Abstract
Objective: To explore the molecular mechanism of umbilical cord blood mesenchymal stem cells (UCBMSCs) in the treatment of advanced osteoarthritis pain. Methods: Normal healthy rats were selected to establish advanced osteoarthritis (OA) model, and the rats were randomly divided into control group, intravenous group, intracavitary group and intrathecal group. The intravenous group received intravenous injection of UCBMSCs, intracavitary group received intra-articular injection of UCBMSCs, and intrathecal group received subarachnoid injection of UCBMSCs. The pain behavior and serum pro-inflammatory factor levels were evaluated before and after treatment. microRNA-29a-3p and FOS mRNA in spinal dorsal horn was detected using qPCR, the phosphorylation of c-fos protein and NR1, NR2B, ERK and PKCg was detected using Western blot, and the level of LncRNA H19 was detected using qPCR. Results: LncRNA H19 was enriched in the exosomes of UCBMSCs. microRNA-29a-3p was the target gene of LncRNA H19, while FOS was the downstream target of microRNA-29a-3p. Pain and inflammation of rats in the intrathecal group improved best, and the phosphorylation levels of c-fos and NR1, NR2B, ERK and PKCg in the spinal dorsal horn of the intrathecal group decreased. LncRNA H19 regulated the central sensitization of astrocytes through microRNA-29a-3p/FOS axis. Conclusion: Intrathecal injection of umbilical cord blood mesenchymal stem cells can improve the pain and central sensitization of advanced osteoarthritis through LncRNA H19/microRNA-29a-3p/FOS axis.
Keywords: Pain of advanced osteoarthritis, central sensitization, umbilical cord blood mesenchymal stem cells, LncRNA H19/microRNA-29a-3p/FOS axis
Introduction
According to International Association for the Study of Pain, pain symptoms caused by somatosensory nervous system diseases or diseases are collectively called Neuropathic pain (NP) [1]. The clinical manifestations of NP are abnormal allergy to external stimuli and harmful response to non-harmful stimuli [2]. NP has a negative impact on patients’ quality of life and limits their normal life [3]. According to statistics, the prevalence rate of NP in general population is 6.9-10% [4]. There are various incentives for NP, which leads to no optimal treatment to improve the pain symptoms of NP patients [5]. Therefore, this article aimed to discuss a safe and effective treatment.
Stem cells have strong regeneration ability and have potential to differentiate into other cell types [6]. Stem cell therapy has broad clinical therapeutic potential, and many studies have shown that stem cells can be used to treat heart disease, osteoarthritis and Crohn’s disease [7-9]. Stem cell therapy has a promising therapeutic prospect for relieving NP pain. Studies have confirmed that mesenchymal stem cells may alleviate NP caused by spinal cord injury. Clinically, there are also reports [11] indicates that stem cell therapy can interfere with the intensity of nerve pain in a certain period of time. Although stem cell therapy has certain therapeutic potential in NP, the mechanism of stem cell action in NP is still unclear.
The expression of c-fos is closely related to pain. The expression of c-fos in frontal cortex, thalamus and periaqueductal gray increased significantly after nerve injury [12], while the expression of c-fos caused changes in nerve activity [13,14], so c-fos can be used as a nerve marker to evaluate pain [15]. Previous studies [16] showed that NP induced the expression of c-fos protein in anterior cingulate cortex of brain, and drug inhibition of c-fos could alleviate the pain degree of NP. Therefore, studying the expression changes of c-fos in NP is helpful to study the mechanism of related treatments in NP.
Pain caused by advanced osteoarthritis (OA) is one of the main types of NP. In this study, the pain model of advanced osteoarthritis in rats was induced by monosodium iodoacetate (MIA), and the therapeutic effects of different injection of umbilical cord blood mesenchymal stem cells (UCBMSCs) on advanced OA pain were compared. At present, there are few studies on stem cell therapy for central sensitization of advanced OA pain. This study will discuss the role of UCBMSCs in central sensitization of advanced OA pain.
Method
Advanced OA pain model
Thirty-five healthy Wistar rats (145-175 g) were selected to adapt to the environment for one week before modeling, with normal drink and food and free movement. The animal experiment was approved by the Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital Ethics Committee, and the experimental operation conformed to animal ethics. Before establishing advanced osteoarthritis (OA) model, rats were anesthetized with 2-4% isoflurane and injected with 50 L MIA into right knee joint. A total of 28 rats were successfully modeled. Twenty-eight rats were randomly divided into control group, intravenous group, intracavitary group and intrathecal group. The intravenous group received intravenous injection of UCBMSCs, the intracavitary group received intra-articular injection of UCBMSCs and the intrathecal group received subarachnoid injection of UCBMSCs. Pain behavior assessment referred to the research of Kimmerling and others [17]. The markers of UCBMSCs including CD105, CD44, CD34 and CD45 were detected by flow cytometry. The levels of inflammatory factors containing IL-1/IL-2/IL-6/IL-10/MCP-1/TNF-α were detected by ELISA. The rats were anesthetized with 2% isoflurane until they stopped breathing after all pain behavioral evaluations. The neurons of spinal dorsal horn were extracted, and the RNA and protein levels were detected by qPCR and Western blot.
