Abstract
Background
A high prevalence of COVID‐19 associated pulmonary aspergillosis (CAPA) has been reported, though histopathological evidence is frequently lacking. To assess the clinical significance of Aspergillus species in respiratory samples of mechanically ventilated COVID‐19 patients, we implemented routine screening for Aspergillus in tracheal aspirate (TA).
Patients/methods
From all adult COVID‐19 patients admitted to the intensive care unit (ICU), TA samples were collected twice a week for Aspergillus screening by PCR and or culture. Bronchoalveolar lavage (BAL) sampling was performed in patients with a positive screening result if possible. Clinical information was obtained from the electronic patient record and patients were categorised according to the recently published consensus case definition for CAPA.
Results
Our study population consisted of 63 predominantly (73%) male patients, with a median age of 62 years and total median ICU stay of 18 days. Aspergillus species were present in TA screening samples from 15 patients (24%), and probable CAPA was diagnosed in 11 (17%) patients. Triazole resistance was detected in one patient (14%). Concordance between TA and BAL was 86%, and all TA culture positives were confirmed in BAL. We were able to withhold treatment in three of fifteen patients with positive screening (20%) but negative BAL results.
Conclusions
Positive culture, molecular detection and or antigen detection of Aspergillus species do not equal infection. Until we understand the clinical relevance of Aspergillus species detected in respiratory samples of COVID‐19 patients, minimal‐invasive screening by TA is a feasible method to monitor patients. Positive screening results should be an indication to perform a BAL to rule out upper airway colonisation.
Keywords: Aspergillus, COVID‐19, COVID‐19 associated pulmonary aspergillosis, intensive care unit, invasive fungal infections, invasive pulmonary aspergillosis, screening
1. INTRODUCTION
Soon after the COVID‐19 pandemic emerged worldwide a high prevalence of COVID‐19 associated pulmonary aspergillosis (CAPA) was reported in patients admitted to the intensive care unit (ICU). 1 , 2 , 3 , 4 Histopathological evidence for CAPA has been described in a number of cases. 5 , 6 , 7 Furthermore, in recent years, severe influenza requiring mechanical ventilation has been recognised as an independent risk factor for invasive pulmonary aspergillosis (IPA) in immunocompetent patients. 8 However, histopathological evidence for CAPA is missing in most reports. 9 Furthermore, a recent summary of available autopsy findings in a series of seven cases did not show signs of invasive aspergillosis (IA) in patients with positive Aspergillus cultures and or positive bronchoalveolar lavage (BAL) galactomannan (GM). 10 Neither was IA reported in a meta‐analysis of pulmonary histopathology reports of 129 COVID‐19 patients, 11 but standard histological stains for fungi were not always performed. Hence, the true prevalence of invasive infection in COVID‐19 patients remains to be elucidated. Also, neither the benefit of (pre‐emptive) antifungal treatment nor the consequences of withholding treatment with antifungals are known yet. 4 , 12
Given that at the moment dexamethasone is the cornerstone of treatment for patients with COVID‐19 admitted to hospital requiring oxygen, 13 , 14 more COVID‐19 patients may be put at risk for IA. Diagnosing CAPA is difficult, because most patients do not fulfil EORTC/MSGERC or AspICU criteria 1 , 2 , 3 , 4 , 15 , 16 and CT‐scans usually reveal severely damaged lungs because of COVID‐19. 2 , 4 Hence, in clinical practice, Aspergillus positive respiratory cultures from COVID‐19 patients cause the following clinical dilemma: are we dealing with colonisation or infection? Although, excess mortality has been observed in patients with COVID‐19 with suspicion of Aspergillus in the lower respiratory tract. 7 , 17 , 18 During the COVID‐19 pandemic diagnosing CAPA was further complicated by concerns about the risk of SARS‐CoV‐2 infection in healthcare workers by aerosol forming diagnostic procedures such as BAL sampling. 19
Currently the optimal diagnostic workflow for CAPA is unknown, but recently a CAPA consensus definition has been proposed. 20 To fully comprehend the diagnostic value and dilemmas of Aspergillus screening we initiated systematic screening for Aspergillus in COVID‐19 patients admitted to the ICU of our academic medical centre. In this study, we evaluated our CAPA screening programme after one month of screening for Aspergillus species by tracheal aspirate (TA). Our goals were to assess the value of TA as a screening sample, to compare different diagnostic tests and to characterise patients with CAPA. By doing so, we wanted to optimise the diagnostic workflow.
2. MATERIALS AND METHODS
2.1. Study setting and patient population
From all adult patients with PCR confirmed COVID‐19, admitted to the ICU of the Leiden University Medical Center (LUMC), TA samples were routinely collected twice a week for bacterial culture, SARS‐CoV2 PCR and Aspergillus screening (culture and Aspergillus PCR). Few BAL procedures were performed at first, but BAL samples were obtained more frequently from mid‐April. In comparison to BAL sampling, TA sampling is a minimal‐invasive method to collect a specimen from the lower respiratory tract. Because TA samples can be contaminated with micro‐organisms from the upper respiratory tract, confirmation by BAL sampling was performed in patients with a positive screening result if deemed possible and clinically indicated. Treatment was initiated in patients with a positive culture or PCR of BAL and in patients with a positive culture and PCR of TA if no BAL was performed. The diagnostic workflow is depicted in Figure 1.
FIGURE 1.
