Abstract
Global spread of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is still ongoing. Before an effective vaccine is available, the development of potential treatments for resultant coronavirus disease 2019 (COVID‐19) is crucial. One of the disease hallmarks is hyper‐inflammatory responses, which usually leads to a severe lung disease. Patients with COVID‐19 also frequently suffer from neurological symptoms such as acute diffuse encephalomyelitis, brain injury and psychiatric complications. The metabolic pathway of sphingosine‐1‐phosphate (S1P) is a dynamic regulator of various cell types and disease processes, including the nervous system. It has been demonstrated that S1P and its metabolic enzymes, regulating neuroinflammation and neurogenesis, exhibit important functions during viral infection. S1P receptor 1 (S1PR1) analogues including AAL‐R and RP‐002 inhibit pathophysiological responses at the early stage of H1N1 virus infection and then play a protective role. Fingolimod (FTY720) is an S1P receptor modulator and is being tested for treating COVID‐19. Our review provides an overview of SARS‐CoV‐2 infection and critical role of the SphK‐S1P‐SIPR pathway in invasion of SARS‐CoV‐2 infection, particularly in the central nervous system (CNS). This may help design therapeutic strategies based on the S1P‐mediated signal transduction, and the adjuvant therapeutic effects of S1P analogues to limit or prevent the interaction between the host and SARS‐CoV‐2, block the spread of the SARS‐CoV‐2, and consequently treat related complications in the CNS.
Keywords: ACE2, COVID‐19, cytokine storm, immuno‐modulators, SARS‐CoV‐2, sphingosine 1‐phosphate
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- AKT
protein kinase B
- APP
amyloid precursor protein
- CNS
central nervous system
- ERK
extracellular signal‐regulated kinase
- FTY720
Fingolimod
- HIV
human immunodeficiency virus
- MFS
major facilitator superfamily
- MODS
multiorgan dysfunction
- NF‐kB
nuclear factor kappa B
- NLRP3
NOD‐like receptor protein 3
- PI3K
phosphatidylinositol 3‐kinase
- PKC
protein kinase C
- PLC
phospholipase C
- S
spike
- S1P
sphingosine 1‐phosphate
- S1PR1
S1P receptor 1
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- SGPL1
S1P lyase type 1
- SphK
sphingosine kinase
- TMPRSS2
transmembrane protease serine 2
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is a severe acute respiratory disease caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2). It is likely to overtake the Spanish flu as being the most severe pandemic. 1 Common symptoms of COVID‐19 include fever, fatigue, cough, conjunctivitis, and dyspnea. In some severe cases, acute respiratory distress syndrome (ARDS) can be noted, which is characterized by acute pulmonary inflammation and increased pulmonary permeability due to alveolar capillaries oedema. 2 SARS‐CoV‐2 infection also can damage other organs, such as the liver, kidneys, heart, and intestines, even causing multiple organ dysfunction syndrome (MODS). 3 Thrombotic complications are evident in some severe COVID‐19 patients. 4 , 5 Moreover, patients reported ischaemic stroke, acute disseminated encephalomyelitis, and brain damage. 6 There are also some patients who reported neurological and neuropsychiatric dysfunctions. 7
Although there are many reports of neurological symptoms in COVID‐19 patients, such as headache, nausea and vomiting, 8 stroke, 2 isolated Guillain‐Barré syndrome, 9 and demyelination. 10 However, it is not clear whether the neurological symptoms are transported via synapses to the nervous system or are the result of passing through the blood–brain barrier (BBB) endothelial cells and inflammatory cells reaching the nervous system. In response to SARS‐CoV‐2 infection, immune cells will be recruited from the circulation to the infected tissues, and these cells will release cytokines in large quantities, which will cause a "cytokine release storm". 2 It is also accompanied by an increased production of a variety of inflammatory mediators including lipid factors, chemokines, cytokines, and extracellular matrix proteins. Overcontrolled immune responses can worsen the condition of COVID‐19 patients. 3
Sphingolipids, biologically active signalling molecules, play an important role in both physiological and pathological conditions (viral infections). 11 , 12 Sphingosine 1‐phosphate (S1P) contributes to determining cell fate through the interaction between ceramide and sphingosine. 13 S1P is a kind of lipid mediator produced by the metabolism of sphingomyelin where is extracellular located. Main functions of SIP include regulating cell–cell and cell–matrix adhesion and affecting cellular migration and differentiation. 14 , 15 S1P also plays a variety of roles in various disease processes, including inflammation and cancer. 16 S1P has five different cell surface G protein (Gi/o, G12/13, and Gq)‐coupled receptors, termed S1PR1‐5 have been described, which are related to cannabinoid and lysophosphatidic acid receptors. 17 Most cells of vertebrates express S1P receptor subtypes, different types of cells express different subtypes. 14 , 18 S1P can act as an intracellular mediator or as a receptor‐ligand. S1P receptors (S1PRs) bind to S1P with high affinity to induce cellular responses. S1P can be transported across the membrane and released from the intracellular environment to the extracellular environment. 2 S1PRs exhibit unique G protein coupling characteristics with S1PR1 being only coupled to the Gi/o family. S1PR2 can be coupled to the families of Gi/o, G12/13 and Gq, causing the activation of small GTPases such as Rac, Rho, and Ras. 19 Among these five receptors of S1P, S1PR1‐3 are mainly expressed in the immune system, central nervous system (CNS) and vascular endothelium. S1PR4 is mainly expressed in lymphatic tissues, while S1PR5 is mainly expressed in the CNS and natural killer cells. 19 , 20 Emerging studies have suggested that the activation of S1PR1 plays an important role in the process of newborn neuroblasts moving from the neurogenic niches to the olfactory bulb. 2 , 21 S1PRs are highly expressed in the olfactory epithelium. 22 , 23 Netland and colleagues 24 have proved that SARS‐CoV has the ability to reach the brain through the olfactory bulb and then causes transneuronal spread to the brain.
Sphingosine‐1‐phosphate binds to the S1PRs on the outer membrane of the cell, activating the Ras/Raf/ERK signal pathway in order to stimulate DNA synthesis, promote cell growth and cell survival. 25 The Ras/Raf/ERK extracellular signal promotes the production of S1P through sphingosine kinase (SphK), which actually forms a positive feedback loop and blocks the pro‐apoptotic function of ceramide. 26 Rho GTPase can be coupled to S1PRs to activate the Hippo signalling pathway and also activate the Hippo signalling related transcriptional effectors. 27 S1P has been proved to regulate the release of free calcium ion in the endoplasmic reticulum 28 (Figure 1). Studies have shown that lacking S1P leads to increased vascular leakage, and S1P signalling is essential for angiogenesis and development. 29 S1P has also been reported to perform their functions through other G protein‐coupled receptor‐independent pathways, such as Histone Deacetylase, 30 TRAF2 31 and Prohibitin 2. 32 SphK and S1P lyase contribute to lymphocyte transport through S1P gradient, playing a role in host inflammatory responses. 33 , 34 Sphingosine analogue FTY720 used for the treatment of multiple sclerosis has been extensively studied. 35 Olfactory epithelial cells have relatively high expression and activity of S1P lyase. 22 S1PR1 analogues AAL‐R 36 and RP‐002 36 inhibit pathophysiological responses in the early stage of H1N1 virus infection. Compared with the antiviral drug oseltamivir, RP‐002 is more effective in reducing lung injury, while a combination therapy being more effective than single treatment. 36 The combination of S1PR1 agonist CYM‐5442 and antiviral drug oseltamivir can provide a maximum protection against acute lung injury. 37 The use of S1PR agonists has been proved to be effective in reducing acute lung injury caused by viral infection. We propose that S1PR agonists may serve as a new therapeutic approach for treating COVID‐19.
FIGURE 1.
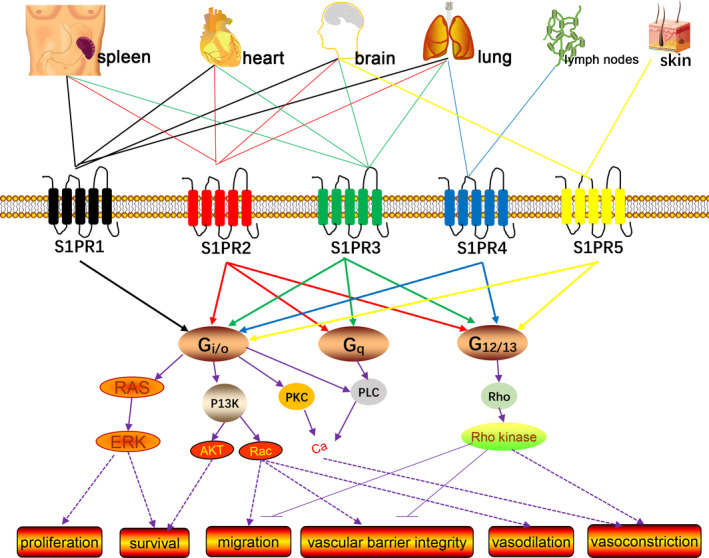
Schematic diagram describing the signalling pathways transduced by S1PRs. S1P activates the S1PRs, S1PR1‐5, which transmits various intracellular signals according to the coupled Gα subunit of the heterotrimeric G protein. S1PR1 is coupled with Gi/o, while S1PR2 and S1PR3 are coupled with Gi/o, Gq and G12/13; S1PR4 and S1PR5 are coupled with Gi/o and G12/13. The signalling through Gi/o is related to the activation of Ras/ERK to promote proliferation and survival, improve survival rate through PI3K/Akt; through PI3K/Rac promote migration, enhance vascular barrier integrity and vasodilation; increase intracellular Ca2+ through PKC or PLC. Signalling through Gq mainly activates the PLC pathways. Signalling through G12/13 can promote the activation of Rho/Rho kinase, thereby inhibiting migration, reducing the integrity of the vascular barrier and inducing vasoconstriction. ERK, extracellular signal‐regulated kinase; PI3K, phosphatidylinositol 3‐kinase; AKT, protein kinase B; PKC, protein kinase C; PLC, phospholipase C
2. SEARCH METHODOLOGY
The literature review was done through the systematic search for current findings mainly from Google Scholar and the National Centre for Biotechnology Information – PubMed database. The following defined words were used as a search strategy to obtain relevant information: (SARS‐CoV‐2; sphingosine 1‐phosphate), (SARS‐CoV‐2; Pathogenic mechanisms), (SARS‐CoV‐2; Nervous system; Clinical features), (COVID‐19; vaccine), and (COVID‐19; Therapeutic strategy or treatment strategy). Supplementary information on vaccine development was obtained from the World Health Organization (WHO) website.