Separation and screening of exosomes
Cell suspension was centrifuged at 1*105 g for 8 h. After that, the cells were cultured in 1640 medium (containing 10% FBS) at 37°C with 5% CO2. After the cells were cultured to 80~90% coverage rate, the supernatant of culture medium was collected and exosomes were extracted. Exosome was centrifuged at 1*105 g for 1.5 h, and the sediment of exosome in lower layer was collected. PBS buffer was added to the exosome precipitate and the buffer was blown repeatedly. Exosome markers TSG101 and CD63 were detected by flow cytometry.
Culture and transfection of astrocytes in vitro
Astrocytes were purchased from ATCC cell bank. The cells were cultured in 1640 medium (Hyclone Company) at 37°C with 5% CO2, and after the cells were cultured to 80~90% coverage rate, subsequent experiments were carried out. Cells with 1*105 were inoculated into the 6-well plate and cultured with medium without FBS. The microRNA-29a-3p inhibitor, FOS siRNA, NC siRNA and NC inhibitor vectors were purchased from Beijing Tiangen company (China). Lipofectamine 2000 transfection kit (Invitrogen, USA) was applied for transfection.
qPCR
Total RNA in astrocytes or spinal dorsal horn of rats was extracted by Trizon reagent (Invitrogen, California, USA). The mRNA expression of microRNA-29a-3p and FOS mRNA was quantified by TaqMan One Step RT-qPCR kit (Solarbio, Beijing, China). The reaction system and reaction procedure referred to the kit instructions. U6 and GAPDH were used as control genes. RNA expression was quantitatively analyzed by QuantStudio™ 7 Flex real-time fluorescence quantitative PCR (Applied Biosystems, USA). The relative expression of RNA was normalized by 2-ΔΔCt method. Primers were obtained from Beijing Tiangen Company.
Western blot
RIPA lysis buffer was used to lyse cells. The lysate was centrifuged for 20 min and then precipitate was discarded. A total of 50 mL supernatant was taken for determining the concentration of protein by BCA kit (Thermo Fisher). SDS-PAGE electrophoresis was used to separate proteins. The filter membrane was polyvinylidene fluoride membrane (EMD millipore company). The primary antibodies of protein were added and placed at 4°C overnight. Then the polyvinylidene fluoride membrane was washed by PBS three times. The secondary antibody was added onto the polyvinylidene fluoride membrane at room temperature for 1h. Finally, ECL luminescent solution was used for visualization treatment. The internal reference protein was β-actin. The antibodies such as c-fos (1:1000), NR1 phosphorylation (1:1000), NR2B phosphorylation (1:1000), PKCγ phosphorylation (1:1000), ERK phosphorylation (1:1000) and β-actin (1:1000) were purchased from Abcam Company (Shanghai, China).
Double luciferase reporter gene
Starbase3.0 and Targetscan 7.2 were applied to analyze the pairing relationship among RNA sequences. pmirGLO vector were constructed with LncRNA H19 or FOS wild type (containing microRNA-29a-3p binding site) and LncRNA H19 or FOS mutant (without microRNA-29a-3p binding site), respectively. The above vectors were transfected into cells respectively, and the luciferase intensity was detected by dual luciferase reporter gene detection system (Promega).
RIP
Wild type LncRNA H19 (with microRNA-29a-3p binding site) and mutant LncRNA H19 (without microRNA-29a-3p binding site) with MS2 hairpin structure were constructed. The above vectors were transfected into the cells respectively, and after 48 h, they were immunoprecipitated by Magna RIPTM RNA binding protein immunoprecipitation kit (Milipore Company). Then RNA was purified, and the enrichment degree of microRNA-29a-3p was quantified by qPCR.
Statistical analysis
The experimental measurement data were expressed as mean ± standard error. KS test confirmed that the data conformed to normal distribution. One-way ANOVA was used to compare the data among multiple groups, and Dunnett-t was used for pair-wise comparison afterwards. The data between the two groups were compared by independent sample t-test. When P<0.5, there are statistical differences between groups.
Results
UCBMSCs improved the advanced OA pain and inflammatory
In this study, the pain behavior of rats in each group was evaluated by Von frey abosolute threshold, change of disability analysis score, poor weight bearing and body weight. Figure 1 showed that compared with the control group, the threshold of Von frey abosolute in rats treated with UCBMSCs increased, while the disability analysis score and load-bearing difference decreased. Among them, the threshold of Von frey abosolute in group 3 was the largest, while the disability analysis score and load-bearing difference were the smallest. There was no significant difference in body weight among groups. Advanced OA pain is often accompanied by severe inflammatory reaction. Therefore, serum inflammatory factors (IL-1/IL-2/IL-6/IL-10/MCP-1/TNF-α) were used to evaluate the inflammatory response of rats in each group. Figure 2 showed that compared with the control group, the levels of serum IL-1, IL-2, IL-6, MCP-1 and TNF-α in rats treated with umbilical cord blood mesenchymal stem cells decreased while IL-10 increased, among which the levels of IL-1, IL-2, IL-6, MCP-1 and TNF-α in group3 were the lowest, while IL-10 was the highest. The above results indicated that UCBMSCs can improve pain and inflammation in advanced OA.
