Abstract
Sepsis-induced myocardial dysfunction (SIMD) is one of the leading causes of death in sepsis. We hypothesized that exosomes released from ECs exposed to bacterial lipopolysaccharides (LPS) have some regulatory effect on cardiomyocytes (CMs). In this study, cultured rat ECs were exposed to 0.5 µg/ml of LPS, and exosomes were isolated from the conditioned medium through ultra-high-speed centrifugation. The exosomes were given to the cultured neonatal rat CMs to test the potential effects and proceed to small RNA sequencing to identify their miRNA expression. We found exosomes from ECs under LPS stimulation (LPS-EC-Exo) enhanced the cell viability and attenuated the injury of CMs. The RNA sequencing depicted the expression of several miRNAs increased in LPS-EC-Exo compared with the exosomes from the control ECs (NC-EC-Exo). Further analysis showed that some miRNAs could promote the survival of CMs by down-regulating the expression of apoptosis-related proteins such as BAK1, P53, and PTEN. This study showed that LPS-EC-Exo has a cardiac protective effect on CMs, which miRNAs may achieve.
Keywords: Sepsis-induced myocardial dysfunction, endothelial cells, exosome, miRNA, cardiomyocytes
Introduction
Sepsis is a critical life-threatening situation that resulted from the severe response of the body to an infection. In sepsis, chemicals such as cytokines are released from the host body to confrontation the infection. When the body’s reaction to the chemicals is out of balance, multiple organ dysfunction could happen.
The heart is one of the most vulnerable target organs for sepsis. Even in the early stages of sepsis, tissue injury occurs in the heart, manifested as microvascular rupture, inflammatory cell infiltration, and focal myocardial necrosis [1,2]. Therefore, sepsis can lead to myocardial dysfunction (SIMD) [3], which is manifested by contractile dysfunction, dilated cardiomyopathy, and decreased ejection fraction [4]. SIMD leads to a significant elevation in mortality [4]. The pathogenesis of SIMD includes myocardial inhibitors, myocardial energy metabolism disorders, oxygen-free radicals damage, cardiomyocyte apoptosis, abnormal gene regulation, cardiac renin-angiotensin system disorder, and autonomic nerve dysfunction, etc. [5].
Endothelial cells (ECs) are located on the blood vessel’s inner side and contact the blood. In sepsis, ECs directly expose to bacterial metabolites or lysates and play a crucial role in the body’s inflammatory response. ECs regulate adjacent cells’ function through paracrine, and exosome transportation is one of the essential paracrine pathways. Exosomes are extracellular vesicles (EVs) with a diameter of 30 to 150 nm [6-8], which contain molecular components of the cells of their origin, including proteins and RNAs [9]. We isolated exosomes from bacterial lipopolysaccharide (LPS)-stimulated rat ECs and used to culture neonatal rat myocardial cells (CMs) to study the biological effects of EC-derived exosomes on CMs.
Material and methods
Ethical approval
The experimental animal procedures conformed to the Guide and Care and Use of Laboratory Animals (the US National Institutes of Health, 1996). The Animal Care and Use Committee of Southwest Medical University approved the experiment.
Primary endothelial cell isolation and cell culture
Male Sprague Dawley rats (8-week-old, bodyweight between 250-300 g) were anesthetized with 120 mg/kg of sodium pentobarbital and sacrificed. The thoracic aorta was harvested under a sterile circumstance, and the adventitia was removed. The artery was cut open, and the vessel’s inner side was digested with 0.1% type I collagenase. Endothelial cells (ECs) were isolated from the cell suspension and cultured in EC medium (ScienCell, Carlsbad, CA, USA) for 5-7 d to obtain primary cells. The anti-von Willebrand factor antibody (Abcam, Cambridge, UK) was used to identify ECs (Figure 1A), and ECs of passage 4-7 were used in subsequent experiments.
Figure 1.
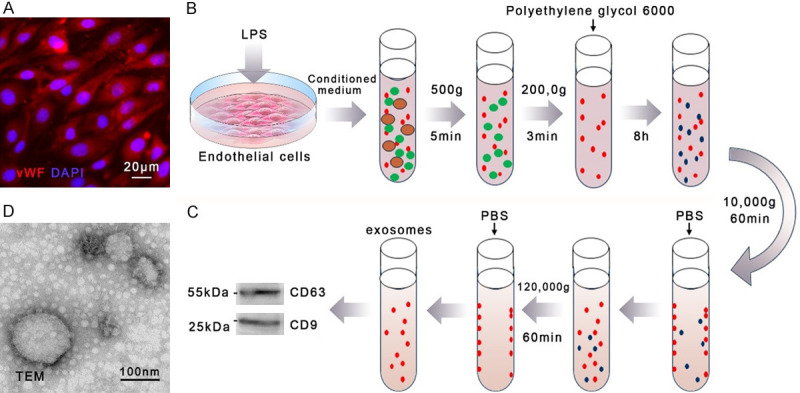
The isolation and identification of rat endothelial cell-derived exosomes. A. Rat endothelial cells (ECs) were isolated from the thoracic aorta and identified with the von Willebrand factor (VWF) antibody. B. ECs were stimulated with 0.5 µg/ml of lipopolysaccharides (LPS) for 6 h. The conditioned medium of ECs was harvested and proceeded to isolated exosomes following the steps demonstrated in the illustration. C. Vesicles were identified with the CD63 and CD9 antibodies. D. The shape and size of endothelial cell-derived exosomes were observed through a transmission electron microscope (TEM). n=5.