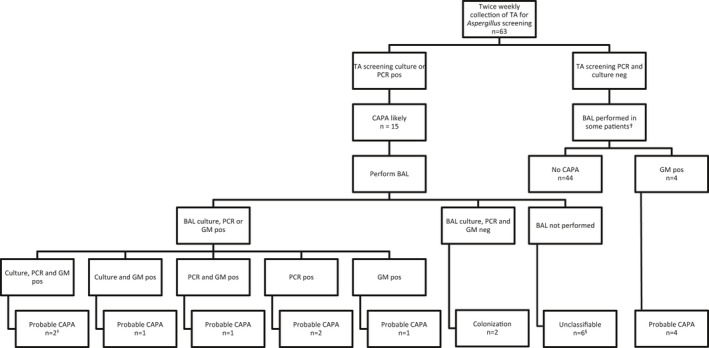
Screening for and classification of COVID‐19 associated pulmonary aspergillosis. TA, tracheal aspirate; BAL, bronchoalveolar lavage; CAPA, COVID‐19 associated pulmonary aspergillosis; GM, galactomannan; pos: positive; neg, negative. Patients were classified according to the CAPA consensus definition.20. †BAL sampling was performed in patients with negative screening results if there was a clinical indication, respiratory deterioration for example. ‡One of the patients with probable CAPA had a positive cytological smear of BAL showing branching hyphae. §In one of the patients branching hyphae were seen in a TA Gram stain
2.2. Study definitions
Categorisation according to CAPA consensus definition
All patients were categorised according to the recently published case definition for CAPA. 20 Tracheobronchitis was not observed. Non‐broncoscopic alveolar lavage was not performed, so patients could not be classified as possible CAPA. Patients not meeting criteria for probable CAPA with negative BAL diagnostics were considered colonised with Aspergillus. If no BAL was performed patients were considered unclassifiable.
2.3. Data collection
We obtained data from the laboratory information system about bacterial culture, SARS‐CoV‐2 PCR, Aspergillus culture and resistance, Aspergillus PCR and GM results between April 1st 2020 and May 11th 2020 (first COVID‐19 peak). These data included TA samples collected for Aspergillus screening as well as other samples collected in this period. From patients with Aspergillus positive culture and or PCR and or GM relevant clinical data about age, sex, underlying illness, diagnosis and follow‐up (dates of first day of illness, hospital admission, ICU admission, discharge and end of follow‐up), treatment (antifungal therapy, antibacterial therapy, treatment with chloroquine or remdesivir, supportive care on ICU), and outcome (death or alive) was obtained from the electronic patient records (EPR). From patients with negative results, we obtained clinical data from the LUMC‐COVID‐19 registration database, as well as all smoking data. Follow‐up was until discharge from hospital, transfer to another hospital or death.
The study was approved by the hospitals’ Institutional Review Board (Medical Ethical Committee Leiden‐Delft‐Den Haag, The Netherlands). Patients were only included if they had consented via an approved opt‐out procedure active in our institution. If patients weren't able to consent because they were intubated, the opt‐out procedure for clinical data for the National Intensive Care Evaluation (NICE) was applied. The screening programme was implemented as temporary routine care.
2.4. Aspergillus culture, PCR techniques and galactomannan
Both TA and BAL samples were inoculated on sheep blood agar, chocolate agar, sheep blood agar with colistin and nalidixic acid and cystine‐lactose‐electrolyte‐deficient (CLED) agar and incubated in an atmosphere enriched with 5% CO2 at 37°C for two days according to standard protocol. In addition, BAL samples were inoculated on Sabouraud dextrose agar with gentamicin (0.04 g/L) and chloramphenicol (0.5 g/L) and incubated at 28°C and 35°C for 10 days. Tracheal aspirate samples were inoculated on Sabouraud agar and were incubated at 35°C for 10 days if they were part of the Aspergillus screening programme.
Triazole resistance screening was performed on Aspergillus fumigatus isolates by a four‐well agar plate with voriconazole (2 mg/L), itraconazole (4 mg/L), posaconazole (0.5 mg/L) and a growth control (VIPcheckTM, Groningen, The Netherlands). Up to four colonies of an isolate were inoculated on Sabouraud agar and after growth of the isolate a suspension of 0.5 ‐ 2.0 McFarland was prepared which was added to the four‐well agar plate. MIC testing was performed on isolates by microbroth dilution (MD) method according to EUCAST by a reference laboratory (Radboudumc, Nijmegen, The Netherlands).
The PathoNostics AsperGenius® PCR assay (PathoNostics, Maastricht, The Netherlands) was performed according to the manufacturer's instructions for detection of Aspergillus fumigatus complex, Aspergillus terreus and Aspergillus species, and TR34/L98H and Y121F/T289A resistance mutations. Samples were diluted in TE buffer and bead beating was carried out. DNA extraction was performed with the MagNA Pure 96, DNA and viral NA small volume kit. Galactomannan testing was performed using the PlateliaTM Aspergillus Ag (Bio‐rad laboratories, Marnes‐la‐Coquette, France). A GM optical density (OD) cut‐off of ≥ 0.5 was deemed positive.
2.5. Statistical analysis
Categorial variables were described as numbers and percentages. Continuous variables were described as median and interquartile range (IQR) or range. Patients with positive and negative Aspergillus culture, PCR or GM results were compared with the Mann Whitney U‐test for numerical data and Chi‐square test or Fisher's exact test for categorical data depending on sample size. A p‐value of less than 0.05 was considered statistically significant. All statistical analyses were performed with SPSS statistics (IBM SPSS Statistics for Windows, version 25.0. Armonk, NY: IBM Corp).
3. RESULTS
3.1. Clinical characteristics
3.1.1. Patient characteristics
From 1 April 1st to May 11th 2020 63 COVID‐19 patients were admitted to the ICU of the LUMC. Median age of the patients was 62 years, 73% were male and median ICU stay was 18 days (range 3‐55). Additional patient characteristics are depicted in Table 1. Classical risk factors for invasive fungal infection 15 were present in three (5%) patients (1 allogeneic stem cell transplantation, 2 organ transplant).
