Dear Editor,
SARS‐CoV2, transmitted through respiratory secretions within close contacts, primarily infects epithelial/endothelial cells lining the respiratory mucosae. Nasopharyngeal swab (NPS), the favoured sample for reverse transcriptase‐polymerase chain reaction (RT‐PCR) retrieves SARS‐CoV2‐infected cells with minimal aerosol formation (Wang et al. 2020; CDC guidelines, 2020). However, NPS collection is somewhat invasive with discomfort, requires medical/technical expertise, and might not be feasible in remote villages, especially in developing countries like India. On the other hand, epithelial cells of the oral mucosa abundantly carry angiotensin converting enzyme‐2 (ACE‐2) receptors that bind SARS‐CoV2 (Huang et al. 2020; Xu et al. 2020). Whole mouth fluid (WMF) is used for diagnosis in many diseases (Azzi et al. 2020; Malamud & Rodriguez‐Chavez, 2011). Its non‐invasive, self‐collectable and low transmission risk makes WMF attractive for diagnosis of Covid‐19 (To et al. 2020). Early and quick detection of SARS‐CoV2 is of prime importance in containing its spread. Currently, most rapid antigen kits are validated for NPS specimens. In this study, we evaluated the utility of a SARS‐CoV2 antigen kit using drooled WMF samples from laboratory‐confirmed SARS‐CoV2 RT‐PCR positive patients.
The study was approved by VHS‐Institutional Ethics Committee (VHS‐IEC/69‐2020). Twenty‐seven RT‐PCR positive (concurrently NPS‐positive) and 10 RT‐PCR negative (5 NPS‐positive and 5 NPS‐negative) WMF samples were selected retrospectively in an anonymous delinked manner. The presence of SARS‐CoV2 antigen was tested using the commercially available NPS rapid antigen test (RAT; SD Biosensor, Korea; Cerutti et al. 2020). Three hundred microliters of free‐flowing WMF was mixed with the extraction buffer and processed as per manufacturer's instructions. Viral copy numbers in NPS samples were calculated using the standard curve equation generated from SARS‐CoV2 RNA standards (Exact Diagnostics, USA). Median and interquartile ranges were calculated using Microsoft excel. Mann–Whitney rank sum test and McNemar's test were performed using VassarStats.
Ten RT‐PCR negative WMF samples were RAT‐negative. Of the 27 RT‐PCR positive WMF samples, 15 (56%) were RAT‐positive. Comparing RAT with RT‐PCR: true positive‐15/27 (56%), true negative‐10/10 (100%), sensitivity‐56%, specificity‐100% and concordance‐78% (p =.0005; McNemar's test). The median and interquartile range of the virus copies in the NPS were statistically higher among the RAT‐positive compared with the RAT‐negative patients (Figure 1; p =.0001; Mann–Whitney rank sum test).
FIGURE 1.
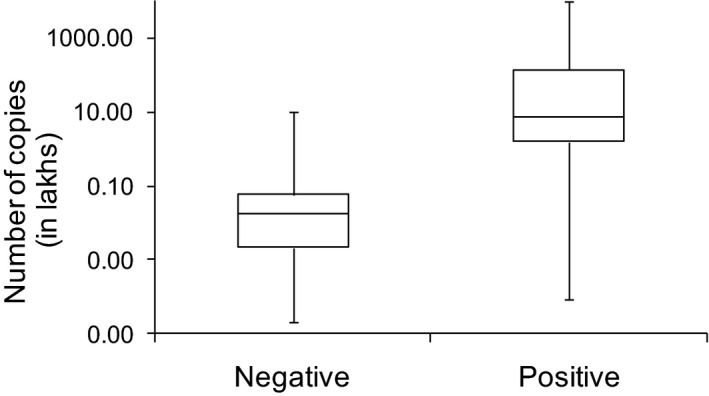
Patients with positive WMF antigen test have higher SARS‐CoV2 copies in their NPS samples. X‐axis denotes negative (n = 12) and positive (n = 15) categories of the WMF antigen test. Y‐axis denotes the number of SARS‐CoV2 copies. The interquartile range shows the 25%–75% range of the virus copies in each category. The error bars depict the minimum and maximum copy numbers in each category
Thus, RAT can detect SARS‐CoV2 antigens in 78% of the cases from WMF in a reliable manner. The significantly higher NPS SARS‐CoV2 burden in the RAT‐positive patients seen in our study is similar to Nagura‐Ikeda et al's report (Nagura‐Ikeda et al. 2020). However, the sensitivity of RAT in their study was only 11.7%, which could be attributed to varying test protocols. We added free‐flowing WMF directly to the extraction buffer, while Nagura‐Ikeda et al dipped a cotton swab in the WMF sample, which was then dipped into the extraction buffer. The latter might retrieve fewer viruses or virus‐infected cells. RT‐PCR, although highly sensitive, also detects dead and/or unpackaged RNA. RAT has moderate sensitivity and detects translated viral proteins. Thus, antigen positivity denotes abundance of proteins and in turn high copy numbers of virus, as shown in the figure. A limitation is the requirement of a diligently collected, free‐flowing, drooled WMF sample without sputum contamination, as thick phlegm/mucous can compromise the lateral flow of the sample across the chromatogram causing false negative results.
