Summary
Bacterial type IV secretion systems (T4SSs) can mediate conjugation. The T4SS from Neisseria gonorrhoeae possesses the unique ability to mediate DNA secretion into the extracellular environment. The N. gonorrhoeae T4SS can be grouped with F-type conjugative T4SSs based on homology. We tested 17 proteins important for DNA secretion by N. gonorrhoeae for protein interactions. The BACTH-TM bacterial two-hybrid system was successfully used to study periplasmic interactions. By determining if the same interactions were observed for F-plasmid T4SS proteins and when one interaction partner was replaced by the corresponding protein from the other T4SS we aimed to identify features associated with the unique function of the N. gonorrhoeae T4SS as well as generic features of F-type T4SSs. For both systems, we observed already described interactions shared by homologs from other T4SSs as well as new and described interactions between F-type T4SS specific proteins. Furthermore, we demonstrate, for the first-time, interactions between proteins with homology to the conserved T4SS outer membrane core proteins and F-type specific proteins and we confirmed two of them by co-purification. The F-type specific protein TraHN was found to localize to the outer membrane and the presence of significant amounts of TraHN in the outer membrane requires TraGN.
Keywords: N. gonorrhoeae, type IV secretion systems, E. coli, F-plasmid, two-hybrid, protein- proteins interactions
Introduction
Type IV secretion systems (T4SSs) are used by bacteria for different functions, the two most common being conjugation and effector protein translocation. These processes play important roles in, respectively, the spread of antibiotic resistance and infection by bacterial pathogens. During conjugation single-stranded DNA is translocated into recipient cells by a contact-dependent process. Neisseria gonorrhoeae is the only bacterium known to use a T4SS to secrete single-stranded DNA into the extracellular milieu (Dillard and Seifert, 2001; Hamilton et al., 2005; Salgado-Pabón et al., 2007).
The most studied T4SS is the P-type system from Agrobacterium tumefaciens, consisting of 12 core proteins named VirB1-VirB11 and VirD4 (Alvarez-Martinez and Christie, 2009). The F-plasmid encodes protein homologs to most of the Vir core proteins (VirB2-VirB10 and VirD4) found in P-type T4SSs, but the F-plasmid additionally encodes several proteins that are conserved only in F-type T4SSs (Lawley et al., 2003). F-type T4SSs have been found on many conjugative plasmids and in genetic islands on the bacterial chromosome (Lawley et al., 2003). The genes encoding the N. gonorrhoeae T4SS proteins are located on a 59 kb genetic island (the Gonococcal Genetic Island, GGI) (Callaghan et al., 2017). 21 genes organized in 4 operons are important for secretion of ssDNA by the N. gonorrhoeae T4SS (Pachulec et al., 2014). The structural T4SS proteins encoded by 17 genes can be divided into three groups: 1. Proteins showing homology to proteins found in most type IV secretion systems, 2. proteins showing homology to proteins conserved only in F-type T4SSs, and 3. proteins specific to the N. gonorrhoeae T4SS or only found in GGI-like T4SSs (Hamilton et al., 2005; Pachulec et al., 2014) (For an overview see Table 1 and Fig.1).
Table 1.
Predicted localization of 17 T4SS proteins essential for DNA secretion by N. gonorrhoeae and the corresponding homologous proteins from the F-plasmid T4SS and P-type T4SSs.
| N. gonorrhoeae T4SS | F-plasmid T4SS | P-type T4SSs |
|---|---|---|
| TraC (CP) | TraC (CP) | VirB4 (CP/IM/PP) |
| TraE (CP/IM/PP) | TraE (CP/IM/PP) | VirB8 (CP/IM/PP) |
| TraG (CP/IM/PP) | TraG (CP/IM/PP) | VirB6 (CP/IM/PP) |
| TraL (CP/IM) | TraL (CP/IM) | VirB3 (CP/IM) |
| TraF (PP) | TraF(PP) | |
| TraH (PP) | TraH (PP) | |
| TraU (PP) | TraU (PP) | |
| TraW (PP) | TraW (PP) | |
| TrbC (PP) | TrbC (PP) | |
| DsbC (PP) | ||
| AtlA (PP/PG) | ||
| LtgX (PP/PG) | Orf169 (PP/PG) | VirB1 (PP/PG) |
| Yag (PP/PG) | ||
| TraB (CP/IM/PP/OM) | TraB (CP/IM/PP/OM) | VirB10 (CP/IM/PP/OM) |
| TraK (PP/OM) | TraK (PP/OM) | VirB9 (PP/OM) |
| TraV (PP/OM) | TraV (PP/OM) | VirB7 (PP/OM) |
| TraN (PP/OM) | TraN (PP/OM) | |
| VirB11 (CP) | ||
CP cytoplasm, IM inner membrane, PP periplasm, PG associated with peptidoglycan, OM outer membrane.
Fig. 1.
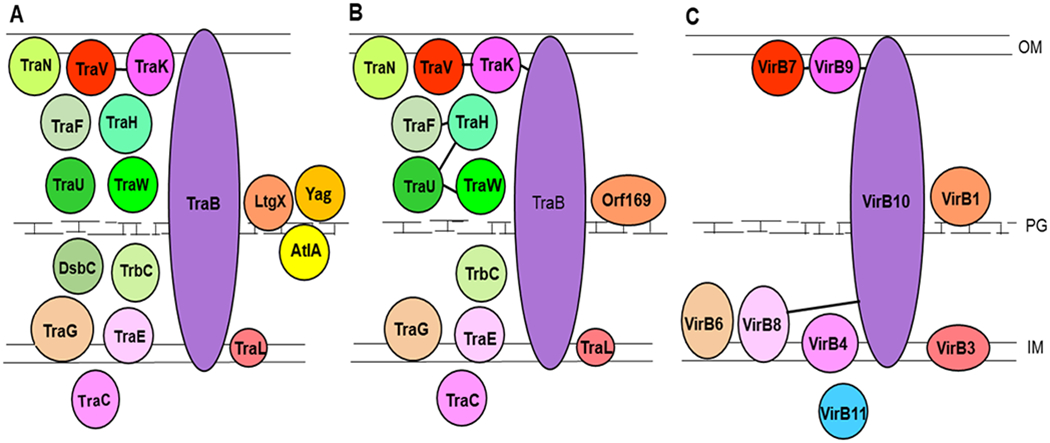
Schematic drawing of T4SSs showing known and/or predicted localization of A. 17 T4SS proteins important for DNA secretion by N. gonorrhoeae, B. homologous proteins from the F-plasmid T4SSs C. homologous proteins from the P-type T4SSs. Previously identified interactions are shown by lines: A. The TraV/TraK interaction described in Ramsey et al. 2014 B. Interactions described in Harris et al. 2001 (Harris et al., 2001) and Harris and Silverman 2004 (Harris and Silverman, 2004). C. Interactions from Das et al. 1997 and Das and Xie 2000. OM, outer membrane, IM, inner membrane, PG, peptidoglycan. Proteins shared between the three systems are shown in red/violet colors while F-type specific proteins are shown in green. Proteins specific to N. gonorrhoeae are shown in yellow while VirB11 found only in the P-type system is shown in blue.
The energy providing ATPases TraC (a VirB4 homolog) is found in F-type T4SSs while F-type T4SSs are missing a homolog to the ATPase VirB11 found in P-type T4SSs (Alvarez-Martines and Christie, 2009). N. gonorrhoeae encodes a TraC homolog required for DNA secretion (Hamilton et al., 2005; Pachulec et al., 2014).