Endothelial derived-exosomes isolation and identification
ECs were seeded in the 100 mm cell culture dish to a confluency of 70% to 80%. Cells were synchronized in serum-free M199 for 12 h. Lipopolysaccharides (LPS) (MilliporeSigma, Burlington, MA, USA) solution was added into the experimental group’s EC culture medium to a final concentration of 0.5 µg/ml. An equal volume of PBS was added into the medium of the control group as a negative control. After 6 hours, the conditioned medium of ECs was collected. The conditioned medium was centrifuged at 500 g for 5 min at 4°C and 2000 g for 30 min to remove cell debris, larger microvesicles, and apoptotic bodies. Polyethylene glycol 6000 (PEG) was added into the supernatant to a final concentration of 8%. The mixture was well mixed to enrich the vesicle and stood for 8 h at 4°C. The hybrid solution was centrifuged at 10,000 g for 60 min at 4°C, and the supernatant containing excess PEG was discarded. Rinse the tube wall with PBS to resuspend exosomes. Centrifuge the suspension again at 120,000 g for 60 min at 4°C and discard the supernatant to further remove the PEG. Finally, the tube’s wall was rinsed with 100 ul of PBS to resuspend the exosomes (Figure 1B). A transmission electron microscope (Hitachi H-9500) was used to detect the shape and size of exosomes. The anti-CD63 antibody (R&D Systems) and anti-CD9 antibody (Abcam) were used to identify the vesicles and their origin of ECs.
Primary cardiomyocytes isolation and cell culture
The heart of the neonatal Sprague Dawley rat was removed in a sterile environment and cut into small pieces. The tissue mass was digested with 0.25% trypsin and 0.1% type II collagenase successively, and the supernatant was collected. The isolated cell collected by centrifuging the cell suspension then cultured in DMEM Complete Medium for 1 d to harvest primary cells. The anti-alpha-actin antibody (Abcam, Cambridge, UK) was used to identify cardiomyocytes (CMs) (Cell Signaling Technology, Danvers, MA, USA) (Figure 2A). Primary CMs were used in subsequent experiments.
Figure 2.
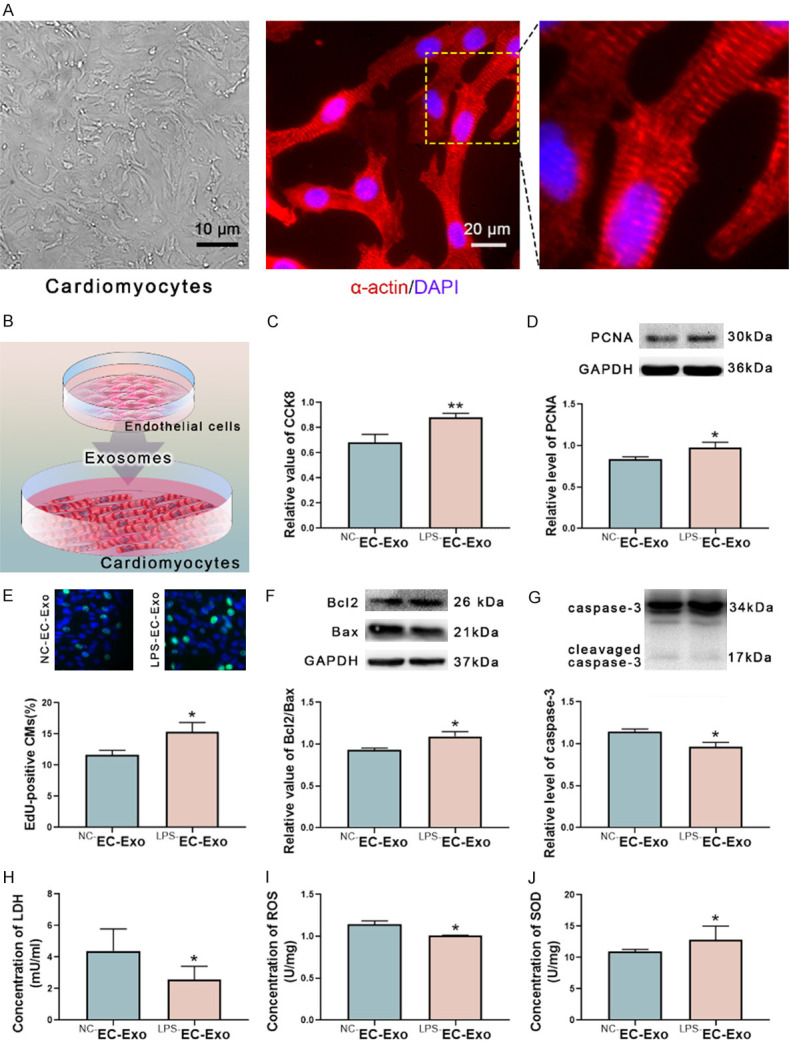
Endothelial cell-derived exosomes alleviate rat cardiomyocytes injury and apoptosis. A. The CMs were harvested from neonatal rats and identified with the α-actin antibody. B. The exosomes derived from ECs (EC-Exo) were isolated from the conditioned medium and added into the medium of CMs to test the biological effects. C. The exosomes from LPS-stimulated ECs (LPS-EC-Exo) increased the cell viability of CMs compared to the exosomes from normal ECs (NC-EC-Exo). D. LPS-EC-Exo elevated the level of proliferating cell nuclear antigen (PCNA) of CMs. E. EdU (5-ethynyl-2’-deoxyuridine) incorporation assay demonstrated that the cell viability of CMs was up-regulated under the LPS-EC-Exo. F. The ratio of Bcl2 and Bax was raised under the induction of LPS-EC-Exo. G. LPS-EC-Exo reduced the level of cleaved caspase-3 of CMs. H. LPS-EC-Exo down-regulated the level of lactate dehydrogenase (LDH) of CMs. I. LPS-EC-Exo decreased the level of reactive oxygen species (ROS) of CMs. J. LPS-EC-Exo enhanced the level of superoxide dismutase (SOD) of CMs. n=7, *P<0.05, **P<0.01.
Incubation of rat cardiomyocytes with endothelial cell-derived exosomes
We seeded 1×105 CMs in the 15.6 mm culture plate with 400 ul DMEM Complete Medium. 1×106 ECs were cultured in the 100 mm culture dish with 5 ml of culture medium, and endothelial-derived exosomes (EC-Exo) were collected as previously described. Then the collected exosomes were resuspended in 100 ul PBS. Finally, the exosomes-containing resuspension was added to the culture medium of CMs and incubated for 24 h (Figure 2B).
