Abstract
Recent evidence indicates that tryptophan metabolites and neurotransmitters are potential mediators of the microbiome–gut–brain interaction. Here, a high-resolution ultra-high performance liquid chromatography-electrospray ionization tandem mass spectrometry (UHPLC-ESI-MS/MS) assay was developed and validated for quantifying 50 neurotransmitters, tryptophan metabolites, and bacterial indole derivatives in mouse serum, feces, and brain. The lower limit of quantitation for the 50 compounds ranged from 0.5 to 100 nmol/L, and sample preparation procedures were adapted for individual compounds to allow quantitation within linearity of the assay with a correlation coefficient >0.99. Reproducibility was tested by intra- and interday precision and accuracy of analysis: intra- and interday precision at the lower limit of quantitation was less than 20% for all compounds, with over two-thirds of the compounds achieving an interday precision below 10%, while the interday accuracy at the lower limit of quantitation ranged from 82.3 to 128.0% for all compounds. The analyte recovery was assessed based on sample-spiked stable-isotope-labeling standards, illustrating a need to consider matrix-specific recovery discrepancies when performing interorgan comparison. Carryover was evaluated by intermittent solvent blank injection. The assay was successfully applied to determining the concentration profiles of neurotransmitter and tryptophan metabolites in serum, feces, and brain of conventionally raised specific pathogen-free (SPF) C57BL/6 mice. Our method may serve as a useful analytical resource for investigating the roles of tryptophan metabolism and neurotransmitter signaling in host–microbiota interaction.
1. Introduction
Recent next-generation sequencing (NGS) efforts have led to explosive growth in knowledge associating the trillions of the commensal micro-organisms to human health.1 One important direction of research is to gauge the neuroactive potentials gut microbiota might hold for modulating the gut–brain axis, either through neuronal or humoral routes, and to discover actionable therapeutic targets toward improved mental health.2,3 Such advances require that the much uncharted biochemical underpinnings of the microbiome–gut–brain axis are deciphered in the first place. Previous evidence derived from animal models4,5 and human cohort studies6 indicates that the gut bacteria–tryptophan/neurotransmitter metabolic network might be a key in orchestrating the gut–brain signaling in response to internal and external stimulus.
l-Tryptophan (Trp) is an essential aromatic amino acid with an indole scaffold by structure. Trp is exclusively obtained from the diet and once administrated, it undergoes three downstream catabolic pathways, including kynurenine (Kyn) pathway, serotonin (5-HT) biosynthesis, and microbial generation of indole derivatives.7 Emerging research showed that the gut microbiota actively regulates these metabolic fluxes to act on the local enteric nervous system (ENS) or to the remote central nervous system (CNS).2 For example, a recent landmark study has identified a signaling cascade through which the gut bacteria can act on the local ENS synthesis of 5-HT, a well-studied monoamine neurotransmitter with multifaceted biological function in mammals.8 Another study further demonstrated that the gut microbiota constantly regulates the maturation of ENS till adulthood through an enteric 5-HT network.9 Importantly, gut bacteria-derived indole compounds have been discovered to not only mitigate enteric inflammation in gut but impact CNS homeostasis and activity. A recent in vivo study of multiple sclerosis showed that indole, indole-3-propionic acid (IPA), and indole-3-carboxaldehyde (I3A) can directly modulate CNS inflammation by activating the aryl hydrogen receptor (AhR) that is expressed in astrocytes.10 A more recent human cohort study associated indole, indole-3-acetic acid (IAA), and skatole with anatomical and functional measures of extended central reward network and measures of diet behaviors and anxiety symptoms.11 The dominating Kyn pathway (accounting for ∼95% of the total Trp catabolic fluxes) with multifaceted CNS effects can also be affected by gut bacteria due to their control over peripheral Trp-Kyn availability.7 In parallel with Trp pathways, numerous bacterial strains such as Lactobacillus rhamnosus and Bifidobacterium breve are known to be able to produce and use neurotransmitters such as γ-aminobutyric acid (GABA) in large quantities in situ and beyond, with demonstrated effects on host neurophysiology.12
Hypothesis testing revolving around how gut microbes modulate brain function via the Trp-neurotransmitter network requires reliable, sensitive, and accurate analytical assays. Trp metabolites and neurotransmitters are structurally diverse, with wide-ranging pKa values and contrasting endogenous concentration levels. In recent years, liquid chromatography tandem mass spectrometry methods have been developed to determine Trp derivatives.13−16 However, most of the existing methods only cover 20 or fewer Trp metabolites while leaving classical neurotransmitters largely unmeasured, and none of the assays tested all relevant sample matrices (i.e., blood, feces, and brain) for advancing the field of the microbiome–gut–brain axis. Importantly, data collected by triple-quadrupole (QqQ) tandem mass spectrometry in selected reaction monitoring (SRM) mode, although sensitive and robust, renders only unit-mass resolution and offers little ion fragmentation details of the transition monitored. This drawback inherently defies novel discovery and may induce potential quantitation biases due to interferences from isobars (with close retention time, similar structure, and fragmentation pattern) when analyzing complex sample matrices. By contrast, high-resolution MS/MS analysis, specifically using quadrupole-orbitrap mass spectrometry in parallel reaction monitoring (PRM) mode not only records the full accurate-mass spectral profile of all fragmentation ion products (<5 ppm) that enables novel discovery and structural elucidation in addition to unambiguous quantitation, but confers comparable or better instrumental sensitivity compared with unit-mass SRM analysis owing to the high resolving power (e.g., up to 140 000 full width at half-maximum (FWHM) for 200 Da using the most basic Q Exactive instrumental model).17 These benefits prompt us to develop a comprehensive, accurate, and sensitive assay using high-resolution mass spectrometry for quantifying Trp catabolites, neurotransmitters, and bacterial indole derivatives with potential neuroactive effects on mammalian hosts. The assay will be validated and applied to targeted metabolomic analysis of mouse serum, feces, and brain and may be a useful analytical resource for advancing the field of microbiome–gut–brain axis.
2. Results and Discussion
2.1. Method Development and Optimization
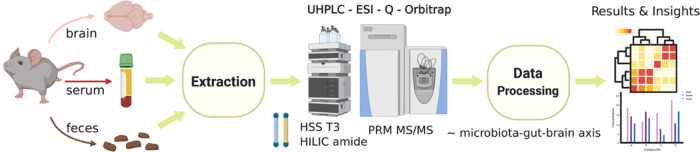
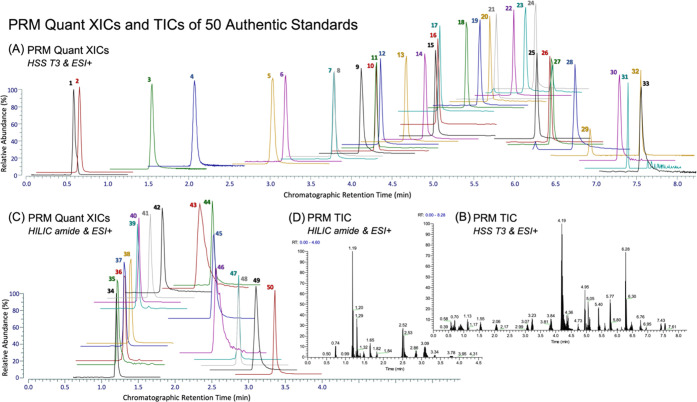
A technical schematic of the high-resolution LC-MS/MS assay is given in Figure 1, which was created by the first author with BioRender.com under a paid subscription (invoice no. 458B55C1-0003). A list of targets was first obtained from a careful literature review, and the authentic chemical standards were purchased as commercially available. Based on the preliminary tests of authentic standards, two complementary chromatographic techniques (i.e., HSS T3 and BEH amide) were applied considering the structural diversity and lipophilicity ranges of the 50 compounds and to minimize ion coelution, with PRM extraction ion chromatograms (XICs) containing at least five points for accurate quantitation. Full scan analysis of authentic standards was first conducted to survey for each compound the chromatographic retention time, ESI polarity, major ion adduct species (selected as precursor ion), and the corresponding accurate m/z. For SIL chemicals, the extent of hydrogen–deuterium exchange was further examined to assess isotope-labeling/chemical purity and the suitability-of-use as internal standards for LC-MS analysis. Then, retention time scheduled PRM MS/MS analysis of select precursor ions was conducted to determine optimal normalized collision energy (NCE) for each compound and to identify major product ions, which were further confirmed by accurate mass (<5 ppm) querying experimental mass spectral databases including mzCloud (www.mzcloud.org, Waltham, MA), metlin (www.metlin.scripps.edu, La Jolla, CA) MoNA (www.mona.fiehnlab.ucdavis.edu, Davis, CA), and hmdb (www.hmdb.ca, Edmonton, Alberta, Canada). PRM transition with the most dominant and stable product ion (relatively independent of NCE, as indicated by ion breakdown curves in mzCloud) was chosen for quantitation, whereas others were kept for confirmatory purposes. Table 1 summarizes the resultant parameters optimized for the 50 analytes and selects SIL internal standards. In addition, Figure 2 shows optimized PRM XICs and total ion chromatograms (TICs) of the 50 compounds.