TABLE 1.
Patient characteristics of COVID‐19 patients with positive and negative Aspergillus culture, PCR or galactomannan results on tracheal aspirate or bronchoalveolar lavage samples
| Aspergillus positive (n = 19) | Aspergillus negative (n = 44) | Total (n = 63) | Significance level, p‐value* | |
|---|---|---|---|---|
| Baseline | ||||
| Age, years (IQR) | 65 (59 ‐ 72) | 61 (55 ‐ 68) | 62 (57 ‐ 71) | .076 a |
| Sex, male (%) | 14 (73.7) | 32 (72.7) | 46 (73.0) | .937b |
| Underlying illness | ||||
| Chronic pulmonary disease or asthma (physician diagnosed), n (%) e | 7 (36.8) | 10 (22.7) d | 17 (27) | .247b |
| Chronic liver disease, n (%) | 0 | 0 | 0 | N/A |
| Diabetes, n (%) e | 5 (26.3) | 10 (22.7) d | 15 (23.8) | 1.000c |
| HIV/AIDS, n (%) | 0 | 0 d | 0 | N/A |
| Malignant neoplasm, n (%) e , f | 2 (10.5) | 3 (6.8) d | 5 (7.9) | .643c |
| Organ transplant, n (%) | 1 (5.3) | 1 (2.3) | 2 (3.2) | .516c |
| Smoking | ||||
| Current smoking, n (%) e | 0 | 3 (6.8) | 3 (4.8) | .537c |
| Stopped smoking, n (%) e | 7 (36.8) | 9 (20.5) | 16 (25.4) | .263c |
| Smoking unknown, n (%) | 7 (36.8) | 22 (50) | 29 (46) | .336b |
| Supportive care on ICU | ||||
| Invasive ventilation, n (%) | 19 (100) | 44 (100) | 63 (100) | 1.000 |
| Prone position, n (%) | 19 (100) | 37 (81.4) | 56 (88.9) | .091c |
| Renal replacement therapy or dialysis, n (%) | 6 (31.6) | 7 (15.9) | 13 (20.6) | .186c |
| Extracorporeal support, n (%) | 1 (5.3) | 0 | 1 (1.6) | .302c |
| Vasoactive drugs, n (%) | 15 (100) | 48 (100) | 63 (100) | 1.000 |
| SARS‐CoV‐2 PCR g | ||||
| SARS‐CoV‐2 PCR conducted during ICU stay, n | 19/19 (100) | 40/44 (90.9) | 59/63 (93.7) | .306c |
| Positive SARS‐CoV‐2 PCR result, n | 18/19 (94.7) | 28/40 (70) | 46/59 (78) | .44c |
| Ct‐value peak load, median (IQR) | 24.3 (22.6 ‐ 28.3) | 27.7 (23.6 ‐ 30) | 25.7 (23.2 ‐ 29.5) | .105 a |
| SARS‐CoV‐2 PCR became negative during ICU stay, n | 8/18 (44.4) | 11/28 (39.3) | 19/46 (41.3) | .729b |
| Outcome | ||||
| Mortality, n (%) | 10 (52.6) | 9 (20.5) | 19 (30.2) | .011b* |
Abbreviations: ct‐value, cycle threshold valueICU, intensive care unit; IQR, interquartile range.
In the Aspergillus negative group the medical history was unknown for asthma (n = 1), diabetes (n = 1), HIV/AIDS (n = 1)) and malignant neoplasm (n = 2).
Unknowns were left out for the statistical test.
Of the patients with a malignant neoplasm, four patients had a solid organ malignancy ≤ 5 years ago and one patient had a haematological malignancy and was recently treated by undergoing an allogenic stem cell transplantation. At presentation in hospital, the patient did not have leukopenia and was not treated with immunosuppressive drugs.
SARS‐CoV‐2 PCR results of tests performed during stay in our hospital.
Mann Whitney U‐test; bChi‐square test; cFischer's exact test.
Statistically significant (p‐value < .05).
During the study period, Aspergillus species were identified in TA screening by culture and or PCR in 15 of 63 (24%) patients. Probable CAPA was diagnosed in 11 of 63 (17%) patients and included four patients who were only BAL GM positive and negative in screening. Patient categorisation is depicted in Figure 1. The clinical characteristics of the patients with positive Aspergillus diagnostics are described in more detail in Table S1. At the time of diagnosis of CAPA CT‐scanning revealed COVID‐19 associated radiographic abnormalities in nine of these patients and cavitation in one patient, who was also diagnosed with a bacterial lung abscess due to Streptococcus anginosus. Six patients with probable CAPA received antifungal treatment. Of the six patients who were treated with antifungals four died, three within one week after treatment initiation and one 23 days after treatment initiation without any signs of antifungal treatment failure as follow‐up samples had become negative. The five untreated patients included three patients with only a positive GM in BAL without other positive Aspergillus diagnostics, one patient positive in screening and only GM positive in BAL and one patient who was screening and BAL PCR positive. No treatment was given because of clinical improvement and or negative follow‐up diagnostics in three patients of whom 2 survived, treatment restrictions in one patient who died shortly thereafter and because of death on the day of BAL results in another patient. One autopsy report was available from a patient with probable CAPA who was only BAL GM positive, but no histological evidence of IA was described. Mortality was significantly higher in patients with probable CAPA (7/11, 63.6%) compared to patients not classified as having an infection (12/52, 23.1%) (p‐value: 0.013). Triazole resistance was detected in one patient (1/7 culture‐positive patients, 14.3%) by VIPcheckTM and MD in a TA screening sample and a T289A/Y12F mutation was detected in two other TA samples from the same patient, but A fumigatus was not isolated from these samples. Bronchoalveolar lavage was not performed in this patient.
