Abstract
Background
Nerve injury-induced changes in gene expression in the dorsal root ganglion (DRG) contribute to neuropathic pain genesis. Eukaryotic initiation factor 4 gamma 2 (eIF4G2) is a general repressor of cap-dependent mRNA translation. Whether DRG eIF4G2 participates in nerve injury-induced alternations in gene expression and nociceptive hypersensitivity is unknown.
Methods
The expression and distribution of eIF4G2 mRNA and protein in mouse DRG after spinal nerve ligation (SNL) were assessed. Effects of eIF4G2 siRNA microinjected through a glass micropipette into the injured DRG on the SNL-induced DRG mu opioid receptor (MOR) and Kv1.2 downregulation and nociceptive hypersensitivity were examined. In addition, effects of DRG microinjection of adeno-associated virus 5-expressing eIF4G2 (AAV5-eIF4G2) on basal DRG MOR and Kv1.2 expression and nociceptive thresholds were analysed.
Results
eIF4G2 protein co-expressed with Kv1.2 and MOR in DRG neurones. Levels of eIF4G2 mRNA (1.7 [0.24] to 2.3 [0.14]-fold of sham, P<0.01) and protein (1.6 [0.14] to 2.5 [0.22]-fold of sham, P<0.01) in injured DRG were time-dependently increased on days 3–14 after SNL. Blocking increased eIF4G2 through microinjection of eIF4G2 siRNA into the injured DRG attenuated SNL-induced downregulation of DRG MOR and Kv1.2 and development and maintenance of nociceptive hypersensitivities. DRG microinjection of AAV5-eIF4G2 reduced DRG MOR and Kv1.2 expression and elicited hypersensitivities to mechanical, heat and cold stimuli in naïve mice.
Conclusions
eIF4G2 contributes to neuropathic pain through participation in downregulation of Kv1.2 and MOR in injured DRG and is a potential target for treatment of this disorder.
Keywords: dorsal root ganglion, eIF4G2, Kv1.2, mu opioid receptor, neuropathic pain
Editor's key points.
-
•
Nerve injury-induced changes in gene expression in the dorsal root ganglion (DRG) contribute to neuropathic pain.
-
•
This study examined the role of eukaryotic initiation factor 4 gamma 2 (eIF4G2) in nerve injury-induced alternations in gene expression and nociceptive hypersensitivity in mouse DRG.
-
•
Nerve injury increased expression of eIF4G2 in injured DRG, and blocking this increase rescued injury-induced downregulation of mu opioid receptors and Kv1.2 in injured DRG and attenuated development and maintenance of injury-induced nociceptive hypersensitivities.
-
•
Increased expression of eIF4G2 contributes to neuropathic pain through downregulation of Kv1.2 and mu opioid receptor expression in injured DRG and may be a target for treatment of this disorder.
Neuropathic pain caused by a lesion or disease of the somatosensory system is a major clinical and socioeconomic problem affecting 7–10% of the world's population.1 In the USA alone, this public health problem translates into billions of US dollars in healthcare expenses and lost productivity.2 Neuropathic pain is characterised by ongoing or intermittent spontaneous pain, often burning in nature, and exaggerated pain in response to noxious stimuli (hyperalgesia) or innocuous stimuli (allodynia).3 Current treatments for these pain hypersensitivities are limited as medications such as NSAIDs and opioids are ineffective and lead to severe side-effects.2
Peripheral nerve injury leads to changes in the expression of pain-associated genes at both transcriptional and translational levels in the first-order sensory neurones of dorsal root ganglia (DRG).4,5 These changes play a key role in neuropathic pain development and maintenance.4,6, 7, 8, 9, 10 For example, peripheral nerve injury downregulates the expression of mu opioid receptor (MOR) and Kv1.2 in injured DRG.6,10, 11, 12, 13, 14, 15, 16 Rescuing this downregulation attenuates nerve injury-induced pain hypersensitivity.6,10, 11, 12, 13, 14, 15, 16 Mimicking their downregulation leads to pain-like hypersensitivity through depolarisation of the resting membrane potential and diminishing current threshold for action potential generation in injured DRG neurones or augmenting MOR-controlled neurotransmitter release from the primary afferents.6,10, 11, 12, 13, 14, 15, 16 Thus, understanding how expression of these pain-associated genes is altered in the DRG after peripheral nerve injury may provide a new avenue for neuropathic pain management.
Eukaryotic initiation factor 4F (eIF4F) is one of the protein complexes involved in the initiation phase of protein translation and is an important effector of post-transcriptional gene regulation.17 The eIF4F complex consists of three subunits: ATP-dependent RNA helicase eIF4A, cap-binding protein eIF4E, and scaffolding protein eIF4G.18 eIF4G comprises three isoforms: eukaryotic initiation factor 4 gamma 1 (eIF4G1), eIF4G2, and eIF4G3.19 eIF4G2 (also known as DAP-5, p97, and NAT1) shares similarities in its C-terminal region (containing the binding sites for eIF4A and eIF3) with eIF4G1.18,20, 21, 22 Unlike eIF4G1, which enhances cap-dependent and cap-independent RNA translation, eIF4G2 acts as a general repressor of cap-dependent translation of most RNAs by forming translationally inactive complexes. Interestingly, eIF4G2 was also shown to promote non-canonical, cap-independent translation initiation, and cap-dependent translation of select mRNAs.23, 24, 25 Previous reports have shown that eIF4G2 plays a critical role in several pathological processes such as cancer, autoimmunity, and neurodegenerative diseases.26,27 Whether eIF4G2 participates in peripheral nerve injury-induced neuropathic pain remains unknown.
In this study, we first examined whether expression of eIF4G2 at mRNA and protein levels was increased in the DRG in a well-characterised mouse model of neuropathic pain induced by unilateral fourth lumbar spinal nerve ligation (SNL) or sciatic nerve chronic constriction injury (CCI). We then determined whether this increase contributed to the development and maintenance of SNL-induced nociceptive hypersensitivities. We finally investigated the mechanisms underlying this process.
Methods
Animals
CD1 mice (7–8 weeks old, male, weight 20–25 g) were obtained from Charles River Laboratories (Wilmington, MA, USA). The mice were group-housed (four per cage) with a standard 12 h light and dark cycle at 23 [2]°C, with food and water available ad libitum. The study was reported in accordance with the guidelines of Animal Research: Reporting of In Vivo Experiments (ARRIVE).28 Sample sizes (eight to 10 mice per group) were similar to those used in previous studies of neuropathic pain.11, 12, 13, 14, 15, 16,29, 30, 31, 32, 33, 34 No missing data and no outliers occurred in this study. Animals were randomly assigned to various treatment groups. All efforts were made to minimise the suffering of animals and the number of animals used. Theexperimenters were blinded to group assignment and treatment conditions during all behavioral tests, including locomotor function test. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Rutgers University and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the International Association for the Study of Pain.
Experimental protocol
Mice were randomised to each model on day 0. Expression of eIF4G2 mRNA and eIF4G2 protein in the DRG and spinal cord was assessed on days 3, 7, and 14 after surgery. The number of eIF4G2-positive neurones was assessed in the DRG on day 7 after surgery. siRNA was microinjected into the injured DRG 5 days before surgery or on day 7 after surgery. Adeno-associated virus 5 (AAV5) was microinjected into unilateral L3 and L4 DRGs. Behavioural nociceptive testing was assessed 1 day before siRNA or AAV5 microinjection, 1 day before surgery, and on days 3–14 after surgery or on weeks 2–8 after microinjection. Expression of eIF4G2, Kv1.2 and MOR in the injured DRG and expression of ERK1 and 2, p-ERK1 and 2 and glial fibrillary acidic protein (GFAP) in the ipsilateral L4 spinal cord were determined on days 5 and 14 after surgery or 8 weeks after microinjection.
Neuropathic pain models
The fourth lumbar (L4) SNL-induced neuropathic pain model was carried out as described.10,12,30 Briefly, mice were anaesthetised with 2–3 vol% [correct?] isoflurane and placed in the prone position. The unilateral L4 spinal nerve was exposed through removal of the L5 transverse process. After exposure and isolation of the L4 spinal nerve, a tight ligation with 7–0 silk thread was made. The ligated nerve was then transected distal to the ligature under a surgical microscope. The skin and muscle were finally closed in layers. The sham group underwent identical procedure except for nerve ligation.
The sciatic nerve CCI-induced neuropathic pain model was carried out as described.10,12,30 In brief, mice were anaesthetised with 2–3 vol% isoflurane. The unilateral sciatic nerve was exposed and loosely ligated with 7–0 silk thread at four sites with intervals of about 1 mm proximal to the trifurcation of the sciatic nerve. Sham group animals were subjected to sciatic nerve exposure and isolation without ligation.
