Abstract
Aims
The SARS‐coV‐2 pandemic continues to cause an unprecedented global destabilization requiring urgent attention towards drug and vaccine development. Thalidomide, a drug with known anti‐inflammatory and immunomodulatory effects has been indicated to be effective in treating a SARS‐coV‐2 pneumonia patient. Here, we study the possible mechanisms through which thalidomide might affect coronavirus disease‐19 (COVID‐19).
Methods
The present study explores the possibility of repurposing thalidomide for the treatment of SARS‐coV‐2 pneumonia by reanalysing transcriptomes of SARS‐coV‐2 infected tissues with thalidomide and lenalidomide induced transcriptomic changes in transformed lung and haematopoietic models as procured from databases, and further comparing them with the transcriptome of primary endothelial cells.
Results
Thalidomide and lenalidomide exhibited pleiotropic effects affecting a range of biological processes including inflammation, immune response, angiogenesis, MAPK signalling, NOD‐like receptor signalling, Toll‐like receptor signalling, leucocyte differentiation and innate immunity, the processes that are aberrantly regulated in severe COVID‐19 patients.
Conclusion
The present study indicates thalidomide analogues as a better fit for treating severe cases of novel viral infections, healing the damaged network by compensating the impairment caused by the COVID‐19.
Keywords: angiogenesis, COVID‐19, endothelium, immune response, inflammation, lenalidomide, SARS‐coV‐2, thalidomide
What is already known about this subject
Severe cases of COVID‐19 infections show an aberrant surge in the host immune response and cytokine storm in lungs.
There is an increased amount of angiogenesis observed in lungs of COVID‐19 patients and the endothelium is heavily affected.
A patient with severe pneumonia‐associated with COVID‐19 has been successfully treated with thalidomide.
What this study adds
This study presents insights into the possible mechanisms by which thalidomide and lenalidomide would suppress the cytokine storm and immune response.
Thalidomide and its derivative lenalidomide modulate expression of several genes and key pathways aberrantly regulated in SARS‐coV‐2 infected tissues.
1. INTRODUCTION
Novel coronavirus, SARS‐coV‐2 has been posing devastating effects on a global scale with a soaring number of infections and an alarming rate of mortality. Along with the tremendous efforts to develop vaccines, repurposing of drugs with known safety and efficacy profiles is one of the viable choices for treatment. Coronavirus disease‐19 (COVID‐19) is clinically very challenging since the novel coronavirus triggers multiorgan turbulence devastating the homeostasis of the human system. Once the SARS‐CoV‐2 virus enters the respiratory tract, there are 4 different stages of the infection from symptoms to multiorgan failures. Phase I begins with the naso‐oral viral entry followed by host immune system alert with active viral replication in the upper respiratory tract (Phase II). In Phase III, a minor cytokine storm occurs in the alveoli, releasing the inflammatory cytokines resulting in leaky blood vessels, which is followed by the second cytokine storm with uncontrolled inflammatory and life‐threatening symptoms, acute respiratory distress syndrome (ARDS), seizure, severe hypoxia and severed organ toxicity (Phase IV). 1 , 2 Manifestation of the biphasic cytokine storm occurs through the activation of a series of cytokines including monocyte chemoattractant protein 1 (CCL2), macrophage inflammatory protein 1α (MIP‐1α), tumour necrosis factor α (TNF‐α), interleukin (IL)‐2R and IL‐6 overwhelming the system leading to indiscriminate damages in multiple organs. 3 , 4 , 5 There is an increased amount of blood vessel growth in the lungs of COVID‐19 patients compared to severe influenza. 6 As COVID‐19 is a multilayer problem, researchers around the world are desperately in search for a drug, which would able to tackle all or few of these COVID‐19 hallmarks.
Thalidomide, a small molecule drug, with many years of history known to cause misery, 7 became a game changer for its multifaceted pharmacological effects such as immunomodulation, anti‐inflammation, antiangiogenesis and antiviral effects. 8 At present, the world needs a smart solution. Thalidomide increases the hope of treating COVID‐19 patients. 9 Chen et al. report successful treatment of SARS‐coV‐2 associated pneumonia with combinatory treatment of thalidomide and a low‐dose glucocorticoid. 10 Two clinical trials, NCT04273581 and NCT04273529 have been registered to check the efficacy of thalidomide in treating COVID‐19 patients. The adverse effects of thalidomide and its analogues are well documented. 11 Extensive information available on thalidomide's mechanisms, its efficacy and safety in haemophagocytic syndrome‐induced cytokine storm 12 and idiopathic pulmonary fibrosis, 13 severe H1N1, and paraquat poisoning lung injury 14 , 15 argues for the possible action of thalidomide on COVID‐19 induced lung effects and cytokine storm. Recent reviews on thalidomide in COVID‐19 treatment endorse the possibility of thalidomide and its analogues for the treatment of COVID‐19 symptoms. 16 , 17
Transcriptome‐based approach to connect diseases with drug responses is a recognized strategy in drug repurposing. 18 With the fast‐growing literature on SARS‐coV‐2 infections, we performed a combined analysis of whole transcriptome signatures of lungs, peripheral blood mononuclear cells (PBMC), bronchoalveolar lavage fluid (BALF) from SARS‐coV‐2 affected patients and A549 cells (transformed adenocarcinoma cells), and compared with the gene expression signatures of A549 cells treated with thalidomide or lenalidomide, haematopoietic cells, and human umbilical vein endothelial cells (HUVEC). We hereby provide possible mechanistic actions of thalidomide in treating the SARS‐coV‐2 pathology. In addition, we suggest that the derivatives of thalidomide, lenalidomide and CC‐220 might also be effective in the treatment of SARS‐coV‐2.
2. METHODS
2.1. Data collection
A total of 16 gene expression datasets including 15 publicly available expression datasets were included in this study. Transcriptomes of SARS‐coV‐2 infected lung tissues matched with healthy control and SARS‐coV‐2 treated A549 cells were obtained from GEO (Accession ID: GSE147507). 19 Transcriptome data of BALF and PBMC were obtained from BIG Data Center (https://bigd.big.ac.cn/; Accession ID: CRA002390). 20 The expression profiles of systemic lupus erythematosus (Accession ID: GSE112087), 21 bone marrow cells treated with lenalidomide (Accession ID: GSE106748), 22 lymphoma cells treated with lenalidomide (Accession ID: GSE60618), 23 CD34‐positive cells treated with pomalidomide (Accession ID: GSE144052), MERS infected PBMC (Accession: GSE1739), 24 lenalidomide‐treated PBMC (Accession ID: GSE84251) and CC‐122 treated lymphoma cells (Accession ID: GSE75420) 25 were procured from GEO and differentially expressed genes (DEGs) were identified using limma. 26 Library of Integrated Network‐Based Cellular Signatures (iLINCS) is a database that contains the gene expression signatures of >21, 000 compounds (http://www.ilincs.org/ilincs/). We obtained the gene expression signatures for A549 cells treated with 10 μM thalidomide for 6 hours (LINCSCP_4683) and 24 hours (LINCSCP_4463), 100 μM lenalidomide for 6 hours (LINCSCP_4650) and 24 h (LINCSCP_4427).
2.2. Transcriptome sequencing of HUVEC treated with thalidomide
Endothelitis is a common sign of COVID‐19 6 and as thalidomide possesses well established vascular and anti‐inflammatory effects, we attempted to explore the effects of thalidomide and its derivatives on endothelium. HUVEC were subjected to 20 μM thalidomide or 20 μM lenalidomide or 20 μM pomalidomide or vehicle control treatment for 8 hours. RNA was isolated using TRIzol method and whole transcriptome sequencing was performed using Illumina HiSeq 2500 platform. The data can be accessed at GEO with the Accession ID GSE118979. The sequence reads were aligned with reference genome of Homo sapiens using TopHat2 (v2.0.8) and then followed by transcript compilation and gene identification was done using Cufflinks (v2.2.0). 27 The DEGs were identified using Cuffdiff program (v2.2.0). 27
2.3. Differential expression and enrichment analysis
The raw counts from the SARS‐coV‐2 transcriptomic profiles were subjected to differential expression analysis by DESEq2 v1.26.0. 28 Subsequently the genes were pre‐ranked using the P‐values from DESeq2 analysis and subjected to pre‐ranked gene set enrichment analysis. 29 Gene sets with false discovery rate < 0.05 were considered to be statistically significant and were visualized using EnrichmentMap plugin 30 of Cytoscape. 31 For drug signatures from iLINCS, differentially expressed genes with P < 0.05 were considered to be statistically significant. Enrichment of kinase perturbation was carried out using Enrichr. 32
2.4. KINOMEscan kinase screening
We analysed our previously published KINOMEscan kinase screening dataset of thalidomide 33 in order to investigate how thalidomide affects the immune system. Kinases whose activities were reduced at least by 60% were considered for further enrichment analysis.