3.1.2. Time to positivity
Screening for IA was initiated at a median of 16 days from first day of illness (FDI) of COVID‐19 (range: 7 ‐ 21). The median duration from FDI and ICU admission to first positive screening result was respectively 18 days (range: 11 ‐ 27) and 8 days (range: 2 ‐ 23), Figure 2. Twelve patients were positive in the first screening sample. Two patients were first positive in the second screening sample and one patient was first positive in the seventh screening sample. Overall 13/15 (87%) patients were found positive in screening within the first 14 days after ICU admission, including 6/7 patients with probable CAPA who were positive in screening. Two patients became positive later, a patient with probable CAPA (positive screening on day 20) who was admitted to the ICU eight days before the screening programme was initiated and a patient who was not classifiable (positive screening on day 23) who was admitted to the ICU in the screening period. Of the seven patients with probable CAPA who were positive in screening, two remained culture and PCR positive and three remained PCR positive in all subsequent TA samples collected during their ICU stay. Of the six patients who were unclassifiable two remained culture and PCR positive. Both colonised patients remained positive in screening. The time to positivity for the different groups is presented in Figure S1.
FIGURE 2.
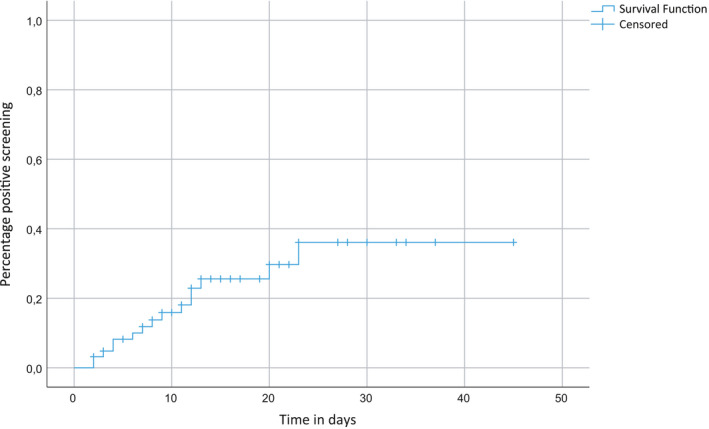
Cumulative incidence of positive screening results from ICU admission. Positive screening result: tracheal aspirate PCR or culture positive. Follow‐up time for patients with negative screening was until last negative culture and or PCR
3.2. Diagnostics
3.2.1. Samples
In total, 421 samples were collected for culture and or PCR which included 366 (86.9%) TA and 55 (13.1%) BAL samples. The median number of samples collected per patient was five (range: 1‐20). A total of 412 (97.9%) samples were cultured, of which 26/412 (6.3%) were positive. Aspergillus PCR was performed on 267/421 (63.4%) samples, of which 49/267 (18.4%) were positive. PCR detected A fumigatus complex (n = 45) A terreus (n = 3) and Aspergillus species (n = 1). Of the remaining samples 215 were negative and three were not interpretable.
3.2.2. Fungal typing and resistance
Aspergillus species were cultured from 26 samples (21 TA and 5 BAL). A fumigatus was cultured most often (n = 26). A. niger (n = 1) and A. flavus (n = 1) were cultured as well. The VIPcheckTM was performed in 23/26 (88%) and 22 A. fumigatus isolates were susceptible in this screening. One isolate was resistant to triazoles and this was confirmed by MD. Microbroth dilution was performed 17 times for 15 A. fumigatus isolates, 1 A. niger isolate and 1 A. flavus isolate, and results for first cultured TA and BAL isolates from culture‐positive patients can be found in Table S1. Cyp‐51 PCR was performed on 28 samples (21 TA and 7 BAL), was successful in 18/28 (64%) samples and could not be interpreted in 10 samples (5 TA and 5 BAL) due to low loads. A T289A/Y121F mutation was detected in 2/18 samples, both from the same patient, but A. fumigatus was not cultured from these samples.
3.2.3. Galactomannan
Galactomannan was performed on 36 BAL samples from 16 patients and was positive (OD cut‐off ≥ 0.5) in 9/19 samples from patients with positive Aspergillus screening in previous TA samples. Positive results were also seen in 7/17 samples from Aspergillus screening negative patients. Using a GM OD cut‐off of ≥ 1 these numbers would have been 8/19 samples and 5/17 samples respectively. PCR was positive in two of these sixteen GM‐positive BAL samples, culture was positive in one sample and both PCR and culture were positive in four samples. The four patients with positive GM as the only positive finding in BAL had negative Aspergillus screening in TA. Details can be found in Table S1. Serum was not routinely collected for GM testing. In the beginning of April, at the start of the screening programme, serum samples from all COVID‐19 patients were tested once, and all were negative (GM OD < 0.5) in spite of positive cultures and or PCR of TA in nine patients (data not shown). Galactomannan was performed on a random subset of 10 TA samples but was weakly positive in three samples (GM OD values: 0.51, 0.66 and 1.36) despite negative PCR and culture, possibly due to the bloody aspect and mucus in the samples and was not performed on this type of sample any further.
3.2.4. TA‐BAL concordance
Concordance of TA and BAL samples could only be analysed in a select group, because mostly BAL sampling was performed if TA screening samples were positive. Bronchoalveolar lavage samples were collected within three days after TA samples for culture and Aspergillus PCR 33 times from 19 different patients for confirmation of a positive TA result, or because there was another clinical indication to perform a BAL. In 22 instances both PCR and culture had been performed on both TA and BAL. Overall concordance between BAL and TA was 19/22 (86%). One additional positive was detected by BAL PCR from a patient with probable CAPA who had been positive in TA screening in earlier samples. No additional positives were detected by BAL culture compared to TA. BAL confirmed all culture‐positive TA samples, but confirmed three of five TA samples that were only PCR positive. Overall TA‐BAL concordance for both PCR and culture is depicted in Table 2 and in Table S2.
