Abstract
It is still not fully understood how to predict the future prognosis of patients at the diagnosis coronavirus disease 2019 (COVID‐19) due to the wide clinical range of the disease. We aimed to evaluate whether severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) viral load could predict the clinical course of pediatric patients. This study was conducted retrospectively with medical records of pediatric patients who were tested for SARS‐CoV2 between April 12 and October 25, 2020 in the University of Health Sciences, Ankara Educating and Training Hospital and Hacettepe University Faculty of Medicine. We evaluated 518 pediatric patients diagnosed with COVID‐19 and classified according to severity as asymptomatic (16.2%), mild (59.6%), moderate (20.2%), and critical/severe (3.9%) cases. We analyzed patients in four groups in terms of ages: <4, 5‐9, 10–14, and 15–17 years. There was no statistically significant difference in terms of ∆C t value among age groups, different gender and the existence of underlying diseases in each disease course. The ∆C t values were relatively lower in the first 2 days of symptoms than after days in all groups. Our study has indicated that children with COVID‐19 have similar amount of viral load in all disease courses irrespective of the age and underlying disease. It should be taken into account that, regardless of the severity of the disease, pediatric patients may have a role in the transmission chain.
Keywords: children, COVID‐19, cycle threshold, PCR, SARS‐CoV‐2, viral load
Research Highlights
Children with COVID‐19 can carry similar amount of viral load in all disease courses irrespective of the age and underlying disease.
The viral load has no prediction utility in terms of the clinical course of children with COVID‐19.
Regardless of the severity of the disease, pediatric patients may have a role in the transmission chain.
1. INTRODUCTION
The whole world is still trying to cope with the coronavirus disease 2019 (COVID‐19) pandemic. For doctors in the clinical setting, the first step for an accurate diagnosis is to suspect the disease according to the symptoms and epidemiologic features of patients and secondly to confirm the diagnosis with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RNA in the nasopharyngeal swabs by reverse transcriptase‐polymerase chain reaction (RT‐PCR). Then, the classification of the clinical course and flow up of the patients to predict the future prognosis and decide for the appropriate treatment are of great importance to find the most appropriate management strategy. However, there is still knowledge gap regarding the prognostic marker for patients with COVID‐19 besides, vaccines and effective therapies. 1 , 2 , 3 Even if, SARS‐CoV‐2 causes less severe disease and progresses better in children than in adults, the clinical manifestations of children's COVID‐19 cases ranged from asymptomatic to critical disease course. 4 It is significant for a pediatrician to provide information and predict which patients are at high risk for the deterioration and have severe or critical disease because of the wide clinical range of COVID‐19.
The standard molecular method for the diagnosis of COVID‐19 is the real‐time RT‐PCR. 5 Real‐time PCR cycle threshold (C t) values represent the number of amplification cycles required for the target gene to exceed a threshold level. 6 It was assumed that C t values are an appropriate surrogate for viral load. 7 Some studies showed the correlations between SARS‐CoV‐2 C t values and clinical outcomes of patients. 3 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 Moreover, the correlation was shown between C t value and the progression to severe disease and even mortality. 9 , 10 Additionally, some parameters, such as increased lactate dehydrogenase, decreased lymphocytes and increased high‐sensitivity troponin showed an association between viral loads in studies. 9 , 11 , 12 , 13
Due to the limited data in pediatric patients regarding the viral load, we aimed to examine whether the diagnostic viral load has any effect or association with disease severity in children.
2. MATERIALS AND METHODS
This study was conducted with medical records of pediatric patients aged under 18 years, who were tested for SARS‐CoV‐2 between April 12 and October 25, 2020, in the University of Health Sciences, Ankara, and Training Hospital and Hacettepe University Faculty of Medicine. This study was approved both by the University of Health Science and the Ankara Educating and Training Hospital Review Board, Ankara, Turkey (No:439).