Figure 1.
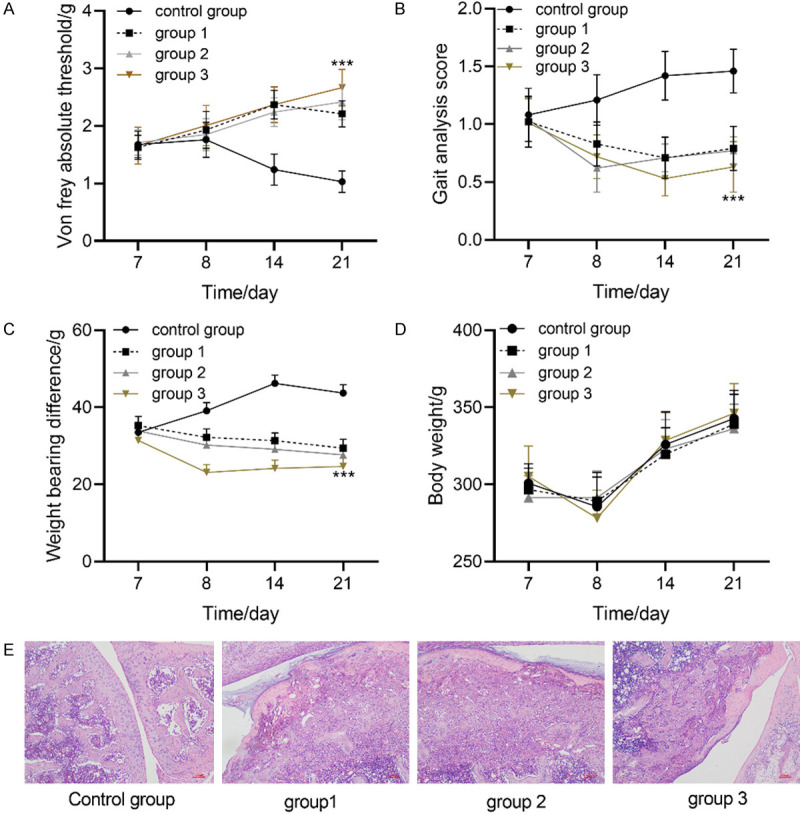
Therapeutic effect of umbilical cord blood mesenchymal stem cells on advanced OA pain. A: Changes of Von frey abosolute threshold in each group. B: Changes of disability analysis scores in each group. C: Change of load-bearing difference of each group. D: Changes of body weight in each group. *** indicates P<0.001. E: The cartilage tissue of osteoarthritis in each group was stained with HE, and the samples were taken after osteoarthritis modeling and before treatment.
Figure 2.
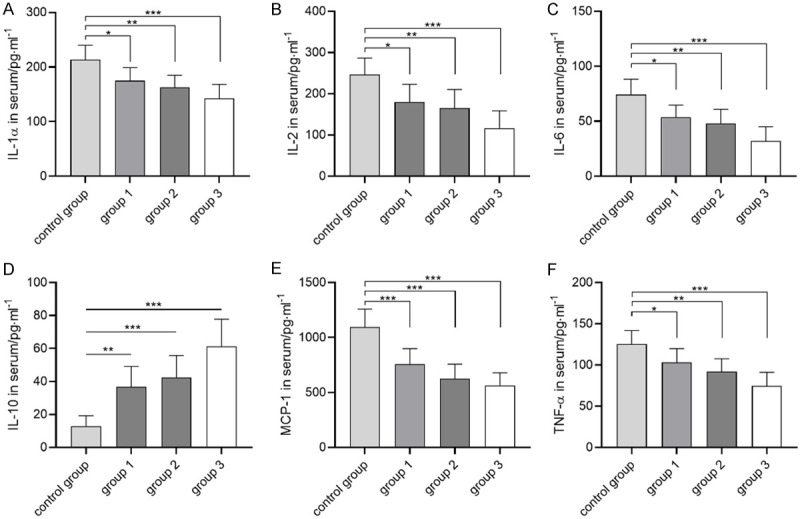
Therapeutic effect of umbilical cord blood mesenchymal stem cells on advanced OA inflammation. A: Changes of serum IL-1 in each group. B: Changes of serum IL-2 in each group. C: Changes of serum IL-6 in each group. D: Changes of serum IL-10 in each group. E: Changes of serum MCP-1 in each group. F: Changes of serum TNF-a in each group. * indicates P<0.05, ** indicates P<0.01, *** indicates P<0.001.
UCBMSCs regulated miR-29a-3p/FOS axis through exosome lncRNA H19 in advanced OA pain
Figure 3A-D showed that CD105, CD44, CD45 and CD34 were analyzed by flow cytometry, and CD105 and CD44 were positive, while CD45 and CD34 were negative. Figure 3E and 3F showed that TSG101 and CD63, the exosome surface markers isolated from mesenchymal stem cells, were analyzed by flow cytometry, and TSC101 and CD63 were positive. LncRNA H19 was highly expressed in exosomes isolated from mesenchymal stem cells. Figure 3H is an electron micrograph of exosomes. Figure 3 showed that lncRNA H19 was highly enriched in exosomes secreted by UCBMSCs.