2.5. Comparative analysis using Toppcluster
The DEGs (Q < 0.05 for transcriptome and P < 0.05 for drug signatures) from all the gene expression profiles were compared for overlapping genes and over‐represented pathways using Toppcluster 34 and the networks were visualized using in Cytoscape. 31
2.6. Identification of protein targets using PharmMapper
For identifying the protein targets of thalidomide, we utilized the PharmMapper server (http://www.lilab-ecust.cn/pharmmapper/). 35 The server identifies possible physiological protein targets of any drug molecule by using a pharmacophore‐based mapping approach. The 3‐dimensional structure of thalidomide was obtained from PubChem and processed on the PharmMapper server choosing only human protein target sets. The top 100 target proteins were selected based on the ranking associated with a fit score (pKd value) for further enrichment analysis using Enrichr. 32
3. RESULTS
3.1. Meta‐analysis of SARS‐coV‐2 affected lung biopsies and PBMCs reveal enrichment of various pathways pertaining to immune response
We reanalysed and compared the SARS‐coV‐2 infected lung biopsies, A549 cells, PBMC and BALF transcriptomic profiles obtained from different studies. SARS‐coV‐2 infection showed a massive surge in inflammatory response, cytokine production and cytokine‐mediated signalling. There was a substantial upregulation of immune response including the processes of haematopoietic development and lymphocyte activation (Figures 1A, 5A, S3A). Activation of viral life cycle and antiviral interferon signalling was observed in infected lungs (Figure 1A) and A549 cells (Figures 1B, S7). Over‐representation of pathways including NOD‐like receptor signalling, MAPK cascade, measles and influenza‐A were seen in the infected lung as identified by gene set enrichment analysis (Figure 1A). Enrichr analysis revealed that many genes upregulated in SARS‐coV‐2 lung are the genes downregulated when SYK was knocked down or inhibited as supported by previous GEO studies (GSE43510, GDS3609 and GSE34176; Figure 1C). Expression to kinase (X2K) analysis showed the possible perturbation of various MAP kinases including ABL1 and JNK1 (Figures S1A, S1B). Human phenotype enrichment analysis shows thrombocytopenia, poor wound healing, abnormality of lymphatic system, serositis and abnormal anticoagulant pathways in SARS‐coV‐2 infected lungs (Figure S5).
FIGURE 1.
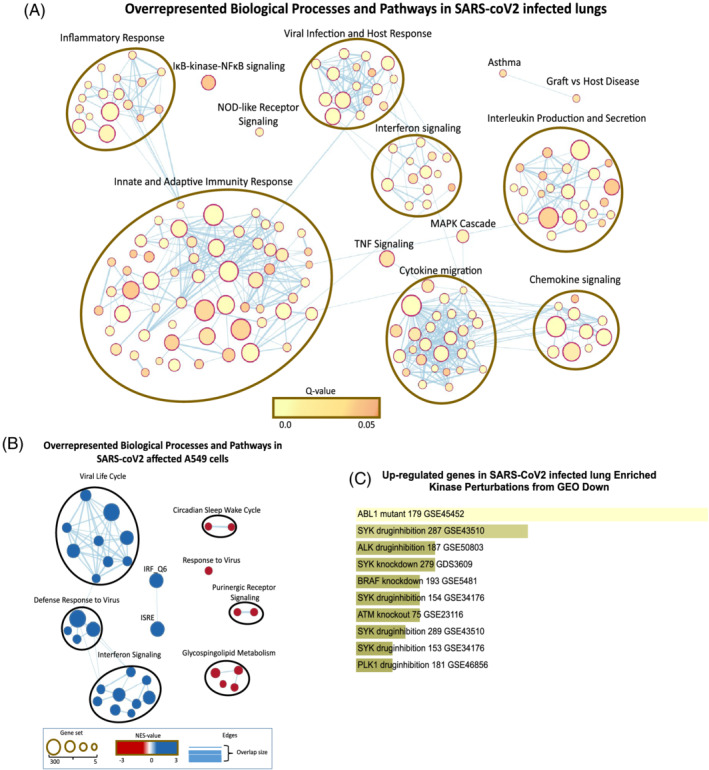
Characteristics of SARS‐coV‐2 infected lungs and A549 cells. Gene set enrichment analysis of genes modulated in (A) SARS‐coV‐2 infected lung and (B) A549 cells. (C) Genes activated in SARS‐coV‐2 overlapping with genes downregulated upon kinase perturbations
FIGURE 5.
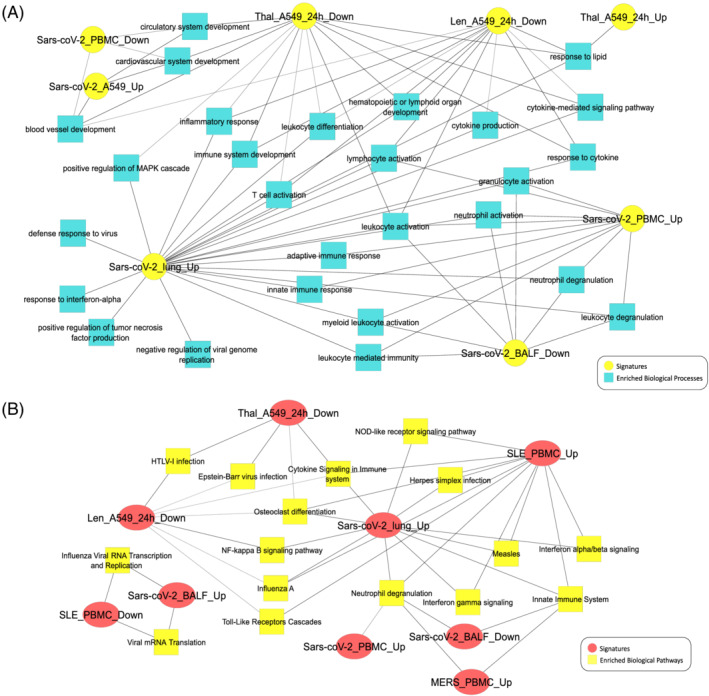
(A) Gene ontology biological process enrichment comparison of gene expression signatures. (B) Biological pathways enriched (false discovery rate < 0.05) in SARS‐coV‐2 infected tissues and their modulation by thalidomide and lenalidomide in A549 cells and lymphoma. The signatures of MERS‐affected and systemic lupus erythematosus patients were used for comparison
3.2. Similarity of SARS‐coV‐2 expression profile with that of systemic lupus erythematosus, lymphoma and multiple myeloma
Comparative enrichment analysis of gene expression profiles for disease‐specific phenotypes revealed the similarity of SARS‐infected tissues with pneumonia, influenza, lymphoma, systemic lupus erythematosus (SLE), multiple myeloma, asthma, auto‐immune diseases, asthma, pneumonia and atherosclerosis while SARS‐coV‐2 PBMC showed exclusive overlap with lymphoma, multiple myeloma and chronic lymphocytic leukemia (Figure 2A). The gene expression profile of SARS‐coV‐2 infected lungs showed high resemblance to that of PBMC from SLE patients (Figure 2B). There was a significant overlap between SARS‐coV‐2 and SLE in upregulated genes involved in immune regulation (Figure S3A), interferon signalling (Figure S7B) and disease‐specific phenotypes including lymphoma and multiple myeloma (Figures S2A, S2B, S2C). Targets of transcription factors, IFN‐sensitive response element and IFN‐regulatory factor (IRF) were upregulated in SARS‐coV‐2 affected lungs similar to upregulation observed in the PBMC of SLE patients (Figures 1B, S1C).
FIGURE 2.
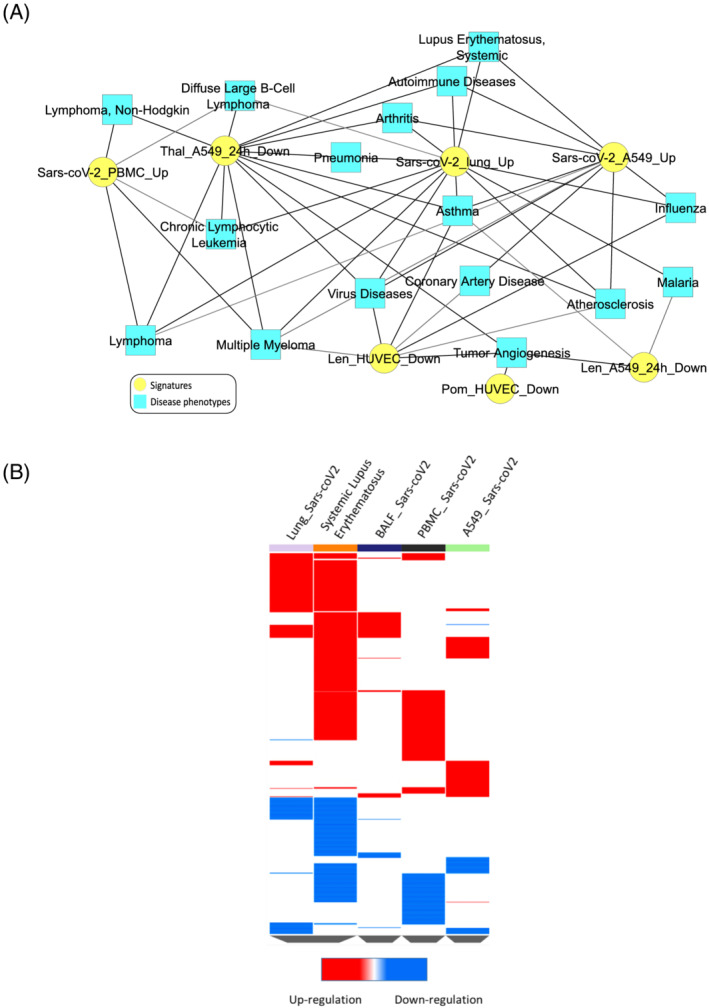
Enrichment of disease‐specific phenotypes. (A) Similarity of SARS‐coV‐2 signatures with disease‐specific phenotypes. (B) Overlapping of SARS‐coV‐2 expression profile of SARS‐coV‐2 affected lung with systemic lupus erythematosus
3.3. Effect of thalidomide on kinases implicated in immune response and MAPK signalling
The kinase screening assay on thalidomide identified key kinases involved in the regulation of immune response. The most kinases affected were LCK and SYK (Figure 3C), critical modulators of T cell receptor signalling. Many LCK substrates, SPI1, TBK1, FOXP3, EGR1, ESR1, IRF1, CBL and STAT1 as well as SYK phosphorylation targets such as OAS1 and MX1 were upregulated in SARS‐coV‐2 lung (Figure s4, S1). Various processes mediating immune response including JUN phosphorylation, IκB phosphorylation, JAK–STAT pathway, leucocyte‐mediated immunity, neutrophil degranulation and activation, B‐cell receptor signalling, and MAPK cascade were found to be affected by thalidomide (Figure 3D). We studied the effects of thalidomide and its derivatives on endothelium and identified the downregulation of several angiogenic genes (Figure 4B and Table 1). PharmMapper results showed strong affinity for LCK, HCK and SYK along with other proteins involved in innate and adaptive immune response (Table 2).