We diagnosed confirmed COVID‐19 cases, according to our national COVID‐19 guidelines which are prepared by Coronavirus Scientific Advisory Board in our country. Suspected cases with positive RT‐PCR against 2019‐nCoV were accepted as confirmed cases. 18 Detection of SARS CoV‐2 RNA in the nasopharyngeal swabs was performed according to the manufacturer's instructions by using a commercial RT‐PCR (The Bio‐Speedy Direct RT‐qPCR SARS CoV‐2 nucleic acid detection kit, Bioeksen, Turkey). The principle of the test is qualitative detection of viral nucleic acid in 40 cycles by RT‐PCR targeting the SARS CoV‐2 specific ORF1ab gene. C t values are inversely related to viral RNA copy numbers. The difference (∆C t) between the sample C t and the positive quality control C t value (C t, sample−C t, ref) was calculated. The SARS‐COV‐2 viral loads of the patients' nasopharyngeal swab samples were estimated with ∆C t values. 10
Data regarding the demographic and clinical characteristics of patients were obtained from the hospital medical records of both hospitals and the records from the Pediatric Infectious Diseases Committee of the hospitals. We categorized the severity of pediatric COVID‐19 cases, based on the clinical characteristics and the results of laboratory examinations and radiologic imaging, as defined by Dong et al. 19
2.1. Statistical analysis
Statistical analyses were performed using SPSS for Windows version 20.0. Descriptive statistics used to define baseline characteristics of cases were mean, median, minimum–maximum, and interquartile ranges (IQRs) for continuous variables and percentages besides numbers for categorical variables. χ 2 and Kruskal–Wallis tests were performed to compare categorical and continuous variables, respectively. The Mann–Whitney U test was used to evaluate non‐normally distributed data. In all the analyses, all tests were two‐tailed and p < .05 was considered significant.
3. RESULTS
3.1. Epidemiological and clinical characteristics
A total of 518 pediatric patients with the diagnosis of COVID‐19 were included in the study. The median (IQR) age of total patients was 11 years (5–14), 48.3% were male and the median day after onset of the symptom to the diagnosis was 1 day, ranging from 0 to 21 days. We analyzed patients in four groups in terms of ages: <4, 5–9, 10–14, and 15–17 years. Most of the patients (31.7%) were between 10 and 14 years old. Fever was present in 50% of cases at any time during the illness. The second most common symptom was cough (34.2% of cases), followed by fatigue or myalgia (22%). Of patients, 8.1% had underlying disease. Patients were classified according to severity, with the percentages of asymptomatic, mild, moderate, and critical or severe cases determined to be 16.2% (n = 84), 59.6% (n = 309), 20.2% (n = 105), and 3.9% (n = 20), respectively. Demographic and clinical characteristics, according to disease severity are summarized in Table 1.
Table 1.
Demographic and clinical data of patients with COVID‐19 accoding to clinical course
| Total (n = 518) | Asymptomatic (n = 84) | Mild (n = 309) | Moderate (n = 105) | Critical/severe (n = 20) | |
|---|---|---|---|---|---|
| Age (years) (median, IQR) | 11 (5–14) | 8 (3–13) | 11 (5–15) | 12 (6–14) | 12 (4–14) |
| Age groups, years | |||||
| <4 | 130 (25.1) | 29 (34.5) | 77 (24.9) | 19 (18.1) | 5 (25) |
| 5–9 | 94 (18.1) | 19 (22.6) | 56 (18.1) | 17 (16.2) | 2 (10) |
| 10–14 | 165 (31.7) | 26 (31) | 88 (27.8) | 44 (41.9) | 9 (45) |
| 15–17 | 129 (24.9) | 10 (11.9) | 90 (29.1) | 25 (23.8) | 4 (20) |
| Male (n, %) | 250 (48.3) | 48 (57.1) | 141 (45.6) | 49 (46.7) | 12 (60) |
| Days after onset (median, IQR) | 1 (0–2) | ‐ | 1 (0–2) | 1 (1–2) | 2 (1.2–3) |
| Underlying disease (n, %) | 42 (8.1) | 1 (1.2) | 21 (6.8) | 10 (9.5) | 10 (50) |
| Symptoms (n, %) | |||||
| Fever | 259 (50) | 0 | 174 (56.3) | 66 (62.9) | 19 (95) |
| Cough | 177 (34.2) | 0 | 113 (36.6) | 55 (52.4) | 9 (45) |
| Dyspnea/tachypnea | 29 (5.6) | 0 | 11 (3.6) | 6 (5.7) | 12 (60) |
| Myalgia/fatigue | 114 (22) | 0 | 76 (24.6) | 26 (24.8) | 12 (60) |
| Sore throat | 106 (20.5) | 0 | 70 (22.7) | 29 (27.6) | 7 (35) |
| Abdominal pain | 39 (7.5) | 0 | 25 (8.1) | 6 (5.7) | 8 (40) |
| Headache | 66 (12.7) | 0 | 47 (15.2) | 11 (10.5) | 8 (40) |
| Diarrhea | 49 (9.5) | 0 | 31 (10) | 13 (12.1) | 5 (25) |
| Vomiting | 27 (5.2) | 0 | 12 (3.9) | 6 (5.7) | 9 (45) |
| Conjunktivitis | 4 (0.8) | 0 | 2 (0.6) | 1 (1) | 1 (5) |
| Loss of smell/taste | 34 (6.6) | 0 | 31 (10) | 3 (2.9) | 0 |
Abbreviations: COVID‐19, coronavirus disease 2019; IQR, interquartile range.
