Abstract
Recent studies have shown the involvement of exosomes in intercellular communication during tumor progression. Circular RNAs (circRNAs) can be packaged into exosomes for extracellular communication, however, the possible effects of exosomal circRNAs in epithelial ovarian cancer (EOC) cells with high metastatic potential have been rarely studied. In this study, we identified exosomal circRNA051239 from high-metastatic ovarian cancer SKOV3.ip cells and subsequently analyzed circRNA051239 levels in both EOC tissues and exosomes derived from plasma and cells by qRT-PCR. A variety of in vitro assays were employed to observe the effects of exosomal circRNA051239 derived from high-metastatic ovarian cancer SKOV3.ip cells on low-metastatic ovarian cancer SKOV3 cells. Bioinformatics analysis and luciferase activity assays were further utilized to confirm the relationship between circRNA051239, miR-509-5p and PRSS3. As a result, circRNA051239 expression was increased in tissues and plasma exosomes from EOC patients. Moreover, si-circRNA051239-Exo (exosomes derived from circRNA051239 knockdown SKOV3.ip cells) inhibited the proliferation, migration as well as invasion of SKOV3 cells. Mechanistically, circRNA051239 functioned as a competitive endogenous RNA (ceRNA) by sponging miR-509-5p to facilitate PRSS3 expression. Exosomal circRNA051239 derived from high-metastatic ovarian cancer SKOV3.ip cells promoted the progression of low-metastatic ovarian cancer SKOV3 cells. Collectively, these outcomes implicated that higher metastatic EOC cells can confer this potential to lower metastatic potential via exosomal circRNA051239, causing enhanced proliferative, migratory and invasive capacities in recipient cells.
Keywords: Epithelial ovarian cancer, circRNA051239, exosomes, metastasis
Introduction
Epithelial ovarian cancer (EOC) is the most prevalent malignancies of the reproductive tract in females, ranking the fifth cancer-associated mortality in females, with a 5-year survival rate of approximately 47% [1]. EOC patients are usually treated by first-line treatment, including optimal cytoreductive surgery and adjuvant chemotherapy [2,3]. However, traditional treatment generally causes chemotherapeutic resistance, ultimately leading to tumor recurrence and even death. Therefore, it is urgent to investigate and to identify novel molecular mechanisms of ovarian cancer tumorigenesis and progression to better control EOC.
Circular RNAs (circRNAs), unlike the better-known linear RNA, are classified as novel endogenous noncoding RNAs, which form covalently closed continuous loops without 5’ caps or 3’ tails [4]. As a novel type of RNA, circRNAs are single-stranded transcripts originating as exons, introns, or intergenic regions with endogenous expression [5,6]. The rapidly developed bioinformatics and high-throughput sequencing have revealed a variety of circRNAs and their properties. Growing evidence implicates the involvement of circRNAs in the pathogenesis cardiovascular disorders [7], neurological diseases [8], as well as tumors [9]. Moreover, circRNAs have been uncovered to affect tumorigenesis as well as metastasis [10]. Current data indicate that circRNAs serve as a ceRNA to regulate tumor proliferation, apoptosis, and angiogenesis [11]. circRNAs have been recently revealed to interact with miRNAs by stably complementary binding and serving as efficient miRNA sponges, thereby posttranscriptional modulating the expression of downstream target genes [12]. Despite the enriched and stable expression of circRNA in exosomes as indicated by accumulative studies [13], it remains undetermined of functions and mechanisms of circRNA in EOC.
Exosomes are membrane vesicles released from various normal and tumor cells with diameters of 30-100 nm [14]. The distribution of exosomes is found in almost all kinds of body fluids, including amniotic fluid, urine, blood, and malignant ascites [15]. Accumulative researches have shown that exosomes are critically involved in intercellular communication by delivering a variety of proteins, lipids, circRNAs, and miRNAs to recipient cells [16]. Exosomes have also been shown to be involved in various disorders, including liver and neurodegenerative diseases [17]. Emerging evidence has demonstrated that tumor-derived exosomes play essential roles in tumor angiogenesis, extracellular matrix remodeling, tumor migration, and metastasis [18,19]. Various proteins, miRNAs or lncRNAs in exosomes are reported to have associations with cancer progression [20]. In addition, the enriched and stable circRNAs in exosomes are considered as promising biomarkers in various cancers [13]. Finally, circRNAs may regulate cell communication through exosomes in some cases by transferring miRNA between different cells [21]. Previous study suggested that exosomes derived from cells with high metastatic potential can promotes invasive growth in low metastatic cancer cells [22]. Nevertheless, the role of exosomal circRNAs in EOC cells with high metastatic potential is still undefined.
Serine protease 3 (PRSS3), a member of the trypsin family of serine proteases, is mainly synthesized in pancreatic acinar cells. PRSS3 has been strongly implicated in tumor growth and metastatic progression of cancers [23]. Ectopic expression of PRSS3 transcripts has also been reported in several epithelial cancer types [24,25], including ovarian cancer [26]. Thus, investigating its molecular mechanism of action might shed light on the development of novel candidate drug targets in cancer.
In the present research, we identified circRNA051239 in tumor exosomes and further focused on the functions as well as underlying mechanisms in EOC cells. Consequently, we detected significantly enhanced expression of circRNA051239 in both ovarian cancer tissues and exosomes from ovarian cancer plasma. Moreover, circRNA051239 was contained in exosomes released by high-metastatic ovarian cancer SKOV3.ip cells, where it promoted proliferation and stimulated migration as well as invasion in low-metastatic ovarian cancer SKOV3 cells. Further mechanistic studies demonstrated that circRNA051239 influenced PRSS3 expression by sponging miR-509-5p. In conclusion, our studies elucidated the mechanism underlying the promoting role of exosomal circRNA051239 on tumor progression in EOC and provided potential therapeutic targets for human EOC.
Materials and methods
Cell culture and transfection
EOC cell lines SKOV3.ip, SKOV3, A2780, CAOV3 and OVCAR3 were obtained commercially from ATCC, which were cultured in RPMI-1640 medium or DMEM containing 10% FBS (Gibco, MA, USA) and 1% penicillin-streptomycin solution at 37°C in a humidified incubator containing 5% CO2. miR-509-5p mimic, miR-509-5p inhibitor, miRNA negative control mimic (NC mimic), miRNA negative control inhibitor (NC inhibitor), circRNA051239 siRNA and control siRNA were synthesized by RiboBio (Guangzhou, China). Lipofectamine 3000 (Invitrogen, USA) was utilized for transfection accordingly, followed by cell collection at 24 h after transfection.
