Summary
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) enters the host cell by binding to angiotensin‐converting enzyme 2 (ACE2) receptor. Other important proteins involved in this process include disintegrin and metalloproteinase domain‐containing protein 17 (ADAM17) also known as tumour necrosis factor‐α‐converting enzyme and transmembrane serine protease 2. ACE2 converts angiotensin II (Ang II) to angiotensin (1–7), to balance the renin angiotensin system. Membrane‐bound ACE2 ectodomain shedding is mediated by ADAM17 upon viral spike binding, Ang II overproduction and in several diseases. The shed soluble ACE2 (sACE2) retains its catalytic activity, but its precise role in viral entry is still unclear. Therapeutic sACE2 is claimed to exert dual effects; reduction of excess Ang II and blocking viral entry by masking the spike protein. Nevertheless, the paradox is why SARS‐CoV‐2 comorbid patients struggle to attain such benefit in viral infection despite having a high amount of sACE2. In this review, we discuss the possible detrimental role of sACE2 and speculate on a series of events where protease primed or non‐primed virus–sACE2 complex might enter the host cell. As extracellular virus can bind many sACE2 molecules, sACE2 level could be reduced drastically upon endocytosis by the host cell. A consequential rapid rise in Ang II level could potentially aggravate disease severity through Ang II‐angiotensin II receptor type 1 (AT1R) axis in comorbid patients. Hence, monitoring sACE2 and Ang II level in coronavirus disease 2019 comorbid patients are crucial to ensure safe and efficient intervention using therapeutic sACE2 and vaccines.
Keywords: COVID‐19, SARS‐CoV‐2 entry, soluble ACE2
Abbreviations
- 6‐HB
six‐helix bundle
- ACE
angiotensin‐converting enzyme
- ACE2
angiotensin‐converting enzyme 2
- ACEIs
angiotensin‐converting enzyme inhibitors
- ADAM17
ADAM metallopeptidase domain 17
- AGT
angiotensinogen
- APA
amino peptidase A
- Ang
angiotensin
- ARBs
angiotensin receptor blockers
- AT1R
angiotensin II receptor 1
- AT2R
angiotensin II receptor 2
- AT4R
Angiotensin IV receptor
- COVID‐19
coronavirus disease 2019
- FP
fusion peptide
- HR
heptad repeats
- MasR
G‐protein‐coupled Mas receptor
- MAPK
mitogen‐activated protein kinases
- MrgD
Mas‐related G protein‐coupled receptor D
- NADPH
nicotinamide adenine dinucleotide phosphate
- RAS
renin–angiotensin system
- TACE
tumour necrosis factor‐α converting enzyme
- TMPRSS2
transmembrane serine protease 2
- RBD
receptor‐binding domain
- RBM
receptor‐binding motif
- ROS
reactive oxygen species
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- SERPIN
serine protease inhibitor
- S protein
spike protein
- VBM
virus‐binding motif
1. INTRODUCTION
The pandemic coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), enters cells via host angiotensin‐converting enzyme 2 (ACE2) receptor. 1 , 2 , 3 , 4 , 5 ACE2 is a key component of renin–angiotensin system (RAS) to maintain delicate balance of health and disease. 6 , 7 It counterbalances the angiotensin II (Ang II)–angiotensin type 1 receptor (AT1R) mediated harmful effects by catalytic cleavage of Ang II. 8 , 9 , 10 , 11 In Ang II rich environment, ACE2 extracellular catalytic domain is shed by disintegrin and metalloproteinase domain‐containing protein 17 (ADAM17) as feedback mechanism. This shed soluble ACE2 (sACE2) retains its catalytic activity and can access Ang II easily. 12 , 13 , 14
During viral entry, virus takes membrane ACE2 (mACE2) inside host cell, reducing mACE2 which impairs the Ang II balance. 3 , 4 , 15 , 16 During infection, binding of viral surface spike (S) glycoprotein to mACE2 also triggers ACE2 shedding. 17 , 18 In some disease conditions and with ageing, ACE2 expression starts to fall and elevated sACE2 is considered a disease biomarker. 19 , 20 , 21 The amount of mACE2 can be correlated with SARS‐CoV‐2 binding and entry; however, very little is known about the role of sACE2 in the overall infection process.
The ACE2‐virus binding and subsequent fusion involve a group of enzymes to prime S protein followed by several conformational rearrangements. 5 , 22 , 23 Several studies have reported the beneficial and preventive role of therapeutic sACE2 in COVID‐19. 21 , 24 , 25 , 26 , 27 , 28 Surprisingly, clinical data suggest that patients with low mACE2 and high sACE2 faced more disease severity and fatality. 25 , 28 This contrasts with the protective role of therapeutic sACE2 claimed in literature. In this perspective, we briefly reviewed the biochemical and structural features of key enzymes and viral S protein in binding and fusion processes. We propose a virus entry mechanism where sACE2–virus complex achieves cellular entry resulting in depletion of both sACE2 and mACE2. This in turn possibly increases Ang II level leading to cytokine storm 29 and other pathological complications. 30
2. RENIN–ANGIOTENSIN SYSTEM AND ACE2
Diversified functions of RAS in different tissues are identified and established so far. 31 , 32 RAS network is a fundamental regulator of many physiological systems and plays a vital role in the pathophysiology of different organ activities. The sole precursor protein, angiotensinogen (AGT), is cleaved by renin at the N‐terminal end and converted into a 10‐amino acid peptide, angiotensin I (Ang I) 33 (Figure 1). AGT belongs to serine protease inhibitor (SERPIN) family. AGT sequences vary but the functional domain, Ang I, sequences are fairly conserved. Ang I is cleaved by ACE to form Ang II. Ang III and IV are formed by sequential removal of N‐terminal amino acid by amino peptidase A and N (APA/N). 11 Ang II is further cleaved by ACE2 to produce Ang (1–7). ACE2 also cleaves Ang I to form Ang (1–9).
FIGURE 1.
