Abstract
Severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) pneumopathy is characterized by a complex clinical picture and heterogeneous pathological lesions, both involving alveolar and vascular components. The severity and distribution of morphological lesions associated with SARS‐CoV‐2 and how they relate to clinical, laboratory, and radiological data have not yet been studied systematically. The main goals of the present study were to objectively identify pathological phenotypes and factors that, in addition to SARS‐CoV‐2, may influence their occurrence. Lungs from 26 patients who died from SARS‐CoV‐2 acute respiratory failure were comprehensively analysed. Robust machine learning techniques were implemented to obtain a global pathological score to distinguish phenotypes with prevalent vascular or alveolar injury. The score was then analysed to assess its possible correlation with clinical, laboratory, radiological, and tissue viral data. Furthermore, an exploratory random forest algorithm was developed to identify the most discriminative clinical characteristics at hospital admission that might predict pathological phenotypes of SARS‐CoV‐2. Vascular injury phenotype was observed in most cases being consistently present as pure form or in combination with alveolar injury. Phenotypes with more severe alveolar injury showed significantly more frequent tracheal intubation; longer invasive mechanical ventilation, illness duration, intensive care unit or hospital ward stay; and lower tissue viral quantity (p < 0.001). Furthermore, in this phenotype, superimposed infections, tumours, and aspiration pneumonia were also more frequent (p < 0.001). Random forest algorithm identified some clinical features at admission (body mass index, white blood cells, D‐dimer, lymphocyte and platelet counts, fever, respiratory rate, and PaCO2) to stratify patients into different clinical clusters and potential pathological phenotypes (a web‐app for score assessment has also been developed; https://r-ubesp.dctv.unipd.it/shiny/AVI-Score/). In SARS‐CoV‐2 positive patients, alveolar injury is often associated with other factors in addition to viral infection. Identifying phenotypical patterns at admission may enable a better stratification of patients, ultimately favouring the most appropriate management. © 2021 The Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Keywords: SARS‐CoV‐2, acute respiratory failure, alveolar injury, vascular injury, COVID‐19
Introduction
Coronavirus disease 2019 (COVID‐19) due to severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) primarily affects the lung [1], with a broad spectrum of clinical manifestations. The severity of symptoms is extremely variable, showing the highest morbidity and mortality in elderly men and in patients with chronic comorbidities [2]. The incidence of acute respiratory distress syndrome (ARDS) among COVID‐19 patients has been reported to range from 47% to 100% in intensive care unit (ICU) patients and from 1% to 68% in overall hospitalized patients [3, 4, 5]. Even though the most severe forms of acute respiratory failure were initially treated as ARDS, the increased awareness of the disease has suggested that acute respiratory failure is characterized in these patients by distinctive clinical features. Indeed, compared with conventional ARDS, SARS‐CoV‐2 acute respiratory failure presents severe hypoxaemia and ventilation/perfusion mismatch, likely due to a downregulation of angiotensin‐converting enzyme‐2 (ACE‐2) secondary to viral endocytosis [6, 7, 8]. However, this aspect is still debated; indeed, other relevant studies reported an increase of ACE‐2 expression after viral infection [9]. In addition, COVID‐19 is often accompanied by a hypercoagulable state, with high levels of fibrinogen, D‐dimers, and an increased risk of thromboembolic complications. Lastly, an abnormal immune response (i.e. ‘cytokine storm syndrome’) with high levels of interleukin (IL)‐2, ‐4, ‐6, and ‐10, and tumour necrosis factor (TNF)‐α occurs. All these distinctive clinical features represent only some examples of the complex clinical scenario characterizing COVID‐19 patients, whose management still remains extremely problematic [10, 11, 12, 13]. Histological hallmarks of ARDS include a spectrum of lung injuries from the hyaline membrane, oedema, and haemorrhage (acute exudative phase) to type 2 pneumocyte hyperplasia, organizing pneumonia, and squamous metaplasia (organizing/proliferative phase) [14, 15]. All these lesions grouped under the generic pattern called diffuse alveolar damage (DAD) were described in deceased COVID‐19 patients as being the major histological findings detected in the lung parenchyma [16].
Consistent vascular lesions, mainly with the features of endothelialitis, thrombotic microangiopathy, and pulmonary intussusceptive angiogenesis, have recently been reported as distinctive pathological features of COVID‐19 pneumopathy [17, 18, 19, 20, 21]. Thus, as clinical features, clear heterogeneous pathological changes have also been reported.
Despite the fact that all the previously mentioned studies have represented an important step forward in our knowledge of SARS‐CoV‐2‐related lesions, the aetiopathogenesis of most of the pathological lung lesions remains unclear. In particular, it is yet to be determined if lung lesions are exclusively related to the viral infection or are conversely more influenced by other factors. Indeed, to date, the distribution of morphological lesions associated with SARS‐CoV‐2 pneumonia and how they relate to clinical data have not been systematically studied.
In the present work, a large number of lung lesions from patients who died from SARS‐CoV‐2 acute respiratory failure were comprehensively analysed and quantified by pathologists. Statistical analyses and machine learning algorithms were then used to ‘phenomap’ patients into prevalent pathological subtypes. The pathological subtypes were analysed in association with clinical, laboratory, molecular, and radiological data with the principal aim of identifying which factors, in addition to SARS‐CoV‐2 infection, may have influenced the presence of the different pathological lesions. An additional exploratory aim was to identify, using the random forest algorithm (RFA), the most discriminative clinical characteristics at hospital admission that might predict SARS‐CoV‐2 pathological phenotypes.
Materials and methods
The present work was a single‐centre prospective study of 26 consecutive COVID‐19 laboratory‐confirmed autopsies performed at the University Hospital of Padua from 23 March to 23 April 2020, according to national and international protocols, as previously described [22]. The Ethics Committee of our Centre was informed about the study (4853/A0/20): the study was approved by our local Clinical Institution Review Board and complied with the Declaration of Helsinki. The diagnosis for COVID‐19 was made according to the WHO interim guidance [23]. Specifically, nasopharyngeal swabs were tested by reverse transcription‐polymerase chain reaction (RT‐PCR) according to international standards. For each patient, the following demographic data and clinical characteristics were recorded: age, gender, body mass index (BMI), comorbidities, sequential organ failure assessment (SOFA) score at hospital admission, ongoing therapies, first positivity for SARS‐CoV‐2, other respiratory pathogens, length of intensive care unit (ICU) or internal medicine ward (IMW) stay, conventional laboratory data and inflammatory parameters, respiratory rate, blood gas analysis (arterial partial pressure of oxygen, PaO2, and carbon dioxide, PaCO2; oxygen saturation), antiviral therapies and anticoagulant therapy during the hospital stay, mode of respiratory support (i.e. conventional O2 therapy; high flow nasal cannula, HFNC; continuous positive airway pressure, CPAP; or invasive mechanical ventilation, IMV), fraction of inspired oxygen (FiO2), and PaO2/FiO2 ratio.