Cell counting kit-8 (CCK-8), LDH-cytotoxicity, reactive oxygen species (ROS) and superoxide dismutase (SOD) activity assay of cardiomyocytes
CMs incubated with EC-Exo were tested for CCK8 cell viability, LDH-cytotoxicity, ROS, and SOD activity according to the manufacturer’s instructions (Boster Biological Technology, China).
Western blot
RIPA buffer was used to lyse cardiomyocytes to extract total protein. The total protein was separated by 10% SDS-PAGE electrophoresis. The levels of PCNA, Bcl2, Bax, cleaved caspase-3, BAK1, P53, and PTEN were detected by western blot with the anti-PCNA antibody (Cell Signaling Technology), anti-Bcl2 antibody (Abcam), anti-Bax antibody (Abcam), anti-caspase-3 antibody (Cell Signaling Technology), anti-BAK1-antibody (Cell Signaling Technology), anti-P53-antibody (Cell Signaling Technology), and anti-PTEN-antibody (Cell Signaling Technology). GAPDH was used as an internal reference. Images were analyzed by Quantity One 4.6 (Bio-Rad, Hercules, CA, USA).
5-ethynyl-20-deoxyuridine (EdU) incorporation assay
Neonatal rat CMs were cultured in 96-well plates to 70% confluency. According to the manufacturer’s instructions, an EdU kit (Ribio, Guangzhou, China) was used to detect the level of nucleic synthesis in CMs. Observe the number of fluorescently labeled CMs under a fluorescence microscope (Olympus TH-4-200) to calculate the proliferation rate.
microRNA sequencing of exosomes and analysis of results
Exosomes extracted from the conditioned medium of ECs were subjected to the miRNA sequencing (Kangchen Biotech, Shanghai, China). The miRNAs with the absolute value of expression fold change >2 and p value <0.05 between the exposed group (LPS-EC-Exo) and the control group (NC-EC-Exo) were considered as differentially expressed miRNAs (DE miRNAs). The expressions of DE miRNAs in the exosomes were validated by quantitative real-time PCR using a miScript II RT Kit, a miScript SYBR PCR kit (Qiagen, Hilden, Germany), and synthetic primers (Ribio). The targets of the DE miRNAs were predicted with TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org) database. The functional clustering of target genes was performed using DAVID v6.8 (Visualization and Integrated Discovery, https://david.ncifcrf.gov/). The signal pathways of target genes may participate in were forecasted using the KEGG Pathway (Kyoto Encyclopedia of Genes and Genomes, https://www.genome.jp/kegg/).
Statistical analysis
All data were expressed as the mean ± SEM. The results were processed with GraphPad Prism 8.0.1. Two-tailed Student’s t-test was used to analyze the data from two groups. Bonferroni-corrected p-value <0.05 was statistically significant.
Results
Morphology and origin identification of endothelial cell-derived exosomes
ECs in the exposed group were stimulated with 0.5 µg/ml of LPS, while ECs in the control group were given an equal volume of PBS. Exosomes were isolated from the conditioned medium of ECs by polyethylene glycol (PEG) separation and ultracentrifugation. The extracted vesicles expressed CD63 and CD9 (Figure 1C). The transmission electron microscope showed that the extract contained many round vesicle structures with a diameter of 30-150 nm (Figure 1D). It is worth noting that when the LPS concentration was gradually increased to 10 µg/ml, exosomes produced by ECs decreased significantly, while the number of apoptotic bodies (ABs) with larger diameters increased markedly.
Exosomes from LPS-exposed ECs increased the viability and proliferation of neonatal rat cardiomyocytes
Cell Counting Kit-8 (CCK-8) assay showed that compared with the exosomes from the normal ECs (NC-EC-Exo), the exosomes from LPS-exposed ECs (LPS-EC-Exo) increased the viability of neonatal rat CMs (Figure 2C). Detection of the level of proliferating cell nuclear antigen (PCNA) and 5-ethynyl-20-deoxyuridine (EdU) incorporation assay showed that LPS-EC-Exo elevated the proliferation of CMs (Figure 2D and 2E).
LPS-EC-Exo alleviated the apoptosis of cardiomyocytes
Western blot showed that the ratio of Bcl2 and Bax (Bcl2/Bax) of CMs raised and the level of cleaved caspase-3 dropped under the action of LPS-EC-Exo, which indicated that LPS-EC-Exo reduced the apoptosis of CMs (Figure 2F and 2G).
LPS-EC-Exo decreased the level of LDH and ROS in cardiomyocytes, while inflated the level of SOD
Compared with NC-EC-Exo, LPS-EC-Exo decreased the levels of lactate dehydrogenase (LDH) and reactive oxygen species (ROS) while increased the level of superoxide dismutases (SOD) in CMs (Figure 2H-J). These indicated that LPS-EC-Exo resisted oxidative stress to CMs.