3.2. The laboratory parameters and ∆C t values of patients
We first analyzed the laboratory parameters and ∆C t value of patients in each clinical course (Table 2). Of the asymptomatic group, the median SARS‐CoV‐2 RNA ∆C t value from nasopharynx samples was 2.4 (IQR: −1.0, 5), while the corresponding median ∆C t value of the mild, moderate and severe or critical groups were 0 (IQR: −2.9, 3.7), 0.9 (−3.5, 4.8), and 1.6 (−1.6, 4.7) respectively. In severe or critical group 70% of patients had increased C‐reactive protein (CRP) rate and 55% had increased lactate dehydrogenase (LDH) rate, with the highest rates among groups. The median procalcitonin rate was 0.04 µg/L (IQR: 0.03–0.07) in asymptomatic group, 0.05 µg/L (IQR: 0.04–0.09) in mild group, 0.05 µg/L (IQR: 0.04–0.89) in moderate group, 0.14 µg/L (IQR: 0.07–2.8) in severe group, 0.14 µg/L (IQR; 0.07‐2.8) in severe/critical group. The lowest lymphocyte (median; 1100 × 106/L (IQR: 500–2400) and thrombocyte (median 199 × 109/L, [IQR: 159–232]) counts were detected in severe/critical group among all groups. We detected the statistically significant differences in increased CRP rates, increased LDH rates, procalcitonin levels, absolute lymphocyte counts and thrombocyte counts among clinical courses of patients, while there were no statistically significant differences in ∆C t value, white blood cell and absolute neutrophil counts (Table 2).
Table 2.
Laboratory data of patients with COVID‐19 accoding to clinical course
| Asymptomatic (n = 84) | Mild (n = 309) | Moderate (n = 105) | Severe/ Critical (n = 20) | p value | |
|---|---|---|---|---|---|
| Delta Ct (median, IQR) | 2.4 (‐1.0, 5) | 0 (‐2.9, 3.7) | 0.9 (‐3.5, 4.8) | 1.6 (‐1.6, 4.7) | >0.05a, b, c |
| Increased CRP (n, %) | 4/62 (6.5) | 34/201 (16.9) | 19/93 (20.4) | 14/20 (70) | 0.001a |
| 0.001b | |||||
| 0.001c | |||||
| Increased LDH (n, %) | 11/43 (25.6) | 26/164 (15.9) | 12/75 (16) | 10/18 (55.6) | 0.02a |
| 0.001b | |||||
| 0.001c | |||||
| Procalcitonin µg/L (median, IQR) | 0.04 (0.03‐0.07) | 0.05 (0.04‐0.09) | 0.05 (0.04‐0.89) | 0.14 (0.07‐2.8) | 0.001a |
| 0.001b | |||||
| 0.001c | |||||
| White blood cell x106/µL (median, IQR) | 5900 (3200‐7800) | 6200 (4500‐8200) | 5900 (4600‐7600) | 7800 (4600‐10800) | >0.05a, b, c |
| ANC x106/µL (median, IQR) | 2400 (1500‐3500) | 3000 (2000‐4700) | 2700 (2000‐4300) | 3500 (1700‐7600) | >0.05a, b, c |
| ALC x106/µL (median, IQR) | 2900 (1900‐4100) | 2100 (1400‐3000) | 2000 (1200‐2900) | 1100 (500‐2400) | 0.001 |
| 0.04 | |||||
| >0.05c | |||||
| Thrombocyte x109/µL (median, IQR) | 283 (232‐334) | 245 (210‐296) | 259 (220‐312) | 199 (159‐232) | 0.001a |
| 0.01b | |||||
| 0.001c |
ALS: Absolute lypmhocyte count. ANS: Absolute neutrophil count. CRP; C‐reactive protein. LDH:Lactate dehydrogenase.
Asymptomatic vs. Severe/Critical.
Mild. vs. Severe/Critical.
Moderate vs. Severe/Critical.