3. PATHOGENIC MECHANISMS OF SARS‐COV‐2 AND POTENTIAL ROLE OF THE SPHK‐S1P‐SIPR PATHWAY
3.1. Non‐nervous system
Approximately 85% sequences are similar between SARS‐CoV‐2 and SARS‐CoV. 38 The SARS‐CoV‐2 sequences are almost the same among different infected persons with the sequence homology exceeding 99.9%. 39 This newly emerged virus has 380 amino acids substitution differences in the receptor binding domain with the virus spike (S) protein of human ACE2 being expressed on the surface. 40 Similar to SARS‐CoV, SARS‐CoV‐2 also finds ACE2 in order to invade cells, suggesting that SARS‐CoV‐2 may target and infect similar cell types as SARS‐CoV did. 41 The ACE2 gene is widely expressed in our body including heart, brain, kidneys, eyes, upper respiratory tract, lungs, gut, vasculature, and liver 42 (Figure 2). It has been gradually established that SARS‐CoV‐2 uses human ACE2 proteins as its receptors in order to enter into the ACE2‐expressing cells and SARS‐CoV‐2 does not use other coronavirus targeting receptors such as dipeptidyl peptidase 4 and aminopeptidase N. 43 Growing evidence confirms that ACE2 is closely associated with lung injury in patients with COVID‐19 44 and could also mediate SARS‐CoV‐2 S‐mediated entry into cells. 45 , 46 , 47 The change of several key amino acid residues leads to the enhancement of hydrophobic interaction and the formation of a salt bridge, making the affinity between SARS‐CoV‐2 and ACE2 much stronger than that of SARS‐CoV. 42 , 48 This can be one of the reasons that current global influence of SARS‐CoV‐2 is far greater than that of SARS‐CoV. Emerging data have suggested that sphingosine can competitively bind to the cell receptor ACE2 with SARS‐CoV‐2 and prevent biological interactions between the viral S protein receptor binding domain and ACE2. 49 This indicates that S1P can interfere with the interaction between the virus and its receptor to prevent viral infection.
FIGURE 2.

Schematic diagram of the main expression of ACE2 in the whole body. ACE2 is expressed in the circumventricular organs of the CNS, upper respiratory tract (goblet and ciliated epithelial cells), the vascular system (endothelial cells, migratory angiogenic cells, and vascular smooth muscle cells), alveolar Type II epithelial cells of the lungs, and pulmonary vasculature, the liver (cholangiocytes and hepatocytes), enterocytes of the gut, kidneys (glomerular endothelial cells, podocytes, and proximal tubule epithelial cells), heart (cardiofibroblasts, cardiomyocytes, endothelial cells, pericytes, and epicardial adipose cells), eyes (pigmented epithelial cells, rod, and cone photoreceptor cells and Müller glial cells)
The SARS‐CoV‐2 S protein has been widely recognized as an important determinant of host tropism, representing a key target for vaccine development. 42 , 45 Both SARS‐CoV 50 and SARS‐CoV‐2 45 S protein can downregulate the expression of ACE2 and induce the shedding of the ACE2 extracellular domain. 51 Subsequently, the function of the renin–angiotensin system can be affected, leading to excessive inflammation and vascular permeability. 45 , 52 In a study of 12 patients with COVID‐19, the level of circulating Ang II levels (linear to viral load) was increased significantly in the COVID‐19 group than the healthy control group. 53 This finding indicates a direct relationship between the down‐regulated expression of ACE2 and systemic renin–angiotensin system imbalance caused by the SARS‐CoV‐2 infection. Sphingosine specifically interacts with ACE2. Strikingly, the ability of binding recombinant ACE2 to the SARS‐CoV‐2 S protein is inhibited significantly after incubating with a low concentration of sphingosine. 49 Amphiphilic sphingosine molecules can bind to the polar and hydrophobic amino acids in the ACE2 pocket, then interacting with the receptor binding domain of the SARS‐CoV‐2 S protein. 54 , 55 , 56 Sphingosine may also interact with other domains of ACE2. 49 The interaction of sphingosine and ACE2 may cause the conformational change of the ACE2 inner S‐binding domain, thereby preventing binding of virus S protein.
During the early stage of infection rapid replication of the SARS‐CoV‐2 may cause epithelial and endothelial cellular apoptosis, vascular leakage, 57 pyroptosis of macrophages and lymphocytes, subsequently triggering the release of pro‐inflammatory cytokines and chemokines. 52 Guan et al 58 found that among 1099 COVID‐19 patients, 82.1% of patients had peripheral blood lymphopenia, indicating pulmonary infiltration of lymphocytes and cell damage through apoptosis. Studies have shown that SARS‐CoV infection leads to increased expression of IFN‐α, β, γ, λ at mRNA level in the lungs, 59 , 60 and SARS‐CoV up‐regulates the expression of IL‐6, TNF‐α, and IL‐12 at the mRNA level of monocyte‐derived macrophages. 61 SARS‐CoV‐2 shows similar cytokine trends to SARS‐CoV. 62 In severe COVID‐19 cases, the serum levels of IL‐2, IL‐1β, IL‐6, IFN‐γ, MIP1A, MCP‐1, and TNF‐α are increased. 63 , 64 After SARS‐Cov‐2 infection, it can trigger the activation of the NOD‐like receptor protein 3 (NLRP3) inflammasome and the secretion of IL‐1b in bone marrow‐derived macrophages, 52 , 65 , 66 which can induce apoptosis and cause the release of inflammatory mediators. 65 Endothelial cell activation is an important part of systemic inflammation and immune response, TNFα can induce the activation of SphK1 and S1P in cells, thereby mediating the activation of endothelial cells and the expression of adhesion molecules. 67 The activation of SphK1 is also related to TNF receptor‐associated factor 2, and locally produces S1P, which mediates the activation of TNFα‐stimulated nuclear factor kappa B (NF‐kB) to promote cell survival and the release of pro‐inflammatory mediators. 31 SphK1 activation can also lead to an increase in the production of IL‐6 induced by lipopolysaccharide and aggravate the inflammatory response. 68 Although SphK2 also has the same ability to phosphorylate sphingosine to S1P as SphK1, 69 SphK2 is considered to be a negative regulator of inflammation. 70 SphK2 has also been confirmed to exert anti‐inflammatory function in human macrophages, and human macrophages are the main regulator of inflammation, its overexpression inhibits the transcriptional activity of NF‐κB and the release of cytokines. 71 The early degradation of SphK2 can cause the activation of macrophages, and the late up‐regulation of SphK2 may be involved in the termination of the production of inflammatory cytokines. 71
In the process of SARS‐CoV‐2 invading cells, both S‐protein and ACE2 are proteolytically modified by host cell proteases. 42 , 72 So host cell proteases also play a key role in the process of SARS‐CoV‐2 invasion. It has been proven that SARS‐CoV‐2 can use a variety of host proteases, 42 including cathepsin L/B, factor X, trypsin, elastase, transmembrane protease serine 2 (TMPRSS2) and furin to trigger S protein and promote cell entry after receptor binding. 73
3.2. Nervous system
In most COVID‐19 cases, the virus spreads along the upper respiratory tract, resulting in limited clinical manifestations. There are also a small number of COVID‐19 cases of SARS‐CoV‐2 reaching the lungs and preferentially affecting type II alveolar cells, leading to serious diseases or possibly entering the brain through some way. Mice transgenic to the SARS‐CoV receptor expressing human ACE2 are highly susceptible to the virus, 74 lung and brain infections were observed in all mice after intranasal inoculation, 75 , 76 confirming that SARS‐CoV infects the brain through olfactory receptor neurons. Although the expression level of human ACE2 in the brain does not exceed 0.1%–1% of the lungs, 24 the level of virus replication in the lungs of these mice is high, but extensive virus replication has also been detected in the brain, SARS‐CoV may use a transneuronal/synaptic pathway that uses axon transport in the brain, 75 , 77 which is also true for SARS‐CoV‐2. Studies showed that SARS‐CoV‐2 enters the endoplasmic cavity in the early and late stages in non‐neuronal cells; therefore, it may be directed to the vesicular axon pathway of neurons. 78 , 79 ACE2 is encoded in neurons, oligodendrocytes and astrocytes, and is also found in the substantia nigra, posterior cingulate cortex, middle temporal gyrus, and ventricles. 80 There are regional differences in the degree of infection in the brain, some areas of the brain (such as the cerebellum) are uninfected, while other areas (such as the thalamus, cerebrum, and brainstem) are severely infected. 24 , 75 There is about a 60‐h delay from nasal infection to the detection of SARS‐CoV in the olfactory bulb. 79 Within these 60 h, the virus may replicate and accumulate in different olfactory epithelial cells, because its subsequent transport to other parts of the brain requires a relatively short time. 79 Considering the similarities between SARS‐CoV and SARS‐CoV‐2, the same is true for SARS‐CoV‐2.
