ACE2 is the primary receptor for SARS‐CoV‐2. We demonstrate that lower airway expression of ACE2 is increased in older adults and males. Lower ACE2 expression in epithelial cells also occurs in people with asthma and is associated with reduced furin and increased ADAM‐17 expression. This may partly explain the relative sparing of people with asthma from severe COVID‐19.
See related Editorial

Keywords: bronchial asthma, chronic obstructive pulmonary disease, coronavirus disease, COVID‐19, pandemic, SARS‐CoV‐2, viral infections
ABSTRACT
Background and objective
COVID‐19 is complicated by acute lung injury, and death in some individuals. It is caused by SARS‐CoV‐2 that requires the ACE2 receptor and serine proteases to enter AEC. We determined what factors are associated with ACE2 expression particularly in patients with asthma and COPD.
Methods
We obtained lower AEC from 145 people from two independent cohorts, aged 2–89 years, Newcastle (n = 115) and Perth (n = 30), Australia. The Newcastle cohort was enriched with people with asthma (n = 37) and COPD (n = 38). Gene expression for ACE2 and other genes potentially associated with SARS‐CoV‐2 cell entry was assessed by qPCR, and protein expression was confirmed with immunohistochemistry on endobronchial biopsies and cultured AEC.
Results
Increased gene expression of ACE2 was associated with older age (P = 0.03) and male sex (P = 0.03), but not with pack‐years smoked. When we compared gene expression between adults with asthma, COPD and healthy controls, mean ACE2 expression was lower in asthma patients (P = 0.01). Gene expression of furin, a protease that facilitates viral endocytosis, was also lower in patients with asthma (P = 0.02), while ADAM‐17, a disintegrin that cleaves ACE2 from the surface, was increased (P = 0.02). ACE2 protein expression was also reduced in endobronchial biopsies from asthma patients.
Conclusion
Increased ACE2 expression occurs in older people and males. Asthma patients have reduced expression. Altered ACE2 expression in the lower airway may be an important factor in virus tropism and may in part explain susceptibility factors and why asthma patients are not over‐represented in those with COVID‐19 complications.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- ADAM
a disintegrin and metallopeptidase
- AEC
airway epithelial cell
- ANOVA
analysis of variance
- ARDS
acute respiratory distress syndrome
- BDP
beclomethasone dipropionate
- COPD
chronic obstructive pulmonary disease
- COVID
coronavirus disease
- CTL
cathepsin‐L
- FEV1
forced expiratory volume in 1 s
- GINA
Global initiative for Asthma
- GOLD
Global Initiative for Chronic Obstructive Lung Disease
- HC
healthy control
- HSPA
heat shock protein
- ICS
inhaled corticosteroid
- IL
interleukin
- PCR
polymerase chain reaction
- PI4KB
phosphatidylinositol 4‐kinase beta
- qPCR
quantitative PCR
- SARS‐CoV
severe acute respiratory syndrome‐associated coronavirus
- TMPRSS
transmembrane serine protease
INTRODUCTION
In December 2019, an outbreak of a novel respiratory illness resulting in severe disease and respiratory failure emerged in Wuhan, China, and has since spread to become the major pandemic in living memory, now known as coronavirus disease (COVID)‐19. 1 The virus responsible was rapidly identified as a previously unknown beta‐coronavirus, closely related to severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV) and is now identified as SARS‐CoV‐2. 2 , 3 The virus binds to the angiotensin‐converting enzyme 2 (ACE2) receptor and requires the transmembrane serine protease 2 (TMPRSS2) to cleave the viral spike protein in order to enter a cell. 3 This step appears to be facilitated by endosomal proteases such as cathepsin‐L (CTL) and enhanced by the protein furin 4 , 5 ; then, the virus enters the host cell by endocytosis.
COVID‐19 has had a devastating impact, now infecting more than 30 million and is responsible for at least 100 000 000 deaths worldwide (World Health Organization (WHO) reports). 6 Infection begins in the upper respiratory tract and while the majority of people experience a mild disease course, 10–15% have more severe disease with progression to infection of the lower airways, resulting in pneumonia, acute respiratory distress syndrome (ARDS) and death. 7 , 8 People with chronic respiratory disease, especially asthma and chronic obstructive pulmonary disease (COPD), are usually at heightened risk of complications from acute respiratory viral infections. 9 This is, however, not clearly the case with COVID‐19, while there appears to be a heightened risk for active smokers, 10 , 11 those with asthma do not appear to be over‐represented. Age and male sex appear to be associated with worse outcomes, and children generally experience only mild illness. 12 , 13 , 14 One factor that may predispose to more severe disease and pneumonia is the level of expression of the ACE2 receptor in airway epithelial cells (AEC) of the lower respiratory tract allowing infection to spread more easily from the upper airway, resulting in a more intense lower airway immune response and more severe clinical disease.
We sought to determine the expression of ACE2 and other genes that are crucial for viral entry in primary human AEC in a cohort of healthy individuals and in people with asthma and COPD.
METHODS
Participants
Subjects for this study had all been previously and independently recruited to provide lower airway samples for studies of chronic asthma or COPD and comprise two independent cohorts based in Newcastle and Perth, Australia (for more details, see Appendix S1 in Supplementary Information). The study was approved by the Hunter New England Human Research Ethics Committee (Reference No. 05/08/10/3.09), Perth Children's Hospital Ethics Committee (RGS1470) and St John of Gods Human Ethics Committee (SJOG#901), and all participants provided written informed consent.
