Abstract
Background
Many arguments suggest that neutrophils could play a prominent role in COVID‐19. However, the role of key components of neutrophil innate immunity in severe forms of COVID‐19 has deserved insufficient attention. We aimed to evaluate the involvement of neutrophil elastase, histone‐DNA, and DNases in systemic and multi‐organ manifestations of COVID‐19.
Methods
We performed a multicenter study of markers of neutrophil innate immunity in 155 cases consecutively recruited in a screening center, local hospitals, and two regional university hospitals. The cases were evaluated according to clinical and biological markers of severity and multi‐organ manifestations and compared to 35 healthy controls.
Results
Blood neutrophil elastase, histone‐DNA, myeloperoxidase‐DNA, and free dsDNA were dramatically increased, and DNase activity was decreased by 10‐fold, compared with controls. Neutrophil elastase and histone‐DNA were associated with intensive care admission, body temperature, lung damage, and markers of cardiovascular outcomes, renal failure, and increased interleukin‐6 (IL‐6), IL‐8, and CXCR2. Neutrophil elastase was an independent predictor of the computed tomography score of COVID‐19 lung damage and the number of affected organs, in multivariate analyses. The increased blood concentrations of NE and neutrophil extracellular traps were related to exacerbation of neutrophil stimulation through IL‐8 and CXCR2 increased concentrations and increased serum DAMPs, and to impaired degradation of NETs as a consequence of the dramatic decrease in blood DNase activity.
Conclusion
Our results point out the key role of neutrophil innate immunity exacerbation in COVID‐19. Neutrophil elastase and DNase could be potential biomarkers and therapeutic targets of severe systemic manifestations of COVID‐19.
Keywords: COVID‐19, DNase, innate immunity, myeloperoxidase, neutrophil extracellular traps (NETs)
Blood levels of neutrophil elastase and histone‐DNA are associated with severe and systemic and multi‐organ manifestations of COVID‐19. Increased blood concentrations of neutrophil elastase and neutrophil extracellular traps are related to exacerbation of neutrophil stimulation through activated IL‐8/CXCR2 pathway. Neutrophil elastase and DNase could be potential biomarkers and therapeutic targets of severe systemic manifestations of COVID‐19.

Abbreviations: COVID‐19, coronavirus disease 2019; CXCR2, C‐X‐C motif chemokine receptor 2; ds, double‐stranded; NETs, neutrophil extracellular traps; NF, nuclear factor; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Abbreviations
- COVID‐19
coronavirus disease 2019
- CXCR2
C‐X‐C motif chemokine receptor 2
- ds
double‐stranded
- NETs
neutrophil extracellular traps
- NF
nuclear factor
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) produces thromboembolic events and multi‐organ manifestations, including heart, liver, and kidneys. 1 , 2 They result in part from the excessive involvement of innate immunity 3 and a cytokine storm produced by lymphocyte and macrophage activation. 4 , 5 , 6 Arguments suggest that neutrophils also play a prominent role in the severe and life‐threatening forms of the disease. 6 , 7 , 8 , 9 However, the involvement of neutrophils in systemic and multi‐organ outcomes of COVID‐19 has deserved insufficient attention. 8
Neutrophils have an arsenal of defensive strategies that include the release of antimicrobial granules and neutrophil elastase (NE), and the formation of neutrophil extracellular traps (NETs). 6 , 8 NETs are histone‐DNA components of dying neutrophils involved in the host defense against pathogens. 6 , 8 A study reported that two markers of NETs, cell‐free DNA and myeloperoxidase (MPO)‐DNA, were increased in hospitalized COVID‐19 patients compared with 30 controls and were correlated with C‐reactive protein, D‐dimer, lactate dehydrogenase, and absolute neutrophil count. 9 Another study from our group showed that MPO‐DNA level is increased in the early phase of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection, in ambulatory cases. 10 The homeostasis of circulating NETs is dependent on NE and DNase 1 and DNase 11L3. 6 Excessive NE and NETs in blood produce vascular and tissue lesions in acute viral pneumonia, which resemble those observed in COVID‐19 infection. 6 , 11 , 12 Despite these evidences, whether NE, DNases, and histone‐DNA are involved in the multi‐visceral manifestations of COVID‐19 has not been evaluated to date.
The present study is the first one, which studied NE, total DNase activity, and histone‐DNA in COVID‐19 cases recruited in screening centers, local hospital, and regional hospital in order to evaluate them according to disease severity and multi‐organ manifestations. We found that NE was an independent predictor of the multi‐organ injury produced by COVID‐19. The release of NE and NETs was related to neutrophil activation by serum damage‐associated molecular patterns (DAMPs) and IL‐8/CXCR2 pathway.
2. MATERIALS AND METHODS
2.1. Recruitment of patients and clinical characteristics of the study population
The NICO study (Neutrophil innate Immunity in COvid‐19) was registered by the Institutional Review Board of Clinical Research (DRCI) of the University Hospital of Nancy (No. 2020PI087) and approved by the Ethical Committee. We performed a cross‐sectional consecutive study of 155 positive patients recruited after informed consent in a screening center, local hospitals, and departments of medicine and intensive care units of two university hospital centers in the highest pandemic period of COVID‐19 in France. We followed the EQUATOR and BRISQ guidelines for the reporting clinical and biological data and the use of biological specimens. The infection was characterized by RT‐PCR on nasal swab specimens. The 34 consecutive ambulatory subjects attended the screening center in the first week of the disease, as previously described. 10 We recruited an additional 121 patients hospitalized for COVID‐19 in medical departments of local hospitals of the region of Tours (n = 43) and Regional University Hospitals of Marseille and Nancy (n = 78). Among them, 13 were treated in intensive care units. The median of the delay between the onset of symptoms and blood sampling was 7 days (IQR: 4–13) in medical departments and 13 days (IQR: 4–14) in intensive care units. We reported the clinical characteristics and outcomes of hospitalized patients listed in Table S1. The scoring of lung damage was evaluated by computed tomography in 5 grades (CT score), as described. 13 The control group included healthy subjects, with no reported contact with infected cases and negative for COVID‐19 screening.
2.2. Blood sampling and biological assessment
The sera were used after completion of biochemical testing ordered by the clinician. The remaining samples were stored in the same conditions among groups, at −20°C in the 24 h following blood withdrawal. Routine biochemical markers were assayed in Cobas 8000 analyzers (Roche Diagnostics, Switzerland) and Atellica (Eschborn, Germany).
2.3. Measurement of elastase, MPO‐DNA, DNA‐histone complexes, dsDNA, and cytokines in serum
Circulating levels of histone‐DNA complexes were measured in serum by the Cell Death Detection ELISA Kit (Roche Diagnostics, Sigma‐Aldrich, USA), as described previously. 14 Values were reported in 450 nm absorbance units. DNA‐MPO complexes were measured by replacing the anti‐histone antibody with a rabbit polyclonal antibody against myeloperoxidase (Merck, Darmstadt, Germany). NE concentration was determined using the Neutrophil Elastase Human ELISA Kit (Invitrogen, Carlsbad, USA). Values were reported in ng/ml. Cell‐free dsDNA was quantified using the Quant‐iT PicoGreen® dsDNA Assay Kit, after subtraction of background (Invitrogen). IL‐6, IL‐8, and CXCR2 were assayed by ELISA Kits from Reagentbio Ireland Ltd, Dublin, Ireland, IL‐1β by Invitrogen, HMGB1 by Novus Biologicals (Centennial, USA), and TNF‐α by ELISA Kit from R&D Systems Inc. (Minneapolis, USA).