3.3. The demographic data and ∆C t values of patients with COVID‐19
We then evaluated the ∆C t value of patients in each clinical course and also according to age groups (<4 years, 5–9 years, 10–14 years, and 15–17 years). There was no statistically significant difference among age groups in each clinical course; asymptomatic, mild, moderate, and severe or critical, in terms of ∆C t value (p = .21, p = .69, p = .21, and p = .41, respectively) (Figure 1). Additionally, there was no statistically significant difference in different gender (p = .61, p = .85, p = .30, and p = .30, respectively) and the presence of the underlying disease in each clinical course (asymptomatic, mild, moderate, and severe or critical, respectively), in terms of ∆C t value (p = .64, p = .94, p = .47, and p = .43, respectively) (Figure 2). Further, we evaluated the ∆C t values of patients who were in the first 2 days and after the 2 days of symptom onset. The ∆C t values were relatively lower in the first 2 days of symptoms than after days in all groups; mild, moderate, and severe or critical (p = .70 and p = .47 respectively) (Figure 3).
Figure 1.
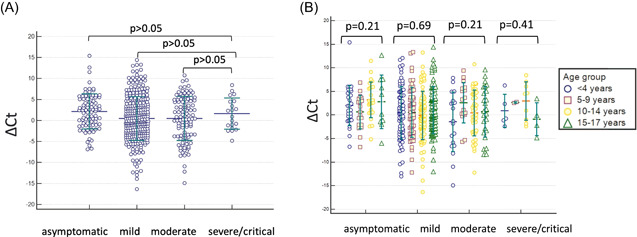
(A) The admission severe acute respiratory syndrome coronavirus 2 (SARS CoV‐2) ∆C t value of patients in each clinical course. There was no statistically significant difference between severe/critical group and others (p > .05). (B) ∆C t value of patients in each clinical courses according to age groups using Kruskal–Wallis test. There was no statistically significance difference among age groups in each clinical course (p > .05)
Figure 2.
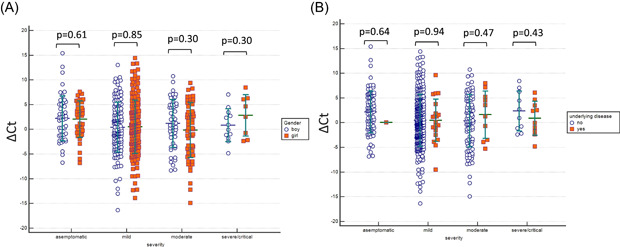
(A) The admission SARS CoV‐2 ∆C t value of patients in different gender (p > .05). (B) ∆C t value of patients in with and without underlying disease (p > .05). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Figure 3.
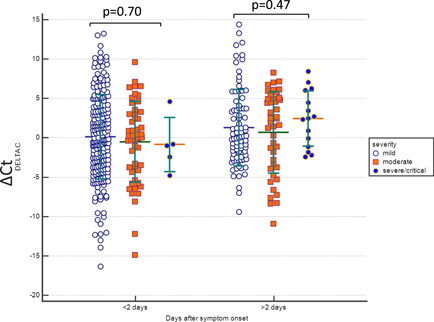
The admission SARS CoV‐2 ∆C t value of patients with mild, moderate, and severe/critical disease course, according to days after symptom onset. (p > .05). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
4. DISCUSSION
Our findings demonstrate that the ∆C t which was assumed to be inversely related to viral load were similar in all clinical courses and in all age groups in children with COVID‐19 in contrast to some previous reports, in which they reported that C t values were associated with disease severity 9 , 17 and even mortality. 20 , 21 On the other hand, limited number of studies reported any difference in median Ct values of groups with different symptom statuses, such as asymptomatic, presymptomatic, atypical, and typical symptoms. 15 , 22 Moreover, it was shown that no obvious difference in viral load and disease severity or overall survival in adults. 2 , 8 Studies in children are limited to small population with conflicting results about the comparability of viral load in children with COVID‐19, in spite of well‐defined cohorts of adult studies. It was reported that children with asymptomatic SARS‐CoV‐2 infection had lower levels of virus than symptomatic children. 23 , 24 Zachariah et al. 25 suggested symptomatic infants have higher NP viral loads at presentation but develop less severe disease as compared to older children and adolescents. However, some studies reported no age correlation with viral load in children. 26 , 27 Although the viral load of SARS‐CoV‐2 might be a useful marker for assessing disease severity and prognosis in adults, there is no such kind of relation between viral load and disease severity in children with COVID‐19 according to the finding of the present study. To the best of our knowledge, our study is also one of the few studies that evaluate the viral load in different clinical courses in a large pediatric population in the English literature.