miRNAs expression difference between LPS-EC-Exo and NC-EC-Exo
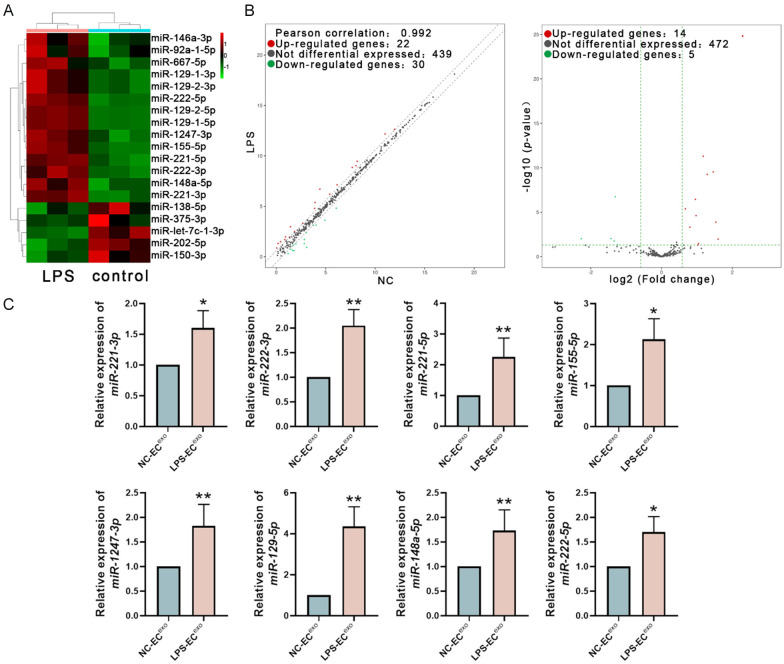
Detected and compared the miRNA expression profiles of NC-EC-Exo and LPS-EC-Exo by small RNA sequencing. miRNAs with an absolute expression fold change >2 and p-value <0.05 between two groups were considered as the differential expressed miRNAs. In LPS-EC-Exo, 15 miRNA expression levels are higher than in NC-EC-Exo, and 4 miRNA expression levels are lower than in NC-EC-Exo (Figure 3A, 3B; Table 1). The top 8 highly expressed miRNAs in LPS-EC-Exo (miR-221-3p, miR-222-3p, miR-221-5p, miR-155-5p, miR-1247-3p, mir-129-5p, miR-148a-5p, and miR-222-5p) were detected by real-time quantitative PCR to confirm their expression differences between the groups. The PCR results were consistent with the sequencing data (Figure 3C).
Figure 3.
The analysis of miRNAs Carried by EC-Exo. A. The endothelial cell-derived exosomes were subjected to miRNA sequencing. The differential expressed miRNAs between NC-EC-Exo and LPS-EC-Exo (absolute value of the expression fold change >2.0, P<0.05) were identified. B. The differential expression of miRNAs in NC-EC-Exo and LPS-EC-Exo was shown in the volcano plots. C. The expression of the 8 miRNAs up-regulated in LPS-EC-Exo was verified with real-time quantification PCR. The results of qPCR confirmed the data of miRNA sequencing. n=7, *P<0.05, **P<0.01.
Table 1.
The detailed information of the differential expressed miRNAs
| Annotation | DE Statistic | Counts per million reads (CPM) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Mature_ID | Length | Seed | Mature_Seq | Regulation | Fold_Change | p_value | LPS | NC |
| rno-miR-221-3p | 23 | GCUACA | AGCUACAUUGUCUGCUGGGUUUC | up | 1.595974069 | 4.12268E-06 | 12.6296558 | 11.95522 |
| rno-miR-222-3p | 21 | GCUACA | AGCUACAUCUGGCUACUGGGU | up | 2.262590717 | 5.09634E-12 | 12.17364154 | 10.99567 |
| rno-miR-221-5p | 23 | CCUGGC | ACCUGGCAUACAAUGUAGAUUUC | up | 2.44701923 | 5.54174E-10 | 9.466560617 | 8.175535 |
| rno-miR-155-5p | 23 | UAAUGC | UUAAUGCUAAUUGUGAUAGGGGU | up | 1.941562851 | 3.60596E-07 | 9.020271954 | 8.063054 |
| rno-miR-1247-3p | 21 | GGGAAC | CGGGAACGUCGAGACUGGAGC | up | 1.965058526 | 2.4416E-05 | 7.129555063 | 6.154983 |
| rno-miR-129-5p | 21 | UUUUUG | CUUUUUGCGGUCUGGGCUUGC | up | 4.887359275 | 1.66167E-25 | 6.698536894 | 4.409482 |
| rno-miR-148a-5p | 23 | AAGUUC | AAAGUUCUGAGACACUCUGACUC | up | 1.744113276 | 0.000486805 | 6.207993957 | 5.4055 |
| rno-miR-222-5p | 21 | GCUCAG | GGCUCAGUAGCCAGUGUAGAU | up | 2.75305286 | 2.93239E-10 | 5.381283427 | 3.920251 |
| rno-miR-129-1-3p | 19 | AGCCCU | AAGCCCUUACCCCAAAAAG | up | 2.893477567 | 0.000131722 | 2.970139124 | 1.437335 |
| rno-miR-129-2-3p | 22 | AGCCCU | AAGCCCUUACCCCAAAAAGCAU | up | 2.893477567 | 0.000131722 | 2.970139124 | 1.437335 |
| rno-miR-92a-1-5p | 23 | GGUUGG | AGGUUGGGAUUUGUCGCAAUGCU | up | 2.035078562 | 0.049602557 | 1.971381937 | 0.946297 |
| rno-miR-146a-3p | 21 | CCUGUG | ACCUGUGAAGUUCAGUUCUUU | up | 2.062720704 | 0.034621139 | 1.477210296 | 0.432662 |
| rno-miR-667-5p | 24 | GGUGCU | CGGUGCUGGUGGAGCAGUGAGCAC | up | 3.029066665 | 0.010661818 | 0.548503655 | -1.05037 |
| rno-let-7c-1-3p | 22 | UGUACA | CUGUACAACCUUCUAGCUUUCC | down | 0.404485731 | 1.79452E-07 | 3.61831496 | 4.924154 |
| rno-miR-138-5p | 23 | GCUGGU | AGCUGGUGUUGUGAAUCAGGCCG | down | 0.395627314 | 0.01767291 | -0.363997435 | 0.973789 |
| rno-miR-375-3p | 22 | UUGUUC | UUUGUUCGUUCGGCUCGCGUGA | down | 0.207440959 | 0.009642982 | -0.727851487 | 1.541376 |
| rno-miR-202-5p | 19 | UCCUAU | UUCCUAUGCAUAUACUUCU | down | 0.371070492 | 0.009487856 | -0.730950453 | 0.699284 |
| rno-miR-150-3p | 19 | UGGUAC | CUGGUACAGGCCUGGGGGA | down | 0.423606483 | 0.046385033 | -1.217325179 | 0.021878 |
MiRNAs in the pink background were up-regulated under the induction of LPS, and the miRNAs in the green were down-regulated.