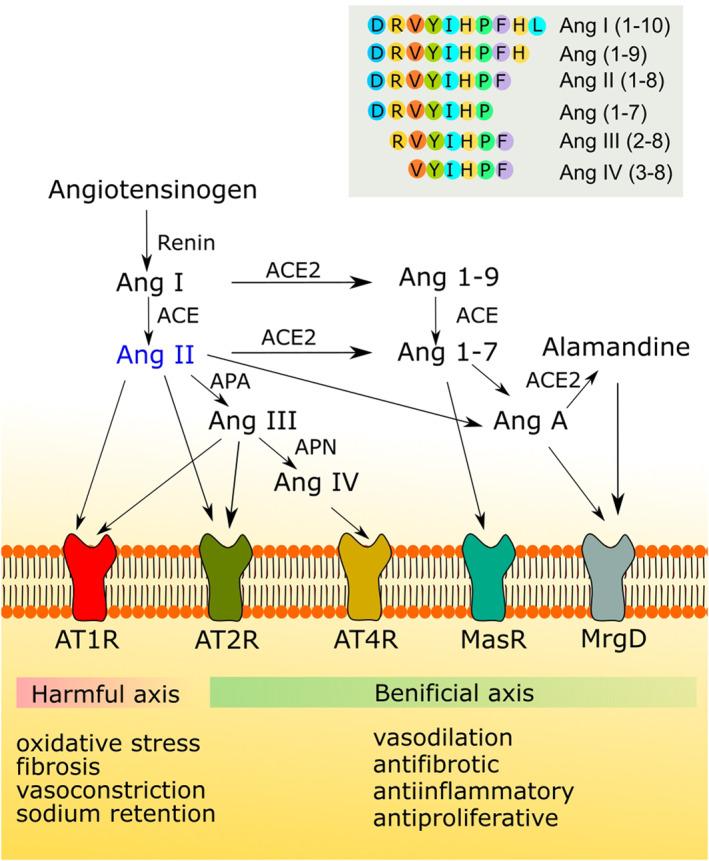
Renin–angiotensin system (RAS) in health and disease. Ang II, being the key molecule, can mediate AT1R‐mediated harmful events. ACE2 can balance the effect of excess Ang II by catalysing it to Ang 1–7. ACE2 can also catalyse Ang I in two‐step process to produce Ang 1–7. Different derivatives of Ang II can confer protective role through AT2R, AT4R, MasR and MrgD receptors. Amino acid configuration of major peptides in the RAS system are shown inset. ACE, angiotensin‐converting enzyme; ACE2, angiotensin‐converting enzyme 2; Ang I, angiotensin I; Ang II, angiotensin II; AT1R, angiotensin II receptor type I; AT2R, angiotensin II receptor type II; AT4R, angiotensin IV receptor; MasR, Mas receptor; MrgD, Mas‐related G protein‐coupled receptor D
AT1R is activated by binding Ang II which forms Ang I–(ACE)–Ang II–AT1R axis. 9 This axis is responsible for vasoconstriction, inflammation, myocardial fibrosis, proliferations, hypertrophy and oedema in different organs and tissues. 34 The second axis is started by the Ang II–(ACE2)–Ang (1–7)–MasR which acts as an antagonist of the Ang I–Ang II–AT1R axis (Figure 1). The third axis is comprised of Ang I–(ACE2)–Ang (1–9)–(ACE)–Ang (1–7)–MasR. Ang II–(APA)–Ang III–(APN)–Ang IV–AT4R can be represented as a fourth axis. 8 , 35 Ang II is sometimes decarboxylated to produce Ang A. Ang A can be cleaved again by ACE2 to generate alamandine which then activates MrgD‐mediated pathway. Through axes two to four, the vasodilator, anti‐inflammatory, anti‐hypertrophic pathways are activated. Production of alamandine also antagonizes the first axis (Figure 1). 8
ACE2 is a transmembrane zinc‐containing metalloenzyme predominantly located in kidney, heart, intestines and lungs. 4 , 10 , 36 ACE2 counteracts the action of ACE by converting a vasoconstrictor peptide, Ang II into a vasodilator Ang (1–7). 36 , 37 , 38 , 39 The ectodomain of ACE2 contains extracellular N‐terminal peptidase that converts Ang I, Ang II and Ang A into Ang (1–9), Ang (1–7) and alamandine, respectively. 10 , 35 , 38 , 39 The catalytic affinity of ACE2 for Ang II is 400 times higher than for Ang I. 40 The C‐terminal domain contains a homologue of a renal protein, collectrin, that regulates amino acid transportation to the cell surface. 10 These distinct enzymatic mechanisms of ACE2 help defend against cancer, inflammatory and cardiovascular diseases. ACE and ACE2 counterbalance each other and their ratio tightly regulates the overall RAS network. 8 , 10
Raised Ang II activates AT1R and triggers downstream cascades. One of these cascades terminates in the initiation of p38, MAPK and ADAM17/ADAM10 phosphorylation by NADPH oxidase 2‐mediated reactive oxygen species formation. 41 , 42 Phosphorylation boosts the enzymatic activity of ADAM17 which facilitates ACE2 ectodomain shedding with a reduction of mACE2. This ensures easy access of ACE2 to Ang II, which stabilizes the AT1R‐mediated pathological effects in a positive feedback cycle. 43 , 44 Elevated Ang II was also found to internalize mACE2 via AT1R‐receptor‐mediated lysosomal degradation. 45
In addition to its normal functions, ACE2 also contains virus‐binding motifs (VBMs) where the receptor‐binding domain (RBD) of S protein binds and facilitates host membrane fusion. 46 , 47 ACE2 acts as receptor for several viruses, including SARS‐CoV‐2 and became a major focus of scientific research. 5 , 12 , 46 Ectodomain shedding does not interfere the VBM, retains both virus‐binding and enzymatic activities and can bind S protein in the extracellular floating state. 12 , 13 , 14
3. MOLECULAR INTERACTION BETWEEN SARS‐CoV‐2 SPIKE PROTEIN AND HOST CELL
3.1. S protein–ACE2 binding dynamics
The SARS‐CoV‐2 S protein is an envelope glycoprotein that creates the distinctive 'corona shape' after which the virus family is named. The spike glycoprotein is a large polypeptide consisting of S1 (residue 13–685) and S2 (residue 686–1273) subunits (Figure 2a). 22 S1 and S2 both contain multiple domains and motifs that play vital roles in attachment, fusion and entry of virus into target cells, and are potential target for the development of neutralizing antibodies, inhibitors and vaccines. Three S1/S2 heterodimers assemble to form a trimer spike protruding from the viral envelope. 48 , 49 Three S2 units form a central core and S1 units sit like a hat on the top with RBDs. The domains of ACE2 and TMPRSS2 are also shown in Figure 2a.
FIGURE 2.
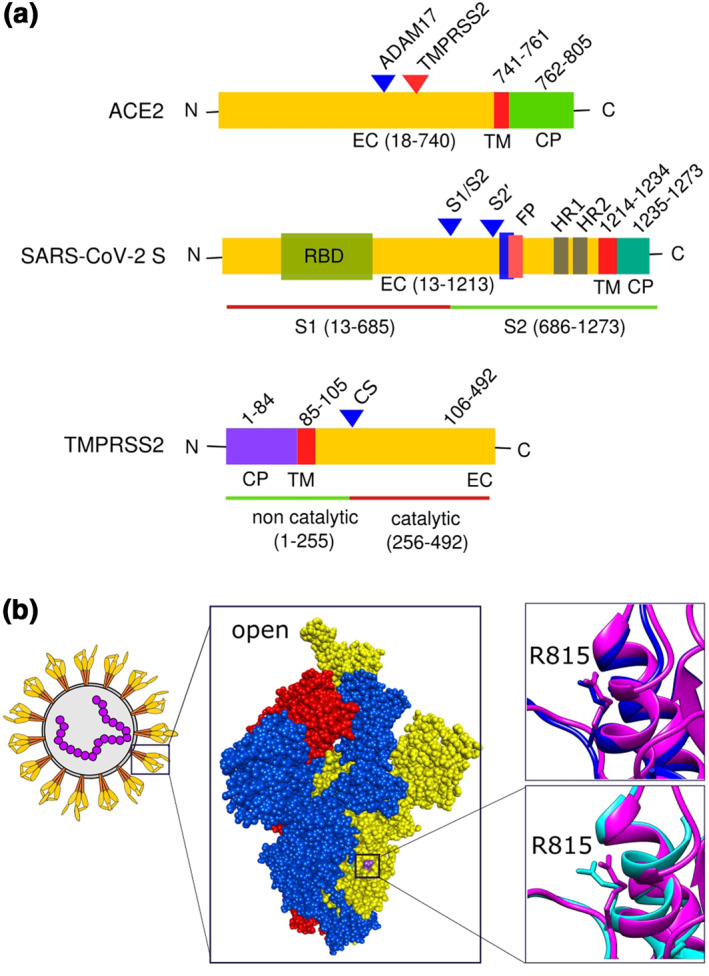
Domains of important proteins and ACE2 binding mediated virus spike protein conformational shift: (a) domains of ACE2, SARS‐CoV‐2 spike protein and TMPRSS2; (b) spike protein trimer of SARS‐CoV‐2 (PDB ID 6XM3) with one monomer open (yellow), and two closed (blue and red). Two superimposed images shown to depict S2′ cleave site residue arginine 815. Closed (blue, PDB ID 6ZGE) and open (pink, PDB ID 6XM3) spike protein monomer superimposed both without ACE2 bound (right top). Open no‐ACE2‐bound spike protein (pink, PDB ID 6XM3) was superimposed on open with ACE2 bound (cyan, PDB 7A96) (right bottom). Discovery Studio Visualizer v17.2.0 and Chimrea v1.13.1 were used to visualize and superimpose S proteins, respectively. Inkscape v0.92.3 was used to compile the figure. CP, cytoplasmic; CS, autocatalytic site; EC, extracellular; FP, fusion peptide; HR, heptad repeat; TM, transmembrane
S1 subunit contains two structurally independent N‐terminal (S1‐NTD) and C‐terminal (S1‐CTD) domains. The S1‐NTD (residue 14–105) contributes to the spike trimer interface. S1‐CTD contains two subdomains: a core structure (RBD, residue 319–541) and a receptor‐binding motif (RBM, residue 437–508) (Figure 2a). 50 A five‐stranded antiparallel β‐sheet is present in RBD. The two‐stranded antiparallel β‐sheet along with two ridges forms a concave‐shaped RBM which recognizes host receptor. Like SARS‐CoV, SARS‐CoV‐2 uses ACE2 membrane receptor to bind S protein for host cell entry with the help of host proteases. 51 , 52 , 53 , 54 Subdomain I of ACE2 in NTD contains several VBMs on the outer surface of ACE2. 50