FIGURE 3.
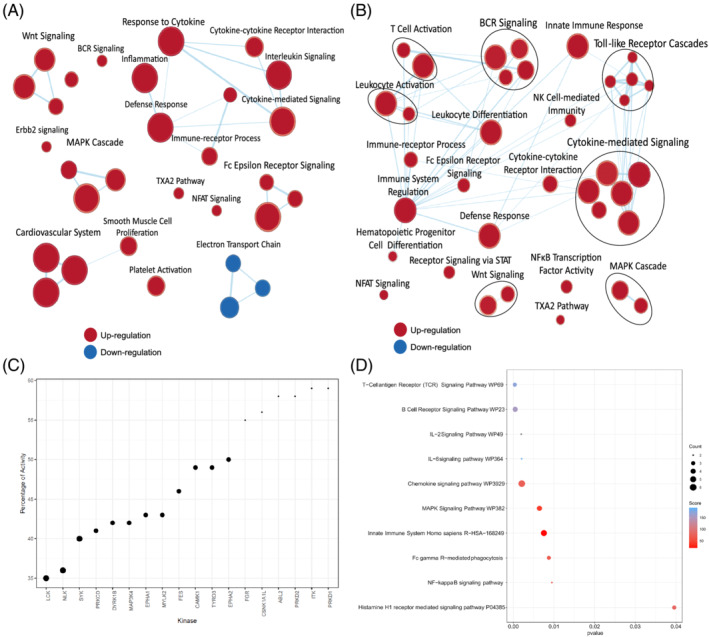
Effect of thalidomide and lenalidomide on immune system. Gene set enrichment analysis network of gene expression profiles of (A) thalidomide and (B) lenalidomide‐treated A549 cells. (C) Activities of kinase involved in immunomodulation and MAPK signalling in the presence of thalidomide. (D) Biological pathways enriched by kinases affected by thalidomide
FIGURE 4.
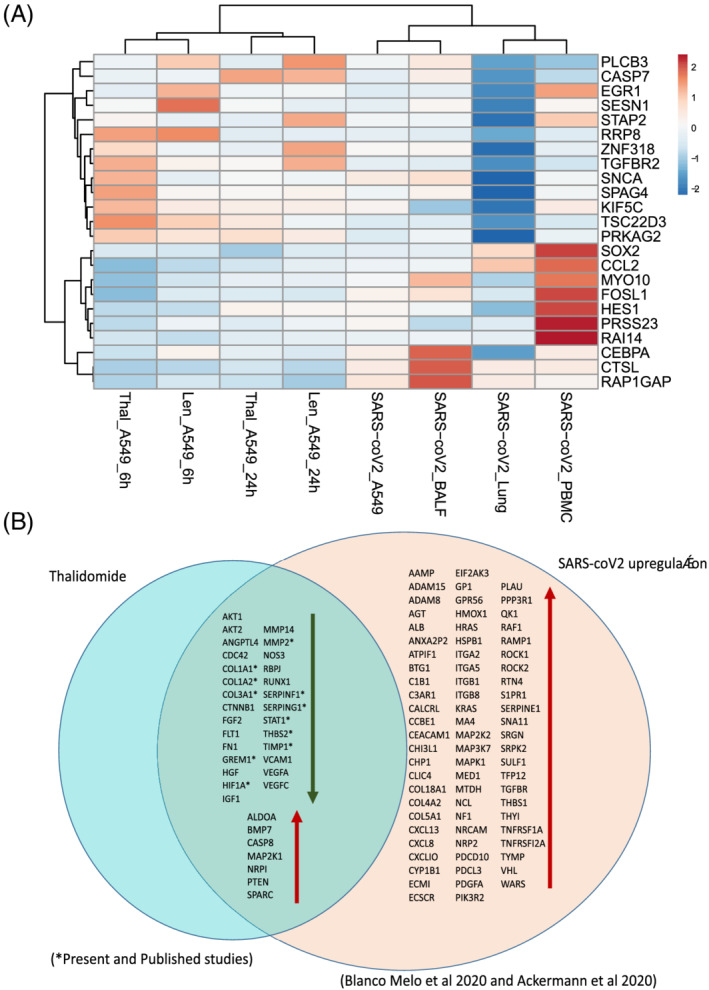
(A) Heatmap depicting the fold change of expression of inflammation and immunomodulatory genes implicated in SARS‐coV‐2 infected tissues and their regulation under thalidomide and lenalidomide treatment (log2FC > 2; false discovery rate < 0.05 for disease signatures). (B) Upregulation of genes implicated in angiogenesis in SARS‐coV2 lung 6 and the modulation of those genes under thalidomide treatment from this study and published studies
TABLE 1.
Genes overexpressed in SARS‐coV‐2 lungs and thalidomide's effects on gene expression
| Gene | SARS‐coV2‐lung | Thalidomide or lenalidomide treatment |
|---|---|---|
| AKT1, AKT2 | Upregulation | Signalling—Down 36 , 37 |
| ANGPTL4 | Upregulation | Down 38 |
| CDC42 | Upregulation | Down 39 |
| COL1A1* | Upregulation | Down 15 |
| COL1A2* | Upregulation | Down 40 |
| MMP2 | Upregulation | Down 41 |
| THBS2* | Upregulation | Down (present study) |
| VEGFA | Upregulation | Down 42 , 43 |
| VEGFC | Upregulation | Down 42 , 43 |
| FGF2* | Upregulation | Down 7 , 43 |
| FLT1 | Upregulation | Down 44 |
| FN1 | Upregulation | Down 45 |
| HIF1A | Upregulation | Down 46 |
| IGF1 | Upregulation | Down 7 |
| MMP14 | Upregulation | Down 41 |
| RBPJ | Upregulation | Down 47 |
| TIMP1 | Upregulation | Down 36 , 37 |
| VCAM1 | Upregulation | Down 38 |
| IFN‐γ | Upregulation | Down 39 |
| IL‐6 | Upregulation | Down 15 |
| HGF | Upregulation | Down 39 |
| IL‐10 | Upregulation | Down 41 |
| IL‐1β | Upregulation | Down 48 |
| CTNNB1* | Upregulation | Down 41 , 43 |
| MCP1* | Upregulation | Down 41 , 43 |
| NF‐κB* | Upregulation | Down 7 , 41 |
| TNF‐α | Upregulation | Down 49 , 50 , 51 , 52 |
| GREM1* | Upregulation | Down (present study) |
| STAT1* | Upregulation | Down 45 |
| NOS3 | Upregulation | Down 53 , 54 |
| CASP8 | Upregulation | Down 40 |
| MAP 2 K1 | Upregulation | Down 46 |
| PTEN | Upregulation | Up 36 |
| IL‐2 | Upregulation | Up 55 |
| BMP7 | Upregulation | Up 36 |
| MIP‐ α | Upregulation | Up/down 48 , 52 |
| SPARC | Upregulation | Up 56 |
| NRPI | Upregulation | Up 57 |
Downregulation of genes/pathways observed in the present study as well.
TABLE 2.
