Abstract
Background
The recent studies highlighted the critical role of exosomes in the regulation of inflammation. Here, we investigated the dynamic biogenesis of the exosomes in the rat model of asthma.
Results
Our finding showed an increase in the expression of IL-4 and the suppression of IL-10 in asthmatic lung tissues compared to the control samples (p < 0.05). Along with the promotion of IL-4, the protein level of TNF-α was induced, showing an active inflammatory status in OVA-sensitized rats. According to our data, the promotion of asthmatic responses increased exosome biogenesis indicated by increased CD63 levels and acetylcholine esterase activity compared to the normal condition (p < 0.05).
Conclusion
Data suggest that the stimulation of inflammatory response in asthmatic rats could simultaneously increase the paracrine activity of pulmonary cells via the exosome biogenesis. Exosome biogenesis may correlate with the inflammatory response.
Keywords: Asthma, Exosome biogenesis, CD63, Inflammatory cytokines, Rats
Background
Asthma is a pathologic state of bronchopulmonary tissue with a considerable socioeconomic burden on health care systems and is mainly characterized by chronic inflammation and profound structural remodeling [1]. Based on the released statistics, the prevalence of asthma is high in either developing or industrialized countries [2]. Histopathological changes are integral to the reduction of the Th2/Th1 ratio and accumulation of immune cells in the pulmonary niche [3]. Due to the sustained immune cell infiltration and activation, it has been shown the normal and physiological activity of multiple cell types with distinct functions are altered within the pulmonary tissue [1, 4]. Monitoring early-stage asthmatic changes in the pulmonary niche could help us to predict the asthma progression and prevent further harmful changes in the function of the respiratory tract. In addition to morphological and functional alterations, the detection of paracrine activity and the release of different factors to the airway conduits are also helpful to monitor underlying pathologies [5]. The disproportion inflammatory response in the severe asthma process may comprise various intercellular communication facilitated by signaling molecules such as exosomes [5]. Exosomes, ranging from 30 to 150 nm size nanoparticles, are released from each cell in response to different stimuli under physiological and pathological conditions [6, 7]. These nano-sized particles harbor both genomic and proteomic elements which could reflect the time-lapse changes of the original cells [8, 9]. In addition to the fundamental role of exosomes in cell homeostasis and the removal of exhausted biomolecules, it is noteworthy that the promotion of asthmatic changes could alter the basal dynamic exosome biogenesis in surface respiratory epithelial cells [10]. Along with these changes, different cell types, either recruited immune cells or local bronchopulmonary cells, could release various amounts of exosomes upon initiation of asthmatic changes to regulate in cell-to-cell communication or to signal the occurrence of pathological conditions [10, 11]. Previously, our research group showed enhanced exosome biogenesis and secretion from human mesenchymal stem cells (MSCs) incubated in a culture medium supplemented with diabetic sera, showing the potency of inflammatory conditions to modulate exosome biogenesis [12]. Commensurate with these descriptions, there are some experiments related to the application of stem cell derived exosomes in the alleviation of asthma [9, 10]. It has been suggested that exosomes can transfer specific bioactive factors, which control the intensity and persistence of inflammation by delivering cargo to the target cells [7, 13–15].
Along with the above-mentioned facts, the current study is a preliminary study to analyze the potency of asthma in the modulation of the exosome biogenesis and secretion in the rat model. We hypothesize that the promotion of asthma could alter total exosome biogenesis in an asthmatic niche. Results from the current experiment could help us to predict the occurrence and intensity of immunological response in the asthmatic niche by monitoring exosome biogenesis. In this case, exosomes are suitable as diagnostic tools that could be used as novel asthma-related biomarkers.
Methods
Animals and ethical issues
All animals were treated according to the previously delivered guidelines (NIH Publication No. 85-23, revised 1996). Experimental protocols were locally reviewed and approved by the ethics committee of Tabriz University of Medical Sciences (No: IR.TBZMED.VCR.REC.1398.308).
Experimental protocol
Eight-week-old male Wistar rats (n = 16; weighing 200–250 g) were subjected to the current experiment. Animals were kept under the standard condition (20–22 °C with a relative humidity of 50 ± 10%). Animals had free access to water and food. Rats were randomly allocated into two groups as follows: control and asthmatic rats (each in 8).
Sensitization and challenge with OVA
On this basis, we followed the previous protocol that lasts over 32 ± 1 days [16–18]. Briefly, rats received 1 mg OVA (Sigma-Aldrich) via an intraperitoneal route on days 1 and 8. OVA was dissolved in 1 ml normal saline solution containing 200 mg aluminum hydroxide followed by the inhalation of 4% aerosolized OVA via a nebulizer apparatus (Model: CX3, Omron Co., Netherland) from days 14 to 32 ± 1. The inhalation protocol was performed for 5 minutes daily in a whole-body inhalation exposure chamber (Dimensions 30 × 20 × 20 cm3). In the control group, normal saline was used as a control vehicle using a similar protocol. After completion of asthma induction, rats from each group were sacrificed humanly and pulmonary tissues were taken soon after rats’ scarification.