Radiological evaluation
To reduce the risk of infection spread, at the beginning of the current pandemic computed tomography (CT) scans in our tertiary centre were seldom performed and COVID‐19 patients were mainly assessed with chest X‐rays at diagnosis and follow‐up. Thus, for all patients, the only imaging available was chest X‐ray.
All radiographs were evaluated by an experienced radiologist in thoracic imaging (CG) using a previously published and validated composed COVID‐19 chest X‐ray score (i.e. COVID‐19 chest X‐ray score – CARE) [24]. In brief, the score was based on the subdivision of each lung in three areas (i.e. upper area, from the apices to the superior margin of the hilum; middle area, from the upper to the lower margin of the hilum; lower area, from the lower margin of the hilum to the costophrenic angle) and a three‐grade score describing, separately, the extension of ground glass (i.e. hazy opacity not obliterating bronchi and vessels) and consolidations (i.e. area of attenuation obscuring airways and vessels). The occurrence of additional findings such as pleural effusion, pneumothorax, pneumomediastinum, and subcutaneous emphysema was also recorded.
Autopsy and histological assessment
Autoptic examinations were carried out with a postmortem interval ranging from 24 hours to 6 days. Lungs were removed en bloc and fixed in 10% buffered formalin. Sixteen tissue blocks from the airways and each lung (three blocks per lobe) were sampled. In selected cases, special stains [e.g. Gram, periodic acid–Schiff (PAS), and Grocott's methenamine silver stains] were performed to highlight other concomitant microorganisms. Two expert lung pathologists (FC and FP) performed the histological evaluation blindly. For each section, several parameters were examined, as follows: (1) trachea, bronchi, and bronchioli: acute and chronic inflammation, presence of squamous metaplasia; (2) alveolar spaces: oedema, blood extravasation, fibrin, hyaline membranes, organizing pneumonia, squamous metaplasia, pneumocyte type II hyperplasia, and inflammatory cells, distinguishing acute (neutrophils) and chronic (macrophages, monocytes, and lymphocytes); (3) alveolar wall: chronic and acute inflammation, interstitial fibroblasts, capillary inflammation, and microthrombi. We define the degrees of capillary inflammation as (a) mild capillary inflammation (only neutrophil margination above baseline), (b) moderate inflammation (neutrophil margination above baseline with at least two back‐to‐back neutrophils), or (c) severe (frank capillaritis with destruction of capillary walls, blood extravasation, and neutrophilic karyorrhexis) [25, 26]; (4) vessels: microthrombi and large thrombi; and (5) pleura: inflammatory infiltrates, fibrosis, and fibrin deposition. In each case, a total of 27 histological parameters were separately quantified in 16 slides, distinguishing each lobe of the right and left lung. A total of 432 pathological features were evaluated for each patient. The presence and the extension of each parameter were scored in a semi‐quantitative manner from 0 to 3 (0: absent; 1: present, focal, in <25% of the section; 2: present, ranging from 25% to 50%; 3: present, diffuse, in >50%). All other additional findings were also carefully reported. Based on the presence and severity of some histological parameters, evaluated in all slides of each case, patients were categorized in a prevalent histological phenotype. Pathological alveolar injury (AI) prevalent phenotype was defined as when the median scores of hyaline membranes, organizing pneumonia, pneumocyte type 2 hyperplasia, and squamous metaplasia were at least as twice as high as the median scores of vascular lesions (microthrombi, large thrombi, vasculitis, and capillary inflammation). We considered prevalent vascular injury (VI) phenotype when the median scores of vascular lesions were double those of AI. A mixed phenotype was defined as when lesions of both AI and VI were equally present (similar median score of the two different types of lesion).
Molecular viral assessment
Total RNA was extracted from about 30 mg of frozen lung tissues using an RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) with a sample homogenization in a TissueLyser System (Qiagen) according to the manufacturer's instructions. The quantity and quality of prepared RNA were examined spectrophotometrically (NanoDrop One/OneC Microvolume UV–Vis Spectrophotometer; Thermo Fisher Scientific, Waltham, MA, USA). Quantitative determination of SARS‐CoV‐2 was performed using the CDC 2019‐Novel Coronavirus (2019‐nCoV) Real‐Time RT‐PCR Diagnostic Panel [https://www.fda.gov/media/134922/download] following the manufacturer's protocol.
The reaction conditions were 10 min at 50 °C, then 95 °C for 3 min, followed by 45 cycles of denaturation at 95 °C for 3 s, then annealing and extension at 55 °C for 30 s. To correct for sampling variability, we used human RNAse P (PRORP) as a reference to normalize the viral load using the comparative cycle threshold (CT) method (ΔCt) that transforms the CT into relative loads (ratios of viral target to human target).
Statistical analysis
All clinical, radiological, and pathological data were recorded in an electronic database. Data are presented as medians (with first and third quartiles) for continuous variables and as percentages for categorical variables. Due to repeated measurements, comparisons among groups were based on generalized linear models (GLMs), with variance inflated using the Huber–White sandwich estimator [27]. GLM family was the Gamma family for continuous response variables with a logarithmic link and quasi‐binomial family for the binary response. To further account for multiple testing, P values were adjusted using the Benjamini–Hochberg procedure [27]. Agreement between pathologists was assessed by using Cohen's kappa. The final dataset comprised more than 1.5 million data points coming from the 26 patients.
Derivation of the AVI score
Data on pathological findings were summarized using robust principal component analysis (RPCA) based on projection pursuits [28]. Since the database had a wide number of variables and a relatively small number of patients, we used more conservative and robust statistical methods for the analysis than the more familiar multilinear regression analysis, which enabled us to separate important features from noisy data. Sparse robust principal components were derived using a grid search algorithm in the plane and with sparseness constraints derived using a grid search and tradeoff product optimization [29]. The PCA loadings were used to characterize the AVI score. The entire analysis was blinded with respect to the operator‐based classification of phenotypes into AI or VI subtypes. Subsequently, after unblinding, a cut‐off for the score was performed using standard ROC analysis to discriminate between the operator‐based classification of phenotypes. The distribution of AVI scores was estimated using a Gaussian kernel and optimal bandwidth selection [30].