Biological processes and pathways enriched by Pharmmapper predicted protein targets of thalidomide, lenalidomide and pomalidomide
| Pharmmapper identified protein targets of thalidomide and enriched biological processes and pathways | |||
|---|---|---|---|
| Enriched biological pathways and processes | Overlap* | Adjusted P‐value** | Genes |
| Innate immune system Homo sapiens R‐HSA‐168249 | 36/807 | 1.19E‐10 | ITK; GSK3B; SRC; CTSV; CTSS; EGFR; MAPK8; CTSL; AKT2; CTSK; ABL1; CASP1; AKT1; MAPK1; JAK3; HRAS; MAP 2 K1; HSP90AA1; SYK; PDPK1; FGG; MAPK14; PTK2; IL2; MAPK10; HCK; LCK; KIT; MAPKAPK2; BTK; MDM2; PRKCQ; BPI; TEK; RAF1; FGFR2 |
| B‐cell receptor signalling pathway WP23 | 13/97 | 7.96E‐10 | GSK3B; MAP 2 K1; SYK; PDPK1; MAPK14; MAPK8; LCK; BTK; RAC2; AKT1; MAPK1; RAF1; HRAS |
| MAPK signalling pathway | 24/295 | 7.42E‐13 | HSPA8; MAP 2 K1; IGF1; MAPK14; EGFR; TGFBR1; IGF1R; MAPK10; MAPK8; AKT2; CASP3; KIT; APKAPK2; KDR; RAC2; AKT1; MAPK1; TEK; RAF1; HRAS; MET; HSPA1B; FGFR2; HSPA1A |
| Interleukin signalling pathway Homo sapiens P00036 | 9/86 | 3.40E‐06 | GSK3B; PDPK1; AKT2; MAPKAPK2; AKT1; MAPK1; RAF1; JAK3; IL2 |
| Interferon‐γ signalling pathway Homo sapiens P00035 | 4/28 | 1.09E‐03 | MAPK10; MAPK8; MAPK1; MAPK14 |
|
Inflammation mediated by chemokine and cytokine signalling pathway Homo sapiens P00031 |
9/188 | 1.05E‐03 | ROCK1; PDPK1; AKT2; AKT1; MAPK1; RAF1; ITGAL; IL2; RHOA |
| IL‐3 signalling pathway WP286 | 9/49 | 2.60E‐08 | HCK; MAP 2 K1; MAPK8; SYK; SRC; AKT1; MAPK1; RAF1; HRAS |
| T‐cell receptor signalling pathway | 15/101 | 6.87E‐12 | ITK; GSK3B; MAP 2 K1; PDPK1; MAPK14; IL2; RHOA; ZAP70; LCK; AKT2; AKT1; MAPK1; PRKCQ; RAF1; HRAS |
| Melanoma | 12/72 | 2.16E‐10 | MAP 2 K1; CDK6; AKT2; MDM2; AKT1; MAPK1; IGF1; RAF1; HRAS; MET; EGFR; IGF1R |
| Measles | 14/138 | 3.11E‐09 | HSPA8; GSK3B; IL2; MAPK10; MAPK8; CDK6; AKT2; CASP3; CDK2; AKT1; RAB9A; JAK3; HSPA1B; HSPA1A |
| Osteoclast differentiation | 13/127 | 9.88E‐09 | MAP 2 K1; SYK; MAPK14; TGFBR1; MAPK10; MAPK8; LCK; AKT2; CTSK; BTK; AKT1; MAPK1; PPARG |
| Chemokine signalling pathway | 15/190 | 1.77E‐08 | ITK; GSK3B; MAP 2 K1; ROCK1; SRC; PTK2; RHOA; HCK; AKT2; RAC2; AKT1; MAPK1; RAF1; HRAS; JAK3 |
| Pharmmapper identified protein targets of lenalidomide and enriched biological processes and pathways | |||
|---|---|---|---|
| Enriched biological pathways and processes | Overlap | Adjusted P‐value | Genes |
| Innate immune system Homo sapiens R‐HSA‐168249 | 22/807 | 7.02E‐05 | GSK3B; ITK; HSP90AA1; PDPK1; SRC; FGG; MAPK14; PTK2; CTSS; MAPK10; LCK; AKT2; CTSK; MAPKAPK2; KIT; BTK; ABL1; MAPK1; BPI; TEK; RAF1; HRAS |
| Toll‐like receptors cascades Homo sapiens R‐HSA‐168898 | 8/140 | 7.97E‐04 | MAPK10; CTSK; MAPKAPK2; BTK; MAPK1; bpi; MAPK14; CTSS |
| Adaptive immune system Homo sapiens R‐HSA‐1280218 | 15/762 | .016 | GSK3B; ITK; PDPK1; SRC; KIF11; CTSS; ZAP70; LCK; AKT2; CTSK; kit; BTK; MAPK1; RAF1; HRAS |
| B cell activation Homo sapiens P00010 | 7/57 | 7.43E‐06 | MAPK10; RAC2; BTK; MAPK1; RAF1; MAPK14; HRAS |
| T cell receptor signalling pathway | 11/101 | 3.16E‐08 | ITK; GSK3B; ZAP70; PDPK1; LCK; AKT2; MAPK1; RAF1; MAPK14; HRAS; RHOA |
| Fc epsilon RI signalling pathway | 9/68 | 1.13E‐07 | MAPK10; PDPK1; AKT2; RAC2; BTK; MAPK1; RAF1; MAPK14; HRAS |
| IL‐17 signalling pathway | 10/93 | 1.15E‐07 | MAPK10; GSK3B; HSP90AA1; MMP13; MMP1; CASP3; MMP3; MAPK1; MAPK14; MMP9 |
| MAPK signalling pathway | 15/295 | 4.32E‐07 | MAPK14; MAPK10; AKT2; CASP3; MAPKAPK2; KIT; KDR; RAC2; MAPK1; TEK; RAF1; HRAS; MET; HSPA1B; HSPA1A |
| TNF signalling pathway | 7/110 | 1.65E‐04 | MAPK10; AKT2; CASP3; MMP3; MAPK1; MAPK14; MMP9 |
| Natural killer cell mediated cytotoxicity | 7/131 | 4.24E‐04 | ZAP70; LCK; CASP3; RAC2; MAPK1; RAF1; HRAS |
| Interferon‐γ signalling pathway Homo sapiens P00035 | 3/20 | 7.82E‐03 | MAPK10; MAPK1; MAPK14 |
| T‐cell antigen receptor signalling pathway | 8/90 | 1.10E‐05 | ITK; ZAP70; PDPK1; LCK; MAPK1; RAF1; MAPK14; HRAS |
| TGF‐β signalling pathway WP366 | 9/132 | 1.80E‐05 | MMP12; MMP1; SRC; MAPK1; RAF1; MAPK14; met; PTK2; RHOA |
| IL‐5 signalling pathway WP127 | 4/40 | 1.82E‐03 | GSK3B; BTK; MAPK1; RAF1 |
| IL‐2 signalling pathway WP49 | 4/42 | 2.10E‐03 | LCK; MAPK1; RAF1; HRAS |
| IL‐3 signalling pathway WP286 | 4/49 | 3.46E‐03 | SRC; MAPK1; RAF1; HRAS |
| Neutrophil‐mediated immunity (GO:0002446) | 26/487 | 1.21E‐10 | CANT1; GSTP1; PYGL; CTSS; PLAU; MAPK1; CTSG; LTA4H; HSP90AA1; ACE; MME; NME2; RNASE3; MMP8; MAPK14; MMP9; RHOA; APRT; BST1; ADAM17; IMPDH1; IMPDH2; BPI; ALDOA; HSPA1B; HSPA1A |
| Neutrophil degranulation (GO:0043312) | 24/479 | 1.94E‐09 | HSP90AA1; CANT1; MME; GSTP1; NME2; RNASE3; PYGL; MMP8; MAPK14; MMP9; CTSS; RHOA; APRT; BST1; PLAU; IMPDH1; IMPDH2; MAPK1; CTSG; BPI; LTA4H; ALDOA; HSPA1B; HSPA1A |
| Neutrophil activation involved in immune response (GO:0002283) | 24/483 | 1.93E‐09 | HSP90AA1; CANT1; MME; GSTP1; NME2; RNASE3; PYGL; MMP8; MAPK14; MMP9; CTSS; RHOA; APRT; BST1; PLAU; IMPDH1; IMPDH2; MAPK1; CTSG; BPI; LTA4H; ALDOA; HSPA1B; HSPA1A |
| Regulation of inflammatory response (GO:0050727) | 10/166 | 1.70E‐04 | ACE2; BST1; PDE2A; PLA2G2A; NR1H4; NR1H3; PPARG; TEK; PPARA; MAPK14 |
| Myeloid leucocyte differentiation (GO:0002573) | 5/50 | 3.14E‐03 | GLO1; kit; PPARG; MAPK14; MMP9 |
| Myeloid leucocyte mediated immunity (GO:0002444) | 9/20 | .04 | ADAM17; ace |
| Cellular response to cytokine stimulus (GO:0071345) | 13/456 | 5.15E‐03 | GSK3B; HSP90AA1; MME; DAPK1; MAOA; MMP1; PDE2A; MMP3; MMP9; RHOA; CASP3; KIT; PIM1A44D61A45A44:D67 |
| Pharmmapper identified protein targets of pomalidomide and enriched biological processes and pathways | |||
|---|---|---|---|
| Enriched biological pathways and processes | Overlap | Adjusted P‐value | Genes |
| T‐cell receptor signalling pathway | 15/101 | 9.64E‐12 | ITK; GSK3B; MAP 2 K1; PDPK1; MAPK14; IL2; RHOA; ZAP70; LCK; AKT2; AKT1; MAPK1; PRKCQ; RAF1; HRAS |
| MAPK signalling pathway | 22/295 | 3.40E‐11 | MAP 2 K1; IGF1; MAPK14; EGFR; TGFBR1; IGF1R; MAPK10; MAPK8; AKT2; CASP3; KIT; MAPKAPK2; KDR; RAC2; AKT1; MAPK1; TEK; RAF1; HRAS; MET; HSPA1B; HSPA1A |
| Neutrophil‐mediated immunity (GO:0002446) | 29/487 | 6.52E‐11 | CDA; GPI; CANT1; ROCK1; GSTP1; ITGAL; CTSS; PLAU; MAPK1; CTSG; LTA4H; HSP90AA1; ACE; MME; NME2; MMP8; MAPK14; MMP9; RHOA; APRT; BST1; ADAM17; FABP5; IMPDH1; IMPDH2; BPI; ALDOA; HSPA1B; HSPA1A |
| T‐cell antigen receptor signalling pathway | 12/90 | 4.23E‐09 | ITK; ZAP70; MAP 2 K1; MAPK8; PDPK1; LCK; AKT1; MAPK1; PRKCQ; RAF1; MAPK14; HRAS |
| Osteoclast differentiation | 13/127 | 1.26E‐08 | MAP 2 K1; SYK; MAPK14; TGFBR1; MAPK10; MAPK8; LCK; AKT2; CTSK; BTK; AKT1; MAPK1; PPARG |
| B‐cell activation Homo sapiens P00010 | 21/94 | 1.49E‐08 | MAPK10; MAP 2 K1; MAPK8; SYK; RAC2; BTK; MAPK1; RAF1; MAPK14; HRAS |
| Chemokine signalling pathway | 15/190 | 2.41E‐08 | ITK; GSK3B; MAP 2 K1; ROCK1; SRC; PTK2; RHOA; HCK; AKT2; RAC2; AKT1; MAPK1; RAF1; HRAS; JAK3 |
| IL‐3 signalling pathway WP286 | 9/49 | 3.05E‐08 | HCK; MAP 2 K1; MAPK8; SYK; SRC; AKT1; MAPK1; RAF1; HRAS |
| B‐cell receptor signalling pathway | 10/71 | 3.74E‐08 | GSK3B; MAP2 K1; SYK; AKT2; RAC2; BTK; AKT1; MAPK1; RAF1; HRAS |
| IL‐17 signalling pathway | 11/93 | 4.19E‐08 | MAPK10; GSK3B; HSP90AA1; MAPK8; MMP13; MMP1; CASP3; MMP3; MAPK1; MAPK14; MMP9 |
| Influenza A | 14/171 | 4.60E‐08 | FDPS; GSK3B; MAP 2 K1; MAPK14; MAPK10; MAPK8; DDX39B; AKT2; CASP1; AKT1; MAPK1; RAF1; HSPA1B; HSPA1A |
| Viral carcinogenesis | 15/201 | 4.62E‐08 | SYK; SRC; HDAC8; RHOA; CCNA2; KAT2B; CDK6; CASP3; CHEK1; MAPKAPK2; CDK2; MDM2; MAPK1; HRAS; JAK3 |
| Regulation of inflammatory response (GO:0050727) | 15/166 | 1.01E‐07 | PDE2A; PLA2G2A; NR1H4; XIAP; NR1H3; MAPK14; IL2; ACE2; BST1; HCK; CASP1; PPARG; TEK; PPARA; PPARD |
