Abstract
Epidemiological data shows a discrepancy in COVID‐19 susceptibility and outcomes with some regions being more heavily affected than others. However, the factors that determine host susceptibility and pathogenicity remain elusive. An increasing number of publications highlight the role of Transmembrane Serine Protease 2 (TMPRSS2) in the susceptibility of the host cell to SARS‐CoV‐2. Cleavage of viral spike protein via the host cell's TMPRSS2 enzyme activity mediates viral entry into the host cell. The enzyme synthesis is regulated by the TMPRSS2 gene, which has also been implicated in the entry mechanisms of previously reported Coronavirus infections. In this review, we have investigated the pathogenicity of SARS‐CoV‐2 and disease susceptibility dependence on the TMPRSS2 gene as expressed in various population groups. We further discuss how the differential expression of this gene in various ethnic groups can affect the SARS‐CoV‐2 infection and Coronavirus disease (COVID)‐19 outcomes. Moreover, promising new TMPRSS2 protease blockers and inhibitors are discussed for COVID‐19 treatment.
Keywords: coronavirus, COVID‐19, differential gene expression, SARS‐CoV‐2, TMPRSS2, TMPRSS2 blockers, TMPRSS2 inhibitors, viral entry
Highlights
1. Entry of SARS‐CoV‐2 into a host cell depends on host protease TMPRSS2.
2. TMPRSS2 gene has localized expression throughout the human body but highly expressed in cells of the respiratory tract (primary target of SARS‐CoV‐2 in humans), gastrointestinal tract, kidneys and prostate.
3. Differences in expression of TMPRSS2 gene in the respiratory among different population groups can be a basis for discrepancy observed in COVID‐19 susceptibility and disease outcomes.
4. Drugs based on the inhibition or blockage of TMPRSS2 protease are undergoing clinical trials as a therapeutic option.
1. INTRODUCTION
In early December 2019, hospitals located in Wuhan (the capital of Hubei Province, China) started reporting patients with pneumonic infections “of unknown origin” and related it to a kind of coronavirus infection. 1 On January 10, 2020, the first whole‐genome sequence of the novel coronavirus was published, which indicated the novel coronavirus to produce symptoms similar to the earlier reported SARS‐like viral infections. 2 Later in January 2020, human‐to‐human transmissibility of the virus in the pneumonia outbreak was reported. 3 The coronavirus responsible for this disease condition was later termed SARS‐CoV‐2 and the associated disease was named coronavirus disease or COVID‐19, by the World Health Organization (WHO) and International Committee on Taxonomy of Viruses (ICTV) on February 12, 2020. 4 Since the reporting of first known COVID‐19 cases in Wuhan, SARS‐CoV‐2, the causative agent of COVID‐19 has spread to 191 countries and has infected more than 90.29 million people around the world claiming more than 1.93 million lives. The United States and India lead the world in the total number of confirmed cases and deaths. 5 SARS‐CoV‐2 primarily infects the respiratory and gastrointestinal tract, 6 however, the main target organ seems to be the lungs. 7 The reported symptoms include fatigue, muscular pain, difficulty in breathing, dry cough, sore throat, diarrhea, etc. with fever and dry cough being the most common of all the symptoms. 8 It is noteworthy to mention that COVID‐19 shares all the symptoms with other viral diseases except breathing difficulties and diarrhea. 6 Phylogenetic studies based on the analysis of open reading frame 1a and 1b (ORF1a/1b), nucleocapsid (N) and Spike (S) genes imply that SARS‐CoV‐2 is a new virus that jumped independently from an animal to a human host. 9 The COVID‐19 pandemic has a zoonotic origin which could have been triggered, in a broader context, by consumption of bushmeat, contact with wild animals, or environmental destruction caused by human activities. 10 Currently, it is believed that SARS‐CoV‐2 originated in bats and then probably passed onto Malayan pangolins as intermediate hosts before infecting humans. 11 , 12 However, there is no clear indication of the involvement of Malayan pangolins intermediary role in SARS‐CoV‐2 spread to humans and the virus could directly have jumped from bats to humans. 12 On the basis of the RNA‐dependent RNA polymerase (RdRp) gene, SARS‐CoV‐2 shares 99% similarity with BtCoV/4991, a horseshoe bat coronavirus strain. 12 As we understand, the RdRp gene is a conserved retroviral gene and thus is used for the evolutionary classification of viruses. 13 On the basis of whole‐genome, SARS‐CoV‐2 bears 80% similarity with SARS‐CoV and 50% similarity with MERS‐CoV, both of them also being coronaviruses that were responsible for major viral outbreaks in the recent past. 14 , 15 To infect humans, spike (S) protein anchored in the envelope of SARS‐CoV‐2 is critical for the virus to bind and infect its host. The receptor‐binding domain (RBD) present in the S‐protein of SARS‐CoV‐2 is also similar to SARS‐CoV, suggesting a mechanism of infection shared by both viruses. SARS‐CoV‐2 binds to the peptidase domain of angiotensin‐converting enzyme‐2 abbreviated as the ACE2 of host cell before fusing and entering the host cell. 7 Moreover, like SARS‐CoV and MERS‐CoV, SARS‐CoV‐2 is also dependent on host protease TMPRSS 2 for its priming and entry into the host cells. 16 This protease has been shown to be crucial for SARS‐CoV‐2 infection and disease outcomes as cells expressing higher levels of TMPRSS2 yield higher SARS‐CoV‐2 cells have a higher viral damage. 17
In a short period of time, SARS‐CoV‐2 has virtually engulfed the whole world and caused high mortality. 5 The condition is further exacerbated as social and economic aspects of life around the globe have been heavily affected due to high infection rates and social distancing measures implemented in response to it. 18 These social distancing measures can slow down but cannot stop the spread of SARS‐CoV‐2 unless a significant population of the world has contracted the virus. 19 In addition to the destruction caused by SARS‐CoV‐2, the nonavailability of viable drugs like antivirals against this new disease makes the condition challenging to deal with. 20 In the review, we have evaluated the role of TMPRSS2 protease in SARS‐CoV‐2 infection and how differences in respiratory expression of this protease can explain the associated discrepancy in SARS‐CoV‐2 infection and COVID‐19 outcomes. Moreover, therapeutic interventions that are based on the inhibition and blocking of TMPRSS2 are also elucidated.
1.1. Mechanism of SARS‐CoV‐2 infection and role of TMPRSS2 protease
Coronaviruses (Coronaviridae family) belong to a group of positive‐sense single‐stranded RNA viruses 21 that cause a variety of respiratory, nervous system, and enteric infections in various animal species including humans. 22 These viruses are subdivided at the level of the genus into four types called Alpha‐(α), Beta‐(β), Gamma‐(γ) and Delta‐(δ) coronaviruses. 23 SARS‐CoV‐2 is a member of β‐ coronaviruses, which have four lineages (A–D) with SARS‐CoV as well as SARS‐CoV‐2 belonging to the B lineage, which contains roughly 200 sequenced viral genomes. 24 To be able to infect, a virus has to first bind to the host surface and subsequently initiate a complex entry mechanism. For coronaviruses in B‐lineage, interaction with the host cell involves attachment of viral spike (S) protein with a specific host cell receptor 25 followed by protease‐mediated cleavage of S‐protein that allows viral entry into the host cells. 24 Both attachment and cleavage have been found to be crucial for lineage B β‐coronaviruses to circumvent species‐specific barriers and infect human cells. 24 , 25 Structural analysis of the S‐protein through cryogenic electron microscopy has revealed that it is a trimeric protein, with each ‐mer containing two functional subunits namely, S1 and S2. The S1 subunit is necessary for recognition of receptors on the surface of a susceptible host cell while the S2 subunit is responsible for fusion of the virus with the cell membrane of the host cell. The S1 subunit also contains an important region called RBD that helps S‐protein in binding with ACE2. 26 The RBD domain contains all the information necessary for host receptor binding as well as folding independently from the rest of the viral S‐protein. The distal S1 domain is cleaved when it binds with the hACE2 receptor. 27
TMPRSS2 is a serine protease that is vital for the SARS‐CoV‐2 infectivity because it causes proteolytic activation and intake of SARS‐CoV‐2 into the host cells. It has been shown that a higher number of SARS‐CoV‐2 viral particles are isolated from the TMPRSS2 expressing cells than non‐expressing cells. 28 Moreover, respiratory cells lacking TMPRSS2 protease are shown to have reduced lung pathology following SARS‐CoV and MERS‐CoV infections. This has been demonstrated by infecting TMPRSS2 knockout and TMPRSS2 proficient mouse models (wild type or WT) with both coronaviruses. The WT batch of mice lost considerable body weight and had observable damaged lungs few days after being infected with SARS‐CoV and MERS‐CoV. These symptoms were not observed, postinfection, in mutant mice. Moreover, as the SARS‐CoV‐2 infection in the lungs results in lung damage followed by a local immune response characterized by the release of cytokines and chemokines, 29 immunological assays of these mutant mice also showed lower lung immunopathology and weakened cytokine and or chemokine mediated immune response as compared with the mutants. 17 This study provided interesting data, which, if proven for humans, could help explain the reasons for higher susceptibility of some population groups to show poor prognosis for COVID‐19 compared with other less susceptible groups. In this review, we explored the currently available data on the role of TMPRSS2 protease in SARS‐CoV‐2 infection and differences in TMPRSS2 gene expression across various population groups, and the associated susceptibility to SARS‐CoV‐2 disease outcomes.
