Growth in the bile salt component deoxycholate (DOC) results in a profound change in the Campylobacter jejuni proteome that is reflected in phenotypic alterations including increased virulence, antibiotic resistance, and nutrient transport. A significantly induced protein was Cj0025c, which shares sequence similarity with TcyP cystine transporters from other bacteria. We show that Cj0025c is required for cystine uptake and deletion of cj0025c results in a sulfur-starved proteome and loss of wild-type phenotypes including human cell invasion, motility and helical morphology.
Keywords: Bacteria, Virulence, Metabolomics, Mass Spectrometry, Pathogens, Bacterial pathogenesis, Campylobacter jejuni, Cystine, Nutrient transport, Sulfur
Graphical Abstract
Highlights
-
•
Growth in the bile salt deoxycholate (DOC) induces virulence proteins in C. jejuni.
-
•
A putative symporter Cj0025c is associated with DOC growth and cystine transport.
-
•
Deletion of cj0025c results in loss of cystine transport and a sulfur starved proteotype.
-
•
Cj0025c is required for wild-type virulence phenotypes including human cell invasion.
Abstract
Campylobacter jejuni is a major cause of food-borne gastroenteritis. Proteomics by label-based two-dimensional liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) identified proteins associated with growth in 0.1% sodium deoxycholate (DOC, a component of gut bile salts), and system-wide validation was performed by data-independent acquisition (DIA-SWATH-MS). LC-MS/MS quantified 1326 proteins (∼82% of the predicted C. jejuni proteome), of which 1104 were validated in additional biological replicates by DIA-SWATH-MS. DOC resulted in a profound proteome shift with 512 proteins showing significantly altered abundance. Induced proteins were associated with flagellar motility and antibiotic resistance; and these correlated with increased DOC motility and resistance to polymyxin B and ciprofloxacin. DOC also increased human Caco-2 cell adherence and invasion. Abundances of proteins involved in nutrient transport were altered by DOC and aligned with intracellular changes to their respective carbon sources. DOC increased intracellular levels of sulfur-containing amino acids (cysteine and methionine) and the dipeptide cystine (Cys-Cys), which also correlated with reduced resistance to oxidative stress. A DOC induced transport protein was Cj0025c, which has sequence similarity to bacterial Cys-Cys transporters. Deletion of cj0025c (Δcj0025c) resulted in proteome changes consistent with sulfur starvation, as well as attenuated invasion, reduced motility, atypical morphology, increased antimicrobial susceptibility and poor biofilm formation. Targeted metabolomics showed Δcj0025c could use known C. jejuni amino and organic acid substrates commensurate with wild-type. Medium Cys-Cys levels however, were maintained in Δcj0025c relative to wild-type. A toxic Cys-Cys mimic (selenocystine) inhibited wild-type growth, but not Δcj0025c. Provision of an alternate sulfur source (2 mm thiosulfate) restored Δcj0025c motility. Our data confirm that Cj0025c is a Cys-Cys transporter that we have named TcyP consistent with the nomenclature of homologous proteins in other species.
Campylobacter jejuni is a Gram-negative, helical, and microaerophilic bacterium with polar flagella that confer motility. The organism is a common cause of food- and water-borne disease (1) and human infections are generally associated with consumption of poorly prepared poultry, because C. jejuni is largely considered a commensal in avian species, although this concept has also been challenged (2, 3, 4). Human disease often requires only low numbers of infecting cells (500–800 cells; a lower infective dose than Salmonella or pathogenic E. coli (5)) and symptoms can range from mild, non-inflammatory diarrhea through to abdominal cramps, vomiting and inflammation. C. jejuni is self-limiting in most clinical cases but has been linked to the development of Guillain-Barré Syndrome (GBS), an acute, debilitating immune-mediated disorder of the peripheral nervous system. Approximately 40% of GBS patients show serological evidence of a prior C. jejuni infection (6).
The “human-chicken paradigm” of why C. jejuni causes disease in humans but not poultry remains poorly understood. Colonization in both hosts has been linked with several C. jejuni traits, of which flagellar motility appears to be the most crucial (7, 8). Weakly motile C. jejuni are poor chicken colonizers and attenuated for human intestinal epithelial cell adherence and invasion. Many other factors, including chemotaxis and protein glycosylation, are also related to infection of both hosts (9, 10, 11, 12). Human pathogenesis involves adherence to epithelial cells, intracellular invasion, and toxin production (13). Colonization of chickens and human epithelia is facilitated by the fibronectin (Fn)-binding proteins CadF (14, 15) and FlpA (16), and many other ‘virulence’ factors have been described, including the surface-exposed glycoprotein adhesin JlpA (17, 18) and the PEB antigens (19, 20). Invasion and cytotoxicity appear largely mediated by the secreted Campylobacter invasion antigens (CiaBC, flagellin-like FlaC, FspA (21, 22, 23)) and the “cytolethal distending” toxin subunits (CdtABC; (24)).
C. jejuni can persist in a range of diverse and potentially stress-inducing environments, including different temperatures, osmolarities, iron and other trace element limitation, presence of toxic compounds and diverse nutrient availabilities. C. jejuni grows preferentially between 37–42 °C (human and poultry body temperatures) but can survive at temperature extremes (e.g. 4 °C; facilitating contamination of >75% of supermarket chicken products (25, 26)). The ability of C. jejuni to utilize host-specific nutrients is considered a major factor in infection. C. jejuni is largely thought to be asaccharolytic, although some strains can catabolize fucose (27). Amino and organic acids (28, 29, 30) are primary carbon sources and C. jejuni preferentially utilizes (in order) serine (Ser), aspartate (Asp), glutamate (Glu) and proline (Pro) (31). Some strains possess a γ-glutamyltranspeptidase (GGT) that enables glutamine (Gln) use (32, 33), whereas others contain an asparaginase (AnsB) that converts extracellular asparagine (Asn) to Asp (32). Ser uptake is mediated by the SdaC transporter and is converted to pyruvate via the l-Ser dehydratase SdaA (34, 35). Pro is the least preferred amino acid and following uptake by the PutP transporter (35) is converted to Glu via 1-pyrroline-5-carboxylate (catalyzed by PutA). Glu and Asp uptake are mediated by several transporters including those encoded by the PEB locus and the C4-dicarboxylate transporters, DcuA, DcuB and DctA, which also transport fumarate and succinate (36). Other organic acids catabolized by C. jejuni include malate, pyruvate and acetate (for which no specific transporters have been identified), citrate (likely transported by the Cj0203 citrate transporter homolog), lactate and α-ketoglutarate (KG), which are transported by the LctP and KgtP permeases, respectively (37).
Sulfur is an important element required for cysteine (Cys) and methionine (Met) biosynthesis, and for the formation of Fe-S clusters in proteins such as SdaA, ferredoxin, Nap nitrate reductase and aconitase (AcnB). C. jejuni is auxotrophic for Cys, despite being unable to utilize Cys as a sole carbon source, and this phenotype cannot be rescued by addition of sulfate as a sulfur source (38). C. jejuni lacks several enzymes in the assimilatory sulfur biosynthesis pathway, as well as typical bacterial ABC-type transport proteins specific for sulfate (38). Although sulfate cannot be used, alternative sulfur sources, including hydrogen sulfide and thiosulfate, allow C. jejuni growth (38, 39). Exogenous glutathione (GSH, a Glu-Cys-Gly tripeptide), can be acted upon by GGT-producing strains to produce Glu as a carbon source and Cys-Gly that can be imported by the Campylobacter peptide transporter CptA (33, 38) as a source of Cys. Additional dipeptides that provide alternative Cys sources include Gly-Cys and Glu-Cys, however transporters for these are currently unknown, and additional Cys-containing dipeptides, including cystine (Cys-Cys) have not, to our knowledge, been tested.
Here we performed proteomics of the C. jejuni response to growth in the presence of the bile salt constituent 0.1% sodium deoxycholate (DOC) using a label-based LC-MS/MS discovery set and global validation by data-independent acquisition (DIA-SWATH-MS). Nutrient transporters were significantly altered by growth in DOC, consistent with in vivo triggers for colonization requiring specific carbon sources. DOC induced a virulence-associated phenotype by increasing motility, antibiotic resistance, and adherence and invasion of human epithelial cells. The product of the cj0025c gene was significantly induced by DOC. Multi-omics and functional analyses confirmed that Cj0025c is a Cys-Cys transporter, required for several wild-type virulence attributes including motility, biofilm formation and cell invasion.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
For discovery-based quantitative proteomics by tandem mass tag (TMT) labeling and LC-MS/MS of C. jejuni growth in 0.1% DOC, 4 biological replicates (control and DOC growth) were processed in tandem. Two additional biological replicates were generated for the DIA-SWATH-MS validation set. Analysis of C. jejuni NCTC11168 wild-type (WT) compared with a Δcj0025c mutant were carried out as above, except 5 biological replicates were analyzed by label-based discovery. Samples for the 2 technical approaches were processed independently of each other. Prior to TMT labeling, equal aliquots from each biological replicate were split to form an internal technical replicate. For targeted metabolomics, 3 biological replicates were prepared in triplicate extractions (9 replicates in total). MS data have been deposited to the ProteomeXchange Consortium via PRIDE with the data set identifier PXD017934.
Bacterial Strains and Growth Conditions
C. jejuni NCTC11168 (WT) and NCTC11168 Δcj0025c (cj0025c deleted in NCTC11168 and DNA were sourced from the Campylobacter Resource Facility and provided as a gift by Prof. Brendan Wren) were prepared and grown as previously described (40). Δcj0025c was grown in the presence of kanamycin (Km; 30 μg/ml) for selection, and 25 μg/ml trimethoprim (all antibiotics Sigma, St. Louis, MO) used for all strains when grown in Mueller-Hinton (MH) medium. Briefly, cells were grown from glycerol stocks in MH broth (Oxoid, Basingstoke, UK) for 48 h at 37 °C under microaerophilic conditions (5% O2, 10% CO2, 85% N2). Cells were sub-cultured into fresh MH broth (in the presence or absence of 0.1% DOC; Sigma) at an initial OD600 of 0.1 and grown until late exponential phase. Cells were collected by centrifugation before lyophilization. For culture supernatant levels of C. jejuni WT and Δcj0025c amino acids, organic acids and sulfur-containing amino acids, cells were also grown in defined MCLMAN medium, supplemented with 10 mm carbon sources and/or 0.8 mm cystine (41).
Western Blotting
Proteins were transferred from SDS-PAGE gels to polyvinylidenedifluoride (PVDF) membrane for 1 h at 400 mA (4°C). Membranes were blocked overnight at 4°C in 5% bovine serum albumin (BSA) in 20 mm Tris, 150 mm NaCl, 0.1% Tween-20, pH 7.4. Membranes were probed with either a 1:1000 CadF-specific or a 1:1000 JlpA-specific rabbit anti-serum overnight at 4°C (18, 42). Proteins were detected using a 1:1000 dilution of goat-anti-rabbit immunoglobulin, followed by incubation with SupersignalTM West Pico Chemiluminescent substrate according to the manufacturer's instructions (Thermo Scientific, Waltham MA) and visualized using a ChemiDOCTM MP Imaging System (Bio-Rad, Hercules CA).
Quantitative PCR (qPCR)
Quantitative PCR (qPCR) was performed as previously described (12). Primers for cj0025c were as follows: forward 5′-CTAATGCTTTCACAAGCTCA-3′ and reverse 5′-TCCTGAGACATTGCTTGCTG-3′. All reactions were performed in technical duplicate. Quantification of gene expression was performed using the 2ΔΔCT method, on a total of n = 3 biological replicates with 16S primers as per (12) used as the housekeeper gene.