Phenomapping
Descriptive analysis of the association between presentation of all clinical data of patients and pathological findings was based on GLMs, with choice of link and variance functions tailored to the type of response (Gaussian functions for continuous variables and binomial functions for binary variables). Survival functions were estimated using a Kaplan–Meier estimator, and multivariable survival models were based on the Cox proportional hazard model. Then to ‘phenomap’ the clinical variables that were mostly associated with pathological phenotypes [31], a random forest algorithm (RFA) was implemented [32]. The choice of an RFA model was suggested by the high k/n ratio, where k is the number of candidate variables and n is the number of observations in the samples. RFA was implemented optimizing over the number of trees (6600), a number of k/3 variables randomly selected as candidates for splitting a node, an average number of five data points in a terminal node, and weighted mean‐squared error as splitting rule. Missing data were iteratively imputed using the Ishwaran algorithm [33]. The relative importance of each clinical variable against the score derived via RPCA was measured by the minimal depth of variables: the smaller the minimal depth, the greater the impact that the variable has on sorting observations, and therefore on the forest prediction. The mean of the minimal depth distribution was used as an analytic threshold for evidence of variable impact, with variables with a minimal depth lower than this threshold being considered as important in the forest prediction [34]. A representative tree was then extracted from the forest to clinically phenomap the pathological patterns. Extraction was based on the Euclidean distance d2 based on closeness to prediction [35]. All analyses were made using the R software [36] with the pcaPP [29] and the randomForestSRC [37] libraries. Machine learning analysis was reported according to the EQUATOR draft guidelines [38].
Results
Clinical data
The median age of the 26 patients was 82 years (Q1–Q3: 76–88 years); 42% were women. Table 1 summarizes the main clinical/radiological data. The estimation of disease duration from symptom onset or hospitalization to patient death was 8.5 days (95% CI 7–13) and 5 days (95% CI 4–7), respectively (supplementary material, Figure S1). A high percentage of patients (14 out of 26 patients, 53.8%) had comorbidities, with the most common being hypertension; 96% of patients had two or more comorbidities, including cardiovascular disease, chronic renal failure, diabetes, dyslipidaemia, dementia, chronic obstructive pulmonary disease, and cancer. Eight (31%) and 14 (54%) patients received therapeutic and prophylactic anticoagulant therapy, respectively, while nine (39%) received antibiotics due to secondary bacterial infections. Seven patients (27%) were admitted to the ICU and were invasively ventilated [tidal volume ≤ 6 ml/kg of ideal body weight (IBW) and driving pressure ≤ 14 cmH2O], with a median length of 6 days (Q1–Q3: 5–14). Nineteen (73%) patients were not invasively ventilated. Specifically, 13 subjects (50%) received O2 therapy, either through a nasal cannula or facial mask, or HFNC [median length of 6 days (Q1–Q3: 4–7)]; six (23%) underwent CPAP or non‐invasive ventilation through a helmet [median length of 4 days (Q1–Q3: 2.5–5)].
Table 1.
Clinical and radiological data of the study cohort.
| N° | Results* | N° | Results* | ||
|---|---|---|---|---|---|
| Clinical characteristics | Laboratory data | ||||
| Age, years | 26 | 76/82/88 | WBC (109/L) | 25 | 4.1/5.5/13.2 |
| Males | 15 | 58% | LY (109/L) | 23 | 0.64/0.80/0.98 |
| Females | 11 | 42% | LY (%) | 23 | 6.4/12.2/17.0 |
| BMI | 20 | 23/25/30 | Platelet count (109/L) | 24 | 143/167/220 |
| SOFA score | 18 | 4.0/5.5/7.0 | D‐dimer (ng/ml) | 20 | 296/474/994 |
| Comorbidity | 26 | 96% (25) | Fibrinogen (mg/dl) | 11 | 4.6/5.3/5.4 |
| Fever (°C) | 21 | 38/38/39 | IL‐6 (μg/ml) | 9 | 41/120/298 |
| Cough | 26 | 62% (16) | Ferritin (ng/ml) | 18 | 577/1162/1814 |
| Shortness of breath | 26 | 88% (23) | |||
| Dyspnoea | 26 | 81% (21) | Respiratory parameters | ||
| Anticoagulant therapy | 26 | Respiratory rate (bpm) | 22 | 22/24/28 | |
| Never | 15% (4) | SpO2 (%) | 26 | 89/93/97 | |
| Prophylactic | 54% (14) | pH | 25 | 7.4/7.4/7.5 | |
| Therapeutic | 31% (8) | PaO2 (mmHg) | 25 | 55/68/90 | |
| Antiviral therapy | 26 | 38% (10) | PaCO2 (mmHg) | 25 | 30/33/40 |
| Antibiotic therapy | 26 | 96% (25) | PaO2/FiO2 | 25 | 74.9/104.4/205.7 |
| Microbiology for SARS‐COV‐2 | 26 | 100% (26) | IMV | 26 | 27% (7) |
| Superimposed infections | 26 | 35% (9) | Length of IMV (days) | 7 | 5/6/14 |
| IMW patients | 26 | 54% (14) | |||
| IMW length of stay (days) | 14 | 2.5/5/7 | Radiological score † | ||
| ICU patients | 26 | 46% (12) | Median global score | 26 | 4 |
| ICU length of stay (days) | 12 | 3.2/5.0/8.0 |
BMI, body mass index; ICU, intensive care unit; IL‐6, interleukin 6; IMV, invasive mechanical ventilation; IMW, internal medicine ward; LY, lymphocytes; SOFA, sequential organ failure assessment; WBC, white blood cells.
Data are expressed as first quartile/median/third quartile for continuous variables and as percentages (absolute numbers) for categorical variables.
Radiological assessment of 97 chest X‐rays of 26 patients.
Radiological findings
A total of 97 chest X‐rays were assessed. During the entire time of hospitalization, all patients except one showed both ground glass and consolidation after at least one chest X‐ray. The highest global score (i.e. 36) was found in four patients. The median scores were 2 (Q1–Q3: 1–2.75) and 0 (Q1–Q3: 0–5) for ground glass and consolidation, respectively. The median global score was 4 (Q1–Q3: 1–6) (Table 1). The ground glass score was significantly higher in the left parenchyma than on the right side (p < 0.001), whereas no statistically significant differences emerged for the consolidation and the global score (supplementary material, Table S1).
Viral molecular findings
RNA was extracted from frozen lung tissue samples obtained from all patients included in the study population. The quantity and quality of RNA samples were adequate for the RT‐qPCR analysis. In particular, mean (± SD) RNA concentration was 241 ± 193 ng/μl; mean (± SD) A260/280 and A260/230 ratios were 2.09 ± 0.03 and 1.52 ± 0.55, respectively.