Structural studies of the outer membrane core complex (OMCC) of two conjugative P-type T4SSs have been published (Chandran et al., 2009; Fronzes et al., 200; Low et al., 2014). The OMCC consists of three proteins that can form a double membrane-spanning complex. The hub protein VirB10 inserts into both the outer and the inner membrane, spans the periplasm and has a short N-terminal end in the cytoplasm (Chandran Darbari and Waksman, 2015). The two other proteins in the OMCC, VirB7, and VirB9 are associated with the outer membrane (Chandran et al., 2009; Low et al., 2014). Structural (Hu et al. 2019) and two-hybrid (Harris et al. 2001) data suggest that the F-plasmid T4SS has a similar OMCC consisting of the VirB10 homolog TraBF, the VirB7 homolog TraVF, and the VirB9 homolog TraKF. TraLF, TraEF, and TraGF from the F-plasmid are proteins associated with the inner membrane with some homology to proteins from the P-type T4SSs (Lawley et al., 2003) (Table 1 and Figure 1B). The N. gonorrhoeae GGI encodes homologs of TraBF, TraVF, TraKF, as well as TraGF, TraLF, and TraEF homologs (Table 1, Fig. 1A). Mutational analyses have shown that all of these proteins are important for DNA secretion by the N. gonorrhoeae T4SS (Hamilton et al., 2001; Hamilton et al. 2005; Pachulec et al., 2014).
In addition to the proteins found in most other type IV secretion systems, F-type T4SSs have a group of periplasmic or peripheral membrane proteins (TraWF, TraUF, TraHF, TraFF, TrbCF, and TraNF) that has been linked to assembly and extension of the conjugation pilus (Arutynov and Frost 2013). Although N. gonorrhoeae does not have pilus-dependent DNA secretion, the TraWN, TraUN, TraHN, TraFN, TrbCN, and TraNN homologs encoded by the N. gonorrhoeae GGI are essential for DNA secretion (Hamilton et al., 2001; Hamilton et al., 2005; Pachulec et al., 2014).
F-type T4SSs generally encode periplasmic thiol-oxidoreductases that promote disulfide bond formation in the periplasm (Hemmis and Schildbach, 2013; Pachulec et al., 2014). Some plasmids with F-type T4SSs such as the F-plasmid encode the protein TrbB which has a redox-active site and a TraF protein without a redox-active site, while others such as the N. gonorrhoeae GGI and the plasmid R27 encode a periplasmic DsbC (disulfide bond) homolog often in combination with a TraF-like protein, both proteins having redox-active sites (Elton et al., 2005; Hemmis and Schildbach, 2013). DsbCN and TraFN are both essential for DNA secretion by the N. gonorrhoeae T4SS (Hamilton et al., 2005).
Lytic transglycosylases capable of peptidoglycan degradation are believed to play a role in the assembly of transport complexes in the cell envelope (Koraimann, 2003). Three of the proteins that are important for DNA secretion by N. gonorrhoeae AtlAN, YagN, and LtgXN are thought to be associated with the peptidoglycan layer (Kohler et al., 2007; Pachulec et al., 2014; Dillard and Seifert, 2001). AtlAN and LtgXN are both lytic transglycosylases (Kohler et al., 2007). LtgXN shows homology to Orf169, a lytic transglycosylase from the F-plasmid T4SS, while AtlAN is specific to the N. gonorrhoeae T4SS (Kohler et al., 2007).
The T4SS from N. gonorrhoeae shows some amino acids sequence similarity to the T4SS from the F-plasmid but the sequence identity is generally low, typically around 25% (see Table 2) (Hamilton et al., 2005; Ramsey et al., 2014), and while other F-type T4SSs are involved in contact-dependent DNA secretion, the T4SS from N. gonorrhoeae carries out contact-independent DNA secretion. Comparing the two systems could potentially be used to define generic features of F-type T4SSs as well as giving information about the specific features associated with the unique function of the N. gonorrhoeae T4SS.
Table 2.
Bioinformatics and literature data used to predict the localization of the N. gonorrhoeae T4SS proteins.
| Signal peptide |
Topology prediction TOPCONS |
Amino acid identity‡ | Experimental evidence for localization (N. gonnorhoeae protein) | Experimental evidence for localization (F-plasmid protein) | |||
|---|---|---|---|---|---|---|---|
| SignalP | TOPCONS | ||||||
| Core complex proteins |
TraBN | No | No | 1 TM N-In, C-out |
24% | (Ramsey et al., 2014) | (Hu et al. 2019) |
| TraKN | Yes | Yes | No TM | 21% | (Ramsey et al., 2014) | (Hu et al. 2019) | |
| TraVN | Yes | Yes | No TM | 18% | (Ramsey et al., 2014). | (Hu et al. 2019) | |
| Other proteins with homology to both F and Vir proteins | TraLN | No | No | 2 TM N-In, C-In † |
(Lawley et al. 2003) | ||
| TraEN | No | No | 1 TM N-In, C-out |
21% | (Arutyunov et al., 2010) | ||
| TraGN | No | No | 8 TM N-Out, C-Out |
(Kohler et al., 2013). | (Firth and Skurray 1992) | ||
| TraCN | No | No | No TM | 24% | (Hu et al. 2019) | ||
| Proteins with homology to proteins
from F-like T4SSs (Hamilton et al., 2005) |
TraWN | Yes | Yes | No TM | 26% | (Ramsey et al., 2012). | (Arutyunov et al., 2010) |
| TraUN | Yes | Yes | No TM | 34% | (Arutyunov et al., 2010) | ||
| TrbCN | No | Yes | No TM | 25% | (Lawley et al. 2003) | ||
| TraNN | Yes | Yes | 1 TM C-In |
(Klimke et al. 2005) | |||
| TraFN | Yes | Yes | No TM | 26% | (Arutyunov et al., 2010) | ||
| TraHN | Yes | Yes | No TM | 23% | (Arutyunov et al., 2010) | ||
| LtgXN | No | Yes | No TM | (Kohler et al., 2007) | Orf169 has a signal sequence | ||
| DsbCN | Yes | Yes | No TM | ||||
| Proteins specific for GGI like T4SS or for the N. gonorrhoeae T4SS (Pachulec et al., 2014) | AtlAN | No | No | No TM | (Kohler et al., 2007) | ||
| YagN | No | No | No TM | Proposed peptidoglycan association (Pachulec et al., 2014) | |||
TM, Transmembrane helix, N/C-In, N/C terminal end of the protein in the cytoplasm, N/C-Out N/C terminal end in the periplasm.
For TraLN the 6 different predictions shown by the TOPCONS webserver differed concerning the localization of the N-terminal end while all predictions indicated a cytoplasmic localization of the C-terminus.
Amino acid identity between the protein from N. gonorrhoeae, and the corresponding protein from the F-plasmid. The amino acid identity is only calculated for the F-plasmid proteins used in this study.
We have compiled sequence-based and published localization information for the N. gonorrhoeae T4SS, and experimentally determined outer membrane localization of TraHN. Only a limited number of studies explore the Tra protein interaction network we have therefore systematically tested 17 proteins important for DNA secretion by N. gonorrhoeae for protein-protein interactions using bacterial two-hybrid systems. To determine if the identified interactions are likely to be specific to the N. gonorrhoeae T4SS or general for F-type T4SSs, we tested the corresponding proteins from the F-plasmid for interactions. Interactions of particular interest were confirmed by co-purification. We have shown cross-system interchangeability of homologous T4SS proteins from the two systems in several cases using both two-hybrid and co-purification approaches, and present interaction models for both systems.
Results
TraHN localizes to the outer membrane dependent on TraGN
To use bacterial two-hybrid systems correctly it is important to know the cellular localization of the proteins.