In humans, the cells co‐expressing ACE2 and protease (such as TMPRSS2) are not only upper respiratory tract epithelial cells. 81 After querying the previously published large amounts of RNA‐seq data from the entire olfactory mucosa of macaques, marmosets, and humans, 82 , 83 it was found that all olfactory mucosa samples expressed almost all genes related to SARS‐CoV‐2 entry. Immunostaining of human olfactory neuroepithelial biopsy tissues confirmed the expression of ACE2 protein, revealing the expression of ACE2 in horizontal basal cells and sustentacular cells, while there is no ACE2 protein expressed in olfactory sensory neurons. 82 The olfactory epithelium is a pseudo‐layered epithelium composed of basal cells (ie stem cells), sensory neurons, supporting cells. 84 In human olfactory epithelium, sustentacular cells and olfactory stem cells may be the direct targets of SARS‐CoV‐2. And sustentacular cells have the highest expression frequency of ACE2 85 , 86 (2.9%), although the frequency is slightly lower than that observed in respiratory cilia (3.6%) and secretory cells (3.9%). 82 Similarly, all horizontal basal cell subtypes express ACE2, but olfactory horizontal basal cells (0.8%) have a lower frequency of expression than respiratory horizontal basal cells (1.7%). 82 Although mature neurons of olfactory receptors do not present or present ACE2 at low levels, 86 SARS‐CoV‐2 may enter and damage stem cells and the sustentacular cells in the nasal epithelium, which are necessary for normal olfactory function or regeneration of impaired neurons. 2 But SARS‐CoV‐2 virus must first invade the non‐neural olfactory epithelial cells which expressing high ACE2, then be transmitted to mature olfactory receptor neurons which expressing low ACE2, and finally be transported along the olfactory axons to the brain. TMPRSS2 transcript is expressed in the lower layers of the olfactory bulb epithelium and sustentacular cells, including stem cells and immature olfactory receptor neurons, single‐cell RNA‐Seq data set and database analysis, as well as subsequent in situ hybridization and immunochemistry experiments, showed that most TMPRSS2 is expressed in sustentacular cells, but the expression level in mature olfactory receptor neurons is very low. 82 , 86 , 87 Although the expression level of SARS‐CoV‐2 entry‐related genes in the olfactory epithelium is lower than that in the respiratory epithelium isolated from the nasal mucosa, it was found that the co‐expression levels of ACE2 and TMPRSS2 in human olfactory epithelial sustentacular cells were similar to those observed in the rest of the non‐nasal respiratory tract. 82 , 88 The co‐expression of TMPRSS2 and ACE2 with their higher expression levels make sustentacular cells more susceptible to SARS‐CoV‐2 infection 86 (Figure 3).
FIGURE 3.
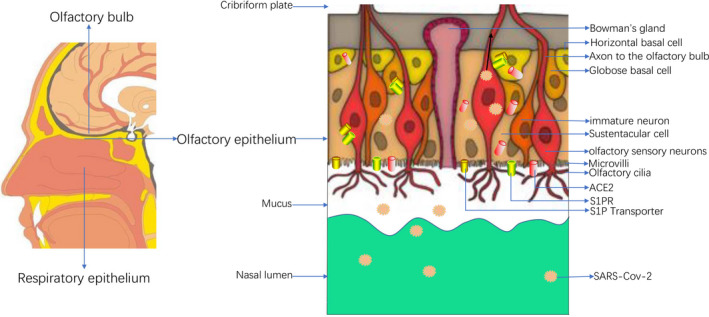
Left: Schematic diagram of the distribution of olfactory bulb, olfactory epithelium, and respiratory epithelium in the nasal cavity: The sagittal view of the human nasal cavity, the arrow points out the position of the olfactory bulb, olfactory epithelium, and respiratory epithelium. Right: Anatomical diagram of the olfactory epithelium and the main known cell types, schematic diagram of SARS‐Cov‐2 invasion of the olfactory epithelium. The olfactory epithelium is mainly composed of olfactory cilia, microvilli, olfactory sensory neurons, sustentacular cells, immature neurons, globular basal cells, horizontal basal cells, Bowman's glands. The expression of ACE2 receptor, S1PR, and S1P transporter is shown in supporting cells and neuronal cells. Some of these cells are the direct targets of SARS‐CoV‐2. The reduced nutritional support caused by damaged sustentacular cells infected by SARS‐CoV‐2 may lead to impaired basal cell renewal
In addition to the olfactory axon pathway, SARS‐CoV‐2 may also be directly transmitted from non‐neuroolfactory epithelial cells to the cerebrospinal fluid surrounding the olfactory nerve bundle, located near the cribriform plate. 79 Once SARS‐Cov‐2 enters the cerebrospinal fluid, it can reach most areas of the brain, including the medulla oblongata. More severe pneumonia can cause systemic hypoxia, which can cause brain damage. Factors that cause damage include peripheral vasodilation, hypercapnia, accumulation of toxic compounds, anaerobic metabolism and hypoxia. These may lead to neuron swelling and brain oedema, and ultimately to nervous system damage. 89
Sphingosine‐1‐phosphate can promote the olfactory ensheathing cells' proliferation in vivo and in vitro through the S1PR1/RhoA/Yes‐associated protein pathway, thereby promoting the projection of the olfactory nerve layer and axons, and the growth of olfactory neurons to the olfactory bulb. 23 S1P plays a key role in stem cells proliferation and olfactory sensory neuron function. S1PRs show different tissue expressions, and S1PRs are highly expressed in the olfactory epithelium both in vitro and in vivo. 23 S1P lyase is an essential enzyme for S1P degradation, it generates S1P chemical gradient and promotes lymphocyte trafficking. 90 S1P lyase is also highly expressed in the CNS, including the cerebral cortex, cerebellum, arachnoid lining cells, choroid plexus, specific neurons of the olfactory bulb, the spinal cord, trigeminal nerve ganglion, midbrain, and hindbrain. 90 Olfactory neurons are one of the tissues that express S1P lyase most strongly around the olfactory bulb. 90
Sphingosine‐1‐phosphate proved to be a powerful neurogenesis stimulant. 91 S1PRs are expressed in different neural progenitor stem cells and different neural cell types. 92 The ATP‐binding cassette transporter has been confirmed as an S1P transporter for various types of normal cells and tumor cells. 16 One of the Major Facilitator Superfamily (MFS) named spinster homologue 2, which hasn't a typical ATP‐binding motif, was identified as promoting S1P output in the heart and brain. 93 MFS domain‐containing 2b (MFSd2b) is also an S1P transporter, which is essential for exporting bioactive lipids from endothelial cells in the brain. 94 The study found that by deleting the gene encoding S1P lyase type 1 (SGPL1) in mice, S1P could accumulate in the brain and then determined the activation of microglia and IL‐6 secretion; this process was related to defective autophagy and was caused by S1P/S1PR2 axis mediated. 95 Van Doorn et al 96 demonstrated that reactive astrocytes cultured in MS lesions and under pro‐inflammatory conditions strongly enhanced the expression of S1P receptors 1 and 3. S1PRs can regulate the release of pro‐inflammatory chemokines in astrocytes and microglia. FTY720 blocked the transcription of chemokines CXCL10, CXCL5/LIX, and monocyte chemotactic protein 1 in astrocytes, and decreased CXCl7 released by microglia. 97
4. SYMPTOMS AND COMPLICATIONS
Generally, the severity of symptoms depends on the patient's immune system status and age, the mortality rate is higher in individuals with weakened immune function and those older than 70 years. 98 The main risk factors for patients with SARS‐Cov‐2 infection include pre‐existing respiratory diseases, hypertension, cardiovascular disease, diabetes, and chronic obstructive pulmonary disease. 99
4.1. Non‐nervous symptoms and complications
The symptoms of each SARS‐Cov‐2 infection patient are different, from asymptomatic infection in mild patients to respiratory failure in severe patients. 100 SARS‐CoV‐2 attacks the lower respiratory tract and upper respiratory tract, the common symptoms of COVID‐19 patients are similar to the common cold and flu symptoms, 100 ranging from mild respiratory diseases, ground‐glass opacity to pneumonia, RNA anaemia, and ARDS. 98 In the early stages of COVID‐19, the increase in the levels of systemic and local blood chemokines and cytokines will trigger the individual's immune response and eventually clear the infection. SARS‐CoV‐2 causes tissue damage, triggers inflammation, and host cell death. 67
A cohort study reported that about 50%–75% of people with positive results of throat swab from nucleic acid tests have no clinical symptoms, most of the remaining people have minor flu‐like symptoms, there are also a small number of people have difficulty breathing, severe interstitial pneumonia, ARDS, and MODS. 100 The common symptoms of SARS‐Cov‐2 infection are cough, fever, fatigue, mild breathing difficulties, headache, sore throat, and conjunctivitis. 64 , 101 A few cases have symptoms of gastrointestinal involvement, including diarrhoea, nausea, and vomiting. 100 Most patients with clinical symptoms and almost all critically ill patients suffer from one or more coexisting underlying diseases, such as diabetes, and cardiovascular diseases, hypertension. 102 , 103 SARS‐CoV‐2 also has a high affinity for the cardiovascular system, which can lead to acute cardiovascular‐related diseases such as congestive heart failure, cerebral medullary cardiopulmonary dysfunction, and other chronic heart diseases. 104 , 105 , 106 The main cause of death in patients with COVID‐19 is respiratory failure, occurring mostly in older people, people with weakened immunity, and patients with other potential health problems. 43 , 107
4.2. Nervous system symptoms and complications
Severe acute respiratory syndrome coronavirus 2 infection primarily affects the respiratory system; therefore, the clinical symptoms of COVID‐19 are predominantly respiratory. But, it is not uncommon for the nervous system to be affected. If it is not detected and treated early, it can lead to serious complications. Under normal circumstances, neurological complications appear in severely ill patients, and sometimes they may even appear earlier than respiratory symptoms, or many COVID‐19 patients only have neurological symptoms.