In all patients, positivity for SARS‐CoV‐2 was confirmed in lung tissues by RT‐qPCR, showing median (Q1–Q3) values of N1 ΔCt of 1.2 (−3.4 to 6.8), ranging from −9.3 to 13.7 (supplementary material, Figure S2).
Pathological findings
Microscopic evaluation of lungs revealed lesions in different anatomic areas with a heterogeneous distribution. The evaluation of all slides showed good inter‐observer agreement (K value between 0.6 and 1). A statistically significant difference in lobe involvement was observed for some parameters. In lung parenchyma, the highest grades of the pathological parameters of AI were more frequently assigned to the lower lobes. The vascular bed showed various lesions from different grades of capillary inflammation, including the most severe form of neutrophilic capillaritis which was mainly detected in the right lungs (p = 0.001), to microthrombi and, less frequently, macrothrombi. There was no preferential distribution of microthrombi and macrothrombi in the parenchyma. Other types of lesions were also detected in 14 cases (54%), including infections (nine cases: seven bacterial and two fungal, morphologically compatible with Aspergillus spp.), neoplasms (three cases: two squamous cell carcinomas and one malignant solitary fibrous tumour), and aspiration pneumonia (two cases). Some of the infections were unknown before autopsy and were also detected by using special stains (e.g. Gram, Grocott, PAS). Large airway acute or chronic inflammation was observed in all cases. Pleura was frequently affected by inflammation and fibrous reaction, without a significant difference in lung/lobe distribution. From the comparison between cases with associated signs of infectious diseases (fungal or bacterial) and pure COVID‐19 cases, pleural involvement was more frequently detected in the second group (39% versus 26%, p = 0.02). Detailed descriptions of the airways, parenchyma, vascular bed, and pleural lesions are reported in supplementary material, Tables S3–S6, respectively. According to the categorization of prevalent histological features, the AI phenotype was present in four cases (16%), the VI phenotype in 11 cases (42%), and the mixed phenotype in the remaining 11 cases (42%).
Based on the above pathological findings, data were synthesized using RPCA. The first RPCA dimension explained 64.67% of the total variance, with higher loadings in absolute values determined by hyaline membrane, intra‐alveolar blood/fibrin and neutrophils, type 2 pneumocyte hyperplasia, intra‐alveolar macrophages/lymphocytes, oedema, organizing pneumonia, intra‐alveolar squamous metaplasia, capillary inflammation, pleural inflammation, and thrombi (supplementary material, Table S2). The first RPCA dimension was thus used to derive a synthetic score to characterize each patient with respect to the most important variables. Such a score was challenged against a blinded classification by the pathologists who classified patients as ‘prevalent AI’, ‘prevalent VI’, or ‘mixed’ phenotype. Standard ROC analysis showed that the score, called ‘AVI score’, discriminated well between prevalent AI and prevalent VI patients. Positive AVI‐score values identified prevalent AI phenotype, while negative AVI‐score values identified prevalent VI phenotype. The mixed phenotype shared AVI‐score values from both phenotypes (Figure 1).
Figure 1.
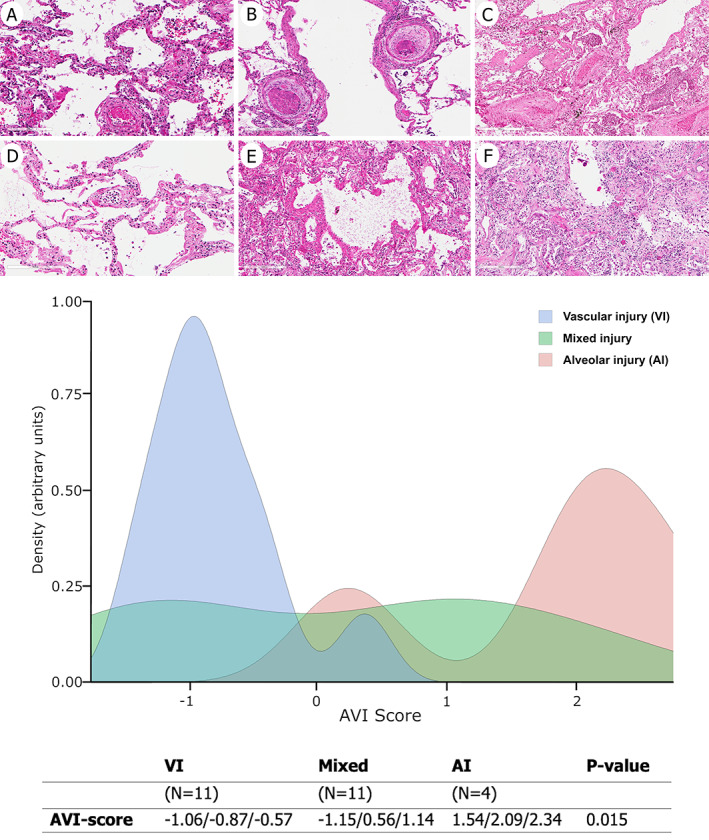
ROC analysis indicates a cut‐off of about zero (−0.034) with an overall accuracy of 0.97 (95% CI 0.91–1.00), sensitivity 0.91 (95% CI 0.74–1.00), and specificity equal to 1.00 to discriminate between AI and VI patients. The mixed phenotype is not characterized by specific ranges of the AVI score. Data in the table are medians (I, III quartile). P value refers to the overall difference among phenotypes. At the top of the figure, example histopathological sections of case 1 (VI phenotype, A, D), case 11 (mixed phenotype, B, E), and case 5 (AI phenotype, C, F) with the most representative lesions in both upper (A–C) and lower lobes (D–F) are shown. (A) Capillaritis and microthrombus (H&E stain). (B) Microthrombi (H&E stain). (C) Hyaline membrane with squamous metaplasia (H&E stain). (D) Neutrophilic margination and capillary inflammation (H&E stain). (E) Diffuse hyaline membrane (H&E stain). (F) Organizing pneumonia (H&E stain). Original magnification: (A, B) ×200; (C–F) ×100.