Dorsal root ganglion microinjection
DRG microinjection was carried out as described.10,12,30 Briefly, a midline incision was performed in the lower lumbar back region and the unilateral L4 DRG (for siRNA) or L3 and L4 DRGs (for AAV5) exposed after removing the corresponding articular processes. AAV5 (1 μl per DRG, 4×1012 GC ml−1) or siRNA (1 μl per DRG, 40 μM; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA; catalog # sc-35170 for eIF4G2 siRNA and catalog # sc-44230 for negative control siRNA) was microinjected into the exposed DRG through a glass micropipette connected to a Hamilton syringe using a dissection microscope. Injected DRGs were stained with blue dye to confirm successful injection. After microinjection, the pipette was kept in place for 10 min before removal. The surgical field was irrigated with sterile saline and iodophor and closed with wound clips. None of the mice showed abnormal locomotor activities after DRG microinjection. Injected DRGs were also stained with haematoxylin and eosin to examine whether their structural integrity was maintained and whether they contained visible leucocytes.
Behavioural tests
Mechanical, thermal, and cold nociceptive tests, conditioned place preference (CPP) tests, and locomotor function tests were carried out as described.10,12,30 To minimise intra- and inter-individual variability in behavioural outcome measurements, animals were acclimatised for at least 2 days before experiments.
Paw withdrawal frequencies (PWFs) in response to calibrated von Frey filament stimuli (0.07 and 0.4 g; Stoelting Co., Wood Dale, IL, USA) were measured. Briefly, mice were placed in a Plexiglas chamber on an elevated mesh screen. Each von Frey filament was applied to the plantar side of a hind paw for ∼1 s, and this was repeated 10 times for each hind paw at 5 min intervals. A quick withdrawal of the paw was regarded as a positive response. Paw withdrawal response in each of these 10 applications was recorded as a percentage withdrawal frequency [(number of paw withdrawals in 10 trials) × 100 = % response frequency].
Paw withdrawal latencies (PWLs) in response to noxious heat stimuli were examined with a Model 336 Analgesic Meter (IITC Life Science Inc., Woodland Hills, CA, USA). Briefly, mice were placed in an individual Plexiglas chamber on a glass plate. A beam of light from a hole of the light box through the glass plate was applied on the middle of the plantar surface of each hind paw. A quick lift of the hind paw automatically turned off the light. The duration of the light beam was defined as the PWL. Each paw was tested three times with 5 min intervals between each trial. A cut-off time of 20 s was used to avoid tissue damage.
PWLs to noxious cold (0°C) were examined by placing mice in an individual Plexiglas chamber on the cold aluminium plate with continuous temperature monitoring. The length of time between placement of the mouse on the plate and the first sign of paw flinching and jumping was defined as the PWL. Each trial was repeated three times at 10 min intervals for the paw on the ipsilateral side. A cut-off time of 20 s was used.
For the CPP test, an apparatus with two Plexiglas chambers (same size, different visual and textured cues) connected by a 4 cm wide × 6 cm high removable door (MED Associates Inc., St. Albans, VT, USA) was used. Time spent in each chamber was monitored by photobeam detectors and automatically recorded in MED-PC IV CPP software. Preconditioning was performed 7 weeks after viral microinjection. Mice were allowed to explore both chambers for 30 min with the internal door opened every day for 3 days. On the fourth day, the mice were placed into one chamber with full access to both chambers for 15 min. The duration of time spent in each chamber was recorded. The mice that spent >720 or <180 s in any chamber were excluded from further testing. The conditioning protocol was then performed for the following 3 days with the internal door closed. The mice were first injected intrathecally with saline (5 μl) specifically paired with one conditioning chamber in the morning for 30 min. About 6 h later, the mice were then injected intrathecally with lidocaine (0.4%, 5 μl) and placed in another chamber for 30 min. Injection order of saline and lidocaine was switched every day. On the test day, at least 20 h after conditioning, the mice were placed in one chamber with free access to both chambers. The time spent in each chamber was recorded for 15 min. Difference scores for chamber preference were calculated by subtracting preconditioning preference time from test time spent in the lidocaine-paired chamber.
Locomotor function, including placing, grasping, and righting reflexes, was examined after the above-described behavioural tests. (1) Placing reflex: the hind limbs were placed slightly lower than the forelimbs, and the dorsal surfaces of the hind paws were brought into contact with the edge of a table. Whether the hind paws were placed on the table surface reflexively was recorded. (2) Grasping reflex: after the animal was placed on a wire grid, whether the hind paws grasped the wire on contact was recorded. (3) Righting reflex: when the animal was placed on its back on a flat surface, whether it immediately assumed the normal upright position was recorded. Each trial was repeated five times at 5 min intervals and the scores for each reflex were recorded based on counts of each normal reflex.
Neuronal culture and transfection
Dorsal root ganglion neuronal cultures were prepared as described.13,14 Briefly, bilateral DRGs collected from 4-week-old CD1 mice were treated with enzyme solution (5 mg ml−1 dispase, 1 mg ml−1 collagenase type I in Hanks' balanced salt solution [HBSS] without Ca2+ and Mg2+; Thermo Fisher Scientific Inc., Waltham, MA, USA) at 37°C for 20 min. After trituration and centrifugation, dissociated neurones were resuspended in cold Neurobasal Medium (Thermo Fisher Scientific) containing 10% fetal bovine serum (JR Scientific, Woodland, CA, USA) and 1% penicillin and streptomycin (Quality Biological, Gaithersburg, MD, USA), and were plated onto poly-d-lysine-coated (Sigma, St. Louis, MO, USA) six-well plates. Neurones were cultured at 37°C in a 95% O2, 5% CO2 atmosphere. On the second day, 2–10 μl of AAV5 (titre ≥1× 1012 GC ml−1) was added to each well. For siRNA transfection, siRNA was diluted to a concentration of 100 nM and transfected into cultured neurones by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Three days later, neurones were harvested for Western immunoblot analysis.
RNA extraction and quantitative real-time reverse transcription–polymerase chain reaction
Total RNA extraction and quantitative real-time reverse transcription–polymerase chain reaction (RT–PCR) assays were carried out as described.14,32,34 Briefly, unilateral L4 DRG from four mice were pooled to achieve enough RNA. Total RNA was extracted using the miRNeasy kit with on-column digestion of genomic DNA (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, and then reverse-transcribed using ThermoScript reverse transcriptase (Invitrogen) using oligo (dT) primers. The template was amplified in a Bio-Rad CFX96 real-time PCR system using the following primers for eIF4G2 mRNA (forward: 5′-CTCCTCAATGTGGGTGTAGAGT-3′, reverse: 5′-AGCGAGCTATACTTTGGCTCTT-3′) or for Tuba1a (forward: 5′- TGTGGATTCTGTGGAAGGCG-3′; reverse: 5′- AAGCACACATTGCCACATACAA-3′). Each sample was run in triplicate in a 20 μl reaction containing 250 nM forward and reverse primers, 10 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), and 20 ng of cDNA. PCR was carried out with an initial 3 min incubation at 95°C, followed by 39 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and a final 5 min incubation at 72°C. Ratios of ipsilateral-side mRNA levels to contralateral-side mRNA levels were calculated using the ΔCt method (2−ΔΔCt). Data were normalised to Tuba1a, an internal control shown to be stable after peripheral nerve injury.29, 30, 31
Plasmid construction and virus production
To construct a plasmid expressing eIF4G2 protein, the full-length sequences of eIF4G2 mRNA from mouse DRG was reverse-transcribed using the SuperScript III One-Step RT-qPCR System with Platinum Taq High Fidelity DNA Polymerase (ThermoFisher Scientific) and amplified by PCR with gene-specific primers (forward: 5′-ATAGAATTCGCCACCGTGGAGAGTGCGATTG; reverse: 5′-GCGTCTAGATTAGTCAGCTTCTTCCTC). The resulting segment was inserted into the pro-viral plasmid at the EcoRI and XbaI restriction sites. The recombinant clones were verified using DNA sequencing. For packaging of AAV5 particles, HEK-293 cells were transfected with the pro-viral plasmid expressing eIF4G2 or Gfp using a PEI Transfection method with pHelper. Three days later, the transfected cells were collected and AAV5 particles were purified using the AAV-pro Purification Kit (Takara Bio, Mountain View, CA, USA).
Western immunoblotting
Protein extraction and Western immunoblotting were carried out as described.14,32,34 Briefly, unilateral L4 DRG from four mice or unilateral L3 and L4 DRGs from two mice were pooled to achieve enough protein. DRG or spinal cord samples were homogenised and cultured neurones ultrasonicated on ice with a lysis buffer (10 mM Tris, 1 mM phenylmethylsulfonyl fluoride, 5 mM MgCl2, 5 mM ethylene glycol tetraacetic acid [EGTA], 1 mM ethylenediaminetetraacetic acid [EDTA], 1 mM dithiothreitol, 40 μM leupeptin, 250 mM sucrose). After centrifugation at 4°C for 15 min at 1000×g, the supernatant was collected. After protein concentration was measured, samples were heated for 5 min at 99°C and loaded onto a 15% stacking plus 7.5% separating sodium dodecyl sulphate (SDS)–polyacrylamide gel (Bio-Rad Laboratories). Proteins were electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories). Membranes were first blocked with 3% non-fat milk in Tris-buffered saline containing 0.1% Tween-20 for 1 h and then incubated overnight at 4°C with the following primary antibodies: rabbit anti-eIF4G2 (1:1000; Invitrogen), rabbit anti-phosphorylated extracellular signal-regulated kinase 1 and 2 (p-ERK 1 and 2) (1:1000; Cell Signaling Technology Inc., Beverly, MA, USA), rabbit anti- ERK 1 and 2 (1:1000; Cell Signaling Technology), mouse anti-GFAP (1:1000; Cell Signaling Technology), rabbit anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:3000; Santa Cruz Biotechnology), rabbit anti-MOR (1:1000; Neuromics, Edina, MN, USA), or mouse anti-Kv1.2 (1:500, NeuroMab, Davis, CA, USA). Proteins were detected by horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (1:3000; Bio-Rad Laboratories) and visualised using peroxide reagent and luminol plus enhancer reagent (Clarity Western ECL Substrate; Bio-Rad Laboratories) with exposure using a ChemiDoc XRS System with Image Lab software (Bio-Rad Laboratories). The intensity of bands was quantified by densitometry using Image Lab software (Bio-Rad Laboratories), and was normalised to GAPDH.