Bronchoalveolar lavage fluid (BALF) preparation
After pectoral incision, BALF was collected immediately by five consecutive 1 mL instillations of sodium chloride 0.9% by a catheter connected to each trachea [4].
Pathological evaluation
To investigate histological changes, lung samples from each group were collected and fixed in a 10% buffer formalin solution (Merck, Germany). Specimens were passaged by serial concentrations of EtOH (70-100%). Then, 4-μm thick sections were prepared and loaded on glass slides. All sections were stained by hematoxylin-eosin (H&E) solution [19]. To confirm the asthmatic condition, we followed the specific pathological changes as follows: leukocyte infiltration, bronchus-associated lymphoid tissue (BALT) hyperplasia, peribronchial cuffing, and emphysema.
Real-time PCR analysis
The transcription levels of IL-4 and IL-10 were quantitatively determined by real-time PCR assay according to previously published methods [16, 20]. In brief, 40 mg lung sample was lysed for total RNA extraction using a total RNA extraction mini kit (YTA, Iran). The concentration of total RNA was measured by a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE 19810 USA). The first-strand cDNA was synthesized from total RNA using a cDNA Synthesis Kit (YTA, Iran). To measure relative mRNA levels, the SYBR Green master mix (YTA, Iran) and Rotor-Gene 6000 instrument (Corbett Life Science, Australia) were used. All samples were measured in triplicate. The relative transcript level of each target gene was normalized to the GAPDH gene using the 2−∆∆Ct method. The list of primer sequences was enlisted in Table 1.
Table 1.
The list of primers used for real-time PCR analysis
| Gene | Primer sequence (5'-3') | |
|---|---|---|
| Forward | Reverse | |
| IL-4 | CTGTCACCCTGTTCTGCTTTCT | CTGGTACAAACATCTCGGTGCA |
| IL-10 | TGAGAATAAAAGCAAGGCAGTGG | GTAGGCTTCTATGCAGTTGATGA |
| GAPDH | TTG CCA TCA ACG ACC CCT TCA | AGC ACC AGC ATC ACC CCA TTT |
Enzyme-linked immunosorbent assay (ELISA)
We used an ELISA to determine the protein level of TNF-α in bronchoalveolar lavage fluid (BALF) using Rat TNF-α ELISA kit (Cat. no: EK0526). Briefly, we incubated each well of 96-well plates with 100 μl BAL samples from each group and kept at 37 °C for 90 min. Then, the solution was discarded and 100 μl working solution was added and incubated at 37 °C for 60 min. The solution was discarded and washed twice in phosphate-buffered saline (PBS). After PBS wash, 100 μl ABC working solution was added to each well and incubated at 37 °C for 30 min. Then, wells were washed 5 times in PBS, and 90 μl chromogen substrate 1,2,4,5- tetramethyl benzene (TMB) was poured into each well and kept for 20 min. To stop the reaction, 100 μl stop solution was added to each well. The absorbance was recorded after 30 min using a microplate reader (Biotek) at 450 nm.
Western blotting
The protein levels of CD63, a protein involved in exosomes biogenesis [21], were evaluated by western blotting analysis. For this purpose, 50 mg of lung tissue were lysed in RIPA lysis buffer supplemented with a protease inhibitor cocktail (Sigma Aldrich) and then homogenized with an electric homogenizer. The lysates were obtained by centrifugation at 12,000g for 20 min at 4 °C. In brief, 100 μg of each sample lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.2-μm polyvinylidene difluoride (PVDF, 249 Millipore) membranes. After blocking with 5% skim milk (Gibco), membranes were incubated with primary anti-human CD63 antibody (Cat no: sc-5275) overnight at 4 °C followed by incubated with secondary antibody (goat anti-mouse IgG-HRP: DB9571, Iran) 1 h at 37 °C. Immunoblotting bands were visualized using a chemiluminescence (ECL) detection system according to the manufacturer’s instructions (Roche). β-actin (Cat no: sc-47778) was kept as a housekeeping protein for normalization. Relative protein levels were calculated using NIH ImageJ software ver. 1.44p.
Exosome isolation
After treatment, BAL samples were collected and after removing debris at 300 g for 10 min, exosomes were isolated using Exo-spin™ kit (EX01-8, Cell Guidance Systems) according to the manufacturer’s guidelines. The proper volume (1/2 ratio) of Exo-spin™ buffer was mixed with each sample and kept overnight at 4 °C. Next, the mixtures were centrifuged at 16,000×g for 1 h using the provided filter columns. Exosome pellets were suspended in 100 μl PBS and kept at 4 °C.
