Abstract
Introduction
The ongoing COVID‐19 pandemic has led to devastating repercussions on health care systems worldwide. This viral infection has a broad clinical spectrum (ranging from influenza‐like disease, viral pneumonia, and hypoxemia to acute respiratory distress syndrome requiring prolonged intensive care unit stays). The prognostic impact of measuring viral load on nasopharyngeal swab specimens (by reverse transcriptase polymerase chain reaction [RT‐PCR]) is yet to be elucidated.
Methods
Between March 3 and April 5, 2020, we conducted a retrospective study on a cohort of COVID‐19 patients (mild or severe disease) who were hospitalized after presenting to the emergency department (ED) and had at least one positive nasopharyngeal swab during their hospital stay. We led our study at the University Hospitals of Strasbourg in the Greater East region of France, one of the pandemic's epicenters in Europe.
Results
We have collected samples from a cohort of 287 patients with a confirmed diagnosis of COVID‐19 who were included in our study. Nearly half of them (50.5%) presented a mild form of the disease, while the other half (49.5%) presented a severe form, requiring mechanical ventilation. Median (interquartile range) viral load on the initial upper respiratory swab at admission was 4.76 (3.29–6.06) log10 copies/reaction. When comparing survivors and nonsurvivors, this viral load measurement did not differ according to subgroups (p = 0.332). Additionally, we have found that respiratory viral load measurement was predictive of neither in‐hospital mortality (adjusted odds ratio [AOR] = 1.05, 95% confidence interval [CI] = 0.85 to 1.31, p = 0.637) nor disease severity (AOR = 0.88, 95% CI = 0.73 to 1.06, p = 0.167).
Conclusion
Respiratory viral load measurement on the first nasopharyngeal swab (by RT‐PCR) during initial ED management is neither a predictor of severity nor a predictor of mortality in SARS‐CoV‐2 infection. Host response to this viral infection along with the extent of preexisting comorbidities might be more foretelling of disease severity than the virus itself.
Keywords: COVID‐19, mortality, nasopharyngeal swabs, severity, viral load
INTRODUCTION
In December 2019, a devastating pandemic caused by a novel coronavirus, SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus 2) started to spread around the world. 1 As of July 1, 2020, this emerging virus has infected more than 10 million people resulting in over 500,000 deaths worldwide. The clinical spectrum of this viral infection is broad. Once infected, the patients are, for a majority, asymptomatic or only slightly symptomatic presenting influenza‐like disease. A certain number develops a mild disease and may require hospitalization (viral pneumonia, hypoxemia). A minority suffers from critical complications such as acute respiratory distress syndrome (ARDS) requiring prolonged mechanical support in the intensive care unit (ICU). 2 , 3 In such challenging times of crisis, health care systems were overwhelmed. Hence, strategies of rationing medical resources were followed. These enforced vital protocols were guided by extreme clinical anticipation and accurate triage where patients are addressed according to severity and chances of survival. 4 Several recent studies have investigated predictive factors of in‐hospital mortality and severity in COVID‐19 patients. Light was shed on clinical, biochemical risk factors, and other demographic characteristics such as comorbidities (hypertension, obesity) and age. 5 , 6 Moreover, in the context of other emerging coronavirus infections, the measurement of viral load on respiratory specimens has proven to be indicative of active virus replication and was used to assess disease progression and effectiveness of treatment. 7
The pathophysiology of this emerging viral infection is yet to be fully understood. We know so far that an immune response dysfunction and an imbalance in the host response seem to determine disease progression along with the risk of developing critical complications. 8 , 9 Sparse studies have focused on initial viral load in respiratory specimens as a marker of critical disease progression. 10 In addition, viral load on nasopharyngeal swabs (by reverse transcriptase polymerase chain reaction [RT‐PCR]) appears to be a risk factor for intubation and in‐hospital mortality. 11 Thus, viral load could be a useful marker of severe disease and may be used to identify patients eligible for more aggressive treatment or early ICU admission and to characterize populations for future therapeutic studies.
We conducted this study on a cohort of COVID‐19 patients (mild or severe disease) who were hospitalized after presenting to the emergency department (ED) at the University Hospitals of Strasbourg (France). Our main outcome was to investigate the viral load in nasopharyngeal swab upon presentation to the ED as a severity and disease progression parameter of SARS‐CoV‐2 infection.
METHODS
Study population and settings
This monocentric study was conducted in the ED of the University Hospitals of Strasbourg. This medical institution has an inpatient bed base of 2500 and its ED receives and treats about 75,000 patients yearly. It is located in the Greater East region of France, one of the most impacted regions in Europe by the pandemic, with nearly 3500 deaths and more than 12,000 infected patients as of end of June 2020. However, during the pandemic, the ED witnessed a paradoxal 40% decrease in the number of patient visits.
