Abstract
Although vaccination campaigns are currently being rolled out to prevent coronavirus disease (COVID‐19), antivirals will remain an important adjunct to vaccination. Antivirals against coronaviruses do not exist, hence global drug repurposing efforts have been carried out to identify agents that may provide clinical benefit to patients with COVID‐19. Itraconazole, an antifungal agent, has been reported to have activity against animal coronaviruses. Using cell‐based phenotypic assays, the in vitro antiviral activity of itraconazole and 17‐OH itraconazole was assessed against clinical isolates from a German and Belgian patient infected with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Itraconazole demonstrated antiviral activity in human Caco‐2 cells (EC50 = 2.3 µM; 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide assay). Similarly, its primary metabolite, 17‐OH itraconazole, showed inhibition of SARS‐CoV‐2 activity (EC50 = 3.6 µM). Remdesivir inhibited viral replication with an EC50 = 0.4 µM. Itraconazole and 17‐OH itraconazole resulted in a viral yield reduction in vitro of approximately 2‐log10 and approximately 1‐log10, as measured in both Caco‐2 cells and VeroE6‐eGFP cells, respectively. The viral yield reduction brought about by remdesivir or GS‐441524 (parent nucleoside of the antiviral prodrug remdesivir; positive control) was more pronounced, with an approximately 3‐log10 drop and >4‐log10 drop in Caco‐2 cells and VeroE6‐eGFP cells, respectively. Itraconazole and 17‐OH itraconazole exert in vitro low micromolar activity against SARS‐CoV‐2. Despite the in vitro antiviral activity, itraconazole did not result in a beneficial effect in hospitalized COVID‐19 patients in a clinical study (EudraCT Number: 2020‐001243‐15).
Keywords: 17‐OH itraconazole, Caco‐2 cells, in vitro, itraconazole, SARS‐CoV‐2, VeroE6‐eGFP cells
Highlights
Itraconazole exerted in vitro low micromolar activity against SARS‐CoV‐2
Remdesivir demonstrated potent antiviral activity, confirming validity of the assay
Itraconazole has since shown no efficacy (study in hospitalized COVID‐19 patients)
1. INTRODUCTION
The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) disease (COVID‐19) has been declared a global pandemic. 1 , 2 SARS‐CoV‐2 is a beta‐coronavirus, which are enveloped viruses containing single‐strand, positive‐sense RNA. 3 Since COVID‐19 emerged in humans in late December 2019, 4 aside from a significant economic loss, more than one million deaths have been reported worldwide, driving urgently the need to identify potential vaccines and therapies.
Given that therapeutic options for antiviral treatment of SARS‐CoV‐2 remain limited, research has initially focused on repurposing available drugs that have demonstrated antiviral activity against coronaviruses. As part of such an initiative, itraconazole was identified as a potential candidate. Itraconazole has been described previously to have activity in an in vitro screen using a luciferase reporter‐expressing recombinant murine betacoronavirus, 5 as well as against a feline alphacoronavirus that causes feline infectious peritonitis. 6 Furthermore, in vitro and in vivo activity has been described against other respiratory viruses, such as influenza A and human rhinovirus. 7 , 8
Itraconazole is a member of the triazole group of broad‐spectrum antifungals, 9 with a well‐established efficacy and safety profile. 8 , 9 , 10 , 11 , 12 The primary mechanism of its antifungal action is the inhibition of ergosterol biosynthesis, by acting on the oxysterol‐binding protein (OSBP). 13 , 14 , 15 , 16 Since ergosterol is closely related to cholesterol, it has been suggested that also in mammalian cells, itraconazole acts as a cholesterol trafficking inhibitor resulting in disruption of the cholesterol‐enriched membranes and thus inhibition of virus replication. 6 , 8 , 17 However, a role in interferon priming has also been suggested as a contributing factor to the antiviral activity. 18
In an attempt to identify therapeutic options for treating COVID‐19 patients, the in vitro antiviral activities of itraconazole, and its metabolite 17‐OH itraconazole were investigated in Caco‐2 and VeroE6‐eGFP cells infected with SARS‐CoV‐2, isolated from COVID‐19 patients.