Clinical–pathological correlations
A positive value of AVI score (consistent with a more representative presence of AI) was directly correlated with endotracheal intubation and the length of invasive mechanical ventilation, longer hospital and ICU stay, duration of illness from symptom onset, and lower tissue viral quantity (p < 0.001) (Figure 2 and supplementary material, Tables S7–S10). A positive AVI score was also associated with the presence of other pathological lesions, such as aspiration pneumonia, other infections, neoplasms, or all three (p < 0.001) (Figure 3 and supplementary material, Table S11). A negative value of AVI score (consistent with a more representative presence of VI) was not related to any clinical data. Neither positive nor negative value of AVI score was correlated with any radiological findings. More than 40 clinical covariates were analysed to characterize the AVI‐score distribution among patients, deriving a synthetic ‘phenomap’ of the underlying pathological aspects. Supplementary material, Table S12 presents anonymized data for the 26 patients. The regression RFA showed good performance (98.7% of explained variance at 10‐fold cross‐validation) using variables recorded at the time of hospital admission: BMI, white blood cells (WBC), D‐dimer, lymphocyte (both absolute and percentage) count, platelet count, fever, respiratory rate, and PaCO2 (Figure 4A). Instead, radiological variables showed no significant correlation and were excluded from further analysis. The combination of the above parameters at the time of admission could lead to the identification of clinical clusters associated with different pathological phenotypes and different AVI scores. For instance, patients with a low respiratory rate, unusual hypercapnia, and lymphocytopenia at admission are potentially at higher risk of developing a ‘prevalent AI’ phenotype (positive AVI score) (Figure 4B).
Figure 2.

Correlation of AVI score with (A) length of intubation (R 2 0.63), (B) ICU stay (R 2 0.72), (C) hospital stay (R 2 0.49) and symptom duration (R 2 0.67), and (D) viral quantity (R 2 0.49 for AI, 0.01 for VI, and 0.33 for mixed). Log(2−ΔCt) represents the SARS‐CoV‐2 relative loads (ratios of viral target to human target) transformed to logarithmic scale for graphical representation.
Figure 3.
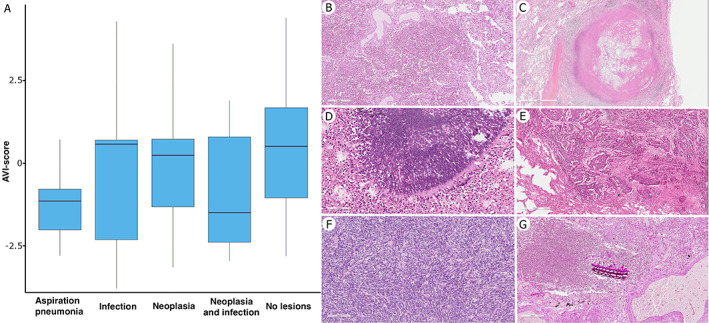
(A) Correlation of AVI score with other pathological lesions. (B–G) On the right, the most representative histological sections of the pathological lesions detected: inflammatory exudate within the intra‐alveolar space resulting in lobar pneumonia (B, H&E stain); necrotizing granuloma (C, H&E stain); pulmonary aspergillosis (D, H&E stain); squamous cell carcinoma (E, H&E stain); lung metastasis of pleural malignant solitary fibrous tumour, previously diagnosed (F, H&E stain); and aspiration pneumonia with inhaled foreign body (G, PAS stain). Original magnification: (D) ×200; (F, G) ×100; (B, E) ×50; (C) ×15.
Figure 4.
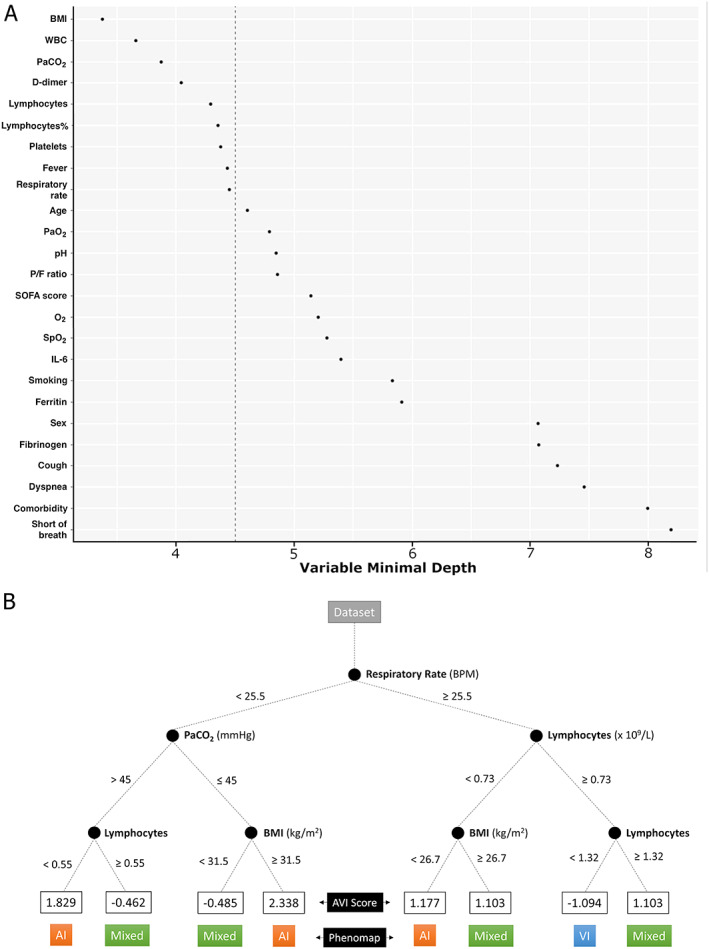
Variables characterizing AVI score. (A) A random forest‐based model was used to obtain the most important variables able to sort patients based on the previous robust principal component analysis (RPCA). The relative importance of each clinical variable against the score derived via RPCA was measured by the minimal depth of variables: the smaller the minimal depth, the more important the variable. Important variables are reported in rank order with the most important at the top. The vertical dashed line indicates a threshold corresponding to the maximal–minimal depth for important variables (left side of the panel). Variables exceeding this value are considered unimportant. (B) Phenomapped representation of the random forest algorithm (pruned version) to show how patients were classified to a specific AVI score based on the most important clinical characteristics. The final node number represents values of the AVI score for specific patterns of covariates.
To facilitate the interpretation and use of the phenotypic representation, a web‐app has been developed and made available at https://r-ubesp.dctv.unipd.it/shiny/AVI-Score/.
Discussion
In this study of 26 consecutive patients who died from SARS‐CoV‐2 acute respiratory failure, we identified three major phenotypes: (1) phenotype with a prevalent DAD (AI); (2) phenotype with prevalent vascular lesions (VI); and (3) phenotype with mixed DAD and VI (Figure 1). This evidence was obtained following a thorough analysis of multiple lung samplings to carefully capture all lesions and to avoid underestimating or missing some important tissue alterations [39].
