Abstract
Data regarding the immunological memory and long‐time kinetics of immunoglobulin (IgG) against viral nucleoprotein (NP) and spike protein S1 receptor‐binding domain (S1RBD) of Severe Acute Respiratory Syndrome‐associated Coronavirus 2 (SARS‐CoV‐2) are lacking. All consecutive COVID‐19 patients admitted to our Clinic between March 1, 2020, and May 1, 2020, who were tested at hospital admission for anti‐S1RBD and anti‐NP IgG were enrolled. Serum samples were tested for anti‐SARS‐CoV‐2 antibodies with the use of two commercially available enzyme‐linked immunosorbent assays. Results are expressed as optical density measurements at 450 nm (OD450). Overall, 111 patients were included; the median (q1–q3) age was 57 (49–73) years, 59 (53%) males. According to disease severity, 31 (28%), 47 (42%), and 33 (30%) patients were considered affected by mild/moderate, severe, and critical SARS‐CoV‐2 infection, respectively. During hospitalization, patients with the critical disease showed a higher peak value of both anti‐NP (median OD450: 3.66 vs. 3.06 vs. 3.00 respectively, p = .043) and anti‐S1RBD IgG (median OD450: 2.33 vs. 1.6 vs. 0.91, respectively, p < .001). By testing 48 subjects 6 months or above from discharge, a significant decrease of anti‐NP IgG was observed (r: −0.5838; p < .0001), whereas anti‐S1RBD IgG showed only a modest reduction (r: −0.1507; p = .0647). Accordingly, 10 (21%) and 2 (4%) patients had a negative serological status for anti‐NP and anti‐S1RBD IgG, respectively; no association with clinical severity was found. IgGs against SARS‐CoV‐2 persisted several months after discharge, regardless of disease severity, suggesting that vaccination could be a valid strategy to fight the pandemic.
Keywords: anti‐S1RBD, COVID‐19, SARS‐CoV‐2, serology
Highlights
‐Antibodies against SARS‐CoV‐2 persisted several months after the disease.
‐ Infection severity apparently did not affect IgG seroconversion.
‐ SARS‐CoV‐2 vaccination could be a valid strategy to fight the pandemic.
1. INTRODUCTION
In early 2020, the entire world and the scientific community faced the spread of Severe Acute Respiratory Syndrome‐related CoronaVirus‐2 (SARS‐CoV‐2), identified for the first time in Wuhan (China). The rapid human‐to‐human transmission through respiratory droplets of this new coronavirus led to the Coronavirus Disease‐2019 (COVID‐19) outbreak, 1 causing over 79 million reported cases and more than 1.8 million deaths worldwide, 2 particularly affecting older and frailer subjects. 3
SARS‐COV‐2 mainly infects pulmonary type II alveolar and endothelial cells and uses its surface spike glycoproteins to bind the angiotensin‐converting enzyme 2 receptor 4 causing a series of pulmonary and extrapulmonary clinical manifestations, 5 including the occurrence of thromboembolic events. 6
Adaptive immune response in the course of infection elicits the production of antibodies directed against viral proteins and, in particular, the SARS‐CoV‐2 nucleoprotein (NP) and spike protein S1 receptor‐binding domain (S1RBD), the latter endowed with neutralizing activity, thus, preventing viral attachment to its receptor and entry into the host cells. The necessary time to obtain anti‐spike conversion from immunoglobulin (IgG) negative to IgG positive is about 14 days 7 although the time and the level of antibody production might depend on either the illness severity or the patient's comorbidities. 8 Regardless, there are still little data about the duration of SARS‐CoV‐2 antibody response. Deepening the knowledge of antibodies kinetic after the recovery from COVID‐19 disease is pivotal to predict if an effective immunity is actually achievable through the vaccination to stop the pandemic.
Herein, serum samples from patients who experienced COVID‐19 were tested during the hospitalization and about 6 months after discharge with the aim to verify if residual antibody levels were still detectable.