Human tissue
Thirty 30 EOC patients along 10 healthy volunteers without any malignancy were enrolled in this study from Peking University People’s Hospital (PUPH) from January 2018 to January 2019. All EOC patients were not subject to any preoperative therapy. After tumor excision, fresh tissue was immediately placed into liquid nitrogen, and further stored at -80°C. In terms of plasma exosome isolation, 5 ml of total blood was extracted from every patient, followed by centrifugation at 3000 rpm for 20 min and storage of plasma supernatant at -80°C for further assay. The study gained approval from Ethics Committee of PUPH, and written informed consent was signed before specimen collection.
Isolation of exosomes from medium and plasma
Cell culture was centrifuged for 5 min at 500×g for discarding cells from the samples. The collected supernatant was transferred to a new polycarbonate tube, followed by centrifugation at 2000×g for 10 min to discard cells as well as other debris. Afterwards, the supernatant was then centrifuged at 10,000×g for 30 min for eliminating shed microvesicles, followed by supernatant collection and filtering with a 0.22-μm membrane filter (Merck Millipore). After centrifugation at 100,000×g for 70 min at 4°C, exosomes were collected and resuspended in 1×PBS, followed by storage at -80°C. The isolation of exosomes from human plasma specimens was performed using ExoQuick solution accordingly (System Biosciences, SBI, CA, USA). In brief, after centrifugation of plasma specimens at 3000×g for 15 min, 63 μl of ExoQuick Exosome Precipitation Solution was mixed with 250 μl of plasma sample. After incubation at 4°C overnight and centrifugation at 1500×g for 30 min at 4°C, PBS was used to resuspend exosome.
Transmission electron microscopy (TEM)
Exosome suspensions were fixed with 4% paraformaldehyde (PFA) as well as 5% glutaraldehyde in 0.1 M PBS (pH 7.4) at 4°C for TEM assay. Briefly, exosome sample was dropped on a carbon film-coated transmission electron microscope grid, followed by incubation at room temperature (RT) for 5 min and subsequent observation by TEM (JEOL, Tokyo, Japan) at 100 KV.
PKH26 labeling of exosomes and exosome uptake into recipient cells
Exosomes isolated from culture medium of SKOV3.ip cells were labeled using a PKH26 red fluorescent labeling kit (Umibio, China) in line with standard protocols. To examine the uptake of exosomes into recipient SKOV3 cells, exosomes were incubated with PKH26 dye for 10 min, followed by incubation with SKOV3 cells at 37°C for 24 h. After rinsing with PBS for three times and fixing with 4% PFA at RT for 30 min, fixed cells were rinsed with PBS for three times, followed by nuclear staining by DAPI for 10 min (Invitrogen). Confocal laser scanning microscopy was applied to detect the cellular uptake and internalization of exosomes.
Microarray analysis
After purifying exosomes from SKOV3.ip and SKOV3 cells, total RNA was isolated by TRIzol LS (Thermo, USA) in accordance with the standard instruction. Specimen preparation as well as microarray hybridization were conducted based on standard protocols. In brief, after digestion of total RNA by RNase R, random priming was used to amplify and transcribe the enriched circRNAs into fluorescent cRNA (Arraystar Super RNA Labeling Kit), followed by hybridization of labeled cRNAs onto Arraystar Human circRNA Array V2 (8x15K, Arraystar). Agilent Scanner G2505C was used for array scanning after washing the slides, and Agilent Feature Extraction software was subsequently utilized for analyzing the array images.
Bioinformatics analysis
GO and KEGG pathway analysis was utilized to predict the functions of differentially expressed (DE) circRNAs between two groups, followed by prediction of circRNA/microRNA targets using miRanda and TargetScan software packages.
Luciferase reporter assay
The 3’-UTR of circRNA051239 and PRSS3 containing miR-509-5p binding sites were subjected to PCR amplification, followed by cloning into the pmirGLO luciferase vector (Promega, USA) to generate the wild-type 3’-UTR luciferase reporters (circRNA051239 WT or PRSS3 WT). The mutant 3’-UTR luciferase reporters (circRNA051239 Mut or PRSS3 Mut) with mutations in several key bases were prepared by the same approach. For the luciferase reporter assay, after inoculating in a 96-well plate, EOC cells were cotransfected with WT or mutant 3’-UTR luciferase reporters and miR-509-5p mimics or negative controls using Lipofectamine 3000. Cells were collected after 48 h, followed by luciferase activity assessment by Luciferase Reporter System (Promega). Each experiment was performed three times.
Nuclear-cytoplasmic fractionation
The cytoplasm and nucleus of SKOV3 cells were isolated with Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). The analysis of extracted RNA from the cytoplasmic and nuclear fractions was conducted by RT-qPCR. GAPDH and U6 were utilized as an internal control for cytoplasm and nucleus, respectively.
Fluorescence in situ hybridization analysis (FISH)
Cy5-labeled circRNA051239 (5’-TGTCCATTAGCTCCTTGGAGGTACAGGACAGC-3’) (BersinBio, Guangzhou, China) was utilized for investigating circRNA051239 localization in ovarian cancer cells. FISH was conducted by a Fluorescence in Situ Hybridization Kit (Geneseed, Guangzhou, China) based on relevant protocol. Cells were fixed with 4% PFA for 20 min, rinsed with PBS as well as permeabilized with 0.5% Triton X-100 for 10 min. In situ hybridization was performed overnight at 37°C with a FISH probe in the hybridization solution. Subsequently, the slide was washed with SSC buffer. Cell nuclei were stained with DAPI (BOSTER, China). ZEISS LSM800 Confocal Microscope (Carl Zeiss AG, Germany) was employed to capture images.
Cell counting kit-8 assay (CCK-8)
Cell proliferation was determined by CCK-8 assay (Dojindo, Kumamoto, Japan). First, 3000 cells were inoculated into 96-well plates, added with 10 μL of CCK-8 reagent after 24 h and subsequent culture at 37°C. At different time points, the OD level was determined using a microplate reader at 450 nm.
5-EdU incorporation assay
An EdU assay kit (RiboBio) was utilized for cell proliferation. First, 10,000 treated EOC cells were inoculated into 96-well plates overnight. Edu solution (25 μM) was added and incubated for 24 h. EOC cells were subsequently fixed with 4% formalin at RT for 2 h, permeabilized by 0.5% Triton X-100 for 10 min. After incubation with Apollo staining solution (RiboBio) for 20 min and subsequent incubation in DAPI (200 μL), cells were imaged with a fluorescence microscope.
Wound-healing assay
Cells were inoculated into 6-well plates in complete medium and cultured at 37°C. After reaching 95% confluence, cells were scraped with a 100-mL sterile pipette tip and rinsed with PBS twice for discarding the suspended cells. Images were photographed in three defined fields at 0 h and 24 h.