| IL‐5 signalling pathway WP127 | 8/40 | 1.06E‐07 | GSK3B; MAP 2 K1; SYK; BTK; AKT1; MAPK1; RAF1; IL2 |
| TNF signalling pathway | 11/110 | 1.99E‐07 | MAPK10; MAP 2 K1; CASP7; MAPK8; AKT2; CASP3; MMP3; AKT1; MAPK1; MAPK14; MMP9 |
| Interleukin signalling pathway Homo sapiens P00036 | 9/86 | 3.96E‐06 | GSK3B; PDPK1; AKT2; MAPKAPK2; AKT1; MAPK1; RAF1; JAK3; IL2 |
| ACE inhibitor pathway WP554 | 5/17 | 5.33E‐06 | ACE2; ACE; CTSG; REN; NR3C2 |
| Natural killer cell mediated cytotoxicity | 10/131 | 7.39E‐06 | ZAP70; MAP 2 K1; SYK; LCK; CASP3; RAC2; MAPK1; RAF1; ITGAL; HRAS |
| Fc γ R‐mediated phagocytosis | 8/91 | 2.57E‐05 | HCK; MAP 2 K1; SYK; AKT2; RAC2; AKT1; MAPK1; RAF1 |
| Toll‐like receptor signalling pathway | 8/104 | 6.29E‐05 | MAPK10; MAP 2 K1; MAPK8; AKT2; CTSK; AKT1; MAPK1; MAPK14 |
| Interferon‐γ signalling pathway Homo sapiens P00035 | 4/20 | .001 | MAPK10; MAPK8; MAPK1; MAPK14 |
|
Inflammation mediated by chemokine and cytokine Signalling pathway Homo sapiens P00031 |
9/188 | .001 | ROCK1; PDPK1; AKT2; AKT1; MAPK1; RAF1; ITGAL; IL2; RHOA |
| JAK–STAT signalling pathway | 8/162 | 0.001 | AKT2; PIM1; AKT1; RAF1; HRAS; JAK3; IL2; EGFR |
| Toll‐like receptor signalling pathway (GO:0002224) | 7/86 | 0.001 | CTSL; CTSK; FGG; MAPKAPK2; BTK; NR1H4; CTSS |
Overlap = the number of genes enriched in the category vs the total no of proteins contributing to the particular pathway/biological process.
Adjusted P‐value denotes the P‐value obtained after multiple testing.
3.4. Upregulated pathways and gene ontology biological processes in SARS‐coV‐2 infection suppressed by thalidomide and lenalidomide
Various genes aberrantly expressed in SARS‐coV‐2 affected lungs are known targets of thalidomide (Table 1). SARS‐coV‐2 infected lungs, PBMC and A549 cells showed significant upregulation of expression of genes involved in inflammation, cytokine signalling, MAPK signalling and activation of cells mediating the immune response whereas BALF exhibited a slightly different immune profile where leucocyte and neutrophil activation was suppressed (Figure 5A). Comparison of differentially expressed genes of all the signatures yielded interesting results. Many of the processes upregulated in SARS‐coV‐2 infected tissues were suppressed by thalidomide and lenalidomide in A549 cells and endothelial cells (Figures 3A,B, 5A). Thalidomide‐treated A549 cells showed suppression of key genes including SYK, JUN, PIK3CA and HLA genes implicated in immune response (Figure S3A,B). Thalidomide and lenalidomide downregulated various proinflammatory and angiogenic genes aberrantly expressed in SARS‐cov‐2 infected lungs including CCL2 and TSC22D3 which are NF‐κB modulators in A549 cells (Figure 4A,B). Thalidomide and lenalidomide treatment resulted in significant suppression of cytokine response, angiogenesis, inflammation, Fc Epsilon receptor signalling and MAPK cascade (Figure 3A). In addition, lenalidomide downregulated STAT1 expression, leucocyte differentiation, TLR signalling as well as IRF activation (Figures 3B, S4B). Many genes implicated in NOD‐like receptor signalling overexpressed in SARS‐coV‐2 were suppressed by lenalidomide in A549 and lymphoma cells (Figure S4C). B‐cell receptor signalling was activated in SARS‐coV‐2 affected PBMC whereas T‐cell activation was observed in SARS‐coV‐2 lungs (Figure S3B). Translation of viral mRNA was exclusively observed in SARS‐coV‐2 infected BALF whereas genes implicated in viral entry and life cycle were upregulated in SARS‐coV‐2 infected lungs, BALF and A549 cells. Genes involved in viral entry and type I interferon signalling were downregulated in thalidomide‐treated A549 cells and lenalidomide‐treated lymphoma, A549 and HUVEC (Figures S7A, S7C). Several aberrantly expressed genes involved in these disease phenotypes were downregulated in thalidomide and lenalidomide treated A549 and endothelium (Figure 2).
4. DISCUSSION
SARS‐coV‐2 infection causes surge in a number of pathways related to inflammation, cytokine signalling, leucocyte and lymphocyte activation, innate and adaptive immune response marking the phenomenon of cytokine storm. As the whole immune system is affected during the SARS‐coV‐2 infection, immunomodulators would be highly beneficial in treating the symptoms. A COVID‐19 patient with pneumonia was treated successfully with thalidomide and low‐dose glucocorticoid. There was a significant decrease in the inflammatory cytokines including IL1‐, IL‐6 and IFN‐γ and increase in the CD4 + and CD8 + T cells and NK cells. Thalidomide reduced the severity of many COVID‐19 symptoms such as lung lesions, exudation due to its pleiotropic effects on the human system. 10 Haemophagocytic syndrome, a hyperinflammatory disorder is another condition in which cytokine storm occurs. It is frequently present with extranodal natural killer/T cell lymphoma (ENKTL). Thalidomide was effective in suppressing the cytokine storm through inhibition of NF‐κB based transcription of IFN‐γ and TNF genes 12 and thalidomide along with P‐Gemox was highly effective in treating ENKTL patients in a Phase II clinical trial. 58 Comparison of SARS‐coV‐2 expression profiles with drug signatures through enrichment analysis revealed striking actions of thalidomide and lenalidomide in A549 and endothelial cells. The results suggest that thalidomide and lenalidomide could reverse the devastating effects of SARS‐coV‐2 infections on immune system. We selected A549, an adenocarcinomic human alveolar basal epithelial cell line to test our hypothesis that thalidomide would be effective against the cytokine storms. The A549 cell line is an appropriate model for testing cytokine storm targeting drugs since a previous study established this model by infecting the cells with influenza A/H1N1 virus (PR‐8) or nonstructural protein 1 plasmid to test the mechanisms behind inflammatory cytokines/chemokines mediated cytokine storm 59 Studies have utilized A549 cells to show the effects of thalidomide on lung fibrosis. 36 , 42 , 60 A limitation of this study is that only 978 genes called landmark genes are profiled in the iLINCS drug signatures. However, the profiles are highly reproducible and represent the whole transcriptome. 18 , 61 Our models of A549 and HUVEC effectively capture the effects of thalidomide in lungs as well as endothelium.
It is also emerging that SARS‐coV‐2 infections perturb vascular plexus significantly and there is a substantial increase in the growth of new blood vessels and evidence of intussusceptive angiogenesis with overexpression of angiogenesis and hypoxia genes in the lungs of COVID‐19 patients. 6 Cytokine storm and atherosclerosis are tightly connected in SARS‐coV‐2, 62 which is consistent with our analysis revealing the enrichment of atherosclerosis in the SARS‐coV‐2 signatures (Figure 2A). Thalidomide is a renowned modulator of vascular system, and it is known to transcriptionally or functionally target various genes (Table 1) upregulated genes in the lungs of COVID‐19 patients. 6 , 19 As SARS‐coV‐2 infection has a huge impact on the haematopoietic system 63 affecting the myeloid cell maturation, we reanalysed the effects of thalidomide and its derivatives on PBMC, bone marrow cells as well as lymphoma cells. Thalidomide and lenalidomide exhibited attenuation of cytokine signalling and inflammation in addition to its anti‐angiogenic action (Figure 3A). The drugs affected most of the pathways upregulated in SARS‐coV‐2 affected lungs and PBMC (Figures 1A, 3A) in A549 cells, mandating direct investigations in SARS‐coV‐2 infected models.