TABLE 2.
Concordance of sequentially collected tracheal aspirate and bronchoalveolar lavage samples
| BAL PCR/culture | ||||
|---|---|---|---|---|
| TA PCR/culture | +/+ | ± | ‐/‐ | % TA finding confirmed in BAL |
| +/+ | 3 a | 0 | 0 | 100 |
| +/− | 0 | 3 b | 2 c | 60 |
| −/− | 0 | 1 d | 13 e | 93 |
Abbreviations: −/−, PCR and culture negative; +/−, PCR positive and culture negative; +/+, PCR and culture positive; BAL, bronchoalveolar lavage; CAPA, COVID‐19 associated pulmonary aspergillosis; GM, galactomannan; TA, tracheal aspirate.
Samples from three patients with probable CAPA.
Samples from three patients with probable CAPA; one patient was culture positive in a later collected BAL sample.
Samples from two patients; 1 colonisation and 1 sample from a patient later diagnosed with probable CAPA based on positive GM in a later collected BAL sample.
Follow‐up sample from a patient with probable CAPA who was TA PCR positive in an earlier sample.
Samples from eight patients; 3 negative and 5 patients with probable CAPA. Of the patients with probable CAPA, three patients were only BAL GM positive but negative in all TA screening sample, one patient was TA PCR positive in an earlier sample and only BAL GM positive and one patient was PCR positive in earlier TA and BAL samples.
3.2.5. PCR‐culture concordance
For 255 samples results were available for both PCR and culture on the same sample, and overall concordance was 225/255 (88.2%). Altogether, PCR detected 28/255 (11.0%) additional positive samples (22 TA and 6 BAL) and missed two culture positives (1 TA and 1 BAL). The additional PCR‐positive samples were from patients who were classified as probable CAPA (n = 6), colonisation (n = 2 and unclassifiable CAPA (n = 3). The samples positive in culture only were a TA sample from a patient who was not classifiable and a BAL sample from a patient with probable CAPA. From this last patient three BAL samples were collected simultaneously and Aspergillus was detected in 2/3 samples, one by culture and one by PCR.
3.2.6. Time to diagnostic result
Aspergillus was detected by PCR and culture in TA screening samples of six patients. All were PCR and culture positive in the same screening sample, except for one patient with probable CAPA. In this patient the first positive screening result by culture was in a sample collected 17 days after the sample of the first positive screening by PCR and after five sequential PCR‐positive and culture‐negative TA samples.
4. DISCUSSION
Whether CAPA is a clinical entity that requires routine screening, invasive diagnostics and treatment is yet unknown. One month of systematic screening yielded positive findings for Aspergillus in 24% of COVID‐19 patients admitted to the ICU. Most patients (87%) were found positive in screening within 14 days after ICU admission. Probable CAPA was diagnosed in 17% of patients, all of whom did not have EORTC/MSGERC host factors for IPA. A large number of samples, 421 in total, were collected for culture and or PCR with a median of five samples per patient. Most isolates were susceptible to triazoles. Concordance between culture and PCR was high (88.2%). We were able to withhold treatment in three of fifteen patients with positive screening (20%) because of negative BAL results, of whom two improved clinically and one died, which demonstrates the added value of BAL sampling. Overall mortality was 30% and mortality was significantly higher in patients with probable aspergillosis (64%) compared to patients not classified as having an infection (23%). Establishing the cause of death was not possible, because mostly autopsies were not performed. These numbers are comparable to other studies in which excess mortality is seen in patients with COVID‐19 with suspicion of Aspergillus in the lower respiratory tract as well, ranging from 44% (30‐day mortality) to 71%. 7 , 17 , 18 To what extent Aspergillus infections contribute to mortality in ICU patients with COVID‐19 and whether antifungal treatment can reduce mortality remains uncertain.
The observed CAPA prevalence in this cohort is within the range reported by other clinical centres from different parts of the world who have reported CAPA prevalence percentages in ICU patients ranging from 3.8 ‐ 35%, 21 , 22 but prevalence numbers should be interpreted with caution as clinical and mycological evidence of CAPA varied per study. Currently, the optimal diagnostic workflow for CAPA is unknown and still under investigation as described in recent literature. 9 , 17 , 22 , 23 , 24 Optimal diagnostics depend both on feasibility for the treating physician and laboratory as well as the diagnostic value. Regarding feasibility, TA samples can be safely and relatively easily obtained from intubated COVID‐19 ICU patients. Fungal culture is a simple and cheap method and prolonged incubation did not seem necessary in our cohort, with all cultures becoming positive within three days (data not shown). With regard to the diagnostic value, TA screening is not sufficient based on the current CAPA consensus definition for which bronchoscopy for inspection of the trachea and BAL sampling or non‐bronchoscopic lavage is required, 20 but may be a screening step to reduce the number of patients in whom invasive BAL sampling is required.