Bioinformatic analysis of the differential expressed miRNAs
TargetScan and miRanda databases were used for the bioinformatics analysis to the top 8 miRNAs (miR-221-3p, miR-222-3p, miR-221-5p, miR-155-5p, miR-1247-3p, mir-129-5p, miR-148a-5p and miR-222-5p).
Gene Ontology (GO) analysis was performed with DAVID 6.8 to forecast the biological process (BP), cellular component (CC), and molecular function (MF) that the DE miRNAs may participate in. The result showed that the DE miRNAs had a high possibility of affecting cell metabolism. We evaluated the potential target genes of the DE miRNAs. We found that many of them were related to cardiomyocytes’ function, such as regulating heart contraction, mo dulating heart rate, and cell communication involved in cardiac conduction (Figure 4A). Pathway analysis was performed on KEGG to predict the signal pathways that the DE miRNAs may participate in (Figure 4B). Twenty-one genes regulate cardiomyocytes’ biological behavior through four pathways, including Adrenergic Signaling in Cardiomyocytes, Hypertrophic Cardiomyopathy, Arrhythmogenic Right Ventricular Cardiomyopathy, and Dilated Cardiomyopathy (Table 2).
Figure 4.
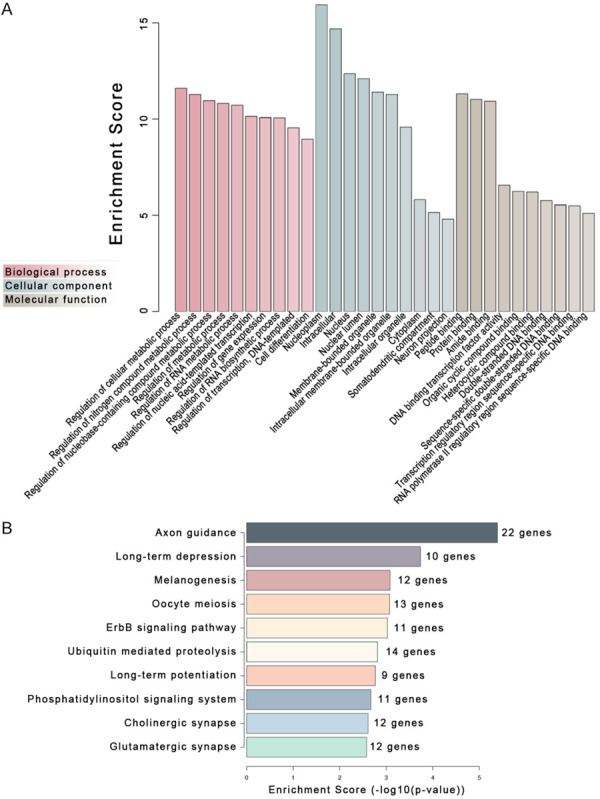
Eight miRNAs up-regulated in the exosomes from LPS-stimulated ECs were subjected to gene ontology (GO) and Pathway analysis to predict their roles and signaling pathways they potentially involved. A. The top 10 entries in the biological process, cellular component, and molecular function according to GO analysis. B. The top 10 signaling pathways predicted by KEGG analysis.
Table 2.
Bioinformatics analysis showed the candidate genes may regulate the biological behavior of cardiomyocytes in four pathways
| ID | Term | Count | Genes |
|---|---|---|---|
| rno04261 | Adrenergic signaling in cardiomyocytes | 14 | Atp1b1, Atp1b4, Atp2b2, Atp2b4, Cacnb4, Creb3l2, Gnai2, Gnai3, Mapk3, Myl3, Plcb1, Ppp2r5e, Ryr2, Scn4b |
| rno05410 | Hypertrophic cardiomyopathy (HCM) | 8 | Cacnb4, Igf1, Itga6, Itgb1, Lmna, Myl3, Prkaa1, Ryr2 |
| rno05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 7 | Cacnb4, Ctnna3, Itga6, Itgb1, Lmna, Ryr2, Tcf7 |
| rno05414 | Dilated cardiomyopathy | 7 | Cacnb4, Igf1, Itga6, Itgb1, Lmna, Myl3, Ryr2 |
The miRNAs highly expressed in LPS-EC-Exo have extensive regulatory effects on cardiomyocytes. They play essential roles in physiological processes such as cardiac growth induced by physical exercise [10-13], embryonic-stem-cell-derived CMs maturation [14,15]. They also take vital roles in pathophysiological processes such as viral myocarditis [16,17], myocardial hypertrophy [18-20], and dilated cardiomyopathy [21] (Table 3A). Notably, several miRNAs have been reported to have anti-apoptotic effects on cardiomyocytes, including miR-221-3p, miR-221-5p, miR-155-5p, and miR-222-5p (Figure 5A).
Table 3.
Cardiac effects of miRNAs highly expressed in LPS-EC-Exo
| A. Cardioprotective miRNAs | |
|
| |
| DE miRNA | Effects |
|
| |
| miR-221-5p | Attenuates LPS induced CM injury [24] |
| Reduces hypoxia-induced CM apoptosis [25,26] | |
| Enhances CM survival and improves cardiac function after MI [27] | |
| miR-222-5p | Improves myocardial survival [28] |
| Necessary for cardiac growth induced by physical exercise [10-13] | |
| Decreases virus-induced CM apoptosis [16] | |
| miR-221/222 clusters | Induce embryonic-stem-cell-derived CMs maturation [14,15] |
| Balance inflammatory response of viral myocarditis [23] | |
| miRNA-155-5p | Improves myocardial survival [28] |
| Improves the survival rate of mice with acute viral myocarditis [17] | |
| miRNA-148a-5p | Prevents the occurrence of dilated cardiomyopathy [21] |
| Improves the survival rate of mice with viral myocarditis [17] | |
|
| |
| B. Pathological Cardiac miRNAs | |
|
| |
| miRNA-155-5p | Induces pathological myocardial hypertrophy [18-20] |
| Induces CM apoptosis in MI [29-32] | |
| miRNA-221-3p | Participates in H2O2 induced myocardial injury [33] |
| miRNA-129-5p | Inhibits CM proliferation [34] |
Abbreviations and acronyms: CM cardiomyocyte, MI myocardial infarction.