TraHF has been implicated in F-pilus extension (Arutynov and Frost 2013), however, without a pilus, the Neisseria TraHN likely plays a distinct role. Since the localization of TraHN was unknown, we epitope-tagged TraHN with a triple FLAG tag at the C-terminus and examined its subcellular localization in N. gonorrhoeae. When traHN-FLAG3 was expressed from the native site, no TraHN-FLAG3 was detected by western blot. Therefore, traHN-FLAG3 was expressed using an inducible promoter from a distant site on the gonococcal chromosome. TraHN-FLAG3 was detectable in that strain upon induction. Cell fractions containing outer membrane, total membrane, or soluble protein were examined, and TraHN-FLAG3 was found in the outer membrane and total membrane fractions only (Fig. 2A). This localization pattern matched that of known outer membrane protein LtgA and was distinct from that of known inner membrane protein SecY and known soluble protein CAT (Fig. 2A).
Fig. 2. Western blots showing TraHN -FLAG3 presence and localization in various N. gonorrhoeae strains.
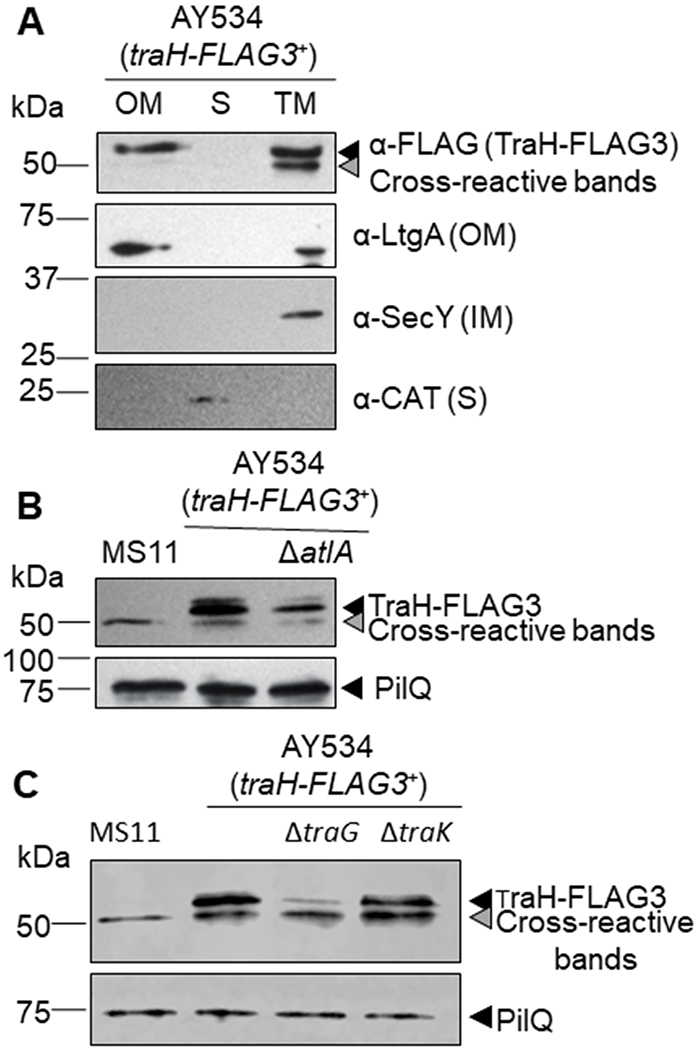
traHN-FLAG3+ strains have the tagged TraH protein expressed from the trpB-iga complementation site under the control of the anhydrotetracycline promoter (PATC) A. OM, outer membrane, S, soluble (periplasmic and cytoplasmic), TM, total membrane (outer and inner membranes). TraHN-FLAG3 was detected using anti-FLAG. Antibodies against LtgA (OM), SecY (IM), and chloramphenicol acetyl-transferase (CAT) (S) were used as fractionation controls. B. traHN-FLAG3 induced in atlAN deletion strain. MS11 is a wild type N. gonorrhoeae strain. C. traHN-FLAG3 induced in traGN and traKN deletion strains.
The traHN,traGN, and atlAN genes are in an operon separate from the one encoding most of the structural proteins of the gonococcal T4SS (Pachulec et al., 2014). The transcript is found at significantly higher levels than that encoding the other T4SS structural proteins, and the translation of TraHN and TraGN, and possibly also AtlAN, is controlled by an RNA switch (Ramsey et al., 2015). The coregulation of these proteins suggested that they might work together for assembly of part of the T4SS. We hypothesized that perhaps TraHN requires the lytic transglycosylase AtlAN to make an opening in the cell wall for TraHN to pass through and that perhaps AtlAN might need TraGN in the inner membrane to access the periplasm.
We used an atlAN deletion mutant to test the necessity of atlA for TraH localization. Outer membrane preparations were examined for TraHN-FLAG3 by western blot. Contrary to our hypothesis, the deletion of atlAN did not significantly reduce TraHN-FLAG3 in the outer membrane (Fig. 2B).
Next, we tested the requirement for co-transcribed TraGN, as well as the structural protein TraKN, for TraH localization to the outer membrane. A traGN deletion strain showed significantly reduced TraHN-FLAG3 in the outer membrane, whereas the deletion of traKN had no effect (Fig. 2C). Thus TraGN, independent of AtlAN and TraKN, is needed for TraHN to be present at significant levels in the gonococcal outer membrane.
To obtain information about the localization of other N. gonorrhoeae T4SS proteins we performed bioinformatic analyses as described in experimental procedures and compiled data from the literature. The outcome is summarized in Table 2.
The BACTH and BACTH-TM systems
In the bacterial adenylate cyclase two-hybrid system (BACTH) (Karimova et al., 1998) the proteins of interest fused with the two fragments (T18 and T25) from the catalytic domain of Bordetella pertussis adenylate cyclase and interaction between the proteins result in functional complementation between T18 and T25 leading to cAMP synthesis and transcriptional activation of the lactose operon. The BACTH-TM system (Ouellette et al., 2014) inserts a transmembrane helix between the proteins of interest and the T18 and T25 fragments of the adenylate cyclase. While the BACTH system requires the proteins of interest to be located in the cytoplasm or the inner membrane (Karimova et al., 1998; Karimova et al., 2005) the BACTH-TM system enables the study of protein interactions in the periplasm (Ouellette et al., 2014). A combination of the BACTH and the BACTH-TM systems can be used to study interactions between inner membrane proteins with a cytoplasmic domain and a periplasmic protein.
Interactions: N. gonorrhoeae genes cloned into the BACTH and the BACTH-TM system vectors
All proteins were fused with both T18 and T25 fragments. The BACTH vectors pUT18C and pKT25 put the T18 and the T25 fragments in the N-terminal end of the protein while pUT18 (Karimova et al., 2001) and p25N (Claessen et al., 2008) put the T18 and the T25 fragment in the C-terminal end of the protein. The BACTH-TM vectors pUTM18C (Ouellette et al., 2014) and pKTM25 (this study) add a transmembrane domain, and the T18 or T25 fragment in the N-terminal end of the proteins.
According to respectively previous work (Ramsey et al., 2014) and our bioinformatics analysis (Table 2), transmembrane proteins TraBN and TraEN are likely to have their N-terminals in the cytoplasm, thus we cloned them in the BACTH vectors pUT18C and pKT25. For the transmembrane proteins TraGN and TraLN, respectively, experimental evidence (Kohler et al., 2013) and bioinformatics analyses indicate that the C-terminal ends of the proteins are likely to be in the cytoplasm while the N-terminal ends are likely to be in the periplasm (Table 2). The genes encoding TraGN and TraLN were therefore cloned in pUT18 and p25N.