A number of hydrophilic residues of both RBD and VBM form a compact link of H‐bond and salt bridge interactions that facilitate the binding of S protein to ACE2. This binding does not interfere with the enzymatic activity of ACE2. 55 The virus–receptor attachment is dominated by polar interactions facilitated by the hydrophilic residues. 50 SARS‐CoV‐2 RBD has a higher affinity for ACE2 compared to SARS‐CoV; on the other hand, the paradox is that the affinity of its S protein to ACE2 is similar or lower than SARS‐CoV S protein. 22 , 51 The reasons for this opposing behaviour are the dynamic state of SARS‐CoV S protein RBD mostly stay in standing‐up position. In contrast, the SARS‐CoV‐2 RBD can stay in lying‐down state 51 making the RBD inaccessible to ACE2 23 and host immune cells as this position is angled towards the core of the trimer. 22 However, structural studies show that SARS‐CoV‐2 S proteins RBD go through hinge‐like conformational rearrangements. 22 , 23 , 50 , 55 , 56 , 57 It transiently hides itself from receptor binding which is thought to be less stable and exhibits spontaneous standing‐up and lying‐down positions. 22 , 57 It can be speculated that four different scenarios can be adopted by a trimer: (a) all RBDs of a trimer are in lying‐down position, (b) one RBD in open position, (c) two RBDs in open positions, and (d) all three RBDs are in standing‐up positions. Structural studies of Benton et al. 23 demonstrated the aforementioned four possibilities by structural biology experiments. However, as like many coronaviruses, S protein contains S1/S2 cleavage site, proteolysis at this site triggers the affinity of RBD towards ACE2. 22 , 23 , 51 , 56 , 57 , 58
3.2. Proteolysis of S protein
S protein of some viruses are primed at S1/S2 site by proteolytic action during packaging once inside cell; however, some are often primed later in entry process, sometimes after receptor bindings. 23 , 56 Proteolysis facilitates the membrane fusion process. 55 Coronaviruses use host proteases in four different phases of virus infection cycle; (a) furin‐like protein convertases during packaging or during cell entry, (b) extracellular proteases, that is, elastase after exocytosis, (c) cell surface proteases like TMPRSS2 just after RBD binds to receptor, and (d) lysosomal protease like cathepsin L, B after endocytosis. However, sometimes, receptor bindings and change in adjacent pH perhaps facilitate proteolysis. 55 , 58 SARS‐CoV‐2 S protein possesses additional four amino acids (PRRA) at S1/S2 junction that chemically changes the site to polybasic or multibasic site. This polybasic site creates a unique cleavage site (RRAR) for furin proteases that is absent in SARS‐CoV. 5 , 22 , 57 Proteolytic cleavage at S1/S2 junction results in the unstable up conformations of all three RBDs that can be more exposed for ACE2 binding. It also facilitates shedding of S1 subunit and initiates refolding of S2. 57 Conversely, in case of standing up of single RBD, cleavage at S1/S2 site dose not separate the subunits fully that remain attached together through non‐covalent interactions. 55 Even though S1/S2 cleavage is not mandatory for cell entry, having a polybasic cleavage site could expand SARS‐CoV‐2 tropism and/or transmissibility and/or altered pathogenicity compared to other coronaviruses. 5 , 23 , 57
3.3. Restructuring of S protein
RBD binding with ACE2 results in a substantial structural rearrangement from a metastable prefusion conformation. 22 , 23 , 57 , 59 RBD domain changes its positions after binding by a rigid‐body rotation. The centre of mass of the domain shifts from trimer axis along with the NTD‐ and RBD‐associated subdomains. This structural rearrangement changes the position of NTDs of all the three S1 of the trimers. Interestingly, binding of ACE2 with two or all RBDs does not produce significant changes in average RBD position. 23 In addition, ACE2 binding with one standing‐up RBD induces a more standing‐up conformation than fully closed conformation of other RBDs suggesting the possibility of binding more ACE2s to other RBDs. The rearrangements of the RBD and NTD domains facilitate the trimeric ring of S1 append to the S2 core and twist the S2 structures. The arginine 815 of S2′ junction is extended outside when ACE2 is bound to the open RBD, but not in non‐bound case (Figure 2b). This alteration eases the S2′ and makes the cleavage site accessible to proteases. The attached S1 trimer is required to be primed at S2′ cleavage site (upstream of S2 fusion peptide) for the further rearrangements for host cell fusion. 23
3.4. Proteolysis at S2′ site
Studies have demonstrated the proteolytic role of furin protease, TMPRSS2 and cathepsins at S2′ cleavage site. 5 , 49 , 60 Several studies on SARS‐CoV and MERS‐CoV along with influenza viruses revealed that activation of S protein and subsequent attachment and fusion with host cell membrane involve multiple cleavage of events at S1/S2 junction and S2′ location. 47 , 52 , 54 TMPRSS2 showed more activity on SARS‐CoV‐2 S protein in vitro compared to cathepsin L and furin. 53 Furin is a membrane‐associated protease predominantly expressed in the Golgi bodies of all cells, which can reach plasma membrane as well and generally been internalized and targeted back to the trans‐Golgi network. 61 The predominant location of furin lies in cytoplasm and its presence on extracellular matrix helps SARS‐CoV‐2 to convert S protein conformation to fuse with host cell membranes. 53 Cathepsins, in contrast, are classically found in lysosomes and endosomes and take part in several degradative and antigen‐presenting progressions. 62 As both furin and cathepsins predominantly work in cytoplasm, it is postulated that serine protease TMPRSS2 is mainly involved in S1/S2 and S2′ cleavage; however, in the absence of TMPRSS2, viruses take advantages of the presence of furins and cathepsins on extracellular matrix to facilitate fusions to host membrane. In addition, extracellular presence of furins may accelerate the membrane attachment by exposing S2′ cleavage site to TMPRSS2 for subsequent irreversible cleavage at that site.