Phenotypic Assays
Motility was assessed as described previously (40) using semi-solid MH medium with 0.4% agar. Plates were inoculated with 2 μl of an overnight biphasic culture (OD600 0.5) and incubated for 48 h at 37 °C under microaerophilic conditions and motility measured by diameter of bacterial spread. For testing thiosulfate (TS) effects, 2 mm TS (Sigma) was added to the medium. Antibiotic sensitivities were measured using Etest® strips for polymyxin B, ciprofloxacin, erythromycin, and amoxacillin/clavulanic acid (Biomérieux, France). A 1 ml culture grown as above was spread on MH agar plates and an Etest® strip added to the center. Plates were incubated for 48 h at 37 °C under microaerophillic conditions. Resistance against oxidative stress was measured by exposure to 5 mm H2O2 for 30 min (12). Cultures were serially diluted and spread onto agar plates, incubated for 48 h at 37 °C under microaerophilic conditions and colony forming units (cfu) enumerated. Biofilm formation was assessed as per (12) with minor modifications; briefly, overnight cultures were diluted to an OD600 of 0.1 in 24 well, flat-bottom cell culture plates containing fresh Brain-Heart Infusion (BHI; Oxoid) medium or BHI supplemented with 5% (v/v) chicken exudate (‘juice’; (43)). Plates were incubated for 48 h under microaerophilic conditions. Non-adherent, non-biofilm cells were removed by washing with 1 ml of sterile PBS. 1.2 ml of fresh medium supplemented with 0.01% (w/v) 2,3,5-triphenyltetrazolium chloride (TTC) was added to each well and further incubated at 37 °C under micraerophilic conditions for 72 h. TTC was removed and the wells air dried. Bound dye was dissolved using 20% acetone/80% ethanol and the absorbance A500 of the solution measured. Human Caco-2 cell adherence and invasion assays were performed using the gentamicin protection method as described (12). Data are from n = 6 replicates.
Selenocystine Inhibition Assay
Selenocystine (Se-Cys-Cys) inhibition assay was performed as described (44). Briefly, cultures were normalized to OD600 of 1 and spread across MCLMAN agar to generate a bacterial lawn. After being allowed to dry, a sterile 10 mm diameter paper disk was placed in the center and 10 μl of 50 mm Se-Cys-Cys (Sigma) added to the disk. Plates were incubated under microaerophilic conditions for 48 h and the zone of inhibition measured.
Lipid A Analysis
Lipid A extraction was performed as per (45). Briefly, cells were solubilized in 4:3 70% isobutyric acid/1 M ammonium hydroxide (Sigma) and boiled at 100 °C for 30 min. Insoluble material was removed by centrifugation for 15 min at 2000 × g, following which supernatants were diluted 1:1 with ultrapure water and lyophilized. Extracts were washed with methanol twice before being reconstituted in 2:1:0.25 chloroform/methanol/water. Aliquots were spotted directly onto stainless steel targets 1:1 with the matrix norharmane (10 mg/ml [Sigma] in 2:1:0.25 chlorofom/methanol/water). Mass spectra were generated on a Bruker UltrafleXtreme (Bruker Daltonics, Billerica, MA) operated in reflectron mode across a mass range of 1000 to 2500 m/z, and a laser frequency of 2000 Hz using 40% global intensity.
Scanning Electron Microscopy (S.E.)
Cells were transferred to 12 well culture plates and allowed to settle onto Nunc Thermanox™ coverslips (Thermo Scientific) for 2 h. Cells were gently washed with 1× phosphate-buffered saline (PBS) 3 times, fixed with 2.5% glutaraldehyde for 1 h and then post-fixed in 1% osmium tetroxide (Sigma), dehydrated with hexamethyldisilazane, mounted on S.E. imaging stubs, and finally sputter coated with 1.5 nm gold. Cells were imaged with a Zeiss Sigma high definition field emission gun (HD-FEG) S.E. microscope at 10,000 × magnification.
Targeted Metabolomics by LC-MS/MS
Metabolites and MS parameters (parent, product ion m/z [transitions], retention time, collision energy and declustering potential) are summarized in supplemental Table S1. Cells (n = 9 samples from 3 biological replicates) were lysed in ultrapure water by 6 rounds of 30 s beadbeating. For assay of extracellular metabolites, aliquots from culture supernatants were collected at 0, 4, 24, 48 and 72 h growth and filtered through 0.22 μm polyethersulfone (PES) filters, following collection of intact cells by centrifugation, and stored at −30 °C. 25 μl of each sample was added to 75 μl of extraction buffer (0.1% formic acid in 80:20 ethanol/water (v/v)), mixed with vortexing, and incubated at 4 °C for 2 h. Solutions were vortexed then centrifuged at 14,000 × g at 4 °C for 15 min. 50 μl of supernatant was lyophilized and metabolites were resuspended in HPLC-grade water, and diluted as necessary. LC-MS/MS was performed as per (12, 46). Metabolites were loaded onto either a Luna Phenyl-Hexyl column (50 mm × 1 mm x 5 μm particle size) (Phenomenex, Torrance CA) or a Synergi Polar-RP (reversed phase), 80 Å column (50 mm × 1 mm × 4 μm particle size) (Phenomenex) using a Nexera UHPLC system (Shimadzu, Kyoto, Japan). Loading and elution were as previously described (12). Metabolites were eluted into a Q-TRAP 5500 mass spectrometer (SCIEX, Framingham MA) operated in targeted MRM mode. Data files were imported into Skyline (v. 4.1.0.18169), and peak areas manually integrated. Statistical analysis was performed in Metaboanalyst (v.4.0) (47).
Quantitative Proteomics by LC-MS/MS
Peptides were prepared as described previously (12) from cells reconstituted in 100 mm HEPES, pH 7.5 and lysed by 6 rounds of 30 s beadbeating. Proteins were precipitated in 1.8:2:2 water/methanol/chloroform, collected by centrifugation, washed twice with methanol, and solubilized in 8 m guanidine-HCl, 100 mm HEPES, pH 7.6. Reduction and alkylation were performed with 10 mm dithiothreitol (DTT) and 20 mm iodoacetamide (IAA) for 1 h each, respectively. Proteins were digested with sequencing grade modified trypsin (Promega, Madison WI), at a protein/protease ratio of 30:1 overnight at 37 °C. Peptides were desalted by solid phase extraction using hydrophilic-lipophilic balance (HLB) cartridges (Waters, Bedford MA) according to the manufacturer's instructions. After this step, samples for DIA-SWATH-MS were lyophilized (see below). For label-based analysis, samples were labeled with 6-plex TMT (Thermo Scientific) according to the manufacturer's instructions. Excess label was quenched with hydroxylamine hydrochloride, after which samples were pooled and desalted using HLB cartridges, as above. TMT labeled samples and pooled, unlabeled samples used to generate SWATH libraries (see below), were fractionated offline by hydrophilic interaction liquid chromatography (HILIC) prior to LC-MS/MS. Samples were reconstituted in 90% acetonitrile (MeCN), 0.1% trifluoroacetic acid (TFA) and loaded onto a 20 cm × 320 μm column packed in-house with 3 μm TSK-amide 80 particles (TOSOH Bioscience, King of Prussia PA) in 100% buffer B (90% MeCN, 0.1% TFA) at 10 μl/min for 10 min using an Agilent 1200 LC system (Agilent Technologies, Santa Clara, CA). Peptides were eluted by adjusting the mobile phase to 70% buffer A (0.1% TFA) over a 40 min linear gradient at 6 μl/min, and collected in 1 min fractions. Adjoining fractions were pooled to a final 6–12 fractions and lyophilized. TMT labeled peptide fractions were separated by reversed phase chromatography over a 90 min gradient as described (12) into a Q Exactive™ HF Hybrid Quadrupole Mass Spectrometer (QE-HF; Thermo Scientific). The QE-HF was configured to perform one full scan MS experiment (scan range 300 to 1650 m/z, resolution of 60000, automatic gain control [AGC] of 3e6 and a maximum ion injection time [IT] of 50 msec) with the top 15 precursors in fulfilment of the selection criteria (charge state 2–4, minimum intensity greater than 9.2e4, dynamic exclusion window of 40 s) selected for MS/MS (scan range 200 to 2000 m/z with a fixed first mass of 110 m/z, resolution of 15,000, AGC of 1e6, maximum IT of 50 msec, isolation window 1.4 m/z, normalized collision energy [NCE] set as 29).
Processing of Mass Spectrometry Files
Data files from TMT experiments were processed in Proteome Discoverer (v. 2.2; Thermo Scientific) and searched against the UniProt C. jejuni NCTC11168 genome database (UP000000799; organism ID 192222; release May 24, 2018 last modification; 1623 proteins (48)) with the SequestHT algorithm. Search parameters included full-trypsin digest with a maximum 2 missed cleavages and carbamidomethyl (C) as a fixed modification; and with variable modifications, oxidation (M), TMT-6plex (peptide N-term, K), and using precursor and fragment ion tolerances of 20 ppm. Data analysis was performed as per (12) with minor modifications. Briefly, peptide level false discovery rate (FDR) was established using Percolator (v. 2.08.01). Rank 1 peptide spectral matches (PSMs) conforming to this 1% peptide FDR were exported, and reporter intensities normalized to the total reporter ion intensity across all channels. Peptides with ambiguous protein assignments were removed from further analysis. Reporter signals for included PSMs were averaged to yield peptide-level quantitation and data from peptides representing each protein were averaged to determine protein-level quantitation. Proteins with a minimum of 2 unique identified peptides were imported into MetaboAnalyst (v 4.0) for statistical analysis by one sample t-tests performed on the log2 transformed protein-level ratios from all available biological replicates. Using the settings unpaired analysis and equal variance, statistical significance was determined using a p value cut-off set to p = 0.05. A minimum protein difference corresponding to a log2 fold change of ±(0.6) (∼±1.5-fold; 0.1% DOC) and ±(0.5) (∼1.4-fold; Δcj0025c) was required for a protein to be considered as altered in abundance, and the values for all proteins in each set were arrayed by volcano plot to highlight those statistically significantly changing. For further stringency, proteins with fold-change values close to the cut-off were only included when validation by DIA-SWATH-MS was available. Data were averaged from all biological replicates in which quantitation was achieved (>2 peptides) and proteins were included only if quantified in a minimum of 2/4 (0.1% DOC) and 2/5 (Δcj0025c) biological replicates. Missing values were not imputed. All peptide identifications are contained within Supplementary Data files 11168DOC_Peptide and 11168cj0025c_Peptide.
Validation by DIA-SWATH-MS
DIA-SWATH-MS was performed as per (12). Briefly, unlabeled peptides were reconstituted in 3% MeCN, 1% FA and loaded onto a 40 cm × 75 μm pulled column packed in-house with 3 μm ReproSil-Pur 120 C18-AQ material maintained at a column temperature of 45 °C at 200 nL/min in 98% buffer A (1% FA) using an Ekspert 425 nanoLC system (SCIEX). Peptides were eluted by altering the mobile phase to 28% buffer B (80% MeCN, 1% FA) over a 146 min linear gradient into a TripleTOF® 6600 mass spectrometer (SCIEX). For generation of the C. jejuni NCTC11168 spectral library, duplicate biological preparations of peptides from the WT controls were analyzed. The 6600 was configured to perform one full scan MS experiment (scan range 350 to 1500 m/z, accumulation time 50 msec) with a maximum of 80 precursors in fulfilment of the selection criteria (charge state 2–5, intensity >400, dynamic exclusion window of 20 s) selected for MS/MS in high sensitivity mode (scan range 100–1500 m/z, dynamic collision energy [DCE] set as true, mass tolerance of 20 ppm). For DIA-SWATH-MS, the instrument was configured to perform one full scan MS experiment (scan range 350 to 1500 m/z, accumulation time 50 msec) followed by 80 variable SWATH windows over the mass range 350 to 1500 m/z (accumulation time 25 msec). Window size was determined using the SWATH variable window calculator tool (SCIEX) from summed total ion chromatograms (TIC) of library-generated samples. SWATH library data files were processed in Protein Pilot (v. 5.0) and searched against the UniProt C. jejuni NCTC11168 proteome (as above) using the Paragon algorithm. Search parameters were sample set as identification; trypsin digestion, maximum 2 missed cleavages; Cys alkylation, IAA; instrument, TripleTOF 6600; search effort as thorough ID, and using a detected protein threshold of 0.05. The group file was then imported into PeakView (v. 2.2.0.11391) using a total protein number corresponding to a global protein level FDR of 1%. SWATH files were processed against the resulting library using the following settings; a peptide confidence threshold of 95%, 1% FDR, a minimum of 6 peaks per peptide selected for highest intensity across a retention time tolerance of 4 min, and an extracted ion chromatogram (XIC) width/mass tolerance of 20 ppm. Acquisitions were aligned by manually selecting a set of high-scoring peptides and generating a retention time standard curve in Peakview that was then applied to all samples. Peptide areas were summed to yield a total protein area that was used for relative quantitation.