Gene expression by qPCR
Detailed description of the quantitative PCR (qPCR) is provided in Table S1 (Supplementary Information). Results were calculated using 2−ΔΔCt (where Ct is the threshold cycle) relative to the mean ΔCt of the healthy control group as described previously. 15
Immunohistochemistry and immunofluorescence
Formalin‐fixed paraffin‐embedded endobronchial biopsies from subjects of the Newcastle cohort were taken from the third‐ to fourth‐generation airways. They were sectioned and rehydrated for immunofluorescence and immunohistochemistry analysis. Detailed methods are provided in Appendix S1 (Supplementary Information).
Statistical analysis
Data were analysed using Stata software version 15 (StataCorp., College Station, TX, USA) or with GraphPad Prism 8 (San Diego, CA, USA). Results are reported as mean (SD) or median (interquartile range), unless otherwise stated. Continuous measures were analysed using Student's t‐test or one‐way analysis of variance (ANOVA) as appropriate. Spearman's correlation coefficients were calculated for univariate association between continuous variables. Linear regression models were used to assess the associations between the variables of interest and ACE2 expression. We initially examined demographic variables alone (bivariate associations and then a multivariable model), and then the clinical characteristics (presence of asthma and COPD) for which the sample was enriched; we also assessed models that included cardiovascular disease, hypertension and diabetes as simple bivariate regression models as well as a model including demographic variables. Due to the relatively low sample size, rather than fitting a full multivariable model and risk over‐fitting, we used a Random Forest to identify the important variables from the collective set. Non‐parametric bootstrapping was used to assess the reliability of results by fitting the model on each of the 50 different samples taken with replacement from the original sample. Regression analyses were performed using R version 4.0.0 (R Core Team (2020), Vienna, Austria).
RESULTS
We assessed gene expression in primary bronchial epithelial cells (pBEC) from participants of the Newcastle cohort (Table 1) and details provided by disease group are summarized separately (Table 2).
Table 1.
Participants of the Newcastle cohort
| Number | 116 |
| Age | 62 (13.5) |
| Sex | Male 49 (42%), female 67 (58%) |
| Smoking status | Never 48 (41%), ex‐smoker 59 (51%), current 9 (8%) |
| BMI | 29.6 (6.6) |
| Asthma | 39 (33.6%) |
| COPD | 38 (33%) |
| Atopy | 23/92 (33%) |
| Cardiovascular disease | 8 (7%) |
| Hypertension | 22 (19%) |
| Diabetes | 12 (10%) |
| Malignancy | 9 (8%) |
| Gastro‐oesophageal reflux | 55 (47%) |
| Prescribed regular inhaled corticosteroids | 55 (47%) |
| Prescribed regular oral corticosteroids | 5 (4%) |
| Prescribed regular ACE or ARB inhibitor | 12 (10%) |
| Prescribed regular proton pump inhibitor | 23 (20%) |
Fourteen subjects had no documented allergy by either skin prick test or radioimmunosorbent assay.
ACE, angiotensin‐converting enzyme; ARB, angiotensin II receptor blocker; BMI, body mass index; COPD, chronic obstructive pulmonary disease.
Table 2.
Clinical details by disease groups in the Newcastle cohort
| Healthy | Asthma | COPD | Analysis | |
|---|---|---|---|---|
| Number | 39 | 39 | 38 | NA |
| Age years (mean and SD) | 64 (55.1, 71.5) | 53.2 (14.8) | 68.2 (8.2) | P = 0.8 |
| Sex, male:female | 11:28 | 16:23 | 16:22 | P = 0.7 |
| Pack‐years smoked (mean and SD) | 0 | 0 | 38.6 (34.9) | P < 0.001 |
| FEV1pp (median and interquartile range) | 90 (83.5, 130.1) | 64.5 (49.8, 86.3) | 46.5 (40, 64) | P < 0.001 † |
| Regular ICS (%) | 0 | 39 (100%) | 17 (42%) | P = 0.001 |
| ICS, median BDP/day, (median and interquartile range) | 0 | 1600 (800, 2000) | 1600 (800, 2000) | P = 0.8 ‡ |
| Atopy | 0 | 19 (49%) | 4 (10.1%) | P < 0.001 |
| Airway eosinophils, % total cell count (median and interquartile range) | 1.0 (0.25, 1.6) | 2.5 (1, 6.9) | 0.9 (0.5, 10.1) | P = 0.002 § |
| GINA | ||||
| GINA step 3 | 16 | |||
| GINA step 4 | 3 | |||
| GINA step 5 | 20 | |||
| TH2 high disease | NA | 28 (72%) | NA | |
| GOLD | ||||
| GOLD 2 | 15 | |||
| GOLD 3 | 19 | |||
| GOLD 4 | 3 |
Pack‐years smoked, 1 packet of 25 cigarettes or equivalent per day for 1 year. ICS dose is expressed as equivalent BDP in mcg per day, where 1 μg of BDP is equivalent to 1 mcg budesonide, 0.5 mcg fluticasone propionate and 0.4 mcg ciclosenide. TH2 high disease was defined as evidence of either atopy or elevated airway eosinophils (2.75% of the total cell count).
FEV1 was significantly lower in participants with COPD compared to both healthy controls (P < 0.001) and participants with asthma (P = 0.01). FEV1 was lower in participants with asthma compared to healthy controls (P < 0.001) (Kruskal–Wallis with Dunn's comparison).
ICS dose was different between those with asthma and COPD.
Participants with asthma had higher airway eosinophils compared to healthy controls (Kruskal–Wallis with Dunn's comparison).
BDP, beclomethasone dipropionate; COPD, chronic obstructive pulmonary disease; FEV1, forced expiratory volume in 1 s; FEV1pp, FEV1 percentage of the predicted value; ICS, inhaled corticosteroid; GINA, Global initiative for Asthma; GOLD, Global Initiative for Chronic Obstructive Lung Disease.