2. MATERIALS AND METHODS
2.1. Study design
All consecutive COVID‐19 patients admitted to the Clinic of Infectious Disease of University Hospital of Bari (Italy) between March 1, 2020 and May 1, 2020, who were tested for anti‐ SARS‐CoV‐2 antibodies at the time of hospitalization were enrolled in this retrospective cohort study. Diagnosis of COVID‐19 was confirmed according to real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) testing (targeting E‐gene, RdRP‐gene, and N‐gene) on the throat and nasopharyngeal swabs performed at the hospital's laboratory with the protocol previously reported by the WHO. 9
For each patient, serial serum samples were collected every 7–10 days during hospitalization. Six months after discharge, all patients who survived were recalled to be retested for anti‐SARS‐CoV‐2 antibodies.
Patients' clinical features (comorbidities, signs, and symptoms at onset, therapies) and laboratory findings (total number of leucocytes, lymphocytes, and platelets, platelet to lymphocytes ratio, and total T CD4+ count) were obtained from clinical records. Patients were classified according to disease severity 10 as:
Mild/moderate, if they had clinical signs of pneumonia (fever, cough, dyspnea, and fast breathing) but no signs of severe pneumonia, including SpO2 ≥ 90% on room air.
Severe, if they had signs of pneumonia (fever, cough, dyspnea, fast breathing) plus one of the following: Respiratory rate > 30 breaths/min; severe respiratory distress; or SpO2 < 90% on room air.
Critical, if they had a diagnosis of acute respiratory distress syndrome was made.
2.2. Antibody detection
Qualitative enzyme‐linked immunosorbent assays (ELISA) were used to detect SARS‐CoV‐2 antibodies in serum samples. Anti‐nucleocapsid protein (NP) and anti‐S1RBD IgG of SARS‐CoV‐2 were detected using a SARS‐CoV‐2 NP IgG ELISA Kit and SARS‐CoV‐2 S1RBD IgG ELISA Kit (ImmunoDiagnostics Limited), respectively. Both kits are two‐step incubation immunoassays that use recombinant nucleocapsid protein (NP) and spike protein S1 receptor‐binding domain (S1RBD) of SARS‐CoV‐2 precoated well plates and an anti‐Human IgG‐horseradish peroxidase (HRP) conjugated antibody. After adding an HRP substrate solution, a color reaction develops whose intensity is proportional to the amount of antibodies captured inside the wells. Results are expressed as optical density (OD450) measurements using an absorbance microplate reader at 450 nm.
According to the manufacturer's instructions, the cutoff for seropositivity was set at an OD450 > 0.582 for anti‐NP IgG and > 0.300 for anti‐S1RBD IgG, respectively; in turn, all samples below these thresholds were considered negative.
2.3. Statistical analysis
All data were anonymized and collated on an electronic database.
Descriptive statistics were produced for demographic, clinical, and laboratory characteristics of cases. Mean and standard deviation were obtained for normally distributed variables, and median and interquartile range (q1–q3) for nonnormally distributed variables, and absolute number, and percentages for categorical variables.
The distribution between groups (according to disease severity) of different variables was analyzed by univariable parametric or nonparametric tests, Kruskal–Wallis or Mann–Whitney U Test (where appropriate) for continuous variables and with Pearson's χ 2 test (Fisher's exact test where appropriate) for categorical variables, according to data distribution.
Basic‐ and Fit‐plot graphics (with fractional polynomial prediction models) were produced to show the trend of IgG serum levels over days after SARS‐CoV‐2 infection; correlations were assessed using a Spearman's rank correlation coefficient (r).
In all cases, p < .05 was considered statistically significant. Statistical analysis was performed using STATA “Special Edition” version 16.1 (STATA Corp.).
3. RESULTS
3.1. General characteristics of the study population
Overall, 111 patients were included; median (q1–q3) age was 57 (49–73) years, 59 (53%) males.