Figure 3.
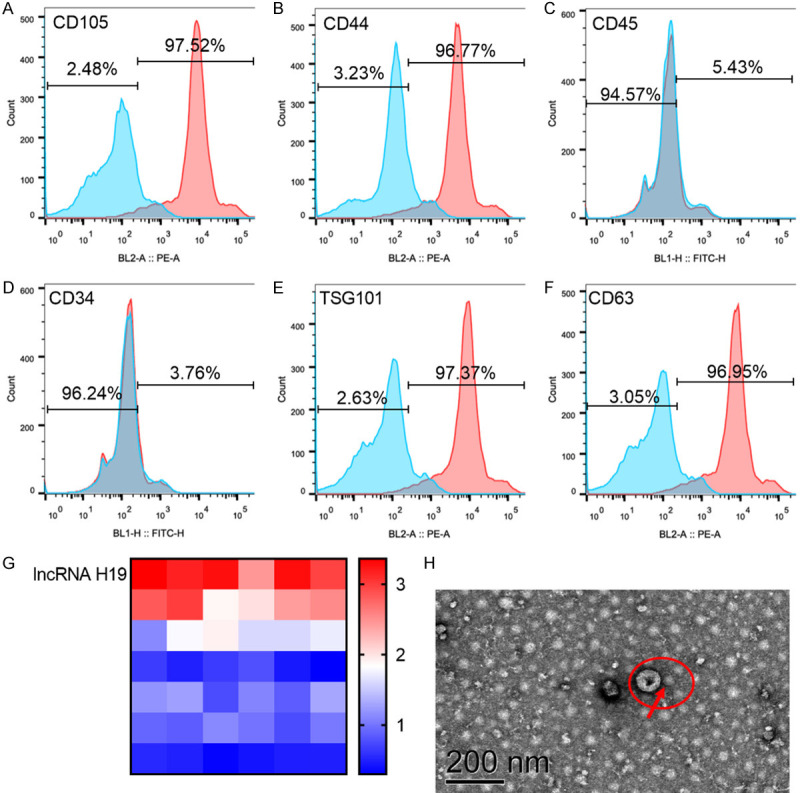
Determination of mesenchymal stem cells and exosomes. A-D: Flow cytometry was used to analyze the surface markers CD105, CD44, CD45 and CD34 of mesenchymal stem cells, in which CD105 and CD44 were positive, while CD45 and CD34 were negative. E, F: Exosome surface markers TSG101 and CD63 isolated from mesenchymal stem cells were analyzed by flow cytometry, and TSC101 and CD63 were positive. G: LncRNA H19 was highly expressed in exosomes isolated from mesenchymal stem cells. H: Electron micrograph of exosome, with the scale of 200 nm.
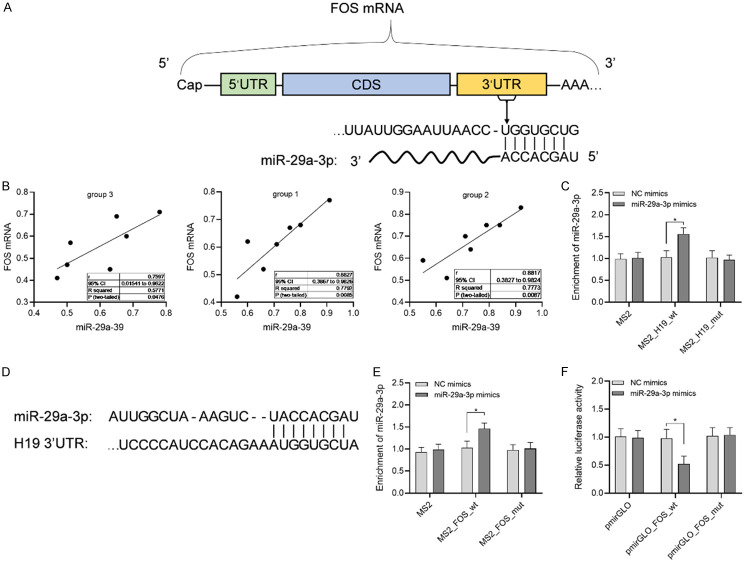
To study the role of LncRNA H19 in the regulation of advanced OA pain, this research applied starbase3.0 and Targetscan7.2 to predict the downstream targets of LncRNA H19. Figure 4 showed that microRNA-29a-3p was the downstream target gene of LncRNA H19, while FOS was the downstream target gene of microRNA-29a-3p, and microRNA-29a-3p was positively correlated with FOS.
Figure 4.
Molecular mechanism of umbilical cord blood mesenchymal stem cells in the treatment of advanced OA pain. A: Predicted binding sites of microRNA-29a-3p and FOS mRNA. B: Pearson analyzed the correlation between microRNA-29a-3p and FOS mRNA. C: Double luciferase reporter gene verified the targeting relationship between microRNA-29a-3p and FOS mRNA. D: Predicted binding site of LncRNA H19 and microRNA-29a-3p. E: microRNA-29a-3p was enriched in LncRNA H19. F: Double luciferase reporter gene verified the targeting relationship between microRNA-29a-3p and LncRNA H19.