Because the BACTH-TM system (Ouellette et al., 2014) had not previously been used for studying periplasmic proteins we cloned TraHN, TraUN, TraNN, TraKN, TraVN, and AtlAN in both the BACTH vectors pUT18C and pKT25 and the BACTH-TM system vectors pUTM18C and pKTM25 to be able to compare the results obtained with the two systems. LtgXN, YagN, DsbCN, TraWN, TraFN, and TrbCN were cloned only in the BACTH-TM system vectors (Table 3). For TraHN, TraUN, TraNN, TraKN, LtgXN, YagN, DsbCN, TraWN, TraFN, and TrbCN, the sequences encoding the signal peptide were detected with SignalP, or in the case of TrbCN and LtgXN with TOPCONS, and removed from the sequences before cloning. For TraVN the first 19 amino acids including the assumed lipobox (Ramsey et al. 2014) were removed before cloning. For AtlAN, and TraCN we chose to put the T18 and T25 fragments at both ends of the protein, thus these proteins were cloned in all four BACTH vectors pUT18C, pUT18, pKT25 or p25N (Table 3).
Table 3.
Interactions between proteins from the N. gonorrhoeae T4SS.
| Transmembrane/cytoplasmic proteins | Periplasmic/outer membrane proteins cloned in both the BACHT and the BACHT-TM system | Periplasmic proteins cloned only in the BACHT-TM system | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T18TraCN | TraCNT18 | T18TraBN TT18 |
T18TraEN | TraGNT18 | TraLNT18 | T18AtlAN | TM18AtlAN | AtlANTT18 | T18TraHN | TM18TraHN | T18TraKN | TM18TraKN | T18TraNN | TM18TraNN | T18TraUN | TM18TraUN | T18TraVN | TM18TraVN | TM18DsbCN | TM18LtgXN | TM18TraFN | TM18TraWN | TM18TrbCN | TM18YagN | ||
| Transmembrane/cytoplasmic proteins | T25TraCN | − | + | − | − | − | − | |||||||||||||||||||
| TraCNT25 | + | − | − | − | − | − | ||||||||||||||||||||
| T25TraBN | − | − | + | + | − | − | − | − | − | − | + | − | + | − | − | − | − | − | + | + | − | − | + | w | − | |
| T25TraEN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| TraGN T25 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| TraLNT25 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Periplasmic/outer membrane proteins cloned in both the BACHT and the BACHT-TM system | T25AtlAN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||
| TM25AtlAN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| AtlANT25 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| T25TraHN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| TM25TraHN | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | |||
| T25TraKN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | |||||||||
| TM25TraKN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| T25TraNN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| TM25TraNN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| T25TraUN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| TM25TraUN | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |||
| T25TraVN | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | |||||||||
| TM25TraVN | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |||
| Periplasmic proteins cloned only in the BACHT-TM system | TM25DsbCN | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |||||||||
| TM25LtgXN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||||
| TM25TraFN | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | ||||||||||
| TM25TraWN | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | ||||||||||
| TM25TrbCN | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | ||||||||||
| TM25YagN | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||||
+, −, and w indicate respectively interaction, no interaction, and weak interaction. Space indicates that the interaction was not tested. The placement of T18 and T25 relative to the protein name indicates N or C-terminal fusion. T18 or T25 indicate that the gene encoding the protein was cloned into the BACTH vectors. TM18 or TM25 indicates that the gene encoding the protein was cloned into the BACTH-TM vectors.
Combinations of N. gonorrhoeae proteins tested
TraBN, TraGN, TraEN, and TraLN are believed to be transmembrane proteins with potential interaction partners in the cytoplasm, the inner membrane, the periplasm and in the case of TraBN also the outer membrane. Due to the possible interactions of these proteins with proteins in several cellular compartments, we tested all possible combinations of these proteins cloned in the BACTH system with all other proteins cloned in the BACTH or the BACTH-TM system by co-transformation into E. coli BTH10. Functional complementation was assayed as described in the method section. (for an overview of the tested combinations of plasmids see Table 3).
For the proteins cloned only in the BACTH-TM system (LtgXN, YagN, DsbCN, TraWN, TraFN, and TrbCN) we tested for interactions between proteins cloned in BACTH-TM vectors and for interactions between transmembrane proteins cloned in BACTH-vectors and periplasmic proteins cloned in BACTH-TM vectors (Table 3). A similar test was done for the six proteins cloned in both the BACTH and the BACTH-TM system i.e., TraKN, TraVN, TraUN, TraHN, TraNN, and AtlAN (Table 3).
As controls, we tested for interactions between the periplasmic proteins cloned in both the BACTH and the BACTH-TM systems (Table 3). Cloning the periplasmic proteins in the BACTH and the BACTH-TM vectors should result in protein expression in different cellular compartments, therefore no interactions should be observed between periplasmic proteins cloned in the two different vector systems. As expected, we did not observe any interactions between a periplasmic protein cloned in a BACTH vector and a periplasmic protein cloned in a BACTH-TM vector (Table 3).
For TraCN, we tested for both a TraCN / TraCN interaction and interactions with transmembrane proteins (Table 3).
The detected interactions
To examine if the interactions observed for the N. gonorrhoeae T4SS proteins were specific for the N. gonorrhoeae T4SS proteins or could potentially be general for F-type T4SS proteins, we analyzed interactions among the corresponding F-plasmid proteins using the protein-adenylate fusions summarized in Table 4.
Table 4.
Interactions between T4SS proteins encoded by the F-plasmid.
| Transmembrane/cytoplasmic proteins | Periplasmic proteins | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T18TraCF | TraCFT18 | T18TraBF | T18TraEF | TM18TraFF | TM18TraHF | T18TraKF | TM18TraKF | TM18TraUF | T18TraVF | TM18TraVF | TM18TraWF | TM18TrbCF | ||
| T25TraCF | − | + | − | − | − | − | − | − | − | − | − | − | − | |
| TraCFT25 | − | + | − | − | − | − | − | − | − | − | − | − | − | |
| T25TraBF | − | + | + | − | + | − | − | + | − | − | − | + | + | |
| T25TraEF | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| Periplasmic proteins | TM25TraFF | − | − | + | − | − | − | − | − | − | − | − | − | − |
| TM25TraHF | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| T25TraKF | − | − | − | − | − | − | − | − | − | + | − | − | − | |
| TM25TraKF | − | − | + | − | + | − | − | − | + | − | − | − | + | |
| TM25TraUF | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| T25TraVF | − | − | − | − | − | − | + | − | − | − | − | − | − | |
| TM25TraVF | − | − | − | − | − | − | − | − | − | − | w | − | + | |
| TM25TraWF | − | − | − | − | + | − | − | − | + | − | w | − | + | |
| TM25TrbCF | − | − | + | − | + | − | − | − | + | − | + | + | + | |
+, − and w indicate respectively interaction, no interactions, and weak interaction. The placement of T18 and T25 relative to the protein name indicates N or C-terminal fusion. T18 or T25 indicate that the gene encoding the protein was cloned into the BACTH vectors. TM18 or TM25 indicates that the gene encoding the protein was cloned into the BACTH-TM vectors.
The observed interactions could be classified into three groups. Group 1 consists of interactions observed both with the N. gonorrhoeae T4SS proteins and with the corresponding proteins from the F-plasmid (Fig. 3A). The OMCC protein homologs TraB and TraV participate in the majority (8/10) of these interactions The following interaction partners were observed: TraB (TraB, TraW, TraK, TraE, and TrbC), TraV (TraK, TraV, and TraW). The two additional shared interactions were TraC/TraC and TraW/TrbC. The TM-TrbCN/TraBN, TM-TraVF/TM-TraVF, and TM-TraVF/TM-TraWF are seen as weak interactions in Fig. 3A. β-galactosidase measurements showed that the signal was approximately 4, 5, and 8 times above the background level for the 3 interactions, respectively (data not shown).
Fig. 3.