Between March 3 and April 5, 2020, all adult patients hospitalized for COVID‐19 were retrospectively included upon admission to the ED of a French teaching hospital. Patients were managed following current guidelines without specific therapeutic intervention. 12 All patients had at least one nasopharyngeal swab where RT‐PCR was positive. Patients who had no positive swab during their hospital stay or received outpatient care were excluded. Those who received palliative therapy or limitation of therapeutic effort upon admission to the ED were also excluded from the study.
Viral load measurement
Upon presentation to the ED, all patients underwent nasopharyngeal swab sampling on which the viral load was measured. Extraction was performed on the eMAG/eSTREAM platform (bioMérieux, Marcy l'Etoile, France) and was followed by amplification on the LightCycler 480 II system (Roche Diagnostics, France). Real‐time RT‐PCR targeted the polymerase gene using the Flo2 and Flo4 primer and probe sets. 13 Quantification was performed using successive dilutions of the CoV_IP transcript to allow the correspondence between the number of cycles and the viral load in copy/reaction.
Data collection
We retrospectively sourced the data from patients’ electronic medical records and then standardized it in a case report file. The collected data included epidemiologic, clinical, and biochemical details. Date of symptom onset was also recorded. Patients’ current treatments and medical history including cardiovascular, diabetes, renal, cancer, and hematologic diseases were collected. Overweight was defined by a body mass index superior to 30 kg/m2. Standard biochemical parameters, such as hemoglobin, platelets, C‐reactive protein (CRP), total leukocytes, and total lymphocytes, were also gathered.
Ethics
This study was approved by the local ethics committee of the University of Strasbourg in France (reference CE 2020‐35). In light of the clinical and epidemiologic context, oral consent for the use of medical data could not be obtained for all patients, but was systematically confirmed by patients’ relatives and after the critical stage by patients themselves or their relatives in case of death.
Data analysis
Continuous variables are presented as median (interquartile range [IQR]) of the distribution and were compared using nonparametric Wilcoxon rank‐sum tests. Categorical variables are described as counts and proportions and were compared using Pearson's chi‐square tests or Fisher's exact tests depending on theoretical numbers. Missing values were handled using multiple imputations by chained equations. 14 , 15 Factors associated with death or severity were identified using multivariable logistic regression models. All the variables with clinical relevance or with a p‐value of <0.2 in univariable analysis were included in the models. No further variable selection procedures were subsequently carried out. We presented the multivariable model based on clinical expertise. Multicollinearity was assessed by using the variance inflation factor (VIF) and ensuring that no factor had a VIF greater than 2. Goodness of fit for the logistic regression models was assessed by using the Hosmer‐Lemeshow test. Results are presented as adjusted odds ratios (AORs) with their 95% confidence intervals (CIs). Receiver operating characteristic (ROC) curve analysis was established to evaluate the predictive performance of viral load on death or severity. Area under the curve (AUC) was computed with its 95% CI. A p‐value of <0.05 was considered as statistically significant. All the analyses were performed using R software version 3.6.0. (R Core Team, 2019. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria; https://www.R‐project.org/).
RESULTS
Clinical characteristics of the study population
During the study period, 5421 patients presented to the ED; 1617 patients were suspected of COVID‐19 and thus received a nasopharyngeal swab. In fine, a total of 577 COVID‐positive patients were hospitalized following their visit to the ED, 154 (26.7%) of whom were in intensive care. We managed to include, after obtaining consent, a total of 287 patients with a confirmed diagnosis of COVID‐19 (Figure 1). The majority of these patients were male (65.8%, 95% CI = 60.3% to 71.3%), the median (IQR) age at time of diagnosis was 63 (50–73) years and one‐third of them were overweight (36.6%, 95% CI = 31% to 42.2%). Regarding medical history, over one‐third of patients (37%) had high blood pressure and nearly 20% of them had a history of diabetes. Upon admission to the ED, median (IQR) CRP level was 86 (30–166) mg/L and median creatinine level was 68.7 µmol/L. The respiratory viral load on the first nasopharyngeal swab was 4.76 (3.29–6.06) log10 copies/reaction. Overall patient characteristics are summarized in Table 1.
FIGURE 1.