Immunohistochemistry
Single- or double-labelled immunofluorescence histochemistry was performed as described.11,16,31 After mice were deeply anaesthetised with isoflurane, they were transcardially perfused with 50–100 ml of 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). The DRG was harvested, post-fixed, and cryoprotected in 30% sucrose in 0.1 M phosphate buffer at 4°C overnight. Tissue was sectioned at a thickness of 20 μm on a cryostat. For single labelling, every fourth section was collected (at least three to four sections per DRG). After being blocked for 1 h at 37°C in phosphate-buffered saline (PBS) containing 10% goat serum and 0.3% Triton X-100, sections were incubated alone with rabbit anti-eIF4G2 (1:200; Invitrogen) overnight at 4°C. Sections were then incubated with goat anti-rabbit IgG conjugated to Cy3 (1:200; Jackson ImmunoResearch, West Grove, PA, USA) for 2 h at room temperature. Control experiments included substitution of normal mouse serum for the primary antiserum and omission of the primary antiserum. For double labelling, seven sets of sections (at least one to two sections per set) from naïve DRGs were collected from each tissue by grouping every seventh serial section. The seven sets were incubated overnight at 4°C with primary rabbit anti-eIF4G2 and one each of the following primary antibodies or reagents, respectively: chicken anti-β III tubulin (1:200; EMD Millipore, Billerica, MA, USA), mouse anti-glutamine synthetase (GS; 1:200; Sigma-Aldrich), mouse anti-NF200 (1:100; Sigma-Aldrich), biotinylated isolectin B4 (IB4, 1:100; Sigma-Aldrich), mouse anti-calcitonin gene-related peptide (CGRP; 1:50; Abcam, Cambridge, UK), guinea pig anti-MOR (1:1000; EMD Millipore) and mouse anti-Kv1.2 (1:200; NeuroMab, Davis, CA, USA). Sections were then incubated for 1 h at 37°C with a mixture of goat anti-rabbit IgG conjugated with Cy3 (1:500, Jackson ImmunoResearch, West Grove, PA, USA) and donkey anti-mouse IgG conjugated with Cy2 (1:500, Jackson ImmunoResearch), donkey anti-guinea pig IgG conjugated to Cy2 (1:500, Jackson ImmunoResearch), donkey anti-chicken IgG conjugated to Cy2 (1:500, Jackson ImmunoResearch), or FITC-labelled avidin D (1:200, Sigma-Aldrich). Fluorescent photomicrographs were captured using a DMI 4000 fluorescence microscope (Leica Microsystems, Mannheim, Germany) with a DFC365 FX camera (Leica) and analysed using NIH ImageJ software.
Statistical analysis
All data are expressed as mean (standard error of the mean [SD]). Data distribution was assumed to be normal based on previous studies.11, 12, 13, 14, 15, 16,29, 30, 31, 32, 33, 34 Results from the behavioural tests, RT–PCR, Western immunoblot, and immunohistochemistry were analysed using one-way or two-way analysis of variance (anova) or paired or unpaired Student's t-tests. When anova showed a significant difference, pairwise comparisons between means was performed using the post hoc Tukey method. Data were analysed by SigmaPlot 12.5 (SYSTAT Software, San Jose, CA, USA). All probability values were two tailed, and a P value <0.05 was considered statistically significant.
Results
Expression and distribution of eIF4G2 in doral root ganglion
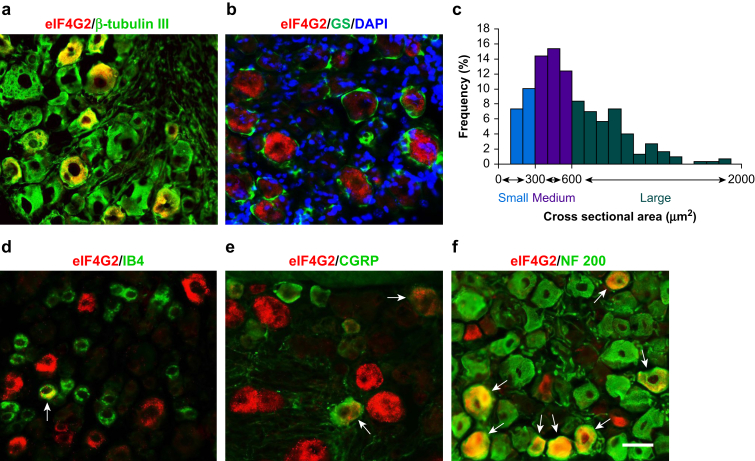
To determine the role of eIF4G2 in neuropathic pain, we first examined its cellular expression in DRG. Double labelling of eIF4G2 and β-tubulin III (a specific neuronal marker) or glutamine synthetase (GS, a marker for satellite glial cells) was carried out. eIF4G2 co-expressed with β-tubulin III in some cells (Fig. 1a) but was not detected in GS-labelled cells (Fig. 1b), indicating that eIF4G2 is expressed predominantly in DRG neurones. About 34% (92 in 269) of β-tubulin III-labelled neurones were positive for eIF4G2. Cross-sectional area analysis of neuronal somata showed that ∼41% of eIF4G2-labelled neurones were large (>600 μm2 in area; Fig. 1c), 42% were medium (300–600 μm2 in area; Fig. 1c), and 17% were small (<300 μm2 in area; Fig. 1c). A DRG subpopulation analysis revealed that ∼13% of eIF4G2-positive neurones were labelled by IB4 (a marker for small non-peptidergic neurones; Fig. 1d), 14% by CGRP (a marker for small DRG peptidergic neurones; Fig. 1e), and 79% by NF200 (a marker for medium and large neurones and myelinated Aβ fibres; Fig. 2f).
Fig 1.
Distribution of eIF4G2 protein in lumbar dorsal root ganglion (DRG) of naïve mice. (a, b) Double-label immunofluorescent staining revealed co-localisation of eIF4G2 (red) with β-tubulin III (green; a), but not with glutamine synthetase (GS; green; b), in the cytoplasm of DRG cells. (b) Cellular nuclei were labelled by 4′,6-diamidino-2-phenylindole (DAPI; blue). (c) Size distribution of eIF4G2-positive neuronal somata in naïve DRG. Large, 41%; medium: 42%; small: 17%. (d–f) Double-labelled immunofluorescent staining (arrows) of eIF4G2 (red) with isolectin B4 (IB4; green), calcitonin gene-related peptide (CGPR; green) or neurofilament 200 (NF200; green) in DRG neurones. n=3 mice. Scale bar, 50 μm. eIF4G2, eukaryotic initiation factor 4 gamma 2.
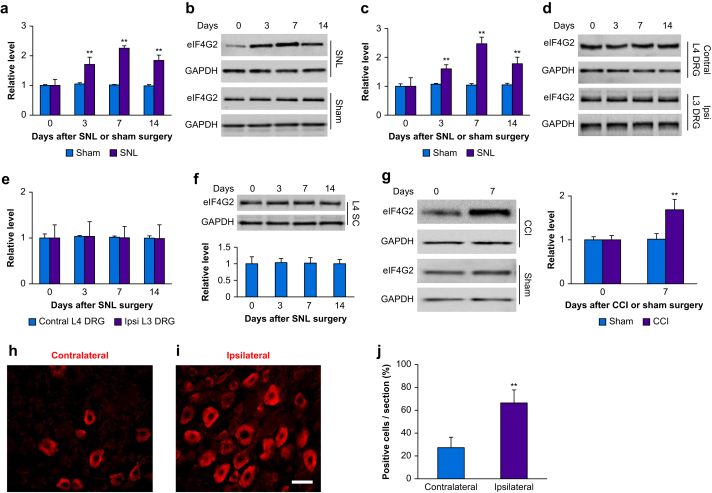
Fig 2.