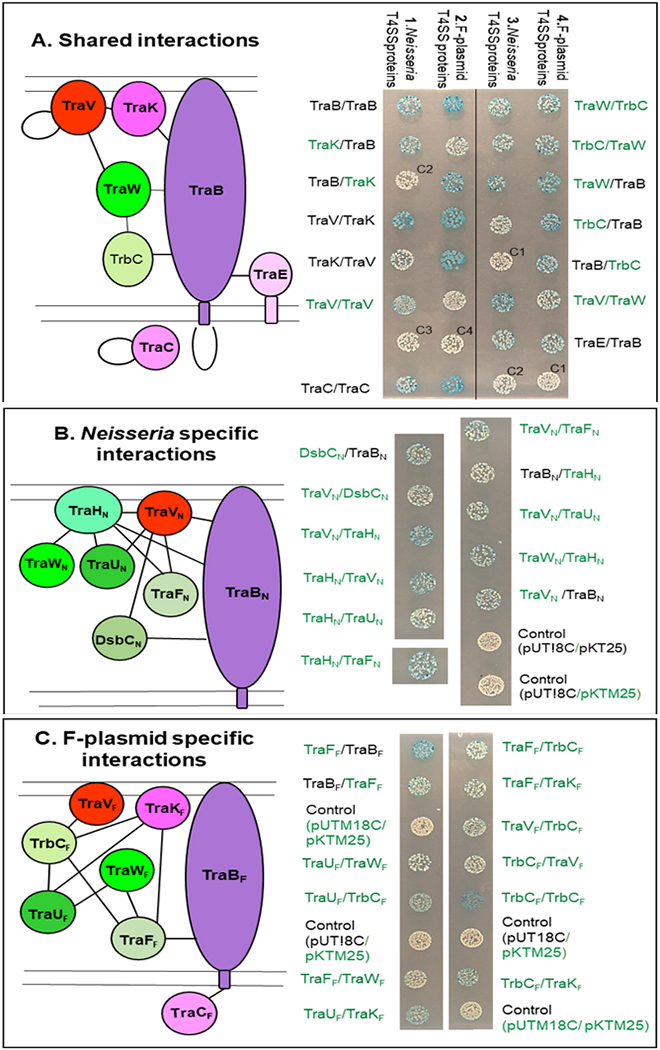
Bacterial 2-hybrid interactions. Left side: schematic drawings showing the observed interactions and the supposed cellular localization of the proteins. Right side: Colonies of E. coli BTH101 transformants carrying plasmids encoding the proteins indicated in the order T18/T25. Protein names in green indicate that the gene encoding the protein was cloned in a BACTH-TM system vector, black names indicate that the gene was cloned in the BACTH system vector. C1, C2, C3, and C4 are vector controls; respectively pUT18C/pKT25, pUTM18C/pKTM25, pUTM18C/pKT25 and pUT18C/pKTM25. A. Interactions observed both between proteins from the N. gonorrhoeae T4SS and the between the corresponding proteins from the F-plasmid. B. Interactions observed only between proteins from the N. gonorrhoeae T4SS. C. Interactions observed only between proteins from the F-plasmid.
Group 2 comprises 10 interactions observed only with periplasmic or membrane-spanning proteins from N. gonorrhoeae, not with the F-plasmid homologs (Fig. 3B). TraVN or TraHN are involved in 9 out of the 10 interactions. The following interaction partners were observed for TraVN and TraHN respectively: TraVN (TraHN, TraFN, TraBN, TraUN, and DsbCN), TraHN (TraBN, TraUN, TraFN, and TraWN). The last Neisseria specific interaction observed was between DsbCN and TraBN. Interactions involving DsbCN were only tested with proteins from N. gonorrhoeae since no relevant homolog is encoded by the F-plasmid.
Group 3 comprises 11 interactions observed only with proteins from the F-plasmid (Fig. 3C). The main proteins involved in the F-plasmid specific are TrbCF, TraFF., TraUF,and TraKF, TraFN has a redox-active site that is missing in TraFF indicating different roles for the two proteins. The interaction observed for TrbCF, TraFF and TraUF are respectively: TrbCF (TraVF, TraFF, TraUF, TraKF and TrbCF), TraFF (TraBF, TraWF and TraKF) and TraUF (TraWF and TraKF ). The last F-plasmid specific interaction is between TraCF and TraBF
Some of the studied proteins were able to interact with a relatively larger number of other proteins, i.e., TraBN and TraBF are involved in 8 and 7 interactions respectively, TraVN is involved in 8 interactions, TrbCF is involved in 7 interactions, and TraKF is involved in 5 interactions (Fig. 3). Some proteins with intrinsic tendency to interact with any protein, so-called ‘‘sticky proteins’’, can give rise to false positives in two-hybrid screens (Battesti and Bouveret, 2012); however, all the proteins tested in this study showed selectivity with regards to interaction partners. Whether these interactions would be formed if several, possibly competing interaction partners were present at the same time is, however, a question that cannot be addressed by two-hybrid studies.
In some cases, for instance, for the TraBN/ TraEN interaction, an interaction was only observed with one of the two possible combinations of T18 and T25. This has been observed in previous two-hybrid analyses, one possible reason being the different copy numbers of the T18 and T25 plasmids (Battesti and Bouveret, 2012).
No interactions were detected for AtlAN, LtgXN, YagN, TraGN, TraNN, TraLN, and TraHF.
Confirmation of selected interactions by co-purification
Since two-hybrid systems can give both false-negative and false-positive results, we aimed to further validate our findings with other methods. For many of the interactions observed in this study, some evidence for the interaction between the studied proteins or homologs from other T4SSs can be found in the literature (Casu et al., 2016; Chandran et al., 2009; Das et al., 1997; Das and Xie, 2000; Ding et al., 2002; Fronzes et al., 2009; Gilmour et al., 2001; Harris et al., 2001; Harris and Silverman, 2004; Hu et al., 2019; Low et al., 2014; Oliveira et al., 2016; Ramsey et al., 2014; Shala-Lawrence et al., 2018; Sivanesan et al., 2010). However, only limited evidence exists for interactions between OMCC proteins homologs (TraB, TraV, and TraK) and F-type specific periplasmic proteins (Arutyunov et al., 2010), and the function of the F-type specific periplasmic proteins is poorly understood. Therefore, we chose to concentrate on the confirmation of interactions between the OMCC protein homolog TraV and the periplasmic proteins TraW and TrbC. For TraVF and TraVN expression, the N-terminal lipobox was omitted. Instead, the proteins were equipped with an N-terminal pelB sequence for periplasmic expression and a C-terminal His-tag. For TraWN, the whole reading frame was expressed without a His-tag. TraWN was retained on Ni-NTA beads in the presence but not in the absence of TraVN-His, confirming that TraWN interacts with TraVN (Fig 4 and for further details Fig. S4). We were, however, unable to pull-down TraWF with TraVF-His. TrbCF was expressed without a His-tag to confirm the F-plasmid specific interaction between TrbCF and TraVF by co-purification. Only a construct without the signal sequence gave a high level of expression and only this protein was used. for pull-down experiments with His-tagged TraVF. TrbCF was found to co-purify with TraVF (Fig. 4 and Fig. S4). Two cysteine residues are found towards the C-terminal end of TrbCF, giving potentially different folding and protein interactions for TrbCF expressed in the cytoplasm compared to TrbCF expressed in the periplasm. E. coli Origami-2(DE3) is a protein expression strain with mutations in both the thioredoxin reductase (trxB) and glutathione reductase (gor) genes. These alterations enhance disulfide bond formation in the cytoplasm. Similar results were obtained for TrbCF expressed in the E. coli BL21(DE3) and E. coli Origami-2(DE3) indicating that disulfide bond formation is not important for the pull-down result observed.
Fig. 4.