Figure 5.
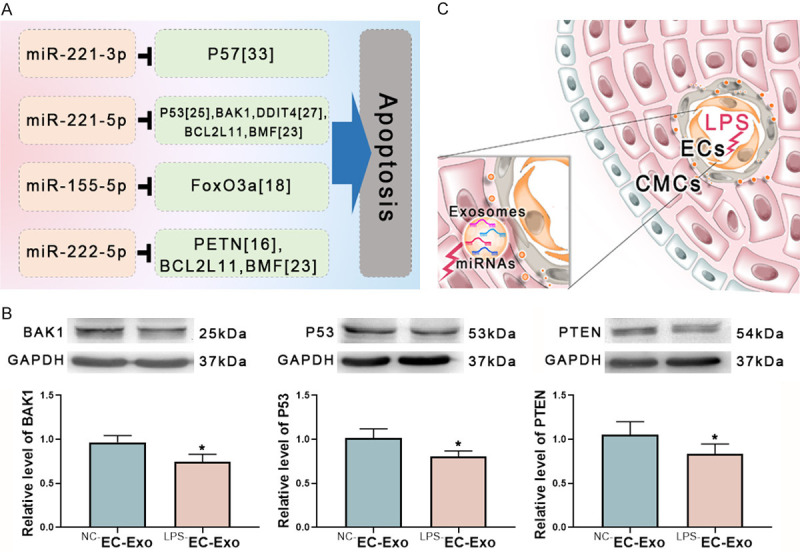
miRNAs prevent cardiomyocytes injury through the predicted targets. A. Several differentially expressed miRNAs in LPS-EC-Exo have shown anti-apoptosis roles in myocardiocytes by targeting cell death-related genes. B. The presumed apoptosis-related genes were tested in the cardiomyocytes stimulated with EC-Exo. LPS-EC-Exo attenuated the level of BAK1, P53, and PTEN. C. The possible mechanism of LPS-EC-Exo protecting cardiomyocytes. In the early sepsis stage, LPS enters the blood circulation and stimulates ECs to express specific miRNAs. Then the differential expressed miRNAs are delivered outside ECs through exosomes and exert a cardiac protective effect. EC-Exo may play an important role in preventing myocardial dysfunction in the early stages of sepsis. n=7, *P<0.05.
Expression of apoptosis-related genes in CMs stimulated by LPS-EC-Exo
We detected the potential targets of the differentially expressed miRNAs shown in Figure 5A after stimulating cardiomyocytes with EC-derived exosomes. Compared with NC-EC-Exo, LPS-EC-Exo down-modulated the level of BAK1, P35, and PTEN, which are apoptosis-related genes in cardiomyocytes (Figure 5B).
Discussion
Sepsis is a life-threatening host response imbalance caused by an infection. Sepsis-induced myocardial dysfunction (SIMD) is one of the critical mortality causes of septic shock. SIMD is often difficult to correct and ultimately leads patients to death. In this study, we found that exosomes released by LPS-stimulated endothelial cells exerted cardiac protective effects.
We found that the exosomes from LPS (0.5 µg/ml) stimulated ECs (LPS-EC-Exo) enhanced neonatal rat cardiomyocytes’ viability and reduced the apoptosis. Meanwhile, LPS-EC-Exo led a decline in LDH and ROS, and a rise in SOD in CMs. These phenomena indicated that LPS-EC-Exo possessed cardiac protective substances. Notably, we also found that in the presence of a higher concentration (10 µg/ml) of LPS, microvesicles (MVs) produced by endothelial cells are mainly apoptotic bodies, while exosomes are markedly reduced. A similar phenomenon has been observed when adipocytes were stimulated with LPS [22]. We deem that at high concentrations of LPS, due to the apoptosis of endothelial cells, the protective effect of EC-derived exosomes on cardiomyocytes weakens or disappears. Therefore, the protective effect of EC-Exo on cardiomyocytes may only exist in the early stages of sepsis.
We detected the miRNAs carried by exosomes and found that the expression profiles of miRNAs of LPS-EC-Exo and NC-EC-Exo were significantly different. The top 8 miRNAs highly expressed in LPS-EC-Exo (miR-221-3p, miR-222-3p, miR-221-5p, miR-1555p, miR-1247-3p, mir-129-5p, miR-148a-5p, and miR-222-5p) were analyzed with bioinformatic tools for target gene prediction, functional enrichment, and pathway forecasting. Some predicted target genes were marked in databases (TargetScan and miRanda) as involved in cardiomyocyte function regulation.
Most DE miRNAs on LPS-EC-Exo are beneficial for the survival of cardiomyocytes (Table 3A). It can explain the reason why LPS-EC-Exo improved the vitality of CMs and reduces myocardial injury markers. Some miRNAs highly expressed in LPS-EC-Exo possess anti-apoptosis roles in cardiomyocytes (Figure 5A). For example, miRNA-221/222 clusters help cardiomyocyte survival and differentiation in most cases. In a viral myocarditis model, miRNA-221/222 clusters play an antiviral role and balance inflammatory response [23]. The clusters also induce the maturation of embryonic-stem-cell-derived cardiomyocytes [14,15]. miRNA-221-5p attenuates LPS induced cardiomyocyte damage by regulating the NF-κB and JNK pathways [24]. Under hypoxic conditions, miRNA-221-5p protects the heart muscle by inhibiting apoptosis [25,26]. In a rat model of myocardial infarction (MI), miRNA-221-5p enhances myocardial cell survival and improves cardiac function after MI [27]. miRNA-222-5p also improves myocardial survival [28]. miRNA-222-5p is necessary for cardiac growth induced by exercise [10-13]. In viral myocarditis, miRNA-222-5p exerts myocardial protection by inhibiting apoptosis [16]. miRNA-155-5p has a double-sided effect on the heart. In some cases, it is beneficial for cardiac survival [28]. It improves the survival rate of mice with acute viral myocarditis [17]. miRNA-148a-5p also has myocardial protective effects. It prevents dilated cardiomyopathy [21] and enhances mice’s survival rate with viral myocarditis [17]. In this study, we found the exosomes from LPS-stimulating ECs (LPS-EC-Exo) attenuated the level of BAK1, P53, and PTEN in rat cardiomyocytes. These proteins are essential to initiate apoptosis. Their reduction can explain the mechanism of the cardioprotective effect of LPS-EC-Exo.