TMPRSS2 contains a type II transmembrane domain, a receptor class A domain, a scavenger receptor cysteine‐rich domain and a protease domain, and belongs to the trypsin‐like serine protease family (Figure 2a). They can interact with other proteins on cell surface, several soluble proteins, matrix components and proteins on other cells or particles. 63 , 64 , 65 Serine proteases are well characterized and known to take part in many cellular, physiological, molecular and pathological processes. Activation of protease domain occurs by autocatalytic cleavage and released from the rest of the part of TMPRSS2. 66 The presence of Ca2+ binding site might be involved in autocatalysis, activation and release of serine protease domain like many serine zymogens. Release of active serine protease can float, reach and act on distant tissues known as trans technique. 67 , 68 , 69 In addition, activation of AT1R receptors raise Ca2+ concentrations in the cytoplasm through AT1R–Gαq/11–PLC–IP3–IP3R axis. 70 Calcium signalling triggers when Ca2+ ions are released from intracellular stores and/or it enters cells through plasma membrane ion channels. 71 Sudden increase of extracellular concentration of Ca2+ may activate several calcium‐dependent enzymes on outer membrane. As mentioned above, the presence of Ca2+‐binding sites 68 on TMPRSS2 are perhaps involved in Ca2+‐dependent autocatalysis and release of activated serine proteases in the extracellular space. There is evidence of released serine proteases in circulatory systems too. 63 , 64 , 72 Cleavage at S2′ location by TMPRSS2 is very important for virus entry as this event can prime and expose fusion peptide at ready to fuse state. 23 , 58 Inhibitors of TMPRSS2 mostly block viral entry into host cells. 5 , 60 , 65
4. SARS‐CoV‐2 UPTAKE AND FUSION PROCESS
4.1. Membrane fusion process
Upon cleavage at S2′ cleavage site, remaining S2 subunit is detached (partially or fully) from S1. 5 , 23 , 46 , 60 , 73 The S2 subunit contains a hydrophobic fusion peptide (residue 816–837) and two heptad repeat regions, HR‐1 (residue 912–984), and HR‐2 (residue 1163–1213). In addition, S protein contains transmembrane (residue 1214–1234) and cytoplasmic (residue 1235–1273) domains. 50 S2′ cleavage site is present just adjacent to fusion peptide; consequently, cleavage at this site exposes the internal fusion peptide along with dramatic conformational changes in S2 subunits. This conformational change exposes and facilitates HR‐1 and HR‐2 to interact with each other to form a six‐helix bundle structure helping host membranes and the virus to fuse. 74 The general mechanism of SARS‐CoV‐2 binding to mACE2 and cellular entry under normal physiological condition is shown in Figure 3.
FIGURE 3.
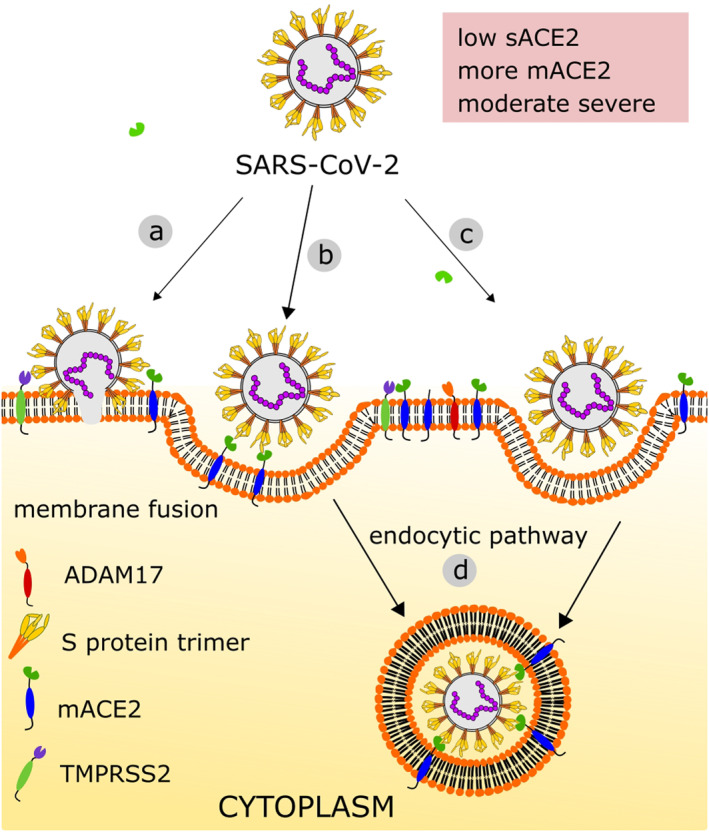
SARS‐CoV‐2 entry in normal physiological condition: (a) virus enter by membrane fusion, (b) through receptor‐dependent endocytosis, (c) through receptor‐independent endocytosis, (d) virus is uptake by host membrane‐bound vesicle where mACE2 is lost. When patient is non‐comorbid, virus can take its entry reducing surface ACE2; however, the Ang II‐mediated severity is thought to be moderate due to the high expression and regeneration of mACE2 and immune capacity of host. ACE2, angiotensin‐converting enzyme 2; Ang II, angiotensin II; mACE2, membrane ACE2; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
4.2. Uptake of virus (endocytic route)
Clathrin‐ and non‐clathrin‐mediated endocytic route of virus entry has also been observed for SARS‐CoV‐2. 75 , 76 , 77 , 78 , 79 These processes are protease independent and facilitate entry at membrane surface in the absence of membrane‐bound or extracellular proteolytic enzymes. After endocytosis, several options are available; budding of clathrin‐coated vesicle, flotillin‐1‐associated endocytosis, caveolae, macropinocytosis, clathrin‐independent carrier and glycosylphosphatidylinositol‐anchored protein‐enriched endosomal compartment. The virus passes through the endocytic pathways towards cell core reducing pH in the endosome. 24 , 76 The acidic environment activates several endosomal proteases like CatB and CatL. These cysteine proteases then act on S protein to facilitate the fusion as mentioned above. 49 , 80 , 81
5. PROPOSED MECHANISM OF sACE2 IN VIRAL ENTRY, SEVERITY AND THERAPEUTICS
5.1. Virus–sACE2 complex mediated entry, RAS imbalance and distant infection
Severe COVID‐19 patients often have underlying health issues, like diabetes, cardiovascular disease, hypertension, obesity, chronic lung disease, are more likely to need ventilation and die. 19 , 20 , 27 Besides, sACE2 has been reported as a biomarker in several diseases. 82 In a study with two large international cohorts comprising more than 5000 elderly with atrial fibrillation, the level of sACE2 was tightly coupled to age, male sex, cardiovascular disease and diabetes. 21 ACE2 ectodomain or sACE2 can bind viral S protein in the similar fashion as mACE2 does, indicating native sACE2 can bind S protein in extracellular space with same affinity. 23 , 27 COVID‐19 comorbid patients have high sACE2, yet are still prone to severity. Based on the present molecular and structural data of virus–ACE2 interaction mechanism, we extend the following possible events of sACE2‐mediated COVID‐19 severity, especially in comorbid patients (Figure 4).