RESULTS
DOC Induces a C. jejuni Proteome Associated with Altered Virulence and In Vivo Growth
Label-based quantitative proteomics post-trypsin digest was applied to identify C. jejuni NCTC11168 proteins associated with growth in 0.1% DOC. TMT labeling and LC-MS/MS provided a “discovery” data set that identified and quantified 1326 proteins (with ≥ 2 peptides per protein) from a minimum of 2/4 biological replicates and which represent 81.7% of the C. jejuni NCTC11168 predicted proteome encoded by 1623 genes (Fig. 1A, supplemental Data S1). System-wide validation was performed using additional biological replicates that were subjected to trypsin digest and DIA-SWATH-MS, which enabled the independent relative quantitation of 1112 proteins (representing 68.5% of the predicted proteome; supplemental Data S1), with 8 identified in the validation set alone (1104 proteins therefore identified in both discovery and validation cohorts). The label-based and DIA-SWATH-MS data sets showed a Pearson correlation of r = 0.8327 (Fig. 1B), indicating strong alignment between the discovery and validation approaches. Log2 (fold change) data were converted to n-fold change for subsequent visualization and analysis and the complete data sets are shown in supplemental Fig. S1.
Fig. 1.
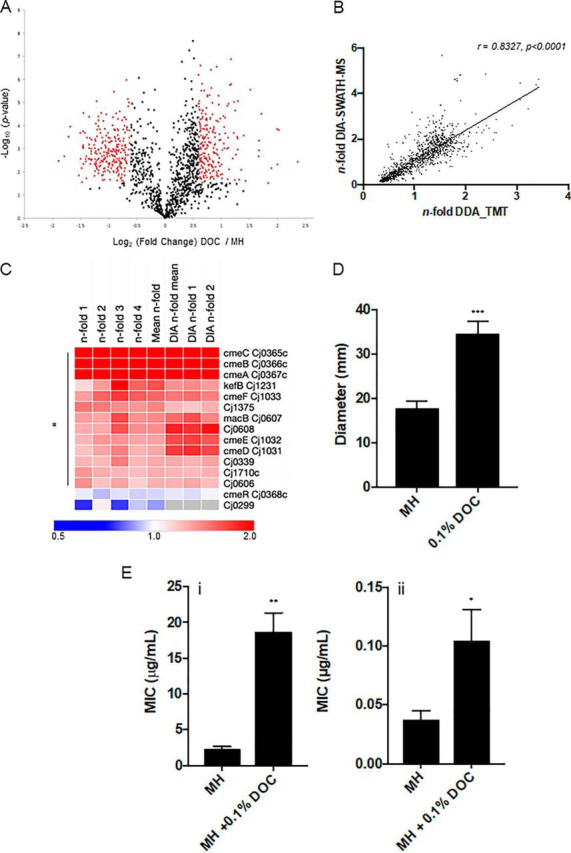
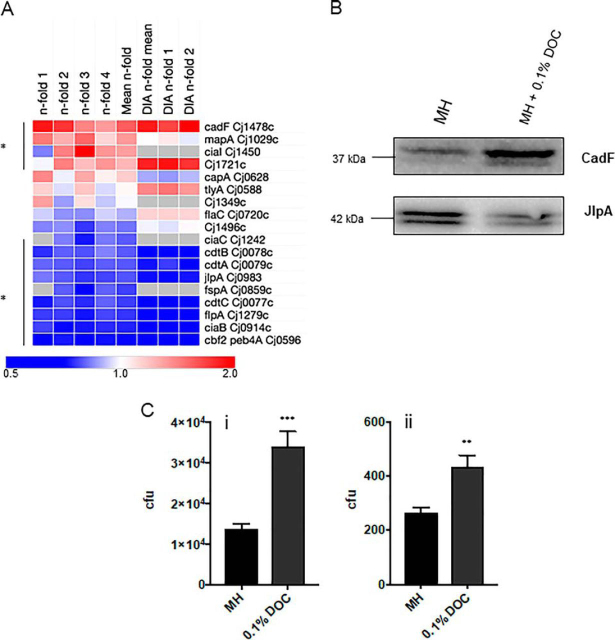
DOC induces a large proteome shift in C. jejuni NCTC11168 and influences phenotypes associated with virulence.A, Volcano plot for cells grown in 0.1% DOC compared with control; x axis represents averaged Log2(DOC/control), y axis represents log10(p value). Significantly differentially abundant proteins are highlighted in red (p < 0.05); B, Correlation plot based on n-fold changes observed in the discovery set (n-fold DDA_TMT) and validation set (n-fold DIA-SWATH-MS), Pearson correlation r = 0.8213 (p < 0.0001) determined from 1104 aligned proteins; C, Heat map of C. jejuni proteins associated with antibiotic resistance and efflux ordered by largest mean n-fold change (label-based discovery; top). Data from each of 4 label-based replicates (n-fold 1–4) and DIA-SWATH-MS validation (‘DIA’ mean of 2 biological replicates [DIA n-fold 1–2]) are shown. Values are gray where the protein was not identified by DIA-SWATH-MS (* denotes proteins significantly altered in abundance); D, Increased motility associated with growth in 0.1% DOC based on colony diameter in mm on semi-solid agar (*** p < 0.001); E, Antibiotic resistance assays for (i) polymyxin B, (ii) ciprofloxacin (** p < 0.005, * p < 0.05).
0.1% DOC induced a profound shift in the C. jejuni proteome with 512 proteins (38.6% of the total identified proteome) considered altered in abundance (244 present at significantly induced, and 268 at significantly reduced, abundance). Two proteins (Cj0879c and Cj1171c) with a conflicting fold change as determined by DIA-SWATH-MS compared with label-based analysis (opposite change in abundance or no change in abundance) were removed. We employed STRINGdb, which identified 3 major functional clusters in the up-regulated protein set (supplemental Fig. S2A); antibiotic resistance (Fig. 1C), flagellar motility and nutrient acquisition. We validated these data by conducting motility and antibiotic resistance assays in the presence or absence of 0.1% DOC. DOC resulted in an ∼2-fold increase in C. jejuni motility (Fig. 1D) and significantly increased resistance to the antimicrobial peptide polymyxin B (8.01-fold, p < 0.005) and the fluoroquinolone, ciprofloxacin (2.8-fold, p < 0.05) (Fig. 1E), consistent with elevated abundances of CmeABC, CmeDEF and other efflux-associated proteins. No difference was observed for amoxicillin+clavulanic acid or erythromycin (not shown).
Clusters associated with virulence factors were identified in both the up- and down-regulated protein data sets (Fig. 2A; supplemental Fig. S2A, S2B). We further validated the discovery and DIA-SWATH-MS data for CadF (1.87-fold in label-based and 2.09-fold in DIA-SWATH-MS) and JlpA (0.63-fold and 0.40-fold, respectively) by Western blotting using anti-CadF and anti-JlpA anti-sera (Fig. 2B). We next examined how DOC influences C. jejuni adherence to, and invasion of, human epithelial Caco-2 cells (Fig. 2C) by using gentamicin protection assays. These data show that C. jejuni “primed” by growth in DOC are significantly more adherent to epithelial cells and capable of increased invasion. This is consistent with previous reports (49), but also suggests some “virulence-associated” proteins may not be as crucial in this process as previously thought, whereas others (e.g. CadF) are validated as virulence determinants (Fig. 2A).
Fig. 2.
C. jejuni NCTC11168 growth in DOC influences abundance of virulence-associated proteins and virulence phenotypes.A, Heat map of known C. jejuni virulence-associated proteins ordered by largest mean n-fold change (label-based discovery; top). Data from each of 4 label-based replicates (n-fold 1–4) and DIA-SWATH-MS validation (“DIA” mean of 2 biological replicates [DIA n-fold 1–2]) are shown. Values are gray where the protein was not identified in a biological replicate and/or by DIA-SWATH MS (* denotes proteins significantly altered in abundance); B, Western blotting using (upper) anti-CadF and (lower) anti-JlpA serum confirming changes in their DOC abundance (JlpA appears as two major bands as it contains a single glycosylation site in NCTC11168 (addition of 1406 Da (18)) (loading controls are shown in Suppl. Fig. S3); C, (i) Adherence to and (ii) invasion of Caco-2 cells for C. jejuni passaged in 0.1% DOC (*** p < 0.001, ** p < 0.005).
Changes to Nutrient Transport Proteins Reflect DOC-induced Metabolic Remodeling
STRINGdb highlighted nutrient transport as a major functional cluster associated with growth in DOC (Fig. 3A). Examination of the proteins contained within this cluster, the down-regulated protein set, and other characterized C. jejuni nutrient transporters (Fig. 3B) suggested that DOC induced metabolic remodeling reflected in altered abundances of specific transporters and therefore potentially uptake of their associated nutrients. LC/MS-MS targeted metabolomics (Fig. 3C; supplemental Data S2) detected significant increases in intracellular α-ketoglutarate (KG, transported by KgtP [2.60-fold and 2.27-fold in label-based and DIA-SWATH-MS, respectively]), Ser (SdaC [2.46-fold/2.47-fold]), Pro (PutP [2.60-fold in DIA-SWATH-MS, only identified in 1 replicate by label-based LC-MS/MS]), lactate (Lac, LctP [2.24-fold/2.52-fold]), succinate (Suc, DctA [2.57-fold/2.27-fold]/DcuB [1.69-fold/2.67-fold]/DcuA [no significant difference]) and Asp (DctA/DcuB and others). We were unable to measure citrate (Cj0203 [2.35-fold/2.40-fold]). Increased intracellular levels of alanine (Ala, hypothesized to be transported by Cj0903c [1.97-fold/1.49-fold]), Glu (associated with the Peb cluster Cj0920c-Cj0922c [Cj0920c, 1.70-fold/2.18-fold; Cj0921c, down-regulated at 0.34-fold/0.13-fold; and Cj0922c, no change in abundance), Asn and Gln were detected, whereas carbon sources with unknown transporters including pyruvate, were present at significantly decreased intracellular abundances (Fig. 3C, supplemental Fig. S4).
Fig. 3.