To our knowledge, this is the first study that has used a combination of robust machine learning techniques to objectively identify pathological phenotypes and factors, which, in addition to SARS‐CoV‐2, may influence their occurrence. The generated AVI score grouped lung lesions in a spectrum identifying vascular injury for negative values and alveolar damage for positive values. The association between AVI score and clinical data showed that a positive value of the score was significantly related to a longer duration of the disease (whether calculated from the onset of symptoms or from the hospital stay), longer ICU stay, tracheal intubation, and prolonged invasive mechanical ventilation, independently of other factors. Generally, patients affected by SARS‐CoV‐2 acute respiratory failure are more hypoxic and usually require invasive mechanical ventilation and a longer hospital stay. Prolonged mechanical ventilation is an important risk factor for ventilator‐associated pneumonia, aspiration, and superimposed infections, and all of these elements could have contributed to the development of a prevalent AI, generating a vicious circle [40, 41].
We also detected a more extensive AI in patients with associated other lesions: neoplasms, aspiration pneumonia, and other infections – the last two also found in patients not treated by prolonged mechanical ventilation. The risk of developing AI in patients with infectious pneumonia is higher in hospitalized patients, particularly in cases with viral, fungal, or mixed infections [42]. The pathogenetic mechanisms at the basis of AI in these pathological lesions are complex, often related to a dysregulation of innate or acquired immune response [43]. Thus, our observations suggest that the development of AI is not exclusively related to SARS‐CoV‐2 infection. Supporting this, the detection of a lower tissue viral quantity in patients with more extensive AI is noteworthy. Such findings are in agreement with earlier observations, obtained in a multicentre study with our contribution, which showed an association of AI with longer disease duration and less frequent tissue virus detection [44]. Based on this, a speculated timeline for the disease may involve viral‐related lung damage and significant vascular injury in the early stages, whereas in the advanced stages the damage may be mainly influenced by other factors (such as prolonged disease duration or mechanical ventilation, superinfections, etc.) responsible for maintaining the inflammatory state with a prevalent pathological appearance of DAD.
In our study, negative AVI score (which means more extensive VI) was not correlated to any clinical outcome data, including all coagulation parameters. This was not an unexpected finding in our case series, considering that the majority of patients had received anticoagulant therapy due to comorbidities or prophylaxis. The absence of any correlation between disease duration, type of clinical management, and other pathological lesions may suggest that VI is a peculiar feature of the COVID‐19 pneumopathy. Indeed, in the literature, considerable evidence indicates the important role of vascular damage in the aetiopathogenesis of the disease [17, 18, 19, 20, 21, 45].
In COVID‐19, the specificity of pathological lung lesions, particularly vascular injury, becomes even more interesting when we compare the characteristics observed in our case series with those of prior severe global pandemics – SARS, MERS, and H1N1 influenza. Patterns of lung injury, including DAD in exudative and/or organizing phases, were identified in MERS and SARS [46, 47, 48, 49, 50, 51]. However, the heterogeneous pattern of vascular lesions frequently detected in SAR‐CoV2 infection was never reported in any of these infections. Particularly, Ackermann et al reported that several vascular lesions, including endothelialitis, thrombosis, and intussusceptive angiogenesis, were detected in COVID‐19 patients more frequently compared with severe influenza virus infections [21].
No associations between AVI score and radiological findings were detected. Although radiological imaging plays a crucial role in diagnosing and monitoring patients with SARS‐CoV‐2 pneumopathy [52, 53, 54], our results showed that chest X‐rays only marginally affect the AVI score. This controversial finding might be due to the severity of the disease in our population and the low specificity of the applied technique. Although chest X‐ray has been shown to be a reliable tool for diagnosing and monitoring patients with COVID‐19 [55, 56, 57], further studies using CT scans are highly recommended to obtain more accurate characterizations of the disease.
We also built a model based on RFA in an attempt to identify clinical, laboratory, or functional ‘biomarkers’ that, at hospital admission, could predict the development of distinct pathological lesions. It is noteworthy that WBC, lymphocyte and platelet counts, BMI, PaCO2, fever, respiratory rate, and D‐dimer were the most important features able to stratify patients into different clinical clusters and ‘potential’ pathological phenotypes with the aim of allowing more individualized management. For instance, in patients at risk of developing a ‘prevalent’ AI phenotype, more attention should be given to optimize mechanical ventilation to ensure a lung‐protective ventilatory strategy, avoiding ‘high’ tidal volumes or transpulmonary pressure, preventing patient self‐inflicted lung injury, atelectasis, barotrauma, and pneumothorax during both invasive and non‐invasive partial ventilatory support [58]. Finally, a proper antibiotic stewardship should be established with the aim of minimizing any potential risk factor for superimposed infections [59]. On the contrary, in patients at risk of developing a potential VI phenotype, the primary aim is to optimize thrombotic prophylaxis. Specifically, thromboprophylaxis should be considered for all hospitalized COVID‐19 patients [60, 61]. This model, if validated in a large multicentre case series, could be highly informative for more appropriate management of COVID‐19 patients.
The present study has limitations. First, the sample size was relatively small and from a single centre. It should be noted, however, that this is one of the largest single‐centre case series in which all the patients received protocolized medical treatments, respiratory support [62], and standardized lung sampling methodology and analysis. Furthermore, the statistical techniques were among those best suited for dealing with a limited number of patients and a large number of covariates, also including the different treatments. However, the risk of instability in the estimates could not be excluded; for example, there could have been a potential underestimation of the rate of ‘pure’ AI, which was observed in a few cases. Nevertheless, both the clinical and the pathological observations, as derived from the statistical models, do agree well with the current knowledge about COVID‐19 pathology. Second, some clinical features could be affected by the numerous therapeutic treatments administered to patients. Accordingly, all clinical information subsequent to the patients' admissions was excluded from the phenomapping and used only for the exploratory analysis.
Finally, all pathological parameters were analysed only on haematoxylin and eosin‐stained slides using a scoring system that is observer‐dependent and subjective. However, all the evaluations were blindly performed by two pathologists, showing good inter‐observer agreement and, overall, were confirmed by machine learning analysis. The present study could represent a good starting point to promote the use of a deep learning approach in digitized slides stained both with haematoxylin and eosin and immunohistochemistry for a more precise definition of the phenotypical characteristics of the inflammatory infiltrate.
