Abstract
A type of coronavirus disease called COVID‐19 is spreading all over the globe. Researchers and scientists are endeavoring to find new and effective methods to diagnose and treat this disease. This article presents an automated and fast system that identifies COVID‐19 from X‐ray radiographs of the chest using image processing and machine learning algorithms. Initially, the system extracts the feature descriptors from the radiographs of both healthy and COVID‐19 affected patients using the speeded up robust features algorithm. Then, visual vocabulary is built by reducing the number of feature descriptors via quantization of feature space using the K‐means clustering algorithm. The visual vocabulary train the support vector machine (SVM) classifier. During testing, an X‐ray radiograph's visual vocabulary is sent to the trained SVM classifier to detect the absence or presence of COVID‐19. The study used the dataset of 340 X‐ray radiographs, 170 images of each Healthy and Positive COVID‐19 class. During simulations, the dataset split into training and testing parts at various ratios. After training, the system does not require any human intervention and can process thousands of images with high precision in a few minutes. The performance of the system is measured using standard parameters of accuracy and confusion matrix. We compared the performance of the proposed SVM‐based classier with the deep‐learning‐based convolutional neural networks (CNN). The SVM yields better results than CNN and achieves a maximum accuracy of up to 94.12%.
Keywords: artificial intelligence, chest X‐ray radiograph, COVID‐19, feature descriptors, medical image processing
1. INTRODUCTION
COVID‐19 is a pathogen that has a high fatality rate, exponential propagation ability, and the lack of definite medical treatment. The COVID‐19 is a beta‐coronavirus with a 70% similarity in genetic sequence to SARS. It is different from MERS and SARS‐CoV. COVID‐19 is the member of the family of coronaviruses that infect humans and highly contagious. Radiography of the chest/lungs uses an X‐ray to help diagnose the cause of several lungs disorders including COVID‐19. Figure 1 shows the sample X‐ray radiographs of Healthy and Positive COVID‐19 cases.
FIGURE 1.
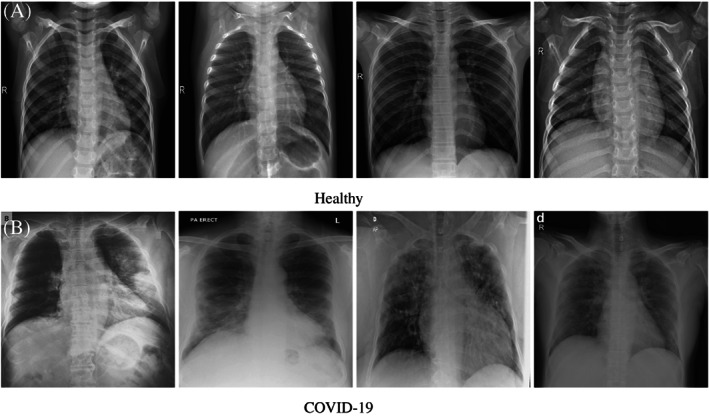
X‐ray radiographs of Healthy and COVID‐19 cases
Early and correct detection of coronavirus can play a decisive factor in saving human lives. A visual inspection of a radiograph by the radiologist can identify the presence of the COVID‐19. However, this requires manual examination that is time‐consuming and not possible for a very large number of images. This work describes an intelligent system to determine the absence or presence of COVID19. The underlying assumption of the study is that COVID‐19 affects the lungs most that exhibit certain characteristics (features) in the lungs tissues. Image processing and machine learning algorithms can detect the COVID‐19 using the X‐ray radiographs of the chest. As a supervised machine learning‐based system, the predetermined Healthy and COVID‐19 radiographs are used to train a classifier. After training, the system can be used to test an unknown radiograph. There are three main stages in the proposed system. In the first stage, feature descriptors are extracted from the radiographs. Then, in the next step, the K‐means clustering algorithm is applied to reduce a large number of feature descriptors into K clusters. In the third stage, either we train the classifier (SVM) using labeled images or identify unlabeled images using an already trained classifier. The main contributions of this work are summarized as follows:
Describe the architecture of an intelligent and automated system to detectCOVID‐19.
Implementation of a fast system that can process thousands of images in a few minutes with high accuracy.
Determination of optimal parameters for speeded up robust features (SURF), K‐means clustering, and SVM algorithms.
Detail statistics of results with analysis to provide a benchmark for future research.
2. RELEVANT WORK
Machine learning is used in a wide range of imaging and text analysis applications, such as cancer or abnormality detection in breast, 1 , 2 , 3 detection of fractures, 4 and detection of crimes from tweets. 5 In the work of Sha et al, 2 they used deep learning convolutional neural network (CNN) to detect breast cancer from mammogram images. Optimized feature extraction is achieved using the Grasshopper algorithm. 6 The method yields 96% sensitivity and 93% specificity rates.