Characterization of exosomes
Exosomes were subjected to transmission electron microscopy (TEM) and flow cytometry analysis. For TEM analysis, 20 μl exosome suspension were fixed with glutaraldehyde 1%, and then loaded on the carbon grids at room temperature and were allowed to dry. After twice washing, exosomes were stained with uranyl acetate 1% for 10 min at room temperature and were visualized using the TEM system (Philips BioTwin, CM100, The Netherlands) at 80 kV. For flow cytometry analysis, 200 μl exosome suspension was incubated with the primary anti-CD63 antibody (Cat no: sc-5275) for 2 h at 4 °C. Next, secondary anti-mouse IG-FITC (cat no: 406001, BioLegend) was added and incubated at 37 °C for 1 h. Samples were subjected to a BD FACSCalibur system and analyzed by FlowJo software (version 7.6.1).
Quantification of exosomes
To quantity the number of exosomes in BAL samples, we performed the acetylcholine esterase assay (AChE activity) using a commercial cholinesterase kit (Cat no. BXC080, Iran) according to the company’s guidelines. In brief, 100 μl of BAL samples were mixed with 500 μl R1 buffer (pyrophosphate + potassium hexacyanoferrate) and kept for 5 min at room temperature. Then, 20 μl R2 buffer (S-butyryl thiocholine iodide) was added, and optical density was obtained at 405 nm by three different intervals using a microplate reader system (BioTek). AhCE activity was measured by the following formula: Activity (U/l) = 65,800 × ΔAbs/min.
Data analysis
All quantitative results were analyzed using a student t-test and presented as mean ± SEM. Statistical significance was set at p values of < 0.05.
Results
Pathological changes were indicated in the asthmatic pulmonary niche
To confirm asthmatic changes, we performed histopathological analysis on pulmonary tissues obtained from the control and asthmatic lungs. Data revealed, massive leukocyte infiltration, bronchus-associated lymphoid tissue (BALT) hyperplasia, peribronchial cuffing, and emphysema in lung tissues of asthmatic rats compared to the control rats, indicating the efficiency of our protocol to induce asthmatic change established (Fig. 1).
Fig. 1.
Histopathological examination of asthmatic pulmonary tissue using H&E staining. Asthmatic changes were indicated by prominent interstitial pneumonia (arrows), focal hemorrhagia (arrows heads), and emphysema. The pathological remodeling coincided with the peribronchial cuffing, bronchial smooth muscle hypertrophy, and BALT hyperplasia. These features highlight the efficiency of our protocol in the induction of asthmatic changes using OVA.
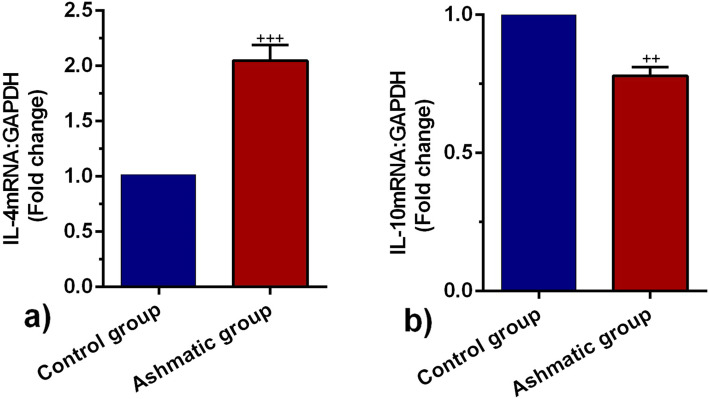
IL-4 and IL-10 expression was changed in the asthmatic lung tissue
To observe the inflammatory status of asthmatic lungs, we monitored the expression of two interleukins IL-4 and IL-10 by real-time PCR assay. According to our data, the expression of IL-4 and IL-10 were changed significantly in the asthmatic group as compared to healthy rats (p < 0.05; Fig. 2a, b). In this regard, we showed enhanced expression of IL-4 coincided with suppression of IL-10 in asthmatic rats as compared with the control group (from p < 0.001 to p < 0.01 respectively) (Fig. 2a, b). The increase of IL-4 and suppression of IL-10 is a valuable tool to show the promotion of asthma in rats sensitized to OVA.
Fig. 2.
Measuring the transcription of IL-4 a and IL-10 b mRNA in the lung tissues of control and asthmatic group (n = 8). Bars represent the mean ± SEM. Statistical differences between control and asthmatic group: ++; p < 0.01 and +++; p < 0.001
Level of TNF-α was increased in the asthmatic BALF
We also monitored the protein levels of TNF-α in the asthmatic discharge along with the gene expression of IL-10 and IL-4. ELISA showed that the level of TNF-α in BALF was prominently increased in the asthmatic rats in comparison with the control rats (p < 0.001; Fig. 3). This data showed that the levels of TNF-α increases with asthma progression.
Fig. 3.