Figure 1.
Technical schematic of high-resolution LC-MS/MS quantitation of 50 neurotransmitters and tryptophan metabolites of significance to the mammalian microbiome–gut–brain axis.
Table 1. List of the Compounds of Analysis with Chromatographic Retention Time and PRM Transitionsa.
| peak no. | compound | abbrev. | formula | chromat. | RT (min) | SIL ISD | polarity | precursor | precursorMZ | NCE | quant MZ | confirm MZ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | l-arginine | Arg | C6H14N4O2 | HSS T3 | 0.59 | d5-Gln | Pos | [M + H]+ | 175.1190 | 50 | 70.0651 | 116.0706; 60.0556; 130.0975 |
| 2 | l-glutamic acid | Glu | C5H9NO4 | HSS T3 | 0.66 | d5-Gln | Pos | [M + H]+ | 148.0604 | 40 | 84.0444 | 130.04987; 102.0550 |
| 3 | 2-pyrrolidinone | C4H7NO | HSS T3 | 1.55 | d3-Trp | Pos | [M + H]+ | 86.0600 | 40 | 86.0600 | 69.03362; 58.06537 | |
| 4 | nudifloramide | C7H8N2O2 | HSS T3 | 2.07 | d4-Kyn | Pos | [M + H]+ | 153.0659 | 100 | 110.0600 | 135.0551; 80.0493 | |
| 5 | kynurenine | Kyn | C10H12N2O3 | HSS T3 | 3.03 | d4-Kyn | Pos | [M + H]+ | 209.0921 | 30 | 94.0647 | 192.0655; 74.0233 |
| 6 | l-phenylalanine | Phe | C9H11NO2 | HSS T3 | 3.19 | d3-Trp | Pos | [M + H]+ | 166.0863 | 50 | 120.0808 | 131.0491; 149.0597 |
| 7 | 3-hydroxyanthranilic acid | 3-Ohaa | C7H7NO3 | HSS T3 | 3.78 | d4-Kyn | Pos | [M + H]+ | 154.0499 | 70 | 80.0492 | 108.0444; 136.0395 |
| 8 | 2-phenethylamine | PEA | C8H11N | HSS T3 | 3.79 | d3-Trp | Pos | [M + H]+ | 122.0963 | 100 | 105.0699 | 79.0542; 103.0542 |
| 9 | N-methylphenethylamine | NMPEA | C9H13N | HSS T3 | 4.12 | d3-Trp | Pos | [M + H]+ | 136.1121 | 50 | 105.0697 | 103.0541; 79.0541; 53.0388 |
| 10 | l-tryptophan | Trp | C11H12N2O2 | HSS T3 | 4.30 | d3-Trp | Pos | [M + H]+ | 205.0970 | 40 | 146.0599 | 188.0705 |
| 11 | indole-3-acrylic acid | IAcrA | C11H9NO2 | HSS T3 | 4.31 | d3-Trp | Pos | [M + H]+ | 188.0708 | 50 | 118.0646 | 146.0596; 170.0597 |
| 12 | xanthurenic acid | XA | C10H7NO4 | HSS T3 | 4.36 | d3-Trp | Pos | [M + H]+ | 206.0449 | 60 | 178.0497 | 132.0441 |
| 13 | tryptamine | C10H12N2 | HSS T3 | 4.67 | d3-Trp | Pos | [M + H]+ | 161.1073 | 40 | 144.0808 | 117.0699; 143.0730; 115.0542 | |
| 14 | N-methyltryptamine | C11H14N2 | HSS T3 | 4.91 | d3-Trp | Pos | [M + H]+ | 175.1231 | 90 | 144.0807 | 143.0729; 117.0697 | |
| 15 | N-acetylserotonin | NAS | C12H14N2O2 | HSS T3 | 5.03 | d3-Trp | Pos | [M + H]+ | 219.1128 | 80 | 115.0539 | 160.0756 |
| 16 | phenylacetyl l-glutamine | PAG | C13H16N2O4 | HSS T3 | 5.08 | d3-Trp | Pos | [M + H]+ | 265.1183 | 40 | 130.0499 | 84.0444; 91.0542 |
| 17 | hippuric acid | C9H9NO3 | HSS T3 | 5.06 | d3-Trp | Pos | [M + H]+ | 180.0655 | 50 | 105.0335 | 95.0489; 77.0382 | |
| 18 | N-(2-phenylacetyl)glycine | PAA | C10H11NO3 | HSS T3 | 5.41 | d2-IAA | Pos | [M + H]+ | 194.0812 | 50 | 91.0542 | 76.0393 |
| 19 | anthranilic acid | 2AA | C7H7NO2 | HSS T3 | 5.58 | d3-Trp | Pos | [M + H]+ | 138.0550 | 50 | 120.0440 | 92.0495; 65.0386 |
| 20 | p-coumaric acid | C9H8O3 | HSS T3 | 5.74 | d2-IAA | Pos | [M + H]+ | 165.0546 | 50 | 119.0491 | 147.0441; 91.0542 | |
| 21 | N-[3-[2-(formylamino)-5-methoxyphenyl]-3-oxypropyl]-acetamide | AFMK | C13H16N2O4 | HSS T3 | 5.78 | d2-IAA | Pos | [M + H]+ | 265.1183 | 35 | 178.0862 | 136.0757; 114.0550; 188.0706 |
| 22 | indole-3-lactic acid | C11H11NO3 | HSS T3 | 5.99 | d2-IAA | Pos | [M + H]+ | 206.0812 | 50 | 118.0650 | 130.0654; 146.0598; 170.0600 | |
| 23 | indole-3-carboxylic acid | C9H7NO2 | HSS T3 | 6.14 | d2-IAA | Pos | [M + H]+ | 162.0550 | 80 | 116.0493 | 144.0442; 118.0649; 117.0570 | |
| 24 | indole-3-carboxaldehyde | I3A | C9H7NO | HSS T3 | 6.26 | d2-IAA | Pos | [M + H]+ | 146.0596 | 80 | 91.0540 | 118.0648; 117.0570 |
| 25 | melatonin | C13H16N2O2 | HSS T3 | 6.29 | d2-IAA | Pos | [M + H]+ | 233.1287 | 65 | 159.0679 | 174.0914; 131.0730 | |
| 26 | indole-3-acetic acid | IAA | C10H9NO2 | HSS T3 | 6.44 | d2-IAA | Pos | [M + H]+ | 176.0708 | 90 | 130.0649 | 103.0539 |
| 27 | indole-3-ethanol | IEt | C10H11NO | HSS T3 | 6.45 | d2-IAA | Pos | [M + H]+ | 162.0919 | 50 | 144.0813 | 135.0810; 130.0657; 116.0500 |
| 28 | coumarin | C9H6O2 | HSS T3 | 6.77 | d2-IPA | Pos | [M + H]+ | 147.0441 | 90 | 91.0542 | 103.0542; 65.0386 | |
| 29 | indole-3-propionic acid | IPA | C11H11NO2 | HSS T3 | 6.93 | d2-IPA | Pos | [M + H]+ | 190.0863 | 80 | 130.0646 | 55.0183 |