Gene expression of ACE2 and related genes
Expression of the following genes was assessed: ACE2, the human receptor that SARS‐CoV‐2 is known to bind, TMPRSS2, CTL and furin, proteases known to be crucial in cleaving the SARS‐CoV‐2 spike protein and facilitating viral entry. 3 , 4 , 5 , 16 In addition, we examined the expression of genes that may be involved in facilitating SARS‐CoV‐2 entry: TMPRSS11A/D that may also be involved in spike protein cleavage 17 ; a disintegrin and metallopeptidase (ADAM)‐10 and ADAM‐17, which mediate shedding of ACE2 from the cell surface and promote uptake of SARS‐CoV 18 , 19 ; and phosphatidylinositol 4‐kinase beta (PI4KB) that creates a lipid microenvironment at the cell surface required for the entry of SARS‐CoV‐1 as well as other RNA viruses. 20 We also assessed the gene expression of proposed alternate receptors for the SARS‐CoV‐2 notably CD147, heat shock protein (HSPA) 5 or GRP78 as potential alternative binding sites for the virus. 21 , 22
We first determined if there were differences in gene expression between subjects who were healthy controls or had a diagnosis of asthma or COPD (Fig. 1). ACE2 expression was significantly lower in those with asthma (mean difference fold change ddCT with healthy controls, 0.23, P = 0.01). Furin was also lower in subjects with asthma (mean difference: 1.1, P = 0.02), but there were no differences between groups in the expression of TMPRSS2 or CTSL. There were no differences in TMPRSS11A/D. ADAM‐10 was higher in COPD (mean difference: 0.15, P = 0.02) and ADAM‐17 was higher in subjects with asthma (mean difference: 0.18, P = 0.02). The groups were not different in CD147 or PI4KB expression. HSPA5 expression was higher in subjects with asthma (mean difference: 0.3, P = 0.02).
Figure 1.
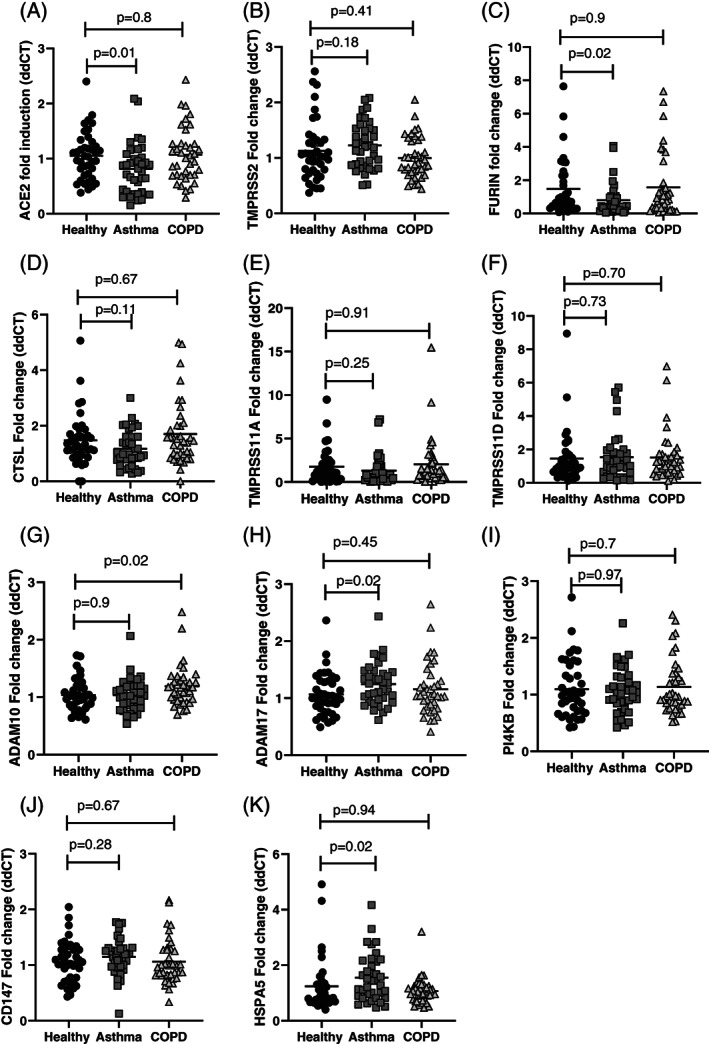
All values are represented individually. The mean and SD are also presented. Differences between the groups were assessed using one‐way analysis of variance (ANOVA), with Dunnett's multiple comparison test. If the ANOVA was significant (P < 0.05), the P‐values are labelled for the group that is different from the healthy controls.
Age and male sex are associated with higher gene expression
Older age was associated with increased ACE2 (Fig. 2), although the correlation was not strong (Pearson's rho = 0.2, P = 0.02). There was no correlation with body mass index (BMI), pack‐years smoked or dose of inhaled corticosteroids (ICS) with any of the genes assessed (data not shown). There were no differences seen in gene expression when analysed separately by obesity (BMI > 30); atopy; history of hypertension, cardiovascular disease or diabetes; use of inhaled or oral corticosteroids; use of ACE inhibitors, ACE2 or proton pump inhibitors (data not shown); and evidence of atopy or airway eosinophil count (data not shown).
Figure 2.