2. METHODS
2.1. Cell culture
Caco‐2 cells (human colon carcinoma cell line; obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen) were cultured in Minimal Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS) with penicillin (100 IU/ml) and streptomycin (100 μg/ml) at 37°C in a 5% CO2 atmosphere. VeroE6‐eGFP (African green monkey kidney cell line; provided by Dr Koen Andries, J&JPRD) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 0.075% sodium bicarbonate, penicillin and streptomycin (100 μg/ml) at 37°C in a 5% CO2 atmosphere. All cell culture reagents were obtained from Sigma‐Aldrich.
2.2. SARS‐CoV‐2 preparation
SARS‐CoV‐2‐FFM1 (strain hCoV‐19/Germany/FrankfurtFFM1/2020) was isolated from a German human‐case sample and cultured in Caco‐2 cells, as previously described. 1 , 19 SARS‐CoV‐2‐Germany stocks were passaged twice in Caco‐2 cells before storage (–80°C).
SARS‐CoV‐2‐Belgium (strain BetaCov/Belgium/GHB‐03021/2020) was recovered from a nasopharyngeal swab taken from a patient returning from Wuhan, China. SARS‐CoV‐2 Belgium stocks were passaged six times in VeroE6‐eGFP cells before storage (–80°C).
2.3. Assessment of antiviral activity
Itraconazole and its metabolite, 17‐OH itraconazole, and remdesivir were synthesized at Johnson & Johnson. GS‐441524, the parent nucleoside of remdesivir, used in studies at KU Leuven was obtained from MedChemExpress. Antiviral activity was assessed by inhibition of virus‐induced cytopathogenic effect (CPE) as described previously. 20 In brief, confluent layers of Caco‐2 cells cultured for 72 h on 96 multi‐well plates (50,000 cells/well) were challenged with SARS‐CoV‐2‐FFM1 at a multiplicity of infection of 0.01. The virus was added together with the compound under investigation and incubated in MEM supplemented with 1% FBS. Itraconazole diluted in MEM without FBS in four‐fold dilutions was added to a concentration range of 0.01–50 μM; 17‐OH itraconazole diluted in MEM without FBS was added in four‐fold dilutions to a concentration range of 0.02–100 μM and remdesivir diluted in MEM without FBS in four‐fold dilutions was added to a concentration range of 0.02–100 μM. Cells were incubated for 48 h; the CPE was then visually scored by two independent laboratory technicians. In addition, CPE was also assessed using a 3‐(4,5‐dimethylthiazol‐2‐yl)−2,5‐diphenyltetrazolium bromide (MTT) assay, performed according to the manufacturer's instructions. Optical densities were measured at 560/620 nm in a Multiskan Reader (MCC/340 Labsystems). Two series of three independent experiments (or one series of three experiments for 17‐OH‐itraconazole), each containing three replicates, were performed. Data were analyzed by four‐parameter curve‐fitting from a dose–response curve using GraphPad Prism (version 7.00) to calculate the EC50 (concentration of the compound that inhibited 50% of the infection) based on visual CPE scoring or the MTT assay.
2.4. Assessment of cell viability
Cell viability in Caco‐2 cells was measured following administration of each of the compounds or metabolites under investigation over the range of concentrations in the absence of virus using the Rotitest Vital (Roth) test according to manufacturer's instructions, as previously described. 21 All assays were performed three times independently in triplicate, this was performed twice (once for 17‐OH‐itraconazole). Data were analyzed by four‐parameter curve‐fitting from a dose–response curve using GraphPad Prism (version 7.00) to calculate the CC50 (cytotoxic concentration of the compound that reduced cell viability to 50%).
2.5. Viral RNA (vRNA) yield reduction assay
Antiviral activity was assessed by inhibition of viral yield in Caco‐2 cells. Confluent layers of Caco‐2 cells cultured for 72 h on 96 multi‐well plates (50,000 cells/well) were challenged with SARS‐CoV‐2‐FFM1 at a multiplicity of infection of 0.01. The virus was added together with the compound under investigation and incubated in MEM supplemented with 1% FBS. Itraconazole diluted in MEM without FBS was added in four‐fold dilutions to a concentration range of 0.01–50 μM; 17‐OH itraconazole diluted in MEM without FBS was added in four‐fold dilutions to a concentration range of 0.02–100 μM and remdesivir diluted in MEM without FBS was added in four‐fold dilutions to a concentration range of 0.02–100 μM. Two series of three independent experiments, each containing two replicates, were performed.