| 30 | indole-3-acetonitrile | IAN | C10H8N2 | HSS T3 | 7.33 | d2-IPA | Pos | [M + H]+ | 157.0760 | 80 | 117.0573 | 130.0651; 90.0464; 89.0386 |
| 31 | 4-methoxyindole | C9H9NO | HSS T3 | 7.40 | d2-IPA | Pos | [M + H]+ | 148.0757 | 100 | 105.0579 | 133.0525; 117.0573; 104.0495 | |
| 32 | methyl indole-3-acetic acid | meIAA | C11H11NO2 | HSS T3 | 7.55 | d2-IPA | Pos | [M + H]+ | 190.0862 | 80 | 130.0652 | 103.0545 |
| 33 | indole | C8H7N | HSS T3 | 7.56 | d2-IPA | Pos | [M + H]+ | 118.0648 | 110 | 91.0537 | 117.0568; 65.0381 | |
| 34 | acetylcholine | ACh | C7H16NO2 | HILIC amide | 1.20 | d13-ACh | Pos | [M + H]+ | 146.1176 | 50 | 87.0441 | 60.0808 |
| 35 | 3-aminopiperidine-2,6-dione | C5H8N2O2 | HILIC amide | 1.21 | d4-5HT | Pos | [M + H]+ | 129.0659 | 40 | 84.0444 | 56.0498 | |
| 36 | 5-hydroxyindole-3-acetic acid | 5-HIAA | C10H9NO3 | HILIC amide | 1.30 | d3-Trp | Pos | [M + H]+ | 192.0655 | 80 | 117.0573 | 118.0647; 91.0537 |
| 37 | 5-hydroxy-Nω-methyltryptamine | me5HT | C11H14N2O | HILIC amide | 1.30 | d4-5HT | Pos | [M + H]+ | 191.1179 | 40 | 160.0758 | 148.0758; 132.0807 |
| 38 | tyramine | C8H11NO | HILIC amide | 1.40 | d4-5HT | Pos | [M + H]+ | 138.0913 | 80 | 103.0542 | 121.0648; 91.0542; 93.0699; 95.0491 | |
| 39 | kynurenic acid | Kyna | C10H7NO3 | HILIC amide | 1.49 | d4-5HT | Pos | [M + H]+ | 190.0499 | 90 | 116.0490 | 162.0547; 89.0381 |
| 40 | serotonin | 5-HT | C10H12N2O | HILIC amide | 1.50 | d4-5HT | Pos | [M + H]+ | 177.1022 | 90 | 115.0540 | 117.0570; 130.0649 |
| 41 | choline | C5H14NO | HILIC amide | 1.66 | d13-ACh | Pos | [M + H]+ | 104.1070 | 110 | 60.0802 | 58.0645 | |
| 42 | nicotinic acid | niacin | C6H5NO2 | HILIC amide | 1.82 | d2-GABA | Pos | [M + H]+ | 124.0393 | 110 | 96.0444 | 80.0495; 78.0338; 53.0386 |
| 43 | l-methionine | Met | C5H11NO2S | HILIC amide | 2.34 | d3-GABA | Pos | [M + H]+ | 150.0589 | 20 | 104.0528 | 56.0495; 133.0318; 61.0106; 102.0550 |
| 44 | l-proline | Pro | C5H9NO2 | HILIC amide | 2.50 | d2-GABA | Pos | [M + H]+ | 116.0706 | 50 | 70.0651 | 68.0495; 116.0706 |
| 45 | histamine | C5H9N3 | HILIC amide | 2.53 | d3-Trp | Pos | [M + H]+ | 112.0869 | 100 | 95.0604 | 68.0495; 81.0447; 54.0338; 67.0417 | |
| 46 | l-tyrosine | Tyr | C9H11NO3 | HILIC amide | 2.57 | d3-Trp | Pos | [M + H]+ | 182.0812 | 60 | 91.0542 | 119.0491; 123.0441 |
| 47 | l-pyroglutamic acid | PCA | C5H7NO3 | HILIC amide | 2.87 | d3-Trp | Pos | [M + H]+ | 130.0497 | 50 | 84.0439 | 102.0546 |
| 48 | Nα-acetyl-l-glutamine | GIcNAc | C7H12N2O4 | HILIC amide | 2.88 | d2-GABA | Pos | [M + H]+ | 189.0870 | 40 | 130.0498 | 129.0659; 172.0603 |
| 49 | trimethylamine N-oxide | TMAO | C3H9NO | HILIC amide | 3.10 | d2-GABA | Pos | [M + H]+ | 76.0757 | 35 | 58.0654 | 59.0732 |
| 50 | γ-aminobutyric acid | GABA | C4H9NO2 | HILIC amide | 3.36 | d2-GABA | Pos | [M + H]+ | 104.0703 | 40 | 87.0437 | 86.0597 |
| ISD | acetylcholine-d13, (N,N,N-trimethyl-d9; 1,1,2,2-d4) | d13-ACh | C7H3D13NO2 | HILIC amide | 1.20 | Pos | [M + H]+ | 159.1992 | 50 | 91.0694 | 91.06938; 69.13753 | |
| ISD | d2-γ-aminobutyric acid | d2-GABA | C4H7D2NO2 | HILIC amide | 3.35 | Pos | [M + H]+ | 106.0832 | 40 | 89.0565 | 89.05647; 88.07254 | |
| ISD | l-glutamine-2,3,3,4,4-d5 | d5-Gln | C5H5D5N2O3 | HSS T3 | 0.67 | Pos | [M + H]+ | 152.1078 | 40 | 135.0810 | 135.08097; 89.0756; 106.1022 | |
| ISD | l-tryptophan-2,3,3-d3 | d3-Trp | C11H9D3N2O2 | HILIC amide | 2.06 | Pos | [M + H]+ | 208.1106 | 40 | 147.0661 | 147.06608; 191.08896; 84.95950 | |
| ISD | serotonin-α,α,β,β-d4 | d4-5-HT | C10H8D4N2O | HILIC amide | 1.52 | Pos | [M + H]+ | 181.1274 | 90 | 118.0732 | 118.07319; 121.08283; 134.09101 | |
| ISD | l-glutamine-2,3,3,4,4-d5 | d5-Gln | C5H5D5N2O3 | HILIC amide | 3.81 | Pos | [M + H]+ | 152.1078 | 40 | 135.0810 | 135.08097; 89.0756; 106.1022 | |
| ISD | l-kynurenine (ring-d4, 3,3-d2) | d4-Kyna | C10H8D4N2O3 | HSS T3 | 2.97 | Pos | [M + H]+ | 213.1172 | 30 | 98.0900 | 98.09003; 198.10228 | |
| ISD | l-tryptophan-2,3,3-d3 | d3-Trp | C11H9D3N2O2 | HSS T3 | 4.34 | Pos | [M + H]+ | 208.1106 | 40 | 147.0661 | 147.06608; 191.08896; 84.95950 | |
| ISD | indole-3-acetic-2,2-d2 acid | d2-IAA | C10H7D2NO2 | HSS T3 | 6.49 | Pos | [M + H]+ | 178.0832 | 90 | 132.0775 | 132.07747; 105.06665 | |
| ISD | indole-3-propionic-2,2-d2 acid | d2-IPA | C11H9D2NO2 | HSS T3 | 6.94 | Pos | [M + H]+ | 192.0988 | 110 | 130.0650 | 130.06495; 56.02437 |
Figure 2.