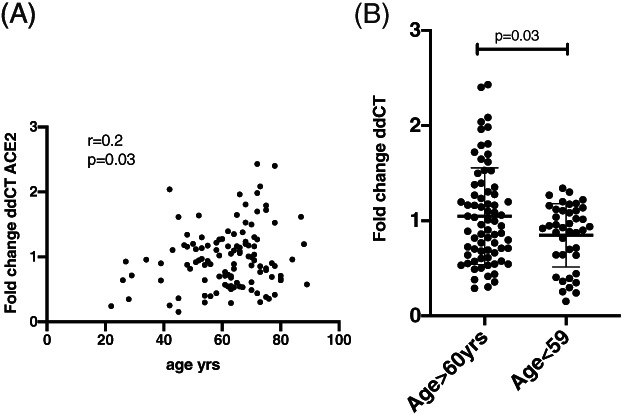
(A) Data from the Newcastle cohort (n = 116), a two‐way comparison between angiotensin‐converting enzyme 2 (ACE2) expression and age. The univariate correlation was measured using Pearson's correlation coefficient with a two‐sided test for significance. (B) Same data with subjects divided by age group. The mean and SD are represented. Differences between the two groups were analysed using an unpaired t‐test.
When we investigated samples from the Perth cohort, there were no differences in ACE2 gene expression between children and adults, but the negative correlation with age still held. We compared ACE2 gene expression between the Perth participants, the Newcastle healthy controls and participants with asthma. While there were no differences between the healthy participant groups, despite the younger age of the Perth healthy controls compared to those from Newcastle, those with asthma still had significantly less ACE2 expression (Fig. 3).
Figure 3.
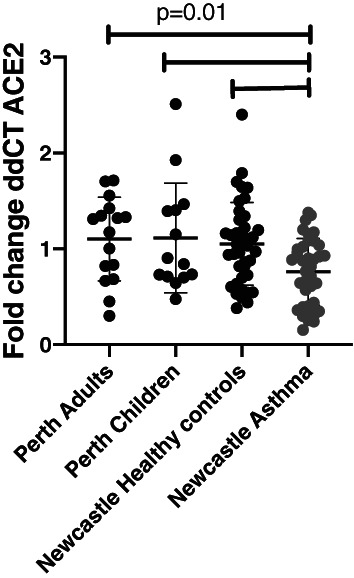
Data represent angiotensin‐converting enzyme 2 (ACE2) gene expression seen in airway epithelial cells (AEC) from Perth adults (n = 16), Perth children (n = 14), Newcastle adult healthy controls (n = 40) and Newcastle asthma (n = 37). Differences between the groups were assessed using one‐way analysis of variance (ANOVA), with Dunnett's multiple comparison test.
We performed a bivariate regression analysis for each variable, adjusting for age, sex, smoking, BMI, asthma, COPD, atopy, hypertension, diabetes, cardiac disease, ICS, prednisone and use of ACE2 inhibitors in all subjects. Age and sex had significant associations with ACE2 expression, and collectively the demographics explained 7.5% of the variation in ACE2 (Table 3). Asthma was also associated with lower ACE2 expression; however, this association was not preserved when adjusting for age, sex and smoking.
Table 3.
Multivariate analysis
| Combined model coefficients | Estimate | SE | Probability > [T] |
|---|---|---|---|
| Intercept | 0.788732 | 0.238309 | <0.01 |
| Age | 0.004731 | 0.003588 | 0.04 |
| Sex | −0.18289 | 0.087999 | 0.04 |
| Pack‐years smoked | 0.002361 | 0.001627 | 0.15 |
| Asthma | −0.158238 | 0.103480 | 0.13 |
ACE2 expression was adjusted for age, sex, smoking, pack‐years smoked and weight. Multiple R‐squared: 0.1185, adjusted R‐squared: 0.08619, degrees of freedom, P = 0.008.
ACE2, angiotensin‐converting enzyme 2.
Expression of ACE2 in the airway epithelium
We stained endobronchial biopsies from five healthy controls, five subjects with asthma and five subjects with COPD, with representative slides displayed (Fig. 4). In comparing slides between healthy controls aged less than 40 years (n = 5) and greater than 65 years (n = 5), there appeared to be a marked difference in ACE2 expression in the epithelium in older donors (Fig. 4A,B). Comparing healthy controls to asthma, there again appeared to be less ACE2 in subjects with asthma (Fig. 4A,C). In healthy controls and subjects with COPD, there was appreciably greater ACE2 observed in the epithelium and subepithelium compared to subjects with asthma (Fig. 4D–F). Most ACE2 expression appeared to be associated with ciliated as well as basal AEC. It was concentrated at the apical end of the AEC, although moderate nuclear and cytoplasmic staining was also observed. The quantification of ACE2 expression (%) in the epithelium with immunohistochemistry staining (IHC) staining indicated less ACE2 expression in subjects with asthma compared to healthy controls and subjects with COPD (Fig. 4G). The percentage (%) ACE2 expression in epithelium was significantly lower in subjects with asthma (median (range): 6.654 (3.955–8.667)) than in subjects with COPD (median (range): 11.084 (9.934–12.136); P = 0.008) and in healthy control (median (range): 9.063 (7.075–12.213); P = 0.05). ACE2 expression (%) in subjects with COPD appeared higher than in controls but failed to reach significance.
Figure 4.