1.1.1. TMPRSS2 protein structure and multiorgan expression
The TMPRSS2 gene located on chromosome 21 expresses a 492 amino acid long cell surface protein of the same name that contains four different domains which include the serine protease (SP) domain, scavenger receptor cysteine‐rich (SRCR) domain, LDL receptor class A (LDLRA) domain and transmembrane (TM) domain. 30 The protein can be divided into a catalytic chain containing and noncatalytic chain containing parts. In the catalytic chain, the amino acid residues HIS 296, ASP 345, and SER 441 provide the catalytic triad binding site, which is involved in catalysis (Figure 1). These amino acid (aa) residues are also involved in the catalytic cleavage of the SARS‐CoV‐2 S‐protein that results in entry of the virus into a host cell. Hence, inhibitors that target these aa residues can be promising drug candidates. 31 The structure of TMPRSS2 including the positions of noncatalytic and catalytic domains, protein domains, and catalytic aa residues is shown in Figure 1.
Figure 1.
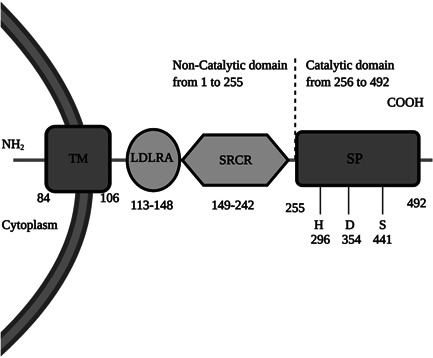
Functional domains of TMPRSS2 protein and their location in the protein sequence. TMPRSS2 contains the noncatalytic chain(1–255) and catalytic chain (256–492). The catalytic chain contains HIS 296, ASP 345, and SER 441 residues, which function as the binding and catalytic sites. ASP (D), HIS (H), LDL receptor class A (LDLRA) domain, scavenger receptor cysteine‐rich (SRCR) domain, SER (S), serine protease (SP) domain, and transmembrane (TM) domain
Serine proteases are usually involved in protein cleavage and are thus important in various physiological processes such as blood coagulation, digestion, tissue remodeling, and programmed cell death. 32 , 33 The TMPRSS2 gene is highly expressed in androgen‐rich environments. 34 Androgen is a hormone that is responsible for the development of male sex characteristics in all vertebrates. 35 Besides androgens, TMPRSS2 activity is also upregulated in the presence of other steroid hormones such as estrogen and glucocorticoids. 36 , 37 TMPRSS2 gene is expressed in a variety of organs, albeit at different levels. The highest level of expression of the gene is reported in the prostate region while lungs also have significantly high expression levels. 38 This high expression could be explained in terms of the fact that TMPRSS2 contains androgen‐regulated elements (AREs) located upstream of transcription start site and first intron. 34 In androgen‐regulated prostate cancer cells, TMPRSS2 is upregulated by androgen hormones while the opposite is true for androgen‐independent prostate cancer cells. Besides this, TMPRSS2 also regulates inflammation by proteolytically activating the protease‐activated receptor‐2 (PAR‐2) in the prostate, whereas in the lungs, TMPRSS2 controls sodium currents in the epithelium by proteolytically cleaving epithelial sodium channels. 39 Still, the exact function of TMPRSS2 protein is not fully understood because the TMPRSS2 gene deletion in mice does not affect their normal growth. 40
1.1.2. SARS‐CoV‐2 respiratory invasion
In humans, SARS‐CoV‐2 primarily enters the respiratory tract to initiate infection. The main route of entry of SARS‐CoV‐2 into the human body is through the nasal epithelium as evidenced by recovery of high virus titers from both symptomatic and asymptomatic patients. 41 Therefore, SARS‐CoV‐2 can spread through both symptomatic as well as asymptomatic individuals. 42 Person‐to‐person viral transmission occurs through respiratory droplets causing invasion of the upper respiratory tract by SARS‐CoV‐2. 43 The host binding receptor ACE2 and the serine protease TMPRSS2 responsible for priming are localized on the surface membrane of nasal epithelial cells. Due to viral attachment, nasal swab samples collected from the upper respiratory tract can be used to detect the virus via an RT‐PCR, 44 and nasal epithelium yields the highest CT values for viral RNA in this region. 41 The bronchial pathway which the virus uses to reach the lungs contain ACE2 and TMPRSS2 protein receptors in the epithelium. 45 The lower respiratory tract, which the virus employs for replication also expresses ACE2 and TMPRSS2, mainly in the alveolar epithelium Type II cells. The viral replication in this region of the respiratory tract can lead to severe conditions such as pneumonia and acute respiratory distress syndrome or ARDS. 46
Both ACE2 receptor and TMPRSS2 proteins are also expressed in a variety of cells in organs besides the respiratory tract such as cholangiocytes, colonocytes, esophageal keratinocytes, gastrointestinal epithelial cells, pancreatic β‐cells, renal proximal tubules, and so on. This broad‐spectrum infectivity explains the ability of the virus to damage organs beyond the immediate exposure of lungs, however, the mechanism by which SARS‐CoV‐2 infects other organs besides lungs is poorly understood. 47
1.1.3. TMPRSS2 protease priming of the SARS‐CoV‐2 S‐protein
Binding of RBD with ACE2 serves as the entry point for the initiation of viral infection. The high binding affinity of S‐protein of SARS‐CoV‐2 with the ACE2 receptor can partly explain the high infection and transmissibility rate than those of the earlier similar coronavirus known as SARS‐CoV. 43 , 48 , 49 Despite high binding affinity, the RBD domain of the viral S‐protein is less than optimum for binding with the host ACE2 receptor compared to the earlier SARS‐CoV 7 suggesting that SARS‐CoV‐2 employs some other technique to bypass this problem. The explanation offered for this high infectivity despite a low affinity for ACE2 protein is that the virus might have evolved through natural selection for optimum ACE2 receptor binding 50 as the high plasticity of the genome allows coronaviruses to adapt specifically to a host. 51 Binding of SARS‐CoV‐2 S‐ protein with ACE2 receptor is then followed by host TMPRSS2‐mediated cleavage of the viral S‐protein. The processing known as priming involves cleavage of the S‐protein at S1/S2 and S2 sites which is essential for the viral fusion with the host cell membrane before entry into the cell. 16 , 28 Besides the TMPRSS2 protein, it has been suggested that SARS‐CoV‐2 can use other proteases such as cathepsin B/L for S‐protein in the absence of TMPRSS2 receptors. However, in the lungs (the primary organ for SARS‐CoV‐2 infection), cathepsin B/L cannot substitute for TMPRSS2 protease activity as the latter is indispensable for viral entry as observed for SARS‐CoV and MERS‐CoV. 52
1.1.4. Temporary loss of smell (anosmia) in COVID‐19 linked to TMPRSS2
One of the most common and distinguishing features of COVID‐19 compared with other viral diseases is the temporary loss of sense of smell, known as anosmia, in many but not all of the symptomatic and asymptomatic patients. 53 , 54 Anosmia could be hypothesized to be linked to the damage caused by SARS‐CoV‐2 to the epithelial layer and subsequent inflammation or impairment of the receptor neurons in the olfactory organ. The latter condition is of particular concern as SARS‐CoV has been found to initially infect olfactory receptors and then proceed onto infecting brains of transgenic mice, bred to express human ACE2 proteins. 55 If the same is true for SARS‐CoV‐2 then the olfactory epithelium (OE) should comprise cells that express ACE2 and TMPRSS2, which should facilitate the viral infection. 56 , 57 To address this, Bilinska et al., 58 analyzed olfactory epithelial cells of mice for expression of ACE2 and TMPRSS2 by RNA sequencing, RT‐PCR analysis, in situ hybridization, Western blot, and immunocytochemistry assays. The researchers found expression of both proteins in the sustentacular OE cells, however, for olfactory receptor neurons (ORNs), ACE2 expression was not detected while TMPRSS2 was only expressed at low levels in mature ORNs only. The preferential target of SARS‐CoV‐2 for sustentacular cells leads to infection and buildup of the viral cells that interfere with the metabolism of these cells. This disturbance in the normal functioning of sustentacular cells could provide an explanation for the loss of olfaction as these cells are important for olfaction because of their role in secretion of odor binding proteins and endocytosis of olfactory binding protein‐odorant complex. 59 This does not however explain whether SARS‐CoV‐2 also the ability has to target brain cells. Further studies are needed to explain whether SARS‐CoV‐2 has the ability to infect ORNs on route to infecting the brain. 58 The investigators, Bilinska et al., 58 reported that the murine OE contains a comparatively denser cluster of ACE2 proteins than respiratory epithelium cells do, which could make OE more susceptible to SARS‐CoV‐2 infection than the respiratory epithelial cells. This is an important finding, which could be replicated in humans to compare levels of ACE2 as well as TMPRSS2 proteins in human OE and respiratory epithelium. If the same is true for human OE as well as for the murine OE, then detecting SARS‐CoV‐2 viral particles in the OE could be more reliable than respiratory epithelium which helps reduce the rate of false‐negative COVID‐19 test results.