2.4. Total deoxyribonuclease (DNase) activity in serum by single radial enzyme diffusion (SRED) assay
DNase activity was quantified on agarose gel containing 0.13 mg.mL−1 DNA from salmon sperm in a buffer containing 100 mM MES pH 6.5, 20 mM MgCl2, 2 mM CaCl2, and SYBR Safe DNA Gel Stain (Invitrogen, Life Technology) as described. 15 , 16 Two microliters of serum was located into wells. Gels were incubated for 24 h at 37°C in a humid chamber. The DNA‐SYBR fluorescence was recorded with a fluorescence scanner.
2.5. Quantification of cell‐bound NETs in neutrophils incubated with patient serum
To determine whether serum of COVID‐19 patients influences cell‐bound NETs, blood from one healthy donor was mixed (1:1 vol/vol) with patient sera prior to PMA treatment and incubated for 6 h, as described. 14 Red cell lysis was achieved by the addition of 1 ml of FACS Lyse reagent (BD Biosciences; 1:10 dilution) for 5 min. Bovine serum albumin (BSA, 1 ml at 2%) in phosphate‐buffered saline (PBS) was then added, and the mixture was centrifuged at 1000 g for 10 min. The pellet resuspended in PBS was analyzed using a Gallios flow cytometer (Beckman Coulter, Villepinte, France). The gating strategy sequentially focused on (i) CD66‐positive cells and then (ii) H3Cit‐positive and MPO‐positive cells.
2.6. Assay for in vitro NET DNA release of neutrophils by patient's serum
Isolated neutrophils were resuspended in RPMI 1640, and 1 × 105 cells were seeded in 96‐well tissue culture plates, as described. 14 Plates were incubated at 37°C to allow cells to adhere. Following stimulation with 100 µl of 0.1 ng/ml PMA for 4 h, wells were incubated with 1 U/ml MNase or DNase 1 (both from Worthington) for 10 min or 10 µl patient or control serum for 6 h, and then 2 mM EDTA was added to stop nuclease activity. The DNA content released in the supernatant was quantified using the Quant‐iT PicoGreen® dsDNA Assay Kit (Invitrogen). The amount of released DNA was considered as 100% in unstimulated neutrophils of healthy donors.
2.7. Statistical analyses
All quantitative variables are shown as the median and interquartile range (IQR, 25–75th percentile) and qualitative variables as percentages and 95% confidence interval (95% CI). Non‐parametric tests were used when data distribution was not normal. Univariate analyses were performed using the chi‐squared test or Fisher's exact test for categorical variables and the Mann‐Whitney U and the Kruskal‐Wallis tests and Spearman's rank correlation for continuous variables. Post hoc pairwise comparisons between subgroups were performed using the Conover test. We subsequently performed receiver operating characteristic (ROC) analysis to look for the optimal thresholds associated with disease outcomes. 17 We subsequently performed receiver operating characteristic (ROC) analysis to look for the optimal threshold associated with disease outcomes, according to DeLong et al. 17 The classification variable used in the ROC analysis was the studied disease outcome. For each ROC analysis, we reported the area under the receiver operating characteristic curve (AUROC) and the associated p‐value. The exact binomial method was applied to estimate the 95% confidence intervals (CIs) of the AUROC. The optimal diagnostic cutoff was defined using the Youden index J. Bias‐corrected and accelerated (BCa) bootstrap interval after 10,000 iterations for the Youden index and its associated values was performed. 18 Clinical and biological criteria were used to define organ injuries (see the Methods section in this article's Online Repository at www.jacionline.org). Bias‐corrected and accelerated (BCa) bootstrap interval after 10,000 iterations for the Youden index and its associated values were performed. 18 In step #2, using multivariable logistic regression analysis, we assessed whether the ROC‐defined threshold defined for elastase (>593 ng/L) was associated with a “number of affected organs ≥2” after accounting for potential confounders. We assessed model discrimination using ROC analysis (AUROC and 95% CI) and model calibration using the Hosmer and Lemeshow goodness‐of‐fit test and the Nagelkerke R2 statistics. 19 In step #3, we assessed the association between elastase level (> or ≤593 ng/L) and patients’ demographics, medical histories, and disease outcomes. A sample size of 7 patients per group was needed to detect an increase ≥200% and 38 patients to detect an increase ≥100% in mean elastase between groups, with 1‐β at 80% and α at 0.05. Statistical analyses were performed using MedCalc, version 19.1 (MedCalc Software, Ostend, Belgium), and Stata SE, version 12.1 (College Station, Texas, USA), based on a two‐sided type I error with an alpha level of 0.05.
3. RESULTS
3.1. NE, MPO‐DNA, histone‐DNA are dramatically increased, and DNase activity was dramatically decreased in COVID‐19 ambulatory and hospitalized patients
The 34 ambulatory cases had a mean age (+/‐SD) of 42+/‐17 years and a sex ratio (M/F) at 0.88. All had at least two symptoms among fever, dry cough, and dyspnea for less than one week. None had a severe form at this step of the disease. The clinical characteristics of the 122 hospitalized cases are reported in Table S1, their biological markers, including IL‐6, IL‐8, CXCR2, TNF‐α, IL‐1β, and HMGB1, are reported in Table S2, and the characteristics of the 35 healthy controls are reported in Table S3. The blood levels of NE, MPO‐DNA, histone‐DNA, and dsDNA of ambulatory and hospitalized COVID‐19 patients are reported in Table S4. They were dramatically higher than those reported in control subjects. In contrast, the global DNase activity was 10‐fold lower in COVID‐19 cases (Figure 1A, B). The serum concentration of NE and NET components was not associated with age and sex among groups, and the serum concentration of α1 antitrypsin (AAT), the main blood inhibitor of NE, was normal (Table S2). The concentration of NE was significantly higher in hospitalized versus ambulatory cases and patients treated in intensive care versus local hospitals and medical departments of university hospitals (Figure 1A). NE was distributed in two clusters of high and lower concentrations in ambulatory cases and patients hospitalized in medical department of regional university hospitals. The clusters with the highest concentrations had similar values than those reported in intensive care units (Figure 1A). As observed for NE and NET components, the concentration of dsDNA was higher in patients admitted in intensive care units (Figure 1B). In addition, the serum concentration of NE and NET components was associated with the number of affected organs (Figure 1C).
FIGURE 1.