Varatharaj et al 7 reported COVID‐19 related neurological and neuropsychiatric complications in 125 patients. Of the 125 patients, 77 cases (62%) had cerebrovascular events, including 57 cases of ischaemic stroke (74%), 9 (12%) cases of cerebral haemorrhage (12%), and 1 (1%) case of CNS vasculitis. Among 125 patients, 39 (31%) had changes in mental status, including 7 (18%) patients with encephalitis and 9 (23%) undiagnosed patients. According to the clinical case definition of psychiatry, the remaining 23 (59%) patients with changed mental status met the definition. Of the 23 neuropsychiatric patients, 10 (43%) had new‐onset psychosis, 6 (26%) had the neurocognitive syndrome, and 4 (17%) had affective disorders. Of the 37 patients with changed mental status, 18 (49%) patients ≤ 60 years old, and 19 (51%) patients > 60 years old, and 13 (18%) of 74 patients with cerebrovascular events ≤ 60 years old, and 61 (82%) patients > 60 years old.
Mao et al 8 reported on two groups of patients with mild and severe conditions. They found that severe patients had less typical coronavirus symptoms, such as dry cough (34.1%) vs. (61.1%) and fever (45.5%) vs. (73%). But, compared with minor cases (45.5%) vs. (30.2%), neurological symptoms in severe cases are significantly more common. The most common neurological symptoms that have been reported are headache and dizziness. There is a prospective case series involving 58 patients that showed a higher incidence of neurological complications, with 49 (84%) patients experiencing neurological symptoms. 108 Agitation was the most common symptom, accounting for 69%, followed by confusion, accounting for 65%. 39 (67%) patients had corticospinal tract signs, and 14 (36%) patients were found to have executive dysfunction syndrome at discharge.
Poyiadji et al 109 reported the first case of acute haemorrhagic necrotizing encephalopathy associated with COVID‐19., which may be due to cytokine storm caused damaged the brain parenchyma and the destruction of the BBB. Zhao et al 110 reported a patient with COVID‐19‐related acute myelitis with fever and body pain, possibly due to cytokine storms and overactive inflammation response. Sharifi et al 111 reported case of intracranial haemorrhage leading to cerebrovascular accidents. Varatharaj, 7 Mao, 8 Helms, 108 and others have also reported similar cases. It is considered that the dysregulation of ACE 2 receptors affects brain self‐regulation, cerebral blood flow, and the sympathetic‐adrenal system, even cause brain haemorrhage. Divani et al 112 reported that stroke rates did increase in areas with a surge in COVID‐19 cases, with a significant increase in the number of strokes presenting with large vessel occlusions in some areas and occurring in younger patients without vascular risk factors.
5. CURRENT METHODS OF TREATING SARS‐COV‐2 INFECTION AND RESEARCH PROGRESS OF VACCINES
Many drugs that have antiviral properties have been proposed for the treatment of SARS‐CoV‐2. But so far, no specific antiviral therapy for human COVID‐19 has been approved. It is now generally accepted that the rationale for treating people with SARS‐Cov‐2 infection is to administer antiviral drugs as soon as possible from the onset of symptoms in order to rapidly reduce viral load and suppress the cytokine response. Through reviewing the literature, we have summarized the current treatment methods for COVID‐19 that are currently in clinical use and are considered for clinical use. Mainly include the following: 1. Use of antiviral drugs; 2. Use of interleukin receptor antagonists; 3. Use of convalescent plasma neutralizing antibodies; 4. Use of monoclonal antibodies. Antiviral drugs are mainly divided into: 1. RNA‐dependent RNA polymerase inhibitors; 2. Viral protease inhibitors; 3. Endosome acidification inhibitors; 4. Late endosome entry inhibitors; 5. Host interferons; 6. Immunomodulator. Antiviral drugs are mainly divided into: 1. RNA‐dependent RNA polymerase inhibitors, a. Remdesivir. It is a kind of adenosine triphosphate nucleoside analogue, and its active metabolite can reduce the production of viral RNA by interfering with virus‐dependent RNA polymerase. 113 A report of 1059 patients showed that the median recovery time of patients receiving remdesivir was significantly reduced by 4 days compared to the median recovery time of patients in the placebo group. 114 In a randomized controlled trial, in patients with symptoms lasting 10 days or less, compared with placebo, the use of redesivir accelerated the clinical improvement time but showed no statistical difference. There is also no difference in virological inhibition 115 ; b. Favipiravir. It can selectively inhibit viral RNA‐dependent RNA polymerase. 116 The effect of treating influenza has been confirmed and approved in China and Japan. 117 Compared with lopinavir/ritonavir, virus clearance time is shortened, radiology is improved significantly, and side effects are less. 118 c. Ribavirin. Research on SARS‐CoV showed that when lobavirin is used in combination with ritonavir, better clinical efficacy can be obtained. 116 A retrospective study by Tong et al 119 did not prove that ribavirin treatment has any clinical effects on SARS‐CoV‐2 infection; 2. Viral protease inhibitors, a. Ritonavir/Lopinavir. Ritonavir/Lopinavir has a better effect on patients infected with SARS‐CoV and is associated with low anti‐SARS‐CoV‐2 activity. 120 But, Lopinavir/Ritonavir was evaluated in randomized controlled trials in severe cases and late‐stage patients (median 13 days after the onset of symptoms), and compared with standard treatment, there was no significant virological or clinical response improvement. 121 b. Nelfinavir. Nelfinavir is currently used to treat HIV infection. It was found in vitro experiments that it is an effective cell fusion inhibitor caused by SARS‐CoV‐2 S‐glycoprotein; 3. Endosome acidification inhibitors. Azithromycin/Hydroxychloroquine/Chloroquine; Azithromycin and Hydroxychloroquine were first demonstrated to be effective in suppressing viral load in a small, non‐randomized clinical trial. 122 But, a larger clinical trial failed to prove the clinical benefit of hydroxychloroquine in the treatment of COVID‐19 and did not get the same results as before. 123 4. Late endosome entry inhibitors, Teicoplanin. Teicoplanin is active against Gram‐positive bacteria and can inhibit bacterial cell wall synthesis. It has been shown to inhibit the MERS‐CoV virus in vitro, affect the replication of MERS‐CoV, and has the potential to treat SARS‐Cov‐2 infection; 124 5. IFN‐β acts as host interferon, and SARS‐CoV‐2 can induce low interferon I–III levels. 125 A randomized controlled trial showed that the future clinical research of dual antiviral therapy based on IFN‐β‐1b is worthy of recognition. 126 Another randomized trial for patients with severe COVID‐19 showed that treatment of SARS‐Cov‐2 infection with IFN‐β‐1a can reduce mortality and allow early patients to be discharged from the hospital. 127 The results of inhaled IFN‐b and IFN‐a‐2b in the treatment of SARS‐Cov‐2 infection are encouraging 128 ; 6. Immunomodulator, Corticosteroids. Although the use of corticosteroids for the treatment of severe respiratory viral infections and respiratory distress syndrome in adults has been controversial, a randomized controlled trial showed that in patients with critical cases of COVID‐19 using ventilators, the use of dexamethasone 6 mg daily for 10 days can reduce the mortality rate by about 12%, while with oxygen support, it can be reduced by 2.9%. 129
Vaccine development is complicated. This is a highly complex science that requires a lot of labour and time. The vaccines developed for SARS‐Cov‐2 mainly include five types: (a) inactivated or attenuated live viruses; (b) subunit vaccines made with partial immune responses to viruses; (c) delivery of messenger RNA to cells Gene vaccines that counteract viral RNA; (d) engineering a part of viral protein to trigger an immune response made viral vector vaccines; (e) vaccines that target the virus reproduction machinery. 67 , 130 , 131 There are hundreds of vaccines currently in development worldwide, and some vaccines show great potential. Oxford University scientists developed a genetically modified adenovirus vaccine, named ChAdOx1 nCoV‐19, which is in the final phase of phase III clinical trials (ClinicalTrials.gov Identifier NCT04324606). The vaccine expresses a S‐glycoprotein that allows entry to human cells through the ACE2 receptor. 67 By identifying this S protein on the ChAdOx1 nCoV‐19 vaccine, the human body can produce an immune response and can prevent the virus from entering human host cells. Researchers have found that ChAdOx1 nCoV‐19 produces similar immune responses in both young and old people, with older people having lower adverse reactions than young people. CanSino Biologics cooperated with Beijing Institute of Biotechnology to develop another vaccine in China. 132 The vaccine was modified to express the SARS‐CoV‐2's S protein using human adenovirus type 5 as vector. 132 This vector has previously been used in the Ebola vaccine Ad5‐EBOV and has shown good safety in clinical trials. 133 It was found in clinical trials that the vaccine can induce neutralizing antibodies and specific T cell responses. 134 The WHO maintains a running list of candidate vaccines in development, and as of December 2nd 2020, there were 51 candidate vaccines in clinical evaluation (Table S1).