1.1.5. Role of TMPRSS2 in previous viral outbreaks
The role of the TMPRSS2 gene in previous major pathogenic viral outbreaks or epidemics of the 21st century is well documented. 60 , 61 The H1N1 influenza pandemic that started in 1918 was among the deadliest epidemics in recorded human history that killed between 50 and 100 million people. 62 Though finding an association between TMPRSS2 and 1918 H1N1 was unlikely to occur at the time of the viral pandemic, it was made possible through a strategic revival of the H1N1 virus and examining the role of TMPRSS2 by infecting TMPRSS2 deficient and proficient mice with the 1918 H1N1 virus. 63 The later outbreaks/epidemics, though being responsible for a far smaller number of causalities, still impacted a significant number of people around the globe. The first major 21st century viral epidemic also caused by a coronavirus termed as severe acute respiratory syndrome virus (SARS‐CoV) was first reported in Foshan city in Guangdong province, China on November 16, 2002. 64 The second major viral outbreak that later became a pandemic caused by H1N1 influenza A virus first emerged in Mexico in April of 2009. 65 , 66 Next, in June 2012, another novel coronavirus later termed as the Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) emerged in Jeddah, Saudi Arabia. 67 Again, in March 2013, another influenza virus, H7N9 emerged in Shanghai and Anhui cities in China. 68 Both viruses that caused these major outbreaks or epidemics can be divided into influenza viruses and coronaviruses. Both H1N1 and H7N9 are influenza‐A viruses that contain hemagglutinin (HA) and neuraminidase (NA) proteins that are essential for host entry and also serve as the basis of classification for these viruses. 69 The HA protein mediates binding of the virus to the sialic acid (SA) residues of the host cell terminus while the NA protein is responsible for the viral release from the SA residues thereby beginning the infection in host cells. 70 SARS‐CoV and MERS‐CoV on the other hand use different proteins and receptors for host cell entry. SARS‐CoV uses spike S‐protein to bind with ACE2 receptor while S‐protein of MERS‐CoV targets dipeptidyl peptidase‐4 (DPP4) for host cell attachment. 71 , 72 In mice models, infection with H1N1 and H7N9 has caused a mortality rate of 20% and 100‐, respectively, while TMPRSS2 knockout has been shown to render H1N1 and H7N9 viruses apathogenic thus, suggesting primal importance of TMPRSS2 in activation of the viruses’ HA protein that leads to viral infection. 63 TMPRSS2 (+) positive cell lines have shown a 100‐fold increase in susceptibility to MERS‐CoV infection than TMRPSS2 (−) negative cell lines. Moreover, treatment of TMPRSS2 containing cell lines treated with camostat (serine protease inhibitor) completely blocks the ability of MERS‐CoV to infect these cells which explains the dependency of MERS‐CoV on TMPRSS2 protease for its infectivity. 73 SARS‐CoV entry into host cells is dependent on cathepsin‐L in addition to TMPRSS2 priming, and the inactivation of these receptors is necessary for complete blockage of SARS‐CoV entry. 74
1.2. Differential TMPRSS2 lung expression and COVID‐19 susceptibility among different population groups
The previous section dealt with a detailed explanation of the SARS‐CoV‐2 infection mechanisms and the role of TMPRSS2 protease in infection. This section deals with the various susceptibility factors noticed among population groups that are associated with differential expression of TMPRSS2.
1.2.1. Age as a susceptibility factor
Progression of COVID‐19 pandemic and hospitalization and mortality rate is highly correlated with population age structure. In the United States, the hospitalization rate due to COVID‐19 has been recorded and published by COVID‐NET, a COVID‐19 case surveillance and data collection program. According to COVID‐NET, the hospitalization rate due to COVID‐19 increases with increasing age. For example, the hospitalization rate for Americans between the age 0–4 was 0.3%, for 50–64 years old, it was 7.4% and 13.8% in individuals ≥65 years of age as of March 30, 2020. 75 Old age also increases the mortality risk for COVID‐19, particularly for those individuals who are aged 80 and above compared with younger members of a population. 76 Indeed, case fatality rate (CFR) estimates from China and Italy have shown that being old increases the risk of dying from COVID‐19 manifold. 77 In China, CFR for 40–49 years old individuals was 0.4% while for those aged above 80 was 14.8% as of February 11, 2020. 78 In Italy, the CFR values estimated until March 30, 2020 were 0.7% for individuals aged between 40 and 49 years while it was 22.7% for individuals aged 80 and above. 79 In the United States, between February 12 and March 16, 2020, about 80% of the COVID‐19 related deaths occurred in individuals who were 65 years or older. 80 Table 1 describes CFR for different age groups in some countries.
Table 1.
Case fatality ratio (CFR) among different population age groups in the named countries
In the case of MERS‐CoV, it was reported that juveniles have higher chances of contracting the virus than adults. 83 For SARS‐CoV‐2, data on infection rates among different age groups is not known. However, down the age group, we see a steady decrease in the number of SARS‐CoV‐2 morbidity and mortality rates as children and infants represent one of the least affected among all population groups. This has been evidenced by the decreased number of children and infants admitted to hospitals due to COVID‐19 related symptoms. 84 On the other hand, younger age groups such as infants and children are highly prone to be admitted into a hospital after viral respiratory infections caused by influenza or syncytial virus which could be associated with an immature immune system, 85 suggesting that there must be other factors at play for the higher probability of worse outcomes in the upper age group.
1.2.1.1. TMPRSS2 expression and its contribution to age susceptibility to COVID‐19
One of the reasons for the decreased infection rate among children and adults as compared to the elderly could be due to decreased expression of ACE2 and TMPRSS2 in organs for which the SARS‐CoV‐2 has a natural tropism in children and adults as compared with the elderly. An experiment in murine models has shown that ACE2 and TMPRSS2 expression in the OE increases with age. 57 OE is a part of the nasal cavity, which then is a component of the upper respiratory tract, 86 acting as the primary route for SARS‐CoV‐2 infection. 45 A preprint study by Schuler et al., 87 has shown that the TMPRSS2 expression in the lung epithelium also increases with age. In the study, lung epithelial cells studied for TMPRSS2 expression included ciliated cells, secretory cells, and alveolar type (AT) 1 and 2 cells while the age groups included infants (up to 2 years old), children (aged between 3 and 17) and adults (aged 54–69) and the results showed that except for AT2 cell types, a significant difference in all other cells was found in the order; infants < children < adults. The study also showed that AT1 and ciliated cells tend to have higher TMPRSS2 expression levels and SARS‐CoV‐2 viral load in severely affected patients while in children and infants, these cells have a lower expression of TMPRSS2 indicating that low levels of TMPRSS2 in the latter two groups have protective effects. In contrast, an earlier report from China highlighted an equal SARS‐CoV‐2 infection risk among children and adults. 88 However, children remain undiagnosed due to the subclinical nature of SARS‐CoV‐2 infection among individuals from this age group which favors the underrepresentation of this group in SARS‐CoV‐2 infection statistics. 84 , 88
1.2.2. Population ethnicity as a susceptibility factor of TMPRSS2
Epidemiological data of COVID‐19 suggests some countries and regions being badly affected by COVID‐19 while others are doing much better. 5 Large genetic studies around the world point to the differences in allele frequencies and protein expression of various genes in geographically distinct groups of people. 89 Based on the TMPRSS2 cumulative genetic expression score (GES), African populations have the lowest observed TMPRSS2 expression levels while East Asians and Admixed Americans show the highest TMPRSS2 expression profile. 90 However, this represents an overall expression profile of TMPRSS2 in different populations while the TMPRSS2 expression profile also varies considerably in lungs as we discuss TMPRSS2 expression in different populations. An increase in the expression of TMPRSS2 in the lungs is expected to increase S‐protein priming, which should thereby make individuals (with higher expression of TMPRSS2) more susceptible to SARS‐CoV‐2 infection and more prone to suffer from worse COVID‐19 outcomes. 91 A comparison of the differences in TMPRSS2 expression among different ethnic populations (including male and female) is shown in Table 2.