UCBMSCs regulated central sensitization mediated by microRNA-29a-3p/FOS axis through exosome lncRNA H19 in advanced OA pain
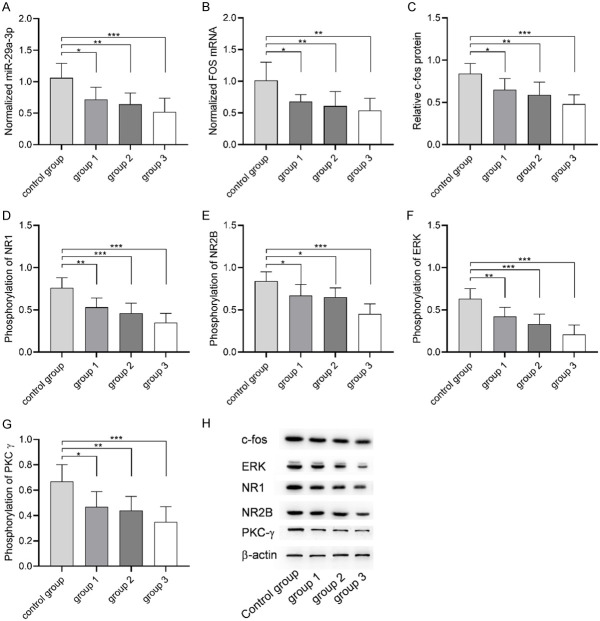
The result 2.2 suggested that microRNA-29a-3p/FOS axis regulated by exosome lncRNA H19 may be the molecular mechanism of UCBMSCs to improve the pain of advanced OA. Therefore, miR-29a-3p and c-fos protein encoded by FOS gene were detected in spinal dorsal horn of each group. Figure 5 showed that compared with the control group, microRNA-29a-3p, FOS mRNA and c-fos protein in spinal dorsal horn of each group were down-regulated after UCBMSCs treatment, and the levels of microRNA-29a-3p, FOS mRNA and c-fos protein in group 3 were the lowest. Phosphorylation of NR1, NR2B, ERK and PKC is directly related to central sensitization. To understand whether UCBMSCs regulate pain through central sensitization, the phosphorylation levels of NR1, NR2B, ERK and PKC in spinal dorsal horn of each group were also detected. Compared with the control group, the phosphorylation levels of NR1, NR2B, ERK and PKCg in spinal dorsal horn of each group treated with UCBMSCs decreased, and the phosphorylation levels of NR1, NR2B, ERK and PKCg in group 3 were the lowest. The above results indicated that UCBMSCs are closely related to central sensitization in advanced OA pain.
Figure 5.
Effects of umbilical cord blood mesenchymal stem cells on microRNA-29a-3p, FOS and central sensitization. A: microRNA-29a-3p expression in spinal dorsal horn of each group. B: FOS mRNA expression in spinal dorsal horn of each group. C: c-fos protein level in each group. D: Changes of NR1 phosphorylation level in spinal dorsal horn of each group. E: Changes of NR2B phosphorylation level in spinal dorsal horn of each group. F: Changes of ERK phosphorylation level in spinal dorsal horn of each group. G: Changes of PKCg phosphorylation level in spinal dorsal horn of each group. H: Western blot. * indicates P<0.05, ** indicates P<0.01, *** indicates P<0.001.
The activation of astrocytes is closely related to central sensitization. The result 2.2 showed that microRNA-29a-3p was the target gene of LncRNA H19, so this part will discuss the influence of LncRNA H19/microRNA-29a-3p axis on astrocytes to study the regulation of this axis on central sensitization of advanced OA pain. Figure 6 showed that exo-LncRNA H19 and microRNA-29a-3p inhibitor could down-regulate microRNA-29a-3p in astrocytes, inhibit FOS mRNA and c-fos protein, and decrease phosphorylation levels of NR1, NR2B, ERK and PKCg. The above results indicated that exo-LncRNA H19 inhibited FOS expression through microRNA-29a-3p and decreased the sensitization activity of astrocytes.
Figure 6.