PRM XICs of quant ions (A, C) and TICs (B, D) of 50 authentic chemical standards (each 5 picomoles on column) under two complementary chromatographic conditions. Compounds of analysis: 1, l-arginine (Arg); 2, l-glutamate (Glu); 3, 2-pyrrolidinone; 4, nudifloramide; 5, kynurenine (Kyn); 6, l-phenylalanine (Phe); 7, 3-hydroxyanthranilate (3-Ohaa); 8, 2-phenethylamine (PEA); 9, N-methylphenethylamine (NMPEA); 10, l-tryptophan (Trp); 11, indole-3-acrylate (IAcrA); 12, xanthurenate (XA); 13, tryptamine; 14, N-methyltryptamine; 15, N-acetylserotonin (NAS); 16, phenylacetyl l-glutamine (PAG); 17, hippurate; 18, N-(2-phenylacetyl)glycine (PAA); 19, anthranilate (2AA); 20, p-coumarate; 21, N-[3-[2-(formylamino)-5-methoxyphenyl]-3-oxypropyl]-acetamide (AMFK); 22, indole-3-lactate; 23, indole-3-carboxylate; 24, indole-3-carboxaldehyde (I3A); 25, melatonin; 26, indole-3-acetate (IAA); 27, indole-3-ethanol (IEt); 28, coumarin; 29, indole-3-propionate (IPA); 30, indole-3-acetonitrile (IAN); 31, 4-methoxyindole; 32, methyl indole-3-acetate (meIAA); 33, indole; 34, acetylcholine (ACh); 35, 3-aminopiperidine-2,6-dione; 36, 5-hydroxyindole-3-acetate (5-HIAA); 37, 5-hydroxy-Nω-methyltryptamine (N-methyl-5HT); 38, tyramine; 39, kynurenate (Kyna); 40, serotonin (5-HT); 41, choline; 42, nicotinate; 43, l-methionine (Met); 44, l-proline (Pro); 45, histamine; 46, l-tyrosine (Tyr); 47, l-pyroglutamate (PCA); 48, Nα-acetyl-l-glutamine (GlcNAc); 49, trimethylamine N-oxide (TMAO); and 50, γ-aminobutyrate (GABA).
2.2. Method Validation
Table 2 summarizes the linearity and intra- and interday precision and accuracy for this assay. Each calibration curve contained at least five consecutive points, and the linearity was determined as linear correlation coefficient >0.99. Except for N-methyl-5HT, all other 49 compounds achieved a linear range with R2 > 0.995. Since orbitrap mass spectrometry operates in such a high resolution, extraction ion chromatogram of the quant ion typically ends up with no noise ions spotted in the background. This means that detection limits could not be defined based on the signal-to-noise ratio that is commonly applied to assessing unit-mass tandem mass spec performances. Instead, the lower limit of quantitation (LLOQ), or the lower end of linear dynamic range (LDR), was defined as both intra- and interday precision <20%. As shown in Table 2, the LDR typically spanned 3 orders of magnitude, with LLOQ ranging from 0.5 to 100 nmol/L (injected 1 μL on column) for all 50 chemicals. The detection limits and linear ranges were comparable or better in comparison with previously reported assays using tandem mass spectrometry.14,15,19 At the higher end of concentration levels, we did not observe any pattern that indicated compound interactions (e.g., competition/saturation) amid the chemical mixtures during our analyses. For method accuracy and precision, rule-of-thumb criteria are 80–120% for accuracy and precision of <20%. We noted that all 50 compounds had excellent interday precision of analysis (<20%, n = 12), with 32 compounds less than 10% at the LLOQ and other 18 less than 20%; for interday accuracy, except for l-methionine (128.0%), indole-3-lactic acid (122.9%), and kynurenic acid (120.2%), other 47 compounds ranged from 82.3 to 116.1%, indicating overall excellent method accuracy. The analyte recovery rates were assessed by spiked SIL internal standards at the start of sample extraction with results summarized in Table S2 and Figure S2. Of note, analyte recovery can be different depending on the biological matrix, as represented by d4-5HT and d2-GABA. This should be carefully addressed in studies aiming at interorgan comparison. In addition, it should be noted that unlike most other compound and/or matrices, d4-5HT reached a mean recovery of 128.6% in the brain, exceeding 100%. This indicated that detection of 5-HT may involve matrix effect, meriting internal standard calibration using its own SIL chemical standards. Carryover was evaluated by examining inserted resuspension solvent blank injections with every 10 injections of samples or standard solutions. Except for occasional cases of Phe (0.027%) and Trp (0.033%) of the previous injection, no observable carryover was found for all other compounds throughout the analysis.
Table 2. Method Validation: Linearity, Intra-, and Interday Precision and Accuracy.
| name | calibration curvea | R2 | linear range (nmol/L) | CV-interdayb (%) | accuracy-interday (%) | CV-day 1 (%) | accuracy-day 1 (%) | CV-day 2 (%) | accuracy-day 2 (%) | CV-day 3 (%) | accuracy-day 3 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PEA | y = 0.0009x – 0.015 | 0.9999 | 5–10 000 | 14.7 | 103.7 | 4.5 | 103.1 | 8.3 | 124.1 | 2.3 | 87.5 |
| 2-pyrrolidinone | y = 0.0014x + 0.02 | 0.9995 | 10–1000 | 8.0 | 105.4 | 12.1 | 97.7 | 12.9 | 116.4 | 13.7 | 107.6 |
| pyroglutamine | y = 0.0002x + 0.0187 | 0.9992 | 100–10 000 | 12.2 | 100.5 | 7.1 | 92.9 | 8.6 | 91.2 | 3.6 | 117.9 |
| 3-ohaa | y = 0.0038x – 1.0377 | 0.9988 | 100–10 000 | 3.7 | 101.3 | 4.7 | 99.3 | 0.6 | 107.0 | 0.9 | 99.1 |
| 4-methoxyindole | y = 0.0004x + 0.0019 | 0.9998 | 50–10 000 | 2.8 | 101.8 | 11.1 | 98.7 | 2.8 | 105.0 | 4.3 | 103.3 |