Transwell migration and invasion assay
Invasion assay was examined using 24-well Matrigel-coated chambers (BD Biosciences, CA, USA), and migration assay was performed similarly using Matrigel-uncoated chambers. A total of 5×103 cells in serum-free RPMI-1640 were seeded into the top chamber. Exosomes were added to the bottom chamber, with complete RPMI-1640 medium as control. After incubating for 24 h, the migrated or invaded cells transferred to the lower chambers were fixed with 4% PFA and were subjected crystal violet, followed by counting and photography of cells under an inverted microscope (Olympus, Japan). All assays were performed in three replicates.
RNA extraction as well as quantitative RT-PCR (qPCR)
TRIzol reagent (Invitrogen) was utilized to extract total RNA from cells as well as tissues, and exosomal RNAs from cells and plasma were isolated utilizing TRIzol LS (Thermo, USA). First-strand cDNA was synthesized by PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). Reverse transcription and qPCR were conducted by RNA-direct SYBR Realtime PCR Master Mix (Toyobo, Japan). GAPDH or U6 were taken as an internal control for circRNAs and miRNAs. 2-ΔΔCt method was utilized for calculating relative expression. All specimens were analyzed in triplicate.
Western blot (WB)
Cells and tissues were lysed in RIPA lysis buffer (Beyotime, Beijing, China) containing protease inhibitor cocktail (Roche, CA, USA). Total protein was separated by 10% SDS-PAGE, transferred to PVDF membranes (Millipore) and blocked with 5% BSA at RT for 1 h. The membranes were reacted with anti-PRSS3 (1:1,000, Abcam), anti-GAPDH, anti-CD63 and anti-CD81 (dilution 1:1000, Abcam) at 4°C overnight. After reacting with corresponding secondary antibodies at RT for 1 h, the membranes were visualized with an ECL kit (Millipore).
Statistical analysis
SPSS 17.0 software package (Chicago, IL, USA) and GraphPad Prism version 6.0 (GraphPad Software) were employed for statistical analysis. Student’s t test, one-way ANOVA as well as chi-square test were used for comparison. Data were shown as mean ± standard deviation (SD). P values < 0.05 implicated statistical significance.
Results
The identification and features of tumor-derived exosomal circRNA051239
In previous study, SKOV3.ip cells were much more tumorigenic and invasive than SKOV3 cells. Thus, we assessed the circRNA expression profile in exosomes from SKOV3.ip and SKOV3 cells using Arraystar Human circular RNA Microarrays. The entire screening workflow was displayed in Figure 1A. The morphology as well as size of extracted exosomes were observed by TEM. Most exosomes were within 100 nm in diameter, consistent with the typical exosomal size (Figure 1B). Moreover, exosomal markers (CD81 and CD63) were also clearly detectable in both groups (Figure 1C).
Figure 1.
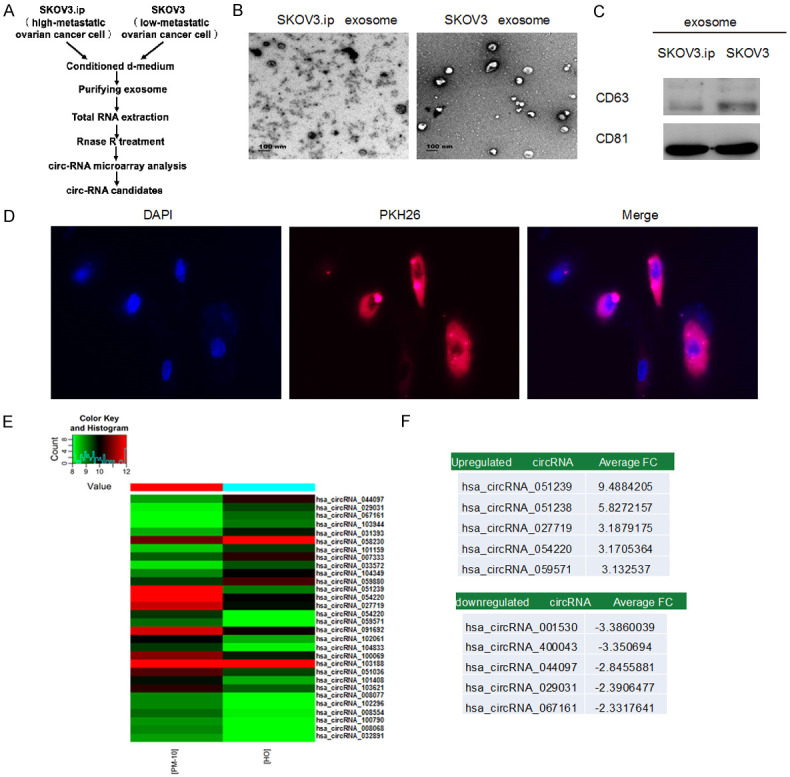
The identification and features of circRNA in tumor exosomes. A. Schematic screening workflow of circRNA candidates. B. TEM image of exosomes isolated from SKOV3.ip (high-metastatic ovarian cancer cell) and SKOV3 (low-metastatic ovarian cancer cell) cell lines. C. Exosomes from culture media were examined by WB (CD63 and CD81). D. SKOV3 cells were treated with 5 μg of PKH26-labeled exosomes (red) derived from SKOV3.ip cells. The nuclei were visualized by DAPI staining. Red: PKH26; Blue: DAPI (nuclei). Confocal microscopy was used to image fluorescent cells. E. Heat map of differentially expressed circRNAs. ‘Red’ and ‘green’ indicates high and low relative expression, respectively. F. The top five upregulated and down-regulated circRNAs.
To investigate the uptake and internalization of ovarian cancer-derived exosomes into recipient cells, SKOV3 cells (low-metastatic potential) were treated with PKH26 red fluorescence-labeled exosomes derived from parental SKOV3.ip (high-metastatic potential). As illustrated in Figure 1D, exosomes from SKOV3.ip cells were significantly absorbed by SKOV3 cells, revealing that exosomes derived from SKOV3.ip cells were more susceptible to be absorbed by SKOV3 cells.
