Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic has become a major public health issue worldwide. Developing and evaluating rapid and easy‐to‐perform diagnostic tests is a high priority. The current study was designed to assess the diagnostic performance of an antigen‐based rapid detection test (COVID‐VIRO®) in a real‐life setting. Two nasopharyngeal specimens of symptomatic or asymptomatic adult patients hospitalized in the Infectious Diseases Department or voluntarily accessing the COVID‐19 Screening Department of the Regional Hospital of Orléans, France, were concurrently collected. The diagnostic specificity and sensitivity of COVID VIRO® results were compared to those of real‐time reverse‐transcriptase quantitative polymerase chain reaction (RT‐qPCR) results. A subset of patients underwent an additional oropharyngeal and/or saliva swab for rapid testing. A total of 121 patients confirmed to be infected and 127 patients having no evidence of recent or ongoing infection were enrolled for a total of 248 nasopharyngeal swab specimens. Overall, the COVID‐VIRO® sensitivity was 96.7% (CI, 93.5%–99.9%). In asymptomatic patients, symptomatic patients having symptoms for more than 4 days and those with an RT‐qPCR cycle threshold value ≥ 32, the sensitivities were 100%, 95.8%, and 91.9%, respectively. The concordance between RT‐qPCR and COVID VIRO® rapid test results was 100% for the 127 patients with no SARS‐CoV‐2 infection. The COVID‐VIRO® test had 100% specificity and sensitivity greater than 95%, which are better than the recommendations set forth by the WHO (specificity ≥ 97%–100%, sensitivity ≥ 80%). These rapid tests may be particularly useful for large‐scale screening in emergency departments, low‐resource settings, and airports.
Keywords: chromatographic techniques, local infection/replication/spread, pathogenesis, research and analysis methods, SARS coronavirus, virus classification
1. INTRODUCTION
At the end of 2019, a pneumonia of initially unknown origin was first reported to the World Health Organization (WHO) Country Office in China. On January 9th, 2020, the Chinese health authorities and the WHO announced the discovery of a novel coronavirus, first named 2019‐nCoV, then officially named severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). This virus, belonging to the coronavirus family but differing from SARS‐CoV‐1 and MERSCoV, is responsible for upper/lower respiratory tract infections known as coronavirus disease 2019 (COVID‐19). The COVID‐19 incubation period is approximately 5.2 days, and the most common onset symptoms are fever, cough, and fatigue. 1 Since SARS‐CoV‐2 emerged in China, it has become a major public health issue worldwide. To date, more than 40 million cases have been detected worldwide, 2 and the pandemic continues to spread unabated.
Minimizing testing delay seems to have the largest impact on reducing onward transmissions, 3 and the availability of highly sensitive and specific tests is essential to quickly identify new cases and contain virus transmission.
Currently, the real‐time reverse‐transcriptase quantitative polymerase chain reaction (RT‐qPCR) assay is the gold standard method to detect SARS‐CoV‐2 RNA in respiratory specimens such as nasopharyngeal or oropharyngeal swabs or bronchoalveolar lavage. 4 However, performing RT‐qPCR is expensive, time‐consuming, and requires special equipment and qualified operators. Faster, cheaper, and easier‐to‐use alternative tools could be represented by novel antigen‐based rapid detection tests or point‐of‐care tests (POCTs). 5
Recently, the WHO approved the first rapid detection test for large‐scale use in low‐ and middle‐income countries, 6 and French health regulation authorities authorized their use in medical settings. 7
Several different POCTs have already been developed, 8 with generally high specificity but variable sensitivity. 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 COVID‐VIRO® (AAZ) is one of the novel immunochromatographic tests designed to detect SARS‐CoV‐2 antigen in nasopharyngeal specimens with that of RT‐qPCR as a reference test is the principal aim of the current study.
2. MATERIALS AND METHODS
2.1. Ethical approval
This study was approved by the Regional North West Ethics and Research Committee. Written informed consent was obtained from each participant.