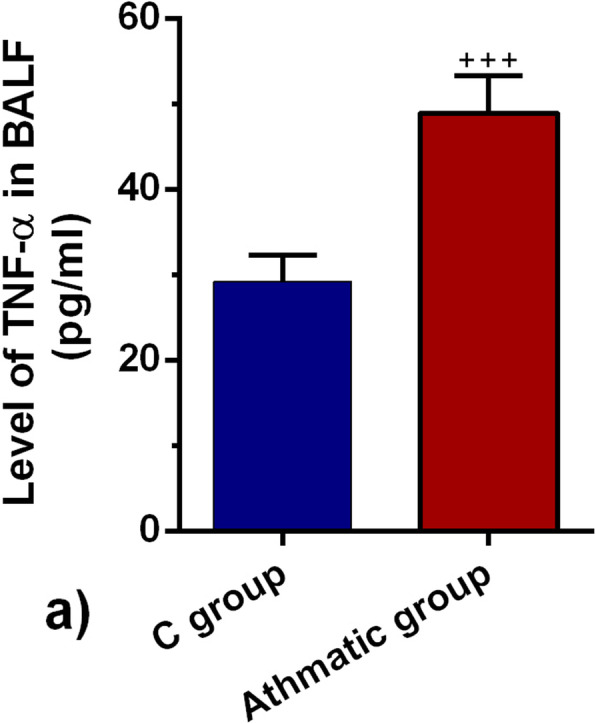
TNF-α in BALF of control and asthmatic rats (n = 8). Bars represent the mean ± SEM. Statistical differences between control and asthmatic groups: +++; p < 0.001
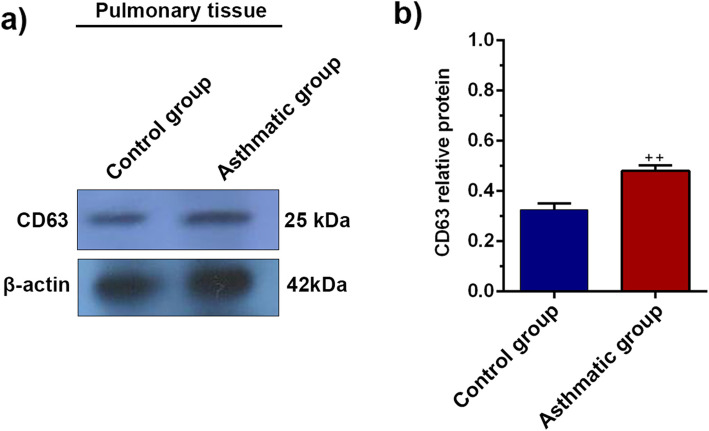
CD63 translation was induced in the lung tissues of asthmatic rats
We also analyzed the expression of CD63 protein in the lung tissues of asthmatic rats using western blotting (Fig. 4a, b). Data showed that the protein level of CD63 was increased in asthmatic rats (~ > 0.5–fold) as compared to control rats (P < 0.01). These data showed enhanced factor synthesis, which correlated with exosome biogenesis and production
Fig. 4.
Western blotting analysis of CD63 protein in lung tissues a. The relative expression of CD63 protein increased in asthmatic lung tissues b. Bars represent the mean ± SEM. Statistical differences between control and asthmatic group: ++; p < 0.01
Enhanced AChE activity was obtained in asthmatic BALF
By monitoring the activity of AChE, we also indirectly showed exosome release to the BALF. Based on data, a significant increase was found in the AChE activity of asthmatic BALF as compared to control samples (P < 0.001, Fig. 5). The data highlighted a prominent AChE activity in asthmatic discharge, which could be related to the paracrine activity of cells exposed to asthmatic changes.
Fig. 5.

Quantification of the number of exosomes secreted into BAL fluid by acetylcholinesterase (AChE) activity assay. Bars represent the mean ± SEM. Statistical differences between control and asthmatic group: +++; p < 0.001
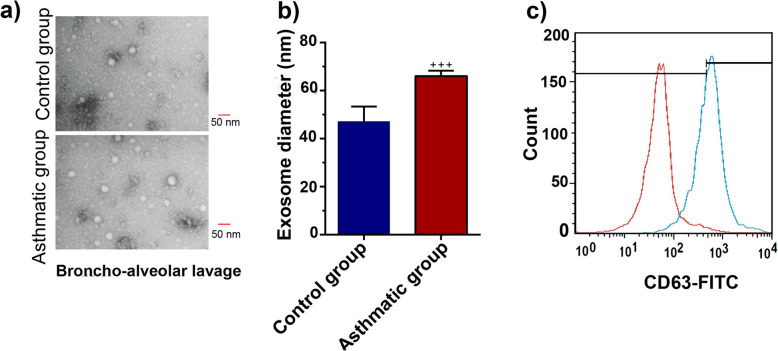
Characterization of purified exosomes
We characterized exosomes released from lung tissue into BALF according to the International Society for Extracellular Vesicles (ISEV) guidelines by TEM and flow cytometry analysis. TEM micrographs indicated a round shape and nano-scale-sized exosomes (Fig. 6a, b). Flow cytometry immunophenotyping confirmed the CD63 marker in purified exosomes (Fig. 6c). Data analysis showed that the diameter of exosomes isolated from asthmatic BAL was significantly increased compared to the control counterparts (p < 0.001; Fig. 6b). It seems that the promotion of an active inflammatory response could alter the production and physiochemical properties of exosomes.