SARS‐CoV‐2 RNA from cell culture supernatant samples was isolated using AVL buffer and the QIAamp Viral RNA Kit (Qiagen) according to the manufacturer's instructions. Absorbance‐based quantification of the RNA yield was performed using the Genesys 10 S UV‐Vis Spectrophotometer (Thermo Scientific). RNA was subjected to OneStep real‐time quantitative polymerase chain reaction (qRT‐PCR) analysis using the Luna Universal One‐Step RT‐qPCR Kit (New England Biolabs) and a CFX96 Real‐Time System, C1000 Touch Thermal Cycler (BioRad). Primers were adapted from the WHO protocol targeting the open reading frame for RNA‐dependent RNA polymerase (RdRp): RdRP_SARSr‐F2 (GTG ARA TGG TCA TGT GTG GCG G) and RdRP_SARSr‐R1 (CAR ATG TTA AAS ACA CTA TTA GCA TA) using 0.4 µM per reaction. Standard curves were created using plasmid DNA (pEX‐A128‐RdRP) harboring the corresponding amplicon regions for RdRP target sequence according to GenBank Accession number NC_045512. 21
Antiviral activity was also assessed by reduction of viral yield in VeroE6‐eGFP cells. VeroE6‐eGFP cells were cultured for 24 h on 96 multi‐well plates (10,000 cells/well) in the absence or presence of compound and infected with SARS‐CoV‐2‐Belgium at a multiplicity of infection of one tissue culture infectious dose (TCID50)/cell, in 200 µl assay medium (DMEM supplemented with 2% FBS and 0.075% sodium bicarbonate). After 2 h at 37°C the cells were washed once and further cultured in 200 µl assay medium containing the same compound concentrations (37°C, 5% CO2) for another 48 h. The viral RNA (vRNA) in the culture supernatant (SN) was extracted using the NucleoSpin kit (Macherey‐Nagel), according to the manufacturer's instructions and quantified by RT‐qPCR performed on a LightCycler96 platform (Roche) using the iTaq Universal Probes One‐Step RT‐qPCR kit (BioRad) with primers and probes specific for SARS‐CoV‐2 obtained from IDTDNA (cat no 10006775). Sequences were identical to the 2019‐nCoV_N1 CDC assay for SARS‐CoV‐2 detection (2019‐nCoV_N1 Forward Primer GAC CCC AAA ATC AGC GAA AT None 500 nM; 2019‐nCoV_N1 Reverse Primer TCT GGT TAC TGC CAG TTG AAT CTG None 500 nM; 2019‐nCoV_N1 Probe FAM‐ACC CCG CAT TAC GTT TGG TGG ACC‐BHQ1). A standard curve was created from serial dilutions of a virus stock with known titer and used to correlate the cycle threshold (C t) values of the experimental samples with absolute virus quantities. One independent experiment, containing two replicates (for itraconazole) or three replicates (17OH‐itraconazole), were performed.
3. RESULTS
3.1. In vitro antiviral activity in Caco‐2 cells
Independent experiments with triplicate measurements were performed with itraconazole (n = 6), 17‐OH itraconazole (n = 3) and remdesivir (n = 6). In Caco‐2 cells, itraconazole resulted in a dose‐dependent inhibition of SARS‐CoV‐2‐FFM1 measured by visual scoring of inhibition of CPE with an EC50 = 1.5 μM (Figure 1; Table 1). Similar findings were obtained using the MTT assay (EC50 = 2.3 μM). The 17‐OH itraconazole metabolite reduced SARS‐CoV‐2 induced CPE with an EC50 value of 1.2 μM and EC50 MTT = 3.6 μM. The positive control, remdesivir, resulted in EC50 values of 0.3 and 0.4 µM in the CPE and MTT assays in Caco‐2 cells, respectively.
Figure 1.