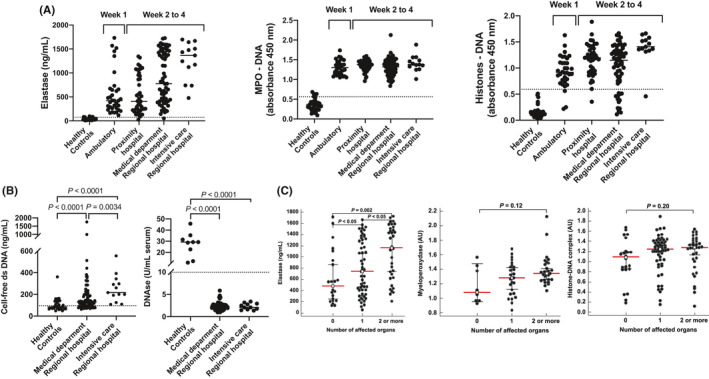
Neutrophil elastase (NE), myeloperoxidase‐DNA (MPO‐DNA), histone‐DNA, and (B) cell‐free dsDNA and total DNase activity in serum of 155 COVID‐19‐positive cases. Symptomatic ambulatory cases (n = 34) were recruited in screening centers and hospitalized patients (n = 122) in local hospitals (n = 43) and in medical departments (n = 65) and intensive care units (n = 13) of two regional university hospitals. The p‐values of NE concentrations between groups were as follows: healthy versus ambulatory <0.0001; healthy versus proximity hospital <0.0001; healthy versus medical department <0.0001; healthy versus intensive care <0.0001; ambulatory versus proximity hospital 0.7870; ambulatory versus medical department 0.0028; ambulatory versus intensive care <0.0001; proximity hospital versus medical department 0.0003; proximity hospital versus intensive care <0.0001; and medical department versus intensive care 0.0239. The p‐values of MPO concentrations between groups were healthy versus ambulatory <0.0001; healthy versus proximity hospital <0.0001; healthy versus medical department <0.0001; healthy versus intensive care <0.0001; ambulatory versus proximity hospital 0.1237; ambulatory versus medical department 0.8589; ambulatory versus intensive care 0.1276; proximity hospital versus medical department 0.1463; proximity hospital versus intensive care 0.5811; and medical department versus intensive care 0.1482. The p‐values of histone_DNA values between groups were healthy versus ambulatory <0.0001; healthy versus proximity hospital <0.0001; healthy versus medical department <0.0001; healthy versus intensive care <0.0001; ambulatory versus proximity hospital 0.0013; ambulatory versus medical department 0.0525; ambulatory versus intensive care <0.0001; proximity hospital versus medical department 0.2740; proximity hospital versus intensive care 0.0.0033; and medical department versus intensive care 0.0003. The thresholds (dotted lines) were evaluated in healthy controls recruited several months before the epidemy. The threshold of NE was estimated at 73 ng/ml and those of MPO‐DNA at 0.562 AU (450 nm absorbance units), histone‐DNA at 0.591 AU, total activity of DNase at 9.94 U/ml, and serum dsDNA at 95.60 ng/ml (C) NE, MPO‐DNA, and histone‐DNA according to the number of organs affected by COVID‐19. Data from (A), (B) and (C) were compared by the Mann‐Whitney test. The dashed lines represent the cutoffs defined by the mean +2 standard deviations for NE, MPO‐DNA, histone‐DNA, cell‐free DNA, and the mean—2 standard deviations for DNase activity
3.2. NE, histone‐DNA, and total DNase activity are associated with severity and multi‐visceral manifestations of COVID‐19
We studied the associations between elastase, myeloperoxidase‐conjugated DNA, and histone‐DNA complex and clinical and biological patients’ characteristics (Table 1). We reported significant correlations of NE with histone‐DNA, MPO‐DNA, and biomarkers of disease severity, SaO2 at hospital admission, leukocytes, neutrophils, neutrophil‐to‐lymphocyte ratio, LDH, markers of cardiovascular and thrombotic risk, including Troponin‐T (cTnT), fibrinogen, and D‐dimer, and markers of renal failure, including urea and creatinine (Table 1 and Figure 2). We also observed a significant association of NE with IL‐6, IL‐8, and CXCR2, but not with TNFα, IL‐1β, and HMBG1 (Figure 3). Significant association of histone‐DNA was reported with the computed tomography score (CT score) of lung damage, 13 markers of cardiovascular and thrombotic risk, 13 (Table 1) including cTnT and fibrinogen, markers of renal failure, including urea and creatinine, and markers of inflammation, including C‐reactive protein, ferritin, and body temperature (Figure S1). In contrast, MPO‐DNA was associated only with leukocytes (p = 0.005) and neutrophils (p = 0.004), and at weaker significance, with D‐Dimer (p = 0.045) and fibrinogen (p = 0.047). Total DNase activity was associated with alkaline phosphatase (PAL) and total bilirubin, while dsDNA was associated only with ferritin (Table 1).
TABLE 1.
Significant correlations between elastase, myeloperoxidase‐conjugated DNA, and histone‐DNA complex, and clinical and biological patients’ characteristics
| Variables | n | Rho | P‐value |
|---|---|---|---|
| Elastase | |||
| hs‐c troponin l (ng/L) | 15 | 0.82 (0.53–0.94) | 0.0003 |
| Fibrinogen (g/L) | 31 | 0.56 (0.25–0.77) | 0.001 |
| Histone‐DNA complex | 121 | 0.55 (0.4–0.66) | <0.0001 |
| Neutrophils (G/L) | 107 | 0.50 (0.34–0.63) | <0.0001 |