Fig. 6.
Transmission electron micrographs of isolated exosomes a. Analyzing the size of exosomes diameter purified from BAL samples of asthmatic and control groups b. Flow cytometry confirmed the expression of CD63 marker on exosomes c. Bars represent the mean ± SEM. Statistical differences between control and asthmatic group: +++; p < 0.001
Discussion
This study was conducted to address the impact of the asthmatic condition on the dynamic biogenesis of exosomes in the rat model. Data showed OVA sensitization promoted asthma-like condition in the rats after 32 days. In addition to pathological changes, the level of inflammatory cytokines was also altered. We found that asthmatic changes increased leukocyte infiltration, which contributes to interstitial bronchopneumonia. Along with these changes, hyperplastic BALT, prominent peribronchial cuffing, and emphysema were also evident in the asthmatic niche [16, 18, 22]. It has been thought that the sustained immune cell recruitment correlated with the increase of IL-4 and suppression of Treg lymphocytes [23, 24]. The secretion of IL-4 triggers central pro-inflammatory responses that regulate eosinophil trans-endothelial migration and IgE production and increases the level of endothelial adhesion molecules such as vascular cell adhesion molecule-1 (VCAM-1). Also, IL-4 can increase mucus secretion, and also accelerate the differentiation of T helper type 2 lymphocytes which leads to cytokine secretion [25, 26]. Conversely, we found that the mRNA level of IL-10 was decreased in lung tissues of asthmatic rats, showing an inhibited Treg activity [27]. Our results share several similarities with previous findings that in asthma conditions, secretion of IL-10 was suppressed, which, in turn, induces the production of many pro-inflammatory cytokines [28, 29]. Besides, ELISA has further confirmed the pro-inflammatory condition in lung tissues of asthmatic rats in that, the production of TNF-a protein in BALF samples was increased. In agreement with previous experiment, it seems that the initiation of asthmatic condition triggers a cascade of cytokine release [30, 31]. TNF-a, a proinflammatory cytokine, plays key roles in the modulation of inflammation in different diseases, like asthma [32], and also in exosome biogenesis [33].
To investigate the dynamic of the exosomal secretory pathway in lung tissues of asthmatic rats, we measured the protein level of CD63. Results from western blotting confirmed that the protein level of CD63 was upregulated in asthmatic lungs. CD63, a tetraspanin protein located on the multivesicular body (MVB) and exosome membranes, is involved in the regulation of exosome biogenesis and loading [34]. Paredes et al. found that exosomes collected from patients BALF with mild allergic asthma abundantly contain CD63 protein compared to those from healthy individuals [35]. Besides, we found that the activity of AChE, an exosome-linked enzyme, was increased in BAL samples of asthmatic rats, indicating an increase in exosome secretion. Our observation showed that asthmatic BALs had a higher level of exosomes in BAL fluid than healthy samples aligns with previous reports [35]. The increased secretion of exosomes may either be due to increased formation of exosomes by airway epithelial cells of asthmatics or amplified immune cells infiltrated in the airways of asthmatics [36]. A growing body of evidence indicates cells under such stress conditions have a higher exosome production capacity. Furthermore, immune cell infiltration is well documented in the airways of asthmatics [37]; those release exosomes, participating in inflammatory responses [38]. Besides, it seems likely that the upregulated expression of IL-4 and TNF-a correlates with an increased level of exosome secretion in asthmatic rats [39, 40]. These results show a close association of the exosome biogenesis and secretion with the synthesis of pro-inflammatory cytokines, especially TNF-a when asthma is induced [5, 13]. The increased exosome biogenesis could correlate with the fact that injured cells tend to find a way to eliminate damaged compounds and unwanted proteins by using exosomes and exocytosis [41, 42]. A study by Takahashi et al., for example, revealed that exosome secretion contributes to supporting cell homeostasis by eliminating damaged DNA molecules from inside cells [43]. Whether asthma can alter the paracrine activity of pulmonary tissues by regulation of exosome secretion needs more investigations. We found that exosome size was increased in asthmatic BALFs, indicating the change and alteration in the exosome biogenesis and sorting machinery. To our knowledge, this finding is preliminary result and there are little details about the effect of asthmatic condition on size of exosomes. Similarly, it was demonstrated that the size of exosomes derived from diabetic MSCs was larger than those of secreted from normal MSCs [12]. Bagheri and co-worker previously showed that the size of exosomes did not alter in injured endothelial cells after exposure to laser irradiation [44]. Previous studies showed that cells may form various subpopulations of MVB/exosomes or respond differently to the insulting conditions [45–47]. Therefore, it can thus be conceivably hypothesized that asthmatic conditions may change MVB loading/biogenesis pathways that, in turn, affect exosomes size [48]. Therefore, additional studies are essential to elucidate whether increased exosome secretion rate in severe asthmatics condition is a way to decrease the intensity of pathological remodeling or it supports the elimination of exhausted intracellular molecules. Collectively, we showed that upon an increase in pro-inflammatory cytokines and damage in asthmatic lung tissues, exosome secretion, biogenesis are stimulated.