According to disease severity, 31 (28%) were considered as mild/moderate COVID‐19, 47 (42%) severe disease, and 33 (30%) as critical SARS‐CoV‐2 infection. General features of patients are resumed in Table 1.
Table 1.
General characteristics of the study population
| Total (n = 111) | Mild/moderate disease (n = 31) | Severe disease (n = 47) | Critical disease (n = 33) | p Value | |
|---|---|---|---|---|---|
| Male sex, n (%) | 59 (53) | 11 (35) | 27 (56) | 21 (64) | .064 |
| Comorbidities, n (%) | (n = 98) | (n = 29) | (n = 42) | (n = 27) | |
| Hypertension | 35 (36) | 4 (14) | 18 (43) | 13 (48) | .012 |
| Obesity (BMI > 30 kg/m2) | 13 (13) | 3 (10) | 5 (12) | 5 (18) | .628 |
| Kidney impairment (CrCl < 30 ml/min) | 6 (6) | 1 (3) | 5 (12) | 0 | .102 |
| Diabetes | 13 (13) | 1 (3) | 10 (24) | 2 (7) | .026 |
| Chronic obstructive pulmonary disease | 19 (19) | 5 (17) | 10 (24) | 4 (15) | .615 |
| Signs and Symptoms at onset, n (%) | (n = 98) | (n = 29) | (n = 42) | (n = 27) | |
| Fever | 81 (83) | 21 (72) | 33 (79) | 27 (100) | .016 |
| Dyspnea | 40 (41) | 2 (7) | 21 (50) | 17 (63) | <.001 |
| Cough | 47 (48) | 18 (62) | 19 (45) | 10 (37) | .155 |
| Laboratory findings at onset, median (q1–q3) | |||||
| Total leucocytes (×103cell/µl) | 5980 (4270–7860) | 4920 (3470–7340) | 6120 (4440–7775) | 6115 (4720–8335) | .281 |
| Total lymphocytes (×103cell/µl) | 939 (690–1321) | 1083 (746–1356) | 1025 (731–1423) | 774 (547–1017) | .010 |
| Total platelets (×103cell/µl) | 190 (151–242) | 191 (159–229) | 178 (152–240) | 197 (143–254) | .977 |
| Platelet to lymphocytes ratio | 0.210 (0.135–0.305) | 0.166 (0.116–0.345) | 0.189 (0.119–0.287) | 0.239 (0.194–0.303) | .054 |
| Total T CD4+ count (cell/µl) | 543 (310–804) | 768 (525–993) | 638 (335–786) | 395 (266–586) | .002 |
| CD4/CD8 ratio | 1.9 (1.21–2.81) | 1.87 (1.15–2.17) | 1.9 (1.21–3.06) | 1.94 (1.3–2.8) | .489 |
| Treatments administered, n (%) | (n = 98) | (n = 29) | (n = 42) | (n = 27) | |
| Use of corticosteroid therapy | 11 (11) | 1 (3) | 2 (5) | 8 (30) | .002 |
| Use of tocilizumab (8 mg/Kg) | 5 (5) | 0 | 0 | 5 (18) | .001 |
| O2 therapy (>10 L/min) | 36 (37) | 0 | 9 (21) | 27 (100) | <.001 |
| Need of noninvasive (NIV) or invasive ventilation (IV) | 21 (21) | 0 | 2 (5) | 19 (70)a | <.001 |
| Survived, n (%) | 103 (93) | 29 (94) | 46 (96) | 28 (87) | .362 |
Abbreviations: BMI, body mass index; CrCL, creatinine clearance; q1–q3, first‐third quartile.
Three cases of invasive mechanical ventilation.