| N-methyl-5HT | y = 0.0005x + 0.0126 | 0.9923 | 20–1000 | 21.5 | 93.3 | 5.9 | 95.6 | 5.3 | 92.5 | 6.0 | 85.1 |
| 5-HIAA | y = 0.196x + 0.8928 | 0.9992 | 0.5–1000 | 5.4 | 93.6 | 4.6 | 90.3 | 4.4 | 89.0 | 14.8 | 95.1 |
| ACh | y = 0.0026x + 0.156 | 0.9982 | 0.5–10 000 | 18.1 | 116.0 | 2.6 | 106.1 | 10.3 | 146.7 | 11.4 | 111.0 |
| AMFK | y = 0.0042x + 0.3512 | 0.9966 | 5–10 000 | 17.4 | 116.1 | 10.6 | 145.2 | 8.6 | 114.2 | 4.9 | 105.1 |
| 2AA | y = 0.0009x – 0.0063 | 0.9999 | 2–10 000 | 11.8 | 114.7 | 11.6 | 124.4 | 7.8 | 106.5 | 5.4 | 128.0 |
| choline | y = 0.0004x + 0.0031 | 0.9984 | 2–10 000 | 10.1 | 108.6 | 11.2 | 123.9 | 8.6 | 101.4 | 4.1 | 109.2 |
| coumarin | y = 0.0024x – 0.0941 | 0.9999 | 20–10 000 | 8.7 | 113.4 | 19.7 | 123.6 | 19.7 | 113.9 | 9.9 | 116.3 |
| hippuric acid | y = 0.0003x – 0.0042 | 0.9996 | 5–10 000 | 7.5 | 107.8 | 10.5 | 102.8 | 13.8 | 110.9 | 4.8 | 117.7 |
| histamine | y = 0.097x – 1.7507 | 0.9986 | 10–500 | 18.7 | 82.3 | 7.7 | 90.2 | 7.1 | 67.6 | 3.2 | 71.4 |
| indole | y = 0.0004x – 0.0103 | 0.9985 | 100–5000 | 8.0 | 97.3 | 6.2 | 106.1 | 2.6 | 95.4 | 1.5 | 87.6 |
| IAA | y = 0.0008x + 0.0419 | 0.9995 | 10–10 000 | 4.8 | 95.0 | 6.7 | 88.0 | 16.0 | 96.5 | 4.6 | 100.6 |
| IAN | y = 0.00004 – 0.0025 | 0.9996 | 100–10 000 | 6.6 | 101.3 | 3.9 | 102.9 | 1.8 | 109.2 | 10.8 | 93.0 |
| IAcrA | y = 0.0005x – 0.0063 | 0.9997 | 5–10 000 | 14.0 | 103.9 | 7.6 | 86.7 | 0.6 | 121.4 | 4.9 | 107.7 |
| I3A | y = 0.0051x + 0.0802 | 0.9994 | 2–10 000 | 2.0 | 102.8 | 11.4 | 104.3 | 1.8 | 104.3 | 9.2 | 102.4 |
| indole-3-carboxylate | y = 0.0001x – 0.0106 | 0.9996 | 100–10 000 | 10.0 | 96.8 | 2.0 | 83.0 | 15.4 | 98.9 | 14.0 | 105.5 |
| IEt | y = 0.0042x + 0.065 | 0.9999 | 5–10 000 | 15.3 | 108.8 | 8.6 | 133.7 | 12.0 | 102.3 | 4.1 | 99.3 |
| indole-3-lactic acid | y = 0.0003x – 0.0174 | 0.9994 | 20–10 000 | 13.0 | 122.9 | 14.9 | 136.7 | 16.0 | 126.3 | 18.0 | 128.8 |
| IPA | y = 0.0011x + 0.0041 | 0.9997 | 20–10 000 | 5.2 | 106.7 | 3.6 | 104.6 | 16.1 | 109.6 | 2.0 | 112.6 |
| Kyna | y = 0.0025x + 0.0448 | 0.9994 | 50–10 000 | 12.4 | 120.2 | 7.1 | 119.3 | 9.4 | 135.2 | 11.2 | 126.1 |
| Kyn | y = 0.0041x – 0.005 | 0.9997 | 10–10 000 | 2.6 | 96.5 | 13.7 | 96.6 | 5.8 | 95.0 | 6.0 | 94.3 |
| Arg | y = 0.003x – 0.1939 | 0.9989 | 5–10 000 | 14.6 | 100.7 | 8.4 | 87.7 | 6.2 | 121.4 | 13.1 | 93.7 |
| Glu | y = 0.0014x + 0.1077 | 0.9985 | 5–10 000 | 8.1 | 93.2 | 3.5 | 85.7 | 11.5 | 99.4 | 12.1 | 87.8 |
| Met | y = 0.00006x – 0.0034 | 0.9984 | 100–10 000 | 14.9 | 128.0 | 5.6 | 135.2 | 8.6 | 134.5 | 5.5 | 142.4 |
| Phe | y = 0.002x – 0.0058 | 0.9999 | 5–10 000 | 4.8 | 94.0 | 2.0 | 91.7 | 6.3 | 93.4 | 3.8 | 97.1 |
| Pro | y = 0.0064x + 0.2074 | 0.9994 | 5–5000 | 3.5 | 99.7 | 16.9 | 94.9 | 4.6 | 100.3 | 3.1 | 103.4 |
| PCA | y = 0.0005x – 0.0163 | 0.9994 | 50–10 000 | 7.1 | 96.6 | 7.7 | 100.5 | 13.2 | 86.4 | 17.6 | 99.7 |
| Trp | y = 0.0012x + 0.0038 | 0.9998 | 2–10 000 | 3.0 | 102.5 | 7.3 | 100.2 | 9.6 | 106.6 | 19.8 | 103.0 |
| Tyr | y = 0.0001x + 0.0164 | 0.9977 | 500–10 000 | 5.3 | 92.7 | 8.9 | 90.2 | 18.8 | 90.8 | 2.6 | 89.6 |
| melatonin | y = 0.0184x + 0.0187 | 0.9995 | 1–1000 | 7.1 | 96.2 | 3.6 | 100.3 | 7.8 | 98.4 | 6.5 | 86.0 |
| meIAA | y = 0.003x – 0.0259 | 0.9999 | 1–10 000 | 8.3 | 112.9 | 15.3 | 118.4 | 8.7 | 111.9 | 4.7 | 121.2 |
| PAA | y = 0.0016x – 0.0188 | 0.9999 | 5–10 000 | 6.9 | 107.8 | 9.1 | 114.7 | 14.9 | 113.6 | 11.0 | 103.0 |
| AFMK | y = 0.0006x – 0.0009 | 1 | 2–500 | 8.7 | 92.4 | 0.8 | 90.7 | 7.6 | 97.1 | 7.2 | 81.8 |
| NAS | y = 0.023x – 0.6326 | 0.9994 | 2–5000 | 8.7 | 100.7 | 2.4 | 111.8 | 2.7 | 90.3 | 3.9 | 100.6 |
| NMPEA | y = 0.0015x + 0.041 | 0.9991 | 5–10 000 | 15.5 | 113.5 | 7.7 | 139.1 | 4.8 | 109.9 | 10.7 | 105.1 |
| nicotinic acid | y = 0.00006x + 0.0003 | 0.9984 | 50–10 000 | 16.1 | 88.8 | 9.6 | 98.1 | 10.6 | 68.8 | 11.9 | 88.4 |
| niacin | y = 0.0021x – 0.0145 | 0.9987 | 10–1000 | 4.8 | 99.9 | 2.0 | 100.6 | 7.0 | 105.5 | 6.3 | 93.7 |
| Nα-acetyl-l-glutamine | y = 0.0007x – 0.0281 | 0.9998 | 20–10 000 | 4.8 | 104.9 | 11.4 | 108.0 | 3.6 | 101.2 | 1.6 | 110.2 |
| GIcNAc | y = 0.0001x – 0.0217 | 0.9961 | 100–10 000 | 18.7 | 106.7 | 4.2 | 81.4 | 5.4 | 122.9 | 6.1 | 122.5 |
| phenylacetyl l-glutamine | y = 0.0001x – 0.0217 | 0.9961 | 100–10 000 | 2.2 | 101.2 | 4.2 | 101.4 | 5.4 | 104.2 | 6.1 | 99.1 |
| PAG | y = 0.0022x + 0.1629 | 0.999 | 20–10 000 | 7.9 | 110.9 | 15.1 | 119.9 | 3.9 | 107.9 | 16.0 | 115.7 |
| 5-HT | y = 0.0002x – 0.00003 | 0.999 | 1–500 | 5.6 | 103.6 | 8.0 | 100.0 | 5.9 | 102.3 | 6.1 | 112.1 |
| TMAO | y = 0.001x – 0.0053 | 0.9997 | 5–500 | 13.2 | 97.7 | 2.8 | 108.9 | 4.2 | 102.7 | 8.5 | 79.2 |
| tyramine | y = 0.0049x + 0.6964 | 0.9959 | 5–10 000 | 8.8 | 91.3 | 10.1 | 82.1 | 15.0 | 95.7 | 12.8 | 87.6 |
| XA | y = 0.0012x – 0.011 | 0.9998 | 5–10 000 | 4.8 | 94.6 | 6.3 | 92.7 | 9.0 | 89.3 | 6.2 | 96.2 |
| GABA | y = 0.0002x + 0.0037 | 0.9998 | 20–10 000 | 6.2 | 91.6 | 1.4 | 89.6 | 7.3 | 87.8 | 8.7 | 88.8 |
The calibration curves listed here are those obtained by plotting ASD-to-ISD peak area ratio (y-axis) over the absolute concentration of targeted analytes (x-axis).