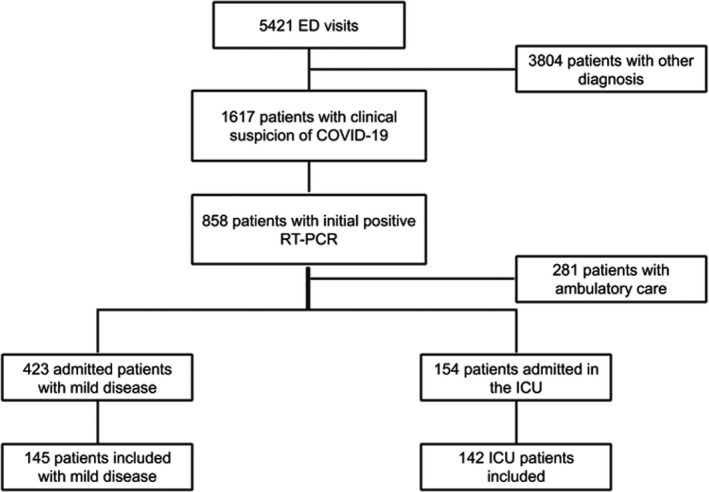
Flowchart of the study. Abbreviations: ICU, intensive care unit; RT‐PCR, reverse transcriptase polymerase chain reaction
TABLE 1.
Clinical characteristics of the study population comparing survivors and nonsurvivors
| Characteristics | Total patients (n = 287) | Nonsurvivors(n = 42) | Survivors (n = 245) | p‐value* |
|---|---|---|---|---|
| Age (y) | 63.1 (50.0–73.0) | 72.5 (64.7–77.9) | 61.4 (47.6–71.6) | <0.001* |
| Sex (men) | 189 (65.8) | 28 (66.7) | 161 (65.7) | 1 |
| Obesity (BMI > 30) | 91 (36.6) | 15 (37.5) | 76 (36.4) | 0.891 |
| Underlying diseases | ||||
| Hypertension | 106 (37.1) | 19 (45.2) | 87 (35.7) | 0.235 |
| Cardiovascular disease | 32 (11.1) | 8 (19.1) | 24 (9.8) | 0.147 |
| Diabetes mellitus | 56 (19.5) | 13 (31.0) | 43 (17.6) | 0.078 |
| Renal insufficiency | 18 (6.3) | 5 (11.9) | 13 (5.3) | 0.209 |
| Dialysis | 5 (1.7) | 3 (7.1) | 2 (0.8) | 0.048* |
| COPD | 8 (2.8) | 3 (7.1) | 5 (2.0) | 0.193 |
| Malignancies | 14 (4.9) | 4 (9.5) | 10 (4.1) | 0.264 |
| Immunotherapy | 8 (2.8) | 3 (7.1) | 5 (2.1) | 0.199 |
| Corticosteroids | 9 (3.2) | 3 (7.1) | 6 (2.5) | 0.269 |
| Laboratory findings in the ED | ||||
| CRP (mg/L) | 86 (30–166) | 122 (63–225) | 74 (25–162) | 0.007* |
| Creatinine (µmol/L) | 68.7 (60.3–82.1) | 75.9 (58.7–111) | 67.9 (61.3–79) | 0.036* |
| Anemia (<10 g/dl) | 17 (6.5) | 8 (20.0) | 9 (4.1) | 0.003* |
| Platelets (×109/L) | 194 (157–252) | 176 (143–229) | 200 (160–254) | 0.152 |
| Total leukocytes (/µL) | 4990 (3275–7745) | 6910 (4640–10,020) | 4810 (3180–7540) | 0.004* |
| Lymphocytes (/µL) | 885 (660–1305) | 750 (500–1020) | 890 (680–1340) | 0.020* |
| Viral load (log10 copies/reaction) | 4.76 (3.29–6.06) | 4.99 (3.58–6.44) | 4.76 (3.21–6.06) | 0.332 |
Data are expressed as median (IQR) or n/N (%), where N is the total number of patients with available data.
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein.
p < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Comparison and correlation between survivors and nonsurvivors
Forty‐two patients (14.6%) died during their hospitalization. Nonsurvivors were significantly older (72.5 years vs. 61.4 years, p < 0.001) than the survivors. Medical history did not differ much in the two groups, aside from more frequent history of dialysis in the nonsurvivor group (p = 0.048). At admission to the ED, nonsurvivors presented significantly higher CRP (122 mg/L vs. 74 mg/L, p = 0.007) and creatinine (p = 0.036) compared to survivors. Nonsurvivors were also more likely to present with anemia (p = 0.003) and lymphopenia (p = 0.02) than survivors. Viral load on the initial respiratory nasopharyngeal swab at admission (4.99 log10 copies/reaction vs. 4.76 log10 copies/reaction, p = 0.332) did not differ between the two subgroups.
We also studied the initial viral load's prognostic performance in the prediction of mortality. The area under the ROC curve was 54.7% (95% CI = 45.4 to 64%; Figure 2).
FIGURE 2.