-
(a)
S proteins of newly entered virus can bind sACE2 in extracellular space to form virus–sACE2 complex. Binding‐induced conformational shifting can allow free floating catalytically active domain of TMPRSS2 or furin to prime S protein for membrane fusion (Figure 4b). Binding of sACE2 to one S protein monomer also facilitates opening of neighbouring S protein monomer for ACE2 binding. Upon priming, the fusion peptide zone of viral S proteins of the complex becomes ready to fuse to host membrane in pH‐dependent mechanism. The virus–sACE2 complex with some free S protein in open state may roll over the membrane through random walk and can bind available ACE2. This could fuse and bring its RNA inside host cell, but post fusion fate of virus‐bound sACE2 needs to be experimentally evaluated.
-
(b)
Endocytosis mediated viral entry can take both receptor‐dependent and ‐independent pathways (Figure 4c,d). In the similar fashion mentioned above, virus partially covered with sACE2 can bind to limited mACE2 present in comorbid patients. This may follow receptor‐mediated endocytosis of virus–sACE2 complex via clathrin‐dependent or ‐independent, cholesterol‐rich caveolae or lipid raft mechanism. In such case, virus–sACE2 complex will get inside the host cell. Again, the replication potential of such internalized complex is still unclear. Although not predominant, macropinocytosis, a receptor‐independent filopodia‐mediated endocytosis process, can also happen and virus–sACE2 can be internalized. 83 , 84
-
(c)
We speculate that primed virus–sACE2 complex is also able to manifest the same host cell entry mechanism in any distant organ once delivered by the circulatory system (Figure 4d). Even cells with no floating proteases and with low or no membrane‐bound proteases can uptake this complex via endocytosis. The coating of sACE2 on virus does not interfere with host immune cells, making it easy to travel to distant organ avoiding immune attack.
FIGURE 4.
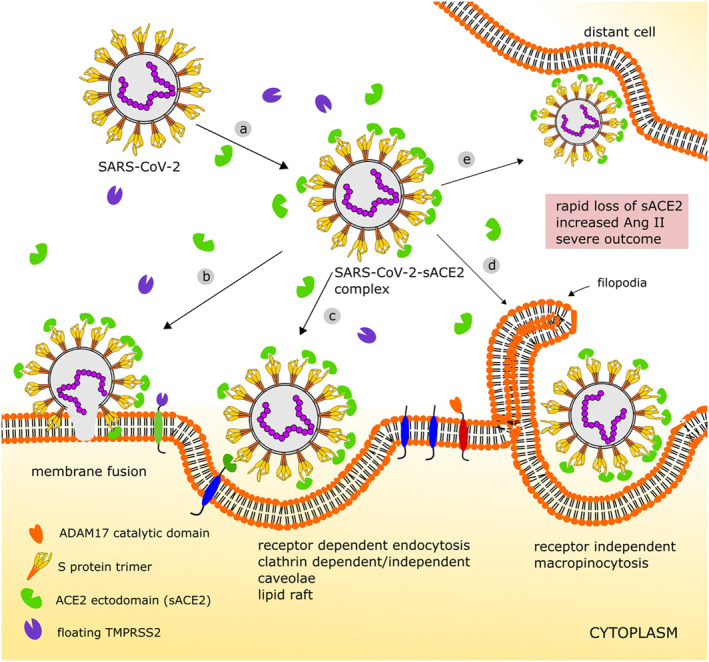
Proposed detrimental role of sACE2 in SARS‐CoV‐2 entry and subsequent disease severity in site of infection and distant cells: (a) in sACE2 abundant condition, virus S protein may partially be covered with circulatory sACE2 forming virus–sACE2 complex and can be primed by floating serine proteases; (b) it can enter its genetic material by membrane fusion; (c) few available mACE2 can bind the virus–sACE2 complex and endocytosis can proceed through several mechanisms; (d) virus–sACE2 complex can be internalized by receptor‐independent macropinocytosis; and (e) the complex can travel to distant cell where serine protease activity is low or absent. In all internalization processes, the virus–sACE2 complex takes sACE2 inside creating rapid crisis of sACE2 and increase of Ang II. This may lead to RAS imbalance and COVID‐19 severity. Ang II, angiotensin II; COVID‐19¸ coronavirus disease 2019; mACE2, membrane ACE2; sACE2, soluble ACE2; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Taken together, in our opinion, this pre‐primed virus–sACE2 complex has potential to create two main harmful effects; (a) if travelled to distant organs where cells lack protease machineries, it can enter cells. This may assist viral spreading from primary infection site to distant organ, especially in comorbid patients to mediate organ failure, and (b) when this virus–sACE2 complex with a large number of bound sACE2 is internalized to host cell, rapid decrease in extracellular sACE2 and mACE2 may create serious imbalance in host RAS homeostasis.
5.2. S protein–ACE2–Ang II triad in using therapeutic sACE2 and vaccination
The published reports claim and hypothesize that therapeutic sACE2 (hACE2) or engineered sACE2 decrease Ang II and block viral entry in treated patients or in cellular experiments. 85 , 86 , 87 , 88 We partially disagree with this claim regarding the reduction of virus entry. As native and therapeutic sACE2 both retain Ang II catalytic activity in addition to virus RBD binding, it is obvious that therapeutic sACE2 can also catalyse Ang II. Experiment with retrovirus pseudo typed with the SARS‐CoV S protein showed successful precipitation of virus both with catalytic‐efficient and ‐deficient variants of sACE2, 89 indicating that sACE2 can catalyse Ang II in its virus‐bound form. However, regarding inhibition of viral entry, as comorbid patients have more sACE2, native sACE2 should alleviate viral entry by covering SARS‐CoV‐2 and preventing cellular entry. In addition, continued Ang II conversion will protect Ang II mediated severe cellular fates. However, the opposite is observed, as comorbid COVID‐19 patients suffer more severe outcome including death.
Zoufaly et al. 86 reported intravenous infusion of recombinant sACE2 to a single patient having comorbidity for 7 days and reported decrease of Ang II and virus in serum. However, they were uncertain whether the reduction of viral load was due to sACE2 or natural course of disease. This case report of only a single patient and lack of control parameters limited the findings. In another report, Monteil et al. 87 used kidney organoids to show that clinical grade sACE2 can block SARS‐CoV‐2 entry in cell lysates. However, they mentioned two limitations, not using lung organoids and their set‐up reflected early‐stage infection only. In addition, the organoids could not mimic the host cells with mACE2/sACE2 for which many questions remain unanswered. In all in vitro experiments, sACE2 was pre‐incubated with SARS‐CoV‐2 and then the complex was incubated with competent cell lines. 85 , 87 , 88 This does not mimic real competition between mACE2 and native or therapeutic sACE2 for virus binding. Thus due to pre‐incubation with sACE2, saturated virus cannot afford to bind mACE2, and subsequent endocytosis is inhibited. We consider the short half‐life of native sACE2, relative low amount and its host cell entry as virus–sACE2 complex might be the reason why comorbid patients fail to gain protection. In case of therapeutic sACE2, additional infusions of sACE2 refill the lost sACE2 and help to balance RAS transiently. In case of comorbid patients, internalized virus–sACE2 replication pattern is unexplained; at least the extracellular elevation of Ang II due to ACE2 down regulation (both mACE2 and sACE2) is thought to worsen the prognosis.