Peripheral nerve injury-induced increase in expression of eIF4G2 mRNA and eIF4G2 protein in injured dorsal root ganglion (DRG). (a) Levels of eIF4G2 mRNA in the ipsilateral L4 DRG on days 3, 7, and 14 after spinal nerve ligation (SNL) or sham surgery. Four unilateral L4 DRGs from four mice were pooled together. n=3 biological repeats (12 mice) per group per time point. Two-way analysis of variance (anova) with repeated measure followed by post hoc Tukey test. Fgroup (1, 16)=174. ∗∗P<0.01 vs the sham group at the corresponding time point. (b, c) eIF4G2 protein expression in the ipsilateral L4 DRG on days 3, 7, and 14 after SNL or sham surgery. (b) Representative Western immunoblots. (c) Statistical summary of the densitometric analysis. Four unilateral L4 DRGs from four mice were pooled together. n=3 biological repeats (12 mice) per group per time point. Two-way anova with repeated measure followed by post hoc Tukey test. Fgroup (1, 16)=33. ∗∗P<0.01 vs the sham group at the corresponding time point. (d–f) Expression of eIF4G2 protein in the contralateral (Control) L4 DRG (d, e), ipsilateral (Ipsi) L3 DRG (d, e) and ipsilateral L4 spinal cord dorsal horn (SC; f) on days 3, 7, and 14 after SNL. Four unilateral L4 DRGs from four mice were pooled together. n=3 biological repeats (DRGs from 12 mice and spinal cord from three mice) per group per time point. One-way anova with repeated measure followed by post hoc Tukey test. Ftime (3, 8)=0.70 for contralateral L4 DRG, 0.01 for ipsilateral L3 DRG and 0.02 for ipsilateral L4 SC. (g) Expression of eIF4G2 in the ipsilateral L3 and L4 DRGs on day 7 after CCI or sham surgery. Two unilateral L3 and L4 DRGs from two mice were pooled together. n=3 biological repeats (six mice) per group per time point. Two-way anova with repeated measure followed by post hoc Tukey test. Fgroup (1, 8)=25. ∗∗P<0.01 vs the sham group at the corresponding time point. (h–j) Neurones labelled by eIF4G2 in the contralateral (h) and ipsilateral (i) L4 DRG on day 7 after SNL. (h, i) Representative immunostaining images. (j) Statistical summary of the number of eIF4G2-labelled neurones. n=3 mice. ∗∗P<0.01 vs the contralateral side by two-tailed unpaired Student's t-test. Scale bar, 50 μm. CCI, chronic constriction injury; eIF4G2, eukaryotic initiation factor 4 gamma 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Time-dependent increase in eIF4G2 in injured dorsal root ganglion after peripheral nerve injury
Next, we examined whether eIF4G2 expression was altered in DRG after peripheral nerve injury. Unilateral SNL increased expression of eIF4G2 mRNA and eIF4G2 protein in the ipsilateral, but not contralateral, L4 DRG (Fig. 2a–f). The levels of eIF4G2 mRNA in the ipsilateral L4 DRG were increased by 1.7-fold (P<0.01), 2.3-fold (P<0.01), and 1.8-fold (P<0.01) on days 3, 7, and 14 after SNL, respectively, compared with those after sham surgery at the corresponding time points (Fig. 2a). Expression of eIF4G2 protein in the ipsilateral L4 DRG was also increased by 1.6-fold (P<0.01), 2.5-fold (P<0.01), and 1.8-fold (P<0.01) on days 3, 7, and 14 after SNL, respectively, as compared with naïve mice (0 day; Fig. 2b and c). Sham surgery did not alter basal expression of eIF4G2 in the ipsilateral L4 DRG (Fig. 2b and c). Neither SNL nor sham surgery changed basal levels of eIF4G2 in the contralateral L4 DRG, ipsilateral (intact) L3 DRG or ipsilateral L4 spinal cord (Fig. 2d–f). Results were similar after CCI. The level of eIF4G2 protein was increased by 1.7-fold in the ipsilateral L3 and L4 DRGs on day 7 after CCI as compared with sham mice at the corresponding time point (P<0.01; Fig. 1g). The number of eIF4G2-labelled neurones in the ipsilateral L4 DRG was 2.4-fold greater than that in the contralateral L4 DRG on day 7 after SNL (P<0.01; Fig. 2h–j).
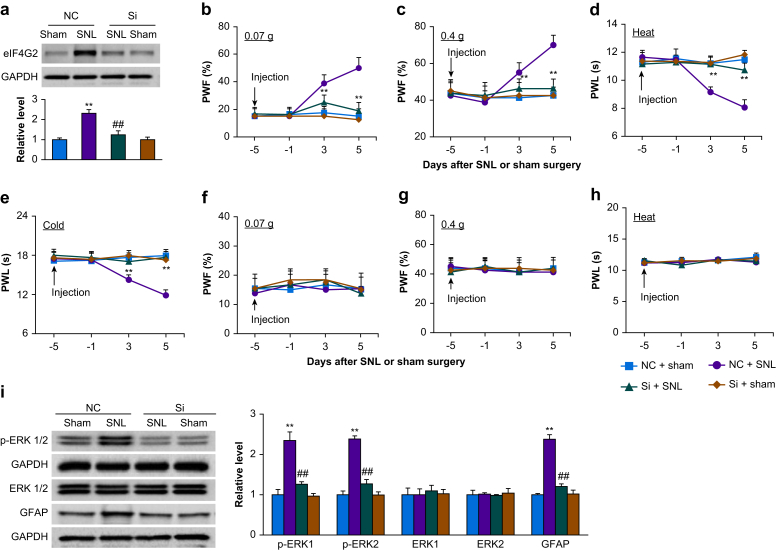
Effect of microinjection of eIF4G2 siRNA on development of nociceptive hypersensitivity
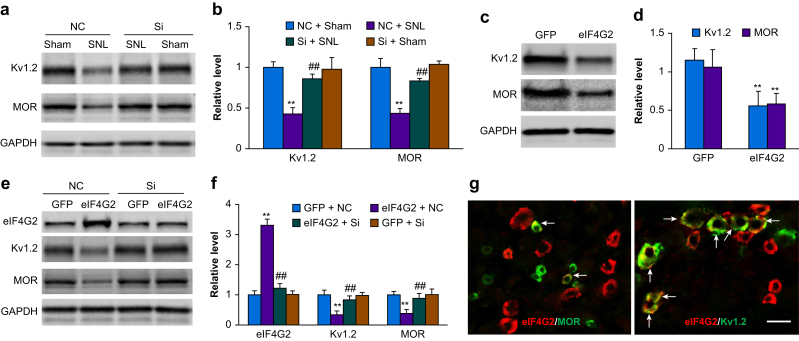
We examined whether the increased eIF4G2 in the injured DRG participated in neuropathic pain. We blocked this increase through microinjection of eIF4G2 siRNA into the ipsilateral L4 DRG 5 days before mice were subjected to either SNL or sham surgery. Negative control siRNA was used as the control. As expected, the level of eIF4G2 protein was significantly increased by 2.6-fold in the ipsilateral L4 DRGs in negative control siRNA-microinjected SNL mice as compared with the negative control siRNA-microinjected sham mice on day 5 after surgery (P<0.01; Fig. 3a). However, this increase was not seen in the eIF4G2 siRNA-microinjected SNL mice (1.2-fold of negative control siRNA-sham, P=0.15; Fig. 3a). Neither siRNA altered basal expression of eIF4G2 in the ipsilateral L4 DRG of sham mice (Fig. 3a).
Fig 3.
Effect of dorsal root ganglion (DRG) microinjection of eIF4G2 siRNA on spinal nerve ligation-induced nociceptive hypersensitivities and dorsal horn central sensitisation during the development period. (a) Expression of eIF4G2 protein in the ipsilateral L4 DRG on day 5 after SNL or sham surgery in mice microinjected with eIF4G2 siRNA (Si) or negative control siRNA (NC). n=3 biological repeats (12 mice) per group. Two-way analysis of variance (anova) with repeated measures followed by post hoc Tukey test. Fgroup (1, 8)=42. ∗∗P<0.01 vs the NC plus sham group. ##P<0.01 vs the NS plus SNL group. (b–h) Effect of micoinjection of eIF4G2 siRNA (Si) or negative control siRNA (NC) into the ipsilateral L4 DRG on PWF to 0.07 g (b and f) and 0.4 g (c and g) von Frey filament stimuli and on paw withdrawal latency (PWL) to heat (d and h) and cold (e) stimuli on the ipsilateral (b–e) and contralateral (f–h) sides at the different days after SNL or sham surgery. n=8 mice per group. Two-way anova with repeated measures followed by post hoc Tukey test. Fgroup (3, 112)=50 for 0.07 g, 14 for 0.4 g, 55 for heat, and 49 for cold. ∗∗P<0.01 vs the NC plus SNL group at the corresponding time points. (i) Effect of microinjection of eIF4G2 siRNA (Si) or negative control siRNA (NC) into the ipsilateral L4 DRG on the expression of p-ERK1 and 2, total ERK1 and 2, and GFAP in the ipsilateral L4 dorsal horn on day 5 after SNL or sham surgery. Left: representative Western immunoblots. Right: statistical summary of densitometric analysis. n=3 mice per group. Two-way anova with repeated measures followed by post hoc Tukey test. Fgroup (1, 8)=30 for p-ERK1, 108 for p-ERK2, 0.50 for ERK1, 0.01 for ERK2, and 128 for GFAP. ∗∗P<0.01 vs the corresponding NC plus sham group. ##P<0.01 vs the corresponding NC plus SNL group. eIF4G2, eukaryotic initiation factor 4 gamma 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; p-ERK, phosphorylated extracellular signal-regulated kinase 1; PWF, paw withdrawal frequency; PWL, paw withdrawal latency.