| Leukocytes (G/L) | 108 | 0.49 (0.33–0.62) | <0.0001 |
| D‐dimer (µg/ml) | 28 | 0.49 (0.13–0.73) | 0.0089 |
| Myeloperoxidase‐conjugated DNA | 121 | 0.45 (0.29–0.58) | <0.0001 |
| LDH (U/L) | 53 | 0.44 (0.18–0.64) | 0.001 |
| Urea nitrogen (g/L) | 87 | 0.34 (0.14–0.52) | 0.0012 |
| C‐reactive protein (mg/L) | 97 | 0.21 (0.01–0.4) | 0.0367 |
| Creatinine (mg/L) | 110 | 0.21 (0.02–0.39) | 0.0261 |
| Platelets (G/L) | 107 | 0.21 (0.01–0.39) | 0.0329 |
| Blood oxygen saturation (%) | 73 | −0.24 (−0.45 to −0.01) | 0.0381 |
| Neutrophil‐to‐lymphocyte ratio | 107 | −0.33 (−0.5 to −0.15) | 0.0004 |
| Myeloperoxidase‐conjugated DNA | |||
| hs‐c troponin l (ng/L) | 15 | 0.68 (0.23–0.89) | 0.007 |
| Elastase (ng/ml) | 121 | 0.45 (0.29–0.58) | <0.0001 |
| Histone‐DNA complex | 121 | 0.42 (0.26–0.56) | <0.0001 |
| D‐dimer (µg/ml) | 28 | 0.38 (0–0.67) | 0.045 |
| Fibrinogen (g/L) | 31 | 0.36 (−0.01to 0.64) | 0.047 |
| Ferritin (µg/L) | 36 | 0.36 (0.03–0.62) | 0.03 |
| LDH (U/L) | 53 | 0.36 (0.10–0.58) | 0.007 |
| C‐reactive protein (mg/L) | 97 | 0.33 (0.13–0.50) | 0.001 |
| Leukocytes (G/L) | 108 | 0.27 (0.08–0.44) | 0.005 |
| Neutrophils (G/L) | 107 | 0.27 (0.08–0.45) | 0.004 |
| γ glutamyltransferase (U/L) | 65 | 0.25 (0–0.48) | 0.04 |
| Total bilirubin (mg/L) | 71 | 0.23 (−0.01 to 0.45) | 0.049 |
| Neutrophil‐to‐lymphocyte ratio | 107 | −0.20 (−0.38 to 0) | 0.04 |
| Histone‐DNA complex | |||
| hs‐c troponin l (ng/L) | 15 | 0.78 (0.44–0.93) | 0.0008 |
| Fibrinogen (g/L) | 31 | 0.64 (0.36–0.82) | <0.0001 |
| Elastase | 121 | 0.55 (0.4–0.66) | <0.0001 |
| Ferritin (µg/L) | 36 | 0.49 (0.19–0.71) | 0.0023 |
| Myeloperoxidase‐conjugated DNA | 121 | 0.42 (0.26–0.56) | <0.0001 |
| LDH (U/L) | 53 | 0.38 (0.11–0.59) | 0.0055 |
| CT score* | 54 | 0.30 (0.02–0.53) | 0.0302 |
| Urea nitrogen (g/L) | 87 | 0.29 (0.08–0.48) | 0.0065 |
| Creatinine (mg/L) | 110 | 0.26 (0.08–0.44) | 0.007 |
| Temperature (°C) | 70 | 0.25 (0.01–0.46) | 0.0372 |
| C‐reactive protein (mg/L) | 97 | 0.21 (0.01–0.4) | 0.0356 |
| ASAT (U/L) | 92 | 0.21 (0–0.4) | 0.0475 |
| Transferrin (g/L) | 10 | −0.83 | 0.0041 |
CT score: computed tomography score based on the visual quantitative evaluation of acute lung inflammatory lesions involving each lobe, which was scored as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%), respectively.
FIGURE 2.
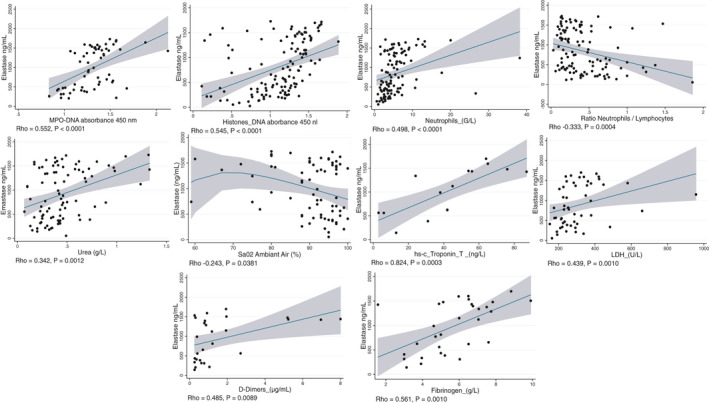
Associations of neutrophil elastase (NE) with components of neutrophil extracellular traps (NETs), clinical features, and biomarkers of severity and multi‐visceral harm of COVID. Significant associations of NE were reported with blood concentration of myeloperoxidase (MPO)‐DNA and histone‐DNA, blood counts of neutrophils, neutrophil/lymphocyte ratio, and blood biomarkers of multi‐visceral harm, including urea, SaO2, troponin‐T, lactate dehydrogenase (LDH), D‐dimer, and fibrinogen. Correlations were assessed by Spearman's rank correlation
FIGURE 3.
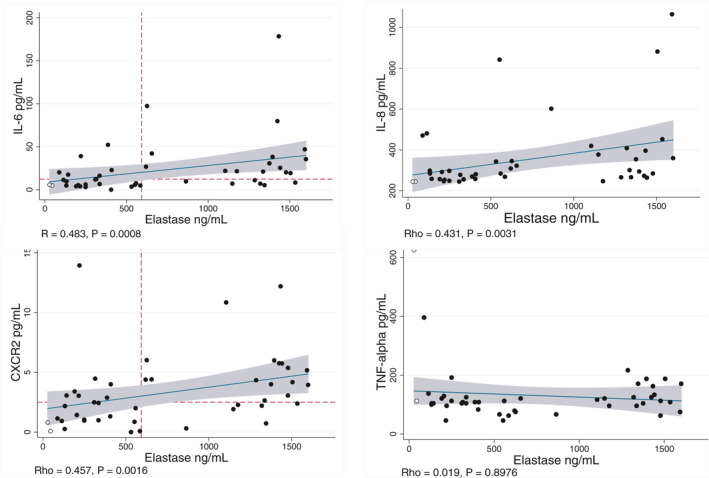
Associations of neutrophil elastase (NE) with cytokines involved in COVID‐19 pathological mechanisms. Significant associations of NE were reported with interleukin‐6 (IL‐6), IL‐8, and neutrophil chemokine receptor CXCR2, but not with TNF‐α. Dotted lines represent the cutoffs reported for multi‐organ damage in receiver operating characteristic (ROC) analyses. These cutoffs were used for the concordance analyses. Correlations were assessed by Spearman's rank correlation
3.3. Diagnostic accuracy of NE, MPO‐DNA, histone‐DNA, dsDNA, DNase activity, and related cytokines for detecting disease outcomes in receiver operating characteristic (ROC) analyses
We performed ROC analyses to assess the diagnostic accuracy of NE, MPO‐DNA, and histone‐DNA for the prediction of disease‐related outcomes (Table S5 and Forest plot in Figure 4). NE had significant thresholds for admission into intensive care units, occurrence of respiratory failure, blood oxygen saturation <85%, and the presence of at least two affected organs (Figure 4). The latter was best predicted with a cutoff at 593 ng/ml of NE. Histone‐DNA had significant thresholds for admission into intensive care units, kidney injury, and blood oxygen saturation <85%. MPO‐DNA had significant thresholds for heart decompensation, kidney injury, respiratory failure, and blood oxygen saturation <85%. The ROC analyses identified also very significant cutoffs for IL‐6, IL‐8, and CXCR2 for admission into intensive care units, heart decompensation, kidney injury, and respiratory failure. In contrast, we did not observe these associations with IL‐1β and TNF‐α.
FIGURE 4.