Two recent papers that used the deep learning (CNN) method to distinguish COVID‐19 from pneumonia and healthy cases using X‐ray radiographs are proposed by References 7 and 8. In his work, 8 uses three types of X‐rays images healthy, pneumonia, and COVID‐19. Their proposed DL‐CRC framework applies data augmentation to the radiograph images to generate synthetic radiographs via zoom and rotation operations. The customized CNN model combines real and synthetic radiographs. The CNN model used in Reference 8 consists of convolution layers for feature extraction and dense layers for classification. The authors report the classification accuracy of 94.61%. The deep learning model of 7 uses ResNet‐101 architecture, a CNN with 101 layers that operates on images of size 224 × 224 × 3. The readymade ResNet‐101 is pretrained to recognize objects from a million images. In their work, [azemin2020] retrained fully connected, softmax, and classification output layers of ResNet‐101 to detect abnormality in chest X‐ray images. The authors report an accuracy of 71.9%.
A study that compares the human experts' evaluation with an artificial intelligence system's using chest radiographs of COVID‐19 is presented by Murphy et al. 9 In this study, radiographs were independently analyzed by six readers (human evaluation) and by the AI system (machine evaluation). This study's AI system is CAD4COVID‐XRay, 10 developed by Thirona (Nijmegen, the Netherlands). The AI system's performance was determined using the receiver operating characteristic curve. The authors conclude that the AI system's performance is comparable with that of human readers.
Some of the researchers used chest CT scans to identify COVID‐19. A deep learning method based on a CNN to detect COVID‐19 from CT scan is presented by Li et al. 11 The system takes as input a series of CT slices and generates a classification prediction of the CT image. The method extracts local and global features from the chest CT exams. A clinical investigation of COVID‐19 pneumonia is presented by Song et al. 12 The study used the chest CT of 51 patients (25 men and 26 women; age between 16 and 76 years from Wuhan, China) with COVID19 diagnosed. A radiologist identifies the chest CT lesions in each patient. The study determines a lung lesion in the CT scan of patients with 5 days or more from the COVID‐19 inception. The authors summarize that fever, cough, and chest CT findings are characteristic of COVID‐19.
Authors of 13 developed a smartphone‐based app AI4COVID‐19 to detect COVID‐19 from the cough sounds of participants. The method uses deep transfer learning to classify four types of coughs, that is, normal, bronchitis, pertussis, and COVID‐19. The overall accuracy of the system is 88.76%.
Our method to identify COVID‐19 relies upon the detection of features and descriptors from a chest radiograph. Some of the well‐known algorithms to extract features or patterns from images are the difference of Gaussians, 14 Laplacian of Gaussians, 15 Harris corner detector, 16 and so forth. Similarly, a variety of algorithms to find descriptors can be found in the literature such as SURF, 17 , 18 Scale Invariant Feature Transform (SIFT) 19 , 20 and discriminative feature description, 21 and so forth.
We employed the support vector machine (SVM) for the classification of X‐ray radiograph. SVM is a supervised machine learning technique that is widely used to analyze and classify data. 22 , 23 , 24 The root of the SVM algorithm goes back to 1963 when it was proposed by Vladimir N. Vapnik and Alexey Ya. Chervonenkis, while the modified version of the algorithm published in 1995 by Corinna Cortes and Vapnik. 22 The earlier version of SVM has the restriction that training data can be separated without errors, while the new version of the algorithm extends it to nonseparable training data. 22
3. SYSTEM ARCHITECTURE AND METHODOLOGY
Figure 2 describes the architecture of the COVID‐19 detection system. There are three major modules of the system training, testing, and classifier. During training, one set of each Healthy and Positive COVID‐19 images are feed to the system. The feature extraction module extracts the local features of the image sets, builds the visual vocabulary (using K‐means clustering), and train the SVM classifier. To find the class of an unknown X‐ray radiograph, the system extracts its feature descriptors, builds the visual vocabulary, and finally predicts the class using the pretrained classifier. The training and testing algorithms of the system are listed in Figure 3.
FIGURE 2.
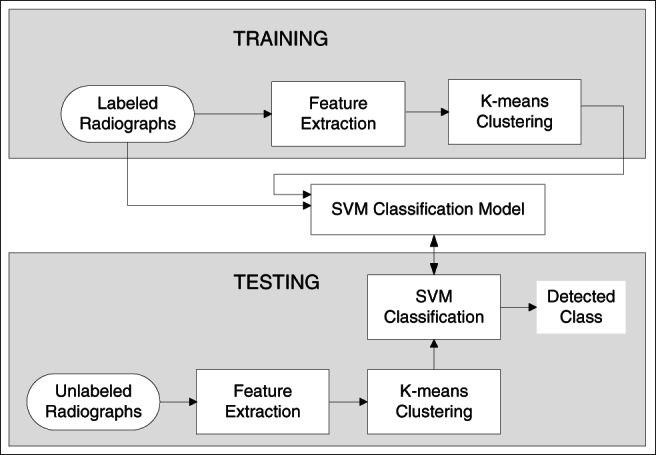
System architecture
FIGURE 3.