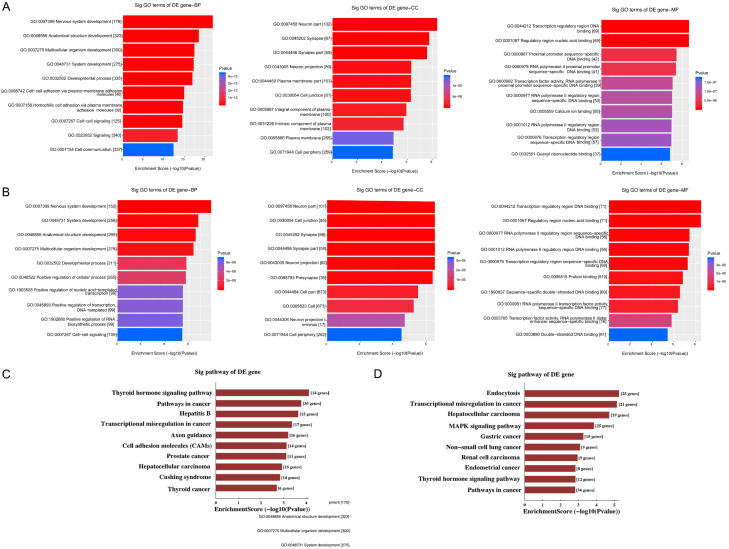
There was obvious dysregulation of exosomal circRNAs in ovarian cancer (Figure 1E). Ten significantly DE circRNAs (five up-regulated and five down-regulated) were selected in ovarian cancer group based on raw microarray signals as well as fold changes (Figure 1F). GO and KEGG pathway analysis demonstrated that these DE circRNAs were related to multiple signaling pathways as well as biological functions (Figure 2A-D).
Figure 2.
GO and KEGG analysis demonstrated that the predicted target genes of these circRNAs were involved in multiple biological processes and signaling pathways. Functional annotation of target genes upregulated (A) and downregulated (B) by circRNAs (GO analysis). Pathways corresponding to the upregulated (C) and downregulated (D) target genes.
qPCR was utilized to assess the levels of 10 circular RNAs in EOC tissues, showing significant up-regulation of circRNA051239 in ovarian cancer tissues than normal controls (Figure 3A and 3B). circRNA051239 was further investigated by first analyzing the levels of exosomal circRNA051239 in plasma from 10 normal controls and 30 EOC patients. As a result, the level of circRNA051239 was significantly increased in plasma exosomes from ovarian cancer patients (Figure 3C).
Figure 3.
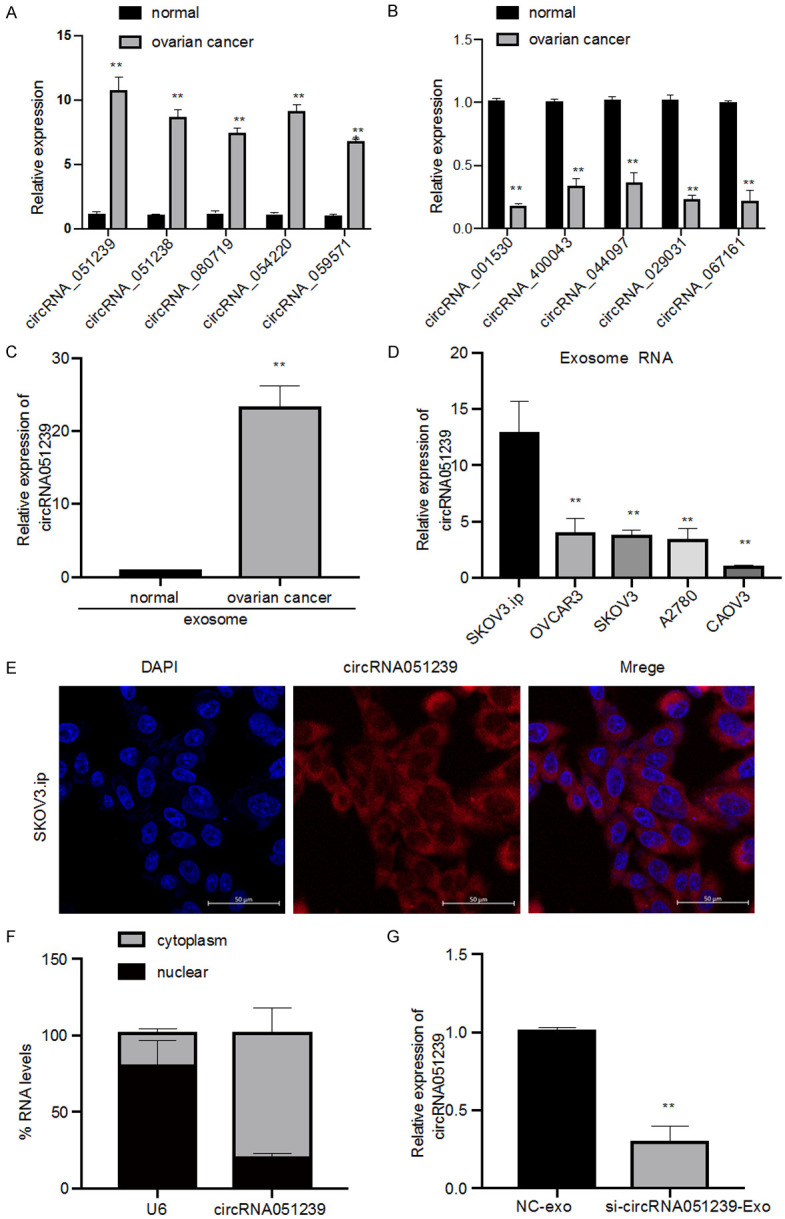
Overexpression of circRNA051239 in EOC tissue and cells. A. Five circRNAs, circRNA051239, circRNA051238, circRNA080719, circRNA054220, and circRNA059571, were validated in EOC tissues by qRT-PCR. B. Five circRNAs, circRNA001530, circRNA400043, circRNA044097, circRNA029031, and circRNA067161, were validated in EOC tissues by qRT-PCR. C. The expression levels of exosomal circRNA051239 in plasma samples from 10 pairs of normal controls and 30 EOC patients by qRT-PCR. D. The relative expression levels of the exosomal circRNA051239 in EOC cell lines (SKOV3.ip, SKOV3, CAOV3, OVCAR3 and A2780) by qRT-PCR. E. Localization of circRNA051239 in SKOV3.ip cells by FISH. F. The subcellular location of circRNA051239. G. The expression of circRNA051239 was examined in SKOV3 cells after treatment with NC-Exo and si-circRNA051239-Exo derived from SKOV3.ip cells by qRT-PCR. Results were shown as mean ± SD. *P < 0.05, **P < 0.01.