Before the CAPA consensus definition was published, an expert opinion on IAPA in ICU patients proposed a CAPA definition, awaiting further histopathological studies, in which the entry criterion was pulmonary infiltrates with at least one of the following: positive GM in serum > 0.5 or BAL ≥ 1, positive Aspergillus culture of BAL (or if BAL is not performed sputum or TA), or cavitation in an area of pulmonary consolidation patients. 23 Bartoletti and colleagues categorised intubated COVID‐19 patients according to this CAPA definition. 7 Probable CAPA was diagnosed in 27.7% patients, whilst by applying the AspICU criteria 17.6% were diagnosed with putative IPA. 7 They concluded that the use of the newly proposed CAPA criteria may allow earlier diagnosis than AspICU, 7 but it may also lead to overtreatment. One of the difficulties is that most COVID‐19 ICU patients have extensive pulmonary infiltrates. Also, Aspergillus is considered a core component of the basal oral microbiome, 9 and false‐positive BAL GM results occur in patients without pulmonary aspergillosis due to various causes. 25 By applying a GM cut‐off value of one, still 32% false‐positive results were seen even in a population including patients with classical host risk factors for IA. 26 By applying the CAPA consensus definition 20 four cases of probable CAPA were found that were only BAL GM positive and had been missed in screening. These patients were not (or only shortly) treated, because IA was not suspected because of spontaneous clinical improvement in one patient who survived, because of treatment restrictions in another patient who died with no evidence of IA in autopsy reports and because of negative follow‐up diagnostics in 2 patients who died. It is our suspicion that the BAL GM was false‐positive in these cases, but CAPA cannot be completely ruled out in the two patients that died without autopsies.
False‐positive GM results can occur in patients colonised with Aspergillus, patients infected or colonised with other fungi such as penicillium, contamination of food and in the past false‐positive results were seen in patients treated with co‐amoxicillin/clavulanic acid and piperacillin‐tazobactam or gluconate‐containing solutions containing GM antigens. 26 , 27 , 28 However, these reasons do not explain the positive results in our cohort. In our validation, TA GM resulted in false positives as well possibly due to the bloody aspect and mucus in the samples. For this reason, we would not advise to perform TA GM. Besides, TA is not validated for the GM assay. The role of BAL GM testing in this setting needs further evaluation.
Concordance between culture and PCR was high (88.2%) for both TA and BAL, and most discordant samples were samples that were PCR positive and culture negative. PCR has the advantage of a rapid time to results and possibly a higher sensitivity compared to culture. However, culture was positive within two days in the majority of patients and hence the increased speed of diagnostics compared to culture is highly dependent on PCR logistics. The added value of PCR for Aspergillus screening on TA remains uncertain. Of the eight patients found positive only by PCR, four were shown to be probable CAPA in BAL samples and two received antifungal treatment, whereas six were left untreated of whom four survived without antifungal treatment suggesting positive Aspergillus PCR on TA may reflect colonisation or contamination of the upper airways. On the other hand, two patients with probable CAPA who were treated with antifungals were only PCR positive in screening and in subsequent BAL samples as well and one patient with probable CAPA had several PCR‐positive samples before becoming culture positive suggesting it may precede CAPA diagnosis by other tests. PCR of BAL is included in the new CAPA consensus definition and a single‐positive Aspergillus PCR in BAL (ct‐value < 36 cycles) is considered mycological evidence for probable CAPA. 20 Before the CAPA consensus definition was available, we also considered it likely for patients with a positive BAL PCR to have CAPA and treated patients accordingly. PCR is included in the revised EORTC/MSGERC, 15 but not in the AspICU criteria. 16 Gangneux and colleagues have proposed two different adaptions of the AspICU criteria by adding i) PCR on TA or ii) PCR TA in combination with blood biomarkers (GM or PCR). 29 By incorporating PCR on TA into the algorithm the amount of patients with putative CAPA increased from 9/45 to 15/45. By incorporating PCR on TA and blood biomarkers into the algorithm the percentage of putative patients subsequently decreased from 9/45 to 7/45. The authors were in favour of the latter algorithm, because it allowed further categorisation of the patients with a better relevance than the regular AspICU classification and had better specificity than PCR on TA alone and this is in accordance with our cohort in which several patients with only a positive PCR on TA were considered colonised, were not treated and had a favourable outcome. In our cohort blood GM was not of added value because it lacked sensitivity, but it was tested in a limited population.
Previous studies on diagnosis of CAPA used various definitions for CAPA and thus sensitivities and specificities of the various diagnostic tests for IA in COVID‐19 patients cannot be reliably compared between studies and cannot be extrapolated from studies on IA in other settings. In general, sensitivity of culture is limited, around 50% 30 and a positive culture cannot discriminate between infection and colonisation, 31 but specificity does increase in immunocompromised patients. 32 In a prospective pilot study including 44 haematological and non‐haematological ICU patients, categorised according to AspICU criteria, BAL culture, PCR and GM (OD cut‐off 1) and serum GM (OD cut‐off 0.5) were compared for putative IPA. 33 Sensitivity (range) for BAL PCR, BAL GM and serum GM was 44% (13.7 ‐ 78.8), 55.6% (21.2 ‐ 86.3) and 33.3% (7.5‐70.1) respectively. Specificity (range) for BAL PCR, BAL GM and serum GM was 94.3% (80.8 ‐ 99.3), 94.3% (80.8 ‐ 99.3) and 97.1% (85.1‐99.9) respectively.
In our centre patients were screened for Aspergillus twice weekly and a BAL was performed if screening was positive, or if there was a clinical indication for a BAL. Although a BAL is a high risk procedure with formation of aerosols which some guidelines advise against, 19 we consider it necessary in the management of COVID‐19 patients with positive Aspergillus screening results. By applying our diagnostic workflow we were able to withhold treatment in three of fifteen patients with positive screening (20%). These patients were only PCR positive in screening. Of these patients two improved clinically and were discharged from the ICU and one patient died. Although TA is considered to be a sample from the lower respiratory tract there is more chance of contamination by tube colonisation with micro‐organisms from the oropharyngeal cavity. A recently published paper proposed a screening and diagnostic algorithm for CAPA that is similar to our proposed workflow. 24 In the algorithm‐positive Aspergillus sputum or TA culture and or clinical deterioration should be an indication to perform a BAL.