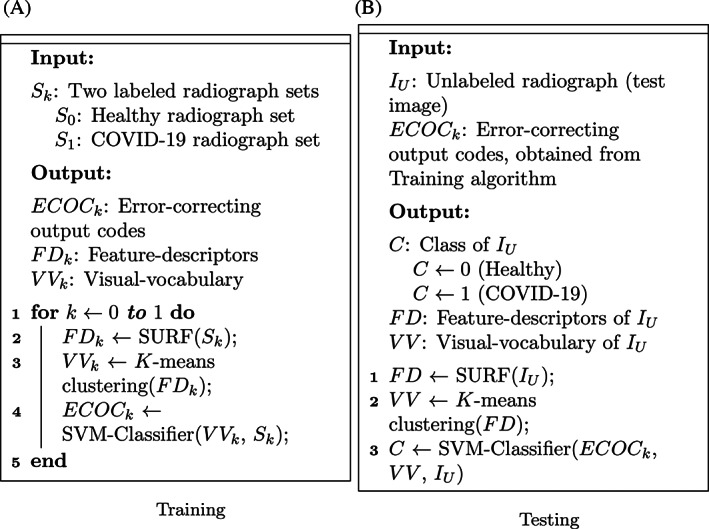
Algorithms
The system uses the following three main steps to classify and identify the radiographs.
Extraction of feature descriptors.
Clustering of feature descriptors.
Classification of radiographs.
The following subsections give the details of the above steps.
3.1. Extraction of feature descriptors
A feature refers to a significant point, that is, a point of interest in an image. A feature descriptor is a set of values that describes the image patch around the point of interest. Local features extractors, for example, SURF, SIFT, and so forth, detects patterns or structures such as a corner, curvature, or edges in a grayscale image via local minima/maxima of some function. We used the SURF algorithm to detect the local features of radiographs. SURF offers a computationally efficient approximation of the second‐order Gaussian derivatives via a set of integral images. For the detection of feature points, instead of relying on perfect Gaussian derivatives, the computation is based on simple two‐dimensional box filters. SURF employs a scale‐invariant blob detector that relies on the determinant of the Hessian matrix for selection of scale selection. The SURF algorithm returns the following information for a feature descriptor:
Spatial coordinates, that is, (x,y) of the feature point.
The scale at which the feature point is detected.
Strength of the feature point.
Sign of the Laplacian operator. This value must be an integer −1, 0, or 1.
The orientation of the feature point in radian.
Figure 4 shows a few feature points (detected using the SURF algorithm) of two classes of radiographs. The image is cropped and enlarged to give a clear and closer view of the pixels that encompass the feature points. Feature points are shown as + sign in green color inside a circle. The large circle size is due to zoom operation.
FIGURE 4.
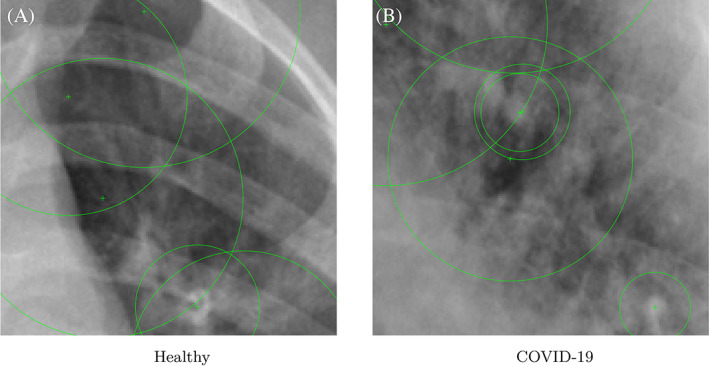
Feature points of two X‐ray radiographs shown as + sign [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Clustering of feature descriptors
The system detects a very large number of feature descriptors from chest radiographs. This big feature descriptor‐vocabulary needs to be reduced to some manageable size. The clustering groups the feature descriptors of similar characteristics in the same group. We group the descriptors obtained from all the radiographs into 500 clusters using the K‐means algorithm. The clusters are mutually exclusive and smaller in number compared to the number of descriptors. The center of each cluster represents a visual‐word. The centers of all the clusters of all the radiographs build the visual‐vocabulary. The following are the main steps of a general K‐means clustering algorithm:
Randomly choose centroid of each of the K clusters.
Allocate ith pixel of the radiograph image to a cluster that minimizes the Euclidean distance between the ith pixel and the centroid.
Recompute the centroid of each cluster by averaging all the pixels in the cluster.
Repeat steps 2 and 3 until convergence is achieved, that is, any pixel does not change its centroid.
3.3. Classification of images
We used SVM for the classification of chest radiographs. Initially, the SVM classifier is trained for two labeled (known) radiograph sets of Healthy and Positive COVID‐19. During the testing, the system is inquired to find the class of an unlabeled (unknown) test radiograph. The SVM classifier compares the feature descriptors of the test radiograph with the visual vocabulary of the classifier. The class of the test radiograph is predicted based on the optimal match.
A binary SVM constructs an optimal hyperplane in high‐dimensional space that separates data into two classes labeled as −1 and 1. The optimal hyperplane is one that maximizes the margin‐width between two classes. The equation of hyperplane H can be written as follows.
| (1) |
where b ∈ R is called the bias and w ∈ R n is referred to as the weight vector. Let sample data are a set of points x i with their classes y i ∈ [1,−1]. Then, for every sample i = 1,2,…,m, the following conditions are satisfied:
| (2) |
| (3) |
Various standard solutions exist to solve the above problem of finding (b,w) such as 25 , 26 , 27 .
4. EVALUATION MATRICES
Let X be one of the two radiograph classes (Healthy or Positive COVID‐19). Then, the following parameters are used to measure the performance of our system:
True negative (TN) are those radiographs that correctly classified as not of class X.