We further observed the behavioural responses of SNL or sham mice after prior microinjection of siRNA. Consistent with previous reports,23,32 SNL led to long-term mechanical allodynia, thermal hyperalgesia, and cold allodynia on the ipsilateral side in negative control siRNA-microinjected mice (Fig. 3b–e). The PWFs in response to 0.07 and 0.4 g von Frey filament stimuli applied to the ipsilateral hind paw were significantly increased on days 3 and 5 after SNL as compared with pre-surgery baseline values (P<0.01; Fig. b and c). The PWLs of the ipsilateral hind paw in response to heat and cold were markedly reduced on days 3 and 5 after SNL (P<0.01; Fig. 3d and e). Microinjection of eIF4G2 siRNA into the ipsilateral L4 DRG largely blocked the SNL-induced increases in PWFs to 0.07 g (25% [5.3%] vs 38% [6.4%] on days 3 and 18% [6.4%] vs 50% [7.6%] on day 5, P<0.01; Fig. 3b) and 0.4 g (46% [5.2%] vs 55% [5.3%] on day 3 and 46% [5.2%] vs 70% [5.3%] on day 5, P<0.01; Fig. 3c) von Frey filament stimuli. The SNL-induced decreases in PWLs to heat (11 [0.5] s vs 9 [0.4] s on day 3 and 11 [0.5] s vs 8 [0.6] s, P<0.01; Fig. 3d) and cold (17 [0.9] s vs 14 [0.7] s on day 3 and 18 [0.9] s vs 12 [0.8] s on day 5, P<0.01; Fig. 3e) stimuli on days 3 and 5 after SNL. DRG microinjection of neither siRNA affected basal paw responses to mechanical, heat, or cold stimuli on the contralateral side of SNL mice, and on either side of sham mice during the observation period (Fig. 3b–h). Both SNL and sham mice with DRG microinjection of either siRNA displayed normal locomotor activity (Supplementary Table S1).
We also examined whether prior microinjection of eIF4G2 siRNA into the DRG affected SNL-induced dorsal horn central sensitisation as indicated by increases in phospho-ERK1 and ERK2 (a marker for neuronal hyperactivation) and GFAP (a marker for astrocyte hyperactivation) in the dorsal horn during the development period. In line with previous studies,23,32 levels of p-ERK1 and 2 (but not total ERK1 and 2) and GFAP increased in the ipsilateral L4 dorsal horn on day 5 after SNL in the negative control siRNA-microinjected SNL mice, but not in sham mice (P<0.01; Fig. 3i). These increases were absent in eIF4G2 siRNA-microinjected SNL mice (1.24 [0.07]-fold vs 2.32 [0.21]-fold for p-ERK1, 1.27 [0.11]-fold vs 2.39 [0.08]-fold for p-ERK2 and 1.21 [0.07]-fold vs 2.38 [0.13]-fold for GFAP, P<0.01; Fig. 3i). Neither siRNA altered basal expression of p-ERK1 and 2, total ERK1 and 2 or GFAP in the dorsal horn of sham mice (Fig. 3i). Taken together, these results indicate that increased eIF4G2 in the injured DRG may be required for development of SNL-induced nociceptive hypersensitivity and dorsal horn central sensitisation.
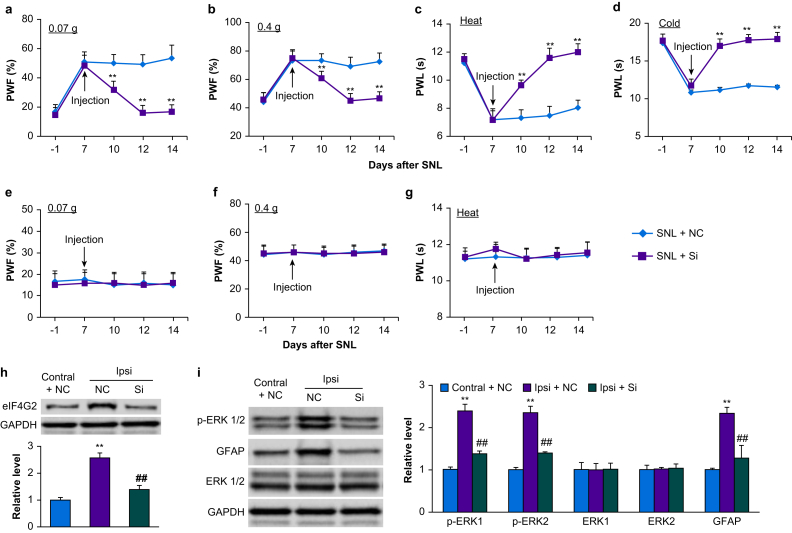
Effect of microinjection of eIF4G2 siRNA on maintenance of nociceptive hypersensitivity
To examine the role of increased DRG eIF4G2 in the maintenance of SNL-induced nociceptive hypersensitivity, we microinjected eIF4G2 siRNA into the ipsilateral L4 DRG starting on day 7 after SNL, when SNL induced nociceptive hypersensitivity peaks. Ipsilateral mechanical, thermal, and cold hypersensitivities were completely developed by this time point (P<0.01, day 7 vs day –1; Fig. 4a–d). However, these hypersensitivities were reduced in the eIF4G2 siRNA-microinjected mice compared with negative control siRNA-microinjected mice on days 10, 12, and 14 after SNL (P<0.01, negative control siRNA-SNL vs eIF4G2 siRNA-SNL; Fig. 4a–d). Neither siRNA changed basal responses to mechanical and thermal stimuli on the contralateral side of SNL mice (Fig. 4e–g). In the ipsilateral L4 DRG, SNL increased expression of eIF4G2 protein by 2.6-fold compared with the contralateral L4 DRG on day 14 after SNL in negative control siRNA-microinjected SNL mice (P<0.01; Fig. 4h), but this increase was almost entirely absent in eIF4G2 siRNA-microinjected mice (P<0.01, negative control siRNA-SNL vs eIF4G2 siRNA-SNL; Fig. 4h). Moreover, SNL-induced increases in p-ERK1 and 2 (but not total ERK1 and 2) and GFAP in the ipsilateral L4 dorsal horn on day 14 after SNL from negative control siRNA-microinjected SNL mice were not seen in eIF4G2 siRNA-microinjected mice (P<0.01, negative control siRNA-SNL vs eIF4G2 siRNA-SNL; Fig. 4i). This evidence suggests that increased DRG eIF4G2 also plays a role in the maintenance of SNL-induced nociceptive hypersensitivities and dorsal horn central sensitisation.
Fig 4.
Effect of dorsal root ganglion (DRG) microinjection of eIF4G2 siRNA on spinal nerve ligation-induced nociceptive hypersensitivities and dorsal horn central sensitisation during the maintenance period. (a–g) Effect of microinjection of eIF4G2 siRNA (Si) or negative control siRNA (NC) into the ipsilateral L4 DRG on PWF to 0.07 g (a and e) and 0.4 g (b and f) von Frey filament stimuli and on paw withdrawal latency (PWL) to heat (c and g) and cold (d) stimuli on the ipsilateral (a–d) and contralateral (e–g) sides at the different days after SNL surgery. n=12 mice per group. Two-way analysis of variance (anova) with repeated measures followed by post hoc Tukey test. Fgroup (1, 110)=267 for 0.07 g, 75 for 0.4 g, 410 for heat, and 505 for cold on the ipsilateral side. ∗∗P<0.01 vs the NC plus SNL group at the corresponding time points. (h) Levels of eIF4G2 protein in the contralateral (Contral) and ipsilateral (Ipsi) L4 DRG on day 14 after SNL in mice post-microinjected with eIF4G2 siRNA (Si) or negative control siRNA (NC). n=3 biological repeats (12 mice) per group. One-way anova with repeated measures followed by post hoc Tukey test. Fgroup (2, 6)=86. ∗∗P<0.01 vs the NC plus SNL group on the contralateral side. ##P<0.01 vs the NC plus SNL group on the ipsilateral side. (i) Effect of microinjection of eIF4G2 siRNA (Si) or negative control siRNA (NC) into the ipsilateral L4 DRG on expression of p-ERK1 and 2, total ERK1 and 2 and GFAP in the contralateral (Contral) and ipsilateral (Ipsi) L4 dorsal horn on day 14 after SNL. Left: representative Western immunoblots. Right: statistical summary of densitometric analysis. n=3 mice per group. One-way anova with repeated measures followed by post hoc Tukey test. Fgroup (2, 6)=103 for p-ERK1, 133 for p-ERK2, 0.03 for ERK1, 0.09 for ERK2, and 370 for GFAP. ∗∗P<0.01 vs the corresponding NC plus SNL group on the contralateral side. ##P<0.01 vs the corresponding NC plus SNL group on the ipsilateral side. eIF4G2, eukaryotic initiation factor 4 gamma 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; p-ERK, phosphorylated extracellular signal-regulated kinase 1; PWF, paw withdrawal frequency; PWL: paw withdrawal latency; SNL, spinal nerve ligation.