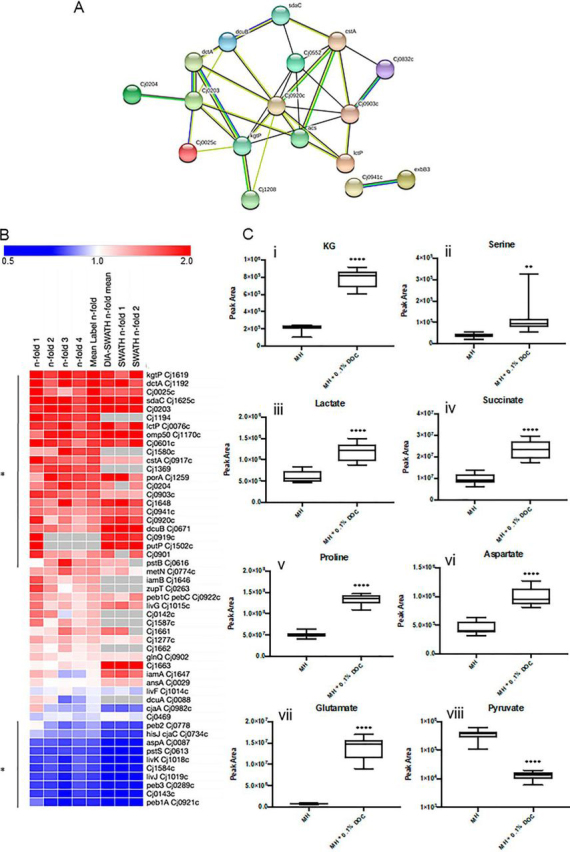
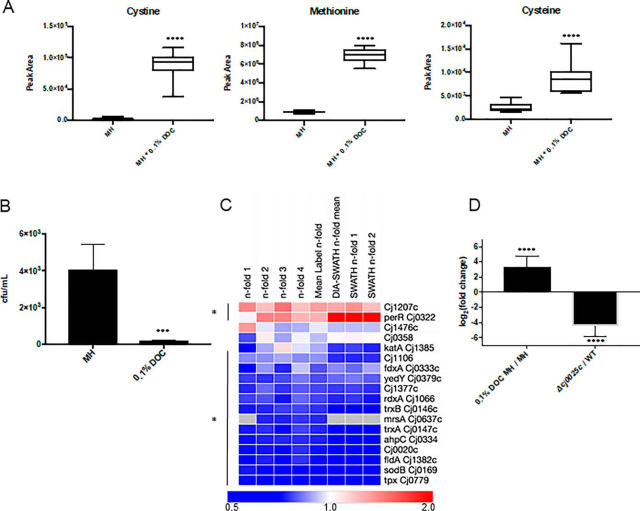
C. jejuni NCTC11168 growth in DOC is associated with changes in nutrient acquisition proteins that correlate with abundance of intracellular nutrients.A, STRINGdb cluster of nutrient transport proteins significantly elevated in abundance during growth in 0.1% DOC; B, Heat map of C. jejuni nutrient transport proteins ordered by largest mean n-fold change (label-based discovery; top). Data from each of 4 label-based replicates (n-fold 1–4) and DIA-SWATH-MS validation (“DIA” mean of 2 biological replicates [DIA n-fold 1–2]) are shown. Values are gray where the protein was not identified in a biological replicate and/or by DIA-SWATH MS (* denotes proteins significantly altered in abundance); C, (i-viii) Comparative intracellular abundances of known C. jejuni carbon sources following growth in MH medium with or without 0.1% DOC and measured by targeted LC-MS/MS metabolomics (**** p < 0.0001, ** p < 0.005).
Metabolomics also showed significantly increased intracellular levels of sulfur-containing compounds, including Cys, Met and the dipeptide Cys-Cys (Fig. 4A), as well as homoCys, biotin and thiamine (supplemental Fig. S5A–S5C). Conversely, we observed decreased acetyl-CoA, which we linked to decreased intracellular levels of both CoA and acetate, another C. jejuni carbon source for which no transporter is known (supplemental Fig. S5D–S5F). The organism is thought to acquire sulfur via Cys or Cys-containing dipeptides (including Cys-Gly that is transported by CptA [Cj0204; 2.02-fold/1.68-fold in 0.1% DOC]; Fig. 3B). Elevated intracellular sulfur is associated with susceptibility to oxidative stress (50). We observed poor survival of C. jejuni in the presence of 5 mm H2O2 for cells previously exposed to 0.1% DOC (Fig. 4B). This phenotype also correlated with reduced abundance of several antioxidant proteins, including thiol peroxidase (0.32-fold/0.17-fold), superoxide dismutase SodB (0.33-fold/0.15-fold) and alkylhydroperoxide reductase AhpC (0.44-fold/0.15-fold) (Fig. 4C).
Fig. 4.
Increased intracellular sulfur-containing amino acids in C. jejuni NCTC11168 grown in DOC correlate with reduced resistance to oxidative stress and increased abundance of the Cj0025c putative sodium:dicarboxylate transporter.A, Comparative intracellular abundances of sulfur-containing amino acids in MH medium with or without 0.1% DOC measured by targeted LC-MS/MS metabolomics (**** p < 0.0001); B, Growth in 0.1% DOC reduces resistance against oxidative stress (cells were exposed to 5 mm H2O2 for 30 min, serial dilutions were plated onto MH agar for CFU enumeration; *** p < 0.001); C, Heat map of C. jejuni antioxidant proteins ordered by largest mean n-fold change (label-based discovery; top). Data from each of 4 label-based replicates (n-fold 1–4) and DIA-SWATH-MS validation (“DIA” mean of 2 biological replicates [DIA n-fold 1–2]) are shown. Values are gray where the protein was not identified in a biological replicate and/or by DIA-SWATH MS (* denotes proteins significantly altered in abundance); D, qPCR showing increased cj0025c gene expression in 0.1% DOC and removal of cj0025c expression in Δcj0025c C. jejuni.
A Putative Sodium:Dicarboxylate Transporter (Cj0025c) is Induced by DOC and Shares Sequence Similarity with the Bacterial Cystine Transporter TcyP
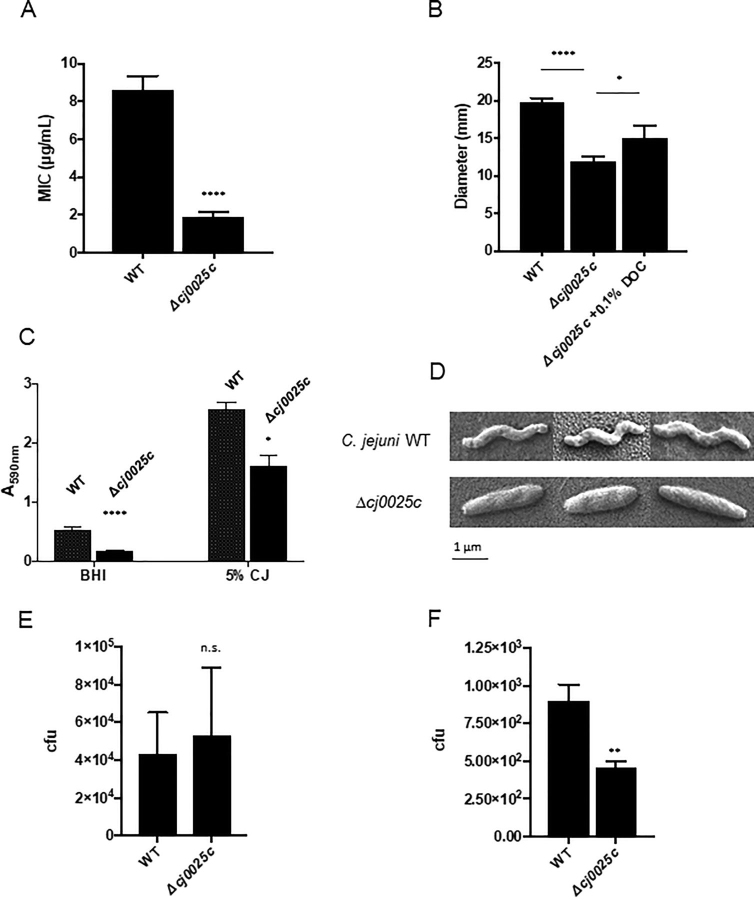
A significantly induced protein in 0.1% DOC was the product of the cj0025c gene, which is currently annotated as a “putative sodium:dicarboxylate transporter” (2.55-fold/1.97-fold; Fig. 3B). A BLAST search revealed Cj0025c shares 34.9% sequence identity with TcyP from Bacillus subtilis (supplemental Fig. S6). TcyP is a Cys-Cys transporter required for sulfur uptake (44, 51). qPCR confirmed that the increased abundance of Cj0025c was consistent with transcription of cj0025c during DOC growth (10.63-fold increased cj0025c expression; Fig. 4D). A deletion mutant of cj0025c (Δcj0025c) was produced in C. jejuni NCTC11168 and loss of gene expression confirmed by qPCR (-22.20-fold decrease; Fig. 4D).
Δcj0025c is Attenuated for Cys-Cys Uptake Compared with Wild-type C. jejuni
Given the correlation between DOC-induced Cj0025c abundance and increased intracellular Cys-Cys, we next tested whether deletion of cj0025c influenced Cys-Cys uptake by monitoring culture supernatant levels in defined medium using targeted metabolomics over a time-course of C. jejuni growth from 0–72 h. Culture supernatant Cys-Cys levels were maintained at ∼100% of uninoculated medium in both wild-type (WT) and Δcj0025c after 4 h growth. By 24 h however, medium Cys-Cys was maintained to ∼57% of the control for Δcj0025c, whereas only ∼27% for WT. Furthermore, at both 48 and 72 h, Cys-Cys was almost, or completely, depleted from the medium in the WT yet maintained at ∼43–47% of the control in Δcj0025c (Fig. 5A). Although this provides strong evidence that Cj0025c is involved in Cys-Cys uptake, we also needed to confirm that Cys-Cys was not being converted to Cys in the medium. Metabolomics confirmed that no change in medium Cys levels occurred in either WT or Δcj0025c and that Cys was maintained at the levels of the uninoculated control in the presence of Cys-Cys (Fig. 5B). Additionally, no changes were observed for extracellular Met (Fig. 5C).
Fig. 5.
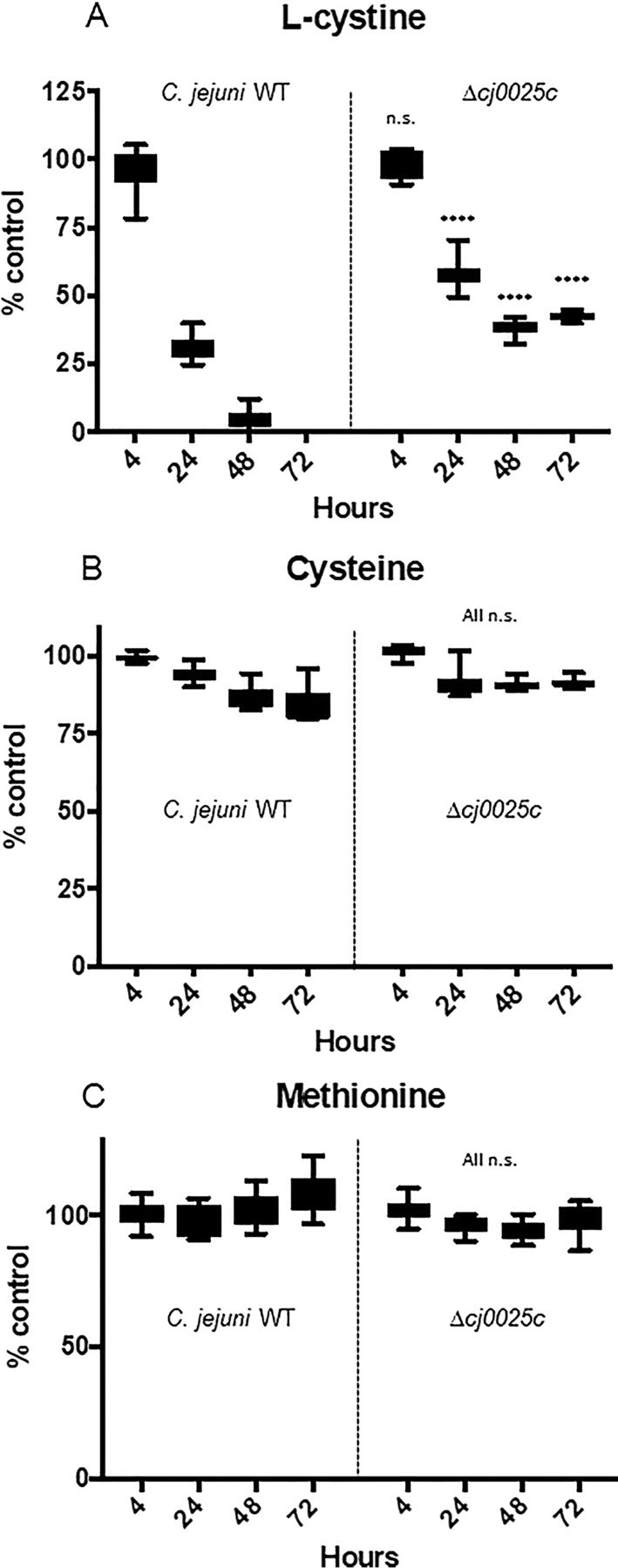
Culture supernatant levels of sulfur-containing amino acids for C. jejuni NCTC11168 WT and Δcj0025c grown in MCLMAN medium across 0–72 h growth. (A) l-cystine; (B) cysteine; (C) methionine (**** p < 0.0001, n.s. not significant). Data expressed at each time point as a % of the uninoculated control (% control). Uninoculated controls were incubated identically and measurements for each amino acid taken at the same time-points, no significant changes were observed (data not shown).