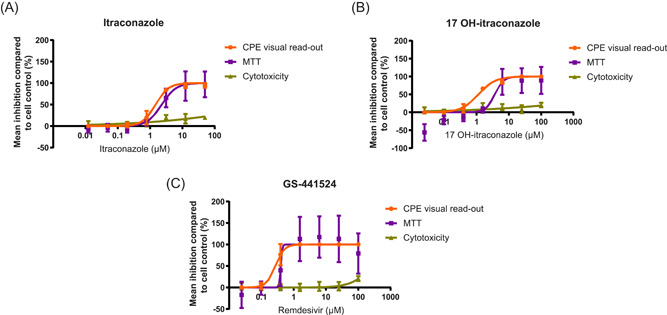
Effect of either (A) itraconazole, (B) 17‐OH itraconazole, or (C) remdesivir on SARS‐CoV‐2 replication in CPE assays and on viability of Caco‐2 cells. Mean percent inhibition for each readout across two series of three independent experiments (itraconazole, remdesivir) or three independent experiments (17‐OH itraconazole) with triplicate measurements are plotted. The error bars represent the standard deviation. Orange represents CPE visual read‐out; purple represents MTT assay; and green represents cytotoxicity. (A) EC50 by visual scoring of inhibition of CPE = 1.5 μM; EC50 by MTT assay = 2.3 μM (B) EC50 by visual scoring of inhibition of CPE = 1.2 μM; EC50 by MTT assay = 3.6 μM (C) EC50 by visual scoring of inhibition of CPE = 0.3 μM; EC50 by MTT assay = 0.4 μM. CPE, cytopathogenic effect; EC50, concentration of the compound that inhibited 50% of the infection; MTT, 3‐(4,5‐dimethylthiazol‐2‐yl)−2,5‐diphenyltetrazolium bromide; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Table 1.
EC50 and CC50 values for the effect of itraconazole, 17‐OH itraconazole and remdesivir on SARS‐CoV‐2 replication and the viability of Caco‐2 cells
| Assessment | Itraconazole | 17‐OH itraconazole | Remdesivir |
|---|---|---|---|
| Antiviral activity | |||
| EC50 by visual scoring of inhibition of CPE, µM | 1.5 | 1.2 | 0.3 |
| EC50 by MTT assay, µM | 2.3 | 3.6 | 0.4 |
| Cell viability | |||
| CC50, µM | >50 | >100 | >100 |
Abbreviations: CC50, cytotoxic concentration of the compound that reduced cell viability to 50%; CPE, cytopathogenic effect; EC50, concentration of the compound that inhibited 50% of the infection; MTT, 3‐(4,5‐dimethylthiazol‐2‐yl)−2,5‐diphenyltetrazolium bromide; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Minimal cytotoxicity was seen in Caco‐2 cells with both itraconazole (CC50 > 50 μM) and 17‐OH itraconazole (CC50 > 100 μM) (Figure 1). For remdesivir, CC50 values greater than 100 μM were observed in Caco‐2 cells.
3.2. In vitro vRNA yield reduction
The in vitro effect of itraconazole and remdesivir (Caco‐2 cells) or GS‐441522 (VeroE6‐eGFP cells) on SARS‐CoV‐2 RNA yield reduction was assessed in Caco‐2 cells and VeroE6‐eGFP cells. A concentration of 6.25 µM and 3.1 µM of itraconazole and its main metabolite 17‐OH itraconazole, respectively, resulted in an approximately 2‐log10 reduction in vRNA levels (as a measure of the number of virus particles in the culture SN) in Caco‐2 cells. This reduction was observed at all higher doses, in the absence of toxicity. However, remdesivir proved more potent (a reduction of ~3 log10 at 6.25 µM) (Figure 2).
Figure 2.
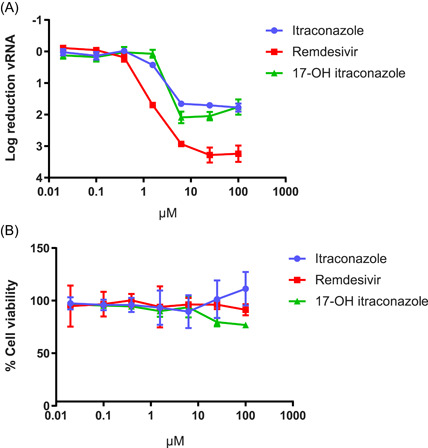
Effect of itraconazole, 17‐OH itraconazole or remdesivir on SARS‐CoV‐2 vRNA yield and viability in Caco‐2 cells. (A) Mean differences in vRNA in the supernatant between untreated cultures and treated cultures at 48 h postinfection with SARS‐CoV‐2‐FFM1 of three independent experiments each containing two replicates is shown. Error bars represent the standard deviation. (B) Mean viability of the cells, based on three independent experiments each containing three replicates is shown. Error bars represent the standard deviation. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; vRNA, viral RNA
In VeroE6‐eGFP cells, a concentration of 1 µM itraconazole resulted in an approximate 1‐log10 reduction in vRNA levels (Figure 3). This reduction was also observed at higher concentrations. As in Caco‐2 cells, GS‐441524 proved more potent in reducing vRNA than itraconazole. At a concentration of 3.7 µM, GS‐441524 reduced vRNA load by greater than 4 log10 to undetectable levels. At concentrations of 10 µM and higher, 17‐OH itraconazole resulted in limited activity, but VeroE6‐eGFP cell viability was markedly affected (Figure 3).