Table 2.
Difference in body TMPRSS2 expression level difference between different ethnicities 90
| Ethnic population | TMPRSS2 expression difference (male) | TMPRSS2 expression difference (female) |
|---|---|---|
| Africans versus Europeans | p < .0001 | p < .0001 |
| Africans versus Americans | p < .0001 | p < .0001 |
| Africans versus East Asians | p < .0001 | p < .0001 |
| Africans versus South Asians | p < .0001 | p < .0001 |
| Europeans versus Americans | p = .084 | p = .97 |
| Europeans versus East Asians | p = .03 | p = .95 |
| Europeans versus South Asians | p = 1 | p < .93 |
| Americans versus East Asians | p = 1 | p = 1 |
| Americans versus South Asians | p = .053 | p = .66 |
| East Asians versus South Asians | p = .015 | p = .55 |
In a recent study by Irham et al., 91 lung expression of TMPRSS2 gene was assessed in population groups from different continents. European and American populations were shown to have higher expression levels of TMPRSS2 than East Asian populations. Notably, the researchers found four TMPRSS2 variants to be associated with higher expression of the gene, and these variants responsible for upregulation of TMPRSS2 were found in higher frequency in European and American populations while the lowest frequency of these variants was found in East Asian populations. Earlier in two distinct patient cohorts, two of these identified variants (rs2070788 and rs383510) made patients more susceptible to infection by influenza virus infection A(H7N9). 92 Another study by Russo et al., 93 identified intergenic lung tissue variant of eQTL (a locus that explains a fraction of the genetic variance of a gene expression phenotype), that is, rs35074065 that is responsible for upregulation of TMPRSS2 and downregulation of the MX‐1 (Interferon‐induced GTP‐binding protein) gene. So, individuals having this eQTL variant are expected to have increased vulnerability to SARS‐CoV‐2 infection and decreased cellular immune response against the virus. In the study, the researchers reported the lowest frequency of this eQTL variant among East Asians while the highest frequency was found in Ashkenazi Jews among the total analyzed population groups (Table 3). Asselta et al., 94 also found rare alleles of two haplotypes associated with higher expression of TMPRSS2 in lungs to be present in higher frequency in the Italian cohort than the East Asian population against whom they were compared. It is also noteworthy to mention here that one of the allele variants of a haplotype responsible for higher TMPRSS2 expression is associated with increased risk to Influenza A virus H7N9 and H1N1 subtype infection and severe outcomes, respectively. 92 Following are some of the susceptibility factors of COVID‐19 disease that encompass TMPRSS2.
Table 3.
The gnomAD database (based on WGS data) annotated allele frequency of eQTL variant rs35074065 in different population groups 93
| Population | Allele frequency |
|---|---|
| Africans/African Americans | 0.12 |
| Latino/Admixed Americans | 0.26 |
| Ashkenazi Jews | 0.47 |
| East Asians | 0.0 |
| Unassigned populations | 0.34 |
| Finnish | 0.34 |
| Non‐Finnish Europeans | 0.45 |
| Southern Europeans | 0.45 |
| North‐Western Europeans | 0.46 |
| Estonians | 0.42 |
1.2.3. TMPRSS2 and gender as a susceptibility factor of SARS‐CoV‐2
According to the sex, gender, and COVID‐19 project, a database for COVI‐19 disaggregation between sexes in the world population, show that more men, irrespective of age, die from COVID‐19 than women, 95 a common factor observed for earlier viral infections such as SARS‐CoV and MERS‐CoV. 96 , 97 The discordancy in death rate is the opposite in India which appears to be an exception to the general trend around the world with more women dying from COVID‐19 than men. 98 If a general trend of the mortality rate is taken into account, then the question arises as to why more men die from COVID‐19 than women? One probable reason for higher infection rates among men could be increased exposure to SARS‐CoV‐2 as more males work outside than females. 99 Another possible explanation for the discrepancy in mortality rate could be the different levels of ACE2 and TMPRSS2 expression between males and females. Although a preprint study conducted on exome and SNP array sample data representing Italy's population found ACE2 to have no association with higher mortality rates among men, differences in expression of TMPRSS2 in bronchial epithelial cells found between males and females could explain severe COVID‐19 outcomes in men. 94 In the study, males were found to express higher levels of TMPRSS2 in bronchial epithelium cells as compared with age‐adjusted females, while TMPRSS2 expression in the lung samples of both sexes exhibited no difference. These findings are interesting as the bronchial epithelium has been proposed to be a preferential target site for SARS‐CoV‐2 infection in humans. Secretory cells in the bronchial epithelium, in particular, are shown to primarily express ACE2 and TMPRSS2. 100 Severe outcomes of SARS‐CoV‐2 infection are related to the alveolar damage caused due to the viral invasion of ciliated host epithelial cells in the bronchi and the bronchioles as well as Type I and II pneumocytes. 101 A preprint study by Song et al., 102 found significantly higher expression of ACE2 and TMPRSS2 in Type II pneumocytes in males as compared with females.
1.2.3.1. Smoking as a contributing factor for higher TMPRSS2 expression in men
It is known that smoking increases TMPRSS2 expression and compared with women, men are more active smokers. 103 , 104 It has been shown that smokers are 1.4 times more likely to be affected by severe COVID‐19 outcomes and 2.4 times more prone to be admitted to ICU due to COVID‐19 than non‐smokers. 105 Analysis of lung expression data in publicly available expression database, that is, Gene Expression Omnibus (GEO) has revealed that smoking increases pulmonary TMPRSS2 expression significantly, however, interestingly, the expression returns to the nonsmoker level when smoking is stopped. This shows that the effect of smoking does not cause a buildup of TMPRSS2 expression but causes a rapid increase, which is more like a switch than a gradual process. 106 Transcriptomic analysis of lung tissue also revealed that the TMPRSS2 expression significantly increases (p = .0002) in smokers as compared to non‐smokers. 104 Smoking as a risk factor for severe COVID‐19 outcomes is however debatable as smokers in China, Korea, and the United States have been shown to be underrepresented among severely affected individuals from COVID‐19. 107
1.2.3.2. Blood androgen‐dependent TMPRSS2 expression as a susceptibility factor
In another study published, 90 differences in TMPRSS2 expression between male and female members, belonging to different ethnic groups were not significant. Although some studies 90 , 94 that point at differences in TMPRSS2 expression might seem to be at odds with each other, the methodology employed was different. In the first study, researchers looked for TMPRSS2 expression localized in lung and bronchial epithelial cells while in the latter, the researchers reported an unlocalized TMPRSS2 expression level in the whole body. Asselta et al. 94 also found the Italian population to express a TMPRSS2 haplotype that is regulated by androgens and is involved in the upregulation of TMPRSS2, so the presence of this haplotype can explain the higher expression associated with poor prognosis among Italian men. 94 The role of TMPRSS2 in prostate cancer is well established, 108 and the cell lines derived from human lungs are regulated by androgens and glucocorticoids and androgen exposure increases TMPRSS2 expression. 109
Contrary to the observed differences in TMPRSS2 expression in males and females by Asselta et al., 94 for the Italian population cohort, Baratchian et al., 106 did not find a statistically different expression of TMPRSS2 between males and females in publicly available lung expression profiles including bronchial epithelia. The observed differences could be attributed to differences in usage of publicly available expression databases and methodologies. Asselta et al. 94 took expression data for ACE2 and TMPRSS2 genes of lungs and bronchial epithelia from the GTEx database (https://gtexportal.org/home/) and GEO repository (https://www.ncbi.nlm.nih.gov/geo/) while genetic data related to the SNPs of both genes were retrieved from the GnomAD repository (https://gnomad.broadinstitute.org/) and compared allele variations and gene expression levels between Italians (men and women) and east Asians. On the other hand, Baratchian et al. 106 obtained publicly available TMPRSS2 expression profiles of lungs and bronchial epithelium from the GEO database only. Table 4 shows the differences in both study methodologies and the outcomes obtained.
Table 4.