Conclusion
Taken together, our data indicate that the exosome secretory pathway concurrently with proinflammatory cytokines production was over-activated in the asthmatic lung tissues. Even so, further scrutiny is essential to uncover the key role of asthmatic-induced over-activation in the dynamic of exosome activities and a better understanding of inflammatory mediated by exosomes to improve the outcome of therapies.
Acknowledgements
Not applicable.
Abbreviations
- AChE
Acetylcholine esterase
- BALF
Bronchoalveolar lavage fluid
- ECL
Chemiluminescence
- ELISA
Enzyme-linked immunosorbent assay
- ISEV
International Society for Extracellular Vesicles
- MSCs
Human mesenchymal stem cells
- MVB
Multivesicular body
- PVDF
Polyvinylidene difluoride
- TMB
Tetramethyl benzene
- VCAM-1
Vascular cell adhesion molecule-1
Authors’ contributions
Conception and manuscript design: J. R. and M. A. Collection of data: A. A. Manuscript writing and analyze data: J. R., M. A., R. K. Made important revisions and confirmed final revision: J. R. and R. R. All authors reviewed and approved the final version of manuscript.
Funding
This study was approved and supported by a grant from the Tuberculosis and Lung Disease Research Center of Tabriz University of medical sciences (No: IR.TBZMED.VCR.REC.1398.308).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jafar Rezaie and Mahdi Ahmadi contributed equally to this work.
Contributor Information
Jafar Rezaie, Email: rezaie.j@umsu.ac.ir.
Mahdi Ahmadi, Email: mahdi59866@gmail.com.
References
- 1.Ahmadi M, Rahbarghazi R, Aslani MR, Shahbazfar AA, Kazemi M, Keyhanmanesh R. Bone marrow mesenchymal stem cells and their conditioned media could potentially ameliorate ovalbumin-induced asthmatic changes. Biomed Pharmacother. 2017;85:28–40. doi: 10.1016/j.biopha.2016.11.127. [DOI] [PubMed] [Google Scholar]
- 2.To T. Stanojevic S, Moores G, Gershon AS, Bateman ED, Cruz AA, Boulet L-P. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health. 2012;12:204. doi: 10.1186/1471-2458-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keyhanmanesh R, Rahbarghazi R, Aslani MR, Hassanpour M, Ahmadi M. Systemic delivery of mesenchymal stem cells condition media in repeated doses acts as magic bullets in restoring IFN-γ/IL-4 balance in asthmatic rats. Life Sci. 2018;212:30–36. doi: 10.1016/j.lfs.2018.09.049. [DOI] [PubMed] [Google Scholar]
- 4.Ahmadi M, Rahbarghazi R, Soltani S, Aslani MR, Keyhanmanesh R. Contributory anti-inflammatory effects of mesenchymal stem cells, not conditioned media, on ovalbumin-induced asthmatic changes in male rats. Inflammation. 2016;39:1960–1971. doi: 10.1007/s10753-016-0431-2. [DOI] [PubMed] [Google Scholar]
- 5.Cañas JA, Sastre B, Rodrigo-Muñoz JM, del Pozo V. Exosomes: a new approach to asthma pathology. Clin Chim Acta. 2019;495:139–147. doi: 10.1016/j.cca.2019.04.055. [DOI] [PubMed] [Google Scholar]
- 6.Zou W, Lai M, Zhang Y, Zheng L, Xing Z, Li T, Zou Z, Song Q, Zhao X, Xia L. Exosome release is regulated by mTORC1. Adv Sci. 2019;6:1801313. doi: 10.1002/advs.201801313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 8.Sancho-Albero M, Navascués N, Mendoza G, Sebastián V, Arruebo M, Martin-Duque P, Santamaria J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J Nanobiotechnol. 2019;17:16. doi: 10.1186/s12951-018-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin K, Wang S, Zhao RC. Exosomes from mesenchymal stem/stromal cells: a new therapeutic paradigm. Biomark Res. 2019;7:8. doi: 10.1186/s40364-019-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cañas JA, Sastre B, Rodrigo-Muñoz JM, del Pozo V. Exosomes: a new approach to asthma pathology. Clin Chim Acta. 2019;495:139–47. [DOI] [PubMed]
- 11.van den Berge M, Tasena H. Role of microRNAs and exosomes in asthma. Curr Opinion Pulmon Med. 2019;25(1):87–93. [DOI] [PubMed]
- 12.Rezaie J, Nejati V, Khaksar M, Oryan A, Aghamohamadzadeh N, Shariatzadeh MA, Rahbarghazi R, Mehranjani MS. Diabetic sera disrupted the normal exosome signaling pathway in human mesenchymal stem cells in vitro. Cell Tissue Res. 2018;374(3):555–65. [DOI] [PubMed]