The impact of mACE2 and sACE2 down regulation in vaccinated individuals can also be explained in the light of the above discussion. All the vaccines under clinical and preclinical trials have RNA‐devoid virus S protein as final derivative to elicit immune response inside our body. 90 SARS‐CoV and SARS‐CoV‐2 S protein alone or expressed in pseudovirus vehicle showed to bind and internalize mACE2 but cannot replicate due to the absence of its RNA. 5 , 12 , 49 , 69 Accordingly, all vaccine ingredients can potentially internalize mACE2 upon binding and trigger mACE2 shedding by ADAM17 to release sACE2. Inactivated or weakened vaccines or vaccines with S protein expressed on pseudovirus or virus‐like particles can bind the available sACE2. Partially saturated vaccine–sACE2 complex can bind to mACE2, and this internalization may reduce the overall ACE2 availability. This will raise Ang II level in the same way as original virus can do and pose additional risk to comorbid and older individuals. Healthy individuals may overcome this vaccine‐induced transient Ang II rise as they do in viral infection. However, with already‐elevated sACE2 level, older and comorbid individual taking vaccine may face sudden Ang II spike mediated harmful effects which need to be carefully observed.
5.3. ACEIs, ARBs and Ang II interaction in COVID‐19 comorbid patients
COVID‐19 severity is more prevalent among patients with the existing comorbidities who use ACE inhibitors (ACEIs) and angiotensin receptor blockers (ARBs). 91 , 92 In the initial phase of COVID‐19, the use of ACEIs and ARBs were thought to be connected with COVID‐19 severity. However, a very large cohort data with 8.2 million participants demonstrated no independent association of using ACEIs and ARBs to COVID‐19 severity. 93 Dalan et al. 91 mentioned that with ageing and diseases the ACE–Ang‐II–AT1R axis takes control over the ACE2–Ang (1–7)–MasR axis. SARS‐CoV‐2 infection further reduces ACE2 and the former axis mediated prognosis takes place. The use of ACEIs and ARBs shift the balance towards beneficial axis as Ang II production is inhibited and AT1R mediated harmful cascade cannot progress. It was thought that the use of ACEIs and ARBs also upregulates ACE2 and could be linked with viral entry‐mediated severity. 91 Critical review of published literature showed no significant overexpression of ACE2 using these drugs. 94 As mentioned earlier, Ang II can mediate lysosomal degradation of mACE2 through AT1R. 45 COVID‐19‐infected comorbid patients with the existing low mACE2 face further reduction of mACE2 by virus‐mediated endocytosis resulting upsurge in Ang II, which in turn triggers another round of mACE2 reduction via AT1R‐mediated lysosomal degradation. Thus the use of ARBs and ACEIs can reduce Ang II formation and AT1R‐mediated mACE2 loss can be stopped. This rescued ACE2 was erroneously considered as ACE2 overexpression as a result of using ARBs and ACEIs. Thus, in comorbid patients, both Ang II and virus are able to downregulate ACE2 shifting the RAS balance to harmful end. The pathological deleterious events are possibly not primarily due to ACE2‐mediated viral entry, rather loss of overall ACE2 and increased Ang II could be the main player behind COVID‐19 catastrophe. Thus, any therapeutic which can reduce the level of Ang II can exert beneficial effects.
6. SUGGESTED FUTURE WORKS
The interconnection between SARS‐CoV‐2, mACE2, sACE2 and Ang II seems to play an important role in COVID‐19 severity in comorbid and older patients. Thus, in vitro and in vivo experiments should be designed to monitor their level. In therapeutic sACE2 and vaccines, virus‐blocking capacity and antibody titre are usually monitored, respectively. However, the level of endocytosis, change of sACE2/mACE2 and Ang II, and extent of filopodia‐driven macropinosome formation need to be evaluated side by side. This will help to formulate safe and effective therapeutic intervention for comorbid group maintaining RAS balance with reduced chance of severity.
7. CONCLUSION
In this review, we propose and discuss possible mechanisms by which virus–sACE2 complex can interact with host cell in comorbid COVID‐19 patients. The interaction can spread virus to distant organ and create sACE2 and mACE2 depletion with negative impact on RAS by increasing Ang II. We further discussed how the mechanism might affect the use of therapeutic sACE2, ACEIs, ARBs and vaccines. These considerations need to be addressed and checked experimentally to understand the precise role of sACE2 in vitro and in vivo. This will hopefully facilitate the design of safe and functional vaccines and therapeutic sACE2 for comorbid people who are at high risk of COVID‐19 infection and severity.
DECLARATION
The first working version of this manuscript has been submitted to SSRN preprint server (https://ssrn.com/abstract=3729704 or http://dx.doi.org/10.2139/ssrn.3729704)
DATA SHARING AND ACCESSIBILITY
Data sharing is not applicable to this article as no new data were created or analysed in this study.
CONFLICT OF INTEREST
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Study concept and design: Mohammad M. Rahman and Asif Ahmed. Literature search and interpretation: Mohammad M. Rahman, Maruf Hasan, and Asif Ahmed. Drafting of the manuscript: Mohammad M. Rahman and Asif Ahmed. Critical revision and editing: Mohammad M. Rahman, Maruf Hasan, and Asif Ahmed. Figure illustration and formatting: Asif Ahmed.
ACKNOWLEDGEMENT
The authors would like to thank Dr. Md. Abdullah Al Kafi (D Card, BSMMU), Assistant Registrar, Department of Cardiology, Sher‐E‐Bangla Medical College Hospital, Barishal, Bangladesh, for his thoughtful intermittent discussion during the review work.
Rahman MM, Hasan M, Ahmed A. Potential detrimental role of soluble ACE2 in severe COVID‐19 comorbid patients. Rev Med Virol. 2021;31(5):e2213. doi: 10.1002/rmv.2213
REFERENCES
- 1. Davidson AM, Wysocki J, Batlle D. Interaction of SARS‐CoV‐2 and other coronavirus with ACE (angiotensin‐converting enzyme)‐2 as their main receptor: therapeutic implications. Hypertension. 2020;76(5):1339‐1349. 10.1161/HYPERTENSIONAHA.120.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferreira‐Duarte M, Estevinho MM, Duarte‐Araujo M, Magro F, Morato M. Unraveling the role of ACE2, the binding receptor for SARS‐CoV‐2, in inflammatory bowel disease. Inflamm Bowel Dis. 2020;26(12):1787‐1795. 10.1093/ibd/izaa249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gross S, Jahn C, Cushman S, Bar C, Thum T. SARS‐CoV‐2 receptor ACE2‐dependent implications on the cardiovascular system: from basic science to clinical implications. J Mol Cell Cardiol. 2020;144:47‐53. 10.1016/j.yjmcc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631‐637. 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamming I, Cooper ME, Haagmans BL, et al. The emerging role of ACE2 in physiology and disease. J Pathol. 2007;212(1):1‐11. 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zamai L. The Yin and Yang of ACE/ACE2 pathways: the rationale for the use of renin‐angiotensin system inhibitors in COVID‐19 patients. Cells. 2020;9(7):1704. 10.3390/cells9071704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hrenak J, Paulis L, Simko F. Angiotensin A/alamandine/MrgD axis: another clue to understanding cardiovascular pathophysiology. Int J Mol Sci. 2016;17(7):1098. 10.3390/ijms17071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mascolo A, Scavone C, Rafaniello C, et al. Renin‐angiotensin system and coronavirus disease 2019: a narrative review. Front Cardiovasc Med. 2020;7:143. 10.3389/fcvm.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Turner AJ. ACE2 cell biology, regulation, and physiological functions. In: Unger T, Ulrike M, Steckelings UM, eds. The Protective Arm of the Renin Angiotensin System (RAS): Functional Aspects and Therapeutic Implications. London, UK: Academic Press; 2015:185‐189. [Google Scholar]