Cj0025c is Not Required for Uptake of Known C. jejuni Carbon Sources
Since there are carbon sources (e.g. pyruvate, malate) for which a transporter has not been identified in C. jejuni, we wished to determine whether Cys-Cys was the sole nutrient transported by Cj0025c. It is also possible that Cj0025c is a secondary transporter for a carbon source imported by other proteins. We measured culture supernatant levels of known carbon sources in C. jejuni over 0–72 h. Our data showed no difference between WT and Δcj0025c uptake for Ser at every time point (supplemental Fig. S7A) and Ser was the most rapidly depleted nutrient for both strains, consistent with this amino acid being the preferred carbon source for C. jejuni. We observed slower uptake of Pro and Glu in Δcj0025c at 24 h (and Asn at 4 h), and more rapid uptake of Asp in Δcj0025c at the same time point; however by 48 h all 4 amino acids were depleted in both strains (supplemental Fig. S7B–E). We saw no uptake of Gln, which is consistent with C. jejuni NCTC11168 lacking a secreted GGT (supplemental Fig. S7F). Culture supernatant levels of the organic acids citrate, fumarate and KG did not differ at any time point (supplemental Fig. S8A–S8C). For pyruvate and malate, we observed slower uptake in Δcj0025c at 24 h and 4 h, respectively; but each nutrient was depleted in both strains by 48 h (supplemental Fig. S8D–S8E). We also confirmed that both WT and Δcj0025c underwent the “succinate shift” where Asp and fumarate are converted within the cell to succinate, which is then exported via DcuB into the medium, for subsequent re-uptake via DctA, which enables succinate use as a primary carbon source (36, 52). Medium succinate peaked at 200–300% the levels seen in uninoculated controls between 4–24 h in both strains (supplemental Fig. S8F). The rate of re-uptake was slower in Δcj0025c (observed at 24 h) but succinate was depleted in both WT and Δcj0025c by 48 h. Finally, we observed reduced depletion of Lac for Δcj0025c compared with WT (supplemental Fig. S8G), with medium Lac maintained at ∼10–20% of the uninoculated control at 48 and 72 h.
The Proteome of Δcj0025c Reflects a Sulfur Starved Phenotype
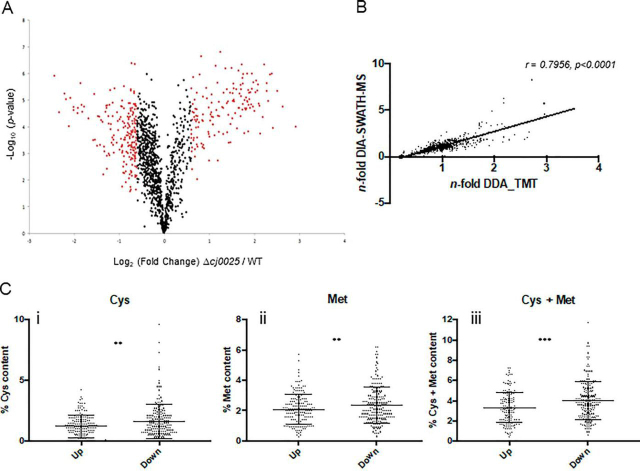
We next hypothesized that if cj0025c encodes a TcyP-like Cys-Cys transporter, then the proteome of Δcj0025c should reflect a sulfur starved phenotype and show depletion of proteins involved in Cys/Met metabolism and biosynthesis, Fe-S cluster proteins and biosynthesis of sulfur-dependent compounds, such as S-adenosylmethionine (SAM), molybdopterins, thiamine, biotin and CoA, as well as processes dependent on these. We conducted label-based discovery and DIA-SWATH-MS validation proteomics as described above. Label-based discovery LC-MS/MS quantified 1349 proteins (≥ 2 peptides per protein; 83.1% of the predicted C. jejuni NCTC11168 proteome) from a minimum of 2/5 biological replicates (Fig. 6A, supplemental Data S3). System-wide validation using additional biological replicates and DIA-SWATH-MS enabled the independent relative quantitation of 983 proteins (60.6% of the predicted proteome; supplemental Data S3). The label-based and DIA-SWATH-MS data showed a Pearson correlation of r = 0.7956 (Fig. 6B; supplemental Fig. S9). Deletion of cj0025c resulted in 371 proteins demonstrating a significant change in abundance (182 elevated and 189 reduced in abundance; Fig. 6A, supplemental Table S2). We observed large increases in abundances of the products of genes from within phase variable gene clusters for which no DIA-SWATH-MS validation was observed (supplemental Fig. S9), suggesting these are of low abundance in the wild-type. Analysis of the “changing” protein data set identified 145 proteins (39.1% of altered proteins; 75 increased in abundance and 70 decreased in abundance) that are involved in sulfur-associated or -dependent processes (supplemental Table S2). We next examined the Cys/Met content of proteins within this data set because bacterial sulfur starvation can result in changed expression of proteins containing high or low content sulfur-containing amino acids (53, 54). Proteins decreased in abundance in Δcj0025c had a statistically significantly higher % Cys, % Met (both p < 0.01) and % Cys+Met (p < 0.005) content than those increased in abundance (Fig. 6C).
Fig. 6.
Deletion of cj0025c alters the C. jejuni NCTC11168 proteome consistent with sulfur starvation.A, Volcano plot for Δcj0025c compared with WT C. jejuni; x axis represents averaged Log2(Δcj0025c/WT), y axis represents log10(p value). Significantly differentially abundant proteins are highlighted in red (p < 0.05); B, Correlation plot based on n-fold changes observed in the discovery set (n-fold DDA_TMT) and validation set (n-fold DIA-SWATH-MS), Pearson correlation r = 0.7956 (p < 0.0001) determined from 983 aligned proteins; C, % sulfur-containing amino acid content for proteins significantly increased (Up) and decreased (Down) in abundance in Δcj0025c compared with WT; (i) cysteine, (ii) methionine, (iii) cysteine + methionine (** p < 0.005, *** p < 0.001).
Finally, we compared the DOC and Δcj0025c data sets and identified 48 proteins with significant and opposite fold changes (15 elevated in DOC and reduced in Δcj0025c [possibly promoted by Cys-Cys uptake/sulfur] and 33 reduced in DOC and elevated in Δcj0025c [possibly repressed by Cys-Cys uptake/sulfur]; supplemental Fig. S10); of these, 23 (47.9%) were associated with sulfur-related functions (supplemental Table S2), indicating enrichment of such proteins compared with the Δcj0025c set alone. There was no association with any functional group in the Cys-Cys/sulfur ‘promoted’ set (although several proteins with large magnitude changes were sulfur-associated [e.g. Cj0908 is a predicted Fe-S cluster protein]), however in the Cys-Cys/sulfur ‘repressed’ set we observed a group of antioxidant proteins (Tpx, SodB, Cj0012c, FldA, TrxB and RdxA), as well as proteins involved in Fe-S cluster repair and sulfur delivery to molybdenum (NifU, IscS, FdhD, ModA, Cj1516 and Cj1517 [MoaD]). These data provide further evidence of a role for Cj0025c in Cys-Cys transport.
Deletion of cj0025c Disrupts Wild-type Virulence Phenotypes
We next phenotypically characterized C. jejuni following deletion of cj0025c. Δcj0025c was attenuated for polymyxin B resistance (Fig. 7A), consistent with significantly reduced abundances of Cme and other efflux associated proteins (supplemental Fig. S11A). Δcj0025c C. jejuni also displayed significantly reduced motility (Fig. 7B). Addition of 0.1% DOC however, restored motility toward WT levels, which suggests Cj0025c is not required for the DOC-associated increase in motility we observed above (Fig. 1D). Δcj0025c was unable to form biofilm at WT levels in two models of biofilm growth - one in BHI medium (3-fold reduction) and the other in medium supplemented with 5% chicken exudate (“juice” [CJ; 1.6-fold reduction in biofilm]), which increases C. jejuni biofilm formation (43) (Fig. 7C). This was consistent with our proteomics data showing significant changes to both SAM-dependent proteins and those involved in signal molecule synthesis and biofilm formation (supplemental Table S2). S.E. revealed a loss of typical C. jejuni helical cell shape, with Δcj0025c displaying a swollen, rod-like appearance (Fig. 7D). Analysis of the proteomics data for shape determinants (supplemental Fig. S11B), indicated that the morphological data were consistent with a significant decrease in abundance of Cj1345c Pgp1 (0.19-fold and 0.04-fold) (55). Despite an increase in the abundance of the phosphoethanolamine (pEtN) transferase EptC that has been shown to modify lipid A, the flagellar protein FlgG and the C. jejuni N-linked glycan (40, 56), we observed no change in the composition or abundance of lipid A variants in Δcj0025c compared with WT (supplemental Fig. S12). Given the reduction of wild-type traits associated with virulence, we tested the ability of WT and Δcj0025c to adhere to, and invade, Caco-2 cells. Δcj0025c showed no significant loss of Caco-2 cell adherence (Fig. 7E), but was attenuated for cell invasion, showing an ∼2-fold reduction in intracellular numbers as determined by gentamicin protection assay (Fig. 7F). These data correlated with the lack of altered protein abundance seen for CadF, but reduced abundances of the Cdt proteins in Δcj0025c (supplemental Fig. S11C).
Fig. 7.
Deletion of cj0025c results in altered C. jejuni NCTC11168 phenotypes associated with virulence.A, Polymyxin B resistance assay (**** p < 0.0001); B, Motility assay based on colony diameter in mm on semi-solid agar (**** p < 0.0001, * p < 0.05); C, Biofilm assays in (left) BHI medium and (right) BHI medium supplemented with 5% chicken exudate (‘juice’ [CJ]; **** p < 0.0001, * p < 0.05); D, Cell morphology assayed by S.E.; E, C. jejuni Caco-2 cell adherence and (F) invasion (** p < 0.005, n.s. not significant).
Addition of a Toxic Cys-Cys Mimic Inhibits Growth of WT But Not Δcj0025c C. jejuni
Our data thus far indicated that Cj0025c encodes a TcyP homolog that is able to transport Cys-Cys as a sulfur source for C. jejuni NCTC11168. We next employed a selenocystine (Se-Cys-Cys) inhibition assay. Se-Cys-Cys is a toxic Cys-Cys analog that inhibits bacterial growth (44) by incorporation of Se into Fe-S cluster proteins. Se-Cys-Cys inhibition plate assays of WT and Δcj0025c, and in medium supplemented with 0.1% DOC, were performed with the hypothesis that DOC elevated Cj0025c protein levels (Fig. 3B) and thus would increase Se-Cys-Cys uptake and increase inhibition. Δcj0025c C. jejuni were 3.5-fold more resistant to Se-Cys-Cys than WT (Fig. 8A). Addition of 0.1% DOC increased the Se-Cys-Cys zone of inhibition ∼2-fold for WT C. jejuni, consistent with the elevated abundance of Cj0025c in DOC growth, while having no influence on the Δcj0025c deletion mutant. Although these data confirm a role for Cj0025c in Cys-Cys transport, Se-Cys-Cys did induce a small zone of inhibition for the Δcj0025c mutant providing some evidence, in addition to our metabolomics assays, that a second, potentially lower affinity, Cys-Cys transporter may be present in C. jejuni. C. jejuni can utilize alternative sulfur sources, including hydrogen sulfide and thiosulfate. We determined whether provision of thiosulfate (TS) could functionally complement Δcj0025c and addition of 2 mm TS resulted in significant recovery of motility (∼1.5-fold increase; p < 0.005) in Δcj0025c (Fig. 8B), without completely restoring this phenotype to WT levels.
Fig. 8.