2.2. Study population
People voluntarily accessing the COVID‐19 Screening Department as well as subjects who tested SARS‐CoV‐2 positive in the previous 5 days and SARS‐CoV‐2 positive patients hospitalized in the Infectious Diseases Department of the Centre Hospitalier Régional (CHR) of Orléans, France, or Drouot Laboratory, Paris, France, from October 12th, 2020, to October 25th, 2020, were included in the study. The diagnosis of SARS‐CoV‐2 infection was confirmed in case of positivity of the specific RT‐qPCR on nasopharyngeal swabs, in accordance with current recommendations. Subjects who recently tested positive for SARS‐CoV‐2 at the COVID‐19 Screening Department were recontacted and retested within 5 days. The inclusion criteria were age ≥ 18 years old and agreement to undergo two concurrent nasopharyngeal swabs for RT‐qPCR and COVID‐VIRO® analysis. Patient age was collected at inclusion, as well as symptom onset date for symptomatic patients. Suggestive symptoms were headache, fatigue, fever, or upper or lower respiratory symptoms. Asymptomatic patients were defined as those not reporting any of these symptoms.
2.3. Case definition
All SARS‐CoV‐2‐positive subjects had confirmed RT‐qPCR positivity within a maximum of 5 days before study sampling and were then retested in parallel with the rapid test.
SARS‐CoV‐2‐negative subjects were patients having a negative RT‐qPCR at the time of inclusion without any previous positive test.
2.4. Specimen collection
Paired nasopharyngeal swabs were obtained for each patient by trained healthcare personnel (nurses, doctors, or biologists). The collection of the two simultaneous samples was always carried out by the same operator.
A polyester‐tipped flexible (viral transport medium tube with swab VTM, Sun‐Trine®) was inserted into two of the nostrils until resistance was felt at the nasopharynx, rotated six times, and withdrawn. After swabbing, the swab applicator was cut off. The first absorbent swab was placed into a vial containing 3 ml of inactivating viral transport media and was immediately transferred to the Virology Unit of the CHR of Orléans Hospital, Orléans, or Drouot Laboratory, Paris, to perform RT‐qPCR. The rapid antigen test was immediately performed on‐site with the second absorbent swab.
An additional oropharyngeal and/or saliva swab specimen was simultaneously collected in a subset of positive patients to determine the diagnostic reliability of these samples in comparison to nasopharyngeal swab specimens. Oropharyngeal specimens were collected on both sides of the tonsillar arches and posterior pharynx. Saliva specimens were collected by swabbing the upper and lower gingiva twice from back to front.
2.5. Real‐time RT‐qPCR assays for the detection of SARS‐CoV‐2 RNA
Nucleic acid extraction was performed with an automated Sample Preparation System MGISP‐960 (MGI). Specific real‐time RT‐qPCR assays targeted three SARS‐CoV‐2 genes, namely, the ORF1ab, S, and N genes (TaqPath Covid‐19 Multiplex RT‐PCR, Thermo Fisher Scientific). Genome amplification was performed using QuantStudio5 (Applied Biosystems). The interpretation of the results was performed according to the manufacturer's instructions. The assay includes an RNA internal extraction control and an amplification control. Samples showing an exponential growth curve and a cycle threshold (C t) value < 37 were considered positive. A unique C t value > 37 was considered negative.
2.6. Rapid antigen test
COVID‐VIRO® (AAZ) is a membrane‐based immunochromatography assay detecting SARS‐CoV‐2 nucleocapsid antigen (N‐protein) in nasopharyngeal samples through monoclonal antibodies. A second monoclonal antibody is conjugated to colloidal gold particles, which are captured in the reaction membrane. The test was performed according to the manufacturer's instructions by mixing nasopharyngeal secretions with 300 µl of dilution buffer in a tube. Then, four drops were added to the appropriate well. When nasopharyngeal secretions cross the strip, passive diffusion allows the solubilized conjugate to migrate with the sample and react with the anti‐SARS‐CoV‐2 antibodies immobilized on the membrane. A control line allows the correct migration of the sample and the reliability of the test to be assessed. Visual interpretation of the results was performed 15 min later (Figure 1).
Figure 1.