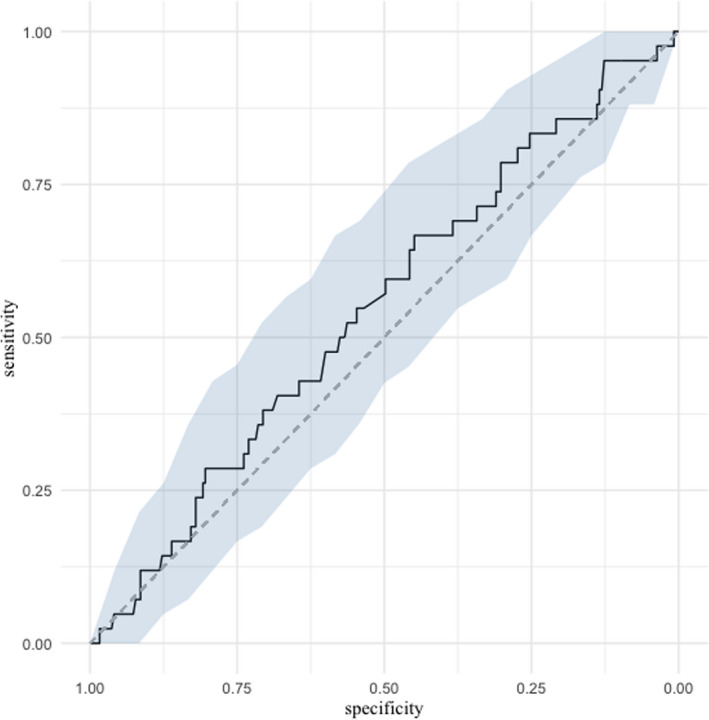
ROC curve presenting the ability of respiratory viral load to predict mortality. ROC, receiver operating characteristics
On multivariable analysis, age over 65 years was a mortality predictor (AOR = 4.70, 95% CI = 1.29 to 17.07, p = 0.019). A previous history of dialysis (AOR = 14.43) and anemia (AOR = 5.14) at baseline was also associated with in‐hospital mortality. However, the viral load on the initial nasopharyngeal swab did not appear to be significantly associated with mortality (AOR = 1.05, 95% CI = 0.85 to 1.31, p = 0.637). Results are summarized in Table 2.
TABLE 2.
Multivariable analysis of factors associated with in‐hospital mortality
| General characteristics | AOR | 95% CI | p value |
|---|---|---|---|
| Age (years) | |||
| <50 | 1 | — | — |
| 50–65 | 1.56 | 0.39–6.27 | 0.532 |
| >65 | 4.70 | 1.29–17.07 | 0.019* |
| Obesity (BMI > 30) | 1.25 | 0.52–3.04 | 0.618 |
| Men | 0.99 | 0.42–2.38 | 0.993 |
| Comorbidities | |||
| Hypertension | 0.79 | 0.33–1.89 | 0.590 |
| Cardiovascular disease | 1.14 | 0.39–3.36 | 0.809 |
| Diabetes mellitus | 1.26 | 0.50–3.19 | 0.626 |
| Renal insufficiency | 0.67 | 0.15–2.98 | 0.596 |
| Dialysis | 14.43 | 1.38–151.14 | 0.027* |
| COPD | 3.14 | 0.63–15.66 | 0.164 |
| Malignancies | 2.81 | 0.60–13.23 | 0.194 |
| Corticosteroids | 1.50 | 0.22–9.99 | 0.678 |
| Immunotherapy | 1.73 | 0.13–22.38 | 0.676 |
| Laboratory findings | |||
| CRP (>100 mg/L) | 1.32 | 0.51–3.40 | 0.567 |
| Creatinine (>90 µmol/L) | 2.15 | 0.83–5.61 | 0.118 |
| Anemia (<10 g/dL) | 5.14 | 1.19–22.19 | 0.032* |
| Platelets (>400 × 109/L) | 0.26 | 0.02–4.09 | 0.339 |
| Lymphopenia (<1500/µL) | 2.95 | 0.56–15.68 | 0.209 |
| Viral load (log10 copies/reaction) | 1.05 | 0.85–1.31 | 0.637 |
Abbreviations: AOR, adjusted odds ratio; BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein.
p < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Comparison and correlation according to disease severity
We found that nearly half of our study population, 145 patients (50.5%), presented with a mild form of the disease, while the other half, 142 patients (49.5%), displayed a severe presentation requiring ICU admission. When comparing those two subgroups, age and overall medical history did not differ (p = 0.159). Patients presenting a severe form of the disease and admitted to the ICU were mostly men (p = 0.009). Time from illness onset to hospital admission did not differ between the two groups (p = 0,23). CRP (42 mg/L vs 153 mg/L, p < 0.001), creatinine (p < 0.001), and platelets (p < 0.008) levels were significantly higher in patients presenting a severe form of the disease. However, viral load on the initial respiratory nasopharyngeal swab at admission was significantly lower in severe patients compared to patients with mild disease (4.97 log10 copies/reaction vs. 4.26 log10 copies/reaction, p = 0.007). Comparison data are summarized in Table 3.