Intra- and interday precision and accuracy at the LLOQ level.
2.3. Sample Analysis
Comprehensive multicompartmental analysis of Trp metabolites and neuroactive chemicals in vivo is crucial to the discovery and validation of gut bacterial mediation of gut–brain interactions. In this study, the validated assay was used to determine the 50 Trp metabolites and neurotransmitters in serum, feces, and brain of conventional specific pathogen-free (SPF) C57BL/6 mice, as a preliminary screening of molecules related to the microbiome–gut–brain axis.
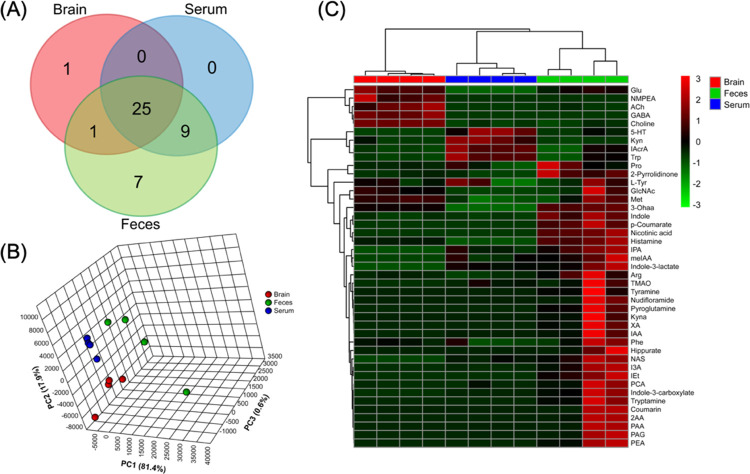
As shown in Figure 3A, a total of 43 compounds were detected and successfully quantified in samples. Among these, 25 metabolites were detected in all three matrices, including an array of gut bacteria-derived indole compounds (i.e., indole, IAA, IAcrA, I3A, and indole-3-lactate) that have been demonstrated as potent inflammation mitigators (e.g., as reactive oxygen species (ROS) scavengers or AhR ligands).10,20 Such prevailing distribution of indole derivatives in the gut–blood–brain system suggests, as an important proof-of-concept, that the gut microbiota may affect the brain function through humoral pathways. Moreover, metabolites of the glutamine–glutamate/GABA cycle, such as GABA, Glu, GlcNAc, and 2-pyrrolidinone, were extensively detected in all three organ compartments. This indicates gut bacterial GABAergic pathways as a potential mechanistic route by which our commensal microbiota modulates the gut–brain signaling.21 The result is consistent with a recently published large cohort study providing the first human population-based evidence associating altered GABA pathways with depression.6 In addition, choline and its derivatives, ACh and TMAO, were detected in all three samples, so were nicotinate (or niacin) and its nicotinamide derivative nudifloramide. For more general interests, amino acid or derivatives of neuroactive potential were found to prevail in the gut–blood–brain systems in large quantities. This includes, but not limit to the aromatic Trp, Phe, Tyr, hippurate along with nonaromatic ones such as Met, Pro, Arg, histamine. By and large, our data indicate several interesting directions of research in the budding field of a microbiome–gut–brain axis, and further studies are warranted, for example, through SIL flux analysis, transporter identification, and mono-association microbiome analysis.
Figure 3.
Tissue-specific distribution of the 50 neurotransmitters and tryptophan metabolites in conventional SPF C57BL/6 mice. (A) Venn diagram of detected compounds; (B) 3D PCA score plot (biological replicates n = 4 for each matrix); and (C) heatmap of metabolite abundance created from sample concentration levels with unit: brain (nmol/g), feces (nmol/g), and serum (μmol/mL).
Some bacterial compounds seem to be strictly compartmentalized in vivo. For example, IPA, methyl IAA, and indole-3-carboxylate were found in serum and feces but not in brain, likely due to lack of transporters across the brain–blood barrier (BBB). Such tissue-specificity or organ compartmentalization can also be seen in Figure 3B, where principal component analysis (PCA) identified a distinct separation among the three sample matrices with PC1 of 81.4% and PC2 of 17.9%. Future studies may consider pooling multiple samples as a quality control (QC) sample to inject intermittently to better delineate natural biological variance and analytical precision in real analysis. The heatmap in Figure 3C illustrates the relative abundances of detected compounds in individual sample matrices, further showing the tissue-specific distribution of these neuroactive molecules.
3. Conclusions
An accurate, precise, and sensitive high-resolution accurate-mass ultra-high performance liquid chromatography-electrospray ionization tandem mass spectrometry (UHPLC-ESI-MS/MS) assay was developed and validated for targeted analysis of 50 neurotransmitters and tryptophan metabolites in mouse serum, feces, and brain. Using high-resolution accurate-mass PRM transitions, the 50 compounds were unambiguously quantified. Sample preparation procedures were kept as simple as can be for consideration of compound coverage and throughput of analysis. Two complementary chromatographic methods were applied, to address the wide-ranging pKa, lipophilicity, and structural diversity of the Trp family molecules and neurotransmitters. Future studies aiming for more extended measurement or discovery of the Trp pathways and bacterial neuroactive molecules may find these chromatographic settings useful as a starting point. With wide linearity, analytical robustness, and sensitivity, this assay is suitable for the routine analysis of Trp catabolites and neurotransmitters in vivo, which shall benefit the elucidation of mechanistic routes through which the gut microbiota mediates mental health.
4. Materials and Methods
4.1. Chemicals and Reagents
LC/MS-grade (Optima) solvents and reagents, including water, acetonitrile (ACN), methanol (MeOH), and formic acid, were purchased from Thermo Fisher Scientific (Waltham, WA). Ammonium formate salt of trace metal purity and dimethyl sulfoxide (DMSO) of ACS reagent grade (>99.9%) were obtained from Sigma-Aldrich (St. Louis, MO). A total of 50 authentic analytical standards, spanning from classical neurotransmitters, tryptophan metabolites to bacterial indole derivatives, were procured from Sigma-Aldrich (St. Louis, MO) and Cayman Chemical (Ann Arbor, MI). Stable-isotope labeled (SIL) internal standards were purchased from Cambridge Isotopes Laboratories Inc. (Tewksbury, MA) and CDN Isotopes (Pointe-Claire, Quebec, Canada).
4.2. Stock Solution Preparation
All chemical standards, including the 50 authentic analytical standards (ASDs) and 8 SIL internal standards (ISDs), were in granular powder forms at delivery to the lab and stored at −80 °C while not in use. The stock solutions of individual standards were made by weighing the original powder to be exact using disposable antistatic microspatula (USA Scientific, Ocala, FL) on a Fisher Scientific analytical balance at 0.1 mg precision (Hampton, NH) and dissolving in MeOH, water, or DMSO to achieve a concentration level of 1.0 mg/mL or less, depending on analyte solubility specified on the Material Safety Data Sheet (MSDS). Molar concentration was converted from mass concentration using a molecular weight that was directly obtained from MSDS distributed by the chemical manufacturers.