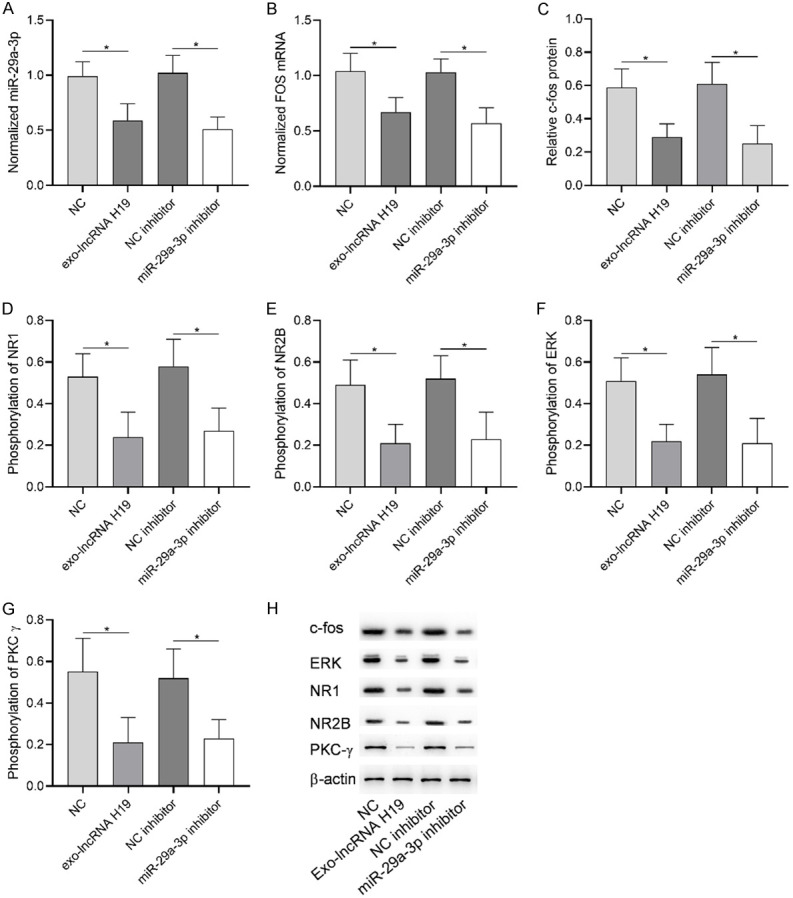
Role of microRNA-29a-3p in advanced OA. A: exo-Lncrna h19 and microRNA-29a-3p inhibitor caused microRNA-29a-3p down-regulation. B: exo-Lncrna H19 and microRNA-29a-3p inhibitor caused FOS mRNA down-regulation. C: exo-Lncrna H19 and microRNA-29a-3p inhibitor caused the down-regulation of c-fos protein. D: The down-regulation of microRNA-29a-3p resulted in the decrease of NR1 phosphorylation level. E: microRNA-29a-3p down-regulated the phosphorylation level of NR2B. F: The down-regulation of microRNA-29a-3p resulted in the decrease of ERK phosphorylation level. G: The down-regulation of microRNA-29a-3p resulted in the decrease of PKCg phosphorylation. H: Western blot. * indicates P<0.05, ** indicates P<0.01.
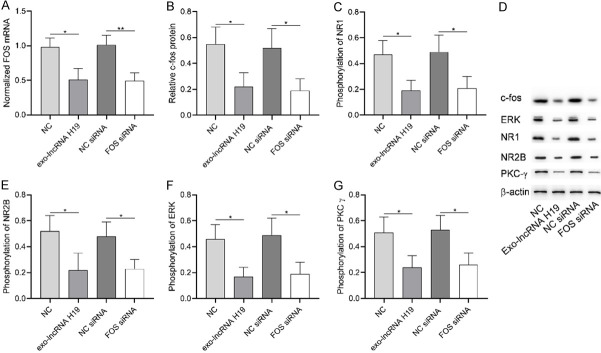
c-fos protein encoded by FOS gene was an important factor in regulating pain. Figure 7 showed that exo-LncRNA H19 could down-regulate FOS mRNA and c-fos protein in astrocytes, and decrease phosphorylation levels of NR1, NR2B, ERK and PKCg. The above results indicated that exo-LncRNA H19 reduced the sensitization activity of central nerve cells by down-regulating FOS and its encoded protein.
Figure 7.
Role of FOS in osteoarthritis pain. A: exo-Lncrna H19 and FOS siRNA caused FOS mRNA down-regulation. B: c-fos protein was down-regulated by exo-Lncrna H19 and FOS siRNA. C: FOS down-regulation caused the decrease of NR1 phosphorylation level. D: Western blot. E: FOS down-regulation caused the decrease of NR2B phosphorylation level. F: FOS down-regulation caused the decrease of ERK phosphorylation level. G: FOS down-regulation caused the decrease of PKCg phosphorylation level. * indicates P<0.05, ** indicates P<0.01.
Discussion
After intervention of UCBMSCs for rats with advanced OA pain, the pain degree has been improved obviously, and the effect of intrathecal injection is the best, which indicates that UCBMSCs can improve pain, and intrathecal injection is more beneficial to improve pain. However, the mechanism of UCBMSCs to improve pain is not clear, so this paper discussed this.
Exosomes secreted by mesenchymal stem cells contain a variety of non-coding RNA, which can be transferred to other cells through vesicles and can regulate the biological functions of cells by targeting downstream genes. Many studies [18-20] have confirmed the authenticity of this mechanism. H19 Imprinted Materially Expressed Transcript (H19) gene is located on chromosome 11, and its coding product is long-chain non-coding RNA (LncRNA) H19. LncRNA H19 is an important regulator of tumors or other diseases. At present, it has been found that LncRNA H19 participates in the occurrence and development of different diseases by adsorbing miRNA or DNA by sponge [21,22]. In this study, it was found that LncRNA H19 was enriched in exosomes secreted by umbilical cord blood mesenchymal stem cells, which may indicate that exosomes LncRNA H19 may play a role in improving pain by UCBMSCs.