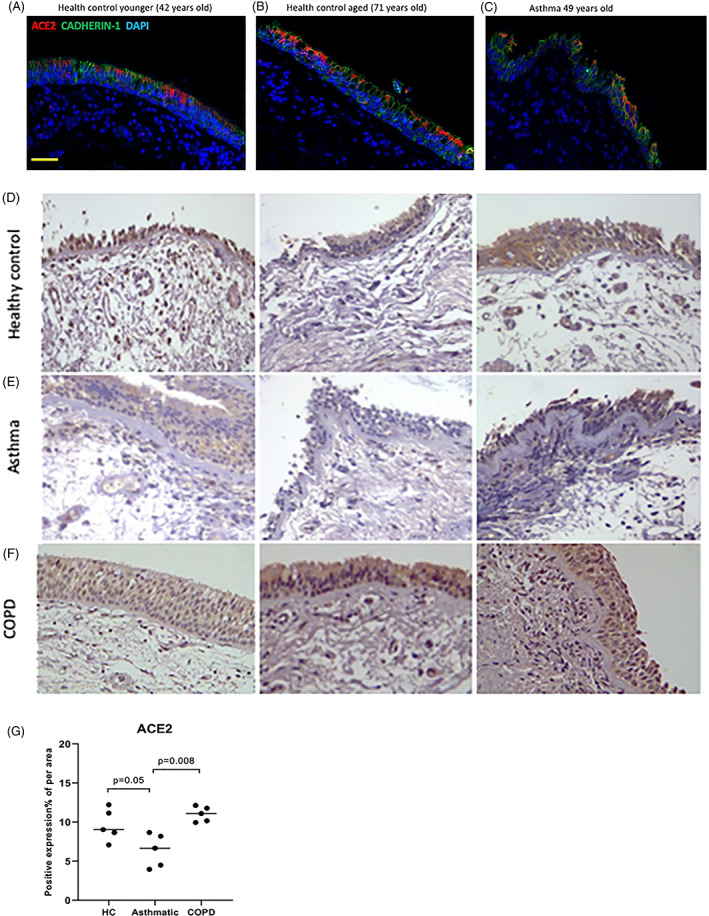
(A–C) Representative immunofluorescent images of formalin‐fixed paraffin‐embedded endobronchial sections showing protein expression of CADHERIN1 (green) and angiotensin‐converting enzyme 2 (ACE2) (red) in healthy controls (HC) (A) under 40 years compared to (B) a HC over 40 years and (C) a participant with asthma (aged 49 years). (D–F) Representative images of endobronchial biopsies stained for ACE2. The images (D) show HC compared to patients with (E) asthma and (F) chronic obstructive pulmonary disease (COPD) (magnification, ×40). (G) The percentage (%) of ACE2 expression measured in the epithelium of HC compared to patients with asthma and COPD. Patients with asthma had significantly lower ACE2 expression compared to HC and COPD patients. There appeared to be a trend for COPD airway cells to have a higher expression compared to HC, this just failed to be significant.
DISCUSSION
The COVID‐19 pandemic represents a unique and critical challenge for the world and a greater understanding of the mechanisms of disease overall and in different patient groups is crucial in determining effective therapeutic responses that so far remain elusive. We found ACE2 gene expression is greater in lower airway cells in older individuals and males. We determined that ACE2 gene and protein levels were lower in subjects with asthma. This was also associated with reduced furin expression and increased ADAM‐17 expression in the epithelium from subjects with asthma, demonstrating a potential mechanism through which IL‐13‐induced airway inflammation may downregulate ACE2 expression.
Viral respiratory tract infections are common triggers of acute asthma, 23 and initially there was great concern that people with asthma would be highly susceptible to SARS‐CoV‐2. Early reports from China 7 and Italy 14 did not suggest that people with asthma were over‐represented in those hospitalized from SARS‐CoV‐2. More recently, the largest cohort review of more than 17 000 hospitalized cases of COVID‐19 from the UK has been pre‐published demonstrating that older age, being male and social deprivation were strongly associated with mortality. 24 While chronic lung disease, including COPD, was associated with mortality, only those with asthma who had required the recent use of oral corticosteroids had increased mortality. 24 We found increased ACE2 was associated with older age and male sex in two independent cohorts in keeping with the epidemiological studies. This suggests that increased lower airway ACE2 expression may be associated with increased risk of more severe disease, and this is consistent with increased ACE2 expression in the small airway epithelium from current smoker and those with COPD. 11 , 25 Asthma alone is not over‐represented in people with COVID‐19 nor is it associated with greater disease severity. This raises the question as to why people with asthma are not at increased susceptibility to SARS‐CoV‐2.
SARS‐CoV‐2 is a highly infectious virus transmitted by airway droplets. 26 Cell entry is the first step necessary and cell tropism is key to infectivity. SARS‐CoV‐2 uses the ACE2 receptor as its point of entry and this receptor has been found to be highly expressed in the upper airway. 27 , 28 While ACE2 is expressed in the lower respiratory tract, in keeping with our results, there are relatively lower levels of expression compared to the upper airway. SARS‐CoV‐2 is able to first infect ciliated columnar epithelial cells and then appears to spread to surrounding secretory cells. 29 ACE2 expression is critical for lower airway infection to occur, and mice deficient in the receptor had greatly reduced SARS‐CoV infection and replication. 30 Thus, it is likely that the level of ACE2 expression in lower airway cells may be an important factor in determining more severe infection.
The entry of SARS‐CoV and SARS‐CoV‐2 into cells is facilitated by the interaction between viral S‐protein with the extracellular domain of the transmembrane ACE2 protein. Endocytosis of the virus then results in downregulation of surface ACE2 expression. 3 , 31 , 32 Once infection is established, SARS‐CoV has been shown to upregulate ADAM‐17 that facilitates further viral endocytosis with ACE2, while knockdown of ADAM‐17 by siRNA prior to infection substantially attenuates SARS‐CoV entry. 33 In 12 selected patients with COVID‐19 and the 6 of these who developed ARDS, circulating angiotensin II levels were markedly elevated, which correlated with viral load as measured by PCR and the degree of pathology. This suggests a link in SARS‐CoV‐2 infection with tissue ACE2 downregulation facilitating the development of ARDS. 34 The spread of infection to the lower airways is reflected in the clinical presentation of the illness, with the first 4–5 days often asymptomatic, followed by dry cough and fever, with viral load peaking at day 10, with those who go onto to develop severe disease, ARDS and severe systemic immune responses 7–10 days after infection. 7 , 35
The biologic importance of reduced ACE2 gene expression and protein levels that we find in asthma is also supported by the observation of reduced expression of furin, a protease that together with TMPRSS2 has been found to be critical in facilitating SARS‐CoV‐2 endocytosis. 16 We also found enhanced expression of ADAM‐17. Phosphorylation of ACE2 enhances its catalytic activity and increases its shedding from the epithelial surface. 36 The role of ADAM‐17 is not well defined in asthma but its presence in AEC has been proposed to be anti‐inflammatory and linked to increased airway remodelling and goblet cell hyperplasia. 37 Enhanced ADAM‐17 activity is a potential mechanism by which ACE2 receptor expression could be downregulated specifically in asthma.