Figure 3.
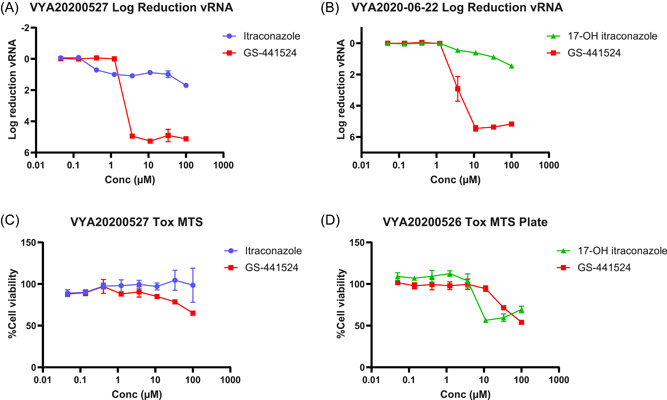
Effect of itraconazole, 17‐OH itraconazole, or GS‐441524 on SARS‐CoV‐2 vRNA yield and viability in VeroE6‐eGFP cells (A) and (B) Mean differences in vRNA in the supernatant between untreated cultures and treated cultures at 48 h postinfection with SARS‐CoV‐2‐Belgium of one independent experiment containing two replicates (A) or three replicates (B) is shown. Error bars represent the standard deviation. (C, D) Mean viability of the cells, based on MTT readout of uninfected cells. Error bars represent the standard deviation. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; vRNA, viral RNA
4. DISCUSSION
While a number of vaccines are available, for the moment there are no antivirals for the prevention of COVID‐19 and only a limited number of antivirals for the treatment of COVID‐19. 22 Given the great unmet need to identify potential treatments for COVID‐19, and the fact that a de novo drug development project will take many years to bring a drug to patients, efforts have been expedited into the in vitro screening of compounds already in the market, or in late‐stage development. In such an effort, itraconazole was identified, with single digit micromolar activity in Caco‐2 cells. Itraconazole was first approved (in the US) in 1992 as an oral treatment for a number of fungal infections in immunocompromised and non‐immunocompromised patients, with a well‐established safety profile. 11
These experiments demonstrated that itraconazole inhibits in vitro SARS‐CoV‐2 replication in Caco‐2 cells, an established cell line for studying SARS‐CoV infections. 1 , 23 The EC50 values for inhibition of virus‐induced CPE formation is in the low micromolar range and in that respect, is only around five‐fold higher than that of remdesivir. The EC50 values that were obtained are in line with recently reported values for itraconazole inhibition of SARS‐CoV‐2 replication in Calu‐3 and VeroE6 cell lines (0.43 and 0.39 μM, respectively). 24 To obtain further evidence for the antiviral potency of the drugs, the effect on vRNA yield in culture was assessed in two cell lines. Itraconazole results in a marked inhibition of virus yield; however, itraconazole was less potent than remdesivir. The use of different SARS‐CoV‐2 strains, virus titers and positive controls in the vRNA yield reduction experiments in Caco‐2 cells and VeroE6‐eGFP cells limits further comparison of these results. In addition, in vitro studies have also demonstrated activity of itraconazole against murine coronavirus (recombinant murine coronavirus, mouse hepatitis virus, EC50 = 7.9 µM) 5 and type I (but not type II) feline coronavirus (EC50 across three strains tested = 0.146–0.597 μM). 6
Itraconazole has documented activity against several enteroviruses, including rhinovirus. 8 , 15 Further, at a dose of 5.7 mg/kg, itraconazole resulted in improved mortality and a lower viral load in mice that had been infected with human influenza (strain PR8M) compared with the control group. 7
As an antifungal agent, itraconazole inhibits fungal ergosterol biosynthesis, through inhibition of a cytochrome P450 enzyme, lanosterol‐14‐alpha‐demethylase. 13 , 14 , 15 , 16 It is also thought to interfere with cholesterol homeostasis and de novo synthesis in the host's cells through inhibition of this same enzyme, thereby influencing the virus or host cell interaction. 17 , 25 , 26 , 27 The antiviral effect of itraconazole may be, at least in part, based on such a mechanism. Itraconazole impairs cholesterol trafficking and blocks late endosomal or lysosomal export of cholesterol to the plasma membrane through NPC intracellular cholesterol transporter 1 (NPC1) leading to cholesterol accumulation in sub‐cellular compartments. 