Of note, subjects with critical COVID‐19 were older than those with severe or mild/moderate disease (median age, 60 vs. 58 vs. 47 years respectively, p < .001), and more frequently affected by hypertension (48% vs. 43% vs. 14% respectively, p = .012). At the time of hospitalization, they also presented more frequently with fever (100% vs. 79% vs. 72%, p = .016), dyspnea (63% vs. 50% vs. 7%, p < .001), low‐median absolute lymphocytes (774 vs. 1,025 vs. 1,083 cell/µl, respectively, p = .010), and CD4 T cells count (395 vs. 638 vs. 768 cell/µl, respectively, p = .002).
3.2. Serologic status and viral clearance during hospitalization
At admission, 70 (62%) and 50 (45%) patients were, respectively, anti‐NP and anti‐S1RBD IgG positive (Table 2). Nevertheless, patients with critical COVID‐19 were more frequently anti‐S1RBD positive at admission, if compared to those experiencing severe and mild/moderate COVID‐19 (64% vs. 46% vs. 23% respectively, p = .004); conversely, no significant difference was observed in the prevalence of anti‐NP positive patients at admission.
Table 2.
Serologic status of patients during hospitalization and 6 months after discharge
| Total (n = 111) | Mild/moderate disease (n = 31) | Severe disease (n = 47) | Critical disease (n = 33) | p Value | |
|---|---|---|---|---|---|
| Serologic status during hospitalization | |||||
| Patients anti‐NP IgG positive at admission, n (%) | 70 (62) | 14 (45) | 33 (69) | 23 (70) | .064 |
| Anti‐NP IgG, median (q1–q3) | |||||
| Time to seroconversion (days) | 14 (11–19) | 14 (10–18) | 13 (10–19) | 17 (11–23) | .296 |
| Peak value during hospitalization (OD450) | 3.27 (1.83–3.91) | 3.00 (1.26–3.71) | 3.06 (1.83–3.94) | 3.66 (2.93–3.95) | .043 |
| Patients Anti‐S1RDB IgG positive at admission, n (%) | 50 (45) | 7 (23) | 22 (46) | 21 (64) | .004 |
| Anti‐S1RDB IgG, median (q1–q3) | |||||
| Time to seroconversion (days) | 18 (13–23) | 16 (12–20) | 17 (12–24) | 19 (16–24) | .532 |
| Peak value during hospitalization (OD450) | 1.52 (0.87–2.29) | 0.91 (0.58–1.34) | 1.6 (1.11–2.03) | 2.33 (1.10–3.09) | <.001 |
| Patients resulted anti‐NP IgG negative at hospital discharge, n (%) | 1 (1) | 0 | 0 | 1 (3) | .320 |
| Patients resulted anti‐S1RBD IgG negative at hospital discharge, n (%) | 2 (2) | 1 (4) | 0 | 1 (3) | .433 |
| Time to viral clearance, days ‐ median (q1–q3) | 22 (14–33) | 24 (14–36) | 22 (14–36) | 22 (15–26) | .711 |
| Serologic status 6 months after discharge | (n = 48) | (n = 14) | (n = 17) | (n = 17) | |
| Anti‐NP IgG | |||||
| Median IgG (q1–q3) value at 6 months of follow‐up (OD450) | 1.09 (0.63–1.69) | 0.70 (0.32–1.73) | 1.38 (0.75–1.65) | 1.11 (0.76–1.32) | .170 |
| Patients resulted anti‐NP IgG negative at 6 months of follow‐up, n (%) | 10 (21) | 5 (36) | 2 (12) | 3 (18) | .243 |
| Anti‐S1RDB IgG | |||||
| Median IgG (q1–q3) value at 6 months of follow‐up (OD450) | 1.03 (0.66–1.82) | 0.89 (0.37–1.00) | 1.48 (0.69–2.78) | 1.40 (0.81–1.82) | .038 |
| Patients resulted in anti‐S1RBD IgG negative at 6 months of follow‐up, n (%) | 2 (4) | 2 (14) | 0 | 0 | .079 |
Abbreviations: anti‐NP, anti‐nucleoprotein; anti‐S1RBD, anti‐spike protein S1 receptor‐binding domain; IgG, immunoglobulin; OD450, optical density 450 nm; q1–q3, first–third quartile.