COVID‐19 coincides with a strong neuro‐endocrine modulation because the disease devastates functions of the organs, and naturally the reciprocal communication between the organs of the endocrine stress system gets a set‐back. 64 ACE2 is expressed along the hypothalamus, pituitary and adrenal axis which is implicated in the stress response and adrenal glands has the highest concentration of virus particles next to lung. 65 A high expression of ACE2 in brain is believed to be the reason for the possible infection of the central nervous system in SARS patients. 66 Chronic elevated stress levels have been reported in SARS and SARS‐coV‐2 patients even long after the outbreak. 10 Notably, thalidomide is also known for its neuro‐endocrine modulation properties. Thalidomide modulates the central nervous system by reducing the generation of proinflammatory cytokines such as IL‐1, IL‐6, IL‐8 and TNF‐α through NF‐κB inhibition. 67 There was a downregulation of genes involved in circadian wake cycle (Figures 1B, S2) including PER3 in the PBMC of COVID‐19 patients, suggesting a reason for the possible sleep disturbances in SARS‐coV‐2 patients. Thalidomide, having a well‐known antiemetic and sedative action on the neuroendocrine axis, would relax the patients, which is supported by the report that thalidomide was effective in treating the anxiety and digestive symptoms in COVID‐19 patients. 10
The anti‐inflammatory properties of thalidomide and its analogues through reduction of IL‐1β, TNF‐α expression and NF‐κB inhibition are well established. 49 SARS‐coV‐2 infections showing elevated NF‐κB signalling and rampage activation of immune response. Unlike other RNA viruses, SARS‐coV‐2 suppresses TNF receptor‐associated factors 3 activation, inhibiting NF‐κB and IRFs, leading to suppression of early proinflammatory and antiviral responses. Whereas later stages of the infection show an enhanced expression of IRF targets in the lungs with an activation of IL‐1, IL‐6 and TNF‐α expression and inhibition of type I interferon signalling. 68 Activation of IRF and IFN‐sensitive response element transcriptional targets in SARS‐coV‐2 affected lungs is in agreement with previous studies reporting the SARS biology. 69 Thalidomide inhibited LCK activity affecting STAT1 phosphorylation, cytokine mediated signalling, NF‐κB signalling, osteoclast differentiation and MAPK signalling through modulation of various upstream activators and downstream effectors. Lenalidomide, in addition, suppressed leucocyte differentiation, TLR signalling along with IRF activation in A549 and lymphoma cells. The effects of thalidomide and lenalidomide observed in our study are consistent with the previous studies where thalidomide and lenalidomide has been shown to inhibit IRF and STAT1 phosphorylation resulting in the downregulation of interferon expression and TLR signalling. 70 , 71
The expression profile of SARS‐coV‐2 infected lungs, PBMC as well as A549 cells show resemblance with profiles of lymphoma, multiple myeloma and SLE (Figure 2A); however, we have focused on SLE as there was a striking similarity of the SLE expression profile and enriched pathways with that of lungs affected by COVID‐19 (Figure 2B). Interestingly, our findings showing the similarity of COVID‐19 infected lung with SLE strongly support a recent study that identified the resemblance between severe cases of COVID‐19 and SLE. 72 We chose PBMC from SLE patients as it also captures the immune activity relatively better, and for better comparison with PBMC from COVID‐19 patients and thalidomide‐treated PBMC. Therefore, drugs that are effective in treating SLE, lymphoma and multiple myeloma might be effective against SARS‐coV‐2 infection. Thalidomide and its derivatives show impressive efficacy in treating multiple myeloma and certain forms of lymphoma. 49 Notably, hydroxychloroquine, an Food and Drug Administration‐approved SLE drug is currently being used in the management of critically ill SARS‐coV‐2 patients. 73 CC‐220, another thalidomide analogue shows very promising results in phase I/II clinical trials against SLE. 74 , 75 CC‐220 through suppression of Ikaros and Aiolos expression, 76 transcription factors that are essential for differentiation of leucocyte and NK cells, thus modulates the innate immune system. As innate immune system pathways are deregulated in SARS‐coV‐2 infected lung and PBMC, further studies are warranted to investigate the efficacy and safety of CC‐220 in treating COVID‐19. 62
Any treatment strategy with thalidomide and its analogues including repurposing thalidomide for COVID‐19, should consider thalidomide‐induced adverse effects including neuropathy and venous thromboembolism. 77 There have been many reports on COVID‐19 patients develop blood clots, 62 a dangerous issue that might be aggravated with the use of thalidomide and lenalidomide. In addition, lenalidomide might cause cytokine release syndrome in chronic lymphocytic leukaemia patients. 78 Therefore, a very careful dosage regimen has to be followed with all these drugs as serious adverse effects have been observed during dose escalation.
5. CONCLUSION
Our study sheds light on the possible mechanisms through which thalidomide and lenalidomide might be effective in the management of SARS‐coV‐2 pathology. Thalidomide and derivatives effectively modulating various aberrantly regulated pathways infections with abundant pharmacological information available make them promising candidates for the treatment of novel coronavirus infections.
CONTRIBUTORS
L.S., S.G., H.S. and S.C. contributed to study design, experiments and data collection while L.S., S.G. and S.C. helped in manuscript preparation.
COMPETING INTERESTS
The authors have none to declare.
Supporting information
FIGURE S1 Upregulated genes mapped to their transcription factors and kinases in (A) SARS‐coV‐2 lung and (B) A549 cells. (C) Transcription factor enrichment of genes upregulated in SARS‐coV‐2 infection.
FIGURE S2 Enrichment of disease‐specific phenotypes. Differentially modulated genes in SARS‐coV‐2 and thalidomide treatment similar to gene expression profiles of (A) lymphoma and (B) systemic lupus erythematosus and (C) multiple myeloma
FIGURE S3 (A) Overexpression of immune response genes in SARS‐coV‐2 tissues and suppression by thalidomide and lenalidomide. (B) B‐cell receptor signalling activated in the PBMC of SARS‐coV‐2 patient and T‐cell activation in SARS‐coV‐2 lungs. (C) Modulation of ERK1 and ERK2 cascade in SARS‐coV2 lung and thalidomide in A549.
FIGURE S4 Activation of (A) inflammation, (B) TLR signalling, (C) NOD‐like signalling, (D) osteoblast differentiation and (E) cytokine signalling in SARS‐coV‐2 and genes targeted by thalidomide and lenalidomide in the signalling pathways
FIGURE S5 The figure illustrates the abnormal human phenotypes enriched in the SARS‐coV‐2 infected lung as identified by Toppcluster (false discovery rate < 0.05)
FIGURE S6 Gene set enrichment analysis identified that the term Circadian sleep wake cycle was significantly enriched in the PBMC of SARS‐coV‐2 patients (false discovery rate = 0.05).
FIGURE S7 Viral life cycle and defines response to virus. (A) Upregulation of pathways pertaining to viral entry, life cycle and antiviral response was observed in SARS‐coV‐2 infected lungs, PBMC, BALF and A549 cells and their modulation by thalidomide and lenalidomide. Overexpression of genes implicated in (B) interferon signalling and (C) type I interferon signalling in SARS‐coV‐2 infected lung and A549.
ACKNOWLEDGEMENTS
This project was partially supported by a grant from University Grant Commission‐Faculty Recharge Programme (UGC‐FRP), Government of India to S.C. The authors gratefully acknowledge Dr. Vijay Ramaswamy, Staff Neuro‐oncologist and Scientist‐Track Investigator, The Hospital for Sick Children, Toronto for critically reviewing the manuscript and providing valuable suggestions.
Sundaresan L, Giri S, Singh H, Chatterjee S. Repurposing of thalidomide and its derivatives for the treatment of SARS‐coV‐2 infections: Hints on molecular action. Brit Jnl Clinical Pharma. 2021;87(10):3835–3850. 10.1111/bcp.14792
Funding information University Grant Commission‐Faculty Recharge Programme (UGC‐FRP), Government of India to S.C
DATA AVAILABILITY STATEMENT
The data generated in this study have been deposited to Gene Expression Omnibus. The datasets that support this study are available in GEO at https://www.ncbi.nlm.nih.gov/geo/, BIG Data Center at https://bigd.big.ac.cn/and iLINCS at http://www.ilincs.org/ilincs/. The appropriate references have been mentioned in the Methods section.
REFERENCES
- 1. Sungnak W, Huang N, Bécavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681‐687. 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420‐422. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L, Liu H, Liu W, et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jie he he Hu Xi Za Zhi. 2020;43(0):E005. [DOI] [PubMed] [Google Scholar]
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):1‐12. 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid‐19. N Engl J Med. 2020;383(2):120–128. 10.1056/NEJMoa2015432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stephens TD, Bunde CJW, Fillmore BJ. Mechanism of action in thalidomide teratogenesis. Biochem Pharmacol. 2000;59(12):1489‐1499. 10.1016/S0006-2952(99)00388-3 [DOI] [PubMed] [Google Scholar]
- 8. Rehman W, Arfons LM, Lazarus HM. The rise, fall and subsequent triumph of thalidomide: lessons learned in drug development. Ther Adv Hematol. 2011;2(5):291‐308. 10.1177/2040620711413165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rismanbaf A. Potential Treatments for COVID‐19; a Narrative Literature Review. Arch Acad Emerg Med. 2020;8(1):e29. http://www.ncbi.nlm.nih.gov/pubmed/32232214 [PMC free article] [PubMed] [Google Scholar]
- 10. Chen C, Qi F, Shi K, et al. Thalidomide Combined with Low‐Dose Glucocorticoid in the Treatment of COVID‐19 Pneumonia. Preprints; 2020. www.preprints.org. Accessed May 20, 2020.