True Positive (TP) are those radiographs that correctly classified as of class X.
False negative (FN) are those radiographs that incorrectly classified as not of class X.
False Positive (FP) are those radiographs that incorrectly classified as of class X.
- Accuracy is the ratio of correctly classified radiographs to the total number of radiographs classified. Mathematically, the following is the equation of percentage accuracy. (A):
(4)
5. EXPERIMENTS
The datasets of 340 X‐ray radiographs used in the experiments belong to the two sources. 28 , 29 Original images are of various resolutions (255 × 249 512 × 512 657 × 657 651 × 6 591 279 × 998). We resize them to 360 × 320 (width × height). Resizing the images also speed up both the training and testing algorithms.
Let there are total N radiographs in the dataset such that N 1 radiographs belong to the Healthy class, while N 2 radiographs are of positive COVID‐19 cases. During each round of the simulation, the dataset is partition into training and testing sets. We randomly partitioned each type of radiograph into two groups such that P percent of the radiographs are taken as the training set, and the remaining (100 − P) percent of radiographs are part of the testing set. Then, at any given value of P, the following is the distribution of radiographs:
| (5a) |
| (5b) |
| (5c) |
| (5d) |
where U 1 is the number of radiographs of Healthy class in the training set, U 2 is the number of radiographs of COVID‐19 class in the training set, V 1 is the number of radiographs of Healthy class in the testing set, V 2 is the number of radiographs of COVID‐19 class in the testing set.
Features are extracted from the training set's radiographs using the SURF algorithm to train the SVM classifier. Let x 1 and x 2 be the number of extracted features of Healthy and COVID‐19 classes. Then, 20% weakest features are discarded from x 1 and x 2 and remaining 80% strongest features, that is, y 1 and y 2 are taken, mathematically,
| (6a) |
| (6b) |
In our experiments, number of Healthy and COVID‐19 radiographs are equal, that is, N 1 = N 2 = 170, consequently, U 1 = U 2 and V 1 = V 2. We take an equal number of features from both the classes to make a unified set of features, that is, x 1 = x 2 and y 1 = y 2. Table 1 provides the details of U 1, U 2, V 1, V 2, x 1, x 2, y 2, y 2, and z at various values of P.
TABLE 1.
Details of radiographs and features at various values of P in the training and test sets (N 1 = N 2 = 170)
| P% | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
|---|---|---|---|---|---|---|---|
| U 1 = U 2 | 34 | 51 | 68 | 85 | 102 | 119 | 136 |
| V 1 = V 2 | 136 | 119 | 102 | 85 | 68 | 51 | 34 |
| x 1 = x 2 | 14960 | 22440 | 29920 | 37400 | 44880 | 52360 | 59840 |
| y 1 = y 2 | 11968 | 17952 | 23936 | 29920 | 35904 | 41888 | 47872 |
| z | 23936 | 35904 | 47872 | 59840 | 71808 | 83776 | 95744 |
The z features are passed to the K‐means clustering algorithm to create 500 mutually exclusive clusters. The center of a cluster is called the visual word. The bag of visual‐words consists of all the visual‐words. Next, the bag of visual‐words is the pass to the SVM classifier for training. Finally, the error‐correcting output codes framework is used to encode radiographs into a visual‐words histogram.
Once the SVM classifier's training is complete, the next step is to evaluate its accuracy using the testing set of radiographs. Note that the classes of radiographs are unknown in the testing set. For each radiograph in the testing set, the system extracts its feature descriptors, group feature descriptors into clusters, builds the visual vocabulary, and finally predicts the radiograph class using the pretrained SVM classifier.
6. CLASSIFICATION USING DEEP‐LEARNING‐BASED CNN
Although this work's main contribution is to build a SVM‐based system to identify positive and negative COVID‐19 cases, we also implemented deep learning‐based CNN to classify COVID‐19 images for comparison. CNN learns feature representations from COVID‐19 radiographs using layers of neurons. Since no pretrained network of COVID‐19 samples is available to us, we trained the deep learning CNN from scratch. Following layers are used to build the CNN:
The input‐layer feed radiographs to a network and applies data normalization.
The convolutional‐layer applies convolutional filters to the radiographs by moving the filters along the rows and columns of images and computing the dot product of the weights and the input and then adding a bias term.
The Batch‐normalization‐layer first normalizes each channel's activations by subtracting the mini‐batch mean and dividing by the mini‐batch SD. Then, the layer shifts the input by learning offset β and scales it by learning scaling factor γ.
The rectified linear unit layer set all the negative input elements to zero.
The fully connected‐layer multiplies the input by a weight matrix and then adds a bias vector.
The softmax‐layer applies a softmax function to the input.
The classification‐layer computes the cross‐entropy loss for multiclass classification problems with mutually exclusive classes.
7. RESULTS
Table 2 and Figure 5 show the accuracy of SVM and CNN‐based methods at various training data values. For each technique, accuracies to detect healthy and positive COVID‐19 radiographs are given separately, along with the mean accuracy (see Equation (4)). In our case, the mean accuracy is the average accuracy of two classes (health and positive COVID‐19) of radiographs at the particular training data value.
TABLE 2.