DRG overexpression of eIF4G2 increases hypersensitivity to nociceptive stimuli
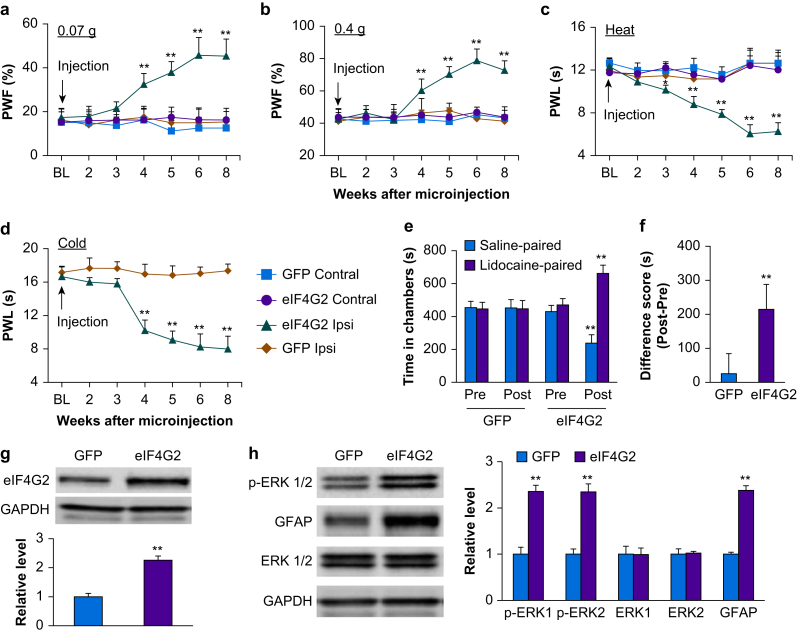
We further examined whether the increased eIF4G2 in injured DRG was sufficient for nerve injury-induced nociceptive hypersensitivity. AAV5 encoding mouse full-length eIF4G2 mRNA (AAV5-eIF4G2) was microinjected into unilateral L3 and 4 DRGs of naïve adult mice. AAV5-Gfp was used as a control. DRG microinjection of AAV5-eIF4G2, but not control AAV5-Gfp, led to marked increases in PWFs in response to 0.07 and 0.4 g von Frey filament stimuli (P<0.01, AAV5-eIF4G2 vs AAV5-Gfp; Fig. 5a and b) and decreases in PWLs in response to heat and cold stimuli (P<0.01, AAV5-eIF4G2 vs AAV5-Gfp; Fig. 5c and d) on the ipsilateral side. These nociceptive hypersensitivities started at 3 or 4 weeks after microinjection and lasted for at least 8 weeks (Fig. 5a–d), consistent with the previously reported 3–4 week lag in AAV5 expression.14,16,29 Neither virus affected locomotor function (Supplementary Table S1) or basal contralateral paw withdrawal responses (Fig. 5a–c). In addition to evoked nociceptive hypersensitivities, DRG microinjection of AAV5-eIF4G2 led to stimulation-independent spontaneous pain indicated by robust preference for the lidocaine-paired chamber (P<0.01, AAV5-eIF4G2 vs AAV5-Gfp; Fig. 5e and f). In contrast, DRG microinjection of AAV5-Gfp did not produce marked preference for either the saline- or lidocaine-paired chamber, evidence of no significant spontaneous pain (Fig. 5e and f). Expression of eIF4G2 protein increased by 2.3-fold in AAV5-eIF4G2-injected DRGs as compared with AAV5-Gfp-injected DRGs 8 weeks after microinjection (P<0.01; Fig. 5g). DRG microinjection of AAV5-eIF4G2 also led to an increase in p-ERK1 and 2 (but not total ERK1 and 2) and GFAP in the ipsilateral L4 dorsal horn (P<0.01, AAV5-eIF4G2 vs AAV5-Gfp; Fig. 5h), suggesting that increased expression of eIFG2 is a sufficient trigger for dorsal horn neuronal and astrocyte hyperactivation. Overall, these findings indicate that increased eIF4G2 in the DRG leads to both spontaneous and evoked nociceptive hypersensitivities, signs of neuropathic pain commonly seen in the clinic.
Fig 5.
Effect of dorsal root ganglion (DRG) eIF4G2 overexpression on nociceptive thresholds and dorsal horn central sensitisation in naïve mice. (a–d) Effect of microinjection of AAV5-eIF4G2 (eIF4G2) or AAV5-Gfp (GFP) into the unilateral L3 and L4 DRGs on PWFs to 0.07 g (a) and 0.4 g (b) von Frey filament stimuli and on paw withdrawal latencies (PWL) to heat (c) and cold stimuli (d) on the ipsilateral (Ipsi) and contralateral (Contral) sides at the different weeks post-viral microinjection. BL, baseline. n=8 mice per group. Two-way analysis of variance (anova) with repeated measures followed by post hoc Tukey test. Fgroup (3, 196)=204 for 0.07 g, 160 for 0.4 g, and 246 for heat. Fgroup (1, 98)=819 for cold. ∗∗P<0.01 vs the AAV5-Gfp group at the corresponding time points on the ipsilateral side. (e and f) Effect of microinjection of AAV5-eIF4G2 or AAV5-Gfp into the unilateral L3 and L4 DRGs on spontaneous ongoing pain as assessed by the CPP paradigm. (e) Time spent in each chamber. (f) Difference scores for chamber preference were calculated by subtracting preconditioning preference time from post-conditioning time spent in the lidocaine-paired chamber. Pre, preconditioning; Post, post-conditioning. n=8 mice per group. Fgroup(3, 56)=71.∗∗P<0.01 vs the corresponding preconditioning by two-way anova with repeated measures followed by Tukey post hoc test (e) or vs the AAV5-Gfp group by two-tailed, independent Student's t test (f). (g) Levels of eIF4G2 protein in the ipsilateral L3 and L4 DRGs 8 weeks after microinjection of AAV5-eIF4G2 (eIF4G2) or control AAV5-Gfp (GFP). n=6 mice per group. ∗∗P<0.01 vs the AAV5-Gfp group by two-tailed independent Student's t test. (h) Effect of microinjection of AAV5-eIF4G2 (eIF4G2) or AAV5-Gfp (GFP) into the unilateral L3 and L4 DRGs on the levels of p-ERK1 and 2, total ERK1 and 2, and GFAP in the ipsilateral L3 and L4 dorsal horn 8 weeks after viral microinjection. Representative Western immunoblots (left panels) and a summary of the densitometric analysis (right graphs) are shown. n=3 mice per group. ∗∗P<0.01 vs the corresponding AAV5-Gfp group by two-tailed, independent Student's t test. AAV5, adeno-associated virus 5; CCP, conditioned place preference; eIF4G2, eukaryotic initiation factor 4 gamma 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; p-ERK, phosphorylated extracellular signal-regulated kinase 1; PWF, paw withdrawal frequency; PWL: paw withdrawal latency.
Increased eIF4G2 downregulates MOR and Kv1.2 expression in injured dorsal root ganglion
We further examined the mechanisms by which increased eIF4G2 in injured DRG participates in SNL-induced nociceptive hypersensitivity. Given that peripheral nerve injury-induced downregulation of MOR and Kv1.2 in injured DRG is known to contribute to neuropathic pain,10, 11, 12, 13,15,16,31,33 and that eIF4G2 functions as a general repressor of cap-dependent translation,20, 21, 22,35 we predicted that increased eIF4G2 contributes to MOR and Kv1.2 downregulation in injured DRG. Consistent with previous reports,10, 11, 12, 13,15,16,31,33 SNL reduced MOR and Kv1.2 expression in the ipsilateral L4 DRG 5 days after SNL in the negative control siRNA-microinjected mice (P<0.01, SNL vs sham; Fig. 6a and b). These reductions were largely reversed in eIF4G2 siRNA-microinjected mice (P<0.01, eIF4G2 siRNA vs negative control siRNA in SNL mice; Fig. 6a and b). Prior microinjection of eIF4G2 siRNA did not change basal expression of MOR and Kv1.2 protein in the DRG of sham mice (Fig. 6a and b). Expression of MOR and Kv1.2 protein in the ipsilateral L3 and L4 DRGs from AAV5-eIF4G2-microinjected mice was decreased by 45% and 51%, respectively, compared with AAV5-Gfp-microinejcted mice at 8 weeks after microinjection (P<0.01, AAV5-eIF4G2 vs AAV5-Gfp; Fig. 6c and d). In addition, cultured DRG neurones co-treated with AAV5-eIF4G2 plus negative control siRNA showed a marked increase in the amount of eIF4G2 and significant decreases in the levels of MOR and Kv1.2 compared with neurones co-treated with AAV5-Gfp plus negative control siRNA (P<0.01; Fig. 6e and f). These effects were nearly abolished in cultured DRG neurones co-treated with eIF4G2-siRNA plus AAV5-eIF4G2 (P<0.01, eIF4G2 siRNA vs negative control siRNA in neurones with eIF4G2 overexpression; Fig. 6e and f), indicating that the decreases in MOR and Kv1.2 expression were specific responses to eIF4G2. Unexpectedly, eIF4G2-siRNA did not alter basal levels of eIF4G2, MOR, and Kv1.2 expression in AAV5-Gfp-transduced cultured DRG neurones (Fig. 6e and f). Finally, double-label immunohistochemistry revealed that ∼52% (38 in 73) of eIF4G2-labelled neurones were positive for Kv1.2 and that ∼24% (26 in 109) of eIF4G2-labelled neurones were positive for MOR in naïve DRG (Fig. 6g).