In conclusion, we found that vascular lesions are an important feature of COVID‐19 pneumonia since they are consistently present in the majority of cases, while AI is related to several factors in addition to SARS‐CoV‐2 infection. The identification of phenotypical patterns associated with clinical characteristics could allow us to stratify COVID‐19 patients into different risk clusters to optimize future management strategies.
Author contributions statement
FC, FP, FF and FL performed the autopsies and histological evaluations. CG provided radiological data. AB, NS, PN, AC, MR, FR, CDV, ACr, RV and MP provided clinical data. DG, LV and GL carried out the statistical data analyses. FC, FP, FF, EC, AB, CG, PN and DG drafted the manuscript. All the authors contributed to study conception, design, and revision of the manuscript.
Supporting information
Figure S1. Kaplan–Meier estimates of disease duration calculated from symptom onset and from hospitalization
Figure S2. Analysis of the SARS‐CoV‐2 Ct values obtained using the CDC 2019‐Novel Coronavirus (2019‐nCoV) Real‐Time RT‐PCR Diagnostic Panel
Table S1. Radiological data obtained from the scoring system
Table S2. Loadings of the principal components analysis (PCA)
Table S3. Pathological findings of the airways
Table S4. Pathological findings of the lung parenchyma
Table S5. Pathological findings of the lung vascular bed
Table S6. Pathological findings of the pleura
Table S7. Distribution of morphological characteristics and AVI scores according to HFNC, CPAP‐NIV, and IMV during ICU stay
Table S8. Distribution of morphological characteristics and AVI score according to duration of IMV
Table S9. Distribution of morphological characteristics and AVI scores according to duration of ICU stay
Table S10. Distribution of AVI score according to duration of disease since hospitalization and symptoms' onset
Table S11. Distribution of morphological characteristics, AVI‐score distribution, and other lesions
Table S12. Anonymized data from 26 patients
Acknowledgements
We wish to thank Judith Wilson for English revision. This research was partially supported by a fellowship from the University of Padua/Intesa San Paolo Vita Bank (2020A08).
No conflicts of interest were declared.
References
- 1. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA 2020; 323: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Porcheddu R, Serra C, Kelvin D, et al. Similarity in case fatality rates (CFR) of COVID‐19/SARS‐COV‐2 in Italy and China. J Infect Dev Ctries 2020; 14: 125–128. [DOI] [PubMed] [Google Scholar]
- 3. Longhini F, Bruni A, Garofalo E, et al. Helmet continuous positive airway pressure and prone positioning: a proposal for an early management of COVID‐19 patients. Pulmonology 2020; 26: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med 2020; 8: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tzotzos SJ, Fischer B, Fischer H, et al. Incidence of ARDS and outcomes in hospitalized patients with COVID‐19: a global literature survey. Crit Care 2020; 24: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. MacLaren G, Fisher D, Brodie D. Preparing for the most critically ill patients with COVID‐19: the potential role of extracorporeal membrane oxygenation. JAMA 2020; 323: 1245–1246. [DOI] [PubMed] [Google Scholar]
- 7. Zarantonello F, Andreatta G, Sella N, et al. Prone position and lung ventilation and perfusion matching in acute respiratory failure due to COVID‐19. Am J Respir Crit Care Med 2020; 202: 278–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santamarina MG, Boisier D, Contreras R, et al. COVID‐19: a hypothesis regarding the ventilation‐perfusion mismatch. Version 2. Crit Care 2020; 24: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li G, He X, Zhang L, et al. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID‐19. J Autoimmun 2020; 112: 102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spiezia L, Boscolo A, Poletto F, et al. COVID‐19‐related severe hypercoagulability in patients admitted to intensive care unit for acute respiratory failure. Thromb Haemost 2020; 120: 998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID‐19 patients reveals IL‐6 and IL‐10 are disease severity predictors. Emerg Microbes Infect 2020; 9: 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arachchillage DRJ, Laffan M. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 2020; 18: 1233–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katzenstein AL, Bloor CM, Leibow AA. Diffuse alveolar damage – the role of oxygen, shock, and related factors. A review. Am J Pathol 1976; 85: 209–228. [PMC free article] [PubMed] [Google Scholar]
- 15. Beasley MB. The pathologist's approach to acute lung injury. Arch Pathol Lab Med 2010; 134: 719–727. [DOI] [PubMed] [Google Scholar]
- 16. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020; 8: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menter T, Haslbauer JD, Nienhold R, et al. Postmortem examination of COVID‐19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020; 77: 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wichmann D, Sperhake JP, Lütgehetmann M, et al. Autopsy findings and venous thromboembolism in patients with COVID‐19: a prospective cohort study. Ann Intern Med 2020; 173: 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet 2020; 395: 1417–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magro C, Mulvey JJ, Berlin D, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID‐19 infection: a report of five cases. Transl Res 2020; 220: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N Engl J Med 2020; 383: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Basso C, Calabrese F, Sbaraglia M, et al. Feasibility of postmortem examination in the era of COVID‐19 pandemic: the experience of a Northeast Italy University Hospital. Virchows Arch 2020; 477: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. World Health Organization (WHO) . Clinical management of severe acute respiratory infection (SARI) when COVID‐19 disease is suspected. WHO. 2020; 2019 (March):12. Available from: https://www.who.int/docs/default‐source/coronaviruse/clinical‐management‐of‐novel‐cov.pdf. [Accessed 22 March 2021].