However, a few DE miRNAs have been reported to be associated with myocardial injury (Table 3B). miRNA-155-5p induces pathological myocardial hypertrophy [18-20]. In the myocardial infarction model of mice, miRNA-155-5p induces cardiomyocyte apoptosis [29-32]. miRNA-221-3p participates in H2O2 induced myocardial damage by inhibiting p57 [33]. miRNA-129-5p inhibits cardiomyocyte proliferation by inhibiting CDK6 [34]. The roles of these miRNAs in myocardial injury caused by LPS needs further study.
In brief, this study found that the exosomes produced by the appropriate concentration of LPS-stimulated ECs to have protective effects on neonatal rat cardiomyocytes. LPS-EC-Exo reduced the apoptosis and injury of CMs (Figure 5C). These protective effects may be the body’s response to avoid sepsis-induced cardiomyocyte dysfunction in the early stages of sepsis and may be achieved by miRNAs carried on exosomes.
Acknowledgements
This study was supported by the National Natural Science Foundation of China, Grant/Award Number: 82070288 and 31900813; The Science & Technology Department of Sichuan Province, Grant/Award Number: 20ZDYF2108 and 2019YJ0409; Office of Science and Technology and Intellectual Property of Luzhou, Grant/Award Number: 2019LZXNYDJ29; Talent development project of The Affiliated Hospital of Southwest Medical University; Collaborative Innovation Center for Prevention and Treatment of Sichuan Province, Southwest Medical University, Grant/Award Number: xtcx2019-18; Education Department of Sichuan Province, Grant/Award Number: 18ZB0637 and 18ZB0641.
Disclosure of conflict of interest
None.
References
- 1.Labugger R, Organ L, Collier C, Atar D, Van Eyk JE. Extensive troponin I and T modification detected in serum from patients with acute myocardial infarction. Circulation. 2000;102:1221–1226. doi: 10.1161/01.cir.102.11.1221. [DOI] [PubMed] [Google Scholar]
- 2.Merx MW, Weber C. Sepsis and the heart. Circulation. 2007;116:793–802. doi: 10.1161/CIRCULATIONAHA.106.678359. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes CJ Jr, Akamine N, Knobel E. Myocardial depression in sepsis. Shock. 2008;30(Suppl 1):14–17. doi: 10.1097/SHK.0b013e3181818617. [DOI] [PubMed] [Google Scholar]
- 4.Blanco J, Muriel-Bombín A, Sagredo V, Taboada F, Gandía F, Tamayo L, Collado J, García-Labattut A, Carriedo D, Valledor M, De Frutos M, López MJ, Caballero A, Guerra J, Alvarez B, Mayo A, Villar J Grupo de Estudios y Análisis en Cuidados Intensivos. Incidence, organ dysfunction and mortality in severe sepsis: a Spanish multicentre study. Crit Care. 2008;12:R158. doi: 10.1186/cc7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takasu O, Gaut JP, Watanabe E, To K, Fagley RE, Sato B, Jarman S, Efimov IR, Janks DL, Srivastava A, Bhayani SB, Drewry A, Swanson PE, Hotchkiss RS. Mechanisms of cardiac and renal dysfunction in patients dying of sepsis. Am J Respir Crit Care Med. 2013;187:509–517. doi: 10.1164/rccm.201211-1983OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawson C, Vicencio JM, Yellon DM, Davidson SM. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J Endocrinol. 2016;228:R57–71. doi: 10.1530/JOE-15-0201. [DOI] [PubMed] [Google Scholar]
- 8.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Tao L, Bei Y, Zhang H, Xiao J, Li X. Exercise for the heart: signaling pathways. Oncotarget. 2015;6:20773–20784. doi: 10.18632/oncotarget.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuttler D, Clauss S, Weckbach LT, Brunner S. Molecular mechanisms of cardiac remodeling and regeneration in physical exercise. Cells. 2019;8:1128. doi: 10.3390/cells8101128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vujic A, Lerchenmuller C, Wu TD, Guillermier C, Rabolli CP, Gonzalez E, Senyo SE, Liu X, Guerquin-Kern JL, Steinhauser ML, Lee RT, Rosenzweig A. Exercise induces new cardiomyocyte generation in the adult mammalian heart. Nat Commun. 2018;9:1659. doi: 10.1038/s41467-018-04083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Bostrom P, Che L, Zhang C, Spiegelman BM, Rosenzweig A. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DS, Chen JH, Lundy DJ, Liu CH, Hwang SM, Pabon L, Shieh RC, Chen CC, Wu SN, Yan YT, Lee ST, Chiang PM, Chien S, Murry CE, Hsieh PC. Defined microRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Rep. 2015;12:1960–1967. doi: 10.1016/j.celrep.2015.08.042. [DOI] [PubMed] [Google Scholar]
- 15.Hu S, Huang M, Nguyen PK, Gong Y, Li Z, Jia F, Lan F, Liu J, Nag D, Robbins RC, Wu JC. Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation. 2011;124:S27–34. doi: 10.1161/CIRCULATIONAHA.111.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Gao X, Hu J, Xie Y, Zuo Y, Xu H, Zhu S. ADAR1p150 forms a complex with dicer to promote miRNA-222 activity and regulate PTEN expression in CVB3-induced viral myocarditis. Int J Mol Sci. 2019;20:407. doi: 10.3390/ijms20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao JL, Lin L. MiR-155 and miR-148a reduce cardiac injury by inhibiting NF-kappaB pathway during acute viral myocarditis. Eur Rev Med Pharmacol Sci. 2014;18:2349–2356. [PubMed] [Google Scholar]