Three other studies have examined ACE2 mRNA expression in epithelial cells from people with asthma. Saheb Sharif‐Askari et al. examined Biobanked Affymetrix (Santa Clara CA, USA) libraries of bronchial epithelial brushings but did not find reduced ACE2 expression in asthma, although they did find an increase in COPD samples. 38 Similar to us, Jackson et al. showed reduced ACE2 expression in nasal samples from children and adults and bronchial epithelial cells from steroid‐naïve mild allergic asthma patients. 39 They demonstrated a negative correlation of ACE2 expression with type 2 airway inflammation, exhaled nitric oxide and nasal epithelial IL‐13 expression. We demonstrate ACE2 expression is seen equally in those with non‐allergic and more severe asthma on regular ICS. Interestingly, it has been shown that IL‐13‐induced epithelial proliferation is mediated by ADAM‐17. 40 There is now evidence that treatment of AEC cultures with IL‐13 reduces the expression of ACE2. 41 This could provide a unifying mechanism by which the effect of IL‐13 on tissue remodelling of the airway epithelium as it occurs in severe allergic disease and asthma may lead to downregulation of ACE2 expression in the lower airways.
In contrast to our findings, Bradding et al. examined bronchial brushings and endobronchial biopsies for the expression of ACE2, TMPRSS2 and furin. 42 Results were available from 356 patients (88 healthy volunteers and 268 patients with asthma (mild to severe)). They did not see any difference between participants with asthma or healthy controls but did find weak, although statistically significant positive, correlations with a TH‐17 gene signature and negative correlation with a TH‐2 gene signature. In contrast, we saw reduced ACE2 expression in patients with asthma but did not find a difference in ACE2 expression with atopy or bronchial lavage eosinophil count. The cells we have measured ACE2 expression in had been cultured without stimulation, as opposed to endobronchial brushings. Our endobronchial biopsies were collected in a similar way.
An important possibility remains that ICS may reduce ACE2 expression. Increased expression of ACE2 and TMPRSS2 was also found in over 300 induced sputum samples from a cohort with severe asthma, although the use of inhaled steroids was associated with lower ACE2 expression. 43 These results however are from induced sputum and the RNA content will have largely come from immune cells and not the airway epithelium as is the case for us. Belgian researchers have seen a similar trend with reduced ACE2 expression in people with COPD or asthma/COPD overlap on regular ICS. 44 While a pre‐print report using in vitro and animal models of ICS in COPD also suggests that they may reduce ACE2 expression. 45 All our participants with asthma were on ICS and 42% of those with COPD. We did not see a negative correlation between ICS dose and ACE2 expression and subjects with airways disease on ICS independent of asthma did not have lower ACE2 expression. Leung et al. demonstrated increased ACE2 expression from resected lung tissue and endobronchial brushings from 26 subjects who were current or former smokers as well as subjects with COPD, suggesting increased expression in COPD. 25 This relationship is likely to be quite complex and observational data in asthma and COPD will inevitably be confounded by ICS use. It is important to note that neither asthma nor ICS use are associated with increased ACE2 expression and on balance the evidence suggests they may be protective. It will take further mechanistic studies to answer these questions. There are now four actively recruiting clinical trials that are assessing if ICS are protective in COVID‐19.
Our study has some weaknesses. Subjects were recruited to study chronic airways disease and were enriched for subjects with moderate to severe asthma and COPD. Our numbers overall were not large enough to give a clear answer with multivariate analysis and a large proportion with both asthma and COPD were on ICS.
We present the largest pathology study that examined ACE2 gene and protein expression from cultured lower airway bronchial epithelial cells enriched with people with asthma and COPD. We have confirmed the presence of ACE2 in samples from children and adults in two independent cohorts and determined with bivariate analysis that age and male sex are associated with increased lower airway ACE2 expression. Our data provide further evidence for the differences observed in RNA expression confirming reduced ACE2 protein levels in subjects with asthma by immunohistochemistry. We are now the second group to find that people with asthma have reduced ACE2 expression in lower airway cells. We propose that this may provide a degree of protection from the complications of SARS‐CoV‐2. The next step is to determine from current epidemiological studies of COVID transmission what is the risk of infection in people with asthma and whether they are relatively protected. We find reduced ACE2 expression associated with reduced furin expression in subjects with asthma and observe increased expression of ADAM‐17. This provides a potential mechanism by which chronic airway inflammation associated with asthma and the action of IL‐13 on the airway epithelium could downregulate ACE2 expression via ADAM‐17 prior to infection. Further studies should now be performed to define this mechanism and determine whether this can be manipulated as a therapeutic target to reduce SARS‐CoV‐2 infection of the airway epithelium.