Classification accuracy of SVM and CNN at various values of a training: testing data ratio
| Training:testing | SVM classification accuracy % | CNN classification accuracy % | ||||
|---|---|---|---|---|---|---|
| Healthy | COVID‐19 | Mean | Healthy | COVID‐19 | Mean | |
| 20:80 | 92.65 | 81.62 | 87.14 | 66.18 | 75.74 | 70.96 |
| 30:70 | 89.92 | 89.08 | 89.50 | 67.23 | 71.43 | 69.33 |
| 40:60 | 90.20 | 95.10 | 92.65 | 75.49 | 73.53 | 74.51 |
| 50:50 | 90.59 | 95.29 | 92.94 | 78.82 | 69.41 | 74.12 |
| 60:40 | 89.71 | 91.18 | 90.45 | 69.12 | 80.88 | 75.00 |
| 70:30 | 92.16 | 96.08 | 94.12 | 80.39 | 76.47 | 78.43 |
| 80:20 | 91.18 | 88.24 | 89.71 | 76.47 | 73.53 | 75.00 |
Abbreviations: CNN, convolutional neural network; SVM, support vector machine.
FIGURE 5.
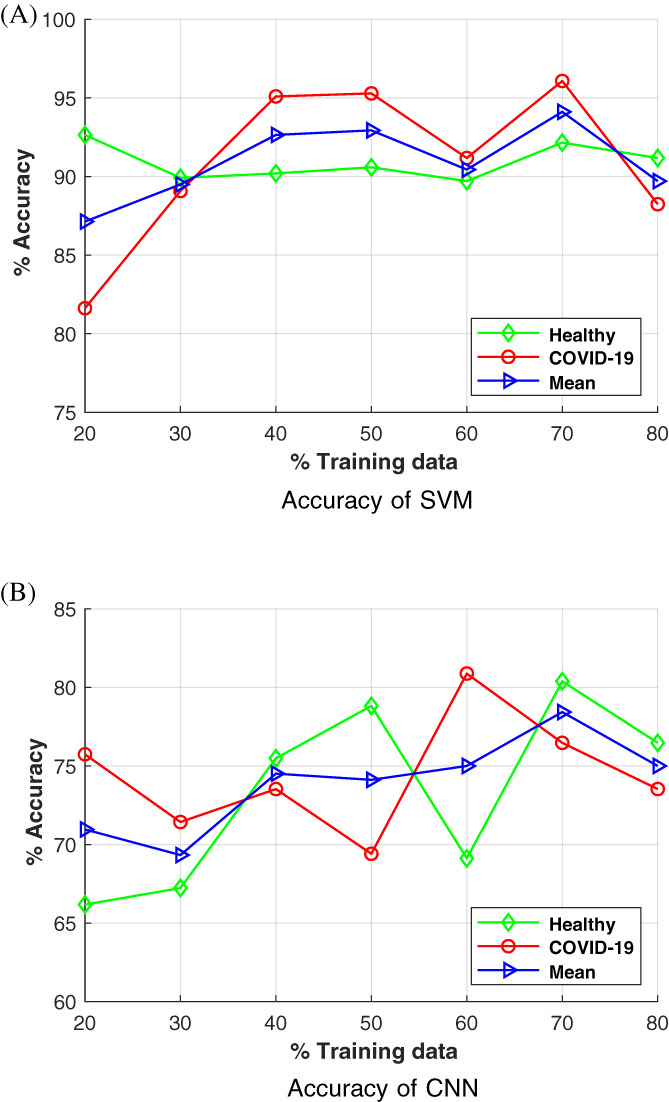
Graph of classification accuracy of support vector machine (SVM) and convolutional neural networks (CNN)‐based methods at various training data values [Color figure can be viewed at wileyonlinelibrary.com]
Figure 6 shows the run‐time for both training and testing modules. There are two graphs (upper and lower) for both CNN and SVM. The upper graph shows the run‐time of the training‐module, while the lower graph shows the runtime of the testing‐module. To test the training‐module, we used only training images. Similarly, to test the testing‐module, we used only testing images. The X‐axis only provides information, how the dataset is divided into training and testing parts. For example, in the dataset of 340 images, at 20% training data, 68 training images are used to compute the testing‐module run‐time. Similarly, at 20% training data, 272 testing images (80% testing data) are used to calculate the testing‐module run‐time.
FIGURE 6.