TABLE 3.
Clinical characteristics of patients with SARS‐CoV‐2 infection by severity of disease
| Characteristics | Total patients (n = 287) |
Mild (n = 145) |
Severe (ICU) (n = 142) | p‐value |
|---|---|---|---|---|
| Age (y) | 63.1 (50.0–73.0) | 61.2 (42.1–74.8) | 65.2 (54.0–72.2) | 0.159 |
| Sex (men) | 189 (65.8) | 85 (58.6) | 104 (73.2) | 0.009* |
| Obesity (BMI > 30) | 91 (36.6) | 31 (29.0) | 60 (42.2) | 0.031 |
| Time from illness onset to hospital admission (days) | 5.5 (1–11) | 5 (1–10) | 7 (2–11) | 0,230 |
| Underlying diseases | ||||
| Hypertension | 106 (37.1) | 49 (33.8) | 57 (40.4) | 0.246 |
| Cardiovascular disease | 32 (11.1) | 17 (11.7) | 15 (10.6) | 0.755 |
| Diabetes mellitus | 56 (19.5) | 26 (17.9) | 30 (21.1) | 0.495 |
| Renal insufficiency | 18 (6.3) | 10 (6.9) | 8 (5.6) | 0.659 |
| Dialysis | 5 (1.7) | 1 (0.7) | 4 (2.8) | 0.349 |
| COPD | 8 (2.8) | 3 (2.1) | 5 (3.5) | 0.700 |
| Malignancies | 14 (4.9) | 5 (3.5) | 9 (6.3) | 0.256 |
| Immunotherapy | 8 (2.8) | 4 (2.8) | 4 (2.8) | 1 |
| Corticosteroids | 9 (3.2) | 7 (4.9) | 2 (1.4) | 0.183 |
| Laboratory findings in the ED | ||||
| CRP (mg/L) | 86 (30–166) | 42 (15–95) | 153 (89–225) | <0.001* |
| Creatinine (µmol/L) | 68.7 (60.3–82.1) | 65.8 (59.8–73.2) | 78(61–103) | <0.001* |
| Anemia (<10 g/dl) | 17 (6.5) | 5 (4.0) | 12 (9.0) | 0.104 |
| Platelets (109/L) | 194 (157–252) | 189 (147–230) | 208 (161–274) | 0.008* |
| Total leukocytes (/µL) | 4960 (3268–7748) | 4050 (2840–5598) | 7030 (4290–9383) | <0.001 |
| Lymphocytes (/µL) | 885 (660–1305) | 1035 (772–1508) | 770 (550–1022) | <0.001* |
| Viral load (log10 copies/reaction) | 4.76 (3.29–6.06) | 4.97 (3.37–6.67) | 4.26 (3.15–5.63) | 0.007* |
Data are expressed as median (IQR) or n/N (%), where N is the total number of patients with available data.
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein; ICU, intensive care unit.
p < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
We also studied the initial viral load's prognostic performance in the prediction of disease severity. The area under the ROC curve was 59.3% (95% CI = 52.7 to 65.9%; Figure 3).
FIGURE 3.
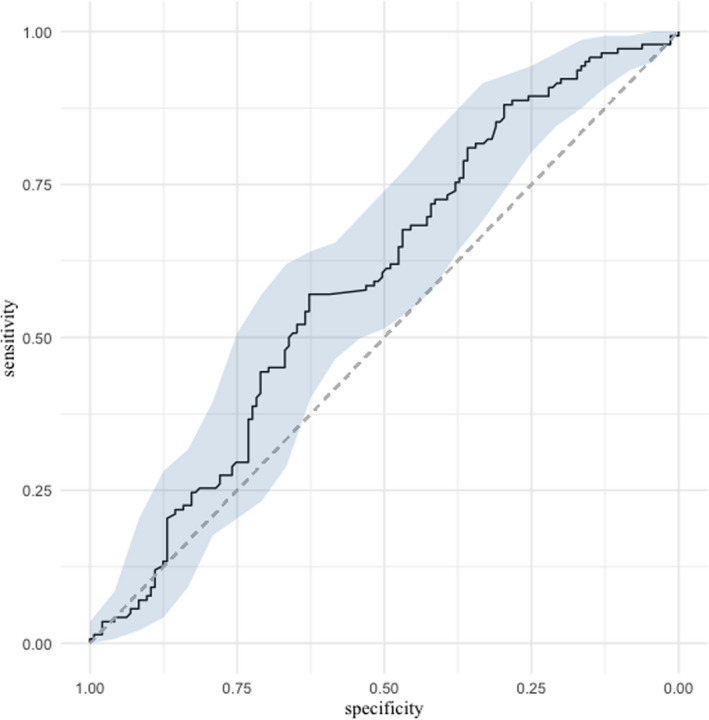
ROC curve presenting the ability of respiratory viral load to predict disease severity (ICU admission). ICU, intensive care unit; ROC, receiver operating characteristics
On multivariable analysis of the factors associated with severity of the disease on admission to the ICU, CRP (AOR = 8.82, p < 0.001), lymphopenia (AOR = 16.59, p < 0.001), and creatinine elevation (AOR = 13.51, p = 0.002) were associated with a critical form of COVID‐19. However, the viral load on the initial nasopharyngeal swab did not appear to be significantly associated with disease severity (AOR = 0.88, 95% CI = 0.73 to 1.06, p = 0.167). Results are summarized in Table 4.