8 , 16 , 25 , 26 , 27 , 28 In turn, elevated cholesterol levels in the endosomal membrane impede fusion of the viral lipid envelope and prevent viral genome transfer into the host cell cytosol. 7 , 29 It has been postulated that such changes in cellular cholesterol are linked with the host's immune response. 30 Disruption of cholesterol biosynthesis has been shown to upregulate type I interferons and thereby accelerate the virus‐induced interferon‐mediated host cell response. 7 , 30 It has also been suggested that itraconazole may have antiviral effects through proteins other than NPC1. 6 In enteroviruses, itraconazole has been shown to inhibit vRNA replication by OSBP, which is responsible for trafficking of cholesterol and phosphatidylinositol‐4‐phosphate between membranes, thus also affecting the membranes of the replication complex. 15 , 16 Since coronaviruses result in the formation of intracellular viral replicative organelles within the endoplasmic reticulum that drive the viral replication cycle, 31 , 32 , 33 it is possible that inhibition of OSBP may cause detrimental changes to replicative organelle membranes. 15 , 16 , 33 , 34 Although it remains to be confirmed how itraconazole may inhibit the replication cycle of coronaviruses, it is conceivable that it acts through multiple mechanisms. 7 , 17
As a lipophilic compound, itraconazole has high bioavailability and extensive distribution throughout the lung, kidney, epidermis and brain. 9 Itraconazole is predominantly metabolized by the cytochrome P450 3A4 isoenzyme system, forming many metabolites. 35 , 36 The main metabolite produced is 17‐OH itraconazole, which also has considerable antifungal activity. 35 Following administration of itraconazole 200‐mg bid, itraconazole and its 17‐OH hydroxy‐metabolite have been shown to reach mean maximal plasma concentrations of 2282 and 3488 ng/ml (~3.2 μM and ~4.8 μM) and a Ctrough of 1855 and 3349 ng/ml (~2.6 and ~4.7 μM), respectively, which is close to the EC50 for inhibition of virus‐induced CPE. Both the parent drug and metabolite have long half‐lives (64 and 56 h, respectively). 11
5. CONCLUSION
Itraconazole and its metabolite, 17‐OH itraconazole show in vitro activity against SARS‐CoV‐2. Based on these in vitro findings, a clinical study in hospitalized COVID‐19 patients was initiated by UZ Leuven in March 2020. 37 This clinical study was ended prematurely because of futility. 38 Other studies in nonhospitalized patients have not been conducted.
CONFLICT OF INTERESTS
Sandra De Meyer, Ellen Van Damme, Christophe Buyck, and Marnix Van Loock are employees and may be stock owners of Johnson & Johnson. Sandra Ciesek received research funding from Janssen for this study. Steven De Jonghe, Dirk Jochmans, Pieter Leyssen, and Johan Neyts are employees from KU Leuven and received funding from Janssen for this study.
AUTHOR CONTRIBUTIONS
Ellen Van Damme, Sandra De Meyer: study design, statistical analysis, manuscript writing, critical review, approval of the version of the manuscript to be published. Denisa Bojkova, Sandra Ciesek, and Jindrich Cinatl: study design, data collection, critical review, approval of the version of the manuscript to be published. Steven De Jonghe, and Dirk Jochmans: data collection, critical review, approval of the version of the manuscript to be published. Johan Neyts: study design, data collection, critical review, approval of the version of the manuscript to be published. Pieter Leyssen: data collection, manuscript writing, critical review, approval of the version of the manuscript to be published. Christophe Buyck: data collection, statistical analysis, critical review, approval of the version of the manuscript to be published. Marnix Van Loock: study design, critical review, approval of the version of the manuscript to be published.