4.3. Animal Rearing and Sample Harvest
Male specific-pathogen-free-grade C57BL/6 mice of ∼7-week-old were purchased from the Jackson Laboratories (Bar harbor, ME, USA) and housed at the University of Georgia animal facility under the following conditions: 22 °C, 40–70% humidity, and a 12:12 h light/dark cycle. Standard pelleted rodent diets and tap water were supplied ad libitum. All mice were observed for 1 week before experimental use. For sample harvest, mice were CO2-sacrificed, and at the time of euthanasia, serum, feces, and brain samples were collected, snap-frozen, and stored at −80 °C prior to analysis. The animals were treated humanely, with all animal procedures approved by the University of Georgia Institutional Animal Care and Use Committee.
4.4. Sample Preparation
Serum, feces, and cerebral cortical brain tissues are selected for targeted analysis due to their relevance to the microbiome–gut–brain axis. The extraction procedures were kept as simple as can be, shunning intricate and selective procedures for considerations of analyte coverage and throughput of analysis.
4.4.1. Blood Serum
Thawed on ice, an exact aliquot of 40 μL of serum sample was used, to which 360 μL of ice-cold ISD-containing MeOH was added, vortexed, and incubated at −20 °C for 30 min. Then, the samples were centrifuged at 15 000g for 10 min to precipitate protein and particulates. Aliquoted supernatants were subsequently dried in a CentriVap vacuum concentrator (Labconco, MO).
4.4.2. Feces
Thawed on ice, ∼20 mg of fecal matter was aliquoted for each sample to a 1.5 mL Eppendorf tube (Hamburg, Germany), which was further filled with ∼30 mg of acid-washed glass beads (Sigma-Aldrich, St. Louis, MO). To every 25 mg of fecal matter, a total of 600 μL of ice-cold ISD-containing MeOH/water (50:50, v/v) solution was added for extraction. The sample was homogenized on a TissueLyzer (Qiagen, Hilden, Germany) for 10 min at 50 Hz and centrifuged at 12 000g for 10 min. Supernatant aliquots were collected and dried in a CentriVap vacuum concentrator (Labconco, MO).
4.4.3. Cerebral Cortical Brain Tissues
Thawed on ice, ∼20 mg of thawed brain tissue of the cortical-hippocampal region was sliced off and placed in a 2 mL screw cap microcentrifuge tube (VWR, Radnor, PA) containing a 5 mm i.d. clean stainless-steel beads (Qiagen, Hilden, Germany). To every 20 mg of brain tissue, 400 μL of ice-cold ISD-spiked MeOH was added. The samples were homogenized on a TissueLyzer (Qiagen, Hilden, Germany) at 50 Hz for 2 min and incubated at −20 °C for 1 h prior to centrifugation at 18 000g for 10 min. The supernatant aliquots of the brain extracts were transferred to a CentriVap vacuum evaporator (Labconco, MO) for dryness.
Upon instrumental analysis, all dried extract aliquots were reconstituted in 98:2 water/acetonitrile and 5:95 water/acetonitrile, respectively, for HSS T3 (reverse phase, C18) and HILIC amide chromatographic analysis. Details of sample extraction, supernatant aliquoting, and solvent resuspension are summarized in Table S1.
4.5. Instrumental Analysis
High-resolution LC-MS/MS analysis was performed using a Thermo Scientific Vanquish UHPLC system coupled to a Q-Exactive mass spectrometer interfaced with a heated electrospray ionization (HESI) source and a hybrid quadrupole-orbitrap mass analyzer. The mass spectrometer was operated at 70 000 mass resolution (FWHM for 200 Da) for full scan analysis (for method development) and 13 500 in parallel reaction monitoring (PRM) mode (for targeted analysis) (Waltham, WA). The instrument was calibrated at least once a week in both ESI positive and negative modes using Thermo Scientific Pierce LTQ Velos ESI ion calibration solutions (Waltham, WA), to ensure optimal and robust instrumental performances throughout the analysis regarding mass accuracy (<5 ppm), ion transfer, ion isolation, and instrumental sensitivity.
Two complimentary chromatographic columns were used, including Waters Acquity UPLC HSS T3 (reverse phase C18, 100 Å, 1.8 μm, 2.1 mm × 100 mm) and Waters Acquity BEH amide (hydrophilic interaction chromatography, i.e., HILIC, 130 Å, 1.7 μm, 2.1 mm × 150 mm) (Milford, MA). For HSS T3, the mobiles phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in ACN (B), with a 15 min gradient: 2% B at 0–1 min; 2–15% B, 1–3 min; 15–50% B, 3–6 min; 50–98% B, 6–7.5 min; 98% B, held at 7.5–11.5 min; 98–2% B, 11.5–11.6 min; 2% B at 11.6–15 min. As of BEH amide, the mobile phases comprised 50:50 (v/v) ACN/water (A) and 15:5:80 (v/v/v) water/MeOH/ACN (B) with both added with 10 mM ammonium formate, and a 11 min gradient was used: 95% B at 0 min; 95–50% B, 0–3.5 min; 50–5% B, 3.5–5.5 min; 5% B, 5.5–6.5 min; 5–95% B, 6.5–6.7 min; and 95% B, 6.7–11 min.
The analyses were conducted in electrospray ionization (ESI) positive mode with sheath gas flow rate set as 60 L/min, aux gas flow rate at 10 L/min; sweep gas flow rate at 1 L/min, spray voltage at 2.75 kV, capillary temperature at 325 °C, and aux gas heater temperature at 400 °C. The mass spectral data for quantitation were acquired in PRM mode, with AGC target set as 2e5, maximum injection time (IT) as 50 ms, and an isolation window of 1.2 m/z for ion fragmentation. The PRM was performed according to an inclusion list containing the elemental composition, (ESI) species, charge numbers, (ionization) polarity, range of retention time for fragmentation, and normalized collision energy (NCE) of higher-energy C-trap dissociation (HCD) that have had optimized for each individual compound.
4.6. Data Processing and Quantitation
The *.RAW data acquired in both full scan and PRM modes can be manually inspected in XCalibur Qual browser 4.1.31.9 (Waltham, WA), and automatic batch peak integration was performed in TraceFinder 4.1 (Waltham, WA). Twelve-point calibration standard solutions of 0.5 nmol/L to 10 μmol/L were made, and calibration curves with at least five consecutive points were generated for each compound, with ASD-to-ISD quant ion peak area ratio being y-axis and ASD-to-ISD amount ratio x-axis. The results of peak area autointegration in TraceFinder were manually inspected in XCalibur and corrected whenever necessary.
4.7. Method Validation
The method validation was carried out following the guidelines in “Bioanalytical method validation: Guidance for industry” by the U.S. Food and Drug Administration (FDA).18
4.7.1. Linear Dynamic Range (LDR)
The LDR was determined by linear regression fitting (R2 > 0.99) between ASD-to-ISD quant ion peak area ratios (y-axis) and ASD-to-ISD amount ratios (x-axis), with the lower end (i.e., lower limit of quantitation, LLOQ) defined as the lowest point tested that had an analytical precision lower than 20%.