- 11. Khalil A, Kamar A, Nemer G. Thalidomide‐Revisited: Are COVID‐19 Patients Going to Be the Latest Victims of Yet Another Theoretical Drug‐Repurposing? Front Immunol. 2020;11:1–7. 10.3389/fimmu.2020.01248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wen H, Ma H, Cai Q, et al. Recurrent ECSIT mutation encoding V140A triggers hyperinflammation and promotes hemophagocytic syndrome in extranodal NK/T cell lymphoma. Nat Med. 2018;24(2):154‐164. 10.1038/nm.4456 [DOI] [PubMed] [Google Scholar]
- 13. Somogyi V, Chaudhuri N, Torrisi SE, Kahn N, Müller V, Kreuter M. The therapy of idiopathic pulmonary fibrosis: what is next? Eur Respir Rev. 2019;28(153):190021. 10.1183/16000617.0021-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu H, Shi X, Ju D, Huang H, Wei W, Dong X. Anti‐Inflammatory Effect of Thalidomide on H1N1 Influenza Virus‐Induced Pulmonary Injury in Mice. Inflammation. 2014;37(6):2091‐2098. 10.1007/s10753-014-9943-9 [DOI] [PubMed] [Google Scholar]
- 15. Li D, Zhang X‐W, Jiang X‐Q, et al. Protective effects of thalidomide on pulmonary injuries in a rat model of paraquat intoxication. J Inflamm. 2015;12(1):46. 10.1186/s12950-015-0093-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goihman‐Yahr M. Proposed use of thalidomide for the cytokine storm of COVID‐19. Clin Dermatol. 2020;38(4):508. 10.1016/j.clindermatol.2020.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dastan F, Tabarsi P, Marjani M, et al. Thalidomide against coronavirus disease 2019 (Covid‐19): A medicine with a thousand faces. Iran J Pharm Res. 2020;19(1):1‐2. 10.22037/ijpr.2020.113369.14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian A, Narayan R, Corsello SM, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171(6):1437‐1452.e17. 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced Host Response to SARS‐CoV‐2 Drives Development of COVID‐19. Cell. 2020;181(5):1036‐1045.e9. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID‐19 patients. Emerg Microbes Infect. 2020;9(1):761‐770. 10.1080/22221751.2020.1747363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oon S, Monaghan K, Ng M, et al. A potential association between IL‐3 and type I and III interferons in systemic lupus erythematosus. Clin Transl Immunol. 2019;8(12):e01097. 10.1002/cti2.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez‐Millan B, Diaz de la Guardia R, Roca‐Ho H, et al. IMiDs mobilize acute myeloid leukemia blasts to peripheral blood through downregulation of CXCR4 but fail to potentiate AraC/Idarubicin activity in preclinical models of non del5q/5q‐ AML. Onco Targets Ther. 2018;7(9):e1477460. 10.1080/2162402X.2018.1477460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gopalakrishnan R, Matta H, Tolani B, Triche T, Chaudhary PM. Immunomodulatory drugs target IKZF1‐IRF4‐MYC axis in primary effusion lymphoma in a cereblon‐dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene. 2016;35(14):1797‐1810. 10.1038/onc.2015.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reghunathan R, Jayapal M, Hsu LY, et al. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6(1):2. 10.1186/1471-2172-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hagner PR, Man HW, Fontanillo C, et al. CC‐122, a pleiotropic pathway modifier, mimics an interferon response and has antitumor activity in DLBCL. Blood. 2015;126(6):779‐789. 10.1182/blood-2015-02-628669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet Mol Biol. 2004;3(1):1‐25. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 27. Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562‐578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545‐15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network‐based method for gene‐set enrichment visualization and interpretation. PLoS ONE. 2010;5(11):e13984. 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498‐2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90‐W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sundaresan L, Kumar P, Manivannan J, et al. Thalidomide and Its Analogs Differentially Target Fibroblast Growth Factor Receptors: Thalidomide Suppresses FGFR Gene Expression while Pomalidomide Dampens FGFR2 Activity. Chem Res Toxicol. 2019;32(4):589‐602. 10.1021/acs.chemrestox.8b00286 [DOI] [PubMed] [Google Scholar]
- 34. Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network‐based dissection of biological systems. Nucleic Acids Res. 2010;38(Web Server):W96‐W102. 10.1093/nar/gkq418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38(suppl_2):W609‐W614. 10.1093/nar/gkq300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun X, Xu Y, Wang Y, Chen Q, Liu L, Bao Y. Synergistic Inhibition of Thalidomide and Icotinib on Human Non‐Small Cell Lung Carcinomas Through ERK and AKT Signaling. Med Sci Monit. 2018;24:3193‐3203. 10.12659/MSM.909977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knobloch J, Schmitz I, Gotz K, Schulze‐Osthoff K, Ruther U. Thalidomide Induces Limb Anomalies by PTEN Stabilization, Akt Suppression, and Stimulation of Caspase‐Dependent Cell Death. Mol Cell Biol. 2008;28(2):529‐538. 10.1128/MCB.00533-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Milanovic D, Sticht C, Röhrich M, Maier P, Grosu AL, Herskind C. Inhibition of 13‐cis retinoic acid‐induced gene expression of reactive‐resistance genes by thalidomide in glioblastoma tumours in vivo. Oncotarget. 2015;6(30):28938‐28948. 10.18632/oncotarget.4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu Y, Li J, Ferguson GD, et al. Immunomodulatory drugs reorganize cytoskeleton by modulating Rho GTPases. Blood. 2009;114(2):338‐345. 10.1182/blood-2009-02-200543 [DOI] [PubMed] [Google Scholar]
- 40. Chen H, Xu H, Luo L, et al. Thalidomide Prevented and Ameliorated Pathogenesis of Crohn's Disease in Mice via Regulation of Inflammatory Response and Fibrosis. Front Pharmacol. 2019;10:1486. 10.3389/fphar.2019.01486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Segarra M, Lozano E, Corbera‐Bellalta M, et al. Thalidomide decreases gelatinase production by malignant B lymphoid cell lines through disruption of multiple integrin‐mediated signaling pathways. Haematologica. 2010;95(3):456‐463. 10.3324/haematol.2009.006395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El‐Aarag B, Kasai T, Masuda J, Agwa H, Zahran M, Seno M. Anticancer effects of novel thalidomide analogs in A549 cells through inhibition of vascular endothelial growth factor and matrix metalloproteinase‐2. Biomed Pharmacother. 2017;85:549‐555. 10.1016/j.biopha.2016.11.063 [DOI] [PubMed] [Google Scholar]
- 43. Rosiñol L, Cibeira MT, Segarra M, et al. Response to thalidomide in multiple myeloma: Impact of angiogenic factors. Cytokine. 2004;26(4):145‐148. 10.1016/j.cyto.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 44. Xiao Y, Miao LL. Expression of PIGF and Its receptor Flt‐1 in Patients with Multiple Myeloma and their Correlation with Chemotherapeutic Efficacy. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016;24(4):1091‐1095. 10.7534/j.issn.1009-2137.2016.04.024 [DOI] [PubMed] [Google Scholar]
- 45. Liang C‐J, Yen Y‐H, Hung L‐Y, et al. Thalidomide inhibits fibronectin production in TGF‐β1‐treated normal and keloid fibroblasts via inhibition of the p38/Smad3 pathway. Biochem Pharmacol. 2013;85(11):1594‐1602. 10.1016/j.bcp.2013.02.038 [DOI] [PubMed] [Google Scholar]
- 46. Badamtseren B, Odkhuu E, Koide N, et al. Thalidomide inhibits interferon‐γ‐mediated nitric oxide production in mouse vascular endothelial cells. Cell Immunol. 2011;270(1):19‐24. 10.1016/j.cellimm.2011.03.018 [DOI] [PubMed] [Google Scholar]
- 47. Ding W, Zeng T, Tao W, et al. Effect of lenalidomide on the human gastric cancer cell line SGC7901/vincristine Notch signaling. J Cancer Res Ther. 2018;14(Supplement):S237–S242. 10.4103/0973-1482.183181 [DOI] [PubMed] [Google Scholar]
- 48. Corral LG, Kaplan G. Immunomodulation by thalidomide and thalidomide analogues. Ann Rheum Dis. 1999;58(Suppl 1):I107‐I113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vallet S, Palumbo A, Raje N, Boccadoro M, Anderson KC. Thalidomide and lenalidomide: Mechanism‐based potential drug combinations. Leuk Lymphoma. 2008;49(7):1238‐1245. 10.1080/10428190802005191 [DOI] [PubMed] [Google Scholar]
- 50. Moreira AL, Sampaio EP, Zmuidzinas A, Frindt P, Smith KA, Kaplan G. Thalidomide exerts its inhibitory action on tumor necrosis factor alpha by enhancing mRNA degradation. J Exp Med. 1993;177(6):1675‐1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klausner JD, Freedman VH, Kaplan G. Thalidomide as an Anti‐TNF‐a Inhibitor: Implications for Clinical Use. Clin Immunol Immunopathol. 1996;81(3):219‐223. 10.1006/clin.1996.0181 [DOI] [PubMed] [Google Scholar]
- 52. Lee E‐S, Kim Y, Kwon H, Bang D, Lee S, Sohn S. Thalidomide upregulates macrophage inflammatory protein‐1? in a herpes simplex virus‐induced Behçet?s disease‐like animal model. Arch Dermatol Res. 2004;296:175‐181 10.1007/s00403-004-0498-8 [DOI] [PubMed] [Google Scholar]