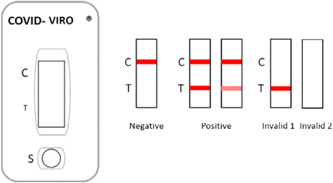
Interpretation of results for COVID‐VIRO® (AAZ, LMB)
2.7. Data analysis
Population characteristics are reported as percentages, mean and median values, standard deviations, and ranges. Data were analyzed in the Infectious Diseases Department.
To determine the diagnostic value of the COVID‐VIRO®, the study population was stratified into two groups:
-
1.
RT‐qPCR‐positive patients were already confirmed at the time of inclusion. A comparison between RT‐qPCR and COVID‐VIRO® results in these patients was used to assess diagnostic test sensitivity.
-
2.
Nonselected symptomatic or asymptomatic patients voluntarily accessed the COVID‐19 Screening Department to detect possible SARS‐CoV‐2 infection. To analyze the data, RT‐qPCR‐positive patients were added to the first group to assess diagnostic test sensitivity. Conversely, RT‐qPCR‐negative patients were selected to quantify the specificity of the rapid test.
The specificity and sensitivity of the COVID‐VIRO® were calculated using the RT‐qPCR results as reference tests, according to the following formulas:
Specificity (%) = 100 × [negative/(negative + positive)], sensitivity (%) = 100 × [positive/(positive + negative)].
The size of the study population was calculated on the basis of a 95% sensitivity with a lower margin of the confidence interval over 91% and a 99% specificity with a lower margin over 95% according to the WHO recommendations for antigenic rapid testing. The student's t‐test was used to compare means.
3. RESULTS
A total of 121 SARS‐CoV‐2‐positive patients and 127 patients with no evidence of recent or ongoing SARS‐CoV‐2 infection were enrolled in the study. A total of 248 couples of nasopharyngeal swabs were analyzed. Of these, 228 were collected in Orléans, and 20 in Paris.
The sex ratio of the study population was 0.9 (117 men and 131 women). The median and mean ages were 38 and 43 years old, respectively (range, 18–96). One patient exhibiting only one positive target (gene S; C t, 36) at RT‐qPCR analysis was considered negative according to the French Society of Microbiology criteria and the extraction kit instructions, and he was therefore included in the group of SARS‐CoV‐2‐negative patients. Ninety‐seven patients were symptomatic, and 24 were totally asymptomatic. No data about household or nonhousehold close contacts were available.
Among the 121 patients diagnosed SARS‐CoV‐2 positive, 17 were hospitalized (14%), and 97 (80.1%) were symptomatic. The median time of symptom duration before sampling was 5 days (mean, 6 days; range, 1–20).
The N gene mean C t value was 25 (range, 15–34) in asymptomatic SARS‐CoV‐2‐positive patients and 27 (range, 13–35) in symptomatic positive patients. The difference was not significant (p > .05). In patients who tested positive within 0 to 4 days from symptom onset, the N gene mean C t value was 25 (range, 13–35), versus 28 (range, 18–33) in those tested within 5 and 7 days and 30 (range, 21–35) in those tested after more than 7 days, showing a constant decrease in viral carriage. Although some RT‐qPCR false‐negative patients had a higher C t value (suggestive of a lower RNA carriage), COVID‐VIRO® was able to detect the antigen in 20 patients who had been reporting symptoms for more than 7 days.
Among the 121 RT‐qPCR‐positive patients, 4 had a negative COVID‐VIRO® result (3.3% false negative). The overall sensitivity of our POCT was estimated at 96.7% (IC, 93.5%–99.9%) (Table 1). Among the 24 asymptomatic patients, no COVID‐VIRO® false‐negative results were reported.
Table 1.
Performance of the COVID‐VIRO® antigenic rapid test in the overall population and in the group of asymptomatic SARS‐CoV‐2‐infected patients
| COVID‐VIRO® | COVID‐VIRO® performances | ||||
|---|---|---|---|---|---|
| Positive | Negative | Sensitivity (%, 95% CI) | Specificity (%) | ||
| 96.7% (93.3%–99.9%) | Positive RT‐PCR | 117 | 4 | ||
| Negative RT‐PCR | 0 | 127 | 100% | ||
| Asymptomatic COVID‐19 + patients | Positive RT‐PCR | 24 | 0 | 100% | |
Table 2 shows the COVID‐VIRO® performances according to the C t value and the delay of symptom onset. COVID‐VIRO® sensitivity was extremely high among patients having Gene N, S or ORF C t values ≥ 32, considering that 34 of 37 patients tested positive □ sensitivity: 91.9% (95% CI, 83.1%–100%).