In severely ill COVID‐19 patients with positive Aspergillus screening results, withholding antifungal treatment based on only moderately validated algorithms is difficult in clinical practice. In our cohort treatment was withheld in five of six patients (2 probable CAPA, 2 colonisation and 1 unclassifiable patient) with positive screening because no clinical signs of CAPA were present of whom four (1 probable, 2 colonisation and 1 unclassifiable patient) had a favourable outcome.
Until there is more clarity about CAPA we will continue screening COVID‐19 patients admitted to the intensive care of the LUMC by culture and PCR on TA. Especially now patients admitted to the intensive care for mechanical ventilation are treated with six mg of dexamethasone (equivalent of 37.5 mg prednisone) for a maximum duration of 10 days. 13 , 14 Prolonged use of corticosteroids is a classical risk factor for IA. 15 If this puts COVID‐19 patients on the ICU at an additional risk for IA remains to be studied. However, our screening in its current extensive form may be simplified for a number of reasons. The large amount of samples pose an extra workload on top of the regular and COVID‐19 associated workload of the laboratory. Since 87% of patients had positive screening results in the first 14 days after ICU admission by PCR or culture, screening might only be necessary in the first two weeks after ICU admission. Finally, screening was done with a combination of both PCR and culture, but screening by culture alone is a possibility. This would cut costs and spare equipment necessary for the PCR. The added value of blood and BAL GM testing seems limited but requires further study.
Due to limited clinical experience with CAPA and new insights during the COVID‐19 pandemic, few BAL procedures were performed in the first months of the pandemic. Because of this clinical decision making could only be based on TA results. We argued that if both TA PCR and culture were positive there was a relevant possibility of actual infection with Aspergillus species and therefore patients were treated as CAPA in case of compatible signs and symptoms, but colonisation cannot be ruled out. Finally, for clinical information about the patients with positive screening and motivation to start antifungal treatment for example we had to rely on information registered in the EPR.
By screening for Aspergillus twice weekly by PCR and culture Aspergillus was detected in respiratory samples of nearly 25% of patients with COVID‐19 admitted to the ICU. Probable CAPA was diagnosed in 17%. Clearly, positive culture, molecular detection (PCR) and/or antigen detection (GM) as evidence of presence of Aspergillus species, do not equal infection. Our findings emphasise that robust and validated definitions and histopathological studies are urgently needed as well as more information on the precise diagnostic performance of serum and BAL GM in this setting. Until we can better comprehend the clinical relevance of Aspergillus species detected in respiratory samples of COVID‐19 patients, minimal‐invasive methods like screening by TA can be performed to monitor patients. Positive screening results should be a trigger to perform a BAL for culture and, if possible, PCR.
AUTHOR CONTRIBUTION
Rebecca van Grootveld: Data curation (lead); Formal analysis (lead); Investigation (lead); Writing‐original draft (equal); Writing‐review & editing (equal). Judith van Paassen: Conceptualization (equal); Writing‐review & editing (equal). Mark G. J. de Boer: Conceptualization (equal); Writing‐review & editing (equal). Eric C. J. Claas: Methodology (equal); Writing‐review & editing (equal). Ed Kuijper: Writing‐review & editing (equal). Martha T. van der Beek: Conceptualization (equal); Methodology (equal); Writing‐original draft (equal); Writing‐review & editing (equal). LUMC‐COVID‐19 Research Group: Data collection
ETHICAL STATEMENT
The study was approved by the hospitals’ Institutional Review Board (Medical Ethical Committee Leiden‐Delft‐Den Haag). The General Data Protection Regulation (GDPR), the Code of conduct for medical research and the Medical Treatment Contracts Act all applied to our study. Patients were only included if they had consented via an approved opt‐out procedure active in our institution. If patients weren't able to consent because they were intubated, the opt‐out procedure for clinical data for the National Intensive Care Evaluation (NICE) was applied. The screening programme was implemented as temporary routine care.
Supporting information
Fig S1
Table S1
Table S2
ACKNOWLEDGMENTS
None
van Grootveld R, van Paassen J, de Boer MGJ, et al. Systematic screening for COVID‐19 associated invasive aspergillosis in ICU patients by culture and PCR on tracheal aspirate. Mycoses. 2021;64:641–650. 10.1111/myc.13259
REFERENCES
- 1. van Arkel ALE, Rijpstra TA, Belderbos HNA, van Wijngaarden P, Verweij PE, Bentvelsen RG. COVID‐19‐associated pulmonary aspergillosis. Am J Respir Crit Care Med. 2020;202(1):132‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koehler P, Cornely OA, Böttiger BW, et al. COVID‐19 associated pulmonary aspergillosis. Mycoses. 2020;63(6):528‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rutsaert L, Steinfort N, Van Hunsel T, et al. COVID‐19‐associated invasive pulmonary aspergillosis. Ann Intensive Care. 2020;10(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alanio A, Dellière S, Fodil S, Bretagne S, Mégarbane B. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID‐19. Lancet Respir Med. 2020;8(6):e48‐e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antinori S, Rech R, Galimberti L, et al. Invasive pulmonary aspergillosis complicating SARS‐CoV‐2 pneumonia: a diagnostic challenge. Travel Med Infect Dis. 2020;101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santana MF, Pivoto G, Alexandre MAA, et al. Confirmed Invasive Pulmonary Aspergillosis and COVID‐19: the value of postmortem findings to support antemortem management. Rev Soc Bras Med Trop. 2020;53:e20200401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartoletti M, Pascale R, Cricca M, et al. Epidemiology of invasive pulmonary aspergillosis among COVID‐19 intubated patients: a prospective study. Clin Infect Dis. 2020;28:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van de Veerdonk FL, Kolwijck E, Lestrade PP, et al. Influenza‐associated aspergillosis in critically ill patients. Am J Respir Crit Care Med. 2017;196(4):524‐527. [DOI] [PubMed] [Google Scholar]