Fig 6.
Role of increased dorsal root ganglion (DRG) eIF4G2 in spinal nerve ligation-induced downregulation of Kv1.2 and mu opioid receptor (MOR) in the injured DRG. (a, b) Expression of Kv1.2 and MOR in the ipsilateral L4 DRG on day 5 after SNL or sham surgery in mice microinjected with eIF4G2 siRNA (Si) or negative control siRNA (NC). Representative Western immunoblots (a) and a statistical summary of the densitometric analysis (b) are shown. n=3 biological repeats (12 mice) per group. One-way analysis of variance (anova) with repeated measures followed by post hoc Tukey test. Fgroup,(3, 8)=29 for Kv1.2 and 25 for MOR. ∗∗P<0.01 vs the corresponding NC plus sham group. ##P<0.01 vs the corresponding NS plus SNL group. (c, d) Levels of Kv1.2 and MOR proteins in the ipsilateral L3 and L4 DRGs 8 weeks after microinjection of AAV5-eIF4G2 (eIF4G2) or control AAV5-Gfp (GFP) in naïve mice. Representative Western immunoblots (c) and a statistical summary of the densitometric analysis (d) are shown. n=3 biological repeats (six mice) per group. ∗∗P<0.01 vs the corresponding AAV5-Gfp group by two-tailed independent Student t test. (e, f) Expression of eIF4G2, Kv1.2 and MOR in mouse cultured DRG neurones transduced or transfected as indicated. GFP, AAV5-Gfp; eIF4G2, AAV5-eIF4G2; NC, control negative siRNA; Si, eIF4G2 siRNA. Representative Western immunoblots (e) and a summary of the densitometric analysis (f) are shown. n=3 biological repeats per group. One-way anova with repeated measures followed by Tukey post hoc test. Fgroup (3, 8)=260 for eIF4G2, 235 for Kv1.2, and 16 for MOR. ∗∗P<0.01 vs the corresponding GFP plus NC group. ##P<0.01 vs the corresponding eIF4G2 plus GFP group. (g) Double-label immunofluorescent staining (arrows) of eIF4G2 (red) with Kv1.2 (green) or MOR (green) in naïve DRG neurones. n=3 mice. Scale bar, 50 μm. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
Long-lasting nociceptive hypersensitivities induced by SNL mimic post-traumatic neuropathic pain frequently seen in the pain clinic. Current treatments for this common and debilitating disorder are limited, at least in part owing to an incomplete understanding of the mechanisms that underlie the development and maintenance of nerve injury-induced pain hypersensitivities. In this study, we show that SNL produced an increase in eIF4G2 expression in the injured DRG. This increase is shown to contribute to development and maintenance of SNL-induced nociceptive hypersensitivities through downregulation of MOR and Kv1.2 protein expression in injured DRG. Our findings present a novel eIF4G2-mediated post-transcriptional mechanism for induction and maintenance of nerve injury-induced pain hypersensitivities.
The present study showed that eIF4G2, a eukaryotic initiation factor implicated in regulation of protein translation, was localised predominantly in DRG neurones, ∼34% of which were labelled by eIF4G2 under normal conditions. Our observations are in line with a previous report that showed distribution of eIF4G2 in the somata and axons of hippocampal neurones.36 Moreover, eIF4G2 mRNA and eIF4G2 protein expression increased in injured DRG, but not in intact (uninjured) DRG, contralateral DRG and the ipsilateral spinal cord dorsal horn, after either SNL or CCI. Consistently, eIF4G2 was detected in a greater number of neurones in injured DRG after SNL. These results suggest that the eIF4G2 gene is activated at the transcriptional level leading to protein translation in injured DRG after peripheral nerve injury. How peripheral nerve injury causes an increase in eIF4G2 mRNA in the DRG is unclear, but this increase is likely triggered by transcription factors and epigenetic modifications or increases in RNA stability.
Increased eIF4G2 in injured DRG is required for peripheral nerve injury-induced nociceptive hypersensitivity. We found that blocking the SNL-induced increase in DRG eIF4G2 through microinjection of eIF4G2 siRNA into the injured DRG nearly abolished SNL-induced nociceptive hypersensitivities to mechanical, heat, and cold stimuli and reduced dorsal horn central sensitisation during both development and maintenance periods. Mimicking the SNL-induced increase in DRG eIF4G2 through overexpression of eIF4G2 led to enhanced nociceptive responses to mechanical, heat, and cold stimuli and increased dorsal horn neuronal and glial activity in mice even in the absence of nerve injury. Given that eIF4G2 siRNA did not affect basal nociceptive responses or locomotor function, our findings provide for a specific and selective role of DRG eIF4G2 in nerve injury-induced nociceptive hypersensitivities. Unexpectedly, eIF4G2 siRNA administration failed to decrease basal expression of eIF4G2 in sham DRGs or in AAV5-Gfp-treated cultured DRGs. However, this siRNA at the same dosage significantly blocked the SNL- or AAV5-eIF4G2-induced increases in DRG eIF4G2. The mechanism by which eIF4G2 siRNA did not affect basal eIF4G2 expression in vivo and in vitro is unknown, but could be attributable to its low basal level of DRG eIF4G2 likely could not be further reduced significantly by its siRNA.
Increased DRG eIF4G2 expression contributes to development and maintenance of peripheral nerve injury-induced nociceptive hypersensitivities likely through downregulation of MOR and Kv1.2 expression in injured DRG. Nerve injury-induced downregulation of Kv1.2 and MOR in the injured DRG plays a key role in the development and maintenance of nerve injury-induced nociceptive hypersensitivity. Kv1.2 is highly expressed in large- and medium-sized DRG neurones.11 Knocking down its expression increased excitability of DRG neurones and produced an augmented responses to mechanical, heat, and cold stimuli in naïve animals.16 Rescuing DRG Kv1.2 expression attenuated nerve injury-induced nociceptive hypersensitivity.11,16 Similarly, MOR knockout mice exhibited enhanced responses to mechanical stimulation of an uninjured paw in addition to increased mechanical allodynia in injured paws of SNL animals.37 Restoring DRG MOR expression blocked MOR-mediated neurotransmitter release from primary afferents and nerve injury-induced nociceptive hypersensitivity.13,15,32 The present work shows that blocking increased DRG eIF4G2 rescued Kv1.2 and MOR expression in injured DRG of SNL mice, and that mimicking the nerve injury-induced increase in DRG eIF4G2 decreased Kv1.2 and MOR expression in naïve DRG. Our in vitro cultured DRG studies further showed regulation of Kv1.2 and MOR expression by eIF4G2 and co-localisation of eIF4G2 with MOR or Kv1.2 in DRG neurones. Given that both Kcna2 (encoding Kv1.2) and Oprm1 (encoding MOR) RNAs contain 5′ cap structures and that eIF4G2 is a repressor of cap-dependent mRNA translation,20, 21, 22,35 increased eIF4G2 likely participates in nerve injury-induced pain hypersensitivity through repressing Kv1.2 and MOR mRNA translation in the injured DRG. It should be noted that other potential mechanisms by which increased DRG eIF4G2 participates in neuropathic pain cannot be ruled out. eIF4G2 also promotes cap-independent translation of B-cell lymphoma 2 (Bcl-2), cyclin-dependent kinase 1 (CDK1), prolyl hydroxylase-domain protein 2 (PHD2), and Down syndrome critical region 1 isoform 4 (DSCR1.4).27,38,39 Whether these targets are involved in eIF4G2 function in neuropathic pain remains to be determined.
Previous studies showed that DNMT1- and DNMT3a-mediated DNA methylation, G9a-mediated histone methylation, transcriptional repressors, FTO-mediated RNA m6A demethylation, and Kcna2 antisense long non-coding RNA are also involved in nerve injury-induced downregulation of Kv1.2 or MOR in injured DRG.6,10,12,13,15,16,33,40,41 It thus appears that multiple mechanisms at both epigenetic and post-translational levels contribute to nerve injury-induced downregulation of the DRG Kcna2 and Oprm1 genes.
Peripheral nerve injury produces pathological alternations in both neurones and glia within the spinal cord dorsal horn.42, 43, 44 Dorsal horn neurones stimulated by increased neurotransmitters/neuromodulators from the injured primary afferents release pro-inflammatory mediators, that cause remarkable activation and gliosis of both microglia and astrocytes within dorsal horn.42, 43, 44 These activated glia in turn release glial factors/mediators that alter neural excitability through modulation of excitatory and inhibitory synaptic transmission, leading to central sensitisation and neuropathic pain.42, 43, 44 We observed that levels of p-ERK1 and 2 and GFAP were markedly increased in dorsal horn on days 5 and 14 after SNL. These increases were attenuated by blocking increased eIF4G2 in injured DRG. Thus, anti-nociception produced by blocking increased DRG eIF4G2 likely results from relieving eIF4G2's repression on DRG Kv1.2 and MOR protein translation, rescuing their downregulation, reducing nerve injury-induced hyperexcitability of injured DRG neurones and decreasing nerve injury-induced increase of neurotransmitter and neuromodulator release from primary afferents. The latter reduces dorsal horn neuronal and glial hyperactivation (i.e. central sensitisation) under neuropathic pain conditions.