To investigate the roles of circRNA051239 in EOC, circRNA051239 expression was determined in a set of EOC cells. Increased levels of circRNA051239 were detected in SKOV3.ip, SKOV3, OVCAR3, A2780, and CAOV3 cells (Figure 3D). Among them, the highest expression of circRNA051239 was observed in SKOV3.ip cells. FISH as well as nuclear-cytoplasmic fractionation analysis were utilized to examine subcellular localization of circRNA051239, showing that cytoplasm was the major location of circRNA051239 (Figure 3E and 3F). Collectively, the above outcomes implicated that circRNA051239 can function as miRNA sponges at the posttranscriptional level.
Exosomal circRNA051239 regulates proliferation, migration as well as invasion in ECO cells
We predicted that exosomal circRNA051239 derived from SKOV3.ip cells was able to alter the biological functions of SKOV3 cells. To establish functions of exosomal circRNA051239, we isolated exosomes from SKOV3.ip cells transfected with circRNA051239 siRNA or NC siRNA, namely, si-circRNA051239-Exo or NC-Exo. The suppression efficiency of si-circRNA051239-Exo in SKOV3 cells was confirmed by qRT-PCR, revealing that si-circRNA051239-Exo suppressed the expression levels of circRNA051239 in SKOV3 cells (Figure 3G) and decreased SKOV3 cell proliferation (Edu assay and CCK-8 assay) in comparison with NC exosomes group (Figure 4A-C). Furthermore, si-circRNA051239-Exo inhibited the invasive and migratory abilities of SKOV3 cells (Figure 5A-F). These findings implicated that exosomal circRNA051239 could modulate proliferation, migration as well as invasion of EOC cells.
Figure 4.
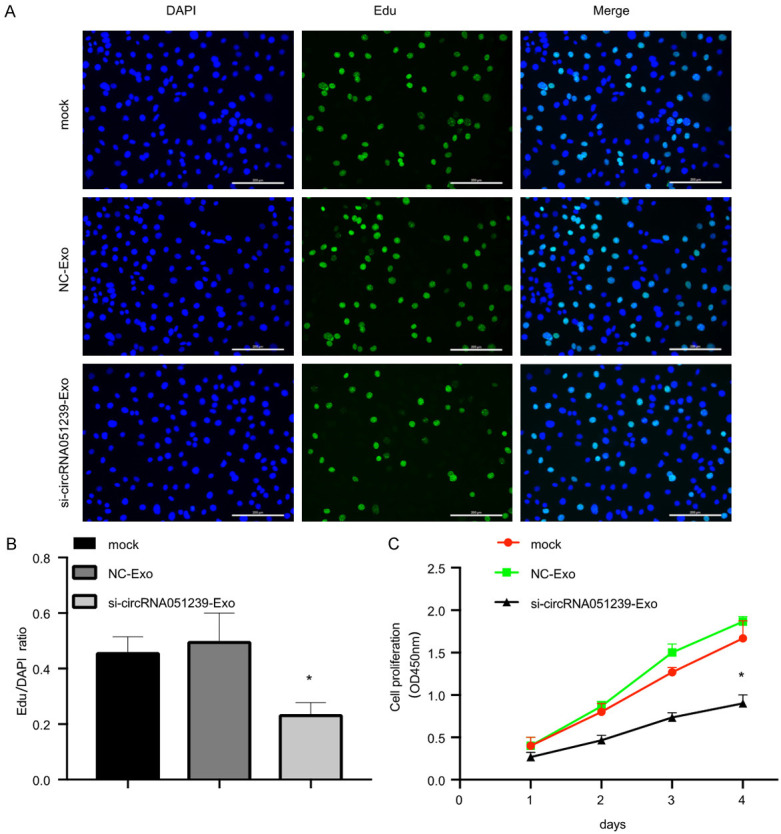
Exosomes derived from circRNA051239 knockdown SKOV3.ip cells suppressed SKOV3 cell proliferation in vitro. A, B. Cell proliferation of SKOV3 cells was observed through EdU assay after treatment with exosomes derived from circRNA051239 knockdown SKOV3.ip cells. C. The proliferation rate of SKOV3 cells was assessed after treatment with exosomes derived from circRNA051239 knockdown SKOV3.ip cells by CCK-8. Data were shown as mean ± SD. *P < 0.05, **P < 0.01.
Figure 5.
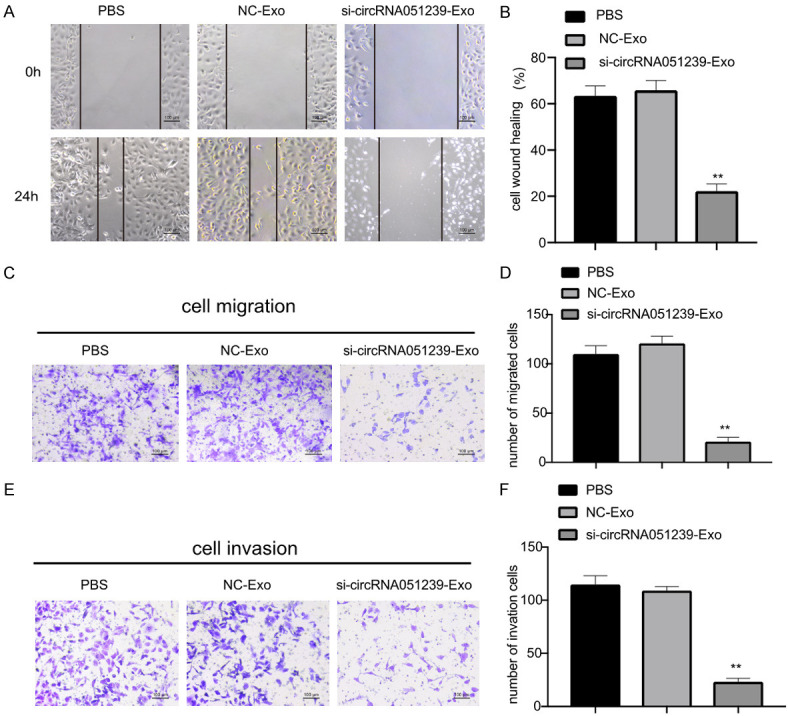
Exosomes derived from circRNA051239 knockdown SKOV3.ip cells suppressed SKOV3 cell migration as well as invasion. A, B. Representative images of the wound-healing assay in SKOV3 cells after treatment with NC-Exo and si-circRNA051239-Exo derived from SKOV3.ip cells at 0 h and 24 h, respectively. C, D. Transwell migration assay was employed for SKOV3 cell migration following treatment with NC-Exo and si-circRNA051239-Exo derived from SKOV3.ip cells. E, F. Invasion of SKOV3 cells after treatment with NC-Exo and si-circRNA051239-Exo derived from SKOV3.ip cells was detected by Transwell invasion assay. Data were shown as mean ± SD. *P < 0.05, **P < 0.01.