6. TARGETING SPHK/S1P/S1PR IN SARS‐COV‐2 INFECTION AND S1P ANALOGUES AS AN ADJUNCTIVE THERAPY IN COVID‐19
Sphingolipid signalling plays an indispensable role in virus replication, immune response activation, and maintenance of vascular system integrity. 67 Edwards et al. 49 proved that exogenous sphingosine can prevent epithelial cells from infecting SARS‐Cov‐2, and it can also prevent SARS‐CoV‐2 from infecting freshly isolated nasal epithelial cells. 0.25 µmol/L sphingosine is sufficient to inhibit SARS‐Cov‐2 infection of human epithelial cells, and 2 µmol/L sphingosine can almost completely inhibit SARS‐Cov‐2 infection. 49 At present, sphingosine has not been found to have side effects or toxicity on cultured experimental cells or human nasal epithelial cells.
Fingolimod is one of the most unique sphingolipid‐based drugs and is currently the main drug candidate for the treatment of COVID‐19. Activated FTY720 is a paradox that acts as both an antagonist and an agonist. 67 FTY720 is a non‐selective agonist of four out of five G‐protein‐coupled S1PRs, namely S1PR1, 3, 4, and 5. 67 Different from naturally occurring S1P, activated FTY720 acts as a selective antagonist of S1PR1 by specifically inducing the internalization and down‐regulation of S1PR1. 135 FTY720 is a kind of sphingosine analogue and also requires SphK for activation and phosphorylation. 136 FTY720 also can act as a substrate and transforms into active state, compared with SphK1, FTY720 is activated by the action of SphK (SphK1,2) isoenzymes and has a higher affinity for SphK2. 137 The potential positive effect of FTY720‐P's antagonism against S1PR1 in the treatment of SARS‐Cov‐2 infection is to inhibit excessive inflammation or "the cytokine storm". 67 FTY720‐P can reduce excessive inflammation causes blood vessel damage. Lymphocytes usually circulate between blood with higher S1P levels and lymphatic tissues with lower S1P levels. Through gradient adjustment, S1P binds to S1PR1, S1PR3, S1PR4, and S1PR5 on lymphocytes. 67 The combination of FTY720‐P with S1PR1 on lymphocytes firstly cause its activation, and then down‐regulation of S1PR1, preventing lymphocytes from infiltrating into the blood from lymphoid tissues, leading to a decrease in peripheral lymphocytes and limiting inflammation. 136 (Figure 4) FTY720 can also be used as a functional antagonist to desensitize the intracellular S1PRs pathway and inactivate the SIPR pathway in potential inflammatory cells. 138 Unlike most immunosuppressants, FTY720 prevents lymphocytes from penetrating into the blood from lymphoid tissues and does not cause cytotoxicity. 139 , 140
FIGURE 4.
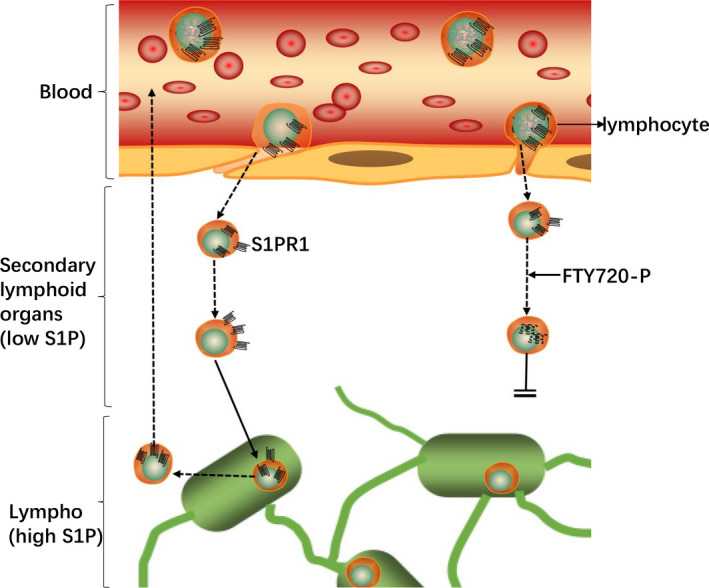
A schematic diagram of the relationship between S1P and lymphocyte transport. Due to the high S1P concentration in circulation, S1PR1 of lymphocytes in circulation mostly internalized. After entering the secondary lymphoid organs with low S1P concentration, they gradually restore the surface expression of S1PR1 and regain the ability to egress from the lymphoid organs toward S1P in circulation. SphK 2 phosphorylates FTY720 and induces the internalization and subsequent degradation of S1PR1, thereby inhibiting the outflow of lymphocytes
As an FDA approved drug, FTY720 has the potential to help intervene and treat the more serious neurological side effects of SARS‐Cov‐2 infection. The FTY720 prodrug (FTY720‐P) can cross the BBB and play a variety of direct roles in the CNS, from neuroprotection to reducing neuroinflammation. 67 FTY720‐P inhibits the invasion of lymphocytes into the CNS by limiting the invasion of lymphocytes in the lymphatic tissue, thereby limiting neuroinflammation. 135 The maintenance of the S1P/S1PR signal can protect vascular endothelial function. 67 FTY720 blocks the inflammatory response mediated by S1P/S1PR1 and has a positive effect on against heart disease. 141 It has been observed to have a protective effect on acute and chronic myocardial damage in mice experiments with ischaemia and hypoxia. 142
Ozanimod is a S1PR agonist that targets S1PR1 and S1PR5, its function is similar to FTY720. When bound to S1PR1 and/or 5, it can lead to the internalization and degradation of S1PRs and the decrease of circulating lymphocytes. 143 Ozanimod does not show side effects like FTY720, such as abnormal cardiac conduction, hypertension, fibrosis, etc., and Ozanimod's half‐life and lymphocyte recovery period are much shorter than FTY720. 143 Ozanimod can competitively bind to SphK2 and prevent the phosphorylation of sphingosine into the active form S1P. 144 SphK2 is a key factor in virus replication and plays an important role in the single‐stranded RNA viruses' replication‐transcription complex. Opaganib's blocking of SphK2 caused a decline in the virus's ability to reproduce and minimizes the possible risk of drug resistance due to virus mutation. 144 In a small group of severely symptomatic COVID‐19 patients treated by Opaganib, Opaganib showed good safety and tolerability, and all the patients' clinical and laboratory examinations were improved. 67
As a selective S1PR1 agonist, AUY954 has been shown in animal experiments to reduce microvascular permeability and inflammation. 29 In contrast, the two S1PR1 antagonists W146 67 and NIBR‐0213 145 showed a loss of capillary integrity in animal experiments.
7. PERSPECTIVES
As SARS‐CoV‐2 infection brings a cytokine storm, another serious complication, namely ARDS, is more likely to occur in elderly and severe cases of COVID‐19 patients. 63 , 146 , 147 Recent studies have pointed out that compared with healthy controls, patients with ARDS have lower serum S1P levels, which is further related to non‐pulmonary organ failure. 148 Lipid species have been shown to be involved in different stages of the virus life cycle and different stages of cell host infection. 2 At present, several drugs that affect lipid metabolism can be used for antiviral treatment, and statins can be used to treat hepatitis C virus infection 149 and the treatment of SARS‐Cov‐2 infection. 150 FTY720 can enter the CNS, where its active metabolite FTY720 phosphate (FTY720‐P) has a pleiotropic neuroprotective effect in the inflammatory microenvironment. 151 FTY720‐P can be used to treat human immunodeficiency virus (HIV) remission that affects virus persistence and T cell function, in neurons exposed to HIV, FTY720‐P reduces the expression of the amyloid precursor protein (APP) gene, thereby reducing APP protein. 151 At present, FTY720 has been reported in the treatment of patients with MS and SARS‐Cov‐2 infection, indicating that the immunomodulatory effect of S1P analogues might be beneficial to reduce mortality. 152 Another patient who has relapsing–remitting MS showed activation of SARS‐Cov‐2 and signs of the hyperinflammatory syndrome after FTY720 was discontinued, 153 indicating that the time of administration may be critical. New S1PRs agonist and S1PRs antagonist drugs have been developed/under development, some of them are undergoing or preparing for clinical trials. S1PR1 and S1PR1 agonists Mayzent and Ozanimod have been approved by the FDA in the past 2 years. 67
8. LIMITATIONS
As an immunomodulator, the effects of S1P analogues on host cells are diverse. We must clarify the potential risks of S1P analogues to the host. First of all, among the existing S1P analogues, only FTY720 is approved for the clinical treatment of multiple sclerosis. 154 FTY720 has broad specificity for S1PR1 and 3–5, and more precise S1P analogues, for instance, CYM5542 or RP‐002, need to be studied to reduce off‐target effects. Second, S1P analogues have shown good therapeutic potential in animal models of pulmonary infections, and their effects on humans have not been studied. Regarding the clinical treatment effect of FTY720 on COVID‐19, clinical trials are currently underway. Third, the effect of S1P analogues on lymphocyte outflow does not involve agonism but is directed at the functional antagonistic activity of S1PR1. 155 It has been reported that both S1P and fingolimod phosphate induced lymphopaenia through agonistic activation of S1PR1 and subsequent internalization of S1P1 in lymphocytes. 154 , 156 , 157 Fourth, due to the lack of S1PR1 on the cell surface, the recirculation of lymphocytes from secondary lymphoid organs to the blood is blocked, 154 because lymphocytes flow out through S1PR1 by a chemotactic response to the S1P concentration gradient. 156 , 158 For S1P, the internalized S1PR1 circulated back to the cell surface within a few hours. 139 But for S1PR1 modulators such as fingolimod phosphate, internalized S1PR1 underwent proteasomal degradation, resulting in a long‐term lack of S1PR1 until it was synthesized de novo. 159 It has a long action time and sometimes may be detrimental to the body. 160 Considering the above factors, we must also consider all potential risks while exploring the huge therapeutic potential of S1P analogues.