TABLE 4.
Multivariable analysis of patients with SARS‐CoV‐2 infection by severity of disease (ICU admission)
| General characteristics | AOR | 95% CI | p‐value |
|---|---|---|---|
| Age (y) | |||
| <50 | 1 | — | — |
| 50–65 | 1.14 | 0.41–3.16 | 0.799 |
| >65 | 0.87 | 0.32–2.39 | 0.784 |
| Obesity | 1.30 | 0.58–2.94 | 0.524 |
| Men | 1.73 | 0.85–3.55 | 0.135 |
| Comorbidities | |||
| Hypertension | 1.95 | 0.80–4.78 | 0.149 |
| Cardiovascular disease | 0.56 | 0.17–1.85 | 0.345 |
| Diabetes mellitus | 1.02 | 0.40–2.62 | 0.962 |
| Renal insufficiency | 0.13 | 0.02–0.75 | 0.023* |
| Dialysis | 36.61 | 0.41–3298.81 | 0.118 |
| COPD | 3.20 | 0.46–22.15 | 0.241 |
| Malignancies | 4.98 | 0.80–31.09 | 0.087 |
| Corticosteroids | 0.17 | 0.01–2.19 | 0.177 |
| Immunotherapy | 5.46 | 0.37–80.19 | 0.217 |
| Laboratory findings | |||
| CRP (>100 mg/L) | 8.82 | 3.36–19.96 | <0.001* |
| Creatinine (>90 µmol/L) | 13.51 | 2.97–61.54 | 0.002* |
| Anemia (<10 g/dl) | 1.55 | 0.34–7.14 | 0.572 |
| Platelets (>400 × 109/L) | 12.11 | 1.26–116.29 | 0.035* |
| Lymphopenia (<1500/µL) | 16.59 | 4.11–66.95 | <0.001* |
| Viral load (log10 copies/reaction) | 0.88 | 0.73–1.06 | 0.167 |
Abbreviations: AOR, adjusted odds ratio; BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein; ICU, intensive care unit.
p < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
DISCUSSION
We studied a cohort of 287 hospitalized patients with a confirmed diagnosis of SARS‐CoV‐2 infection. Infection diagnosis is usually confirmed by RT‐PCR on nasopharyngeal swab on which respiratory viral load measurement is therefore possible, yet not widely used in the management of viral infections. Some authors have suggested that serum viral load correlates with inflammation (IL‐6) and clinical severity. 16 We have analyzed the initial viral loads on nasopharyngeal swabs collected at admission to the ED during the COVID‐19 pandemic. This allowed us to demonstrate that respiratory viral load was neither a risk predictor of infection severity nor mortality. To the best of our knowledge, our study is one of the first of this type, comparing viral load measurement in a population of COVID‐19 hospitalized patients.