Table 2.
Sensitivity of the COVID‐VIRO® antigenic rapid test in comparison to reverse‐transcriptase polymerase chain reaction (RT‐PCR) according to viral carriage and delay from symptom onset
| COVID‐VIRO® | ||||
|---|---|---|---|---|
| Positive | Negative | Sensitivity, % (95% CI) | ||
| Patients with high viral carriage (C t ≤ 28) | Positive RT‐PCR | 65 | 1 | 98.5% (95.5%–100%) |
| Patients with high and moderate viral carriage (C t < 32) | Positive RT‐PCR | 83 | 1 | 98.8% (96.5%–100%) |
| Patients with low viral carriage (C t ≥ 32) | Positive RT‐PCR | 34 | 3 | 91.9% (83.1%–100%) |
| Delay from symptoms onset ≤ 4 days | Positive RT‐PCR | 34 | 2 | 94.4% (87.0%–100%) |
| Delay from symptoms onset > 4 days | Positive RT‐PCR | 60 | 2 | 96.8% (92.4%–100%) |
Table 3 reports the characteristics of the four COVID‐VIRO® false‐negative cases. Three out of four had C t values ≥ 32 and were considered noncontagious, regardless of the date of onset of symptoms.
Table 3.
Characteristics of the four discordant positives reverse‐transcriptase polymerase chain reaction (RT‐PCR) negative COVID‐VIRO®. No patients were hospitalized
| S, N, ORF | |||||
|---|---|---|---|---|---|
| Gene | |||||
| Patient | Age range | Sex | Symptoms | C t, respectively | Delay from symptom onset |
| #1 | 25 | F | Yes | 31, 34, 31 | 5 |
| #2 | 29 | M | Yes | 32, 32, 31 | 1 |
| #3 | 65 | F | Yes | Neg, 32, 32 | 16 |
| #4 | 46 | M | Yes | 32, 28, 28 | 3 |
Twenty positive patients with a previous positive RT‐qPCR tested negative when the second RT‐qPCR was simultaneously performed with the POCT. All of these had a positive COVID‐VIRO® result (mainly weak or very weak line), suggesting that it may still be positive some days after PCR negativization.
Among the 127 patients with no SARS‐CoV‐2 infection, no false‐positive result was observed, as the concordance between RT‐qPCR and COVID‐VIRO® was 100%. COVID‐VIRO® specificity is therefore estimated at 100%.
Additionally, 48 patients with a positive COVID‐VIRO® test on nasopharyngeal swab specimens underwent simultaneous oropharyngeal (34 patients) or saliva swabs (14 patients). COVID‐VIRO® turned positive in 24 of 34 and 0 out of 14 patients on oropharyngeal and saliva specimens, respectively. The sensitivity was 70.6% for oropharyngeal specimens and 0% for saliva specimens.
4. DISCUSSION
This observational study aimed to evaluate the diagnostic performance of a POCT developed to detect SARS‐CoV‐2 antigens from a nasopharyngeal swab directly after sampling and provide the result within 15 min. The diagnostic value of COVID‐VIRO® was determined using RT‐qPCR as the gold standard in a community setting. In the current study, the COVID‐VIRO® sensitivity and specificity were 96.7% and 100%, respectively, with no observed false positives. Although the results should be evaluated after 15 min, positive results almost always appeared within the first five minutes and often within one minute.