- 18.Fan Y, Liu L, Fang K, Huang T, Wan L, Liu Y, Zhang S, Yan D, Li G, Gao Y, Lv Y, Chen Y, Tu Y. Resveratrol ameliorates cardiac hypertrophy by down-regulation of mir-155 through activation of breast cancer type 1 susceptibility protein. J Am Heart Assoc. 2016;5:e002648. doi: 10.1161/JAHA.115.002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Wang J, Chen Q, Wu Q, Deng W, Zhou H, Shen D. Long non-coding RNA cytoskeleton regulator RNA (CYTOR) modulates pathological cardiac hypertrophy through miR-155-mediated IKKi signaling. Biochim Biophys Acta Mol Basis Dis. 2019;1865:1421–1427. doi: 10.1016/j.bbadis.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Seok HY, Chen J, Kataoka M, Huang ZP, Ding J, Yan J, Hu X, Wang DZ. Loss of MicroRNA-155 protects the heart from pathological cardiac hypertrophy. Circ Res. 2014;114:1585–1595. doi: 10.1161/CIRCRESAHA.114.303784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raso A, Dirkx E, Philippen LE, Fernandez-Celis A, De Majo F, Sampaio-Pinto V, Sansonetti M, Juni R, El Azzouzi H, Calore M, Bitsch N, Olieslagers S, Oerlemans M, Huibers MM, de Weger RA, Reckman YJ, Pinto YM, Zentilin L, Zacchigna S, Giacca M, da Costa Martins PA, Lopez-Andres N, De Windt LJ. Therapeutic delivery of miR-148a suppresses ventricular dilation in heart failure. Mol Ther. 2019;27:584–599. doi: 10.1016/j.ymthe.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega FJ, Moreno M, Mercader JM, Moreno-Navarrete JM, Fuentes-Batllevell N, Sabater M, Ricart W, Fernandez-Real JM. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin Epigenetics. 2015;7:49. doi: 10.1186/s13148-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corsten MF, Heggermont W, Papageorgiou AP, Deckx S, Tijsma A, Verhesen W, van Leeuwen R, Carai P, Thibaut HJ, Custers K, Summer G, Hazebroek M, Verheyen F, Neyts J, Schroen B, Heymans S. The microRNA-221/-222 cluster balances the antiviral and inflammatory response in viral myocarditis. Eur Heart J. 2015;36:2909–2919. doi: 10.1093/eurheartj/ehv321. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Wang L, Guo E, Qi Y. Silence of lncRNA CHRF protects H9c2 cells against lipopolysaccharide-induced injury via up-regulating microRNA-221. Exp Mol Pathol. 2019;107:43–50. doi: 10.1016/j.yexmp.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Yu B, Gong M, Wang Y, Millard RW, Pasha Z, Yang Y, Ashraf M, Xu M. Cardiomyocyte protection by GATA-4 gene engineered mesenchymal stem cells is partially mediated by translocation of miR-221 in microvesicles. PLoS One. 2013;8:e73304. doi: 10.1371/journal.pone.0073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q, Zhou Y, Richards AM, Wang P. Up-regulation of miRNA-221 inhibits hypoxia/reoxygenation-induced autophagy through the DDIT4/mTORC1 and Tp53inp1/p62 pathways. Biochem Biophys Res Commun. 2016;474:168–174. doi: 10.1016/j.bbrc.2016.04.090. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Richards AM, Wang P. MicroRNA-221 is cardioprotective and anti-fibrotic in a rat model of myocardial infarction. Mol Ther Nucleic Acids. 2019;17:185–197. doi: 10.1016/j.omtn.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao L, Bei Y, Zhou Y, Xiao J, Li X. Non-coding RNAs in cardiac regeneration. Oncotarget. 2015;6:42613–42622. doi: 10.18632/oncotarget.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang G, Hao F, Hu X. Downregulation of microRNA-155 stimulates sevoflurane-mediated cardioprotection against myocardial ischemia/reperfusion injury by binding to SIRT1 in mice. J Cell Biochem. 2019;120:15494–15505. doi: 10.1002/jcb.28816. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Bei Y, Huang P, Zhou Q, Shi J, Sun Q, Zhong J, Li X, Kong X, Xiao J. Inhibition of miR-155 protects against LPS-induced cardiac dysfunction and apoptosis in mice. Mol Ther Nucleic Acids. 2016;5:e374. doi: 10.1038/mtna.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Huang CX, Rao PP, Cao GQ, Zhang Y, Zhou JP, Zhu LY, Liu MX, Zhang GG. MicroRNA-155 inhibition attenuates endoplasmic reticulum stress-induced cardiomyocyte apoptosis following myocardial infarction via reducing macrophage inflammation. Eur J Pharmacol. 2019;857:172449. doi: 10.1016/j.ejphar.2019.172449. [DOI] [PubMed] [Google Scholar]
- 32.Guo J, Liu HB, Sun C, Yan XQ, Hu J, Yu J, Yuan Y, Du ZM. MicroRNA-155 promotes myocardial infarction-induced apoptosis by targeting RNA-binding protein QKI. Oxid Med Cell Longev. 2019;2019:4579806. doi: 10.1155/2019/4579806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng Q, Liu Y, Huo X, Sun H, Wang Y, Bu F. MicroRNA221-3p contributes to cardiomyocyte injury in H2O2 treated H9c2 cells and a rat model of myocardial ischemia reperfusion by targeting p57. Int J Mol Med. 2018;42:589–596. doi: 10.3892/ijmm.2018.3628. [DOI] [PubMed] [Google Scholar]
- 34.Majumdar G, Raghow R. Trichostatin A induces a unique set of microRNAs including miR-129-5p that blocks cyclin-dependent kinase 6 expression and proliferation in H9c2 cardiac myocytes. Mol Cell Biochem. 2016;415:39–49. doi: 10.1007/s11010-016-2675-4. [DOI] [PubMed] [Google Scholar]