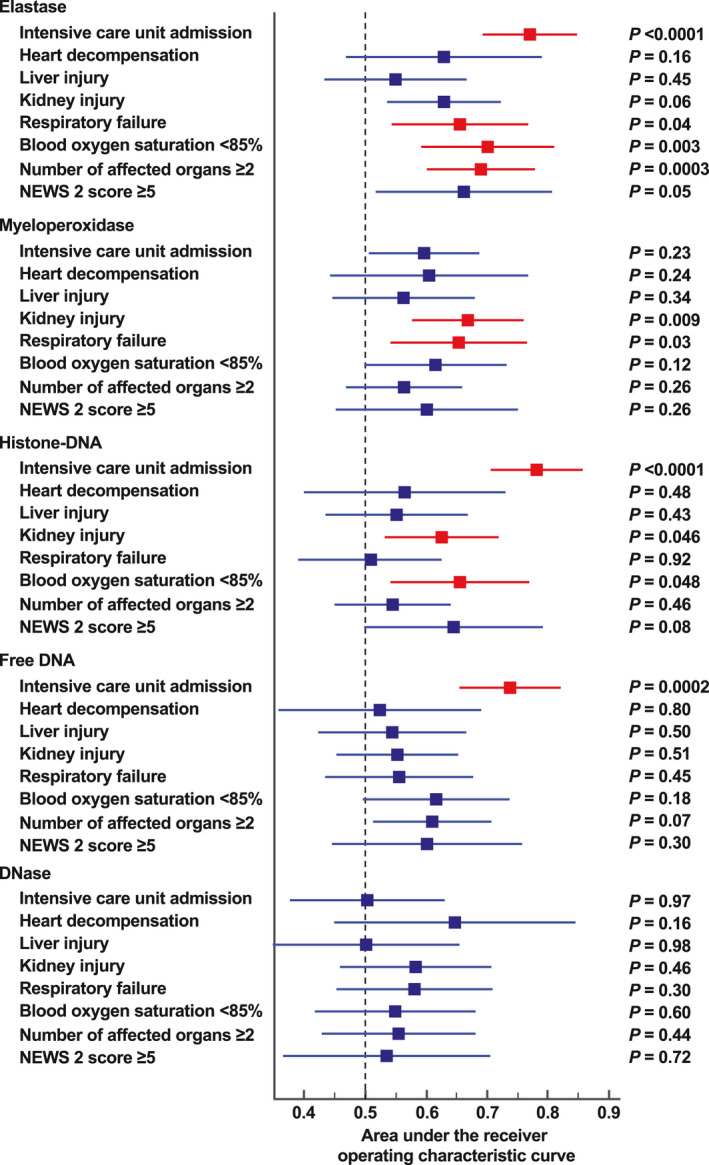
Forest plot reporting the summary of the receiver operating characteristic (ROC) analyses to assess the diagnostic accuracy of neutrophil elastase (NE), myeloperoxidase (MPO)‐conjugated DNA, histone‐DNA, cell‐free dsDNA, and DNase activity for the prediction of disease‐related outcomes. For each biomarker, a ROC analysis was performed using the following classification variables: intensive care unit admission, heart decompensation, liver injury, kidney injury, respiratory failure, blood oxygen saturation <85%, number of affected organs ≥2, and NEWS 2 score ≥5. For each ROC analysis, the summary results were reported using the area under the ROC curve, the 95% confidence interval, and the associated p‐value (Table S3)
3.4. Associations between NE >593 ng/ml and patients’ characteristics and outcomes
In univariate analyses, NE >593 ng/ml was significantly associated with CT score, blood oxygen saturation, respiratory failure, presence of at least two affected organs, and admission into the intensive care unit (Table S6). We retained chronic kidney disease, diabetes, and hypertension as potential confounders in multivariable analyses (Table S7). In the multivariable models that were used to account for the collinearity issue, NE >593 ng/ml was independently associated with CT score and presence of at least two affected organs (Table S8). The optimal multivariable model had an area under the receiver operating characteristics (AUROC) of 0.876 (95% CI, 0.758–0.950) and a percentage of cases correctly classified of 85%. We performed concordance analyses between cutoffs of NE and cytokines in the prediction of least two affected organs. We reported a 70.4% and 72.7% concordance of NE with CXCR2 and IL‐6, respectively (Figure 3).
3.5. Patient sera decrease the retention of NETs and increase the release of dsDNA in isolated control neutrophils
We studied cell‐bound NETs of control neutrophils incubated with patient sera using flow cytometry. We observed a decreased retention of cell‐bound NETs, which reflected an increased release, from cells incubated with patient versus control sera (Figure 5A). We further studied the release of dsDNA from control neutrophils produced by patient sera. We observed an increased release of dsDNA from neutrophils incubated with patient versus control sera (Figure 5B). The inhibition of NE and other serum serine proteases with 10 μM aprotinin had a weak inhibition effect on dsDNA release (Figure 5C). Taken together, these data suggested that the sera from patients increased the release of components of NETs, with a limited role of NE.
FIGURE 5.
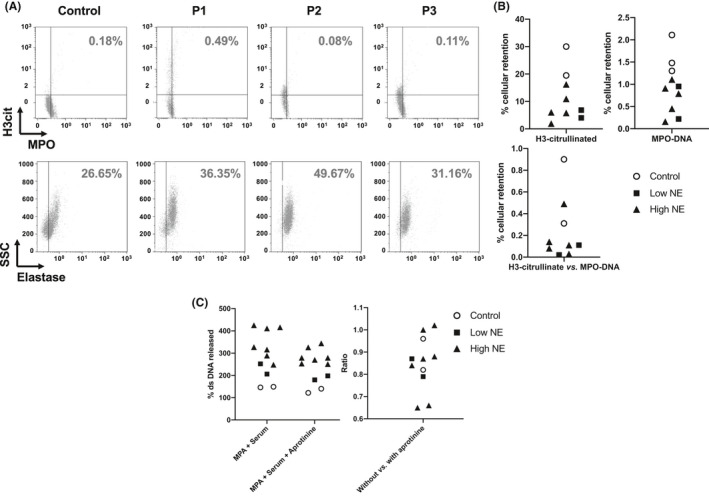
Neutrophil innate immunity is a target of the systemic effects of COVID‐19. (A, B) NET formation and release of neutrophil dsDNA by sera of COVID‐19 patients. (A) Representative data of flow cytometry experiments showing NET detection as citrullinated H3 (H3cit) and myeloperoxidase (MPO) double‐positive neutrophils (top) and NE‐positive neutrophils (bottom), after incubation of neutrophils with sera. (B) Quantitative analysis of NET retention on neutrophils from a healthy donor incubated with control or patient sera. (C) Quantification of DNA release from neutrophils of a healthy donor stimulated with PMA to induce NET formation and subsequently incubated with 10% serum from controls or patients in the absence or in the presence of aprotinin. The amount of released DNA was considered as 100% in unstimulated neutrophils from the donor
4. DISCUSSION
Our study showed that NE, global DNase activity, and components of NETs are involved in the early and later steps of COVID‐19. NE and histone‐DNA were associated with clinical manifestations and biomarkers related to pulmonary damage, cardiovascular manifestations, renal insufficiency, and inflammation. Elastase >593 ng/ml was an independent predictor of multi‐organ injury in multivariate analysis.