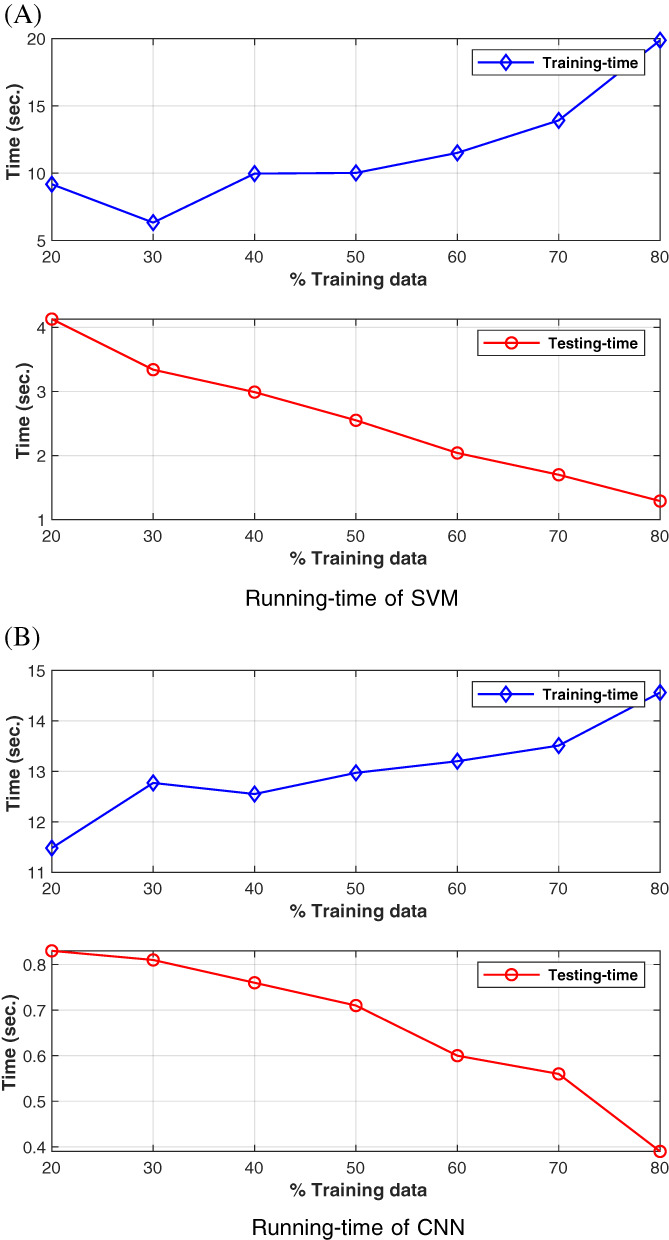
Time plots (training and testing) of support vector machine (SVM) and convolutional neural networks (CNN)‐based methods at various training data values [Color figure can be viewed at wileyonlinelibrary.com]
8. DISCUSSION
Comparative results suggest that SVM‐based method performs better than the deep‐learning‐based CNN method. SVM performs mapping of image features into high‐dimensional feature spaces and then determines the decision boundary, that is, where to draw the best hyperplane that divides the space into two subspaces, that is, healthy and COVID‐19. The CNN algorithm does not provide the optimal division of features and yield low accuracy compared to SVM. In general, deep‐learning‐based methods works better for massive data sets (thousands of images).
Figure 4 shows the features points of Healthy and Positive COVID‐19 radiographs. A closer look at this figure reveals that lesions (cloud like white areas) in the Positive COVID‐19 radiograph are much stronger than the Healthy radiograph. These lesions assist the machine learning algorithm, that is, SVM classifier to distinguish between two classes of radiographs.
Now let analyze some statistics related to confusion matrices (CMs) of Figure 7. The CM in Figure 7B belongs to the SVM method at 50:50 training: testing data, that is, out of 170 radiographs of each class, 85 are randomly chosen for training and the remaining 85 for testing. Statistics (TN, TP, FN, and FP) shown in Figure 7B correspond to testing data. Therefore, the sum of each row is 85. For example, in row 1 of this CM, out of 85 (77 + 8) radiographs, 77 are classified as Healthy while 8 as Positive COVID‐19. It means that out of 85 test radiographs, 77 are correctly classified, while 8 Healthy radiographs are incorrectly classified as Positive COVID‐19. Similarly, in row 2 of the CM, out of 85 (4 + 81) radiographs, 4 radiographs are incorrectly classified as Healthy, while 81 radiographs are correctly classified as Positive COVID‐19. The number of correctly classified radiographs, that is, 77 and 81 are written in the diagonal cells, that is, ., (1, 1) and (2, 2) of the Figure 7B. The accuracy to identify Healthy class is (77/85)*100 = 90.59%, while the accuracy to identify COVID‐19 class is (81/85)*100 = 95.29%. Therefore, the mean accuracy of SVM at 50% training: testing data are (90.59 + 95.29)/2 = 92.94%. The CM of CNN at 50:50 training: testing data ratio is shown in Figure 7C. For CNN: TN = 67, TP = 59, FN = 26, and FP = 18. Therefore, according to Equation (4) mean accuracy is: A = ((67 + 59) ÷ (67 + 59 + 26 + 18)) × 100 = 74.12%. The mean accuracy of SVM is comparatively quite higher than CNN at 50:50 training: testing data ratio.
FIGURE 7.