In summary, we identified an eIF4G2-mediated post-translational mechanism by which nerve injury downregulates expression of Kv1.2 and MOR in injured DRG. Given that blocking the increased eIF4G2 in the DRG impaired nerve injury-induced nociceptive hypersensitivities without affecting locomotor function and acute pain, eIF4G2 may be an essential mediator of neuropathic pain and a potential target for management of this disorder. However, potential unwanted side-effects caused by targeting eIF4G2 should be assessed because of its expression in other tissues.
Authors' contributions
Study conception and design: YXT, ZZ, BZ, SD
Data acquisition and analysis: ZZ, BZ, SD, GH, HZ, SW, SJ, YXT
Drafting figures: ZZ, BZ, TB, AB, YXT
Drafting manuscript: BZ, TB, YXT
Finalising manuscript: YXT
Declarations of interest
The authors declare that they have no conflicts of interest.
Funding
Grants (R01NS094224, R01NS111553, and RFNS113881) from the US National Institutes of Health (Bethesda, MD, USA).
Handling editor: Hugh C Hemmings Jr
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bja.2020.10.032.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cohen S.P., Mao J. Neuropathic pain: mechanisms and their clinical implications. BMJ. 2014;348:f7656. doi: 10.1136/bmj.f7656. [DOI] [PubMed] [Google Scholar]
- 2.Gaskin D.J., Richard P. The economic costs of pain in the United States. J Pain. 2012;13:715–724. doi: 10.1016/j.jpain.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Campbell J.N., Meyer R.A. Mechanisms of neuropathic pain. Neuron. 2006;52:77–92. doi: 10.1016/j.neuron.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laumet G., Garriga J., Chen S.R. G9a is essential for epigenetic silencing of K(+) channel genes in acute-to-chronic pain transition. Nat Neurosci. 2015;18:1746–1755. doi: 10.1038/nn.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S., Marie L.B., Miao X. Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Mol Pain. 2016;12:1–14. doi: 10.1177/1744806916629048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Guo X., Sun L. N(6)-methyladenosine demethylase FTO contributes to neuropathic pain by stabilizing G9a expression in primary sensory neurons. Adv Sci (Weinh) 2020;7:1902402. doi: 10.1002/advs.201902402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang L., Lutz B.M., Bekker A., Tao Y.X. Epigenetic regulation of chronic pain. Epigenomics. 2015;7:235–245. doi: 10.2217/epi.14.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutz B.M., Bekker A., Tao Y.X. Noncoding RNAs: new players in chronic pain. Anesthesiology. 2014;121:409–417. doi: 10.1097/ALN.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S., Bono J., Tao Y.X. Long noncoding RNA (lncRNA): a target in neuropathic pain. Expert Opin Ther Targets. 2019;23:15–20. doi: 10.1080/14728222.2019.1550075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J.Y., Liang L., Gu X. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun. 2017;8:14712. doi: 10.1038/ncomms14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan L., Guan X., Wang W. Impaired neuropathic pain and preserved acute pain in rats overexpressing voltage-gated potassium channel subunit Kv1.2 in primary afferent neurons. Mol Pain. 2014;10:8. doi: 10.1186/1744-8069-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang L., Gu X., Zhao J.Y. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci Rep. 2016;6:37704. doi: 10.1038/srep37704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang L., Zhao J.Y., Gu X. G9a inhibits CREB-triggered expression of mu opioid receptor in primary sensory neurons following peripheral nerve injury. Mol Pain. 2016;12:1–16. doi: 10.1177/1744806916682242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo K., Wu S., Gu X. MBD1 contributes to the genesis of acute pain and neuropathic pain by epigenetic silencing of Oprm1 and Kcna2 genes in primary sensory neurons. J Neurosci. 2018;38:9883–9899. doi: 10.1523/JNEUROSCI.0880-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L., Zhao J.Y., Gu X. Nerve injury-induced epigenetic silencing of opioid receptors controlled by DNMT3a in primary afferent neurons. Pain. 2017;158:1153–1165. doi: 10.1097/j.pain.0000000000000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X., Tang Z., Zhang H. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prevot D., Darlix J.L., Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch A.G., Lorsch J.R. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henis-Korenblit S., Shani G., Sines T., Marash L., Shohat G., Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci U S A. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imataka H., Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imataka H., Olsen H.S., Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka S., Poksay K.S., Arnold K.S., Innerarity T.L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 23.Lee S.H., McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 2006;25:4008–4019. doi: 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nousch M., Reed V., Bryson-Richardson R.J., Currie P.D., Preiss T. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA. 2007;13:374–384. doi: 10.1261/rna.372307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiyama H., Takahashi K., Yamamoto T. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2017;114:340–345. doi: 10.1073/pnas.1617234114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai Y., Xie M. LINC01579 promotes cell proliferation by acting as a ceRNA of miR-139-5p to upregulate EIF4G2 expression in glioblastoma. J Cell Physiol. 2019;234:23658–23666. doi: 10.1002/jcp.28933. [DOI] [PubMed] [Google Scholar]
- 27.Seo J.Y., Jung Y., Kim D.Y. DAP5 increases axonal outgrowth of hippocampal neurons by enhancing the cap-independent translation of DSCR1.4 mRNA. Cell Death Dis. 2019;10:49. doi: 10.1038/s41419-018-1299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Percie du S.N., Hurst V., Ahluwalia A. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. J Physiol. 2020 doi: 10.1113/EP088870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L., Han G., Wu S. Toll-like receptor 7 contributes to neuropathic pain by activating NF-kappaB in primary sensory neurons. Brain Behav Immun. 2020;87:840–851. doi: 10.1016/j.bbi.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L.N., Zou Y., Wu S.G. Fn14 Participates in neuropathic pain through NF-kappaB pathway in primary sensory neurons. Mol Neurobiol. 2019;56:7085–7096. doi: 10.1007/s12035-019-1545-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Gu X., Sun L. Dorsal root ganglion myeloid zinc finger protein 1 contributes to neuropathic pain after peripheral nerve trauma. Pain. 2015;156:711–721. doi: 10.1097/j.pain.0000000000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z., Mao Y., Liang L. The transcription factor C/EBPbeta in the dorsal root ganglion contributes to peripheral nerve trauma-induced nociceptive hypersensitivity. Sci Signal. 2017;10 doi: 10.1126/scisignal.aam5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L., Gu X., Pan Z. Contribution of DNMT1 to neuropathic pain genesis partially through epigenetically repressing Kcna2 in primary afferent neurons. J Neurosci. 2019;39:6595–6607. doi: 10.1523/JNEUROSCI.0695-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan J., Wen J., Wu S. Contribution of dorsal root ganglion octamer transcription factor 1 to neuropathic pain after peripheral nerve injury. Pain. 2019;160:375–384. doi: 10.1097/j.pain.0000000000001405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamanaka S., Zhang X.Y., Maeda M. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 2000;19:5533–5541. doi: 10.1093/emboj/19.20.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kar A.N., MacGibeny M.A., Gervasi N.M., Gioio A.E., Kaplan B.B. Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J Neurosci. 2013;33:7165–7174. doi: 10.1523/JNEUROSCI.2040-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansikka H., Zhao C., Sheth R.N., Sora I., Uhl G., Raja S.N. Nerve injury induces a tonic bilateral mu-opioid receptor-mediated inhibitory effect on mechanical allodynia in mice. Anesthesiology. 2004;100:912–921. doi: 10.1097/00000542-200404000-00022. [DOI] [PubMed] [Google Scholar]
- 38.Marash L., Liberman N., Henis-Korenblit S. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell. 2008;30:447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Bryant J.D., Brown M.C., Dobrikov M.I. Regulation of hypoxia-inducible factor 1alpha during hypoxia by DAP5-induced translation of PHD2. Mol Cell Biol. 2018;38 doi: 10.1128/MCB.00647-17. e00647–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchida H., Matsushita Y., Ueda H. Epigenetic regulation of BDNF expression in the primary sensory neurons after peripheral nerve injury: implications in the development of neuropathic pain. Neuroscience. 2013;240:147–154. doi: 10.1016/j.neuroscience.2013.02.053. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Chen S.R., Laumet G., Chen H., Pan H.L. Nerve injury diminishes opioid analgesia through lysine methyltransferase-mediated transcriptional repression of mu-opioid receptors in primary sensory neurons. J Biol Chem. 2016;291:8475–8485. doi: 10.1074/jbc.M115.711812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H., Alam A., Chen Q. The role of microglia in the pathobiology of neuropathic pain development: what do we know? Br J Anaesth. 2017;118:504–516. doi: 10.1093/bja/aex006. [DOI] [PubMed] [Google Scholar]
- 43.Chen G., Zhang Y.Q., Qadri Y.J., Serhan C.N., Ji R.R. Microglia in pain: detrimental and protective roles in pathogenesis and resolution of pain. Neuron. 2018;100:1292–1311. doi: 10.1016/j.neuron.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji R.R., Donnelly C.R., Nedergaard M. Astrocytes in chronic pain and itch. Nat Rev Neurosci. 2019;20:667–685. doi: 10.1038/s41583-019-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.