- 11. Volpe M, Rubattu S. Angiotensinogen and angiotensins. In: Huhtaniemi I, Martini L, eds. Encyclopedia of Endocrine Diseases. 2nd ed. London, UK: Academic Press; 2019:483‐489. [Google Scholar]
- 12. Heurich A, Hofmann‐Winkler H, Gierer S, Liepold T, Jahn O, Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88(2):1293‐1307. 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mukerjee S, Gao H, Xu J, Sato R, Zsombok A, Lazartigues E. ACE2 and ADAM17 interaction regulates the activity of presympathetic neurons. Hypertension. 2019;74(5):1181‐1191. 10.1161/HYPERTENSIONAHA.119.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zipeto D, Palmeira JDF, Arganaraz GA, Arganaraz ER. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID‐19. Front Immunol. 2020;11:576745. 10.3389/fimmu.2020.576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS‐CoV‐2. Front Cell Infect Microbiol. 2020;10:317. 10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zemlin AE, Wiese OJ. Coronavirus disease 2019 (COVID‐19) and the renin‐angiotensin system: a closer look at angiotensin‐converting enzyme 2 (ACE2). Ann Clin Biochem. 2020;57(5):339‐350. 10.1177/0004563220928361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glowacka I, Bertram S, Herzog P, et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84(2):1198‐1205. 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H, Yang P, Liu K, et al. SARS coronavirus entry into host cells through a novel clathrin‐ and caveolae‐independent endocytic pathway. Cell Res. 2008;18(2):290‐301. 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holman N, Knighton P, Kar P, et al. Risk factors for COVID‐19‐related mortality in people with type 1 and type 2 diabetes in England: a population‐based cohort study. Lancet Diabetes Endocrinol. 2020;8(10):823‐833. 10.1016/S2213-8587(20)30271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanyaolu A, Okorie C, Marinkovic A, et al. Comorbidity and its impact on patients with COVID‐19. SN Compr Clin Med. 2020:1‐8. 10.1007/s42399-020-00363-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wallentin L, Lindback J, Eriksson N, et al. Angiotensin‐converting enzyme 2 (ACE2) levels in relation to risk factors for COVID‐19 in two large cohorts of patients with atrial fibrillation. Eur Heart J. 2020:41:4037‐4046. 10.1093/eurheartj/ehaa697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benton DJ, Wrobel AG, Xu P, et al. Receptor binding and priming of the spike protein of SARS‐CoV‐2 for membrane fusion. Nature. 2020;588(7837):327‐330. 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir Res. 2020;178:104792. 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roshanravan N, Ghaffari S, Hedayati M. Angiotensin converting enzyme‐2 as therapeutic target in COVID‐19. Diabetes Metab Syndr. 2020;14(4):637‐639. 10.1016/j.dsx.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pang X, Cui Y, Zhu Y. Recombinant human ACE2: potential therapeutics of SARS‐CoV‐2 infection and its complication. Acta Pharmacol Sin. 2020;41(9):1255‐1257. 10.1038/s41401-020-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muslim S, Nasrin N, Alotaibi FO, et al. Treatment options available for COVID‐19 and an analysis on possible role of combination of rhACE2, angiotensin (1‐7) and angiotensin (1‐9) as effective therapeutic measure. SN Compr Clin Med. 2020;2(10):1761‐1766. 10.1007/s42399-020-00407-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basit A, Ali T, Rehman SU. Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS‐CoV‐2 spike glycoprotein and potent COVID‐19 therapeutic agent. J Biomol Struct Dyn. 2020:1‐10. 10.1080/07391102.2020.1768150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mahmudpour M, Roozbeh J, Keshavarz M, Farrokhi S, Nabipour I. COVID‐19 cytokine storm: the anger of inflammation. Cytokine. 2020;133:155151. 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kai H, Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evidence and insights into COVID‐19. Hypertens Res. 2020;43(7):648‐654.2020/07/01. 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sparks MA, Crowley SD, Gurley SB, Mirotsou M, Coffman TM. Classical renin‐angiotensin system in kidney physiology. Compr Physiol. 2014;4(3):1201‐1228. 10.1002/cphy.c130040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu CH, Mohammadmoradi S, Chen JZ, Sawada H, Daugherty A, Lu HS. Renin‐angiotensin system and cardiovascular functions. Arterioscler Thromb Vasc Biol. 2018;38(7):e108‐e116. 10.1161/ATVBAHA.118.311282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu H, Cassis LA, Kooi CW, Daugherty A. Structure and functions of angiotensinogen. Hypertens Res. 2016;39(7):492‐500. 10.1038/hr.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zores F, Rebeaud ME. COVID and the renin‐angiotensin system: are hypertension or its treatments deleterious? Front Cardiovasc Med. 2020;7:71. 10.3389/fcvm.2020.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu P, Sriramula S, Lazartigues E. ACE2/ANG‐(1‐7)/Mas pathway in the brain: the axis of good. Am J Physiol Regul Integr Comp Physiol. 2011;300(4):R804‐R817. 10.1152/ajpregu.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Donoghue M, Hsieh F, Baronas E, et al. A novel angiotensin‐converting enzyme‐related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1‐9. Circ Res. 2000;87(5):E1‐E9. 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 37. Wang W, McKinnie SM, Farhan M, et al. Angiotensin‐converting enzyme 2 metabolizes and partially inactivates pyr‐apelin‐13 and apelin‐17: physiological effects in the cardiovascular system. Hypertension. 2016;68(2):365‐377. 10.1161/HYPERTENSIONAHA.115.06892. [DOI] [PubMed] [Google Scholar]
- 38. Keidar S, Kaplan M, Gamliel‐Lazarovich A. ACE2 of the heart: from angiotensin I to angiotensin (1‐7). Cardiovasc Res. 2007;73(3):463‐469. 10.1016/j.cardiores.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 39. Chamsi‐Pasha MA, Shao Z, Tang WH. Angiotensin‐converting enzyme 2 as a therapeutic target for heart failure. Curr Heart Fail Rep. 2014;11(1):58‐63. 10.1007/s11897-013-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS‐CoV‐2 infection. Eur J Intern Med. 2020;76:14‐20. 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu P, Derynck R. Direct activation of TACE‐mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor‐dependent cell proliferation. Mol Cell. 2010;37(4):551‐566. 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scott AJ, O'Dea KP, O'Callaghan D, et al. Reactive oxygen species and p38 mitogen‐activated protein kinase mediate tumor necrosis factor alpha‐converting enzyme (TACE/ADAM‐17) activation in primary human monocytes. J Biol Chem. 2011;286(41):35466‐35476. 10.1074/jbc.M111.277434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Patel VB, Clarke N, Wang Z, et al. Angiotensin II induced proteolytic cleavage of myocardial ACE2 is mediated by TACE/ADAM‐17: a positive feedback mechanism in the RAS. J Mol Cell Cardiol. 2014;66:167‐176. 10.1016/j.yjmcc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 44. Xu J, Sriramula S, Xia H, et al. Clinical relevance and role of neuronal AT1 receptors in ADAM17‐mediated ACE2 shedding in neurogenic hypertension. Circ Res. 2017;121(1):43‐55. 10.1161/CIRCRESAHA.116.310509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deshotels MR, Xia H, Sriramula S, Lazartigues E, Filipeanu CM. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin II type I receptor‐dependent mechanism. Hypertension. 2014;64(6):1368‐1375. 10.1161/HYPERTENSIONAHA.114.03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bradding P, Richardson M, Hinks TSC, et al. ACE2, TMPRSS2, and furin gene expression in the airways of people with asthma‐implications for COVID‐19. J Allergy Clin Immunol. 2020;146(1):208‐211. 10.1016/j.jaci.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. Apr 7. 2009;106(14):5871‐5876. 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. NCBI. Conserved Protein Domain Family: SARS‐CoV‐2_Spike_S1_RBD. https://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=cd21480 [Google Scholar]