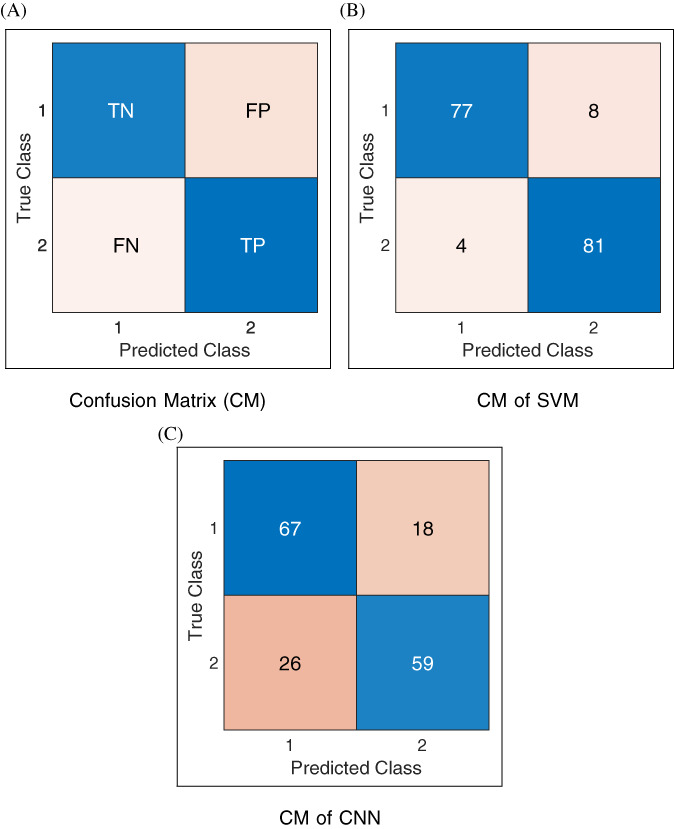
Confusion matrices at 50:50 training:testing: In the labeling of classes, 1 refers to Healthy (COVID‐19 negative), and 2 refers to COVID‐19 positive [Color figure can be viewed at wileyonlinelibrary.com]
Table 2 indicates that the maximum classification accuracy occurs at 70% of the training data. After 70%, the overfitting problem causes a decline in accuracy value. To build a fast and automatic system, we did not apply preprocessing steps such as noise filtering and cropping to remove unwanted parts from the radiographs. These preprocessing steps may improve accuracy but slow down the system speed or need human intervention.
It can be inferred from Figure 6 that with the increasing percentage of training data, the training‐time increases, while the testing‐time decreases. However, testing‐time is much smaller than the training‐time, which is one of the objectives of this research, that is, to devise a fast method to detect COVID‐19. In terms of running time, deep‐learning‐based CNN performs better than the proposed SVM‐based method. However, both approaches are fast and suitable for real‐life systems.
9. LIMITATIONS AND FUTURE WORK
COVID‐19 almost affected the lives of all the people in the world. Due to the topic's importance, we deem it worth investigating the COVID‐10 detection of the system using the well‐established image processing techniques (SURF and K‐mean clustering) and machine learning algorithms (SVM classifier). However, more novel ideas need to explore in the future, and the system needs evaluation with a larger dataset, preferably in thousands of radiographs. Currently, the polymerase chain reaction is the most reliable and standard test to detect and diagnose COVID‐19.
10. CONCLUSIONS
This work presents an image processing and machine learning‐based system to detect COVID‐19 from the chest X‐ray radiographs. The system can process thousands of images in a few minutes and produce results with high accuracy. Initially, radiographs are feed to the system that divides them into training and testing sets. Then, feature descriptors are extracted from the training set using the SURF algorithm. A visual‐vocabulary is constructed by grouping descriptors into clusters using the K‐means clustering algorithm. Feature‐descriptors train the SVM classifier. During the testing phase to determine an unknown test radiograph class, its features are extracted and clustered then sent to the pretrained SVM classifier. The classifier identifies the type of test radiograph (Healthy or COVID‐19 positive). The performance of the system is validated using 340 X‐ray radiographs. The most suitable value of K for the K‐means clustering algorithm is out to be 500. We compared the performance of our SVM‐based method with the deep‐learning‐based CNN method. The proposed SVM‐based system achieves a maximum average accuracy of 94.12% at 70:30 training: testing data ratios. After training, the system does not require any human intervention and can process thousands of images with high accuracy in a few minutes.
CONFLICT OF INTEREST
The author has no relevant financial interests in the manuscript and no other potential conflicts of interest to disclose.
Khan MA. An automated and fast system to identify COVID‐19 from X‐ray radiograph of the chest using image processing and machine learning. Int J Imaging Syst Technol. 2021;31:499–508. 10.1002/ima.22564
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in GitHub at: https://github.com/ieee8023/covid-chestxray-dataset and https://github.com/jurader/covid19_xp.