Differences in the methodology and results obtained
| Methodology | GEO datasets used | Result | References |
|---|---|---|---|
| GTEX and GEO repository | GSE66499 and GSE19804 | Men have significantly higher expression in Bronchial Epithelium (p = .029) but not in lungs | [94] |
| Only GEO | GSE103174, GSE123352, GSE16008, GSE18385, GSE18385, GSE37147, GSE4115, and GSE43696 | No difference in TMPRSS2 expression in lungs and Bronchial Epithelium | [106] |
1.3. Treatment options for COVID‐19 based on targeting TMPRSS2 expression
Considering the primal importance of TMPRSS2 protease in SARS‐CoV‐2 infection and COVID‐19 outcome, designing therapeutic targets of TMPRSS2 protease has been considered. The following section describes some of the therapeutic agents that could be used for COVID‐19 treatment based on decreasing or inhibiting TMPRSS2 expression.
1.3.1. Inhibition of TMPRSS2 androgen exposure as a potential COVID‐19 treatment strategy
Much of the knowledge regarding the role of the TMPRSS2 gene comes from the field of cancer biology. The gene is a diagnostic marker for prostate cancers as it is upregulated while being frequently involved in translocation with the ERG gene. 110 TMPRSS2 contains an androgen receptor (AR), which is involved in transcriptional regulation through binding with androgen hormone. 111 Being androgen‐regulated, androgen deprivation therapy (ADT) is regularly used in cancer patients 112 which can impart its effect by decreasing TMPRSS2 expression. 113 Besides prostate cancer, increasing exposure with androgen has been reported to increase TMPRSS2 expression in the lungs. 114 Building on these premises, it seems logical to hypothesize that inhibiting androgen exposure in lungs should suppress TMPRSS2 activity. For such purposes, AR expression antagonists, AR coregulatory factors, and transcription factors such as ETS1 and SP1, and so on, could be used. 115 Apart from similarities in the TMPRSS2 gene role, the two diseases also share commonalities in risk factors that increase the likelihood of developing severe disease outcomes. Other risk factors of co‐morbidity include alcoholism, diabetes, and hypertension; behavioral traits such as smoking and age >50 years. 116
Recently, it has been suggested that ADT used for prostate cancer patients could also be used in SARS‐CoV‐2 patients. 115 Compounds that are currently used as ADT include blockers of androgen receptors such as bicalutamide and enzalutamide, anti‐gonadotropins and inhibitors of androgen synthesis. 117 ADTs such as estradiol, enzalutamide, genistein, and phytoestrogens have been shown to decrease expression of TMPRSS2 by 1.6‐ to 14‐fold, depending on the cell lines and experimental conditions, and so on, used in these studies. On the other hand, androgen exposure has been found to increase TMPRSS2 expression by 1.4‐ to 20‐fold. 108 Montopoli et al. 115 found prostate cancer patients receiving ADT to be a lower risk of contracting SARS‐CoV‐2 infection. In this in silico study, the researchers analyzed metadata of 9280 SARS‐CoV‐2 infected patients in Veneto, Italy, and found that prostate cancer was associated with increased chances of hospitalization, admission into ICU, and the overall death rate from COVID‐19. However, the increased percentage can also be attributed to the greater testing rate for COVID‐19 in prostate cancer patients as these patients are more frequently hospitalized than the healthy population which raises the issue of sample bias. The researchers then analyzed a sample of ADT (+plus) and ADT (−minus) prostate cancer patients and found that a significantly smaller number of ADT receiving patients contracted SARS‐CoV‐2 infection (only 4 out of 5273 ADT receiving patients). The study provides an important insight into the potential new treatment option for COVID‐19 patients that if successful will help a significant population around the world. Androgens being involved in the regulation of TMPRSS2, are also implicated in severe immune reactions by increasing production of interleukins and transforming growth factor‐β while decreasing antibody production in response to viral infections. 118 The positive association between higher androgen levels and neutrophils produced can also be a cause of cytokine storm that can worsen COVD‐19 related outcomes. 118 , 119 Contrary to the hypothesis and observed association found by Montpoli et al., 115 using the androgen hormone antagonist enzalutamide did not result in a significant decrease of TMPRSS2 expression in the lungs of mice models. 106 Moreover, a study done in Veneto, Italy found no evidence for the role of ADT in protecting patients from the poor prognosis of COVID‐19, as 25% of prostate cancer patients died from COVID‐19 than the 13% average infected Italian males. 120 One of the reasons for the huge differences in results could likely be due to different levels of social distancing practices observed by the two research groups in Veneto, Italy. Moreover, Montopoli et al., 115 did not define the stage of pancreatic cancer in patients that were investigated, while Caffo et al., 120 studied a total of 1949 pancreatic cancer patients all of whom were in the advanced stage of their disease. It is likely that differences in stages of pancreatic cancer could have significantly produced divergent observations made by the two scientists. 120
1.3.2. Therapeutic strategies based on TMPRSS2 protease inhibition
Besides ADT, there have been other treatment options suggested for inhibiting or decreasing the TMPRSS2 expression as a therapeutic target. There are various publications that have indicated a plethora of TMPSS2 inhibitors and among these inhibitors, one of the most promising inhibitors that could be used for COVID‐19 treatment is Camostat Mesylate (CM). The inhibitor has been shown to protect mice from death after lethal injection with SARS‐CoV. In Japan, CM is already being used as a treatment drug, for diseases other than COVID‐19. 93 When mice are given the same concentration of CM as required for humans, the mortality rate has been shown to be reduced by 70%–65%. 121 In Caco‐2 and Vero cell lines, treatment with CM has shown partial inhibition of SARS‐CoV‐2 entry into the respective cell lines. However, when CM is used in combination with the CatB/L inhibitor called E‐64d for the same cell lines, complete viral inhibition is attained, suggesting a combinational approach that acts synergistically in blocking SARS‐CoV‐2 entry completely. 50 , 122 Another compound that is of interest for TMPRSS2 expression inhibition is Bromohexine. 123 The same compound has already been recommended for the treatment of SARS‐CoV and MERS‐CoV infections. 60 In the bronchial and pulmonary epithelial cells, the concentration of bromhexine can become four to six times higher than the in plasma, which is sufficient to suppress TMPRSS2 activity. 123 The compound is also being investigated for treatment of cough and chest congestion associated with pneumonia caused by COVID‐19 in China. 124 Recently, Nafamostat mesylate (INN) has been shown to inhibit SARS‐CoV‐2 entry into the host Calu‐3 human cell lines with 15 times higher efficiency than the CM. 125 A molecular docking study by Sonawane et al., 126 has shown that the three abovementioned TMPRSS2 protein inhibitors strongly interact with the His296, Ser441, and Asp435 TMPRSS2 protein residues present in the catalytic region that results in the inhibitory action of these blockers. The authors also found that among these inhibitors, CM shows the strongest binding with TMPRSS2. Finally, Bestle et al., 127 used aprotinin and two synthetic TMPRSS2 inhibitors called MI‐432 and MI‐1900. Treatment with these inhibitors resulted in a maximum of 20‐ to 35‐, 5‐, and 25‐ to 70‐fold decrease of SARS‐CoV‐2 titers in Calu‐3 cell lines compared to uninfected Calu‐3 cells, respectively. Among the abovementioned TMPRSS2 inhibitors, only CM is currently being investigated as a treatment option for COVID‐19 in a clinical trial (National Library of Medicine, 2020 [NLM] NCT04321096). So, to increase the number of treatment options, the TMPRSS2 inhibitors described in the section should be examined in clinical trials (Figure 2).
Figure 2.