Overall, this easy to use point‐of‐care RAT may be used for the detection of SARS‐CoV2 in WMF samples for the rapid confirmation in symptomatic cases requiring urgent medical/dental care; in posttreatment or postquarantine people to rule out transmission risk; as a self‐test at home; and in small remote medical centres where medical expertise to collect NPS samples and technical expertise for RT‐PCR are often not available. The small group of antigen‐negative people may be confirmed by collecting NPS samples and transporting to higher facilities for RT‐PCR.
AUTHOR CONTRIBUTIONS
Priya Kannian: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Project administration; Supervision; Validation; Writing‐original draft; Writing‐review & editing. Chandra Lavanya: Resources; Supervision; Visualization. Krittika Ravichandran: Investigation; Resources; Visualization. Jayaraman Bagavad Gita: Project administration; Writing‐original draft; Writing‐review & editing. Pasuvaraj Mahanathi: Investigation; Resources; Visualization. Veeraraghavan Ashwini: Investigation; Resources; Visualization. Nagalingeswaran Kumarasamy: Conceptualization; Methodology; Project administration; Supervision; Writing‐review & editing. Gunaseelan Rajan: Conceptualization; Funding acquisition; Project administration; Resources; Supervision; Visualization; Writing‐review & editing. Kannan Ranganathan: Conceptualization; Data curation; Formal analysis; Supervision; Writing‐review & editing. Stephen J. Challacombe: Visualization; Writing‐review & editing. Jennifer Webster‐Cyriaque: Visualization; Writing‐review & editing. Newell W. Johnson: Visualization; Writing‐review & editing.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/odi.13793.
ACKNOWLEDGEMENTS
Funded by intramural funds of Chennai Dental Research Foundation, India. Authors have no conflicts of interest to declare.
Kannian P, Lavanya C, Ravichandran K, et al. SARS‐CoV2 antigen in whole mouth fluid may be a reliable rapid detection tool. Oral Dis. 2021;00:1–2. 10.1111/odi.13793
REFERENCES
- Azzi, L. , Carcano, G. , Gianfagna, F. , Grossi, P. , Gasperina, D. D. , Genoni, A. , Fasano, M. , Sessa, F. , Tettamanti, L. , Carinci, F. , Maurino, V. , Rossi, A. , Tagliabue, A. , & Baj, A. (2020). Saliva is a reliable tool to detect SARS‐CoV‐2. Journal of Infection, 81, e45–e50. 10.1016/j.jinf.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Guidelines (2020). Overview of testing for SARS‐CoV‐2 (COVID‐19). National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases Oct (2020). https://www.cdc.gov/coronavirus/2019‐ncov/hcp/testing‐overview [Google Scholar]
- Cerutti, F. , Burdino, E. , Milia, M. G. , Allice, T. , Gregori, G. , Bruzzone, B. , & Ghisetti, V. (2020). Urgent need of rapid tests for SARS‐CoV2 antigen detection: Evaluation of the SD‐Biosensor antigen test for SARS‐CoV2. Journal of Clinical Virology, 132, 104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N. , Perez, P. , Kato, T. , Mikami, Y. , Okuda, K. , Gilmore, R. C. , Conde, C. D. , Gasmi, B. , Stein, S. , Beach, M. , Pelayo, E. , Maldonado, J. , LaFont, B. , Padilla, R. , Murrah, V. , Maile, R. , Lovell, W. , Wallet, S. , Bowman, N. M. , Byrd, K. M. (2020). Integrated single‐cell atlases reveal an oral SARS‐CoV2 infection and transmission axis. MedrXiv, 2020.10.26.20219089. 10.1101/2020.10.26.20219089 [DOI] [Google Scholar]
- Malamud, D. , & Rodriguez‐Chavez, I. R. (2011). Saliva as a diagnostic fluid. Dental Clinics of North America, 55, 159–178. 10.1016/j.cden.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagura‐Ikeda, M. , Imai, K. , Tabata, S. , Miyoshi, K. , Murahara, N. , Mizuno, T. , Horiuchi, M. , & Kato, Y. (2020). Clinical Evaluation of Self‐Collected WMF by Quantitative Reverse Transcription‐PCR (RT‐qPCR), Direct RT‐qPCR, Reverse Transcription–Loop‐Mediated Isothermal Amplification, and a Rapid Antigen Test to Diagnose COVID‐19. Journal of Clinical Microbiology, 58, e01438–e1520. 10.1128/JCM.01438-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, K.‐W. , Tsang, O.‐Y. , Yip, C.‐Y. , Chan, K.‐H. , Wu, T.‐C. , Chan, J.‐C. , Leung, W.‐S. , Chik, T.‐H. , Choi, C.‐C. , Kandamby, D. H. , Lung, D. C. , Tam, A. R. , Poon, R.‐S. , Fung, A.‐F. , Hung, I.‐N. , Cheng, V.‐C. , Chan, J.‐W. , & Yuen, K.‐Y. (2020). Consistent detection of 2019 novel coronavirus in saliva. Clinical Infectious Diseases, 71, 841–843. 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Xu, Y. , Gao, R. , Lu, R. , Han, K. , Wu, G. , & Tan, W. (2020). Detection of SARS‐CoV‐ 2 in different types of clinical specimens. JAMA, 323, 1843–1844. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Zhong, L. , Deng, J. , Peng, J. , Dan, H. , Zeng, X. , Li, T. , & Chen, Q. (2020). High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. International Journal of Oral Science, 12, 8. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]