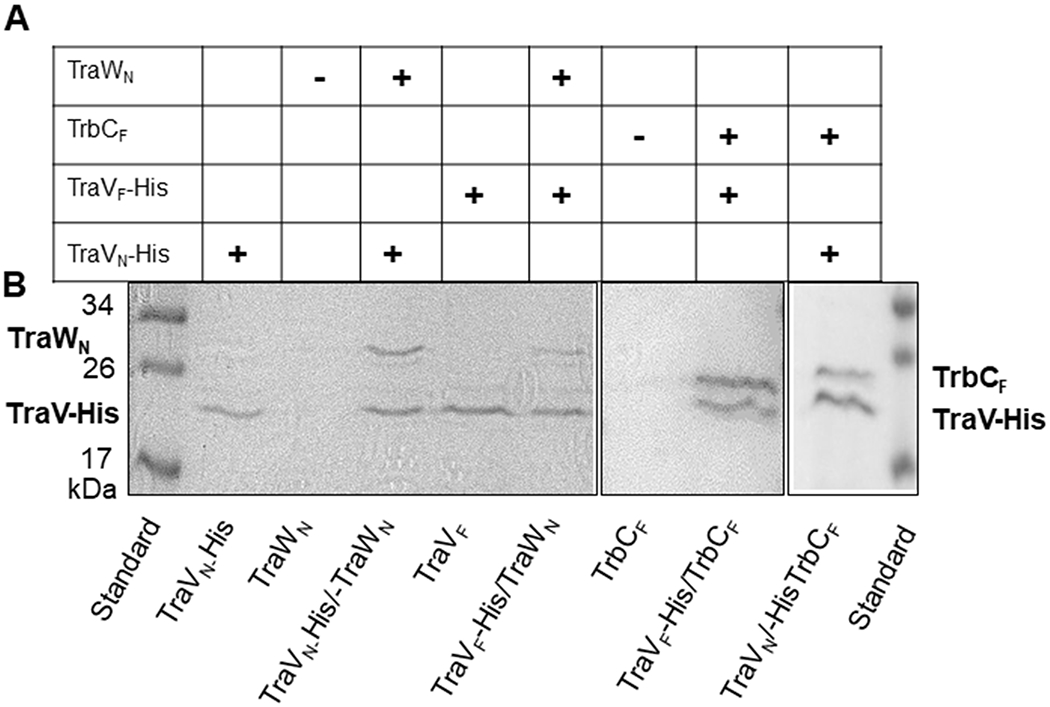
Co-purification of TraWN and TrbCF with TraVN-His and TraVF-His on Ni-NTA beads. A. The predicted outcome of the co-purifications if the proteins interact. B. The samples are separated by SDS-PAGE and visualized with coomassie blue. Samples eluted from Ni-NTA beads with 250 mM imidazole are shown. For the TrbCF the results shown are for TrbCF expressed in E. coli Origami2(DE3), all other proteins were expressed in E. coli BL21(DE3) as described under experimental procedures. The calculated molecular weights are 19.5, 17.2, 28.2, and 21.4 kDa for TraVN, TraVF, TraWN, and TrbCF respectively.
Interactions between proteins from the F-plasmid T4SS and proteins from the N. gonorrhoeae T4SS
For interactions observed with proteins from the F-plasmid T4SS or with proteins from the N. gonorrhoeae T4SS, we examined if one of the proteins could be replaced by the corresponding F or N. gonorrhoeae T4SS proteins giving mixed F/N. gonorrhoeae T4SS interactions (Table 5). For all the interactions shared between the N. gonorrhoeae T4SS and F-plasmid T4SS proteins (Fig. 3A), either one or both proteins could be replaced by the corresponding protein from the other system (Table 5 and Fig. S3). While the TraVN/TraKN and TraVF/TraKF interactions were only observed when the genes encoding the two proteins were cloned in the BACTH system, the two mixed interactions TM-TraVF/TM-TraKN and TM-TraVN/TM-TraKF were observed only for the BACTH-TM clones (Table 5).
Table 5.
Mixed interactions N. gonorrhoeae T4SS / F-plasmid T4SS (periplasmic and membrane-spanning proteins).
| Proteins from the F-plasmid T4SS | Proteins from the N. gonorrhoeae T4SS | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T18TraBF | T18TraCF | TraCFT18 | T18TraEF | TM18TraFF | TM18TraHF | T18TraKF | TM18TraKF | TM18TraUF | T18TraVF | TM18TraVF | TM18TraWF | TM18TrbCF | TM18DsbCN | T18TraBN | T18TraCN | TraCNT18 | T18TraEN | TM18TraFN | TM18TraHN | T18TraKN | TM18TraKN | TM18TraUN | T18TraVN | TM18TraVN | TM18TraWN | TM18TrbCN | ||
| Proteins from the F-plasmid T4SS | T25TraBF | + | + | − | − | + | + | + | + | + | + | + | ||||||||||||||||
| T25TraCF | − | + | + | |||||||||||||||||||||||||
| TraCFT25 | − | − | − | |||||||||||||||||||||||||
| T25TraEF | − | |||||||||||||||||||||||||||
| TM25TraFF | − | − | − | + | − | |||||||||||||||||||||||
| TM25TraHF | − | − | − | − | − | − | ||||||||||||||||||||||
| T25TraKF | − | |||||||||||||||||||||||||||
| TM25TraKF | + | + | + | + | + | |||||||||||||||||||||||
| TM25TraUF | − | − | − | − | − | |||||||||||||||||||||||
| T25TraVF | − | |||||||||||||||||||||||||||
| TM25TraVF | − | − | − | − | − | − | − | − | − | |||||||||||||||||||
| TM25TraWF | − | − | − | + | − | |||||||||||||||||||||||
| TM25TrbCF | − | + | + | + | + | + | + | |||||||||||||||||||||
| Proteins from the N. gonorrhoeae T4SS | TM25DsbCN | − | + | |||||||||||||||||||||||||
| T25TraBN | + | + | + | − | + | − | + | + | + | + | ||||||||||||||||||
| T25TraCN | − | − | ||||||||||||||||||||||||||
| TraCNT25 | − | − | ||||||||||||||||||||||||||
| T25TraEN | − | |||||||||||||||||||||||||||
| TM25TraFN | + | − | − | − | − | |||||||||||||||||||||||
| TM25TraHN | + | − | + | + | − | |||||||||||||||||||||||
| T25TraKN | − | |||||||||||||||||||||||||||
| TM25TraKN | − | − | − | + | + | |||||||||||||||||||||||
| TM25TraUN | − | − | − | − | + | |||||||||||||||||||||||
| T25TraVN | − | |||||||||||||||||||||||||||
| TM25TraVN | + | − | − | − | − | + | − | + | ||||||||||||||||||||
| TM25TraWN | + | − | + | + | + | |||||||||||||||||||||||
| TM25TrbCN | − | − | − | − | − | − | + | |||||||||||||||||||||
+ and − indicate respectively interaction and no interactions. Space indicates that the interaction was not tested. The placement of T18 and T25 relative to the protein name indicates N or C-terminal fusion. T18 or T25 indicate that the gene encoding the protein was cloned into the BACTH vectors. TM18 or TM25 indicates that the gene encoding the protein was cloned into the BACTH-TM vectors.
Also, for groups 2 and 3 containing, respectively, N. gonorrhoeae specific interactions and F specific interactions, several mixed interactions were observed (Table 5, Fig. S1, and S2). TraBN, TraBF, TraVN, TrbCF, and TraKF, which were able to form several non-mixed interactions, were also proficient in forming mixed interactions. Co-purification confirmed mixed interactions between TraVF and TraWN and between TraVN and TrbCF (Fig 4).