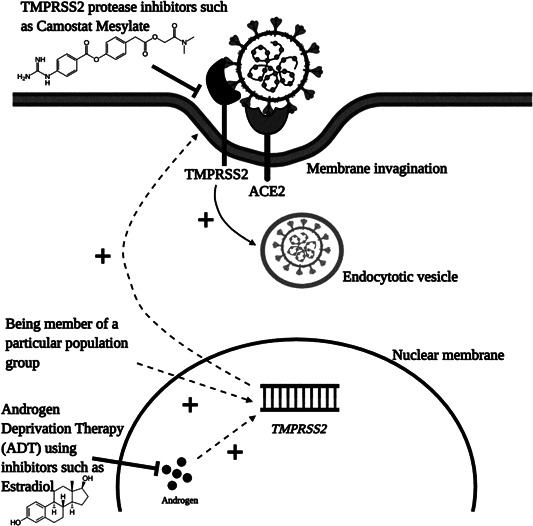
Role of TMPRSS2 protease in SARS‐CoV‐2 in infection. After binding of the SARS‐CoV‐2 S‐protein with the ACE2 receptor, the S‐protein is primed by TMPRSS2 protease that results in the viral entry. Plus (+) sign indicates all the factors that increase TMPRSS2 protease expression and SARS‐CoV‐2 priming. Inhibitor (‐‐‐) sign indicates therapeutic targets that can have inhibitory action against TMPRSS2 protease
2. SUMMARY
The discrepancy in COVID‐19 related severe health deterioration and death rate can be due to many factors including socioeconomic and biological differences. Among the biological differences, TMPRSS2, a gene whose role in prostate cancer is well established is being also investigated for its involvement in the ongoing pandemic, that is, COVID‐19. SARS‐CoV‐2 is dependent upon the expression of this gene as it causes priming of the viral S‐protein, allowing it to enter the host cells. The gene is naturally expressed in the nasal epithelium (entry site of SARS‐CoV‐2) and in the known target organ of the virus (different cell types in lungs). This, together with the studies showing that higher SARS‐CoV‐2 load is obtained from host cells expressing higher levels of TMPRSS2 prompted us to investigate the matter further. In the review, we have analyzed current data on how differences in lung expression of TMPRSS2 among different human population groups affect COVID‐19 outcomes. Indeed, there are studies that link the higher expression of TMPRSS2 with an increase in hospitalization and death rates. However, much of the work done in this area is reporting the association between differential TMPRSS2 expression in different population groups with differences in susceptibility to SARS‐CoV‐2 infection and COVID‐19 disease‐related outcomes. These association studies include analysis of SNP data profiling, TMPRSS2 gene expression data profiling, murine SARS‐CoV‐2 lung infection, and a small number of human lung tissue samples. Though promising, further research is still needed because the studies establish only a correlation rather than evidence. So, it is the need of the hour to elucidate more on the matter and establish a conclusive link between the two. This will help in distinguishing more susceptible population groups from less susceptible, which, therefore, can guide us in designing better treatment strategies in the absence of effective drug or vaccine treatment, the situation in the world as of writing this review. In this context, therapeutic agents targeting TMPRSS2 expression have also been described. Among these, camostat mesylate (CM) is the only inhibitor that is currently being examined for its efficacy in treating COVID‐19 in a clinical trial. There is a need to increase the number of treatment options and so, other TMPRSS2 protease inhibitors like bromhexine, INN, and aprotinin, and so on, should also be investigated in clinical trials. Like basic research about the role of TMPRSS2 in COVID‐19, further research is needed to substantiate the utility of TMPRSS2 inhibitors for treating COVID‐19 patients.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Asim Z. Abbasicontributed up to 50% of the writing content. The main text of the manuscript, illustrations, and legends were prepared. Dania A. Kiyani contributed intellectually up to 10% through discussions, provided details for the gene databases, and helped with references. Syeda M. Hamid also contributed intellectually up to 10% through discussions and helped in the layout and restructuring of the review. Muhammad Saalim contributed up to 10% through proofreading, restructuring, and language proficiency. Ammad Fahim contributed up to 10% through proofreading, restructuring, and provided valuable suggestions regarding the direction of the manuscript. Nasir Jalal proposed the initial idea along with main headings, hypothesis building, and a structured methodical approach to address the question. His percentage contribution was 10%–15% through in‐house peer review, proofreading, the concept for illustrations and legends, and overall guidance throughout the project.
PEER REVIEW STATEMENT
The peer review history for this article is available at https://publons.com/publon/10.1002/jmv.26911
Abbasi AZ, Kiyani DA, Hamid SM, Saalim M, Fahim A, Jalal N. Spiking dependence of SARS‐CoV‐2 pathogenicity on TMPRSS2. J Med Virol. 2021;93:4205–4218. 10.1002/jmv.26911
REFERENCES
- 1. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y‐Z. Novel 2019 coronavirus genome. 2020. http://virological.org/t/novel-2019-coronavirus-genome/319. Accessed January 20, 2021.
- 3. Chan JF‐W, Yuan S, Kok K‐H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y, Geng X, Tan Y, et al. New understanding of the damage of SARS‐CoV‐2 infection outside the respiratory system. Biomed Pharmacother. 2020;127:110195. 10.1016/j.biopha.2020.110195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johns Hopkins Coronavirus Resource Center . Johns Hopkins University of Medicine. COVID‐19 map. 2020. https://coronavirus.jhu.edu/map.html. Accessed January 1, 2021.
- 6. Ali I, Alharbi OML. COVID‐19: disease, management, treatment, and social impact. Sci Total Environ. 2020;728:138861. 10.1016/j.scitotenv.2020.138861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127‐20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khadke S, Ahmed N, Ahmed N, et al. Harnessing the immune system to overcome cytokine storm and reduce viral load in COVID‐19: a review of the phases of illness and therapeutic agents. Virol J. 2020;17(1):1‐18. 10.1186/s12985-020-01415-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1):313‐319. 10.1080/22221751.2020.1725399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kenyon C. Emergence of zoonoses such as COVID‐19 reveals the need for health sciences to embrace an explicit eco‐social conceptual framework of health and disease. Epidemics. 2020;33:100410. 10.1016/j.epidem.2020.100410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui J, Li F, Shi Z‐L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181‐192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brierley L, Fowler A. Predicting the animal hosts of coronaviruses from compositional biases of spike protein and whole genome sequences through machine learning. bioRxiv. 2020. 10.1101/2020.11.02.350439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolf YI, Kazlauskas D, Iranzo J, et al. Origins and evolution of the global RNA virome. mBio. 2018;9(6):e02329‐18. 10.1128/mBio.02329-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren L‐L, Wang Y‐M, Wu Z‐Q, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133:1015‐1024. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwata‐Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93(6):e01815‐e01818. 10.1128/JVI.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kain MP, Childs ML, Becker AD, Mordecai EA. Chopping the tail: how preventing superspreading can help to maintain COVID‐19 control. Epidemics. 2020:100430. 10.1016/j.epidem.2020.100430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roche B, Garchitorena A, Roiz D. The impact of lockdown strategies targeting age groups on the burden of COVID‐19 in France. Epidemics. 2020;33:100424. 10.1016/j.epidem.2020.100424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahdi M, Mótyán JA, Szojka ZI, Golda M, Miczi M, Tőzsér J. Analysis of the efficacy of HIV protease inhibitors against SARS‐CoV‐2's main protease. Virol J. 2020;17(1):1‐8. 10.1186/s12985-020-01457-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Woo PCY, Huang Y, Lau SKP, Yuen K‐Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804‐1820. 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holmes KV. Coronaviruses In: Fields virology. 4th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2001:1187‐1203. [Google Scholar]
- 23. Abebe EC, Dejenie TA, Shiferaw MY, Malik T. The newly emerged COVID‐19 disease: a systemic review. Virol J. 2020;17(1):1‐8. 10.1186/s12985-020-01418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562‐569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237‐261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang Y, Xiao Z, Ye K, et al. SARS‐CoV‐2: characteristics and current advances in research. Virol J. 2020;17(1):1‐17. 10.1186/s12985-020-01369-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science (80‐). 2005;309(5742):1864‐1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 28. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS‐CoV‐2 by TMPRSS2‐expressing cells. Proc Natl Acad Sci USA. 2020;117(13):7001‐7003. 10.1073/pnas.2002589117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS‐CoV‐2 infection. Mayo Clin Proc. 2020:1989‐1999. 10.1016/j.mayocp.2020.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paoloni‐Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics. 1997;44(3):309‐320. [DOI] [PubMed] [Google Scholar]
- 31. Roomi M, Khan Y. Potential compounds for the Inhibition of TMPRSS2. chemrxiv. 2020. [Google Scholar]