The overall epidemiologic data of our population are comparable to what is classically demonstrated in recent literature (elder male patients with cardiovascular comorbidities). 17 , 18 We have shown that age (>65 years) was a predictor of severity supporting what has been demonstrated thus far. 19 We have also found that anemia and prior history of dialysis were predictors of mortality. Our findings align with recent studies about the frequency of acute kidney injury related to SARS‐CoV‐2 and its association with severe forms of the infection, especially in case of preexisting renal failure. 20 High CRP (>100 mg/dL), anemia (<10 g/dL), and lymphopenia (<1500/µL) were predictive of clinical severity (admission to ICU). 21 These elements highlight the role of comorbidities in the progression of the disease as in the case of sepsis; the more fragile and ailing the patient, the more severe SARS‐CoV‐2 infection is. 22
Regarding the respiratory viral load, there was no significant difference between various compared subgroups (survivors vs. nonsurvivors and mild vs. severe infection). Similarly, on multivariate analyses, we did not find any significant difference and the areas under the ROC curve confirmed the viral load's poor prognostic performance in the prediction of both mortality and disease severity. However, other studies have shown better performances of viral load levels in predicting disease outcome and mortality. 23 , 24 One of the hypotheses could be that some patients at admission deteriorated quickly due to the situation in our region, where people were advised, at that time, to stay home unless critical care was needed. This might mean that viral load at admission could already have declined from a higher level formerly attained. 25 Detection of viral nucleic acids by RT‐PCR is considered the criterion standard for the diagnosis of SARS‐CoV‐2. While RT‐PCR is the most sensitive method to detect viruses, a negative nasopharyngeal RT‐PCR result does not completely rule out the presence of COVID‐19 (sensitivity between 83% and 93%). 26 These false‐negative test results could result from inadequate sampling techniques or from a lower viral localization in the respiratory tract at certain stages of the disease. Although SARS‐CoV‐2 RT‐PCR techniques use two genes to ensure detection of potential mutant viral strains presenting genomic modifications in one of two targets, laboratory error or viral genome mutations could also happen explaining negative results. 10
Our results support the theory that host immune response to the virus displays a great influence on the evolution and severity of the disease. This response to infection seems to be more related to the extent of the hosts’ chronic comorbid medical conditions and their impaired physiological reserve than to the level of viral load itself; severity of disease progression hence seems to be more determined by the hosts’ characteristics than the quantifiable parts of the virus. Thus, potential future therapeutic strategies should consider approaching and targeting this disproportionate and dysregulated host response to viral infection. 27
The pathophysiology of this SARS‐CoV‐2–related disease is not yet well established. 28 However, the virus appears to initially target respiratory epithelial cells, endothelial cells, and lung macrophages, resulting in a true local host cell response to the infection. 9
The clinical severity of the disease (leading to ARDS) manifests, at times, brutally at the beginning of the infection and, at other times, more progressively 6 to 10 days after the onset of symptoms. 29 This biphasic evolution is underlined by a cytokine storm, which suggests a dysregulated inflammatory host response to an imbalance between pro‐ and anti‐inflammatory mediators. 30 The systemic inflammatory response syndrome is particularly responsible for activating blood coagulation and, thus, increasing risks of thrombosis in patients with severe SARS‐CoV‐2 infection. 31 These also lead to the recruitment and accumulation of leukocytes in tissues causing ARDS. Although these data shed light on the role and impact of immunologic and molecular physiopathology, further mechanisms remain to be investigated at this stage. 32
As in bacterial sepsis, viral sepsis is also the result of a dysregulated host immune response to infection. 33 A hyperinflammatory “cytokine storm” is sometimes visible at first, yet a secondary anti‐inflammatory phase (CARS syndrome) tries to counterbalance more or less concomitantly the first phase via host‐induced immunosuppression (which may lead to secondary infections). Thus, as is the case in bacterial sepsis, mortality of elder patients with SARS‐CoV‐2 infection is significant probably due to immunosenescence. 34 From a therapeutic perspective, immunotherapies are rapidly expanding in sepsis and could represent an innovative avenue in the treatment of SARS‐CoV‐2 infection.
LIMITATIONS
Our study presents some limitations like its retrospective nature and the relatively small sample size. The results of the multivariable model for in‐hospital mortality should be cautiously interpreted due to a low number of events per variable. Furthermore, our study was led in one center (Strasbourg's University Hospitals), yet it should be noted that it was conducted at the pandemic's epicenter in France. We did not monitor temporal changes in viral load over time (e.g., rate of viral load expansion or viral load clearance); this variation could be assessed as a marker of severity. In addition, viral load could be relevant to other aspects of COVID‐19 like transmissibility and other studies will be needed to address those questions. Finally, given the workload and pressure submerging our health care systems during the pandemic, the overall parameters of the hospital stay could not be exhaustively collected and detailed.
CONCLUSION
In conclusion, respiratory viral load measurement on the first nasopharyngeal swab (reverse transcriptase polymerase chain reaction) during initial ED management was neither a predictor of severity nor a predictor of mortality in SARS‐CoV‐2 infection. Host response to viral infection and the extent of preexisting comorbidities might be more determinant of disease severity and critical progression than the virus load itself.
CONFLICT OF INTEREST
The authors have no potential conflicts to disclose.
AUTHOR CONTRIBUTIONS
All authors contributed to the design of the study and analysis of data, drafting the paper, and approving the final version.