Author contributions
Conceptualization: P.A.B.W., P.S.P., G.K., S.S.S., S.M.S., P.M.H., A.K. Data curation: P.A.B.W., P.S.P., G.K., A.A., E.N.S., L.W.G., M.S.E., A.R., P.V., A.C.‐Y.H., K.L., A.K. Formal analysis: P.A.B.W., L.C., E.N.S., M.S.E., C.O., A.K. Investigation: K.N., A.A., L.C., L.W.G., S.S.S., W.L., N.B., A.R., P.V., A.C.‐Y.H., K.L., T.I. Methodology: P.S.P., G.K., K.N., E.N.S., L.W.G., S.S.S., W.L., M.S.E., A.R., A.C.‐Y.H., K.L., T.I. Project administration: P.A.B.W., K.N., S.M.S., P.M.H., A.K. Resources: P.A.B.W., E.N.S., S.S.S., N.B., S.M.S., A.K. Supervision: P.A.B.W. Software: L.W.G. Validation: K.N., A.A., E.N.S., L.W.G., M.S.E., A.K. Visualization: K.N., L.C. Writing—original draft: P.A.B.W., P.S.P., G.K., K.N., L.C., E.N.S., L.W.G., S.S.S., M.S.E., C.O., N.B., A.R., K.L., T.I., S.M.S., A.K. Writing—review and editing: P.A.B.W., P.S.P., G.K., K.N., A.A., L.W.G., S.S.S., M.S.E., C.O., A.R., T.I., S.M.S., P.M.H., A.K.
Supporting information
Appendix S1 Supplementary methods.
Table S1 Detailed list of genes assessed by PCR.
Visual Abstract ACE2 Expression is elevated in airway epithelial cells from aged and male donors but reduced in asthma.
Acknowledgements
S.S.S. is supported by the Clifford Craig Foundation Launceston General Hospital and the Rebecca L. Cooper Medical Research Foundation. P.M.H. is funded by the Clifford Craig Foundation Launceston General Hospital, a Rebecca L. Cooper Fellowship and grants from the National Health and Medical Research Foundation Council (NHMRC) of Australia (1079187 and 1175134) and SPHERE.
Wark PAB, Pathinayake PS, Kaiko G, et al. ACE2 expression is elevated in airway epithelial cells from older and male healthy individuals but reduced in asthma. Respirology. 2021;26:442–451. 10.1111/resp.14003
Associate Editor: Judith Mak; Senior Editor: Philip Bardin
See related Editorial
REFERENCES
- 1. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020; 382: 727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoffmann M, Kleine‐Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat. Commun. 2020; 11: 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS‐CoV‐2. Proc. Natl. Acad. Sci. U. S. A. 2020; 117: 11727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect. Dis. 2020; 20: 533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395: 1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodriguez‐Morales AJ, Cardona‐Ospina JA, Gutiérrez‐Ocampo E, Villamizar‐Peña R, Holguin‐Rivera Y, Escalera‐Antezana JP, Alvarado‐Arnez LE, Bonilla‐Aldana DK, Franco‐Paredes C, Henao‐Martinez AF et al. Clinical, laboratory and imaging features of COVID‐19: a systematic review and meta‐analysis. Travel Med. Infect. Dis. 2020; 34: 101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loubet P, Samih‐Lenzi N, Galtier F, Vanhems P, Loulergue P, Duval X, Jouneau S, Postil D, Rogez S, Valette M et al. Factors associated with poor outcomes among adults hospitalized for influenza in France: a three‐year prospective multicenter study. J. Clin. Virol. 2016; 79: 68–73. [DOI] [PubMed] [Google Scholar]
- 10. Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, Liu XQ, Chen RC, Tang CL, Wang T et al. Comorbidity and its impact on 1590 patients with Covid‐19 in China: a nationwide analysis. Eur. Respir. J. 2020; 55: 2000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brake SJ, Barnsley K, Lu W, McAlinden KD, Eapen MS, Sohal SS. Smoking upregulates angiotensin‐converting enzyme‐2 receptor: a potential adhesion site for novel coronavirus SARS‐CoV‐2 (Covid‐19). J. Clin. Med. 2020; 9(3): 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du R‐H, Liang L‐R, Yang C‐Q, Wang W, Cao T‐Z, Li M, Guo G‐Y, Du J, Zheng C‐L, Zhu Q et al. Predictors of mortality for patients with COVID‐19 pneumonia caused by SARS‐CoV‐2: a prospective cohort study. Eur. Respir. J. 2020; 55: 2000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host susceptibility to severe COVID‐19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit. Care 2020; 24: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, Cereda D, Coluccello A, Foti G, Fumagalli R et al. Baseline characteristics and outcomes of 1591 patients infected with SARS‐CoV‐2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020; 323: 1574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2̂(−delta CT) method for quantitative real‐time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013; 3: 71–85. [PMC free article] [PubMed] [Google Scholar]
- 16. Bestle D, Heindl MR, Limburg H, van Thuy VL, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O et al. TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS‐CoV‐2 in human airway epithelial cells and provide promising drug targets. bioRxiv [Preprint] posted on 15 April 2020. 10.1101/2020.04.15.042085. [DOI] [Google Scholar]
- 17. Bertram S, Glowacka I, Müller MA, Lavender H, Gnirss K, Nehlmeier I, Niemeyer D, He Y, Simmons G, Drosten C et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin‐like protease. J. Virol. 2011; 85: 13363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia HP, Look DC, Tan P, Shi L, Hickey M, Gakhar L, Chappell MC, Wohlford‐Lenane C, McCray PB Jr. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009; 297: L84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haga S, Nagata N, Okamura T, Yamamoto N, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS‐CoV are candidate antiviral compounds. Antiviral Res. 2010; 85: 551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang N, Ma P, Lang J, Zhang Y, Deng J, Ju X, Zhang G, Jiang C. Phosphatidylinositol 4‐kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike‐mediated cell entry. J. Biol. Chem. 2012; 287: 8457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang K, Chen W, Zhou Y‐S, Lian J‐Q, Zhang Z, Du P, Gong L, Zhang Y, Cui H‐Y, Geng J‐J et al. SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. bioRxiv [Preprint] posted on 14 March 2020. 10.1101/2020.03.14.988345. [DOI] [Google Scholar]