ACKNOWLEDGMENTS
This project has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no.: 101003627. Part of this study work was performed using the “Caps‐It” research infrastructure (project ZW13‐02) that was financially supported by the Hercules Foundation and Rega Foundation, KU, Leuven. This study has been funded in part with Federal funds from the Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, under OTA No. HHSO100201800012C. Medical writing support for the development of this manuscript was provided by Patrick Hoggard of Zoetic Science, an Ashfield company, part of UDG Healthcare plc; this support was funded by Janssen Pharmaceuticals. We thank Lena Stegmann, Winston Chiu, Piet Maes, Robbert Boudewijns, Nguyen Dan Thuc Do, Xin Zhang, Jana Van Dycke, Rana Abdelnabi, Laura Thijs, Peggy Geluykens, Doortje Borrenberghs, and Christel Van den Eynde for technical support for antiviral assays.
Damme EV, De Meyer S, Bojkova D, et al. In vitro activity of itraconazole against SARS‐CoV‐2. J Med Virol. 2021;93:4454–4460. 10.1002/jmv.26917
DATA AVAILABILITY STATEMENT
The data sharing policy of Janssen Pharmaceutical Companies of Johnson & Johnson is available at https://www.janssen.com/clinical-trials/transparency. As noted on this site, requests for access to the study data can be submitted through Yale Open Data Access (YODA) Project site at http://yoda.yale.edu.
REFERENCES
- 1. Hoehl S, Rabenau H, Berger A, et al. Evidence of SARS‐CoV‐2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020;382(13):1278‐1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao J, Forrest JC, Zhang X. A screen of the NIH Clinical Collection small molecule library identifies potential anti‐coronavirus drugs. Antiviral Res. 2015;114:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takano T, Akiyama M, Doki T, Hohdatsu T. Antiviral activity of itraconazole against type I feline coronavirus infection. Vet Res. 2019;50(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schloer S, Goretzko J, Kühnl A, Brunotte L, Luwdig S, Rescher U. The clinically licensed antifungal drug itraconazole inhibits influenza virus in vitro and in vivo. Emerg Microbes Infect. 2019;8(1):80‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shim A, Song JH, Kwon BE, et al. Therapeutic and prophylactic activity of itraconazole against human rhinovirus infection in a murine model. Sci Rep. 2016;6:23110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saag MS, Dismukes WE. Azole antifungal agents: emphasis on new thiazoles. Antimicrob Agents Chemother. 1988;32(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J, Tang JY, Gong R, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17(4):388‐399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ortho‐McNeil‐Janssen . Pharmaceuticals, Inc. Sporanox® Summary of Product Characteristics. 2012. https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/020083s048s049s050lbl.pdf. Accessed December 10, 2020.
- 12. De Doncker P, Pande S, Richarz U, Garodia N. Itraconazole: what clinicians should know? Indian. J Drugs Dermatol. 2017;3(1):4‐10. [Google Scholar]
- 13. Da Silva Ferreira ME, Colombo AL, Paulsen I, et al. The ergosterol biosynthesis pathway, transporter genes, and azole resistance in Aspergillus fumigatus . Med Mycol. 2005;43(Suppl 1):S313‐S319. [DOI] [PubMed] [Google Scholar]
- 14. Lepesheva GI, Waterman MR. Sterol 14α‐demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta. 2007;1770(3):467‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strating JRPM, van der Linden L, Albulescu, et al. Itraconazole inhibits enterovirus replication by targeting the oxysterol‐binding protein. Cell Rep. 2015;10(4):600‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bauer L, Ferla S, Head SA, et al. Structure‐activity relationship study of itraconazole, a broad‐range inhibitor of picornavirus replication that targets oxysterol‐binding protein (OSBP). Antiviral Res. 2018;156:55‐63. [DOI] [PubMed] [Google Scholar]