The pain model of advanced osteoarthritis in rats was induced by MIA, and UCBMSCs were injected intrathecally. qPCR was used to detect the abnormally expressed genes in spinal dorsal horn. The results indicated that microRNA-29a-3p and FOS mRNA were down-regulated after stem cell therapy, suggesting that microRNA-29a-3p and FOS might play a role in pain improvement. In this paper, it was found that microRNA-29a-3p was the target gene of LncRNA H19, and can regulate the FOS expression of its target gene through the binding site by using starbase3.0 and Targetscan7.2. Pearson analysis showed that microRNA-29a-3p was positively correlated with FOS, and the results of double luciferase reporter gene showed that FOS was the downstream target gene of microRNA-29a-3p. In addition, the results of RIP and double luciferase reporter gene also showed that LncRNA H19 could sponge microRNA-29a-3p through its base sequence of 3’UTR segment.
c-fos protein was encoded by FOS gene, which was an important factor in regulating pain. In this paper, we found that c-fos in spinal dorsal horn of rats was significantly down-regulated after UCBMSCs treatment, which may be the reason for pain improvement. NR1, NR2B, PKCγ and ERK are biochemical indicators of NP, and their phosphorylation level is closely related to pain sensitization [23-26]. The phosphorylation levels of NR1, NR2B, PKCγ, ERK in spinal dorsal horn were detected. It was found that the phosphorylation levels of NR1, NR2B, PKCγ, ERK in spinal dorsal horn of rats with advanced OA pain decreased significantly after intervention of UCBMSCs, indicating that the central sensitization of rats with advanced OA pain decreased and the pain symptoms improved. In the central nervous system, the activity of astrocytes is closely related to central sensitization. The expression of microRNA-29a-3p and FOS in astrocytes was regulated, and the phosphorylation levels of NR1, NR2B, PKCγ and ERK in astrocytes were observed. The results indicated that down-regulation of microRNA-29a-3p or FOS resulted in the decrease of phosphorylation levels of NR1, NR2B, PKCγ and ERK in astrocytes.
According to the above results, it is speculated that exocrine LncRNA H19 secreted by UCBMSCs regulates the central sensitization of advanced OA pain through microRNA-29a-3p/FOS axis, thus relieving pain. Although LncRNA H19/microRNA-29a-3p/FOS axis is considered to be the mechanism of central sensitization of UCBMSCs in the treatment of advanced OA pain (even NP), this paper only discussed the model of advanced OA pain. The relationship between LncRNA H19/microRNA-29a-3p/FOS axis and sciatic nerve pain model will be further studied in future studies.
To sum up, we found that exocrine LncRNA H19 secreted by UCBMSCs regulated the central sensitization of pain in advanced osteoarthritis by targeting the microRNA-29a-3p/FOS axis of astrocytes. This finding indicated that microRNA-29a-3p/FOS axis might have potential value in pain target therapy, and it is worth further exploring its clinical application value in the follow-up study.
Acknowledgements
Supported by Sichuan Science and Technology Program (grant number, 2019YFH0073).
Disclosure of conflict of interest
None.
References
- 1.Meacham K, Shepherd A, Mohapatra DP, Haroutounian S. Neuropathic pain: central vs. peripheral mechanisms. Curr Pain Headache Rep. 2017;21:28. doi: 10.1007/s11916-017-0629-5. [DOI] [PubMed] [Google Scholar]
- 2.Cavalli E, Mammana S, Nicoletti F, Bramanti P, Mazzon E. The neuropathic pain: an overview of the current treatment and future therapeutic approaches. Int J Immunopathol Pharmacol. 2019;33:2058738419838383. doi: 10.1177/2058738419838383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gok Metin Z, Arikan Donmez A, Izgu N, Ozdemir L, Arslan IE. Aromatherapy massage for neuropathic pain and quality of life in diabetic patients. J Nurs Scholarsh. 2017;49:379–388. doi: 10.1111/jnu.12300. [DOI] [PubMed] [Google Scholar]
- 4.van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155:654–662. doi: 10.1016/j.pain.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Bouchenaki H, Begou M, Magy L, Hajj R, Demiot C. Pharmacological management of neuropathic pain. Therapie. 2019;74:633–643. doi: 10.1016/j.therap.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Kim SU, de Vellis J. Stem cell-based cell therapy in neurological diseases: a review. J Neurosci Res. 2009;87:2183–2200. doi: 10.1002/jnr.22054. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Niu R, Li W, Lin J, Stamm C, Steinhoff G, Ma N. Therapeutic potential of menstrual blood-derived endometrial stem cells in cardiac diseases. Cell Mol Life Sci. 2019;76:1681–1695. doi: 10.1007/s00018-019-03019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang R, Li W, Zhao Y, Yang F, Xu M. Clinical efficacy and safety of stem cell therapy for knee osteoarthritis: a meta-analysis. Medicine (Baltimore) 2020;99:e19434. doi: 10.1097/MD.0000000000019434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panes J, Garcia-Olmo D, Van Assche G, Colombel JF, Reinisch W, Baumgart DC, Dignass A, Nachury M, Ferrante M, Kazemi-Shirazi L, Grimaud JC, de la Portilla F, Goldin E, Richard MP, Diez MC, Tagarro I, Leselbaum A, Danese S ADMIRE CD Study Group Collaborators. Long-term efficacy and safety of stem cell therapy (Cx601) for complex perianal fistulas in patients with Crohn’s disease. Gastroenterology. 2018;154:1334–1342. e4. doi: 10.1053/j.gastro.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Yousefifard M, Nasirinezhad F, Shardi Manaheji H, Janzadeh A, Hosseini M, Keshavarz M. Human bone marrow-derived and umbilical cord-derived mesenchymal stem cells for alleviating neuropathic pain in a spinal cord injury model. Stem Cell Res Ther. 2016;7:36. doi: 10.1186/s13287-016-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickers ER, Karsten E, Flood J, Lilischkis R. A preliminary report on stem cell therapy for neuropathic pain in humans. J Pain Res. 2014;7:255–263. doi: 10.2147/JPR.S63361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narita M, Ozaki S, Narita M, Ise Y, Yajima Y, Suzuki T. Change in the expression of c-fos in the rat brain following sciatic nerve ligation. Neurosci Lett. 2003;352:231–233. doi: 10.1016/j.neulet.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 13.VanElzakker M, Fevurly RD, Breindel T, Spencer RL. Environmental novelty is associated with a selective increase in Fos expression in the output elements of the hippocampal formation and the perirhinal cortex. Learn Mem. 2008;15:899–908. doi: 10.1101/lm.1196508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragunow M, Faull R. The use of c-fos as a metabolic marker in neuronal pathway tracing. J Neurosci Methods. 1989;29:261–265. doi: 10.1016/0165-0270(89)90150-7. [DOI] [PubMed] [Google Scholar]
- 15.Harris JA. Using c-fos as a neural marker of pain. Brain Res Bull. 1998;45:1–8. doi: 10.1016/s0361-9230(97)00277-3. [DOI] [PubMed] [Google Scholar]
- 16.Takeda R, Watanabe Y, Ikeda T, Abe H, Ebihara K, Matsuo H, Nonaka H, Hashiguchi H, Nishimori T, Ishida Y. Analgesic effect of milnacipran is associated with c-Fos expression in the anterior cingulate cortex in the rat neuropathic pain model. Neurosci Res. 2009;64:380–384. doi: 10.1016/j.neures.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Kimmerling KA, Gomoll AH, Farr J, Mowry KC. Amniotic suspension allograft modulates inflammation in a rat pain model of osteoarthritis. J Orthop Res. 2020;38:1141–1149. doi: 10.1002/jor.24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 2018;17:2411–2422. doi: 10.1080/15384101.2018.1526603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Zou R, Wang Z, Wen C, Zhang F, Lin F. Exosomal KLF3-AS1 from hMSCs promoted cartilage repair and chondrocyte proliferation in osteoarthritis. Biochem J. 2018;475:3629–3638. doi: 10.1042/BCJ20180675. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging (Albany NY) 2019;11:8777–8791. doi: 10.18632/aging.102264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Luo S, Zhang J, Yuan Y, Jiang W, Zhu H, Ding X, Zhan L, Wu H, Xie Y, Song R, Pan Z, Lu Y. lncRNA H19 alleviated myocardial I/RI via suppressing miR-877-3p/Bcl-2-mediated mitochondrial apoptosis. Mol Ther Nucleic Acids. 2019;17:297–309. doi: 10.1016/j.omtn.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu YT, Tang H, Xie WQ, Yao H, Gu WT, Zheng YZ, Shang HB, Wang Y, Wei YX, Wu ZR, Wu ZB. Exosome-transmitted lncRNA H19 inhibits the growth of pituitary adenoma. J Clin Endocrinol Metab. 2019;104:6345–6356. doi: 10.1210/jc.2019-00536. [DOI] [PubMed] [Google Scholar]
- 23.Meng X, Zhang Y, Lao L, Saito R, Li A, Backman CM, Berman BM, Ren K, Wei PK, Zhang RX. Spinal interleukin-17 promotes thermal hyperalgesia and NMDA NR1 phosphorylation in an inflammatory pain rat model. Pain. 2013;154:294–305. doi: 10.1016/j.pain.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Yan H, Li X, Liu J, Cao S, Huang B, Huang D, Wu L. Inhibition of connexin 43 and phosphorylated NR2B in spinal astrocytes attenuates bone cancer pain in mice. Front Cell Neurosci. 2018;12:129. doi: 10.3389/fncel.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao YQ, Yin JB, Wu HH, Ding T, Wang Y, Liang JC, Guo XJ, Tang K, Chen DS, Chen GZ. Contribution of spinal PKCgamma expression to short- and long-lasting pain behaviors in formalin-induced inflamed mice. Pain Physician. 2018;21:E555–E564. [PubMed] [Google Scholar]
- 26.Maruta T, Nemoto T, Hidaka K, Koshida T, Shirasaka T, Yanagita T, Takeya R, Tsuneyoshi I. Upregulation of ERK phosphorylation in rat dorsal root ganglion neurons contributes to oxaliplatin-induced chronic neuropathic pain. PLoS One. 2019;14:e0225586. doi: 10.1371/journal.pone.0225586. [DOI] [PMC free article] [PubMed] [Google Scholar]