To date, several studies have evaluated the diagnostic performance of POCTs in real life, yielding conflicting data. 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 In general, the sensitivity ranged from 60.8% to 79.6%. 13 , 14 , 15 , 16 , 17 , 18 Occasionally, significantly high sensitivity (93.9% and 98.3%) has been reported. 19 , 20 , 21
Concerning the Panbio COVID‐19 Ag Rapid test (Abbott), the manufacturer reported high sensitivity (93.3%; 95% CI, 83.8–98.2) in a highly endemic setting in Brazil, 22 but other independent cohort studies have not confirmed these data. In 257 symptomatic and asymptomatic patients enrolled at the Emergency Department and Primary Health Care Setting in Spain, the overall sensitivity was 73.3%, reaching 86.5% among patients having symptoms for less than seven days. 11 In another multicentric study performed on 200 SARS‐CoV‐2‐positive patients, the POCT sensitivity was 72.6% (95% CI, 64.5%–79.9%) in the Netherlands and 81.0% (95% CI, 69.0%–89.8%) in Aruba. The test sensitivity was as high as 95.2% (95% CI, 89.3%–98.5%) in patients with RT‐qPCR test positivity for C t values < 32. 23
In our study, stratifying patients by the C t value, COVID‐VIRO® sensitivity remained extremely high even for C t values > 32 (96.9%; 95% CI, 91.1%–100%).
Detection of viral RNA in nasopharyngeal samples is not necessarily related to infectiousness. 24 Several factors determine the risk of viral transmission: the viability of a virus, the amount of viral replication estimated by the C t, the presence of respiratory symptoms, the individual's local mucosal immune response to the virus, and the behavior of the infected individual and their close contacts. 25 However, in the present study, the number of viral particles estimated by the C t value did not differ in asymptomatic and symptomatic SARS‐CoV‐2‐infected patients. COVID‐VIRO® appears to be as sensitive as RT‐qPCR to detect infected patients in a limited number of asymptomatic patients.
Available data report that the RNA viral load rapidly decreases after the onset of symptoms, and infectiousness generally declines within 7–10 days. 24 , 26 , 27 , 28 , 29 Stratifying our patients by symptom duration at sampling, we observed a similar decline from Day 1 to Day 14 in the mean C t value enregistered. Only one hospitalized patient aged 91 exhibited a C t value of S, 21; N, 22; ORF, 21 at Day 10. Our POCT was able to detect the antigen both within and 4 days after the onset of symptoms.
Considering discordant results, the analytical performances depend on different factors, including viral load, quality of the specimen, and processing. The two nasopharyngeal swabs were concurrently performed by the same operator, but a greater quantity of secretions and viruses were likely to be more concentrated on the first one. Unfortunately, we cannot know the temporal order in which each swab was performed. Furthermore, waiting one hour between the two samples was infeasible.
Analyzing POCT results on oropharyngeal and saliva specimens of positive patients, we quickly realized that sensitivity dramatically drops. Even if nasopharyngeal swabs are uncomfortable for patients, this specimen should be privileged for the diagnosis of SARS‐CoV‐2 infection, whether obtained through PCR or POCT.
This study has several limitations. First, this study is not a real‐life study, as when PCR‐positive patients returned for the study, the operators knew the status of the patient. Then, operators paid more attention to the results within 15 min. This method allows us to give the real performances of the test done in the best conditions. This could not occur in real life, with many operators and emergency wards where nurses do not necessarily wait enough before reading or do not have the best conditions to read a thin line. Second, the date of symptom onset was reported by patients and may not always be accurate, leading to an inaccurate stratification of patients. Then, the number of asymptomatic patients is rather limited to obtain conclusive data, even if we did not observe any discordance between tests in this group of patients. Finally, we did not perform any parallel comparison with other POCTs.
In France, nurses, pharmaceuticals, and general practitioners have recently been authorized to perform POCTs in medical settings. 7 Data obtained from the current study could reassure health authorities in expanding access to POCTs that, in addition to being quick and easy to use, they are also reliable. In the current setting, unaware people could wait for RT‐qPCR results for several days. During this time, they could easily infect their close contacts. A more rapid diagnosis and the subsequent contact tracing would certainly positively impact the containment of transmission.
In emergency departments, POCTs could also be used to quickly recognize asymptomatic positive patients from negative patients, avoiding SARS‐CoV‐2 nosocomial infection. Furthermore, such tests would likely be useful in low‐ and middle‐income countries and at airports to limit viral spread worldwide.