Exosomal circRNA051239 regulates PRSS3 expression via miR-509-5p
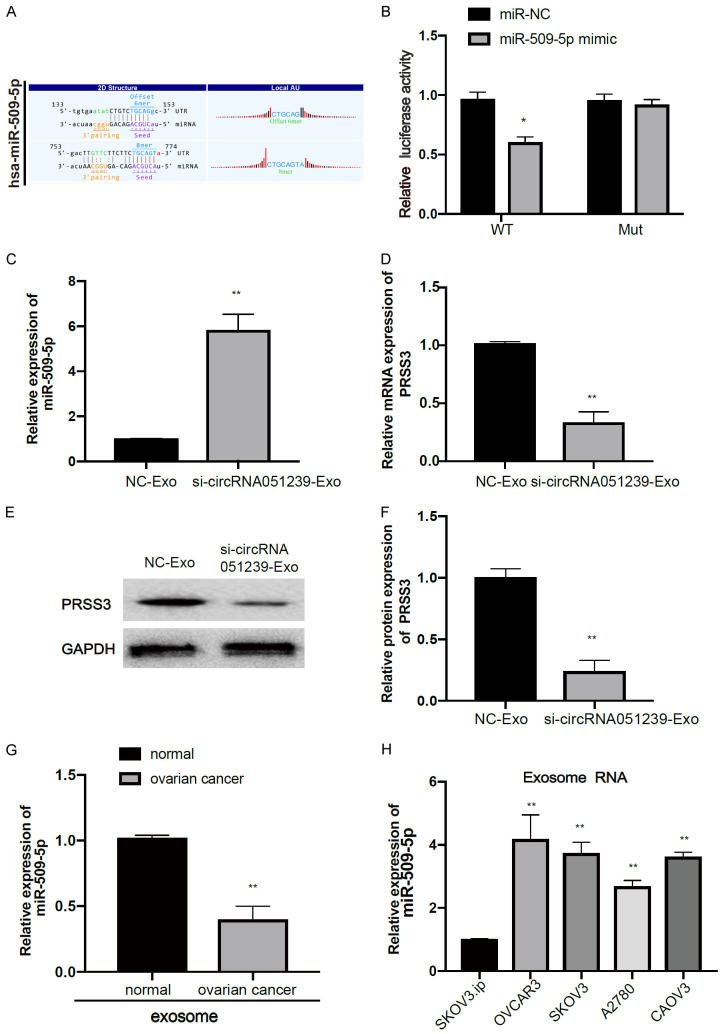
Bioinformatics analysis revealed complementary binding sites of miR-509-5p with circRNA051239 (Figure 6A). To further validate whether miR-509-5p could directly bind with circRNA051239, luciferase reporter assay was performed with WT or mutant circRNA051239 3’-UTR luciferase vector (circRNA051239 WT or circRNA051239 Mut) in SKOV3 cells. Consequently, miR-509-5p mimic treatment significantly attenuated the luciferase activity of WT circRNA051239 (circRNA051239-WT), rather than that of mutant circRNA051239 (circRNA051239-Mut), in both SKOV3 cell lines (Figure 6B).
Figure 6.
CircRNA051239 targeted miR-509-5p as a miRNA sponge. A. Binding sites of miR-509-5p in 3’-UTR of circRNA051239. B. Luciferase reporter assays were performed following transfection with miR-NC or miR-509-5p. C, D. The expression of miR-509-5p and PRSS3 was examined in SKOV3 cells after treatment with NC-Exo and si-circRNA051239-Exo derived from SKOV3.ip cells by qRT-PCR. E, F. Detection of PRSS3 protein expression in SKOV3 cells following treatment with NC-Exo and si-circRNA051239-Exo derived from SKOV3.ip cells by WB. G. qRT-PCR analysis was performed to examine the expression of exosomal miR-509-5p in EOC tissues and normal tissues was examined by qRT-PCR. H. Exosomal miR-509-5p expression was detected in five EOC cell lines (SKOV3.ip, SKOV3, CAOV3, OVCAR3 and A2780). Data were shown as mean ± SD. *P < 0.05, **P < 0.01.
For further validation on the interaction of miR-509-5p with circRNA051239, we found that si-circRNA051239-exo could significantly increase miR-509-5p expression in SKOV3 cells (Figure 6C). si-circRNA051239-Exo/NC-Exo was added to SKOV3 cells to detect PRSS3 expression. Interestingly, si-circRNA051239-Exo also inhibited PRSS3 expression in SKOV3 cells by qPCR and WB (Figure 6D-F). Moreover, miR-509-5p expression was obviously attenuated in plasma exosomes in EOC patients as well as SKOV3.ip cells (Figure 6G and 6H). Thus, circRNA051239 might function as a sponge against miR-509-5p in EOC cells.
Moreover, we exploited the possible target genes of miR-509-5p in EOC. Consequently, PRSS3 was predicted as a direct target of miR-509-5p using TargetScan tool (Figure 7A), validated by luciferase reporter assay. Luciferase activity was markedly attenuated in SKOV3 cells when cotransfected with miR-509-5p mimics and WT PRSS3 vectors (PRSS3 WT) but was unchanged when cotransfected with miR-509-5p mimics and mutant PRSS3 vectors (PRSS3 Mut) (Figure 7B). In addition, we detected the mRNA levels of miR-509-5p and PRSS3 in SKOV3 cells with miR-509-5p mimics-Exo and miR-509-5p Inhibitor-Exo derived from SKOV3.ip cells. qRT-PCR revealed that miR-509-5p expression was elevated in SKOV3 cells after treatment with mimics-Exo and was decreased in SKOV3 cells following miR-509-5p inhibitor treatment (Figure 7C). Additionally, the expression of PRSS3 was decreased in SKOV3 cells following mimics-Exo treatment, but was increased following miR-509-5p inhibitor treatment (Figure 7D). WB data showed that miR-509-5p mimics-Exo significantly decreased PRSS3 protein expression in SKOV3 cells (Figure 7E and 7F), further implicating that PRSS3 was a direct target of miR-509-5p in EOC cells.
Figure 7.