- 32. Nelson PS, Gan L, Ferguson C, et al. Molecular cloning and characterization of prostase, an androgen‐regulated serine protease with prostate‐restricted expression. Proc Natl Acad Sci USA. 1999;96(6):3114‐3119. 10.1073/pnas.96.6.3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rittenhouse HG, Finlay JA, Mikolajczyk SD, Partin AW. Human Kallikrein 2 (hK2) and prostate‐specific antigen (PSA): two closely related, but distinct, kallikreins in the prostate. Crit Rev Clin Lab Sci. 1998;35(4):275‐368. 10.1080/10408369891234219 [DOI] [PubMed] [Google Scholar]
- 34. Lin B, Ferguson C, White JT, et al. Prostate‐localized and androgen‐regulated expression of the membrane‐bound serine protease TMPRSS2. Cancer Res. 1999;59(17):4180‐4184. [PubMed] [Google Scholar]
- 35. Sriram D, Yogeeswari P. Steroids. Medicinal Chemistry. 2nd ed. India: Pearson Education; 2010:437. [Google Scholar]
- 36. Arora VK, Schenkein E, Murali R, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155(6):1309‐1322. 10.1016/j.cell.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Z, Wang Y, Xiao ZG, et al. Nuclear receptor ERRα and transcription factor ERG form a reciprocal loop in the regulation of TMPRSS2:ERG fusion gene in prostate cancer. Oncogene. 2018;37(48):6259‐6274. 10.1038/s41388-018-0409-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stopsack KH, Mucci LA, Antonarakis ES, Nelson PS, Kantoff PW. TMPRSS2 and COVID‐19: serendipity or opportunity for intervention? Cancer Discov. 2020;10(6):779‐782. 10.1158/2159-8290.CD-20-0451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park Y. TMPRSS2 (transmembrane protease, serine 2). Atlas Genet Cytogenet Oncol Haematol. 2010;14(12):1163‐1165. 10.4267/2042/44922 [DOI] [Google Scholar]
- 40. Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2‐encoded protease. Mol Cell Biol. 2006;26(3):965‐975. 10.1128/MCB.26.3.965-975.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou P, Yang X‐L, Wang X‐G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park SW, Cornforth DM, Dushoff J, Weitz JS. The time scale of asymptomatic transmission affects estimates of epidemic potential in the COVID‐19 outbreak. Epidemics. 2020;31:100392. 10.1016/j.epidem.2020.100392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell. 2020;181(4):894‐904. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang H, Lan Y, Yao X, Lin S, Xie B. The chest CT features of coronavirus disease 2019 (COVID‐19) in China: a meta‐analysis of 19 retrospective studies. Virol J. 2020;17(1):1‐17. 10.1186/s12985-020-01418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bertram S, Heurich A, Lavender H, et al. Influenza and SARS‐coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLOS One. 2012;7(4):e35876. 10.1371/journal.pone.0035876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sungnak W, Huang N, Bécavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681‐687. 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID‐19. Nat Med. 2020;26(7):1017‐1032. 10.1038/s41591-020-0968-3 [DOI] [PubMed] [Google Scholar]
- 48. Lei C, Qian K, Li T, et al. Neutralization of SARS‐CoV‐2 spike pseudotyped virus by recombinant ACE2‐Ig. Nat Commun. 2020;11(1):1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS‐CoV‐2. Nat Med. 2020;26(4):450‐452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus. Virol J. 2020;17:131. 10.1186/s12985-020-01402-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hoffmann M, Kleine‐Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Mol Cell. 2020;78:779‐784. 10.1016/j.molcel.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lechien JR, Chiesa‐Estomba CM, De Siati DR, et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild‐to‐moderate forms of the coronavirus disease (COVID‐19): a multicenter European study. Eur Arch Oto‐Rhino‐Laryngol. 2020;277:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moein ST, Hashemian SMR, Mansourafshar B, Khorram‐Tousi A, Tabarsi P, Doty RL. Smell dysfunction: a biomarker for COVID‐19. Int Forum Allergy Rhinol. 2020;10(8):944‐950. 10.1002/alr.22587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol. 2008;82(15):7264‐7275. 10.1128/JVI.00737-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Butowt R, Bilinska K. SARS‐CoV‐2: olfaction, brain infection, and the urgent need for clinical samples allowing earlier virus detection. ACS Chem Neurosci. 2020;11(9):1200‐1203. 10.1021/acschemneuro.0c00172 [DOI] [PubMed] [Google Scholar]
- 57. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single‐cell RNA‐seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019‐nCoV infection. Front Med. 2020:1‐8. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bilinska K, Jakubowska P, Von Bartheld CS, Butowt R. Expression of the SARS‐CoV‐2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Neurosci. 2020;11:1555‐1562. 10.1021/acschemneuro.0c00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Strotmann J, Breer H. Internalization of odorant‐binding proteins into the mouse olfactory epithelium. Histochem Cell Biol. 2011;136(3):357‐369. 10.1007/s00418-011-0850-y [DOI] [PubMed] [Google Scholar]
- 60. Chaipan C, Kobasa D, Bertram S, et al. Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol. 2009;83(7):3200‐3211. 10.1128/JVI.02205-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1‐10. 10.1016/j.biochi.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morens DM, Taubenberger JK, Harvey HA, Memoli MJ. The 1918 influenza pandemic: lessons for 2009 and the future. Crit Care Med. 2010;38(4 suppl):e10‐e20. 10.1097/CCM.0b013e3181ceb25b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tarnow C, Engels G, Arendt A, et al. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol. 2014;88(9):4744‐4751. 10.1128/JVI.03799-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Seto W, Tsang D, Yung R, et al. Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet. 2003;361(9368):1519‐1520. 10.1016/S0140-6736(03)13168-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kelly H, Peck HA, Laurie KL, Wu P, Nishiura H, Cowling BJ. The age‐specific cumulative incidence of infection with pandemic influenza H1N1 2009 was similar in various countries prior to vaccination. PLOS One. 2011;6(8):e21828. 10.1371/journal.pone.0021828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Perez‐Padilla R, De La Rosa‐zamboni D, Ponce de Leon S, et al. Pneumonia and respiratory failure from swine‐origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;361(7):680‐689. 10.1056/NEJMoa0904252 [DOI] [PubMed] [Google Scholar]
- 67. Chowell G, Blumberg S, Simonsen L, Miller MA, Viboud C. Synthesizing data and models for the spread of MERS‐CoV, 2013: key role of index cases and hospital transmission. Epidemics. 2014;9:40‐51. 10.1016/j.epidem.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gao R, Cao B, Hu Y, et al. Human infection with a novel avian‐origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888‐1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 69. CDC . Influenza type A viruses. Influenza (Flu). 2017. https://www.cdc.gov/flu/avianflu/influenza-a-virus-subtypes.htm/. Accessed January 13, 2021.
- 70. Kosik I, Yewdell JW. Influenza hemagglutinin and neuraminidase: Yin–Yang proteins coevolving to thwart immunity. Viruses. 2019;11(4):346. 10.3390/v11040346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci USA. 2020;117(21):11727‐11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang N, Shi X, Jiang L, et al. Structure of MERS‐CoV spike receptor‐binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986‐993. 10.1038/cr.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87(23):12552‐12561. 10.1128/JVI.01890-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86(12):6537‐6545. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory‐confirmed coronavirus disease 2019—COVID‐NET, 14 States, March 1–30, 2020. Morb Mortal Wkly Rep. 2020:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dowd JB, Andriano L, Brazel DM, et al. Demographic science aids in understanding the spread and fatality rates of COVID‐19. Proc Natl Acad Sci USA. 2020;117(18):9696‐9698. 10.1073/pnas.2004911117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. CDC COVID−19 Team Response , Bialek S, Boundy E, et al. Severe outcomes among patients with coronavirus disease 2019 (COVID‐19)—United States, February 12–March 16, 2020. Morb Mortal Wkly Rep. 2020;69(12):343‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention . The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID‐19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145‐151. 10.3760/cma.j.issn.0254-6450.2020.02.003 [DOI] [PubMed] [Google Scholar]
- 79. EpiCentro . Istituto Superiore di Sanità. Infographic Available in English – 13 May 2020 Update in COVID‐19 Integrated Surveillance: Key National Data. 2020. https://www.epicentro.iss.it/en/coronavirus/sars-cov-2-integrated-surveillance-data. Accessed January 13, 2021.
- 80. Bialek S, Boundy E, Bowen V, et al. Severe outcomes among patients with coronavirus disease 2019 (COVID‐19) – United States, February 12–March 16, 2020. Morb Mortal Wkly Rep. 2020;69(12):343‐346. 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salmon A. Why are Korea's COVID‐19 death rates so low? Asia Times. https://asiatimes.com/2020/03/why-are-koreas-covid-19-death-rates-so-low/. Accessed January 11, 2021.