- 22. Chu H, Chan C‐M, Zhang X, Wang Y, Yuan S, Zhou J, Au‐Yeung RK‐H, Sze K‐H, Yang D, Shuai H et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018; 293: 11709–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansbro NG, Horvat JC, Wark PA, Hansbro PM. Understanding the mechanisms of viral induced asthma: new therapeutic directions. Pharmacol. Ther. 2008; 117: 313–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williamson E, Walker AJ, Bhaskaran KJ, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P et al. OpenSAFELY: factors associated with COVID‐19‐related hospital death in the linked electronic health records of 17 million adult NHS patients. medRxiv [Preprint] Posted on 7 May 2020. 10.1101/2020.05.06.20092999. [DOI] [Google Scholar]
- 25. Leung JM, Yang CX, Tam A, Shaipanich T, Hackett TL, Singhera GK, Dorscheid DR, Sin DD. ACE‐2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID‐19. medRxiv [Preprint] Posted on 23 March 2020. 10.1101/2020.03.18.20038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanche S, Lin YT, Xu C, Romero‐Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020; 26: 1470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera‐López C, Maatz H, Reichart D, Sampaziotis F et al.; HCA Lung Biological Network. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020; 26: 681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, Li T, Chen Q. High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020; 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ravindra NG, Alfajaro MM, Gasque V, Wei J, Filler RB, Huston NC, Wan H, Szigeti‐Buck K, Wang B, Montgomery RR et al. Single‐cell longitudinal analysis of SARS‐CoV‐2 infection in human bronchial epithelial cells. bioRxiv [Preprint] Posted on 13 July 2020. 10.1101/2020.05.06.081695. [DOI] [Google Scholar]
- 30. Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus‐induced lung injury. Nat. Med. 2005; 11: 875–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426: 450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dijkman R, Jebbink MF, Deijs M, Milewska A, Pyrc K, Buelow E, van der Bijl A, van der Hoek L. Replication‐dependent downregulation of cellular angiotensin‐converting enzyme 2 protein expression by human coronavirus NL63. J. Gen. Virol. 2012; 93: 1924–9. [DOI] [PubMed] [Google Scholar]
- 33. Haga S, Yamamoto N, Nakai‐Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF‐alpha‐converting enzyme by the spike protein of SARS‐CoV and ACE2 induces TNF‐alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. U. S. A. 2008; 105: 7809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, Wang Z, Li J, Li J, Feng C et al. Clinical and biochemical indexes from 2019‐nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020; 63: 364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020; 382: 1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel VB, Clarke N, Wang Z, Fan D, Parajuli N, Basu R, Putko B, Kassiri Z, Turner AJ, Oudit GY. Angiotensin II induced proteolytic cleavage of myocardial ACE2 is mediated by TACE/ADAM‐17: a positive feedback mechanism in the RAS. J. Mol. Cell. Cardiol. 2014; 66: 167–76. [DOI] [PubMed] [Google Scholar]
- 37. Dreymueller D, Uhlig S, Ludwig A. ADAM‐family metalloproteinases in lung inflammation: potential therapeutic targets. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015; 308: L325–43. [DOI] [PubMed] [Google Scholar]
- 38. Saheb Sharif‐Askari N, Saheb Sharif‐Askari F, Alabed M, Temsah M‐H, Al Heialy S, Hamid Q, Halwani R. Airways expression of SARS‐CoV‐2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol. Ther. Methods Clin. Dev. 2020; 18: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackson DJ, Busse WW, Bacharier LB, Kattan M, O'Connor GT, Wood RA, Visness CM, Durham SR, Larson D, Esnault S et al. Association of respiratory allergy, asthma, and expression of the SARS‐CoV‐2 receptor ACE2. J. Allergy Clin. Immunol. 2020; 146: 203–6.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Booth BW, Sandifer T, Martin EL, Martin LD. IL‐13‐induced proliferation of airway epithelial cells: mediation by intracellular growth factor mobilization and ADAM17. Respir. Res. 2007; 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kimura H, Francisco D, Conway M, Martinez FD, Vercelli D, Polverino F, Billheimer D, Kraft M. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J. Allergy Clin. Immunol. 2020; 146: 80–8.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bradding P, Richardson M, Hinks TSC, Howarth PH, Choy DF, Arron JR, Wenzel SE, Siddiqui S. ACE2, TMPRSS2, and furin gene expression in the airways of people with asthma‐implications for COVID‐19. J. Allergy Clin. Immunol. 2020; 146: 208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peters MC, Sajuthi S, Deford P, Christenson S, Rios CL, Montgomery MT, Woodruff PG, Mauger DT, Erzurum SC, Johansson MW et al. COVID‐19 related genes in sputum cells in asthma: relationship to demographic features and corticosteroids. Am. J. Respir. Crit. Care Med. 2020; 202: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maes T, Bracke K, Brusselle GG. COVID‐19, asthma, and inhaled corticosteroids: another beneficial effect of inhaled corticosteroids? Am. J. Respir. Crit. Care Med. 2020; 202: 8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Finney LJ, Glanville N, Farne H, Aniscenko J, Fenwick P, Kemp SV, Trujillo‐Torralbo M‐B, Calderazzo MA, Wedzicha JA, Mallia P et al. Inhaled corticosteroids downregulate the SARS‐CoV‐2 receptor ACE2 in COPD through suppression of type I interferon. bioRxiv [Preprint] Posted on 15 June 2020. 10.1101/2020.06.13.149039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary methods.
Table S1 Detailed list of genes assessed by PCR.
Visual Abstract ACE2 Expression is elevated in airway epithelial cells from aged and male donors but reduced in asthma.