REFERENCES
- 1. Zhu NA, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berlin DA, Gulick RM, Martinez FJ. Severe covid‐19. N Engl J Med. 2020;383(25):2451–2460. [DOI] [PubMed] [Google Scholar]
- 3. Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID‐19 in the New York City Area. JAMA. 2020;323(20):2052‐2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Truog RD, Mitchell C, Daley GQ. The toughest triage ‐ allocating ventilators in a pandemic. N Engl J Med. 2020;382(21):1973‐1975. [DOI] [PubMed] [Google Scholar]
- 5. Argenziano MG, Bruce SL, Slater CL, et al. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ. 2020;369:m1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID‐19. JAMA Intern Med. 2020;180(8):1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Memish ZA, Al‐Tawfiq JA, Makhdoom HQ, et al. Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome. J Infect Dis. 2014;210:1590–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tay MZ, Poh CM, Renia L, MacAry PA, Ng LF. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng S, Fan J, Yu F, et al. Viral load dynamics and disease severity in patients infected with SARS‐CoV‐2 in Zhejiang province, China, January‐March 2020: retrospective cohort study. BMJ. 2020;369:m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Magleby R, Westblade LF, Trzebucki A, et al. Impact of SARS‐CoV‐2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2020. 10.1093/cid/ciaa851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alhazzani W, Møller MH, Arabi YM, et al. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with coronavirus disease 2019 (COVID‐19). Intensive Care Med. 2020;46(5):854‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benotmane I, Gautier Vargas G, Wendling MJ, et al. In‐depth virological assessment of kidney transplant recipients with COVID‐19. Am J Transplant. 2020;20(11):3162‐3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Buuren S, Groothuis‐Oudshoorn K. mice: multivariate imputation by chained equations in R. J Stat Softw. 2011;45(3):1‐67. [Google Scholar]
- 15. Pedersen AB, Mikkelsen EM, Cronin‐Fenton D, et al. Missing data and multiple imputation in clinical epidemiological research. Clin Epidemiol. 2017;9:157‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X, Zhao B, Qu Y, et al. Detectable serum SARS‐CoV‐2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL‐6) level in critically ill COVID‐19 patients. Clin Infect Dis. 2020;71(8):1937‐1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Y, Koh V, Marimuthu K, et al. Epidemiological and clinical predictors of COVID‐19. Clin Infect Dis. 2020;71(15):786‐792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suleyman G, Fadel RA, Malette KM, et al. Clinical characteristics and morbidity associated with coronavirus disease 2019 in a series of patients in metropolitan Detroit. JAMA Netw Open. 2020;3(6):e2012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grasselli G, Greco M, Zanella A, et al. Risk factors associated with mortality among patients with COVID‐19 in intensive care units in Lombardy, Italy. JAMA Intern Med. 2020;180(10):1345‐1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gabarre P, Dumas G, Dupont T, Darmon M, Azoulay E, Zafrani L. Acute kidney injury in critically ill patients with COVID‐19. Intensive Care Med. 2020;46(7):1339‐1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan X, Huang W, Ye B, et al. Changes of hematological and immunological parameters in COVID‐19 patients. Int J Hematol. 2020;112(4):553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan WJ, Liang WH, Zhao Y, et al. Comorbidity and its impact on 1590 patients with COVID‐19 in China: a nationwide analysis. Eur Respir J. 2020;55(5):2000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shlomai A, Ben‐Zvi H, Glusman Bendersky A, Shafran N, Goldberg E, Sklan EH. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID‐19 patients. Crit Care. 2020;24(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pujadas E, Chaudhry F, McBride R, et al. SARS‐CoV‐2 viral load predicts COVID‐19 mortality. Lancet Respir Med. 2020;8(9):e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He XI, Lau EH, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26(5):672‐675. [DOI] [PubMed] [Google Scholar]
- 26. Kim H, Hong H, Yoon SH. Diagnostic performance of CT and reverse transcriptase‐polymerase chain reaction for coronavirus disease 2019: a meta‐analysis. Radiology. 2020;296(3):E145–E155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horie S, McNicholas B, Rezoagli E, et al. Emerging pharmacological therapies for ARDS: COVID‐19 and beyond. Intensive Care Med. 2020;46(12):2265–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020;382:1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao YM, Xu G, Wang B, Liu BC. Cytokine storm syndrome in coronavirus disease 2019: a narrative review [published online ahead of print]. J Intern Med. 2020. 10.1111/joim.13144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Helms J, Severac F, Merdji H, Anglés‐Cano E, Meziani F. Prothrombotic phenotype in COVID‐19 severe patients. Intensive Care Med. 2020;46(7):1502‐1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science. 2020;369(6504):718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315(8):801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Opal SM, Girard TD, Ely EW. The immunopathogenesis of sepsis in elderly patients. Clin Infect Dis. 2005;41(Suppl 7):S504–S512. [DOI] [PubMed] [Google Scholar]