- 13.Samanta S, Rajasingh S, Drosos N, Zhou Z, Dawn B, Rajasingh J. Exosomes: new molecular targets of diseases. Acta Pharmacol Sinica. 2018;39:501–513. doi: 10.1038/aps.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18:32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdyazdani N, Nourazarian A, Nozad Charoudeh H, Kazemi M, Feizy N, Akbarzade M, Mehdizadeh A, Rezaie J, Rahbarghazi R. The role of morphine on rat neural stem cells viability, neuro-angiogenesis and neuro-steroidgenesis properties. Neurosci Letters. 2017;636:205–212. doi: 10.1016/j.neulet.2016.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Ahmadi M, Rahbarghazi R, Shahbazfar A-A, Baghban H, Keyhanmanesh R. Bone marrow mesenchymal stem cells modified pathological changes and immunological responses in ovalbumin-induced asthmatic rats possibly by the modulation of miRNA155 and miRNA133. General Physiol Biophys. 2018;37:263–274. doi: 10.4149/gpb_2017052. [DOI] [PubMed] [Google Scholar]
- 17.Neamati A, Boskabady MH, Mahdavi-Shahri N, Mahmoudabady M. The preventive effect of Brassica napus L. oil on pathophysiological changes of respiratory system in experimental asthmatic rat. Avicen J Phytomed. 2013;3:56. [PMC free article] [PubMed] [Google Scholar]
- 18.Keyhanmanesh R, Rahbarghazi R, Ahmadi M. Systemic transplantation of mesenchymal stem cells modulates endothelial cell adhesion molecules induced by ovalbumin in rat model of asthma. Inflammation. 2018;41:2236–2245. doi: 10.1007/s10753-018-0866-8. [DOI] [PubMed] [Google Scholar]
- 19.Rahbarghazi R, Keyhanmanesh R, Aslani MR, Hassanpour M, Ahmadi M. Bone marrow mesenchymal stem cells and condition media diminish inflammatory adhesion molecules of pulmonary endothelial cells in an ovalbumin-induced asthmatic rat model. Microvasc Res. 2019;121:63–70. doi: 10.1016/j.mvr.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Oghbaei H, Alipour MR, Hamidian G, Ahmadi M, Ghorbanzadeh V, Keyhanmanesh R. Two months sodium nitrate supplementation alleviates testicular injury in streptozotocin-induced diabetic male rats. Exp Physiol. 2018;103:1603–1617. doi: 10.1113/EP087198. [DOI] [PubMed] [Google Scholar]
- 21.Rezaie J, Rahbarghazi R, Pezeshki M, Mazhar M, Yekani F, Khaksar M, Shokrollahi E, Amini H, Hashemzadeh S, Sokullu SE. Cardioprotective role of extracellular vesicles: a highlight on exosome beneficial effects in cardiovascular diseases. J Cell Physiol. 2019;234:21732–21745. doi: 10.1002/jcp.28894. [DOI] [PubMed] [Google Scholar]
- 22.Yoon S-Y, Hong GH, Kwon H-S, Park S, Park SY, Shin B, Kim T-B, Moon H-B, Cho YS. S-adenosylmethionine reduces airway inflammation and fibrosis in a murine model of chronic severe asthma via suppression of oxidative stress. Exp Mol Med. 2016;48:e236. doi: 10.1038/emm.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YC, Lee KH, Lee HB, Rhee YK. Serum levels of interleukins (IL)-4, IL-5, IL-13, and interferon-γ in acute asthma. J Asthma. 2001;38:665–671. doi: 10.1081/JAS-100107544. [DOI] [PubMed] [Google Scholar]
- 24.Wang K, Zhu X, Zhang K, Zhu L, Zhou F. Investigation of gallic acid induced anticancer effect in human breast carcinoma mcf-7 cells. J Biochem Mol Toxicol. 2014;28:387–393. doi: 10.1002/jbt.21575. [DOI] [PubMed] [Google Scholar]
- 25.Steinke JW, Borish L. Th2 cytokines and asthma—interleukin-4: its role in the pathogenesis of asthma, and targeting it for asthma treatment with interleukin-4 receptor antagonists. Respir Res. 2001;2:66. doi: 10.1186/rr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao H, Tong J, Wang Z, Zhang Z, Chen J. Effect of acupoint application therapy on expression of IL-4 and IFN-gamma in lung tissue of asthma rats. Zhen Ci Yan Jiu. 2007;32:174–178. [PubMed] [Google Scholar]