9. SUMMARY AND CONCLUSIONS
Overall, S1P analogues may enhance the integrity of lung endothelial cells, and help to prevent and treat the nervous system damage caused by SARS‐Cov‐2 infection in the early stage. The cytokine storm induced by SARS‐CoV‐2 is a very serious immunopathology, which may cause the death of COVID‐19 patients. S1P analogues protect against pulmonary infection earlier by suppressing cytokine storms. And the regulation of sphingolipid signalling has shown many beneficial effects, including selective vascular barrier protection, neuroprotection, limiting inflammation, regulation of coagulation, and cardioprotection. The regulation of S1P release or S1P content by S1P transporter, SphK or S1P lyase activity, and S1PRs‐mediated signal transduction may prevent SARS‐CoV‐2 infection or virus transmission. Currently, three S1P‐based drugs FTY720, Opaganib and Ozanimod, which have been approved by the FDA, have been considered for the treatment of COVID‐19 patients, and clinical trials are currently underway, highlighting the potential of targeting SphK‐S1P‐S1PRs to alleviate symptoms in COVID‐19 patients. S1P analogues have great potential in the treatment of COVID‐19, and further research is needed to explore the therapeutic effects and risks of S1P analogues in SARS‐Cov‐2 infected patients.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTION
All authors contributed to the study conception and design. Methodology, software, formal analysis: Yuehai Pan, Shuai Zhao, Jinming Han; writing—original draft preparation: Fan Chen, Fei Gao; writing—review and editing: Fan Chen; all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Supporting information
Tab S1
ACKNOWLEDGEMENTS
None.
Yuehai Pan and Fei Gao contribute equally.
Funding information
This research received no external funding.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available at https://www.doi.org//10.1016/S2215‐0366(20)30287‐X, reference number [reference number] and supplementary materials.
REFERENCES
- 1. Pan Y, Zhao S, Chen F. Letter to the editor: ‘What are the long‐term neurological and neuropsychiatric consequences of COVID‐19?’. World Neurosurg. 2020;144:310‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meacci E, Garcia‐Gil M, Pierucci F. SARS‐CoV‐2 infection: a role for S1P/S1P receptor signaling in the nervous system? Int J Mol Sci. 2020;21(18):6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang B, Zhou X, Qiu Y, et al. Clinical characteristics of 82 cases of death from COVID‐19. PLoS One. 2020;15(7):e0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klok FA, Kruip M, van der Meer NJM, et al. Confirmation of the high cumulative incidence of thrombotic complications in critically ill ICU patients with COVID‐19: an updated analysis. Thromb Res. 2020;191:148‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willyard C. Coronavirus blood‐clot mystery intensifies. Nature. 2020;581(7808):250. [DOI] [PubMed] [Google Scholar]
- 6. Reichard RR, Kashani KB, Boire NA, Constantopoulos E, Guo Y, Lucchinetti CF. Neuropathology of COVID‐19: a spectrum of vascular and acute disseminated encephalomyelitis (ADEM)‐like pathology. Acta Neuropathol. 2020;140(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Varatharaj A, Thomas N, Ellul MA, et al. Neurological and neuropsychiatric complications of COVID‐19 in 153 patients: a UK‐wide surveillance study. Lancet Psychiatry. 2020;7(10):875‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao L, Jin H, Wang M, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whittaker A, Anson M, Harky A. Neurological manifestations of COVID‐19: a systematic review and current update. Acta Neurol Scand. 2020;142(1):14‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zanin L, Saraceno G, Panciani PP, et al. SARS‐CoV‐2 can induce brain and spine demyelinating lesions. Acta Neurochir. 2020;162(7):1491‐1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia‐Gil M, Albi E. Nuclear lipids in the nervous system: what they do in health and disease. Neurochem Res. 2017;42(2):321‐336. [DOI] [PubMed] [Google Scholar]
- 12. Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19(3):175‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Newton J, Lima S, Maceyka M, Spiegel S. Revisiting the sphingolipid rheostat: evolving concepts in cancer therapy. Exp Cell Res. 2015;333(2):195‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cartier A, Hla T. Sphingosine 1‐phosphate: lipid signaling in pathology and therapy. Science. 2019;366(6463):eaar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chakraborty P, Vaena SG, Thyagarajan K, et al. Pro‐survival lipid sphingosine‐1‐phosphate metabolically programs t cells to limit anti‐tumor activity. Cell Rep. 2019;28(7):1879‐1893.e1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T. The role of sphingosine‐1‐phosphate in inflammation and cancer progression. Cancer Sci. 2018;109(12):3671‐3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar A, Saba JD. Regulation of immune cell migration by sphingosine‐1‐phosphate. Cell Mol Biol (Noisy‐le‐grand) (OMICS). 2015;61(2):121. [PMC free article] [PubMed] [Google Scholar]
- 18. Takuwa Y, Okamoto Y, Yoshioka K, Takuwa N. Sphingosine‐1‐phosphate signaling in physiology and diseases. BioFactors. 2012;38(5):329‐337. [DOI] [PubMed] [Google Scholar]
- 19. Mendelson K, Evans T, Hla T. Sphingosine 1‐phosphate signalling. Development. 2014;141(1):5‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pyne NJ, Pyne S. Sphingosine 1‐phosphate and cancer. Nat Rev Cancer. 2010;10(7):489‐503. [DOI] [PubMed] [Google Scholar]
- 21. Alfonso J, Penkert H, Duman C, Zuccotti A, Monyer H. Downregulation of sphingosine 1‐phosphate receptor 1 promotes the switch from tangential to radial migration in the OB. J Neurosci. 2015;35(40):13659‐13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Genter MB, Van Veldhoven PP, Jegga AG, et al. Microarray‐based discovery of highly expressed olfactory mucosal genes: potential roles in the various functions of the olfactory system. Physiol Genomics. 2003;16(1):67‐81. [DOI] [PubMed] [Google Scholar]
- 23. Bao X, Xu X, Wu Q, et al. Sphingosine 1‐phosphate promotes the proliferation of olfactory ensheathing cells through YAP signaling and participates in the formation of olfactory nerve layer. Glia. 2020;68(9):1757‐1774. [DOI] [PubMed] [Google Scholar]
- 24. Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol. 2008;82(15):7264‐7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shu X, Wu W, Mosteller RD, Broek D. Sphingosine kinase mediates vascular endothelial growth factor‐induced activation of ras and mitogen‐activated protein kinases. Mol Cell Biol. 2002;22(22):7758‐7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cuvillier O, Pirianov G, Kleuser B, et al. Suppression of ceramide‐mediated programmed cell death by sphingosine‐1‐phosphate. Nature. 1996;381(6585):800‐803. [DOI] [PubMed] [Google Scholar]
- 27. Yu FX, Zhao B, Panupinthu N, et al. Regulation of the Hippo‐YAP pathway by G‐protein‐coupled receptor signaling. Cell. 2012;150(4):780‐791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolf JJ, Studstill CJ, Hahm B. Emerging connections of S1P‐metabolizing enzymes with host defense and immunity during virus infections. Viruses. 2019;11(12):1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camerer E, Regard JB, Cornelissen I, et al. Sphingosine‐1‐phosphate in the plasma compartment regulates basal and inflammation‐induced vascular leak in mice. J Clin Investig. 2009;119(7):1871‐1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hait NC, Allegood J, Maceyka M, et al. Regulation of histone acetylation in the nucleus by sphingosine‐1‐phosphate. Science. 2009;325(5945):1254‐1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alvarez SE, Harikumar KB, Hait NC, et al. Sphingosine‐1‐phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465(7301):1084‐1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strub GM, Paillard M, Liang J, et al. Sphingosine‐1‐phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration. FASEB J. 2011;25(2):600‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vicente C, Arriazu E, Martínez‐Balsalobre E, et al. A novel FTY720 analogue targets SET‐PP2A interaction and inhibits growth of acute myeloid leukemia cells without inducing cardiac toxicity. Cancer Lett. 2020;468:1‐13. [DOI] [PubMed] [Google Scholar]
- 34. Herz J, Köster C, Crasmöller M, et al. Peripheral T cell depletion by FTY720 exacerbates hypoxic‐ischemic brain injury in neonatal mice. Front Immunol. 2018;9:1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ziemssen T, Medin J, Couto CA, Mitchell CR. Multiple sclerosis in the real world: a systematic review of fingolimod as a case study. Autoimmun Rev. 2017;16(4):355‐376. [DOI] [PubMed] [Google Scholar]
- 36. Matheu MP, Teijaro JR, Walsh KB, et al. Three phases of CD8 T cell response in the lung following H1N1 influenza infection and sphingosine 1 phosphate agonist therapy. PLoS One. 2013;8(3):e58033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao J, Zhu M, Jiang H, Shen S, Su X, Shi Y. Combination of sphingosine‐1‐phosphate receptor 1 (S1PR1) agonist and antiviral drug: a potential therapy against pathogenic influenza virus. Sci Rep. 2019;9(1):5272. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38. Yang X, Chen F. Letter to the editor: "asymptomatic carrier transmission of coronavirus disease 2019 (COVID‐19) and multipoint aerosol sampling to assess risks in the operating room during a pandemic". World Neurosurg. 2020;142:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in china. Cell Host Microbe. 2020;27(3):325‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qian H, Kang X, Hu J, et al. Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature. 2020;582(7813):550‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gheblawi M, Wang K, Viveiros A, et al. Angiotensin‐converting enzyme 2: SARS‐CoV‐2 receptor and regulator of the renin‐angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126(10):1456‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Imai Y, Kuba K, Penninger JM. The discovery of angiotensin‐converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93(5):543‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐292.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Edwards MJ, Becker KA, Gripp B, et al. Sphingosine prevents binding of SARS‐CoV‐2 spike to its cellular receptor ACE2. J Biol Chem. 2020;295(45):15174‐15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Glowacka I, Bertram S, Herzog P, et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84(2):1198‐1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jia HP, Look DC, Tan P, et al. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297(1):L84‐L96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu Y, Cheng Y, Wu Y. Understanding SARS‐CoV‐2‐mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virologica Sinica. 2020;35(3):266‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang K, Gheblawi M, Oudit GY. Angiotensin converting enzyme 2: a double‐edged sword. Circulation. 2020;142(5):426‐428. [DOI] [PubMed] [Google Scholar]