- 9. Verweij PE, Gangneux JP, Bassetti M, et al. Diagnosing COVID‐19‐associated pulmonary aspergillosis. Lancet Microbe. 2020;1(2):e53‐e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flikweert AW, Grootenboers M, Yick DCY, et al. Late histopathologic characteristics of critically ill COVID‐19 patients: Different phenotypes without evidence of invasive aspergillosis, a case series. J Crit Care. 2020;59:149‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Polak SB, Van Gool IC, Cohen D, von der Thüsen JH, van Paassen J. A systematic review of pathological findings in COVID‐19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol. 2020;33(11):2128‐2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fekkar A, Poignon C, Blaize M, Lampros A. Fungal infection during COVID‐19: does aspergillus mean secondary invasive aspergillosis? Am J Respir Crit Care Med. 2020;202(6):902‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. JAMA. WHO rapid evidence appraisal for COVID‐19 therapies (REACT) Working Group. association between administration of systemic corticosteroids and mortality among critically Ill patients with COVID‐19: a meta‐analysis. 2020;324(13):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. RECOVERY Collaborative Group . Dexamethasone in hospitalized patients with Covid‐19 ‐ preliminary report. N Engl J Med. 2020;17:2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the european organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis. 2020;71(6):1367‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blot SI, Taccone FS, Van den Abeele AM, et al. A clinical algorithm to diagnose invasive pulmonary aspergillosis in critically ill patients. Am J Respir Crit Care Med. 2012;186(1):56‐64. [DOI] [PubMed] [Google Scholar]
- 17. White PL, Dhillon R, Cordey A, et al. A national strategy to diagnose COVID‐19 associated invasive fungal disease in the ICU. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dellière S, Dudoignon E, Fodil S, et al. Risk factors associated with COVID‐19‐associated pulmonary aspergillosis in ICU patients: a French multicentric retrospective cohort. Clin Microbiol Infect. 2020;13:S1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wahidi MM, Shojaee S, Lamb CR, et al. The use of bronchoscopy during the coronavirus disease 2019 pandemic: CHEST/AABIP guideline and expert panel report. Chest. 2020;158(3):1268‐1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koehler P, Bassetti M, Chakrabarti A, et al. Defining and managing COVID‐19‐associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect Dis. 2020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamoth F, Glampedakis E, Boillat‐Blanco N, Oddo M, Pagani JL. Incidence of invasive pulmonary aspergillosis among critically ill COVID‐19 patients. Clin Microbiol Infect. 2020;26(12):1706‐1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arastehfar A, Carvalho A, van de Veerdonk FL, et al. COVID‐19 Associated pulmonary aspergillosis (CAPA)‐from immunology to treatment. J Fungi (Basel). 2020;6(2):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verweij PE, Rijnders BJA, Brüggemann RJM, et al. Review of influenza‐associated pulmonary aspergillosis in ICU patients and proposal for a case definition: an expert opinion. Intensive Care Med. 2020;46(8):1524‐1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Armstrong‐James D, Youngs J, Bicanic T, et al. Confronting and mitigating the risk of COVID‐19 associated pulmonary aspergillosis. Eur Respir J. 2020;56(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nguyen MH, Jaber R, Leather HL, et al. Use of bronchoalveolar lavage to detect galactomannan for diagnosis of pulmonary aspergillosis among nonimmunocompromised hosts. J Clin Microbiol. 2007;45(9):2787‐2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farmakiotis D, Le A, Weiss Z, Ismail N, Kubiak DW, Koo S. False positive bronchoalveolar lavage galactomannan: effect of host and cut‐off value. Mycoses. 2019;62(3):204‐213. [DOI] [PubMed] [Google Scholar]
- 27. Swanink CM, Meis JF, Rijs AJ, Donnelly JP, Verweij PE. Specificity of a sandwich enzyme‐linked immunosorbent assay for detecting Aspergillus galactomannan. J Clin Microbiol. 1997;35(1):257‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ansorg R, van den Boom R, Rath PM. Detection of Aspergillus galactomannan antigen in foods and antibiotics. Mycoses. 1997;40(9–10):353‐357. [DOI] [PubMed] [Google Scholar]
- 29. Gangneux JP, Reizine F, Guegan H, et al. Is the COVID‐19 Pandemic a good time to include aspergillus molecular detection to categorize aspergillosis in ICU patients? a monocentric experience. J Fungi (Basel). 2020;6(3):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meersseman W, Lagrou K, Maertens J, Van Wijngaerden E. Invasive aspergillosis in the intensive care unit. Clin Infect Dis. 2007;45(2):205‐216. [DOI] [PubMed] [Google Scholar]
- 31. Ullmann AJ, Aguado JM, Arikan‐Akdagli S, et al. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID‐ECMM‐ERS guideline. Clin Microbiol Infect. 2018;24(Suppl 1):e1‐e38. [DOI] [PubMed] [Google Scholar]
- 32. Perfect JR, Cox GM, Lee JY, et al. The impact of culture isolation of Aspergillus species: a hospital‐based survey of aspergillosis. Clin Infect Dis. 2001;33(11):1824‐1833. [DOI] [PubMed] [Google Scholar]
- 33. Boch T, Reinwald M, Spiess B, et al. Detection of invasive pulmonary aspergillosis in critically ill patients by combined use of conventional culture, galactomannan, 1‐3‐beta‐D‐glucan and Aspergillus specific nested polymerase chain reaction in a prospective pilot study. J Crit Care. 2018;47:198‐203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2