- 49. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11(1):1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell. 2020;181(4):894‐904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci. U. S. A. 2020;117(21):11727‐11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258(1):1‐20. 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 53. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23(6):1–5. 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29‐57. 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221‐224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐292 e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol Sin. 2020;41(9):1141‐1149. 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2020. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffmann M, Kleine‐Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779‐784. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Teuchert M, Berghofer S, Klenk HD, Garten W. Recycling of furin from the plasma membrane. Functional importance of the cytoplasmic tail sorting signals and interaction with the AP‐2 adaptor medium chain subunit. J Biol Chem. 1999;274(51):36781‐36789. 10.1074/jbc.274.51.36781. [DOI] [PubMed] [Google Scholar]
- 62. Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two‐step, furin‐mediated activation of the spike protein. Proc Natl Acad Sci U. S. A. 2014;111(42):15214‐15219. 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1‐10. 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vaarala MH, Porvari K, Kyllonen A, Lukkarinen O, Vihko P. The TMPRSS2 gene encoding transmembrane serine protease is overexpressed in a majority of prostate cancer patients: detection of mutated TMPRSS2 form in a case of aggressive disease. Int J Cancer. 2001;94(5):705‐710. 10.1002/ijc.1526. [DOI] [PubMed] [Google Scholar]
- 65. Afar DE, Vivanco I, Hubert RS, et al. Catalytic cleavage of the androgen‐regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61(4):1686‐1692. [PubMed] [Google Scholar]
- 66. Wilson S, Greer B, Hooper J, et al. The membrane‐anchored serine protease, TMPRSS2, activates PAR‐2 in prostate cancer cells. Biochem J. 2005;388(Pt 3):967‐972. 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Geybels MS, McCloskey KD, Mills IG, Stanford JL. Calcium channel blocker use and risk of prostate cancer by TMPRSS2:ERG gene fusion status. Prostate. 2017;77(3):282‐290. 10.1002/pros.23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. NCBI . Transmembrane protease serine 2 isoform 1 [homo sapiens]. 2020. https://www.ncbi.nlm.nih.gov/protein/NP_001128571.1. Accessed 6 November 2020. [Google Scholar]
- 69. Glowacka I, Bertram S, Muller MA, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85(9):4122‐4134. 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. KEGG . Vascular smooth muscle contraction. 2020. https://www.kegg.jp/kegg-bin/show_pathway?ko04270+K04166. Accessed 4 November 2020. [Google Scholar]
- 71. Bagur R, Hajnoczky G. Intracellular Ca(2+) sensing: its role in calcium homeostasis and signaling. Mol Cell. 2017;66(6):780‐788. 10.1016/j.molcel.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Meyer D, Sielaff F, Hammami M, Bottcher‐Friebertshauser E, Garten W, Steinmetzer T. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem J. 2013;452(2):331‐343. 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]
- 73. Ragia G, Manolopoulos VG. Inhibition of SARS‐CoV‐2 entry through the ACE2/TMPRSS2 pathway: a promising approach for uncovering early COVID‐19 drug therapies. Eur J Clin Pharmacol. 2020;76(12):1623–1630. 10.1007/s00228-020-02963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xia S, Zhu Y, Liu M, et al. Fusion mechanism of 2019‐nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17(7):765‐767. 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bayati A, Kumar R, Francis V, McPherson PS. SARS‐CoV‐2 uses clathrin‐mediated endocytosis to gain access into cells. BioRxiv preprint. 2020. 10.1101/2020.07.13.201509. [DOI] [Google Scholar]
- 76. Glebov OO. Understanding SARS‐CoV‐2 endocytosis for COVID‐19 drug repurposing. FEBS J. 2020;287(17):3664‐3671. 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kiani AK, Dhuli K, Anpilogov K, Bressan S, Bertelli M. Natural compounds as inhibitors of SARS‐CoV‐2 endocytosis: a promising approach against COVID‐19. Acta Biomed 2020;91. 10.23750/abm.v91i13-S.10520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xiu S, Dick A, Ju H, et al. Inhibitors of SARS‐CoV‐2 entry: current and future opportunities. J Med Chem. 2020. 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci. 2020;16(10):1724‐1731. 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Qiu Z, Hingley ST, Simmons G, et al. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike‐mediated entry. J Virol. 2006;80(12):5768‐5776. 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Regan AD, Shraybman R, Cohen RD, Whittaker GR. Differential role for low pH and cathepsin‐mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet Microbiol. 2008;132(3‐4):235‐248. 10.1016/j.vetmic.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ortiz‐Perez JT, Riera M, Bosch X, et al. Role of circulating angiotensin converting enzyme 2 in left ventricular remodeling following myocardial infarction: a prospective controlled study. PloS One. 2013;8(4):e61695. 10.1371/journal.pone.0061695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Freeman MC, Peek CT, Becker MM, Smith EC, Denison MR. Coronaviruses induce entry‐independent, continuous macropinocytosis. mBio. 2014;5(4):e01340‐14. 10.1128/mBio.01340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Caldas LA, Carneiro FA, Higa LM, et al. Ultrastructural analysis of SARS‐CoV‐2 interactions with the host cell via high resolution scanning electron microscopy. Sci Rep. 2020;10(1):16099. 10.1038/s41598-020-73162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guo L, Bi W, Wang X, et al. Engineered trimeric ACE2 binds viral spike protein and locks it in “Three‐up” conformation to potently inhibit SARS‐CoV‐2 infection. Cell Res. 2021;31(1):98–100. 10.1038/s41422-020-00438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zoufaly A, Poglitsch M, Aberle JH, et al. Human recombinant soluble ACE2 in severe COVID‐19. Lancet Respir Med. 2020;8(11):1154‐1158. 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181(4):905‐913 e7. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tada T, Fan C, Chen JS, et al. A soluble ACE2 microbody protein fused to a single immunoglobulin Fc domain is a potent inhibitor of SARS‐CoV‐2. Cell Rep. 2020:108528. 10.1016/j.celrep.2020.108528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moore MJ, Dorfman T, Li W, et al. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin‐converting enzyme 2. J Virol. 2004;78(19):10628‐10635. 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Callaway E. The race for coronavirus vaccines. Nature. 2020;580:576‐577. [DOI] [PubMed] [Google Scholar]
- 91. Dalan R, Bornstein SR, El‐Armouche A, et al. The ACE‐2 in COVID‐19: foe or friend? Horm Metab Res. 2020;52(5):257‐263. 10.1055/a-1155-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ahmadian E, Pennefather PS, Eftekhari A, Heidari R, Eghbal MA. Role of renin‐angiotensin system in liver diseases: an outline on the potential therapeutic points of intervention. Expet Rev Gastroenterol Hepatol. 2016;10(11):1279‐1288. 10.1080/17474124.2016.1207523. [DOI] [PubMed] [Google Scholar]
- 93. Hippisley‐Cox J, Young D, Coupland C, et al. Risk of severe COVID‐19 disease with ACE inhibitors and angiotensin receptor blockers: cohort study including 8.3 million people. Heart. 2020;106(19):1503‐1511. 10.1136/heartjnl-2020-317393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sriram K, Insel PA. Risks of ACE inhibitor and ARB usage in COVID‐19: evaluating the evidence. Clin Pharmacol Ther. 2020;108(2):236‐241. 10.1002/cpt.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]