- 54. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 55. Barnes CO, West AP Jr, Huey‐Tubman KE, et al. Structures of human antibodies bound to SARS‐CoV‐2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828‐842.e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barnes CO, West AP Jr, Huey‐Tubman KE, et al. Structures of human antibodies bound to SARS‐CoV‐2 spike reveal common epitopes and recurrent features of antibodies. bioRxiv. 2020;182(4):828‐842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Labò N, Ohnuki H, Tosato G. Vasculopathy and coagulopathy associated with SARS‐CoV‐2 infection. Cells. 2020;9(7):1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. de Lang A, Baas T, Teal T, et al. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS‐CoV‐infected macaques. PLoS Pathog. 2007;3(8):e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Naz F, Arish M. Battling COVID‐19 pandemic: sphingosine‐1‐phosphate analogs as an adjunctive therapy? Front Immunol. 2020;11:1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type i interferon and inflammatory monocyte‐macrophage responses cause lethal pneumonia in SARS‐CoV‐infected mice. Cell Host Microbe. 2016;19(2):181‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang W, Zhao Y, Zhang F, et al. The use of anti‐inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID‐19): the perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao S, Chen F, Wang D, Han W, Zhang Y, Yin Q. NLRP3 inflammasomes are involved in the progression of postoperative cognitive dysfunction: from mechanism to treatment. Neurosurg Rev. 2020;Sep(12):1‐17. [DOI] [PubMed] [Google Scholar]
- 66. Zhao S, Chen F, Yin Q, Wang D, Han W, Zhang Y. Reactive oxygen species interact with NLRP3 inflammasomes and are involved in the inflammation of sepsis: from mechanism to treatment of progression. Front Physiol. 2020;11:571810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McGowan EM, Haddadi N, Nassif NT, Lin Y. Targeting the SphK‐S1P‐SIPR pathway as a potential therapeutic approach for COVID‐19. Int J Mol Sci. 2020;21(19):7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ebenezer DL, Fu P, Suryadevara V, Zhao Y, Natarajan V. Epigenetic regulation of pro‐inflammatory cytokine secretion by sphingosine 1‐phosphate (S1P) in acute lung injury: role of S1P lyase. Adv Biol Reg. 2017;63:156‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hatoum D, Haddadi N, Lin Y, Nassif NT, McGowan EM. Mammalian sphingosine kinase (SphK) isoenzymes and isoform expression: challenges for SphK as an oncotarget. Oncotarget. 2017;8(22):36898‐36929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pyne NJ, Adams DR, Pyne S. Sphingosine kinase 2 in autoimmune/inflammatory disease and the development of sphingosine kinase 2 inhibitors. Trends Pharmacol Sci. 2017;38(7):581‐591. [DOI] [PubMed] [Google Scholar]
- 71. Weigert A, von Knethen A, Thomas D, et al. Sphingosine kinase 2 is a negative regulator of inflammatory macrophage activation. Biochim Biophys Acta. 2019;1864(9):1235‐1246. [DOI] [PubMed] [Google Scholar]
- 72. McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS‐CoV‐2 and COVID‐19. Pharmacol Res. 2020;157:104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McCray PB Jr, Pewe L, Wohlford‐Lenane C, et al. Lethal infection of K18‐hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81(2):813‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tseng CT, Perrone LA, Zhu H, Makino S, Peters CJ. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J Immunol. 2005;174(12):7977‐7985. [DOI] [PubMed] [Google Scholar]
- 77. Tseng CT, Huang C, Newman P, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin‐converting enzyme 2 virus receptor. J Virol. 2007;81(3):1162‐1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Butowt R, Bilinska K. SARS‐CoV‐2: olfaction, brain infection, and the urgent need for clinical samples allowing earlier virus detection. ACS Chem Neurosci. 2020;11(9):1200‐1203. [DOI] [PubMed] [Google Scholar]
- 80. Divani AA, Andalib S, Biller J, et al. Central nervous system manifestations associated with COVID‐19. Curr Neurol Neurosci Rep. 2020;20(12):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bertram S, Heurich A, Lavender H, et al. Influenza and SARS‐coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7(4):e35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brann DH, Tsukahara T, Weinreb C, et al. Non‐neuronal expression of SARS‐CoV‐2 entry genes in the olfactory system suggests mechanisms underlying COVID‐19‐associated anosmia. Sci Adv. 2020;6(31):eabc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Saraiva LR, Riveros‐McKay F, Mezzavilla M, et al. A transcriptomic atlas of mammalian olfactory mucosae reveals an evolutionary influence on food odor detection in humans. Sci Adv. 2019;5(7):eaax0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Choi R, Goldstein BJ. Olfactory epithelium: cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig Otolaryngol. 2018;3(1):35‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fodoulian L, Tuberosa J, Rossier D, Landis B, Carleton A, Rodriguez I. SARS‐CoV‐2 receptor and entry genes are expressed by sustentacular cells in the human olfactory neuroepithelium. BioRxiv. 2020;23(12):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bilinska K, Jakubowska P, Von Bartheld CS, Butowt R. Expression of the SARS‐CoV‐2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Neurosci. 2020;11(11):1555‐1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Torabi A, Mohammadbagheri E, Akbari Dilmaghani N, et al. Proinflammatory cytokines in the olfactory mucosa result in COVID‐19 induced anosmia. ACS Chem Neurosci. 2020;11(13):1909‐1913. [DOI] [PubMed] [Google Scholar]
- 88. Sungnak W, Huang N, Bécavin C, Berg M. SARS‐CoV‐2 entry genes are most highly expressed in nasal goblet and ciliated cells within human airways. ArXiv. 2020;Mar(13):1‐21. [Google Scholar]
- 89. Ahmad I, Rathore FA. Neurological manifestations and complications of COVID‐19: a literature review. J Clin Neurosci. 2020;77:8‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Borowsky AD, Bandhuvula P, Kumar A, et al. Sphingosine‐1‐phosphate lyase expression in embryonic and adult murine tissues. J Lipid Res. 2012;53(9):1920‐1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ng ML, Yarla NS, Menschikowski M, Sukocheva OA. Regulatory role of sphingosine kinase and sphingosine‐1‐phosphate receptor signaling in progenitor/stem cells. World J Stem Cells. 2018;10(9):119‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Terai K, Soga T, Takahashi M, et al. Edg‐8 receptors are preferentially expressed in oligodendrocyte lineage cells of the rat CNS. Neuroscience. 2003;116(4):1053‐1062. [DOI] [PubMed] [Google Scholar]
- 93. Spiegel S, Maczis MA, Maceyka M, Milstien S. New insights into functions of the sphingosine‐1‐phosphate transporter SPNS2. J Lipid Res. 2019;60(3):484‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kobayashi N, Kawasaki‐Nishi S, Otsuka M, Hisano Y, Yamaguchi A, Nishi T. MFSD2B is a sphingosine 1‐phosphate transporter in erythroid cells. Sci Rep. 2018;8(1):4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Karunakaran I, Alam S, Jayagopi S, et al. Neural sphingosine 1‐phosphate accumulation activates microglia and links impaired autophagy and inflammation. Glia. 2019;67(10):1859‐1872. [DOI] [PubMed] [Google Scholar]
- 96. Van Doorn R, Van Horssen J, Verzijl D, et al. Sphingosine 1‐phosphate receptor 1 and 3 are upregulated in multiple sclerosis lesions. Glia. 2010;58(12):1465‐1476. [DOI] [PubMed] [Google Scholar]
- 97. O'Sullivan SA, O'Sullivan C, Healy LM, Dev KK, Sheridan GK. Sphingosine 1‐phosphate receptors regulate TLR4‐induced CXCL5 release from astrocytes and microglia. J Neurochem. 2018;144(6):736‐747. [DOI] [PubMed] [Google Scholar]
- 98. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. J Autoimmun. 2020;109:102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zheng F, Tang W, Li H, Huang YX, Xie YL, Zhou ZG. Clinical characteristics of 161 cases of corona virus disease 2019 (COVID‐19) in Changsha. Eur Rev Med Pharmacol Sci. 2020;24(6):3404‐3410. [DOI] [PubMed] [Google Scholar]
- 100. Pascarella G, Strumia A, Piliego C, et al. COVID‐19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288(2):192‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Resp Med. 2020;8(5):475‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hu Y, Sun J, Dai Z, et al. Prevalence and severity of corona virus disease 2019 (COVID‐19): a systematic review and meta‐analysis. J Clin Virol. 2020;127:104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li B, Yang J, Zhao F, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID‐19 in China. Clin Res Cardiol. 2020;109(5):531‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Guzik TJ, Mohiddin SA, Dimarco A, et al. COVID‐19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res. 2020;116(10):1666‐1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu PP, Blet A, Smyth D, Li H. The science underlying COVID‐19: implications for the cardiovascular system. Circulation. 2020;142(1):68‐78. [DOI] [PubMed] [Google Scholar]