- 24. Giraudo C, Cavaliere A, Fichera G, et al. Validation of a composed COVID‐19 chest radiography score: the CARE project. ERJ Open Res 2020; 6: 359–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berry GJ, Burke MM, Andersen C, et al. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody‐mediated rejection in heart transplantation. J Heart Lung Transplant 2013; 32: 1147–1162. [DOI] [PubMed] [Google Scholar]
- 26. Wallace WD, Li N, Andersen CB, et al. Banff study of pathologic changes in lung allograft biopsy specimens with donor‐specific antibodies. J Heart Lung Transplant 2016; 35: 40–48. [DOI] [PubMed] [Google Scholar]
- 27. Huber PJ. The behavior of maximum likelihood estimates under nonstandard conditions. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics, Le Cam LM, Neyman J (eds). University of California Press: Berkeley and Los Angeles, 1967; 221–233. [Google Scholar]
- 28. Croux C, Filzmoser P, Oliveira MR. Algorithms for Projection–Pursuit robust principal component analysis. Chemom Intel Lab Syst 2007; 87: 218–225. [Google Scholar]
- 29. Croux C, Filzmoser P, Fritz H. Robust sparse principal component analysis. Dent Tech 2013; 55: 202–214. [Google Scholar]
- 30. Sheather SJ, Jones MC. A reliable data‐based bandwidth selection method for kernel density estimation. J R Stat Soc B Methodol 1991; 53: 683–690. [Google Scholar]
- 31. Braunwald E. Another step toward personalized care of patients with heart failure. Eur J Heart Fail 2015; 17: 988–990. [DOI] [PubMed] [Google Scholar]
- 32. Breiman L. Random forests. Mach Learn 2001; 45: 5–32. [Google Scholar]
- 33. Ishwaran H, Kogalur UB, Blackstone EH, et al. Random survival forests. Ann Appl Stat 2008; 2: 841–860. [Google Scholar]
- 34. Ishwaran H, Kogalur UB, Gorodeski EZ, et al. High‐dimensional variable selection for survival data. J Am Stat Assoc 2010; 105: 205–217. [Google Scholar]
- 35. Banerjee M, Ding Y, Noone AM. Identifying representative trees from ensembles. Stat Med 2012; 31: 1601–1616. [DOI] [PubMed] [Google Scholar]
- 36. R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, 2019. Available at: https://www.R-project.org/. [Accessed 22 March 2021]. [Google Scholar]
- 37. Ishwaran H, Kogalur UB. Fast Unified Random Forest for Survival, Regression, and Classification (RF‐SRC), R package version 2.9.3, 2020. Available at: https://CRAN.R-project.org/package=randomForestSRC. [Accessed 25 March 2021].
- 38. Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res 2016; 18: e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Calabrese F, Pezzuto F, Fortarezza F, et al. Pulmonary pathology and COVID‐19: lessons from autopsy. The experience of European Pulmonary Pathologists. Virchows Arch 2020; 477: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hegeman MA, Hemmes SNT, Kuipers MT, et al. The extent of ventilator‐induced lung injury in mice partly depends on duration of mechanical ventilation. Crit Care Res Pract 2013; 2013: 435236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rezoagli E, Fumagalli R, Bellani G. Definition and epidemiology of acute respiratory distress syndrome. Ann Transl Med 2017; 5: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kojicic M, Li G, Hanson AC, et al. Risk factors for the development of acute lung injury in patients with infectious pneumonia. Crit Care 2012; 16: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med 2008; 358: 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borczuk AC, Salvatore PS, Seshan SV, et al. COVID‐19 pulmonary pathology: a multi‐institutional autopsy cohort from Italy and New York City. Mod Pathol 2020; 33: 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calabrese F, Fortarezza F, Giraudo C, et al. Two sorts of microthrombi in a patient with Coronavirus Disease 2019 and lung cancer. J Thorac Oncol 2020; 15: 1782–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gill JR, Sheng ZM, Ely SF, et al. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med 2010; 134: 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakajima N, Sato Y, Katano H, et al. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Mod Pathol 2012; 25: 1–13. [DOI] [PubMed] [Google Scholar]
- 48. Ng DL, Al Hosani F, Keating MK, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 2016; 186: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franks TJ, Chong PY, Chui P, et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol 2003; 34: 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1953–1966. [DOI] [PubMed] [Google Scholar]
- 51. Hwang DM, Chamberlain DW, Poutanen SM, et al. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol 2005; 18: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Borghesi A, Zigliani A, Masciullo R, et al. Radiographic severity index in COVID‐19 pneumonia: relationship to age and sex in 783 Italian patients. Radiol Med 2020; 125: 461–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fichera G, Stramare R, De Conti G, et al. It's not over until it's over: the chameleonic behavior of COVID‐19 over a six‐day period. Radiol Med 2020; 125: 514–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toussie D, Voutsinas N, Finkelstein M, et al. Clinical and chest radiography features determine patient outcomes in young and middle‐aged adults with COVID‐19. Radiology 2020; 297: E197–E206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cleverley J, Piper J, Jones MM. The role of chest radiography in confirming covid‐19 pneumonia. BMJ 2020; 370: m2426. [DOI] [PubMed] [Google Scholar]
- 56. Rubin GD, Ryerson CJ, Haramati LB, et al. The role of chest imaging in patient management during the COVID‐19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology 2020; 296: 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simpson S, Kay FU, Abbara S, et al. Radiological Society of North America expert consensus statement on reporting chest CT findings related to COVID‐19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA – secondary publication. J Thorac Imaging 2020; 35: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fan E, Beitler JR, Brochard L, et al. COVID‐19‐associated acute respiratory distress syndrome: is a different approach to management warranted? Lancet Respir Med 2020; 8: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Torres A, Niederman MS, Chastre J, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital‐acquired pneumonia and ventilator‐associated pneumonia: guidelines for the management of hospital‐acquired pneumonia (HAP)/ventilator‐associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur Respir J 2017; 50: 1700582. [DOI] [PubMed] [Google Scholar]
- 60. Spyropoulos AC, Levy JH, Ageno W, et al. Scientific and Standardization Committee communication: clinical guidance on the diagnosis, prevention, and treatment of venous thromboembolism in hospitalized patients with COVID‐19. J Thromb Haemost 2020; 18: 1859–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Piazza G, Morrow DA. Diagnosis, management, and pathophysiology of arterial and venous thrombosis in COVID‐19. JAMA 2020; 324: 2548–2549. [DOI] [PubMed] [Google Scholar]
- 62. Pasin L, Sella N, Correale C, et al. Regional COVID‐19 network for coordination of SARS‐CoV‐2 outbreak in Veneto, Italy. J Cardiothorac Vasc Anesth 2020; 34: 2341–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Kaplan–Meier estimates of disease duration calculated from symptom onset and from hospitalization
Figure S2. Analysis of the SARS‐CoV‐2 Ct values obtained using the CDC 2019‐Novel Coronavirus (2019‐nCoV) Real‐Time RT‐PCR Diagnostic Panel
Table S1. Radiological data obtained from the scoring system
Table S2. Loadings of the principal components analysis (PCA)
Table S3. Pathological findings of the airways
Table S4. Pathological findings of the lung parenchyma
Table S5. Pathological findings of the lung vascular bed
Table S6. Pathological findings of the pleura
Table S7. Distribution of morphological characteristics and AVI scores according to HFNC, CPAP‐NIV, and IMV during ICU stay
Table S8. Distribution of morphological characteristics and AVI score according to duration of IMV
Table S9. Distribution of morphological characteristics and AVI scores according to duration of ICU stay
Table S10. Distribution of AVI score according to duration of disease since hospitalization and symptoms' onset
Table S11. Distribution of morphological characteristics, AVI‐score distribution, and other lesions
Table S12. Anonymized data from 26 patients