- 27.Ogawa Y, Duru EA, Ameredes BT. Role of IL-10 in the resolution of airway inflammation. Curr Mol Med. 2008;8:437–445. doi: 10.2174/156652408785160907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung F. Anti-inflammatory cytokines in asthma and allergy: interleukin-10, interleukin-12, interferon-gamma. Mediators Inflamm. 2001;10:51–59. doi: 10.1080/09629350120054518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawano H, Kayama H, Nakama T, Hashimoto T, Umemoto E, Takeda K. IL-10-producing lung interstitial macrophages prevent neutrophilic asthma. Int Immunol. 2016;28:489–501. doi: 10.1093/intimm/dxw012. [DOI] [PubMed] [Google Scholar]
- 30.Vuillemenot BR, Rodriguez JF, Hoyle GW. Lymphoid tissue and emphysema in the lungs of transgenic mice inducibly expressing tumor necrosis factor-α. Am J Respir Cell Mol Biol. 2004;30:438–448. doi: 10.1165/rcmb.2003-0062OC. [DOI] [PubMed] [Google Scholar]
- 31.Mehta AA, Mahajan S. Role of cytokines in pathophysiology of asthma. Iran J Pharmacol Ther. 2006;5:1–14. [Google Scholar]
- 32.Babu KS, Davies DE, Holgate ST. Role of tumor necrosis factor alpha in asthma. Immunol Allergy Clin. 2004;24:583–597. doi: 10.1016/j.iac.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Ye L, Lu H, Chen H, Zhang Y, Huang Y, Zheng JC. TNF-α promotes extracellular vesicle release in mouse astrocytes through glutaminase. J Neuroinflamm. 2017;14:87. doi: 10.1186/s12974-017-0853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torregrosa Paredes P, Esser J, Admyre C, Nord M, Rahman QK, Lukic A, Rådmark O, Grönneberg R, Grunewald J, Eklund A, et al. Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy. 2012;67:911–919. doi: 10.1111/j.1398-9995.2012.02835.x. [DOI] [PubMed] [Google Scholar]
- 36.Hough KP, Wilson LS, Trevor JL, Strenkowski JG, Maina N, Kim Y-I, Spell ML, Wang Y, Chanda D, Dager JR. Unique lipid signatures of extracellular vesicles from the airways of asthmatics. Sci Rep. 2018;8:1–16. doi: 10.1038/s41598-018-28655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker C, Kaegi MK, Braun P, Blaser K. Activated T cells and eosinophilia in bronchoalveolar lavages from subjects with asthma correlated with disease severity. J Allergy Clin Immunol. 1991;88:935–942. doi: 10.1016/0091-6749(91)90251-I. [DOI] [PubMed] [Google Scholar]
- 38.Fujita Y, Yoshioka Y, Ito S, Araya J, Kuwano K, Ochiya T. Intercellular communication by extracellular vesicles and their microRNAs in asthma. Clin Ther. 2014;36:873–881. doi: 10.1016/j.clinthera.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Tkaczyk C, Villa I, Peroneta R, David B, Chouaib S, Mécheri S. In vitro and in vivo immunostimulatory potential of bone marrow–derived mast cells on B-and T-lymphocyte activation. J allergy Clin Immunol. 2000;105:134–142. doi: 10.1016/S0091-6749(00)90188-X. [DOI] [PubMed] [Google Scholar]
- 40.Podbielska M, Szulc ZM, Kurowska E, Hogan EL, Bielawski J, Bielawska A, Bhat NR. Cytokine-induced release of ceramide-enriched exosomes as a mediator of cell death signaling in an oligodendroglioma cell line. J Lipid Res. 2016;57:2028–2039. doi: 10.1194/jlr.M070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann BD, Paine MS, Brooks AM, McCubrey JA, Renegar RH, Wang R, Terrian DM. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008;68:7864–7871. doi: 10.1158/0008-5472.CAN-07-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan B-T, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, Takasugi M, Watanabe S, Kanemaki MT, Obuse C. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagheri HS, Mousavi M, Rezabakhsh A, Rezaie J, Rasta SH, Nourazarian A, Avci ÇB, Tajalli H, Talebi M, Oryan A, et al. Low-level laser irradiation at a high power intensity increased human endothelial cell exosome secretion via Wnt signaling. Lasers Med Sci. 2018;33:1131–1145. doi: 10.1007/s10103-018-2495-8. [DOI] [PubMed] [Google Scholar]
- 45.Edgar JR, Eden ER, Futter CE. Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic. 2014;15:197–211. doi: 10.1111/tra.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willms E, Johansson HJ, Mäger I, Lee Y, Blomberg KEM, Sadik M, Alaarg A, Smith CIE, Lehtiö J, El Andaloussi S, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G, Schiffelers RM, Gucek M, van Balkom BWM. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:18396. doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.