5. CONCLUSION
COVID‐VIRO® (AAZ) was a reliable test for SARS‐CoV‐2 diagnosis. The diagnostic sensitivity and specificity of nasopharyngeal swabs were 96.7% (95% CI, 9%–99.9%) and 100%, respectively. To date, this is the only COVID‐19 antigenic rapid test that meets the WHO's criteria for a screening test (sensitivity ≥ 80%, specificity ≥ 97%–100%). Unfortunately, performing the test on oropharyngeal or salivary samples yields less reliable results.
The results of this study have already been mentioned by the manufacturer in the kit insert. 30
AUTHOR CONTRIBUTIONS
Experimental strategy design: L. Courtellemont, G. Pialoux, T. Prazuck. Experiments: L. Courtellemont, J. Guinard, C. Guillaume, S. Giaché, V. Rzepecki, A. Seve, C. Gubavu, K. Baud, C. Le Helloco, G.N. Cassuto, T. Prazuck. Manuscript writing: L. Courtellemont, L. Hocqueloux, T. Prazuck. Manuscript editing: L. Courtellemont, T. Prazuck.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Courtellemont L, Guinard J, Guillaume C, et al. High performance of a novel antigen detection test on nasopharyngeal specimens for diagnosing SARS‐CoV‐2 infection. J Med Virol. 2021;93:3152‐3157. 10.1002/jmv.26896
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Rothan HA, Byrareddy SN. The epiademiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. J Autoimmun. 2020;109:102433. 10.1016/j.jaut.2020.102433. PMID: 32113704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20:533‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kretzschmar ME, Rozhnova G, Bootsma MCJ, van Boven M, van de Wijgert JHHM, Bonten MJM. Impact of delays on effectiveness of contact tracing strategies for COVID‐19: a modelling study. Lancet Public Heal. 2020;5:e452‐e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gorbalenya AE, Baker SC, Baric RS, et al. Severe acute respiratory syndrome‐related coronavirus: the species and its viruses—a statement of the Coronavirus Study Group. Nat Microbiol. 2020;(5. 536–544. 10.1101/2020.02.07.937862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev. 2020;8, CD013705. 10.1002/14651858.CD013705. Accessed August 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO.com . retrieved 2020[Internet]. Available at https://www.who.int/news/item/28-09-2020-global-partnership-to-make-available-120-million-affordable-quality-covid-19-rapid-tests-for-low-and-middle-income-countries. Accessed October 26, 2020. [Google Scholar]
- 7. Arrêté du 16 octobre modifiant l'arrêté du 10 juillet 2020 prescrivant les mesures générales nécessaires pour faire face à l'épidémie de covid‐19 dans les territoires sortis de l'état d'urgence sanitaire et dans ceux où il a été prorogé – Légifrance JORF no 0253 du octobre 17, 2020.
- 8. Finddx.org . 2020. https://www.finddx.org/covid-19/pipeline/. Accessed December 23, 2020.
- 9. Hirotsu Y, Maejima M, Shibusawa M, et al. Comparison of automated SARS‐CoV‐2 antigen test for COVID‐ 19 infection with quantitative RT‐PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scohy A, Anantharajah A, Bodéus M, Kabamba‐Mukadi B, Verroken A, RodriguezVillalobos H. Low performance of rapid antigen detection test as frontline testing for COVID‐19 diagnosis. J Clin Virol. 2020;129:104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linares M, Pérez Tanoira R, Carrero A, et al. Panbio antigen rapid test is reliable to diagnose SARS‐ CoV‐2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133:104659. 10.1016/j.jcv.2020.104659. Epub October 16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenollar F, Bouam A, Ballouche M, et al. Evaluation of the panbio COVID‐19 rapid antigen detection test device for the screening of patients with COVID‐19. J Clin Microbiol. 2020;59(2):e02589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mak GCK, Lau SSY, Wong KKY, et al. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS‐CoV‐2. J Clin Virol. 2021;134:104712. 10.1016/j.jcv.2020.104712. Accessed December 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lanser L, Bellmann‐Weiler R, Öttl KW, et al. Evaluating the clinical utility and sensitivity of SARS‐CoV‐2 antigen testing in relation to RT‐PCR Ct values. Infection. 