The dramatic increase in blood levels of NE and NETs in ambulatory cases shows their involvement in the early host response to SARS‐CoV‐2, as previously observed for other viral pneumonia. 4 , 6 , 11 , 20 The increase in NE and NETs is still observed in the later phase of the infection, as evidenced in hospitalized cases, in particular those recruited in intensive care unit two weeks after the onset of symptoms. Alveolar epithelial cells from lungs infected with influenza virus stimulate NETosis. 21 However, there is a debate on the detrimental versus protective effects of NETs in acute respiratory distress syndrome (ARDS). 22 , 23 The citrullinated H3 and cell‐free DNA reflect the production of extracellular traps by neutrophils, granulocytes, and monocytes. 23 , 24 The dramatic increase in blood concentration of NE reflects more specifically the activation of neutrophils. 24 , 25 Of note, NE concentration correlates better with alveolar inflammation than neutrophil count, in acute respiratory distress syndrome. 25 , 26
NE, but not histone‐DNA and MPO‐DNA, was an independent predictor of multi‐organ damage in COVID‐19 patients. This result reflects the prominent role of NE in the release of NET components and mechanisms of neutrophil innate immunity. 24 NE is dispensed in tissues and blood by degranulation or release with NETs. 24 , 27 , 28 , 29 The induction of ROS by viral infection activates MPO, which then activates the release of NE. 29 , 30 The highly significant correlation between NE and histone‐DNA and MPO‐DNA illustrates the role of NE in NET formation. Upon neutrophil stimulation, NE translocates to the nucleus, where it participates in histone degradation before it releases with the DNA/chromatin material of NETs. 6 , 30 Importantly, NE associated with NETs remains active and escapes the endogenous anti‐protease activity of AAT. 31 NE can produce tissue damages in lung, heart, liver, vessels, and kidney and exerts prothrombotic effects in viral infection. 6 , 26 , 31 , 32 , 33 , 34 , 35 , 36 , 37 The degradation of extracellular matrix (ECM) components by NE produces the same lung and vascular injuries as those observed in autopsy specimens of COVID‐19 patients. 34 , 35 , 36 , 37 , 38 , 39 The associations of NE and histone‐DNA with SaO2 at hospital admission and CT score of lung damage of COVID‐18 are consistent with their effects in other lung viral infections. 6 , 12 , 40 Their associations with troponin‐T, D‐dimer, and fibrinogen suggest their role in the prothrombotic effects reported in COVID‐19. 39 , 40 , 41 The myocardial injury during COVID‐19 is not clearly understood. 42 , 43 , 44 The mechanisms include direct viral infection, thrombosis, microvascular, and myocardial injury related to reduced oxygen delivery and release of cytokines. 34 , 43 , 45 The increased cTnT associated with NE could be secondary to coronary thrombosis and myocardial infarction and/or myocarditis. 34 , 43 , 44 NE and NETs could also contribute to the association of heart injury through systemic effects in kidney and other organs. 2 , 45 , 46 The associations of NE and histone‐DNA with urea and creatinine suggest their involvement in the acute kidney injury reported in about 30% of COVID‐19 patients. 33 , 47 , 48 , 49
Our study contributes to a better understanding of pathological mechanisms of COVID‐19 by pointing out the key role of the disruption of neutrophil innate immunity during and after viral replication, as summarized in graphical abstract. IL‐6, IL‐8, and TNF‐α account among the cytokines predominantly associated with COVID‐19 severity. 50 We confirmed this association for IL‐6 and IL‐8. Increased IL‐1β concentrations were positively correlated with disease severity in several studies. 51 However, we did not find any association of IL‐1β with outcomes of COVID‐19 severity, as previously reported in a recent meta‐analysis. 52 These results are consistent with a recent large size study, in which IL‐1β was poorly detected and, as a result, had only marginal predictive value of disease severity. 50 IL‐6 and IL‐8 are produced upon NF‐κB activation of infected alveolar macrophages through mechanisms that probably involve Bruton tyrosine kinase (BTK). 53 This is illustrated by the promising clinical effects produced by the BTK inhibitor acalabrutinib in patients with severe COVID‐19. 53 IL‐8 acts as neutrophil‐activating chemokine through its binding to CXCR2, which is a major chemokine receptor of neutrophils. 15 Therefore, the dramatic increase in NE in severe COVID‐19 may be related to neutrophil activation by the IL‐8/CXCR2 pathways. 16 LDH, ferritin, and D‐dimer are highly correlated with NE and could reflect the macrophage activation of COVID‐19. 49 , 54 Conversely, NE and NETs could also play a role in macrophage activation and the processing of pro‐inflammatory cytokines. 3 , 6 , 36 , 37 , 38 We reported a dramatic decrease in global DNase activity in serum, which could participate in NETosis through decreased degradation of circulating chromatin‐DNA fragments (graphical abstract). 55 , 56 , 57 , 58 It is noteworthy that a Dnase1–/– Dnase1 l3–/– mouse model exposed to pathogens produces lung lesions similar to those observed in patients with respiratory distress and/or sepsis and autopsies of COVID‐19 patients. 40 , 59 Consistently, a recent study reported a decreased activity of DNase I in bacterial and viral pneumonia in children. 60
The sera of COVID‐19 patients decreased the cell retention of NETs and increased the release of dsDNA of neutrophils from healthy donors. Similar results have been previously obtained with COVID‐19 serum using the cell dye SYTOX Green to quantify NETs. 9 These results show that the NETosis can be triggered by endogenous stimuli released in blood by injured tissues such as DAMPs, including free dsDNA. 61 HMGB1 seems not to be involved in increased NETosis, as its concentration was not increased in COVID‐19 cases, as previously reported. 62
Our study opens up therapeutic perspectives. The excessive NE activity reported in our study suggests evaluating the use of new generation potent NE inhibitors, including lonodelestat (POL6014), alvelestat, CHF6333, and elafin in COVID‐19. 8 , 27 The dramatic decrease in DNase reported in our study also suggests evaluating the use of recombinant deoxyribonuclease I (dornase alfa) and/or DNase 1L3. 63 , 64 , 65 One expected effect is the release of NE from degraded NETs, with increased free NE in blood and subsequently improved efficacy of NE inhibitors. 30 For this reason, we think that the association of DNase inhibitors with NE inhibitors should be considered in the treatment of COVID‐19. Another therapeutic option to be considered could be the use of inhibitors of CXCR2 to block the neutrophil recruitment and activation. Impairing NE and/or the release of NETs in the early phase of COVID‐19 could produce benefits but also risks related to decreasing the host innate immunity. In contrast, we presume that this strategy could decrease the systemic manifestations of COVID‐19 in the post‐replication phase of the disease.
Our study suffered limitations. We could not systematically report all biomarkers in all patients. The recruitment and cross‐sectional evaluation did not allow us to study the kinetics of NE and NETs by the follow‐up of patients. However, we were able to compare the concentration of NE and NETs in ambulatory symptomatic patients recruited during the first week of COVID‐19 and patients hospitalized during the second to fourth week of the disease. The multicentric recruitment in local hospitals and in regional university hospitals was a strength of our study for evaluating cases with contrasted disease severity. The exhaustive recording of clinical data allowed us to perform multivariate analysis of the association of NE and NETs with severity and multi‐organ damage of COVID‐19, after forced adjustment for medical history.