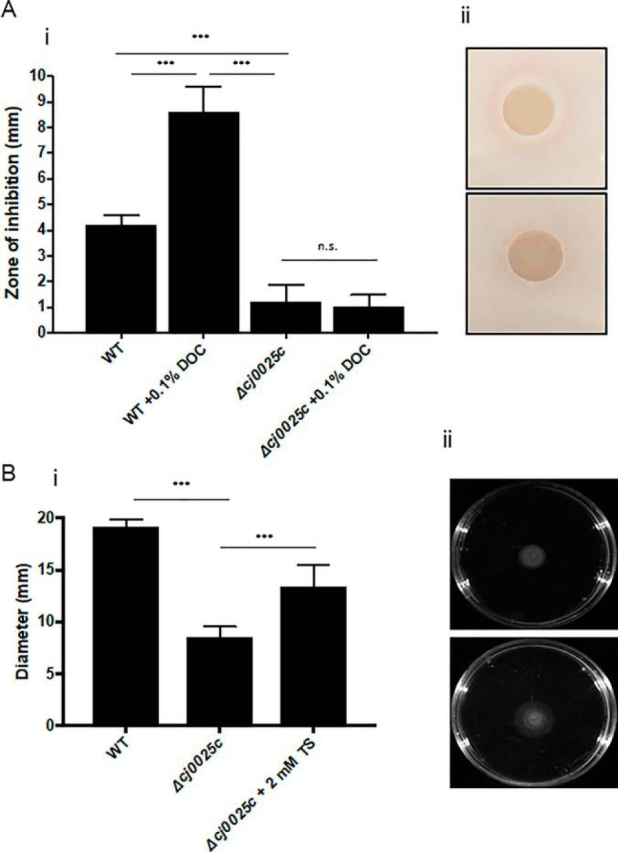
Cj0025c is a TcyP-like Cys-Cys transport protein.A, Selenocystine inhibition assay (i) Se-Cys-Cys inhibits WT C. jejuni and 0.1% DOC increases inhibition consistent with increased abundance of Cj0025c under 0.1% DOC growth (Fig. 3B), whereas Δcj0025c is only slightly inhibited and 0.1% DOC does not influence this phenotype (*** p < 0.001, n.s. not significant), (ii) representative Se-Cys-Cys inhibition plates (upper) C. jejuni WT and (lower) Δcj0025c; B, Provision of an alternative sulfur source (2 mm thiosulfate [TS]) (i) restores Δcj0025c motility toward WT levels, (ii) representative motility plate assay (upper) Δcj0025c, (lower) Δcj0025c + 2 mm TS (*** p < 0.001).
DISCUSSION
C. jejuni encounters a hostile environment upon colonization of the human gut, facing chemical and biological factors that must be overcome to establish infection, including bile salts and the presence of established microflora, which compete for nutrients and employ toxic products to inhibit foreign microorganisms. We examined the proteome response of C. jejuni to the bile salt DOC. DOC induced a proteome response indicative of C. jejuni being primed for epithelial cell colonization that is largely in agreement with transcriptomic studies of DOC growth (49). C. jejuni increased abundance of a functionally undefined transporter, Cj0025c, which we have shown is required for WT uptake of Cys-Cys and for several virulence-associated phenotypes, most likely driven by a requirement for sulfur to be incorporated into small molecules that drive virulence processes, including motility and biofilm formation.
DOC positively influenced virulence traits including resistance to polymyxin B and ciprofloxacin. These data aligned with elevated abundances of proteins related to antibiotic efflux. Antibiotic resistance is largely driven by the tripartite efflux system CmeABC, which has been associated with resistance to bile salts (2 mm choleate) (57). Elevated transcription of cmeABC has also been observed upon bile salt exposure (49). cmeB negative C. jejuni are more susceptible to ciprofloxacin (58), which is consistent with our data showing elevated CmeABC and increased resistance to this fluoroquinolone. Despite this, cmeB mutants did not show any difference with respect to polymyxin B resistance suggesting DOC-associated resistance to this antimicrobial peptide may be independent of CmeABC.
C. jejuni exposed to DOC demonstrated significantly higher Caco-2 cell adherence and invasion. These data support the critical role of the adhesin CadF in C. jejuni pathogenesis (14, 15). DOC-associated increases in invasion were initially difficult to reconcile with the proteomics data, which showed no change or significantly decreased abundances of CiaBC and CdtABC. Previous work examining the transcriptomics response of C. jejuni to DOC indicated that exposure had no effect on cadF expression or adherence (to INT407 cells), but resulted in increased ciaB expression and invasion (49). Cia and Cdt proteins are secreted, whereas our experiments measured their intracellular levels. Protein secretion in C. jejuni is poorly understood but is thought to occur via a type III secretion system (T3SS)-like flagellar export apparatus (30, 60). Proteomics for these and other ‘secretion-associated proteins’ revealed most were elevated in DOC (supplemental Fig. S13A), which suggests increased secretion, rather than reduced expression, may be responsible for intracellular abundances of ‘invasion’ proteins. This agrees with (21) who showed increased DOC-generated secretion of CiaB.
The largest protein cluster that changed in response to DOC was nutrient transporters, suggesting bile salts induce uptake of gut-associated carbon sources and trace elements. C. jejuni likely adapts to the gut by elevating expression of transporters specific for mucin-associated amino acids, such as Ser and Pro, as well as organic acids produced as by-products by resident microflora [28, 29,61]. C. jejuni increased abundance of SdaC and PutP in DOC, resulting in increased intracellular Ser and Pro. sdaC deletion mutants are attenuated for chicken colonization and mouse intestinal model virulence (34, 35), whereas those defective for Pro uptake are attenuated in the mouse alone. Transporters for organic acids were also elevated in DOC, including KgtP, LctP and DctA/DcuB, and these were also correlated with increased intracellular levels of their specific nutrients. Lactate is more abundant in the upper gastrointestinal tract and may act as an environmental cue that allows C. jejuni to determine an optimal niche during infection (62). Conversely, we observed significantly decreased intracellular acetate and pyruvate, two carbon sources for which transporters are yet to be elucidated. C. jejuni also produces pyruvate from both Lac and Ser (28, 29), however increases of these did not result in elevated pyruvate. Proteins responsible for production of pyruvate from Lac and Ser were largely unchanged in DOC (data not shown), however we did observe reduced abundances of phosphoenolpyruvate carboxylase (PckA) and the Cj0073c-Cj0075c membrane lactate dehydrogenase-like complex (supplemental Data S1) (37).
DOC increased intracellular abundances of several sulfur-containing compounds, including Cys, Met and Cys-Cys, although decreased acetyl-CoA was observed. Acetate uptake is linked to acetyl-CoA production for maintenance of the TCA cycle and we observed significantly induced DOC levels of acetyl-CoA synthase (Acs; Suppl. Data S1), suggesting that lower acetate may account for intracellular acetyl-CoA. Acquisition of sulfur is crucial for colonization, with the gut rich in sulfated biomolecules including mucins, free dietary Cys and the bile component taurocholic acid (63). Gut microflora also produce hydrogen sulfide as a by-product of sulfur metabolism. C. jejuni is capable of utilizing 0.5–1 mm hydrogen sulfide, which is within the range encountered in the human gastrointestinal tract; however, beyond ∼2 mm growth is inhibited (38, 64). Host epithelial cells also oxidize microbiota-derived hydrogen sulfide to thiosulfate (and further oxidation in the gut to tetrathionate, although not all C. jejuni [e.g. NCTC11168] can use this sulfur source) which can also act as a sulfur source (38, 65). C. jejuni generally acquires sulfur from free Cys, Cys-containing dipeptides (Cys-Gly, Gly-Cys and Glu-Cys) and for some strains, GSH (29, 38). Specific transporters for most Cys-dipeptides have not been identified. In DOC, we observed elevated abundance of a putative sodium:dicarboxylate symporter, Cj0025c, which shares significant sequence similarity to the TcyP family of bacterial Cys-Cys transporters (44, 51). Deletion of cj0025c attenuated Cys-Cys uptake. We were surprised that low-level Cys-Cys transport was still observed in Δcj0025c. This suggests the presence of a second Cys-Cys transporter, possibly the Cys-Gly transport protein CptA (Cj0204), or the C. jejuni carbon starvation protein A (CstA) that mediates uptake of other di- and tripeptides (66). This hypothesis was strengthened by the elevated abundance of CptA in Δcj0025c. Evidence of a second Cys-Cys transporter was also demonstrated by Se-Cys-Cys assay where, despite significant protection, a small zone of inhibition remained for Δcj0025c. In addition to TcyP, E. coli and B. subtilis contain ATP-binding cassette (ABC) transporters (TcyJKLMN and TcyABC in B. subtilis and TcyJLN in E. coli; (44, 50)) with affinity for Cys-Cys and other Cys-containing compounds. Unlike TcyP, we saw no similar proteins via BLAST search to any of these Tcy ABC transport proteins in C. jejuni (not shown), suggesting Cys-Cys uptake in the absence of cj0025c occurs via an unidentified process.
The Δcj0025c proteome displayed changes associated with sulfur-dependent processes including redox homeostasis, Fe-S clusters, Met and SAM biosynthesis and SAM-dependent proteins, and biosynthesis of sulfur compounds (e.g. CoA, biotin, thiamine, molybdopterins and lipoic acid). C. jejuni (like other bacteria) also responded to sulfur starvation by reducing expression of high %Cys/Met-containing proteins and increasing expression of low %Cys/Met-containing proteins. Some organisms contain large gene clusters encoding proteins with low Cys/Met content (‘sulfur clusters‘), although C. jejuni does not contain such regions (54). Phenotypic characterization of Δcj0025c indicated the attenuation of WT traits. Biofilm formation was reduced in Δcj0025c consistent with a need for sulfur in SAM biosynthesis, the first step in the pathway leading to autoinducer-2 (AI-2) production for quorum sensing. ΔluxS C. jejuni (LuxS catalyzes the final step in AI-2 biosynthesis) are defective for motility at 37 °C (although unaffected at 42 °C), and similar AI-2 mutants in Helicobacter pylori are both defective for motility and show altered expression of flagellar genes (67, 68). Biofilm formation and motility are linked in C. jejuni (69), which may explain the similarly attenuated phenotypes observed here. Motility is a fundamental virulence determinant in C. jejuni and our data show that cj0025c is required for WT motility irrespective of LuxS, which is maintained at WT levels in Δcj0025c (supplemental Data S3). Little is known regarding a link between sulfur and motility in C. jejuni; however, recent work in the Gram-negative pathogen Serratia marcescens (70) has shown that Cys, and thus sulfur, limitation reduces flagellar motility and secretion of phospholipase through the T3SS-like flagellar export apparatus, phenotypes that are correlated with sulfur-directed gene expression. We noted significantly reduced abundances of several proteins associated with flagellar export (supplemental Fig. S13B). Sulfur was also required for optimal cytotoxicity in S. marcescens - a phenotype we observed in Δcj0025c, which was attenuated for Caco-2 cell invasion. Finally, the association of C. jejuni motility with sulfur could be demonstrated by supplementation with 2 mm TS, which rescued Δcj0025c motility.
Helical cell shape is considered a major virulence factor. Two peptidoglycan modifying peptidases, Pgp1 (Cj1345c) and Pgp2 (Cj0906c), are responsible for cell morphology, with pgp1 and pgp2 negative C. jejuni displaying a rod-like appearance (55, 71). Transposon screens for cell shape defective C. jejuni have shown that single nucleotide polymorphisms or insertions/deletions within these 2 genes account for near all rod-like mutants (72, 73). An additional gene associated with curvature (Cjj81176_1105; Cj1087c in NCTC11168) and homologous to the cell shape determinant 1 (Csd1) from H. pylori has also been identified (74). Δcj0025c showed no change to the abundance of Pgp2, whereas Cj1087c was induced and Pgp1 reduced in abundance (supplemental Fig. S11B). Increased abundance of pEtN transferase EptC was observed but could not be linked to any observable change in lipid A. We cannot unequivocally rule out the presence of a mutation in Pgp1, however our proteomics data were based on quantitation of 17 peptides (11 for DIA-SWATH-MS) spanning the full length of the predicted 52 kDa protein (data not shown), suggesting that truncation or sequence modification are not responsible for the phenotype we observed here. Peptidoglycan formation requires biosynthesis of UDP-N-acetylglucosamine (UDP-GlcNAc) and UDP-N-acetylmuramic acid (UDP-MurNAc), both of which receive their acetyl group from the sulfur compound acetyl-CoA; and peptidoglycan can also be modified by O-acetylation mediated by PatAB (75). In DOC-treated C. jejuni, we saw no evidence of a change in cell morphology (not shown), however we did observe significant increases in Pgp2 and PatB abundances, which may help explain why acetyl-CoA was reduced in abundance under these conditions. Therefore, limited acetyl-CoA in sulfur starved Δcj0025c C. jejuni may result in reduced availability of peptidoglycan precursors and/or an inability to acetylate mature peptidoglycan resulting in changes to cell morphology associated with Cj1087c and Pgp1.