According to the scientific report of World Health Organization, transmission can also occur from people who are infected and remain asymptomatic. 28 Additionally, SARS‐CoV‐2 burden in respiratory epithelial cells indicates a risk to transmit this virus, as well. 29 In this situation, we should pay attention the role of children in the spread of COVID‐19 due to the fact that most of the children with COVID‐19 have asymptomatic or mild disease course as in our study.
We also demonstrated that viral load was similar, even in different gender groups and the presence of the underlying disease, besides age and clinical course in pediatric patients. Namely, host factors including underlying disease, gender and age did not impact viral load and the viral load has no effect on the prediction of the clinical course of children. There are many unclear issues regarding the increased risk of severe disease in children with underlying disease, 30 including the age‐related difference in the severity of children with COVID‐19, virus dynamics and host genetic factors that influence the clinical course of the disease. Therefore, identifying the nature of immune system in children may possibly be the key for understanding the COVID‐19 pathogenesis, transmission features and finding the treatment options as well as vaccines.
Similarly, to the literature, we found that children have relatively high viral load in their upper airways, in the early days of acute COVID‐19 24 and asymptomatic patients had similar viral load as severe patients. From an infection control perspective, it is the significant point to identify infected children early and especially in the asymptomatic clinical course for prevention of transmission.
Our study has several limitations. First, we could not perform serial sampling for PCR and viral load due to the retrospective nature of the study. Serial sampling would be better to evaluate viral dynamics and shedding patterns and to determine the transmission potential of children with COVID‐19. Second, we analyzed only the nasopharyngeal swab fluid no other body fluids, such as sputum, blood, feces, and urine. Persistence and clearance of viral RNA from different patient specimens would give information regarding virus transmission dynamics. Finally, we could not evaluate the relationship between viral load with neutralizing antibodies, cytokines, chemokine, or any host immune system functions of the patients. Further virologic and immunological studies are urgently needed in children regarding how they could cope with COVID‐19 better than adults to find the treatment and management strategies of COVID‐19 and to understand the role of children in transmission.
In conclusion, our study has indicated that children with COVID‐19 can carry a similar amount of viral load at all ages irrespective of the clinical course. So, it seems that viral load has no prediction utility in terms of the clinical course of children with COVID‐19. As a result, host factors, such as immune response to virus seem one of the further investigating targets in children to understand the actual disease course.
AUTHOR CONTRIBUTIONS
Kubra Aykac: study design, manuscript writing and review. Burcu Ceylan Cura Yayla: study design, data collection. Yasemin Ozsurekci: discussion and review. Kubra Evren: study design, data collection. Pembe Derin Oygar: data collection. Sibel Lacinel Gurlevik: data collection. Tugce Coskun: data collection. Onur Tasci: data collection. Filiz Demirel Kaya: study design, data collection. Ilknur Fidanci: data collection. Medine Aysin Tasar: data collection. Alpaslan Alp: study design, data collection. Ali Bulent Cengiz: results, discussion. Sevilay Karahan: statistical analysis. Mehmet Ceyhan: results, discussion. All authors read and approved the final manuscript.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Aykac K, Cura Yayla BC, Ozsurekci Y, et al. The association of viral load and disease severity in children with COVID‐19. J Med Virol. 2021;93:3077–3083. 10.1002/jmv.26853
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. B Booth AL, Abels E, McCaffrey P. Development of a prognostic model for mortality in COVID‐19 infection using machine learning. Mod Pathol. 2020:1‐10. 10.1038/s41379-020-00700-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Argyropoulos KV, Serrano A, Hu J, et al. Association of initial viral load in severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) patients with outcome and symptoms. Am J Pathol. 2020;190:1881‐1887. 10.1016/j.ajpath.2020.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID‐19. Lancet Infect Dis. 2020;20:656‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zimmermann P, Curtis N. Coronavirus infections in children including COVID‐19: an overview of the epidemiology, clinical features, diagnosis, treatment, and prevention options in children. Pediatr Infect Dis J. 2020;39:355‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang Y‐W, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID‐19: current issues and challenges. J Clin Microbiol. 2020;58:e00512‐e00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bustin SA, Mueller R. Real‐time reverse transcription PCR (qRT‐PCR) and its potential use in clinical diagnosis. Clin Sci. 2005;109:365‐379. [DOI] [PubMed] [Google Scholar]