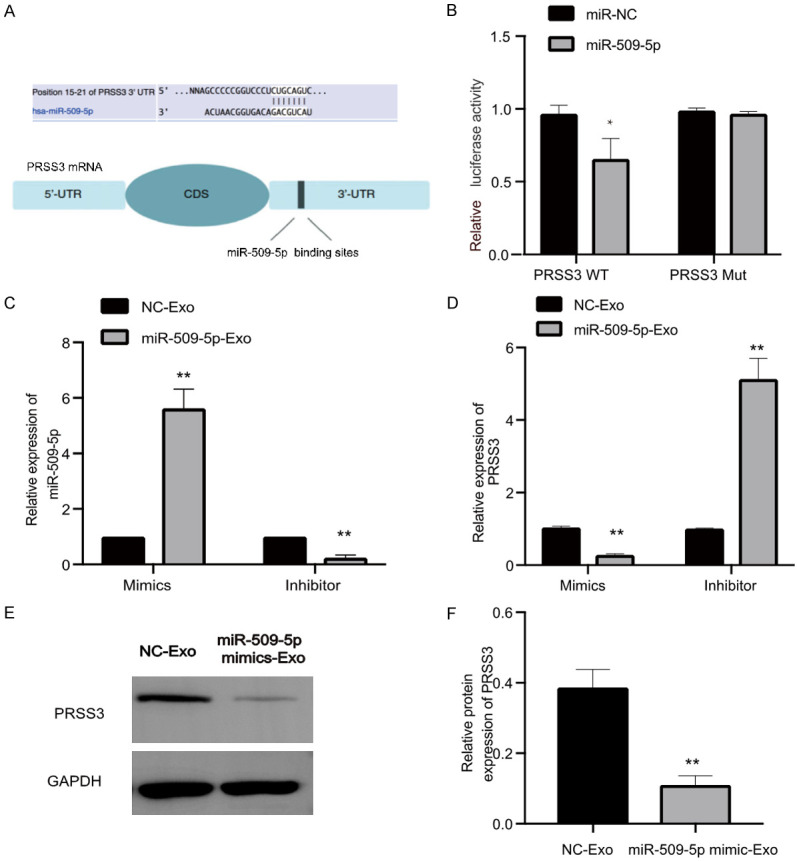
PRSS3 was a target of miR-509-5p. A. Predicted miR-509-5p-binding sites in PRSS3 3’-UTR. B. Relative luciferase activity of SKOV3 cells cotransfected with either WT or Mut PRSS3 3’-UTR reporter along with either miR-509-5p mimic or miR-NC (control). C. Quantification of miR-509-5p expression in SKOV3 cells after treatment with miR-509-5p mimic-Exo or miR-509-5p inhibitor-Exo derived from SKOV3.ip cells by qRT-PCR. D. Relative expression of PRSS3 in SKOV3 cells after treatment of miR-509-5p mimic-Exo or miR-509-5p inhibitor-Exo derived from SKOV3.ip cells by qRT-PCR analysis. E, F. Detection of PRSS3 protein expression in SKOV3 cells following treatment with miR-509-5p mimic-Exo or miR-NC-Exo by WB. Data were hown as mean ± SD. *P < 0.05, **P < 0.01.
Discussion
Ovarian cancer is a major gynecologic cancer worldwide, with complex and multifactorial etiology. Here, microarray analysis identified circRNA051239 from high-metastatic ovarian cancer SKOV3.ip cells and demonstrated the up-regulation of circRNA051239 in ovarian cancer tissue as well as plasma exosomes. Additionally, the study showed that exosomes derived from high-metastatic ovarian cancer cells could transfer circRNA051239 into low-metastatic ovarian cancer cells, which significantly promoted the malignant behaviors of ovarian cancer cells by increasing proliferation, invasion and migration abilities. In addition, further study revealed that exosomal circRNA051239 stimulated the expression of PRSS3 in ovarian cancer by sponging miR-509-5p.
Recently, increasing studies have indicated that dysregulated noncoding RNAs, including miRNAs, lncRNA, and some circRNAs, are expressed in multiple types of cancers, including ovarian cancer. circRNAs participate in many cellular processes. Despite obscure roles of the majority of circRNAs, mounting evidence has revealed that deregulation of circRNAs can act as tumor suppressors or oncogenes, thereby affecting cell proliferation, migration as well as metastasis in human tumors [27,28]. Additionally, circRNAs can sponge miRNAs and function as ceRNAs to regulate gene expression in various types of cells [29,30].
Exosomes are small membranous vesicles with endocytic origin. To date, accumulated evidence has shown that exosomes act as biological delivery vehicles during cell-cell communication due to the various contained lipids, miRNAs, circRNAs and proteins. Many studies suggest that factors in exosomes can promote tumor initiation, development, and progression. Exosomes derived from diverse kinds of donor cells affect cell growth and metastasis, which is suggestive of material exchange between donor and recipient cells [31,32]. In addition, exosomal circRNAs have been identified to promote tumor progression, suggesting that exosomal circRNAs can be potential biomarkers for cancers [33]. Mounting researches have validated the stable and abundant distribution of circRNAs in exosomes. However, the role of exosomal circRNAs in EOC cell lines with different metastatic potential remains insufficiently explored.
circRNA051239 was enriched in tissues of ovarian cancer patients in our study. Ovarian cancer patients have significantly increased levels of plasma exosomal circRNA051239 than normal controls, suggesting exosomal circRNA051239 as a possible diagnostic marker for ovarian cancer. The functional effects of exosomal circRNA051239 were further confirmed in recipient cells. Our results showed that exosomes from circRNA051239 knockdown ovarian cancer SKOV3.ip cells significantly suppressed the invasion, migration and proliferation of recipient SKOV3 cells, suggesting that EOC-derived exosomes could enhance tumor growth and metastasis by transmitting circRNA051239. Hence, our findings suggest that circRNA051239 is directly transported from high-metastatic cells to low-metastatic cells via exosomes and regulates the biological functions of ovarian cancer in vitro.
The function of exosomal circRNAs has been well-investigated in cancers. For example, the tumor-released exosomal circRNA PDE8A might a promising diagnostic and progressive marker in pancreatic cancer, which aggregates invasion via miR-338/MACC1/MET pathway in pancreatic cancer [20]. Additionally, exosomal circRNAs secreted from adipocytes were transferred into recipient tumor cells in vitro and in vivo, thereby promoting tumor growth and decreasing DNA damage via miR-34a inhibition as well as USP7/Cyclin A2 signaling pathway activation in recipient cells [34].
To further explore the effects of circRNA051239 in EOC, we determined the location of circRNA051239 in EOC cells (cytoplasm). Cytoplasmic circRNAs, as well as long noncoding RNAs (lncRNAs) are revealed to act as miRNA sponges to indirectly modulate targeted mRNA expression. Bioinformatics analysis revealed that circRNA051239 shared miR-509-5p response elements with PRSS3. Luciferase reporter assay validated that circRNA051239 functioned as a ceRNA of miR-509-5p. miR-509-5p has been previously uncovered to significantly enhance or suppress proliferation invasion as well as migration of pancreatic cancer cells [35]. In our study, comprehensive bioinformatics along with the luciferase reporter assay confirmed that PRSS3 was a direct functional target of miR-509-5p in EOC cells. Exosomal transportation is a highly reliable cell-cell communication method. These findings implicated that circRNA051239 derived from high-metastatic SKOV3.ip cells enhanced the metastatic abilities of low-metastatic SKOV3 cells by affecting the miR-509-5p-PRSS3 pathway of recipient cells.