In conclusion, our study points out the dramatic increase in NE, DNase activity, and NETs during the first phase of COVID‐19 and their key role in the severity of the acute respiratory distress syndrome and cardiovascular, renal, and inflammatory systemic manifestations in the later step of the disease. They suggest evaluating NE, DNase 1, and NETs as potential therapeutic targets.
CONFLICT OF INTEREST
None of the authors had any financial relationships for themselves and their immediate family/significant others.
AUTHOR CONTRIBUTIONS
JL Guéant had full access to all of the data, supervised the study, and takes responsibility for the integrity and accuracy of the data analysis. JL Guéant and V Regnault conceived and designed the study. JL Guéant, V Regnault, RM Guéant‐Rodriguez, A Oussalah, and P Lacolley acquired the data, analyzed the data, or interpreted the data. JL Guéant involved in main drafting of the manuscript. V Regnault, RM Guéant‐Rodriguez, and A Oussalah involved in partial drafting of specific parts. All authors critically revised the manuscript. RM Guéant‐Rodriguez, Julien Fromonot, and A Oussalah involved in database management. RM Guéant‐Rodriguez and A Oussalah involved in statistical analyses.
Supporting information
Tables S1–S8
Figure S1
ACKNOWLEDGEMENTS
The NICO study was registered by the Institutional Review Board of Clinical Research (DRCI) of the University Hospital of Nancy (No. 2020PI087) and approved by the Ethical Committee of the University Hospital of Nancy. The authors had no conflict of interest to declare.
Rosa‐Maria Guéant‐Rodriguez and Abderrahim Oussalah have Equal contribution.
Funding information
The study was funded by the research project FHU ARRIMAGE and the French PIA project “Lorraine Université d’Excellence,” reference ANR‐15‐IDEX‐04‐LUE.
REFERENCES
- 1. Zhou P, Yang X‐L, Wang X‐G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grasselli G, Zangrillo A, Zanella A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS‐CoV‐2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323(16):1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75:1564‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGonagle D, Sharif K, O'Regan A, Bridgewood C. The role of cytokines including interleukin‐6 in COVID‐19 induced pneumonia and macrophage activation syndrome‐like disease. Autoimmun Rev. 2020;19:102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mo P, Xing Y, Xiao YU, et al. Clinical characteristics of refractory COVID‐19 pneumonia in Wuhan, China. Clin Infect Dis. 2020:ciaa270. [Epub ahead of print]. 10.1093/cid/ciaa270 [DOI] [Google Scholar]
- 6. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18:134‐147. [DOI] [PubMed] [Google Scholar]
- 7. Lagunas‐Rangel FA. Neutrophil‐to‐lymphocyte ratio and lymphocyte‐to‐C‐reactive protein ratio in patients with severe coronavirus disease 2019 (COVID‐19): a meta‐analysis. J Med Virol. 2020;92(10):1733‐1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barnes BJ, Adrover JM, Baxter‐Stoltzfus A, et al. Targeting potential drivers of COVID‐19: neutrophil extracellular traps. J Exp Med. 2020;217:e20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zuo Y, Yalavarthi S, Shi H, et al. Neutrophil extracellular traps in COVID‐19. JCI. Insight. 2020;5: e138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gueant JL, Fromonot J, Gueant‐Rodriguez RM, Lacolley P, Guieu R, Regnault V. Blood Myeloperoxidase‐DNA, a biomarker of early response to SARS‐CoV‐2 infection? Allergy. 2021;76:892‐896. 10.1111/all.14533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cortjens B, de Boer OJ, de Jong R, et al. Neutrophil extracellular traps cause airway obstruction during respiratory syncytial virus disease. J Pathol. 2016;238:401‐411. [DOI] [PubMed] [Google Scholar]
- 12. Twaddell SH, Baines KJ, Grainge C, Gibson PG. The emerging role of neutrophil extracellular traps in respiratory disease. Chest. 2019;156:774‐782. [DOI] [PubMed] [Google Scholar]
- 13. Li K, Fang Y, Li W, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID‐19). Eur Radiol. 2020;30:4407‐4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Popovic B, Zannad F, Louis H, et al. Endothelial‐driven increase in plasma thrombin generation characterising a new hypercoagulable phenotype in acute heart failure. Int J Cardiol. 2019;274:195‐201. [DOI] [PubMed] [Google Scholar]
- 15. Uddin M, Watz H, Malmgren A, Pedersen F. NETopathic inflammation in chronic obstructive pulmonary disease and severe asthma. Front Immunol. 2019;10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Didangelos A. COVID‐19 hyperinflammation: what about neutrophils? mSphere. 2020;5:e00367‐e00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837‐845. [PubMed] [Google Scholar]
- 18. Efron B, Tibhirani RJ. An introduction to the bootstrap. CRC press; 1994. [Google Scholar]
- 19. Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691‐692. [Google Scholar]
- 20. Funchal GA, Jaeger N, Czepielewski RS, et al. Respiratory syncytial virus fusion protein promotes TLR‐4‐dependent neutrophil extracellular trap formation by human neutrophils. PLoS One. 2015;10:e0124082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pillai PS, Molony RD, Martinod K, et al. Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science. 2016;352:463‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bendib I, de Chaisemartin L, Mekontso Dessap A, Chollet‐Martin S, de Prost N. Understanding the role of neutrophil extracellular traps in patients with severe pneumonia and ARDS. Chest. 2019;156:1278‐1280. [DOI] [PubMed] [Google Scholar]
- 23. Granger V, Faille D, Marani V, et al. Human blood monocytes are able to form extracellular traps. J Leukoc Biol 2017;102:775‐781. [DOI] [PubMed] [Google Scholar]
- 24. Kettritz R. Neutral serine proteases of neutrophils. Immunol Rev. 2016;273:232‐248. [DOI] [PubMed] [Google Scholar]
- 25. Lengas A, Poletti V, Pacifico L, di Domizio C, Patelli M, Spiga L. Acute lung inflammation: neutrophil elastase versus neutrophils in the bronchoalveolar lavage—neutrophil elastase reflects better inflammatory intensity. Intensive Care Med. 1994;20:354‐359. [DOI] [PubMed] [Google Scholar]
- 26. Polverino E, Rosales‐Mayor E, Dale GE, Dembowsky K, Torres A. The role of neutrophil elastase inhibitors in lung diseases. Chest. 2017;152:249‐262. [DOI] [PubMed] [Google Scholar]
- 27. Korkmaz B, Moreau T, Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie. 2008;90:227‐242. [DOI] [PubMed] [Google Scholar]
- 28. Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003;5:1317‐1327. [DOI] [PubMed] [Google Scholar]
- 29. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolaczkowska E, Jenne CN, Surewaard BGJ, et al. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat Commun. 2015;6:6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor S, Dirir O, Zamanian RT, Rabinovitch M, Thompson AAR. The role of neutrophils and neutrophil elastase in pulmonary arterial hypertension. Front Med (Lausanne). 2018;5:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakazawa D, Marschner JA, Platen L, Anders HJ. Extracellular traps in kidney disease. Kidney Int. 2018;94:1087‐1098. [DOI] [PubMed] [Google Scholar]