REFERENCES
- 1. Bakkouri I, Afdel K. Multi‐scale CNN based on region proposals for efficient breast abnormality recognition. Multimed Tools Appl. 2018;78(10):12939‐12960. [Google Scholar]
- 2. Sha Z, Hu L, Rouyendegh BD. Deep learning and optimization algorithms for automatic breast cancer detection. Int J Imag Syst Technol. 2020;30(2):495‐506. [Google Scholar]
- 3. Yengec Tasdemir SB, Tasdemir K, Aydin Z. A review of mammographic region of interest classification. WIREs Data Mining and Knowledge Discovery; United States: Wiley; 2020. [Google Scholar]
- 4. Adams M, Chen W, Holcdorf D, McCusker MW, Howe PDL, Gaillard F. Computer vs human: deep learning versus perceptual training for the detection of neck of femur fractures. J Med Imaging Radiat Oncol. 2018;63(1):27‐32. [DOI] [PubMed] [Google Scholar]
- 5. AlGhamdi MA, Khan MA. Intelligent analysis of Arabic tweets for detection of suspicious messages. Arab J Sci Eng. 2020;45(8):6021‐6032. 10.1007/s13369-020-04447-0. [DOI] [Google Scholar]
- 6. Saremi S, Mirjalili S, Lewis A. Grasshopper optimisation algorithm: theory and application. Adv Eng Softw. 2017;105:30‐47. [Google Scholar]
- 7. Azemin MZC, Hassan R, Tamrin MIM, Ali MAM. COVID‐19 deep learning prediction model using publicly available radiologist‐adjudicated chest x‐ray images as training data: preliminary findings. Int J Biomed Imaging. 2020;2020:1‐7. 10.1155/2020/8828855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakib S, Tazrin T, Fouda MM, Fadlullah ZM, Guizani M. DL‐CRC: deep learning‐based chest radiograph classification for COVID‐19 detection: a novel approach. IEEE Access. 2020;8:171575‐171589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murphy K, Smits H, Knoops AJG, et al. COVID‐19 on chest radiographs: a multireader evaluation of an artificial intelligence system. Radiology. 2020;296(3):E166‐E172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Artificial Intelligence to Screen for COVID‐19 on CT and X ray images. Cad4covid‐xray. https://thirona.eu/cad4covid/; 2011.Accessed February 12, 2021.
- 11. Lin L, Qin L, Xu Z, et al. Artificial intelligence distinguishes COVID‐19 from community acquired pneumonia on chest CT. Radiology. 2020;296:200905. 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song F, Shi N, Shan F, et al. Emerging 2019 novel coronavirus (2019‐nCoV) pneumonia. Radiology. 2020;295(1):210‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imran A, Posokhova I, Qureshi HN, et al. AI4covid‐19: AI enabled preliminary diagnosis for COVID‐19 from cough samples via an app. Inform Med Unlock. 2020;20:100378. 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lowe DG. Distinctive image features from scale‐invariant keypoints. Int J Comp Vis. 2004;60(2):91‐110. [Google Scholar]
- 15. Lindeberg T. Feature detection with automatic scale selection. Int J Comp Vis. 1998;30(2):79‐116. [Google Scholar]
- 16. Christopher G. Harris, Mike Stephens, et al. a combined corner and edge detector. Alvey Vis Conf. 1988;15(50):10‐5244. [Google Scholar]
- 17. Bay H, Ess A, Tuytelaars T, Van Gool L. Speededup robust features (surf). Comput Vis Image Underst. 2008;110(3):346‐359. [Google Scholar]
- 18. Bay H, Tuytelaars T, van Gool L. Surf: speeded up robust features. Proceedings of the 9th European Conference on Computer Vision ‐ Volume Part, ECCV'06I. Austria: Berlin, Springer‐Verlag; 2006:404‐417. 10.1007/11744023_32. [DOI] [Google Scholar]
- 19. Ledwich L, Williams S. Reduced sift features for image retrieval and indoor localisation. Australian Conference on Robotics and Automation. Vol 322. Australia: CiteSeer; 2004:3. [Google Scholar]
- 20. Lowe DG. Object Recognition from Local Scale‐Invariant Features. In Proceedings of the 7th IEEE International Conference on Computer Vision, Vol. 2, 1150‐1157; 1999.
- 21. Sarfraz MS, Hellwich O. On head pose estimation in face recognition. In: Ranchordas AK, Araúijo HJ, Pereira JM, Braz J, eds. Communications in Computer and Information Science. Germany: Springer Berlin Heidelberg; 2009:162‐175. [Google Scholar]
- 22. Cortes C, Vapnik V. Support‐vector networks. Mach Learn. 1995;20(3):273‐297. [Google Scholar]
- 23. Cristianini N, Shawe‐Taylor J. An Introduction to Support Vector Machines and Other Kernel‐Based Learning Methods. Cambridge, United Kingdom: Cambridge University Press; 2000. 10.1017/CBO9780511801389. [DOI] [Google Scholar]
- 24. Stoean C, Stoean R. Support Vector Machines and Evolutionary Algorithms for Classification. Switzerland: Springer International Publishing; 2014. 10.1007/978‐3‐319‐06941‐8. [Google Scholar]
- 25. Fan R‐E, Chen P‐H, Lin C‐J. Working set selection using second order information for training support vector machines. Mach Learn Res. 2005;6:1889‐1918. [Google Scholar]
- 26. Shao X, Wu K, Liao B. Single directional SMO algorithm for least squares support vector machines. Comput Intell Neurosci. 2013;2013:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kecman V, Huang TM, Vogt M. In: Wang L, ed. Support Vector Machines: Theory and Applications in Support Vector Machines. Germany: Springer International Publishing; 2005. [Google Scholar]
- 28. Cohen JP, Morrison P, Dao L. Covid‐19 image data collection. arXiv 2003.11597; 2020.
- 29. Nishio M, Noguchi S, Matsuo H, Murakami T. Automatic classification between COVID‐19 pneumonia, nonCOVID‐19 pneumonia, and the healthy on chest x‐ray image: combination of data augmentation methods. Sci Rep. 2020;10(1).1‐6. 10.1038/s41598-020-74539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GitHub at: https://github.com/ieee8023/covid-chestxray-dataset and https://github.com/jurader/covid19_xp.