- 7. Rao S, Manissero D, Steele VR, Pareja J. A narrative systematic review of the clinical utility of cycle threshold values in the context of COVID‐19. Infect Dis Ther. 2020;9:573‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26:672‐675. [DOI] [PubMed] [Google Scholar]
- 9. Huang JT, Ran RX, Lv ZH, et al. Chronological changes of viral shedding in adult inpatients with COVID‐19 in Wuhan, China. Clin Infect Dis. 2020;71(16):2158‐2166. 10.1093/cid/ciaa631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu XS, Sun S, Shi Y, Wang H, Zhao R, Sheng J. SARSCoV‐2 viral load in sputum correlates with risk of COVID‐19 progression. Crit Care. 2020;24:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation between relative nasopharyngeal virus RNA load and lymphocyte count disease severity in patients with COVID‐19. Viral Immunol. 2020:vim.2020.0062. 10.1089/vim.2020.0062 [DOI] [PubMed] [Google Scholar]
- 12. Yuan C, Zhu H, Yang Y, et al. Viral loads in throat and anal swabs in children infected with SARS‐CoV‐2. Emergin Microbe Infect. 2020;9:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS‐CoV‐2. J Infect. 2020;81:e45‐e50. 10.1016/j.jinf.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xia XYW, Wu J, Liu HL, Xia H, Jia B, Huang WX. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID‐19. J Clin Virol. 2020;127:104360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kimball A, Hatfield KM, Arons M, et al. Asymptomatic and presymptomatic SARS‐CoV‐2 infections in residents of a long‐term care skilled nursing facility—King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi F, Wu T, Zhu X, et al. Association of viral load with serum biomakers among COVID‐19 cases. Virology. 2020;546:122‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng S, Fan J, Yu F, et al. Viral load dynamics anddisease severity in patients infected with SARS‐CoV‐2 in Zhejiang province, China, January‐March 2020: retrospective cohort study. BMJ. 2020;369:m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The Coronavirus Scientific Advisory Board (Turkey) . 2020. https://covid19bilgi.saglik.gov.tr/depo/rehberler/COVID-19_Rehberi.pdf
- 19. Dong Y, Mo X, Hu Y, et al. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China. Pediatrics. 2020;145:e20200702.32179660 [Google Scholar]
- 20. Shlomai A, Ben‐Zvi H, Glusman Bendersky A, Shafran N, Goldberg E, Sklan EH. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID‐19 patients. Crit Care. 2020;24:539. 10.1186/s13054-020-03244-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Faíco‐Filho KS, Passarelli VC, Bellei N. Is Higher viral load in SARS‐CoV‐2 associated with death? Am J Trop Med Hyg. 2020;103:2019‐2021. 10.4269/ajtmh.20-0954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS‐CoV‐2 infections and transmission in a skilled nursing facility. New Engl J Med. 2020;382:2081‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han MS, Seong MW, Kim N, et al. Viral RNA load in mildly symptomatic and asymptomatic children with COVID‐19, Seoul, South Korea. Emerging Infect Dis. 2020;26(10):2497‐2499. 10.3201/eid2610.202449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kociolek LK, Muller WJ, Yee R, et al. Comparison of upper respiratory viral load distributions in asymptomatic and symptomatic children diagnosed with SARS‐CoV‐2 infection in pediatric hospital testing programs. J Clin Microbiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zachariah P, Halabi KC, Johnson CL, Whitter S, Sepulveda J, Green DA. Symptomatic infants have higher nasopharyngeal SARS‐CoV‐2 viral loads but less severe disease than older children. Clin Infect Dis. 2020;71:2305‐2306. 10.1093/cid/ciaa608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yonker LM, Neilan AM, Bartsch Y, et al. Pediatric severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2): clinical presentation, infectivity, and immune responses. J Pediatr. 2020;227:45‐52.e5. 10.1016/j.jpeds.2020.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baggio S, L'Huillier AG, Yerly S, et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) viral load in the upper respiratory tract of children and adults with early acute coronavirus disease 2019 (COVID‐19). Clin Infect Dis. 2020:ciaa1157. 10.1093/cid/ciaa1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. World Health Organization . Transmission of SARS‐CoV‐2: imlications for infection prevention precautions. 2020. https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions
- 29. Mason RJ. Pathogenesis of COVID‐19 from a cell biology perspective. Eur Respir J. 2020;55(4):2000607. 10.1183/13993003.00607-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Centers for Disease Control and Prevention . People with certain medical conditions. 2020. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html#children-underlying-conditions
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