4.7.2. Intra- and Interday Accuracy and Precision
To assess assay robustness, calibration standard solutions at three different concentration levels (including LLOQ) were used to determine intra- and interday accuracy and precision of analysis. The intraday tests were conducted using three replicate injections within the same day (n = 3), whereas the interday tests were based on three separate days, each with three replicate injections (n = 12). Precision represented by percent relative standard deviation (RSD%) of ASD-to-ISD peak area ratio replicates was calculated in Microsoft Excel (Redmond, WA) as: RSD% = 100% × σ/μ, where μ is the mean and σ is the standard deviation; the accepted precision of analysis was set at 20%. On the contrary, accuracy (Acc) was calculated by Acc = 100% × experimental value/actual value.
4.7.3. Analytical Recovery and Matrix Effect
The analyte recovery was evaluated by analyzing all spiked SIL internal standards in the three sample matrices.
4.7.4. Carryover
Potential carryover of analysis was assessed by injecting resuspension solvent blanks every 10 samples or calibration standard solutions during analysis. The same set of columns were used for all three biological matrices, which were conducted separately to fully evaluate sample-specific carryover.
Acknowledgments
This work was funded by National Institute of Environmental Health Sciences (NIEHS) under award nos. R01ES024950, R03ES032067, R35ES028366, P30ES010126, and P42ES031007. We especially thank the instrumentation support from the Chemistry and Analytical Core (CAC) of the UNC Superfund Research Program (P42ES031007). The authors would also like to acknowledge partial support from the Chen-Yu Yen and Whay-Ray C. Yen Graduate Fellowship awarded by the UNC Gillings School of Global Public Health (2018-2019).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c05789.
Overview of sample processing for addressing wide dynamic ranges of compounds (Figure S1); barplots of analyte recovery in sample matrices as indicated by SIL standards (Figure S2); sample pretreatment procedures for individual compounds (Table S1); and analyte recovery rates in sample matrices (Table S2) (PDF)
Author Present Address
† Metaorganism Immunity Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892, United States.
The authors declare no competing financial interest.
Supplementary Material
References
- Kau A. L.; Ahern P. P.; Griffin N. W.; Goodman A. L.; Gordon J. I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327. 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan T. G.; Cryan J. F. The Microbiome-Gut-Brain Axis in Health and Disease. Gastroenterol. Clin. North Am. 2017, 46, 77–89. 10.1016/j.gtc.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Groen R. N.; de Clercq N. C.; Nieuwdorp M.; Hoenders H. J. R.; Groen A. K. Gut microbiota, metabolism and psychopathology: A critical review and novel perspectives. Crit. Rev. Clin. Lab. Sci. 2018, 55, 283–293. 10.1080/10408363.2018.1463507. [DOI] [PubMed] [Google Scholar]
- Gao J.; Xu K.; Liu H.; Liu G.; Bai M.; Peng C.; Li T.; Yin Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M.; Nollen E. A. A.; Rohrig U. F.; Fallarino F.; Opitz C. A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discovery 2019, 18, 379–401. 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- Valles-Colomer M.; Falony G.; Darzi Y.; Tigchelaar E. F.; Wang J.; Tito R. Y.; Schiweck C.; Kurilshikov A.; Joossens M.; Wijmenga C.; Claes S.; Van Oudenhove L.; Zhernakova A.; Vieira-Silva S.; Raes J. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- Cervenka I.; Agudelo L. Z.; Ruas J. L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- Yano J. M.; Yu K.; Donaldson G. P.; Shastri G. G.; Ann P.; Ma L.; Nagler C. R.; Ismagilov R. F.; Mazmanian S. K.; Hsiao E. Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F.; Grasset E.; Manneras Holm L.; Karsenty G.; Macpherson A. J.; Olofsson L. E.; Backhed F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 6458–6463. 10.1073/pnas.1720017115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V.; Mascanfroni I. D.; Bunse L.; Takenaka M. C.; Kenison J. E.; Mayo L.; Chao C. C.; Patel B.; Yan R.; Blain M.; Alvarez J. I.; Kebir H.; Anandasabapathy N.; Izquierdo G.; Jung S.; Obholzer N.; Pochet N.; Clish C. B.; Prinz M.; Prat A.; Antel J.; Quintana F. J. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osadchiy V.; Labus J. S.; Gupta A.; Jacobs J.; Ashe-McNalley C.; Hsiao E. Y.; Mayer E. A. Correlation of tryptophan metabolites with connectivity of extended central reward network in healthy subjects. PLoS One 2018, 13, e0201772 10.1371/journal.pone.0201772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J. A.; Forsythe P.; Chew M. V.; Escaravage E.; Savignac H. M.; Dinan T. G.; Bienenstock J.; Cryan J. F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 16050–16055. 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hényková E.; Vranova H. P.; Amakorova P.; Pospisil T.; Zukauskaite A.; Vlckova M.; Urbanek L.; Novak O.; Mares J.; Kanovsky P.; Strnad M. Stable isotope dilution ultra-high performance liquid chromatography-tandem mass spectrometry quantitative profiling of tryptophan-related neuroactive substances in human serum and cerebrospinal fluid. J. Chromatogr. A 2016, 1437, 145–157. 10.1016/j.chroma.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Lefèvre A.; Mavel S.; Nadal-Desbarats L.; Galineau L.; Attucci S.; Dufour D.; Sokol H.; Emond P. Validation of a global quantitative analysis methodology of tryptophan metabolites in mice using LC-MS. Talanta 2019, 195, 593–598. 10.1016/j.talanta.2018.11.094. [DOI] [PubMed] [Google Scholar]
- Chen G.-Y.; Zhong W.; Zhou Z.; Zhang Q. Simultaneous determination of tryptophan and its 31 catabolites in mouse tissues by polarity switching UHPLC-SRM-MS. Anal. Chim. Acta 2018, 1037, 200–210. 10.1016/j.aca.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley L.; Nye L. C.; Grant I.; Andreas N.; Chappell K. E.; Sarafian M. H.; Misra R.; Plumb R. S.; Lewis M. R.; Nicholson J. K.; Holmes E.; Swann J. R.; Wilson I. D. Ultrahigh-Performance Liquid Chromatography Tandem Mass Spectrometry with Electrospray Ionization Quantification of Tryptophan Metabolites and Markers of Gut Health in Serum and Plasma-Application to Clinical and Epidemiology Cohorts. Anal. Chem. 2019, 91, 5207–5216. 10.1021/acs.analchem.8b05884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsein G. E.; Pamir N.; von Haller P. D.; Kim D. S.; Oda M. N.; Jarvik G. P.; Vaisar T.; Heinecke J. W. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J. Proteomics 2015, 113, 388–399. 10.1016/j.jprot.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. Bioanalytical Method Validation: Guidance for Industry; US Food & Drug Administration, 2018.
- Marcos J.; Renau N.; Valverde O.; Aznar-Lain G.; Gracia-Rubio I.; Gonzalez-Sepulveda M.; Perez-Jurado L. A.; Ventura R.; Segura J.; Pozo O. J. Targeting tryptophan and tyrosine metabolism by liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2016, 1434, 91–101. 10.1016/j.chroma.2016.01.023. [DOI] [PubMed] [Google Scholar]
- Wlodarska M.; Luo C.; Kolde R.; d’Hennezel E.; Annand J. W.; Heim C. E.; Krastel P.; Schmitt E. K.; Omar A. S.; Creasey E. A.; Garner A. L.; Mohammadi S.; O’Connell D. J.; Abubucker S.; Arthur T. D.; Franzosa E. A.; Huttenhower C.; Murphy L. O.; Haiser H. J.; Vlamakis H.; Porter J. A.; Xavier R. J. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37 e6. 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj A.; Moro E.; Bistoletti M.; Orlandi V.; Crema F.; Giaroni C. Glutamatergic Signaling Along The Microbiota-Gut-Brain Axis. Int. J. Mol. Sci. 2019, 20, 1482 10.3390/ijms20061482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.