2020. 10.1007/s15010-020-01542-0. Epub ahead of print. PMID: 33185807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Albert E, Torres I, Bueno F, et al. Field evaluation of a rapid antigen test (Panbio™ COVID‐19 Ag Rapid Test Device) for COVID‐19 diagnosis in primary healthcare centers. Clin Microbiol Infect. 2020 Nov 12;S1198‐743X(20):30697‐2. 10.1016/j.cmi.2020.11.004. Epub ahead of print. PMID: 33189872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cerutti F, Burdino E, Milia MG, et al. Urgent need of rapid tests for SARS CoV‐2 antigen detection: evaluation of the SD‐Biosensor antigen test for SARS‐CoV‐2. J Clin Virol. 2020;132:104654. 10.1016/j.jcv.2020.104654. Epub September 29, 2020. PMID: 33053494; PMCID: PMC7522649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nalumansi A, Lutalo T, Kayiwa J, et al. Field evaluation of the performance of a SARS‐CoV‐2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int J Infect Dis. 2020;104:282‐286. 10.1016/j.ijid.2020.10.073. Epub ahead of print. PMID: 33130198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young S, Taylor SN, Cammarata CL, et al. Clinical evaluation of BD Veritor SARS‐CoV‐2 point‐of‐care test performance compared to PCR‐based testing and versus the Sofia 2 SARS Antigen point‐of‐care test. J Clin Microbiol. 2020;59(1):e02338. 10.1128/JCM.02338-20. Epub ahead of print. PMID: 33023911 JCM.02338‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaimayo C, Kaewnaphan B, Tanlieng N, et al. Rapid SARS‐CoV‐2 antigen detection assay in comparison with real‐time RT‐PCR assay for laboratory diagnosis of COVID‐19 in Thailand. Virol J. 2020;17(1):177. 10.1186/s12985-020-01452-5. PMID: 33187528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Porte L, Legarraga P, Vollrath V, et al. Evaluation of a novel antigen‐based rapid detection test for the diagnosis of SARS‐CoV‐2 in respiratory samples. Int J Infect Dis. 2020;99:328‐333. 10.1016/j.ijid.2020.05.098. Epub June 1, 2020. PMID: 32497809; PMCID: PMC7263236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. PANBIO™ COVID‐19 Ag Rapid Test Device . https://www.globalpointofcare.abbott/en/index.html. Accessed December 23, 2020.
- 23. Gremmels H, Winkel BMF, Schuurman R, Ritger NAM, Rodriguez O, et al. Real‐life validation of the Panbio™ COVID‐19 antigen rapid test (Abbott) in community‐dwelling subjects with symptoms of potential SARS‐CoV‐2 infection. Lancet E Clin Med. December 5, 2020. 10.1016/j.eclinm.2020.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walsh KA, Jordan K, Clyne B, et al. SARS‐CoV‐2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81(3):357‐371. 10.1016/j.jinf.2020.06.067. Accessed June 29, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. World Health Organization . Criteria for releasing COVID‐19 patients from isolation. June 17, 2020. https://www.who.int/publications/i/item/criteria-for-releasing-covid-19-patients-from-isolation
- 26. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J. Ho A. SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV viral load dynamics, duration of viral sedding, and infectiousness: a systematic review and meta‐analysis. Lancet Microbe. 2020. 10.1016/S2666-5247(20)30172-5. Accessed November 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jefferson T, Spencer E, Brassey J, Heneghan C. Viral cultures for COVID‐19 infectivity assessment. Systematic review. medRxiv. 2020:20167932, 08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park M, Pawliuk C, Nguyen T, et al. Determining the period of communicability of SARS‐CoV‐2: a rapid review of the literature, 2020. medRxiv. 2020:20163873, 07.28.
- 29. Walsh KA, Spillane S, Comber L, et al. The duration of infectiousness of individuals infected with SARS‐CoV‐2. J Infect. 2020;81(6):847‐856. 10.1016/j.jinf.2020.10.009. Accessed October 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. COVID‐VIRO® Rapid antigenic test. https://www.covid19aaz.com/en/rapide-antigenic-test/. Accessed December 23, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