We were surprised that no Cys uptake was observed in the presence of Cys-Cys. Several Cys transporters have been considered including CjaA and the Paq ABC transport system (Cj0467–0469) (76), however mutants in cjaA and paqP maintained WT growth in the presence of Cys as well as Cys-containing dipeptides (76, 77). Other potential Cys transporters including Peb1A (Cj0921c) and CjaC (Cj0734c) have also been reported to not transport Cys or Cys-Cys. Our data correlate with these observations because we saw no change in abundance of CjaA, CjaC or Cj0469 in DOC (where intracellular Cys and Cys-Cys are strongly increased) or Δcj0025c. Additionally, Peb1A was repressed in DOC, which was also in contrast with elevated intracellular levels of Peb1A substrates, Asp and Glu, however these are transported by other proteins (e.g. DctA) that were induced under the same conditions. The extracellular redox balance is generally in favor of Cys-Cys (78) and we saw no evidence of inter-conversion as Cys-Cys was depleted from the medium without commensurate change in Cys. Cys-Cys drives oxidative stress in E. coli (50) because uptake results in rapid intracellular conversion to Cys that drives the Fenton reaction. Furthermore, over-accumulation of Cys is avoided by secretion of excess Cys into the medium. It is therefore possible that in the presence of Cj0025c, medium Cys levels are maintained because of both the intracellular conversion of Cys-Cys to Cys, release of excess Cys and concurrent Cys uptake resulting in an overall net appearance of extracellular maintenance. Given however, that we observed the same maintenance of medium Cys in Δcj0025c, it appears more likely that Cys import did not occur under these conditions.
It remains possible that nutrients other than Cys-Cys may be imported via Cj0025c. We observed little difference between WT and Δcj0025c C. jejuni for uptake of any of the amino and organic acids tested; although we did observe differences in the rate of uptake for some, and low level preservation of medium Lac in Δcj0025c. Unfortunately, we did not observe the Lac permease LctP in Δcj0025c, therefore we are unable to say whether this difference is a result of effects on that protein or are specific to Cj0025c. Additionally, we were unable to monitor acetate in culture medium and can make no assertions about any role for Cj0025c in acetate transport. Finally, we are also unable to conclude whether Cys-Cys is the sole dipeptide transported by Cj0025c. Given increased abundance of Cj0204/CptA upon cj0025c deletion, it remains plausible that there is some functional redundancy between these two transporters.
DOC induces an in vivo-like response at the proteome level that is reflected in increased virulence phenotypes, including metabolic rearrangement resulting in increased uptake of specific amino and organic acids, as well as sulfur compounds, including Cys-Cys. Cj0025c was induced by DOC and shares sequence identity with bacterial TcyP Cys-Cys transporters. Deletion of cj0025c resulted in C. jejuni that were attenuated for Cys-Cys import, and that display a proteome switch to a sulfur starvation-like response. Δcj0025c C. jejuni were attenuated for virulence phenotypes including motility, cell morphology, antibiotic resistance, and human epithelial cell invasion, consistent with a requirement for sulfur. Resistance to the toxic analog Se-Cys-Cys confirmed that Cj0025c encodes a Cys-Cys transporter that we have named TcyP to be consistent with the nomenclature of homologous proteins in other bacteria.
DATA AVAILABILITY
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via PRIDE with the data set identifier PXD017934.
Acknowledgments
We thank Prof. Brendan Wren (London School of Hygeine and Tropical Medicine) for provision of strains and molecular biology reagents; and Ben Crossett, David Maltby, Angela Connolly and Melanie White for technical advice.
Department of Health | National Health and Medical Research Council (NHMRC) (APP1106878) to Stuart J. Cordwell
Footnotes
This article contains supplemental data.
Funding and additional information—This work was supported in part by the National Health and Medical Research Council (NHMRC) of Australia (Project Grant APP1106878 to S.J.C). L.M., A.L.D. and J.A.C are supported by Australian Government Research Training Program (RTP) Stipends. A.L.D. is additionally supported by the William G. Murrell Postgraduate Scholarship in Microbiology and a University of Sydney Merit Award Supplementary Scholarship. W.P.K. is supported by a University of Sydney/Faculty of Science Postgraduate Award in Microbial Vaccinology. This work was facilitated by access to Sydney Mass Spectrometry, a Core Research Facility of the University of Sydney.
Author contributions—L.M. and S.J.C. designed research; L.M., A.L.D., W.P.K., J.A.C., Z.S.-B., P.N., N.S., and S.J.C. performed research; L.M., N.S., and S.J.C. analyzed data; L.M. and S.J.C. wrote the paper.
Conflict of interest—Authors declare no competing interests.
Abbreviations—The abbreviations used are:
- LC-MS/MS
- liquid chromatography coupled to tandem mass spectrometry
- DOC
- sodium deoxycholate
- DIA-SWATH-MS
- system-wide validation was performed by data-independent acquisition
- GBS
- Guillain-Barré Syndrome
- HILIC
- hydrophilic interaction liquid chromatography.
Present address for Paula Niewold: Department of Parasitology, Leiden University Medical Centre, Leiden University, Leiden, The Netherlands.
Present address for Nestor Solis: Centre for Blood Research, Department of Oral Biological and Medical Sciences, University of British Columbia, Vancouver, Canada.
Supplementary Material
REFERENCES
- 1.Epps S.V.R., Harvey R.B., Hume M.E., Phillips T.D., Anderson R.C., Nisbet D.J. Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health. 2013;10:6292–6304. doi: 10.3390/ijerph10126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermans D., Pasmans F., Heyndrickx M., Van Immerseel F., Martel A., Van Deun K., Haesebrouck F. A tolerogenic mucosal immune response leads to persistent Campylobacter jejuni colonization in the chicken gut. Crit. Rev. Microbiol. 2012;38:17–29. doi: 10.3109/1040841X.2011.615298. [DOI] [PubMed] [Google Scholar]
- 3.Humphrey S., Chaloner G., Kemmett K., Davidson N., Williams N., Kipar A., Humphrey T., Wigley P. Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio. 2014;5:e01364–14. doi: 10.1128/mBio.01364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awad W.A., Hess C., Hess M. Re-thinking chicken-Campylobacter jejuni interaction: a review. Avian Pathol. 2018;47:352–363. doi: 10.1080/03079457.2018.1475724. [DOI] [PubMed] [Google Scholar]
- 5.Black R.E., Levine M.M., Clements M.L., Hughes T.P., Blaser M.J. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 6.Nachamkin I., Allos B.M., Ho T. Campylobacter species and Guillain-Barré Syndrome. Clin. Microbiol. Revs. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo C.D., Taboada E., Nash J.H., Lanthier P., Kelly J., Lau P.C., Verhulp R., Mykytczuk O., Sy J., Findlay W.A., Amoako K., Gomis S., Willson P., Austin J.W., Potter A., Babiuk L., Allan B., Szymanski C.M. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 2004;279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- 8.Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15:456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Lertsethtakarn P., Ottermann K.M., Hendrixson D.R. Motility and chemotaxis in CampylobacterHelicobacter. Annu. Rev. Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korolik V. The role of chemotaxis during Campylobacter jejuni colonisation and pathogenesis. Curr. Opin. Microbiol. 2019;47:32–37. doi: 10.1016/j.mib.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Szymanski C.M., Gaynor E.C. How a sugary bug gets through the day: recent developments in understanding fundamental processes impacting Campylobacter jejuni pathogenesis. Gut Microbes. 2012;3:135–144. doi: 10.4161/gmic.19488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cain J.A., Dale A.L., Niewold P., Klare W.P., Man L., White M.Y., Scott N.E., Cordwell S.J. Proteomics reveals multiple phenotypes associated with N-linked glycosylation in. Campylobacter jejuni. Mol. Cell. Proteomics. 2019;18:715–734. doi: 10.1074/mcp.RA118.001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young K.T., Davis L.M., DiRita V.J. Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 14.Konkel M.E., Garvis S.G., Tipton S.L., Anderson DE, Jr, Cieplak W., Jr Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- 15.Monteville M.R., Yoon J.E., Konkel M.E. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiol. 2003;149:153–165. doi: 10.1099/mic.0.25820-0. [DOI] [PubMed] [Google Scholar]
- 16.Konkel M.E., Larson C.L., Flanagan R.C. Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J. Bacteriol. 2010;192:68–76. doi: 10.1128/JB.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S., Joe A., Lynett J., Hani E.K., Sherman P., Chan V.L. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 2001;39:1225–1236. doi: 10.1111/j.1365-2958.2001.02294.x. [DOI] [PubMed] [Google Scholar]
- 18.Scott N.E., Bogema D.R., Connolly A.M., Falconer L., Djordjevic S.P., Cordwell S.J. Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 2009;8:4654–4664. doi: 10.1021/pr900544x. [DOI] [PubMed] [Google Scholar]
- 19.Pei Z.H., Burucoa C., Grignon B., Baqar S., Huang X.Z., Kopecko D.J., Bourgeois A.L., Fauchere J.L., Blaser M.J. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 1998;66:938–943. doi: 10.1128/iai.66.3.938-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leon-Kempis M.D., Guccione E., Mulholland F., Williamson M.P., Kelly D.J. The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 2006;60:1262–1275. doi: 10.1111/j.1365-2958.2006.05168.x. [DOI] [PubMed] [Google Scholar]
- 21.Rivera-Amill V., Kim B.J., Seshu J., Konkel M.E. Secretion of the virulence associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J. Infect. Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- 22.Dasti J.I., Tareen A.M., Lugert R., Zautner A.E., Gross U. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 2010;300:205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Song Y.C., Jin S., Louie H., Ng D., Lau R., Zhang Y., Weerasekera R., Al Rashid S., Ward L.A., Der S.D., Chan V.L. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol. Microbiol. 2004;53:541–553. doi: 10.1111/j.1365-2958.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- 24.Pickett C.L., Pesci E.C., Cottle D.L., Russell G., Erdem A.N., Zeytin H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 1996;64:2070–2078. doi: 10.1128/iai.64.6.2070-2078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ransom G.M., Kaplan B., McNamara A.M., Wachsmuth I.K. Campylobacter prevention and control: the USDA Food Safety and Inspection Service role and new food safety approaches. In: Nachamkin I., Blaser M.J., editors. Campylobacter. 2nd ed. ASM Press; Washington, D.C: 2000. pp. 511–528. [Google Scholar]
- 26.Bhaduri S., Cottrell B. Survival of cold-stressed Campylobacter jejuni on ground chicken and chicken skin during frozen storage. Appl. Environ. Microbiol. 2004;70:7103–7109. doi: 10.1128/AEM.70.12.7103-7109.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stahl M., Friss L.M., Nothaft H., Liu X., Li J., Szymanski C.M., Stintzi A. L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7194–7199. doi: 10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl M., Butcher J., Stintzi A. Nutrient acquisition and metabolism by Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2012;2:5. doi: 10.3389/fcimb.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofreuter D. Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni. Front. Cell Infect. Microbiol. 2014;4:137. doi: 10.3389/fcimb.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnham P.M., Hendrixson D.R. Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 2018;16:551–565. doi: 10.1038/s41579-018-0037-9. [DOI] [PubMed] [Google Scholar]