Several limitations exist in our research. Firstly, we screened out the circRNA051239 from ovarian cancer cell-derived exosomes, instead of the generally-used tumor tissues, which can cause a smaller individual bias in microarray study. Secondly, our results showed that plasma exosomal circRNA051239 levels in EOC patients were increased than those of normal controls. However, the correlations of exosomal circRNA051239 expression in plasma with tumor grade, stage or prognosis of EOC patients remain unclear. Thus, we should further evaluate the prognostic and diagnostic value of circulating exosomal circRNA051239 in EOC in a large-scale set of clinical samples. Because many circRNAs can be easily detected in clinical practice, we believe that exosomal circRNAs might be proper and reliable biomarkers in EOC.
In our study, circRNA051239 was identified from ovarian cancer cell-derived exosomes. We demonstrated the up-regulation of circRNA051239 expression in tumor tissue as well as plasma exosomes in EOC patients. Exosome-mediated transfer of circRNA051239 can promote ovarian cancer cell proliferation and migration. In addition, exosomal circRNA051239 from high-metastatic cells could impact low-metastatic cells by regulating PRSS3 levels by competing for miR-509-5p in recipient cells, leading to EOC progression. In conclusion, these findings suggest that exosomal circRNAs function as messengers in intercellular communication, which also implicate another possible mechanism for ovarian cancer therapy.
Acknowledgements
This work was supported by Peking University People’s Hospital Research and Development Funds (RDX2019-05); the Peking University Medicine Seed Fund for Interdisciplinary Research (BMU2020MX02); the National Key Research and Development Program of China (Grant No. 2016YFA0201400); and the National Key Technology R&D Program (Grant No. 2015BAI13B06).
Disclosure of conflict of interest
None.
References
- 1.Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 2.Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ, Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, Brenton JD, Chiappinelli KB, Martins FC, Coukos G, Drapkin R, Edmondson R, Fotopoulou C, Gabra H, Galon J, Gourley C, Heong V, Huntsman DG, Iwanicki M, Karlan BY, Kaye A, Lengyel E, Levine DA, Lu KH, McNeish IA, Menon U, Narod SA, Nelson BH, Nephew KP, Pharoah P, Powell DJ Jr, Ramos P, Romero IL, Scott CL, Sood AK, Stronach EA, Balkwill FR. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–679. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroeger PT Jr, Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Curr Opin Obstet Gynecol. 2017;29:26–34. doi: 10.1097/GCO.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 5.Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, Finn SP. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu LP, He YJ, Hou JC, Chen X, Zhou SY, Yang SJ, Li J, Zhang HD, Hu JH, Zhong SL, Zhao JH, Tang JH. The role of circRNAs in cancers. Biosci Rep. 2017;37:1–11. doi: 10.1042/BSR20170750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li TR, Jia YJ, Wang Q, Shao XQ, Lv RJ. Circular RNA: a new star in neurological diseases. Int J Neurosci. 2017;127:726–734. doi: 10.1080/00207454.2016.1236382. [DOI] [PubMed] [Google Scholar]
- 9.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, Sun SH, Yang F, Zhou WP. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng W, Dean DC, Hornicek FJ, Shi H, Duan Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol Cancer. 2019;18:124. doi: 10.1186/s12943-019-1049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Xu J, Xia K, Chang Y, Liu J, Yuan W. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howitt J, Hill AF. Exosomes in the pathology of neurodegenerative diseases. J Biol Chem. 2016;291:26589–26597. doi: 10.1074/jbc.R116.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conigliaro A, Cicchini C. Exosome-mediated signaling in epithelial to mesenchymal transition and tumor progression. J Clin Med. 2019;8:26. doi: 10.3390/jcm8010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li K, Chen Y, Li A, Tan C, Liu X. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144:1486–1495. doi: 10.1002/ijc.31774. [DOI] [PubMed] [Google Scholar]
- 20.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai X, Chen C, Yang Q, Xue J, Chen X, Sun B, Luo F, Liu X, Xiao T, Xu H, Sun Q, Zhang A, Liu Q. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018;9:454. doi: 10.1038/s41419-018-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 23.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc Natl Acad Sci U S A. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diederichs S, Bulk E, Steffen B, Ji P, Tickenbrock L, Lang K, Zänker KS, Metzger R, Schneider PM, Gerke V, Thomas M, Berdel WE, Serve H, Müller-Tidow C. S100 family members and trypsinogens are predictors of distant metastasis and survival in early-stage non-small cell lung cancer. Cancer Res. 2004;64:5564–5569. doi: 10.1158/0008-5472.CAN-04-2004. [DOI] [PubMed] [Google Scholar]
- 26.Ma R, Ye X, Cheng H, Ma Y, Cui H, Chang X. PRSS3 expression is associated with tumor progression and poor prognosis in epithelial ovarian cancer. Gynecol Oncol. 2015;137:546–552. doi: 10.1016/j.ygyno.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Xu H, Sun Y, You B, Huang CP, Ye D, Chang C. Androgen receptor reverses the oncometabolite R-2-hydroxyglutarate-induced prostate cancer cell invasion via suppressing the circRNA-51217/miRNA-646/TGFbeta1/p-Smad2/3 signaling. Cancer Lett. 2020;472:151–164. doi: 10.1016/j.canlet.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Yang B, Ren H, Xiao T, Zhang L, Li L, Li M, Wang X, Zhou H, Zhang W. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10:953. doi: 10.1038/s41419-019-2180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Wang R, Wu Z, Bai P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019;19:328. doi: 10.1186/s12935-019-0994-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Yao Z, Xu R, Yuan L, Xu M, Zhuang H, Li Y, Zhang Y, Lin N. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10:945. doi: 10.1038/s41419-019-2176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12:133. doi: 10.1186/s13045-019-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z, Li S. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38:477. doi: 10.1186/s13046-019-1473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, Yuan W, Sun Z, Ming L. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Li X, Li Y, Wan L, Chen R, Chen F. miR-509-5p inhibits cellular proliferation and migration via targeting MDM2 in pancreatic cancer cells. Onco Targets Ther. 2017;10:4455–4464. doi: 10.2147/OTT.S130378. [DOI] [PMC free article] [PubMed] [Google Scholar]