Overall, the median time to viral clearance, calculated as the number of days between the first positive and the first negative PCR test on a nasopharyngeal swab was 22 (14–33) days; no correlation was found with disease severity (24 vs. 22 vs. 22 days, p = .711) (Table 2).
During hospitalization, in patients with the critical disease compared to those with severe and mild/moderate disease, a higher peak value of both anti‐NP (median OD450: 3.66 vs. 3.06 vs. 3.00, p = .043) and anti‐S1RBD IgG (median OD450: 2.33 vs. 1.6 vs. 0.91, p < .001) was observed. In contrast, the time to obtain the anti‐NP and anti‐ S1RBD seroconversion and viral clearance was not associated with disease severity.
Furthermore, at discharge, only one and two patients continued to be negative for anti‐NP I and anti‐S1RBD IgG, respectively.
3.3. Serologic status after 6 months from discharge
Six months after discharge, 103 (93%) patients survived and were recalled for serological testing. Forty‐eight subjects agreed to provide an additional blood sample for the detection of anti‐SARS‐CoV‐2 antibodies and included 14 (30%) patients who had experienced a mild/moderate disease, 17 (35%) a severe COVID‐19, and 17 (35%) a critical infection during the previous hospitalization (Table 2).
Overall, median OD values for anti‐S1RBD IgG appeared to be higher in patients with the severe disease than in those with mild/moderate and critical disease (median OD450 1.48 vs. 0.89 vs. 1.40 respectively, p = .038); the same association was not observed as regards anti‐NP IgG.
As shown in Figure 1A,B the OD values for anti‐NP IgG significantly decreased over time from diagnosis (r: −0.5838; p < .0001), whereas the OD values for anti‐S1RBD IgG showed a not statistically significant reduction (r: −0.1507; p = .0647) remaining above the threshold of positivity. After stratification for disease severity, a significant decreasing trend was observed for anti‐NP IgG, independently of the clinical picture (Figure 2A), whereas anti‐S1RBD IgG substantially persisted over time and remained above the positivity threshold, although a significant decrease in OD450 was observed in a patient with a critical disease (r:−0.4800; p = .0006; Figure 2B).
Figure 1.
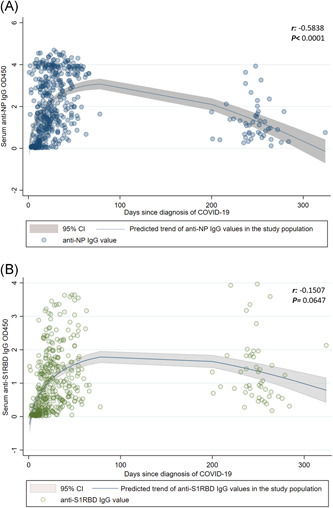
Trend over the months of anti‐NP IgG (A) and anti‐S1RBD IgG (B). IgG, immunoglobulin; NP, nucleoprotein; S1RBD, spike protein S1 receptor‐binding domain
Figure 2.
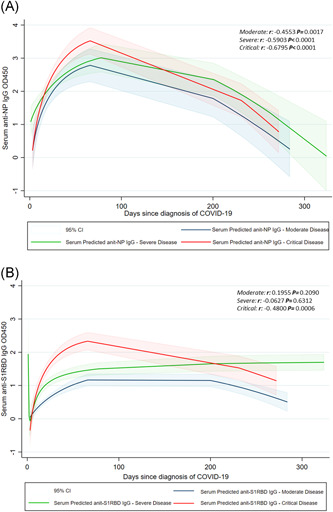
Trend over the months of anti‐NP IgG (A) and anti‐S1RBD IgG (B) according to disease severity. IgG, immunoglobulin; NP, nucleoprotein; S1RBD, spike protein S1 receptor‐binding domain
Accordingly, 10 (21%) and 2 (4%) patients showed a negative serologic status for anti‐NP and anti‐S1RBD IgG, respectively. However, no association was found between COVID‐19 severity and a negative serological test for anti‐NP and anti‐S1RBD IgG after 6 months of follow‐up (Table 2).