Discussion
Evaluation of the BACTH-TM system
Since the BACTH two-hybrid system (Karimova et al., 1998), requires the interaction between the T18 and the T25 fragments of the adenylate cyclase to occur in the cytoplasm it is not optimal for studying periplasmic interactions. Ouellette et al. (Ouellette et al., 2014) developed a modified BACTH system (the BACTH-TM system) where the E. coli OppB transmembrane domain is inserted between the protein of interest and the T18 or T25 fragment, exposing the protein of interest to the periplasmic environment but did not use the system to study interactions between periplasmic proteins. To evaluate the system, we cloned 13 proteins from the N. gonorrhoeae T4SS and 7 proteins from the F-plasmid T4SS system in the BACTH-TM system. For comparison, 6 proteins from the N. gonorrhoeae T4SS and 2 proteins from the F-plasmid T4SS were cloned into both the BACTH and the BACTH-TM systems. For TraHN and TraUN we only observed interactions when the proteins were cloned in the BACTH-TM system (Table 3). Since TraHN and TraUN are proteins rich in cysteine residues (9 and 16 respectively), it seems likely that these proteins are unable to fold correctly in the cytoplasm. For periplasmic proteins cloned only in the BACTH-TM system, we observed interactions between TraW and TrbC from both the N. gonorrhoeae T4SS and the F-plasmid T4SS. In several F-type T4SSs, TrbC proteins are fused to the N-terminus of TraW, and TrbCF and TraWF were found to co-purify, indicating that the TraW/TrbC interaction is important for F-type T4SSs (Shala-Lawrence et al., 2018). These results indicate that BACTH-TM system can be used successfully to study periplasmic protein interactions.
For the peripheral membrane proteins, TraVN and TraKN, an interaction in the cytoplasm after removal of signal sequences have previously been demonstrated using a bacterial two-hybrid system (Ramsey et al., 2014). Since the only cysteine in TraKN and TraKF is in the predicted signal peptide, the TraV/TraK interaction is unlikely to involve a disulfide bridge (Ramsey et al., 2014). TraVN, TraKN, TraVF, and TraKF were cloned in both the BACTH and the BACTH-TM systems. The TraVN/TraKN and the TraVF/TraKF interactions were only observed for the proteins cloned in the BACTH system (Table 3). It is possible that linking the proteins to the inner membrane interferes with the TraV/TraK interaction. We did, however, observe other interactions for TM-TraVN, TM-TraKN, TM-TraVF, and TM-TraKF. (Table 3, Table 4, Fig. 3).
Interactions between F-type specific T4SS periplasmic proteins
Two interaction groups for F-proteins have been defined by yeast two-hybrid screens (Harris and Silverman, 2004; Harris et al., 2001) - one consisting of the three proteins with homology to the outer membrane core proteins TraVF, TraKF and TraBF (Harris et al., 2001) and one consisting of F-specific proteins (TrbBF, TrbIF, TraWF, TraUF, TraHF, and TraFF) (Harris and Silverman, 2004). No interactions were observed between the two interaction groups. Our study using bacterial two-hybrid systems confirmed several of these previously observed interactions (Fig. 3, Table 3). We have not included TrbBF and TrbIF in this study since the N. gonorrhoeae T4SS does not possess a TrbB homolog and the deletion of trbIN has been found not to affect DNA secretion by N. gonorrhoeae (Pachulec et al., 2014). For TraWF, TraHF, TraUF, and TraFF the study carried out by Harris and Silvermann (Harris and Silverman, 2004) demonstrated TraHF/TraFF, TraHF/TraUF, and TraWF/TraUF interactions. For TraHF, we were unable to demonstrate any interaction (Table 3). It is possible that anchoring TraHF to the inner membrane inhibits the formation of a correctly folded TraHF protein. For TraHN we did observe TraHN/TraFN and TraHN/TraUN interactions similar to the interactions observed by Harris and Silverman (Harris and Silverman, 2004) (Fig. 3B). The TraWF/TraUF interaction observed by Harris and Silvermann (Harris and Silverman, 2004) was identified in this study as an F-specific interaction (Fig. 3C). Besides, we observed the TrbCF/TraWF and TrbCF/TrbCF interactions demonstrated by Shala-Lawrence et al. 2018 (Fig. 3A and C) as well as some new interactions between the F-type specific proteins (Fig. 3B and C).
We demonstrate that TraHN is an outer membrane-associated protein in N. gonorrhoeae (Fig. 2). Its F-plasmid homolog TraHF also associates with the outer membrane in the presence of other T4SS proteins (Arutyunov et al.,2010). TrbIF is required for correct TraHF localization (Arutyunov et al., 2010). Gonococci do not require TrbIN for DNA secretion (Pachulec et al., 2014). The transcriptomic study of Remmele, as well as the qRT-PCR results of Ramsey, indicated that the traHN-traGN-atlAN transcript was found at much higher levels than the long transcript containing most other T4SS genes (Remmele CW et al. 2014; Ramsey et al. 2015). Thus, we sought to determine if TraHN might work together with AtlAN or TraGN. TraGN was found to affect the localization of TraHN possibly by stabilizing TraHN or by facilitating the transport of TraHN to the gonococcal outer membrane
A possible biological implication of the interaction between TraV/TraK/TraB and F-type specific proteins
In this study, we observed several interactions between the proposed OMCC protein TraB/TraV/TraK and F-type specific proteins (Fig. 3, Fig. 4, Table 3, 4, and 5). The F-type specific proteins have been assigned a function in pilus assembly/retraction and mating pair stabilization based on mutant studies (Arutyunov and Frost, 2013). Until recently the physical and functional relationship of the T4SS apparatus and the F-pilus has been undefined. However, a recent CryoET study (Hu et al. 2019) indicates that the F-pilus is connected to the T4SS outer membrane complex (OMC). Further, the study indicates that the F pilus nucleates assembly at the outer membrane in a process leading to a structural change in the OMC (Hu et al. 2019). It is tempting to speculate that the F-type specific proteins are involved in this structural change. Although the N. gonorrhoeae T4SS lacks a pilus, a structural change of the OMC mediated by the F-type specific proteins might still be needed to allow for substrate transfer.
Interactions between TraV and F-type specific proteins
TraVF is an outer membrane lipoprotein (Doran et al., 1994). TraHF, TraFF, TraUF, and TraWF have been shown to localize to the outer membrane when in the context of the complete transfer apparatus, probably with TraVF as the anchor protein (Arutyunov et al., 2010). We observed several interactions between TraVN and TraVF and F-type specific periplasmic proteins using BACTH studies (Fig. 3A, B, and C). The TraWN/TraVN and the TrbCF/TraVF interactions were confirmed by co-purification (Fig. 4). The results indicate that TraV can anchor F-type specific periplasmic proteins to the outer membrane both for the N. gonorrhoeae T4SS and the F-plasmid T4SS. The TraV homolog VirB7 is a small lipoprotein that helps to stabilize the outer membrane complex at the outer membrane (Christie, 2016). VirB7 from the A. tumefaciens is only 55 amino acids; however longer forms of VirB7 with additional functions have been described (Christie, 2016). TraVN and TraVF are respectively 193 and 171 amino acids with only the N-terminal part of the proteins showing weak homology to VirB7 (Ramsey et al., 2014). It is, therefore possible that the C-terminal part of TraV could be involved in interactions with F-type specific proteins.