- 31.Guccione E., Leon-Kempis M.D., Pearson B.M., Hitchin E., Mulholland F., van Diemen P.M., Stevens M.P., Kelly D.J. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 2008;69:77–93. doi: 10.1111/j.1365-2958.2008.06263.x. [DOI] [PubMed] [Google Scholar]
- 32.Hofreuter Novik D., V, Galán J.E. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe. 2008;4:425–433. doi: 10.1016/j.chom.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Barnes I.H., Bagnall M.C., Browning D.D., Thompson S.A., Manning G., Newell D.G. Gamma-glutamyltranspeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 2007;43:198–207. doi: 10.1016/j.micpath.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velayudhan J., Jones M.A., Barrow P.A., Kelly D.J. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofreuter D., Mohr J., Wensel O., Rademacher S., Schreiber K., Schomburg D., Gao B., Galán J.E. Contribution of amino acid catabolism to the tissue specific persistence of Campylobacter jejuni in a murine colonization model. PLoS ONE. 2012;7:e50699. doi: 10.1371/journal.pone.0050699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wösten M.M., van de Lest C.H., van Dijk L., van Putten J.P. Function and regulation of the C4-dicarboxylate transporters in Campylobacter jejuni. Front. Microbiol. 2017;8:174. doi: 10.3389/fmicb.2017.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas M.T., Shepherd M., Poole R.K., van Vliet A.H., Kelly D.J., Pearson B.M. Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 2011;13:48–61. doi: 10.1111/j.1462-2920.2010.02307.x. [DOI] [PubMed] [Google Scholar]
- 38.Vorwerk H., Mohr J., Huber C., Wensel O., Schmidt-Hohagen K., Gripp E., Josenhans C., Schomburg D., Eisenreich W., Hofreuter D. Utilization of host-derived cysteine-containing peptides overcomes the restricted sulphur metabolism of Campylobacter jejuni. Mol. Microbiol. 2014;93:1224–1245. doi: 10.1111/mmi.12732. [DOI] [PubMed] [Google Scholar]
- 39.Garvis S.G., Tipton S.L., Konkel M.E. Identification of a functional homolog of the Escherichia coliSalmonella typhimurium cysM gene encoding O-acetylserine sulfhydrylase B in Campylobacter jejuni. Gene. 1997;18:63–67. doi: 10.1016/s0378-1119(96)00631-2. [DOI] [PubMed] [Google Scholar]
- 40.Scott N.E., Nothaft H., Edwards A.V.G., Labbate M., Djordjevic S.P., Larsen M.R., Szymanski C.M., Cordwell S.J. Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 2012;287:29384–29396. doi: 10.1074/jbc.M112.380212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alazzam B., Bonnassie-Rouxin S., Dufour V., Ermel G. MCLMAN, a new minimal medium for Campylobacter jejuni NCTC 11168. Res. Microbiol. 2011;162:173–179. doi: 10.1016/j.resmic.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 42.Scott N.E., Marzook N.B., Deutscher A., Falconer L., Crossett B., Djordjevic S.P., Cordwell S.J. Mass spectrometric characterization of the Campylobacter jejuni adherence factor CadF reveals post-translational processing that removes immunogenicity while retaining fibronectin binding. Proteomics. 2010;10:277–288. doi: 10.1002/pmic.200900440. [DOI] [PubMed] [Google Scholar]
- 43.Brown H.L., Reuter M., Salt L.J., Cross K.L., Betts R.P., van Vliet A.H. Chicken juice enhances surface attachment and biofilm formation of Campylobacter jejuni. Appl. Environ. Microbiol. 2014;80:7053–7060. doi: 10.1128/AEM.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burguière P., Auger S., Hullo M., Danchin A., Martin-Verstraete I. Three different systems participate in L-cystine uptake in Bacillus subtilis. J. Bacteriol. 2004;186:4875–4884. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung L.M., Fondrie W.E., Doi Y., Johnson J.K., Strickland D.K., Ernst R.K., Goodlett D.R. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci. Rep. 2017;7:6403. doi: 10.1038/s41598-017-04793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei R., Li G.D., Seymour A.B. High-throughput and multiplexed LC/MS/MRM method for targeted metabolomics. Anal. Chem. 2010;82:5527–5533. doi: 10.1021/ac100331b. [DOI] [PubMed] [Google Scholar]
- 47.Chong J., Soufan O., Li C., Caraus I., Li S.Z., Bourque G., Wishart D.S., Xia J.G. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parkhill J., Wren B.W., Mungall K., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Jagels K., Karlyshev A.V., Moule S., Pallen M.J., Penn C.W., Quail M.A., Rajandream M.A., Rutherford K.M., van Vliet A.H.M., Whitehead S., Barrell B.G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 49.Malik-Kale P., Parker C.T., Konkel M.E. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J. Bacteriol. 2008;190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chonoles Imlay K.R., Korshunov S., Imlay J.A. Physiological roles and adverse effects of the two cystine importers of Escherichia coli. J. Bacteriol. 2015;197:3629–3644. doi: 10.1128/JB.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lensmire J.M., Dodson J.P., Hsueh B.Y., Wischer M.R., Delekta P.C., Shook J.C., Ottosen E.N., Kies P.J., Ravi J., Hammer N.D. The Staphylococcus aureus cystine transporters TcyABC and TcyP facilitate nutrient sulfur acquisition during infection. Infect. Immun. 2020;88:e00690–19. doi: 10.1128/IAI.00690-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Stel A.X., van de Lest C.H.A., Huynh S., Parker C.T., van Putten J.P.M., Wösten M.M.S.M. Catabolite repression in Campylobacter jejuni correlates with intracellular succinate levels. Environ. Microbiol. 2018;20:1374–1388. doi: 10.1111/1462-2920.14042. [DOI] [PubMed] [Google Scholar]
- 53.Tralau T., Vuilleumier S., Thibault C., Campbell B.J., Hart C.A., Kertesz M.A. Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J. Bacteriol. 2007;189:6743–6750. doi: 10.1128/JB.00889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott C., Hilton M.E., Coppin C.W., Russell R.J., Oakschott J.G., Sutherland T.D. A global response to sulfur starvation in Pseudomonas putida and its relationship to the expression of low-sulfur-content proteins. FEMS Microbiol. Lett. 2007;267:184–193. doi: 10.1111/j.1574-6968.2006.00575.x. [DOI] [PubMed] [Google Scholar]
- 55.Frirdich, E., Biboy, J., Adams, C., Lee, J., Ellermeier, J., Gielda, L.D., DiRita, V.J., Girardin, S.E., Vollmer, W., and Gaynor, E.C., Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni, . PLoS Pathog. 8, e1002602. [DOI] [PMC free article] [PubMed]
- 56.Cullen T.W., Trent M.S. A link between the assembly of flagella and lipooligosaccharide of the Gram negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin J., Sahin O., Michel L.O., Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 2003;71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin J., Michel L.O., Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002;46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alemka A., Corcionivoschi N., Bourke B. Defense and adaptation: the complex inter-relationship between Campylobacter jejuni and mucus. Front. Cell. Infect. Microbiol. 2012;20:15. doi: 10.3389/fcimb.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luethy P.M., Huynh S., Ribardo D.A., Winter S.E., Parker C.T., Hendrixson D.R. Microbiota-derived short-chain fatty acids modulate expression of Campylobacter jejuni determinants required for commensalism and virulence. mBio. 2017;8:e00407–17. doi: 10.1128/mBio.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carbonero F., Benefiel A.C., Alizadeh-Ghamsari A.H., Gaskins H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012;3:448. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol. Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y.W., Denkmann K., Kosciow K., Dahl C., Kelly D.J. Tetrathionate stimulated growth of Campylobacter jejuni identifies a new type of bi-functional tetrathionate reductase (TsdA) that is widely distributed in bacteria. Mol. Microbiol. 2013;88:173–188. doi: 10.1111/mmi.12176. [DOI] [PubMed] [Google Scholar]
- 65.Rasmussen J.J., Vegge C.S., Frokiaer H., Howlett R.M., Krogfelt K.A., Kelly D.J., Ingmer H. Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J. Med. Microbiol. 2013;62:1135–1143. doi: 10.1099/jmm.0.059345-0. [DOI] [PubMed] [Google Scholar]
- 66.He Y., Frye J.G., Strobaugh T.P., Jr, Chen C. Analysis of AI-2/LuxS-dependent transcription in Campylobacter jejuni 81–176. Foodborne Pathog. Dis. 2008;5:399–415. doi: 10.1089/fpd.2008.0106. [DOI] [PubMed] [Google Scholar]
- 67.Shen F., Hobley L., Doherty N., Loh J.T., Cover T.L., Sockett R.E., Hardie K.R., Atherton J.C. In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transulphuration, regulates motility through modulation of flagellar gene expression. BMC Microbiol. 2010;10:210. doi: 10.1186/1471-2180-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Šimunović K, Ramić D., Xu C., Smole Možina S. Modulation of Campylobacter jejuni motility, adhesion to polystyrene surfaces, and invasion of INT407 cells by quorum-sensing inhibition. Microorganisms. 2020;11, 8(1) doi: 10.3390/microorganisms8010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson M.T., Mitchell M.A., Sintsova A., Rice K.A., Mobley H.L.T. Sulfur assimilation alters flagellar function and modulates the gene expression landscape of Serratia marcescens. mSystems. 2019;6(4):e00285–19. doi: 10.1128/mSystems.00285-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frirdich E., Vermeulen J., Biboy J., Soares F., Taveirne M.E., Johnson J.G., DiRita V.J., Girardin S.E., Vollmer W., Gaynor E.C. Peptidoglycan LD-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J. Biol. Chem. 2014;289:8007–8018. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frirdich E., Biboy J., Huynh S., Parker C.T., Vollmer W., Gaynor E.C. Morphology heterogeneity within a Campylobacter jejuni helical population: the use of calcofluor white to generate rod-shaped C. jejuni 81–176 clones and the genetic determinants responsible for differences in morphology within 11168 strains. Mol. Microbiol. 2017;104:948–971. doi: 10.1111/mmi.13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esson D., Mather A.E., Scanlan E., Gupta S., de Vries S.P., Bailey D., Harris S.R., McKinley T.J., Méric G., Berry S.K., Mastroeni P., Sheppard S.K., Christie G., Thomson N.R., Parkhill J., Maskell D.J., Grant A.J. Genomic variations leading to alterations in cell morphology of Campylobacter spp. Sci. Rep. 2016;6:38303. doi: 10.1038/srep38303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esson D., Gupta S., Bailey D., Wigley P., Wedley A., Mather A.E., Méric G., Mastroeni P., Sheppard S.K., Thomson N.R., Parkhill J., Maskell D.J., Christie G., Grant A.J. Identification and initial characterisation of a protein involved in Campylobacter jejuni cell shape. Microb. Pathog. 2017;104:202–211. doi: 10.1016/j.micpath.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwata T., Watanabe A., Kusumoto M., Akiba M. Peptidoglycan acetylation of Campylobacter jejuni is essential for maintaining cell wall integrity and colonization in chicken intestines. Appl. Environ. Microbiol. 2016;82:3284–3290. doi: 10.1128/AEM.02068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Müller A., Thomas G.H., Horler R., Brannigan J.A., Blagova E., Levdikov V.M., Fogg M.J., Wilson K.S., Wilkinson A.J. An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein. Mol. Microbiol. 2005;57:143–155. doi: 10.1111/j.1365-2958.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- 76.Lin A.E., Krastel K., Hobb R.I., Thompson S.A., Cvitkovitch D.G., Gaynor E.C. Atypical roles for Campylobacter jejuni amino acid ATP binding cassette transporter components PaqP and PaqQ in bacterial stress tolerance and pathogen-host cell dynamics. Infect. Immun. 2009;77:4912–4924. doi: 10.1128/IAI.00571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banerjee R. Redox outside the box: linking extracellular redox remodeling with intracellular redox metabolism. J. Biol. Chem. 2012;287:4397–4402. doi: 10.1074/jbc.R111.287995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Uncited reference
- 59.Neal-McKinney J.M., Konkel M.E. The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front. Cell. Infect. Microbiol. 2012;20:31. doi: 10.3389/fcimb.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via PRIDE with the data set identifier PXD017934.