- 53. Tamilarasan KP, Kolluru GK, Rajaram M, Indhumathy M, Saranya R, Chatterjee S. Thalidomide attenuates nitric oxide mediated angiogenesis by blocking migration of endothelial cells. BMC Cell Biol. 2006;7(1):7‐17. 10.1186/1471-2121-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Majumder S, Rajaram M, Muley A, et al. Thalidomide attenuates nitric oxide‐driven angiogenesis by interacting with soluble guanylyl cyclase. Br J Pharmacol. 2009;158(7):1720‐1734. 10.1111/j.1476-5381.2009.00446.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shannon E, Aseffa A, Pankey G, Sandoval F, Lutz B. Thalidomide's ability to augment the synthesis of IL‐2 in vitro in HIV‐infected patients is associated with the percentage of CD4+ cells in their blood. Immunopharmacology. 2000;46(2):175‐179. 10.1016/S0162-3109(99)00169-1 [DOI] [PubMed] [Google Scholar]
- 56. Pellagatti A, Jadersten M, Forsblom A‐M, et al. Lenalidomide inhibits the malignant clone and up‐regulates the SPARC gene mapping to the commonly deleted region in 5q‐ syndrome patients. Proc Natl Acad Sci. 2007;104(27):11406‐11411. 10.1073/pnas.0610477104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang Z, Wen R, Zhang Y, et al. Thalidomide induce response in patients with corticosteroid‐resistant or relapsed ITP by upregulating Neuropilin‐1 expression. Int Immunopharmacol. 2019;72:437‐444. 10.1016/j.intimp.2019.04.041 [DOI] [PubMed] [Google Scholar]
- 58. Huang H, Yan G, Su H, et al. Clinical Outcome of an Multicentre, Randomized, Phase II Clinical Trial for Patients with Extranodal NK/T Cell Lymphoma Treated By P‐Gemox or Aspametdex. Blood. 2019;134(Supplement_1):1569‐1569. 10.1182/blood-2019-123478 [DOI] [Google Scholar]
- 59. Phung TTB, Sugamata R, Uno K, et al. Key role of regulated upon activation normal T‐cell expressed and secreted, nonstructural protein1 and myeloperoxidase in cytokine storm induced by influenza virus PR‐8 (A/H1N1) infection in A549 bronchial epithelial cells. Microbiol Immunol. 2011;55(12):874‐884. 10.1111/j.1348-0421.2011.00396.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin Y‐C, Shun C‐T, Wu M‐S, Chen C‐C. A Novel Anticancer Effect of Thalidomide: Inhibition of Intercellular Adhesion Molecule‐1‐Mediated Cell Invasion and Metastasis through Suppression of Nuclear Factor‐ B. Clin Cancer Res. 2006;12(23):7165‐7173. 10.1158/1078-0432.CCR-06-1393 [DOI] [PubMed] [Google Scholar]
- 61. Cheng L, Li L. Systematic Quality Control Analysis of LINCS Data. CPT Pharmacometrics Syst Pharmacol. 2016;5(11):588‐598. 10.1002/psp4.12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vinciguerra M, Romiti S, Fattouch K, De Bellis A, Greco E. Atherosclerosis as Pathogenetic Substrate for Sars‐Cov2 Cytokine Storm. J Clin Med. 2020;9(7):2095. 10.3390/jcm9072095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020. 10.1101/2020.02.06.20020974 [DOI] [Google Scholar]
- 64. Steenblock C, Todorov V, Kanczkowski W, et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and the neuroendocrine stress axis. Mol Psychiatry. 2020;(8):1‐1617. 10.1038/s41380-020-0758-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lv Q, Yang Q, Cui Y, et al. Effects of taurine on ACE, ACE2 and HSP70 expression of hypothalamic‐pituitary‐adrenal axis in stress‐induced hypertensive rats. Adv Exp Med Biol. 2017;975:871‐886. 10.1007/978-94-024-1079-2_69 [DOI] [PubMed] [Google Scholar]
- 66. Chrousos GP, Kaltsas G. Post‐SARS sickness syndrome manifestations and endocrinopathy: How, why, and so what? Clin Endocrinol (Oxf). 2005;63(4):363‐365. 10.1111/j.1365-2265.2005.02361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jung YJ, Tweedie D, Scerba MT, Greig NH. Neuroinflammation as a Factor of Neurodegenerative Disease: Thalidomide Analogs as Treatments. Front Cell Dev Biol. 2019;7:313. 10.3389/fcell.2019.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID‐19: Immunology and treatment options. Clin Immunol. 2020;215:108448. 10.1016/j.clim.2020.108448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spiegel M, Pichlmair A, Martínez‐Sobrido L, et al. Inhibition of Beta Interferon Induction by Severe Acute Respiratory Syndrome Coronavirus Suggests a Two‐Step Model for Activation of Interferon Regulatory Factor 3. J Virol. 2005;79(4):2079‐2086. 10.1128/JVI.79.4.2079-2086.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Millrine D, Tei M, Gemechu Y, Kishimoto T. Rabex‐5 is a lenalidomide target molecule that negatively regulates TLR‐induced type 1 IFN production. Proc Natl Acad Sci U S A. 2016;113(38):10625‐10630. 10.1073/pnas.1611751113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Millrine D, Miyata H, Tei M, et al. Immunomodulatory drugs inhibit TLR4‐induced type‐1 interferon production independently of Cereblon via suppression of the TRIF/IRF3 pathway. Int Immunol. 2016;28(6):307‐315. 10.1093/intimm/dxw005 [DOI] [PubMed] [Google Scholar]
- 72. Woodruff MC, Ramonell RP, Nguyen DC, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID‐19. Nat Immunol. 2020;21(12):1506‐1516. 10.1038/s41590-020-00814-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Taccone FS, Gorham J, Vincent J‐L. Hydroxychloroquine in the management of critically ill patients with COVID‐19: the need for an evidence base. Lancet Respir Med. 2020;8(6):539‐541. 10.1016/S2213-2600(20)30172-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schafer PH, Ye Y, Wu L, et al. Cereblon modulator iberdomide induces degradation of the transcription factors Ikaros and Aiolos: immunomodulation in healthy volunteers and relevance to SLE. Ann Rheum Dis. 2018;77(10):1516‐1523. 10.1136/annrheumdis-2017-212916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lonial S, van de Donk NWCJ, Popat R, et al. First clinical (phase 1b/2a) study of iberdomide (CC‐220), a CELMoD, in combination with dexamethasone (DEX) in patients (pts) with relapsed/refractory multiple myeloma (RRMM). J Clin Oncol. 2019;37(15_suppl):8006. 10.1200/JCO.2019.37.15_suppl.8006 [DOI] [Google Scholar]
- 76. Mazzurana L, Forkel M, Rao A, et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3−ILC1/NK cell transdifferentiation. Eur J Immunol. 2019;49(9):1344‐1355. 10.1002/eji.201848075 [DOI] [PubMed] [Google Scholar]
- 77. Palumbo A, Palladino C. Venous and arterial thrombotic risks with thalidomide: Evidence and practical guidance. Ther Adv Drug Saf. 2012;3(5):255‐266. 10.1177/2042098612452291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Aue G, Njuguna N, Tian X, et al. Lenalidomide‐induced upregulation of CD80 on tumor cells correlates with T‐cell activation, the rapid onset of a cytokine release syndrome and leukemic cell clearance in chronic lymphocytic leukemia. Haematologica. 2009;94(9):1266‐1273. 10.3324/haematol.2009.005835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Upregulated genes mapped to their transcription factors and kinases in (A) SARS‐coV‐2 lung and (B) A549 cells. (C) Transcription factor enrichment of genes upregulated in SARS‐coV‐2 infection.
FIGURE S2 Enrichment of disease‐specific phenotypes. Differentially modulated genes in SARS‐coV‐2 and thalidomide treatment similar to gene expression profiles of (A) lymphoma and (B) systemic lupus erythematosus and (C) multiple myeloma
FIGURE S3 (A) Overexpression of immune response genes in SARS‐coV‐2 tissues and suppression by thalidomide and lenalidomide. (B) B‐cell receptor signalling activated in the PBMC of SARS‐coV‐2 patient and T‐cell activation in SARS‐coV‐2 lungs. (C) Modulation of ERK1 and ERK2 cascade in SARS‐coV2 lung and thalidomide in A549.
FIGURE S4 Activation of (A) inflammation, (B) TLR signalling, (C) NOD‐like signalling, (D) osteoblast differentiation and (E) cytokine signalling in SARS‐coV‐2 and genes targeted by thalidomide and lenalidomide in the signalling pathways
FIGURE S5 The figure illustrates the abnormal human phenotypes enriched in the SARS‐coV‐2 infected lung as identified by Toppcluster (false discovery rate < 0.05)
FIGURE S6 Gene set enrichment analysis identified that the term Circadian sleep wake cycle was significantly enriched in the PBMC of SARS‐coV‐2 patients (false discovery rate = 0.05).
FIGURE S7 Viral life cycle and defines response to virus. (A) Upregulation of pathways pertaining to viral entry, life cycle and antiviral response was observed in SARS‐coV‐2 infected lungs, PBMC, BALF and A549 cells and their modulation by thalidomide and lenalidomide. Overexpression of genes implicated in (B) interferon signalling and (C) type I interferon signalling in SARS‐coV‐2 infected lung and A549.
Data Availability Statement
The data generated in this study have been deposited to Gene Expression Omnibus. The datasets that support this study are available in GEO at https://www.ncbi.nlm.nih.gov/geo/, BIG Data Center at https://bigd.big.ac.cn/and iLINCS at http://www.ilincs.org/ilincs/. The appropriate references have been mentioned in the Methods section.