- 17. Takano T, Endoh M, Fukatsu H, Sakurada H, Doki T, Hohdatsu T. The cholesterol transport inhibitor U18666A inhibits type I feline coronavirus infection. Antiviral Res. 2017;145:96‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schloer S, Goretzko J, Kühnl A, Brunotte L, Ludwig S, Rescher U. The clinically licensed antifungal drug itraconazole inhibits influenza virus in vitro and in vivo. Emerg Microbes Infect. 2019;8(1):80‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivens T, Eynde CV, AckerAcker, KV, et al. Development of a homogeneous screening assay for automated detection of antiviral agents active against severe acute respiratory syndrome‐associated coronavirus. J Virol Methods. 2005;129(1):56‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Meyer S, Bojkova D, Cinatl J, et al. Lack of antiviral activity of darunavir against SARS‐CoV‐2. Int J Infect Dis. 2020;97:7‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bojkova D, Klann K, Koch B, et al. Proteomics of SARS‐CoV‐2‐infected host cell reveals therapy targets. Nature. 2020;583(7816):469‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu H. Drug treatment options for the 2019‐new coronavirus (2019‐nCoV). Biosci Trends. 2020;14(1):69‐71. [DOI] [PubMed] [Google Scholar]
- 23. Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr H. Treatment of SARS with human interferons. Lancet. 2003;362:293‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schloer S, Brunotte L, Mecate‐Zambrano A, et al. Drug synergy of combinatory treatment with remdesivir and the repurposed drugs fluoxetine and itraconazole effectively impairs SARS‐CoV‐2 infection in vitro. bioRxiv. 2020. 10.1101/2020.10.16.342410 [DOI] [PMC free article] [PubMed]
- 25. Shoemaker CJ, Schornberg KL, Delos SE, et al. Multiple cationic amphiphiles induce a Niemann‐Pick C phenotype and inhibit Ebola virus entry and infection. PLOS One. 2013;8(2):e56265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trinh MN, Lu F, Li X, et al. Triazoles inhibit cholesterol export from lysosomes by binding to NPC1. Proc Natl Acad Sci USA. 2017;114(1):89‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amini‐Bavil‐Olyaee S, Choi YJ, Lee JH, et al. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13(4):452‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Head SA, Shi WQ, Yang EJ, et al. Simultaneous targeting of NPC1 and VDAC1 by itraconazole leads to synergistic inhibition of mTOR signaling and angiogenesis. ACS Chem Biol. 2017;12(1):174‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kühnl A, Musiol A, Heitzig N, et al. Late endosomal/lysosomal cholesterol accumulation is a host cell‐protective mechanism inhibiting endosomal escape of influenza A virus. mBio. 2018;9(4):e01345‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. York AG, Williams KJ, Argus JP, et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163(7):1716‐1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Risco C, de Castro IF, Sanz‐Sánchez L, Narayan K, Grandinetti G, Subramaniam S. Three‐dimensional imaging of viral infections. Annu Rev Virol. 2014;1(1):453‐473. [DOI] [PubMed] [Google Scholar]
- 32. Romero‐Brey I, Bartenschlager R. Membranous replication factories induced by plus‐strand RNA viruses. Viruses. 2014;6(7):2826‐2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neuman BW, Angelini MM, Buchmeier MJ. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22(11):642‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takano T, Wakayama Y, Doki T. Endocytic pathway of feline coronavirus for cell entry: differences in serotype‐dependent viral entry pathway. Pathogens. 2019;8(4):300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peng CC, Shi W, Lutz JD, et al. Stereospecific metabolism of itraconazole by CYP3A4: Dioxoalane ring scission of azole antifungals. Drug Metab Dispos. 2012;40(3):426‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prentice AG, Glasmacher A. Making sense of itraconazole pharmockinetics. J Antimicrob Chemother. 2005;56(Suppl 1):i17‐i22. [DOI] [PubMed] [Google Scholar]
- 37. EUDRACT . COVID‐19: a randomized, open‐label, adaptive, proof‐of‐ concept clinical trial of new antiviral drug candidates against SARS‐CoV‐2. 2020. https://www.clinicaltrialsregister.eu/ctr-search/trial/-001243-15/BE. Accessed December 10, 2020.
- 38. Liesenborghs L, Spriet I, Jochmans D, et al. Itraconazole for COVID‐19: preclinical studies and a proof‐of‐concept pilot clinical study. Lancet. 2021. 10.2139/ssrn.3731461. Accessed February 5, 2021. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sharing policy of Janssen Pharmaceutical Companies of Johnson & Johnson is available at https://www.janssen.com/clinical-trials/transparency. As noted on this site, requests for access to the study data can be submitted through Yale Open Data Access (YODA) Project site at http://yoda.yale.edu.