4. DISCUSSION
The COVID‐19 pandemic is bringing down the global healthcare system, the national economy, and people's social life; therefore, developing strategies to stop the SARS‐COV‐2 spread is essential. The newly developed vaccines are “the light at the end of the tunnel” for the achievement of herd immunity and, consequently, for the end of this pandemic.
However, to reach this goal, the development of an effective and long‐lasting immunological response is a major concern. Therefore, investigating the long‐term serological status of COVID‐19 survivors is pivotal to predict if the world‐wide vaccination will be a successful strategy.
In this study, we showed that although the anti‐NP and anti‐S1RBD response (according to the OD values) was more robust in patients with critical COVID‐19, almost all subjects in our series resulted reactive for SARS‐CoV‐2 antibodies within 18 days of hospitalization. In fact, the detection rate of anti‐NP and anti‐S1RBD was 99% and 98%, respectively. About 6 months later, a progressive decline of IgG values was observed; in particular, almost one out five of patients had a significant reduction of anti‐NP antibodies below the threshold of positivity. Conversely, long‐term persistence of anti‐S1RBD IgG in virtually all patients was demonstrated.
Our data are in line with previous studies showing the seroconversion of SARS‐CoV‐2 antibodies in almost all hospitalized patients within 2 weeks from the admission 7 , 11 , 12 ; these studies also demonstrated higher IgG serum levels in subjects who had a severe clinical picture. 7
In contrast, data concerning the long‐term persistence and levels of anti‐SARS‐CoV‐2 over time are poor. The majority of studies investigating the kinetics of antibodies to SARS‐CoV‐2 are limited to about 40 days after the onset of symptoms 7 , 13 , 14 showing the trend and the rapid increase of IgG around the acute phase of COVID‐19.
However, in a recent study where the long‐term kinetics of neutralizing anti‐SARS‐CoV‐2 antibodies was analyzed, 15 suggested that anti‐S1RBD IgG are durable, with only a modest decline in titers over months post symptoms onset, and that at 5 12 to 8 months post‐COVID‐19 almost all individuals remain positive for S1RBD IgG.
Conversely, other studies indicated that, although anti‐S1RBD IgG can last for more than 6 months after SARS‐CoV‐2 infection, a significant reduction of the antibody levels is observed 16 which possibly presages a progressive loss of a protective antibody response within a few months after SARS‐CoV‐2 infection. 17
In our study a progressive decline of IgG titers was noted; however, a significant reduction of IgG levels only concerned anti‐NP. Interestingly, anti‐S1RBD remained almost stable over time regardless of the severity of the clinical picture, and always above the thresholds of positivity, although lower anti‐S1RBD OD450 values were observed in patients who had suffered from mild/moderate infection.
Accordingly, in a study performed on three family members who had had a mild COVID‐19, the patient with a relatively more severe disease showed a higher level of anti‐S1RBD IgG during hospitalization. Noteworthily, the serum sample of these patients 6 months after discharge was still neutralizing in vitro. 18
In summary, our study adds data to the lacking information on the serological kinetics of survived COVID‐19 patients after a long enough time.
In any case, this study has some limitations. First, this is a retrospective study with only one serum evaluation of IgG levels after discharge. Moreover, the number of patients recalled after hospitalization is quite small, hampering deeper analysis on IgG kinetics according to different clinical pictures. Third, the ELISA test used for antibody dosage was only qualitative and the neutralization assay on IgG serum level was not performed.