- 82. Kayano T, Nishiura H. A comparison of case fatality risk of COVID‐19 between Singapore and Japan. J Clin Med. 2020;9(10):3326. 10.3390/jcm9103326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dighe A, Jombart T, Van Kerkhove MD, Ferguson N. A systematic review of MERS‐CoV seroprevalence and RNA prevalence in dromedary camels: implications for animal vaccination. Epidemics. 2019;29:100350. 10.1016/j.epidem.2019.100350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kelvin AA, Halperin S. COVID‐19 in children: the link in the transmission chain. Lancet Infect Dis. 2020;20:633‐634. 10.1016/S1473-3099(20)30236-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23(1):74‐98. 10.1128/CMR.00032-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sarnat BH. Development of olfaction and taste in the human fetus and neonate. Fetal Neonatal Physiol. 2017:1411‐1420. [Google Scholar]
- 87. Schuler BA, Habermann AC, Plosa EJ, et al. Age‐determined expression of priming protease TMPRSS2 and localization of SARS‐CoV‐2 in lung epithelium. J Clin Invest. 2021;131(1):e140766. 10.1101/2020.05.22.111187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bi Q, Wu Y, Mei S, et al. Epidemiology and transmission of COVID‐19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020;20(8):911‐919. 10.1016/S1473-3099(20)30287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein‐coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285‐291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ortiz‐Fernández L, Sawalha AH. Genetic variability in the expression of the SARS‐CoV‐2 host cell entry factors across populations. Genes Immun. 2020;21(4):269‐272. 10.1038/s41435-020-0107-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Irham LM, Chou W‐H, Calkins MJ, Adikusuma W, Hsieh S‐L, Chang W‐C. Genetic variants that influence SARS‐CoV‐2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem Biophys Res Commun. 2020;529(2):263‐269. 10.1016/j.bbrc.2020.05.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cheng Z, Zhou J, To KK‐W, et al. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J Infect Dis. 2015;212(8):1214‐1221. 10.1093/infdis/jiv246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Russo R, Andolfo I, Lasorsa VA, Iolascon A, Capasso M. Genetic analysis of the novel SARS‐CoV‐2 host receptor TMPRSS2 in different populations. bioRxiv. 2020. 10.1101/2020.04.23.057190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID‐19 severity in Italy. SSRN Electron J. 2020. 10.2139/ssrn.3559608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.The Sex, Gender and COVID‐19 Project, Global Health 50/50. 2020. https://globalhealth5050.org/the-sex-gender-and-covid-19-project/. Accessed January 11, 2020.
- 96. Alghamdi IG, Hussain II, Almalki SS, Alghamdi MS, Alghamdi MM, El‐Sheemy MA. The pattern of Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive epidemiological analysis of data from the Saudi Ministry of Health. Int J Gen Med. 2014;7:417‐423. 10.2147/IJGM.S67061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Karlberg J, Chong DSY, Lai WYY. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am J Epidemiol. 2004;159(3):229‐231. 10.1093/aje/kwh056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Joe W, Kumar A, Rajpal S, Mishra US, Subramanian SV. Equal risk, unequal burden? Gender differentials in COVID‐19 mortality in India. J Glob Heal Sci. 2020;2(1). 10.35500/jghs.2020.2.e17 [DOI] [Google Scholar]
- 99. Xiong D, Zhang L, Watson GL, et al. Pseudo‐likelihood based logistic regression for estimating COVID‐19 infection and case fatality rates by gender, race, and age in California. Epidemics. 2020;33:100418. 10.1016/j.epidem.2020.100418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lukassen S, Chua RL, Trefzer T, et al. SARS ‐CoV‐2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10). 10.15252/embj.20105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6494):1012‐1015. 10.1126/science.abb7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Song H, Seddighzadeh B, Cooperberg MR, Huang FW. Expression of ACE2, the SARS‐CoV‐2 receptor, and TMPRSS2 in prostate epithelial cells. Eur Urol. 2020;78(2):296‐298. 10.1016/j.eururo.2020.04.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cai H. Sex difference and smoking predisposition in patients with COVID‐19. Lancet Respir Med. 2020;8(4):e20. 10.1016/S2213-2600(20)30117-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Saheb Sharif‐Askari N, Saheb Sharif‐Askari F, Alabed M, et al. Airways expression of SARS‐CoV‐2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev. 2020;18:1‐6. 10.1016/j.omtm.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vardavas CI, Nikitara K. COVID‐19 and smoking: a systematic review of the evidence. Tob Induc Dis. 2020;18:20. 10.18332/tid/119324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Baratchian M, McManus J, Berk M, et al. No evidence that androgen regulation of pulmonary TMPRSS2 explains sex‐discordant COVID‐19 outcomes. bioRxiv. 2020. 10.1101/2020.04.21.051201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Patanavanich R, Glantz SA. Smoking is associated with COVID‐19 progression: a meta‐analysis. Nicotine Tob Res. 2020;22(9):1653‐1656. 10.1093/ntr/ntaa082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang X, Dhindsa R, Povysil G, et al. TMPRSS2 transcriptional inhibition as a therapeutic strategy for COVID‐19. Preprints.org. 2020. 10.20944/preprints202003.0360.v2 [DOI]
- 109. Mikkonen L, Pihlajamaa P, Sahu B, Zhang F‐P, Jänne OA. Androgen receptor and androgen‐dependent gene expression in lung. Mol Cell Endocrinol. 2010;317(1):14‐24. 10.1016/j.mce.2009.12.022 [DOI] [PubMed] [Google Scholar]
- 110. Tomlins SA. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644‐648. 10.1126/science.1117679 [DOI] [PubMed] [Google Scholar]
- 111. Lucas JM, Heinlein C, Kim T, et al. The androgen‐regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4(11):1310‐1325. 10.1158/2159-8290.CD-13-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Isbarn H, Boccon‐Gibod L, Carroll PR, et al. Androgen deprivation therapy for the treatment of prostate cancer: consider both benefits and risks. Eur Urol. 2009;55(1):62‐75. 10.1016/j.eururo.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shaw GL, Whitaker H, Corcoran M, et al. The early effects of rapid androgen deprivation on human prostate cancer. Eur Urol. 2016;70(2):214‐218. 10.1016/j.eururo.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor‐dependent prostate cancer growth. Mol Cell. 2007;27(3):380‐392. 10.1016/j.molcel.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Montopoli M, Zumerle S, Vettor R, et al. Androgen‐deprivation therapies for prostate cancer and risk of infection by SARS‐CoV‐2: a population‐based study (N = 4532). Ann Oncol. 2020;31(8):1040‐1045. 10.1016/j.annonc.2020.04.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chakravarty D, Nair SS, Hammouda N, et al. Sex differences in SARS‐CoV‐2 infection rates and the potential link to prostate cancer. Commun Biol. 2020;3(1):374. 10.1038/s42003-020-1088-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Goren A, Vano‐Galvan S, Wambier CG, et al. A preliminary observation: male pattern hair loss among hospitalized COVID‐19 patients in Spain—a potential clue to the role of androgens in COVID‐19 severity. Exp Med. 2020;217:6. 10.1111/jocd.13443 [DOI] [PubMed] [Google Scholar]
- 118. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626‐638. 10.1038/nri.2016.90 [DOI] [PubMed] [Google Scholar]
- 119. Barnes BJ, Adrover JM, Baxter‐Stoltzfus A, et al. Targeting potential drivers of COVID‐19: neutrophil extracellular traps. J ExpMed. 2020;217(6):e20200652. 10.1084/jem.20200652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Caffo O, Zagonel V, Baldessari C, et al. On the relationship between androgen‐deprivation therapy for prostate cancer and risk of infection by SARS‐CoV‐2. Ann Oncol. 2020;31:1415‐1416. 10.1016/j.annonc.2020.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. National Library of Medicine (U.S.) . The Impact of Camostat Mesilate on COVID‐19 Infection. Identifier NCT04321096. 2020. https://clinicaltrials.gov/ct2/show/NCT04321096. Accessed January 13, 2021.
- 122. Padmanabhan P, Desikan R, Dixit N. Targeting TMPRSS2 and Cathepsin B/L together may be synergistic against SARS‐CoV‐2 Infection. PLOS Comput Biol. 2020;16(12):e1008461. 10.26434/CHEMRXIV.12213125.V2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Maggio R, Corsini GU. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS‐CoV‐2 infection. Pharmacol Res. 2020;157:104837. 10.1016/j.phrs.2020.104837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. National Library of Medicine (U.S.) . Evaluating the Efficacy and Safety of Bromhexine Hydrochloride Tablets Combined With Standard Treatment/Standard Treatment in Patients With Suspected and Mild Novel Coronavirus Pneumonia (COVID‐19). Identifier NCT04273763. 2020. https://clinicaltrials.gov/ct2/show/NCT04273763. Accessed January 13, 2021.
- 125. Hoffmann M, Schroeder S, Kleine‐Weber H, Müller MA, Drosten C, Pöhlmann S. Nafamostat mesylate blocks activation of SARS‐CoV‐2: new treatment option for COVID‐19. Am Soc Microbiol. 2020;64(6):e00754‐20. 10.1128/AAC.00754-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sonawane K, Barale SS, Dhanavade MJ. Homology modeling and docking studies of TMPRSS2 with experimentally known inhibitors camostat mesylate, nafamostat and bromhexine hydrochloride to control SARS‐coronavirus‐2. ChemRxiv. 2020. 10.26434/chemrxiv.12162360.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS‐CoV‐2 in human airway epithelial cells and provide promising drug targets. bioRxiv. 2020. 10.1101/2020.04.15.042085 [DOI] [PMC free article] [PubMed] [Google Scholar]