- 33. Doring Y, Libby P, Soehnlein O. Neutrophil extracellular traps participate in cardiovascular diseases: recent experimental and clinical insights. Circ Res. 2020;126:1228‐1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bronze‐da‐Rocha E, Santos‐Silva A. Neutrophil elastase inhibitors and chronic kidney disease. Int J Biol Sci. 2018;14:1343‐1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alam SR, Newby DE, Henriksen PA. Role of the endogenous elastase inhibitor, elafin, in cardiovascular injury: from epithelium to endothelium. Biochem Pharmacol. 2012;83:695‐704. [DOI] [PubMed] [Google Scholar]
- 36. Clancy DM, Henry CM, Sullivan GP, Martin SJ. Neutrophil extracellular traps can serve as platforms for processing and activation of IL‐1 family cytokines. FEBS J. 2017;284:1712‐1725. [DOI] [PubMed] [Google Scholar]
- 37. Alfaidi M, Wilson H, Daigneault M, et al. Neutrophil elastase promotes interleukin‐1beta secretion from human coronary endothelium. J Biol Chem. 2015;290:24067‐24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62:726‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N Engl J Med. 2020;383:120‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bikdeli B, Madhavan MV, Jimenez D, et al. COVID‐19 and thrombotic or thromboembolic disease: implications for prevention, antithrombotic therapy, and follow‐up: JACC state‐of‐the‐art review. J Am Coll Cardiol. 2020;75:2950‐2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Driggin E, Madhavan MV, Bikdeli B, et al. Cardiovascular considerations for patients, health care workers, and health systems during the COVID‐19 pandemic. J Am Coll Cardiol. 2020;75:2352‐2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Babapoor‐Farrokhran S, Gill D, Walker J, Rasekhi RT, Bozorgnia B, Amanullah A. Myocardial injury and COVID‐19: possible mechanisms. Life Sci. 2020;253:117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Doyen D, Moceri P, Ducreux D, Dellamonica J. Myocarditis in a patient with COVID‐19: a cause of raised troponin and ECG changes. Lancet. 2020;395:1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Renu K, Prasanna PL, Valsala GA. Coronaviruses pathogenesis, comorbidities and multi‐organ damage—A review. Life Sci. 2020;255:117839. 10.1016/j.lfs.2020.117839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Libby P. The heart in COVID19: primary target or secondary bystander? JACC Basic Transl Sci. 2020;5(5):537‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng Y, Luo R, Wang K, et al. Kidney disease is associated with in‐hospital death of patients with COVID‐19. Kidney International. 2020. 97:829‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oussalah A, Gleye S, Clerc Urmes I, et al. Long‐term ACE Inhibitor/ARB use is associated with severe renal dysfunction and acute kidney injury in patients with severe COVID‐19: results from a referral center cohort in the northeast of France. Clin Infect Dis. 2020;71(9):2447‐2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng Y, Luo R, Wang K, et al. Kidney disease is associated with in‐hospital death of patients with COVID‐19. Kidney Int. 2020;97:829‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu T, Zhang J, Yang Y, et al. The role of interleukin‐6 in monitoring severe case of coronavirus disease 2019. EMBO Mol Med. 2020;12:e12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Del Valle DM, Kim‐Schulze S, Hsin‐Hui H, et al. An inflammatory cytokine signature helps predict COVID‐19 severity and death. Nat Med. 2020;26:1636‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao Y‐D, Ding M, Dong X, et al. Risk factors for severe and critically ill COVID‐19 patients: a review. Allergy. 2021;76:428‐455. 10.1111/all.14657. [DOI] [PubMed] [Google Scholar]
- 52. Akbari H, Tabrizi R, Lankarani KB, et al. The role of cytokine profile and lymphocyte subsets in the severity of coronavirus disease 2019 (COVID‐19): a systematic review and meta‐analysis. Life Sci. 2020;258:118167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roschewski M, Lionakis MS, Sharman JP, et al. Inhibition of Bruton tyrosine kinase in patients with severe COVID‐19. Sci Immunol. 2020;5(48):eabd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Colafrancesco S, Alessandri C, Conti F, Priori R. COVID‐19 gone bad: a new character in the spectrum of the hyperferritinemic syndrome? Autoimmun Rev. 2020;19:102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takeshita H, Yasuda T, Nakajima T, Hosomi O, Nakashima Y, Kishi K. Mouse deoxyribonuclease I (DNase I): biochemical and immunological characterization, cDNA structure and tissue distribution. Biochem Mol Biol Int. 1997;42:65‐75. [DOI] [PubMed] [Google Scholar]
- 56. Ueki M, Takeshita H, Fujihara J, et al. Caucasian‐specific allele in non‐synonymous single nucleotide polymorphisms of the gene encoding deoxyribonuclease I‐like 3, potentially relevant to autoimmunity, produces an inactive enzyme. Clin Chim Acta. 2009;407:20‐24. [DOI] [PubMed] [Google Scholar]
- 57. Ueki M, Kimura‐Kataoka K, Takeshita H, et al. Evaluation of all non‐synonymous single nucleotide polymorphisms (SNPs) in the genes encoding human deoxyribonuclease I and I‐like 3 as a functional SNP potentially implicated in autoimmunity. The FEBS Journal. 2014;281:376‐390. [DOI] [PubMed] [Google Scholar]
- 58. Keyel PA. Dnases in health and disease. Dev Biol. 2017;429:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jiménez‐Alcázar M, Rangaswamy C, Panda R, et al. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science. 2017;358:1202‐1206. [DOI] [PubMed] [Google Scholar]
- 60. Chen D, Yang XL, Shen ZB, et al. Significance of neutrophil extracellular trap and its markers in the early diagnosis of community‐acquired pneumonia in children. Zhongguo Dang Dai Er Ke Za Zhi. 2019;21:868‐875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Denning NL, Aziz M, Gurien SD, Wang P. DAMPs and NETs in Sepsis. Front Immunol. 2019;10:2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen L, Long X, Xu Q, et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID‐19 patients. Cell Mol Immunol. 2020;17:992‐994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zitter JN, Maldjian P, Brimacombe M, Fennelly KP. Inhaled Dornase alfa (Pulmozyme) as a noninvasive treatment of atelectasis in mechanically ventilated patients. J Crit Care. 2013;28(218):e1‐e7. [DOI] [PubMed] [Google Scholar]
- 64. Pottecher J, Noll E, Borel M, et al. Protocol for TRAUMADORNASE: a prospective, randomized, multicentre, double‐blinded, placebo‐controlled clinical trial of aerosolized dornase alfa to reduce the incidence of moderate‐to‐severe hypoxaemia in ventilated trauma patients. Trials. 2020;21:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fuchs TA, Jimenez‐Alcazar M, Göbel J, Englert H.Engineered DNAse enzymes and use in therapy . 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S8
Figure S1