In conclusion, our study suggested the persistence after several months of antibodies against the S1RBD in all patients, regardless of disease severity; consequently, the vaccination should be a valid strategy to fight the pandemic, if these data are confirmed also in vaccinated subjects.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceptualization: Laura Monno and Annalisa Saracino. Data curation: Paola Laghetti, Eugenio Milano, and Gaetano Brindicci. Formal analysis: Davide Fiore Bavaro; Methodology: Anna Volpe, Paola Laghetti, and Antonella Lagioia; Supervision: Laura Monno. Writing—original draft: Davide Fiore Bavaro and Paola Laghetti. Writing—review and editing: Laura Monno and Annalisa Saracino.
ETHICS STATEMENT
Human samples were collected with the consent of patients and with the approval of the ethics committee of the University Hospital Policlinico (Bari, Italy) (Study Code: 6357). The study was conducted in accordance with the Declaration of Helsinki and national and institutional standards. All patients provided informed consent for the use of their data for research purposes. In any case, data were previously anonymized, according to the requirements set by the Italian Data Protection Code (leg. Decree 196/2003).
Bavaro DF, Laghetti P, Milano E, et al. Anti‐spike S1 receptor‐binding domain antibodies against SARS‐CoV‐2 persist several months after infection regardless of disease severity. J Med Virol. 2021;93:3158–3164. 10.1002/jmv.26878
Davide F. Bavaro and Paola Laghetti contributed equally to this study.
DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. COVID‐19 Situation Reports . 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed January 7, 2021.
- 3. Balena F, Bavaro DF, Fabrizio C, et al. Tocilizumab and corticosteroids for COVID‐19 treatment in elderly patients. Gerontol Geriatr. 2020;68:197‐203. [Google Scholar]
- 4. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson KD, Harris C, Cain JK, Hummer C, Goyal H, Perisetti A. Pulmonary and extra‐pulmonary clinical manifestations of COVID‐19. Front Med (Lausanne). 2020;7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bavaro DF, Poliseno M, Scardapane A, et al. Occurrence of acute pulmonary embolism in COVID‐19—a case series. Int J Infect Dis. 2020;98:225‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients with Novel Coronavirus Disease 2019. Clin Infect Dis. 2020;71(16):2027‐2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee YL, Liao CH, Liu PY, et al. Dynamics of anti‐SARS‐Cov‐2 IgM and IgG antibodies among COVID‐19 patients. J Infect. 2020;81(2):e55‐e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. https://www.who.int/docs/default-source/coronaviruse/uscdcrt-pcr-panel-for-detection-instructions.pdf?sfvrsn=3aa07934_2. Accessed January 7, 2021.
- 10. World Health Organization . Clinical management of COVID‐19. Interim Guidance. 2020. https://apps.who.int/iris/bitstream/handle/10665/332196/WHO-2019-nCoV-clinical-2020.5-eng.pdf?sequence=1&isAllowed=y
- 11. Bao Y, Ling Y, Chen YY, et al. Dynamic anti‐spike protein antibody profiles in COVID‐19 Patients. Int J Infect Dis. 2021;103:540‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science. 2020;370(6521):1227‐1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020;26(6):845‐848. [DOI] [PubMed] [Google Scholar]
- 14. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in Coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X, Lu S, Li H, et al. Viral and antibody kinetics of COVID‐19 patients with different disease severities in acute and convalescent phases: a 6‐month follow‐up study. Virol Sin. 2020;35(6):820‐829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang L, Yang B, Jiang N, et al. Three‐month follow‐up study of survivors of coronavirus disease 2019 after discharge. J Korean Med Sci. 2020;35(47):e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flehmig B, Schindler M, Ruetalo N, et al. Persisting neutralizing activity to SARS‐CoV‐2 over months in sera of COVID‐19 patients. Viruses. 2020;12(12):1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