- 106. South AM, Diz DI, Chappell MC. COVID‐19, ACE2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020;318(5):H1084‐H1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Helms J, Kremer S, Merdji H, et al. Neurologic features in severe SARS‐CoV‐2 infection. N Engl J Med. 2020;382(23):2268‐2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. COVID‐19‐associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology. 2020;296(2):E119‐E120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhao K, Huang J, Dai D, Feng Y, Liu L, Nie S. Acute myelitis after SARS‐CoV‐2 infection: a case report. MedRxiv. 2020;03(16):1‐7. [Google Scholar]
- 111. Sharifi‐Razavi A, Karimi N, Rouhani N. COVID‐19 and intracerebral haemorrhage: causative or coincidental? New Microbes New Infect. 2020;35:100669. In:2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Divani AA, Andalib S, Di Napoli M, et al. Coronavirus disease 2019 and stroke: clinical manifestations and pathophysiological insights. J Stroke Cerebrovasc Dis. 2020;29(8):104941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sheahan TP, Sims AC, Graham RL, et al. Broad‐spectrum antiviral GS‐5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396):eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19 ‐ final report. N Engl J Med. 2020;383(19):1813‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID‐19: a randomised, double‐blind, placebo‐controlled, multicentre trial. Lancet. 2020;395(10236):1569‐1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hung IF. Treatment of coronavirus disease 2019. Curr Opin HIV AIDS. 2020;15(6):336‐340. [DOI] [PubMed] [Google Scholar]
- 117. Shiraki K, Daikoku T. Favipiravir, an anti‐influenza drug against life‐threatening RNA virus infections. Pharmacol Ther. 2020;209:107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cai Q, Yang M, Liu D, et al. Experimental treatment with favipiravir for COVID‐19: an open‐label control study. Engineering (Beijing). 2020;6(10):1192‐1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tong S, Su Y, Yu Y, et al. Ribavirin therapy for severe COVID‐19: a retrospective cohort study. Int J Antimicrob Agents. 2020;56(3):106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chan JF, Chan KH, Kao RY, et al. Broad‐spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect. 2013;67(6):606‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Cao B, Wang Y, Wen D, et al. A trial of lopinavir‐ritonavir in adults hospitalized with severe Covid‐19. N Engl J Med. 2020;382(19):1787‐1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID‐19: results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sarma P, Kaur H, Kumar H, et al. Virological and clinical cure in COVID‐19 patients treated with hydroxychloroquine: a systematic review and meta‐analysis. J Med Virol. 2020;92(7):776‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Baron SA, Devaux C, Colson P, Raoult D, Rolain JM. Teicoplanin: an alternative drug for the treatment of COVID‐19? Int J Antimicrob Agents. 2020;55(4):105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036‐1045.e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hung IF, Lung KC, Tso EY, et al. Triple combination of interferon beta‐1b, lopinavir‐ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID‐19: an open‐label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695‐1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rahmani H, Davoudi‐Monfared E, Nourian A, et al. Interferon β‐1b in treatment of severe COVID‐19: a randomized clinical trial. Int Immunopharmacol. 2020;88:106903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhou Q, Wei X‐S, Xiang X, et al. Interferon‐a2b treatment for COVID‐19. MedRxiv. 2020;11:1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with Covid‐19 ‐ preliminary report. N Engl J Med. 2020;17:NEJMoa2021436 [Google Scholar]
- 130. Lundstrom K. Coronavirus pandemic‐therapy and vaccines. Biomedicines. 2020;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gao Y, Yan L, Huang Y, et al. Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science. 2020;368(6492):779‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Taxt AM, Grødeland G, Lind A, Müller F. Status of COVID‐19 vaccine development. Tidsskr Nor Laegeforen. 2020;140(13):14. [DOI] [PubMed] [Google Scholar]
- 133. Wu L, Zhang Z, Gao H, et al. Open‐label phase I clinical trial of Ad5‐EBOV in Africans in China. Hum Vacc Immunother. 2017;13(9):2078‐2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhu FC, Li YH, Guan XH, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type‐5 vectored COVID‐19 vaccine: a dose‐escalation, open‐label, non‐randomised, first‐in‐human trial. Lancet. 2020;395(10240):1845‐1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Grassi S, Mauri L, Prioni S, et al. Sphingosine 1‐phosphate receptors and metabolic enzymes as druggable targets for brain diseases. Front Pharmacol. 2019;10:807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Obinata H, Hla T. Sphingosine 1‐phosphate and inflammation. Int Immunol. 2019;31(9):617‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Billich A, Bornancin F, Dévay P, Mechtcheriakova D, Urtz N, Baumruker T. Phosphorylation of the immunomodulatory drug FTY720 by sphingosine kinases. J Biol Chem. 2003;278(48):47408‐47415. [DOI] [PubMed] [Google Scholar]
- 138. Teijaro JR, Walsh KB, Long JP, et al. Protection of ferrets from pulmonary injury due to H1N1 2009 influenza virus infection: immunopathology tractable by sphingosine‐1‐phosphate 1 receptor agonist therapy. Virology. 2014;452–453:152‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Stepanovska B, Huwiler A. Targeting the S1P receptor signaling pathways as a promising approach for treatment of autoimmune and inflammatory diseases. Pharmacol Res. 2020;154:104170. [DOI] [PubMed] [Google Scholar]
- 140. Huwiler A, Zangemeister‐Wittke U. The sphingosine 1‐phosphate receptor modulator fingolimod as a therapeutic agent: recent findings and new perspectives. Pharmacol Ther. 2018;185:34‐49. [DOI] [PubMed] [Google Scholar]
- 141. Premer C, Kanelidis AJ, Hare JM, Schulman IH. Rethinking endothelial dysfunction as a crucial target in fighting heart failure. Mayo Clin Proc Innov Qual Outcomes. 2019;3(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang F, Xia Y, Yan W, et al. Sphingosine 1‐phosphate signaling contributes to cardiac inflammation, dysfunction, and remodeling following myocardial infarction. Am J Physiol Heart Circ Physiol. 2016;310(2):H250‐H261. [DOI] [PubMed] [Google Scholar]
- 143. Scott FL, Clemons B, Brooks J, et al. Ozanimod (RPC1063) is a potent sphingosine‐1‐phosphate receptor‐1 (S1P1) and receptor‐5 (S1P5) agonist with autoimmune disease‐modifying activity. Br J Pharmacol. 2016;173(11):1778‐1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. French KJ, Zhuang Y, Maines LW, et al. Pharmacology and antitumor activity of ABC294640, a selective inhibitor of sphingosine kinase‐2. J Pharmacol Exp Ther. 2010;333(1):129‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bigaud M, Dincer Z, Bollbuck B, et al. Pathophysiological consequences of a break in S1P1‐dependent homeostasis of vascular permeability revealed by S1P1 competitive antagonism. PLoS One. 2016;11(12):e0168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Resp Med. 2020;8(4):420‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Int Med. 2020;180(7):934‐943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zhao J, Tan Y, Wang L, Su X, Shi Y. Serum sphingosine‐1‐phosphate levels and Sphingosine‐1‐Phosphate gene polymorphisms in acute respiratory distress syndrome: a multicenter prospective study. J Transl Med. 2020;18(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 149. Heaton NS, Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19(7):368‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Fedson DS, Opal SM, Rordam OM. Hiding in plain sight: an approach to treating patients with severe COVID‐19 infection. MBio. 2020;11(2):e00398‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Geffin R, Martinez R, de Las PA, Issac B, McCarthy M. Fingolimod induces neuronal‐specific gene expression with potential neuroprotective outcomes in maturing neuronal progenitor cells exposed to HIV. J Neurovirol. 2017;23(6):808‐824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Foerch C, Friedauer L, Bauer B, Wolf T, Adam EH. Severe COVID‐19 infection in a patient with multiple sclerosis treated with fingolimod. Mult Scler Relat Disord. 2020;42:102180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gomez‐Mayordomo V, Montero‐Escribano P, Matías‐Guiu JA, González‐García N, Porta‐Etessam J, Matías‐Guiu J. Clinical exacerbation of SARS‐CoV2 infection after fingolimod withdrawal. J Med Virol. 2020;93(1):546‐549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Park SJ, Im DS. Sphingosine 1‐phosphate receptor modulators and drug discovery. Biomol Ther. 2017;25(1):80‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Matloubian M, Lo CG, Cinamon G, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427(6972):355‐360. [DOI] [PubMed] [Google Scholar]
- 156. Brinkmann V, Cyster JG, Hla T. FTY720: sphingosine 1‐phosphate receptor‐1 in the control of lymphocyte egress and endothelial barrier function. Am J Transplant. 2004;4(7):1019‐1025. [DOI] [PubMed] [Google Scholar]
- 157. Thangada S, Khanna KM, Blaho VA, et al. Cell‐surface residence of sphingosine 1‐phosphate receptor 1 on lymphocytes determines lymphocyte egress kinetics. J Exp Med. 2010;207(7):1475‐1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Chiba K. New therapeutic approach for autoimmune diseases by the sphingosine 1‐phosphate receptor modulator, fingolimod (FTY720). Yakugaku zasshi. 2009;129(6):655‐665. [DOI] [PubMed] [Google Scholar]
- 159. Oo ML, Thangada S, Wu MT, et al. Immunosuppressive and anti‐angiogenic sphingosine 1‐phosphate receptor‐1 agonists induce ubiquitinylation and proteasomal degradation of the receptor. J Biol Chem. 2007;282(12):9082‐9089. [DOI] [PubMed] [Google Scholar]
- 160. Subei AM, Cohen JA. Sphingosine 1‐phosphate receptor modulators in multiple sclerosis. CNS Drugs. 2015;29(7):565‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab S1
Data Availability Statement
The data that support the findings of this study are openly available at https://www.doi.org//10.1016/S2215‐0366(20)30287‐X, reference number [reference number] and supplementary materials.