The TraB-TraE interaction
Due to the high divergence of the primary sequence, some VirB8 homologs have been identified only upon structural analysis (Goessweiner-Mohr et al., 2013). A bioinformatic study placed TraEF in a universally present group of VirB8 homologs (Guglielmini et al., 2014) and secondary structure predictions also indicate that TraEF and TraEN are VirB8 like proteins (Goessweiner-Mohr et al., 2013). CryoEM of a P-type T4SS from the conjugative R388 plasmid shows that the inner membrane complex consists of the N-terminal part of VirB10 in connection with a set of other inner membrane-associated proteins including VirB8 (Low et al., 2014). Interactions between VirB8 homologs and VirB10 homologs have been demonstrated using the BACTH system (Casu et al., 2016) as well as other two-hybrid systems (Das and Xie, 2000; Ding et al., 2002). TraEF has been shown to associate with the inner membrane (Arutyunov et al., 2010) and is essential for conjugation (Lawley et al., 2003), but the function of the protein is unknown. We observed the interaction between TraB and TraE for both proteins from the N. gonorrhoeae T4SS and proteins from the F-plasmid (Fig. 3A) and a mixed TraEN/TraBF interaction (Table 5 and Fig. S3). This result indicates that in addition to inner membrane localization, TraE shares with VirB8 the ability to interact with the VirB10 homolog TraB.
Cross-system interchangeability of T4SS proteins
In this study, we observed several interactions between proteins from the F-plasmid T4SS and proteins from the N. gonorrhoeae T4SS system, indicating a high degree of cross-system interchangeability of homologous T4SS proteins despite low sequence homology (Table 5, Fig. S1, S2 and S3). This phenomenon has been observed in several other studies (Carraro et al., 2017; Casu et al., 2016; Gillespie et al., 2015; Gordon et al., 2017). For P-type T4SS there are indications for cross-system interchangeability between VirB8, VirB10, and VirB5 homologs (Casu et al., 2016; Gillespie et al., 2015; Gordon et al., 2017; Schmidt-Eisenlohr et al., 1999). In nature, this cross-system interchangeability might be important for bacteria carrying more than one T4SS (Gillespie et al., 2015). With regards to F-type T4SSs, it is not unusual for multidrug-resistant Enterobacteria to carry both IncF and IncA/C plasmids (Rayamajhi et al., 2011; Silva et al., 2015). Like IncF plasmids, IncA/C plasmids encode F-type T4SSs (Harmer and Hall, 2015). An interesting example of crosstalk occurs between an IncA/C plasmid and Salmonella genomic island 1 (Carraro et al., 2017). While the IncA/C plasmid encodes an F-type T4SS, the Salmonella genomic island 1 only encodes homologs of TraN, TraH, and TraG with amino acid identity between 37% and 78% to the plasmid proteins (Carraro et al., 2017). The Tra subunits of the genomic island can complement their plasmid counterpart in mutant studies; however, the outcome of the conjugation is shifted towards the spread of the genomic island rather than the IncA/C plasmid (Carraro et al., 2017). The presence of an IncF plasmid increases the conjugation rate of co-residing IncA/C plasmids by an unknown mechanism (Gama et al., 2017). Our data support cross-system interchangeability between F-type T4SS proteins. This interchangeability might be a way different co-residing conjugative plasmids can interact in processes that could influence the spread of antibiotic resistance.
In conclusion, our results indicate that the T4SSs from the F-plasmid and N. gonorrhoeae share an overall architecture, especially with regards to conserved T4SS protein homologs (Fig 3A). However, interactions between F-type specific proteins and between F-type specific proteins and conserved T4SS proteins (TraV, TraK, and TraB) exhibit more variation between systems (Fig. 3B, C). We present maps of the protein interactions that build these two F-type T4SSs and demonstrate that multiple protein components are likely interchangeable within these interaction networks.
Experimental procedures
Bacterial strains and growth conditions
All bacterial strains are listed in supplementary material Table S1. N. gonorrhoeae MS11 was grown on GC chocolate agar plates with VCAT (EO labs.) at 5% CO2 at 37°C or in GCBL liquid medium containing 0.042% NaHCO3 and Kellogg’s supplement (Kellogg et al., 1963) with aeration at 37°C. E. coli were grown in LB medium (Bertani, 1951) at 37°C or 30°C. Antibiotics were used at the following concentrations: ampicillin (100 µg ml−1), kanamycin (50 µg ml−1), and streptomycin (50 µg ml−1).
Construction of plasmids
Chromosomal DNA from N. gonorrhoeae MS11 was isolated from liquid overnight cultures using the GenElute Bacterial Genomic DNA kit from Sigma following the recommendation of the manufacturer except that approx. 8-9 ml of overnight culture were used for each preparation (rather than 1.5 ml) to compensate for a low OD600 in the overnight cultures. N. gonorrhoeae genes were amplified with N. gonorrhoeae MS11 chromosomal DNA as a template using primers 3- 43, 67, 68, 71, and 72 (supplementary material Table S2). Genes encoded by the F-plasmid were PCR amplified with cell lysates of E. coli JM101 as a template using primers 44-64 and 75 - 78 (Table S2). The PCR products were digested with restriction enzymes cutting the restrictions sites underlined in Table S2 and cloned into pKT25 (Karimova et al., 2001), pKTM25 (this study), p25N (Claessen et al., 2008), pUT18C (Karimova et al., 2001), pUTM18C (Ouellette et al., 2014) or pUT18 (Karimova et al., 2001) digested with the same restriction enzymes. Cloning into pCOLADuet-1 and pET22b were done with the primers indicated in supplementary material Table S1 and S2 using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs.) according to the manufacturer’s instruction All plasmids are listed in supplementary material Table S1. The structure of all plasmids was confirmed by DNA sequencing. The sequences of the N. gonorrhoeae MS11 genes were found to match the sequence derived from Accession no. CP003909.
Construction of N. gonorrhoeae mutants, subcellular fractionation, and western blotting
The traHN gene from N. gonorrhoeae strain MS11 was cloned into pMR100 to add the 3x-FLAG tag in-frame with the TraHN coding sequence, creating the intermediate pAY25. The resulting traHN-FLAG3 gene was subcloned from pAY25 into pMR68 to place it under transcriptional control by the anhydro-tetracycline inducible promoter and locate it between gonococcal genes iga and trpB. The resulting plasmid, pAY27, was then used to insert the traHN-FLAG3 construct onto the gonococcal chromosome in wild-type N. gonorrhoeae strain MS11 or its derivatives lacking traKN (MR535, Ramsey et al. 2014), traGN (PK186, Kohler PL et al. 2013), or atlAN (PK127, Kohler PL et al. 2007). To FLAG3-tag TraHN at the native locus, a fragment of DNA downstream of the traH native site was cloned in pAY25, creating pAY28. pAY28 was used to transform MS11, generating AY529. Gonococci were transformed as previously described (Ramsey et al. 2015). The expression of TraHN-FLAG3 was induced with 0.2 ng/ml anhydro-tetracycline. Subcellular fragmentation and western blots were performed essentially as described before (Ramsey et al. 2014, Ramsey et al. 2015). For the western blots approximately 5 µg protein from each fraction was subject to SDS-PAGE.
Bioinformatic analyses
For in silico localization studies, we used the SignalP 4.1 Server with the default setting for Gram-negative bacteria (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011), the TatP 1.0 server (http://www.cbs.dtu.dk/services/TatP/) (Bendtsen et al., 2005) and TOPCONS server (http://topcons.cbr.su.se/pred/reference/) (Tsirigos et al., 2015). Amino acid identity between the protein from N, gonorrhoeae and the corresponding protein from the F-plasmid were calculated using the Needleman-Wunsch global alignment algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Determination of interactions and measurements of β-galactosidase activity
The initial interaction screening was done after the co-transformation of T18 and T25 encoding plasmids into E. coli BTH101, plating on LB agar plates with appropriate antibiotics, 0.5 mM IPTG and 40 µg ml−1 X-gal and incubated 40 - 48 hours at 30°C. In the absence of functional complementation between T18 and T25, the colonies are white, while they are blue when functional complementation occurs. As a negative control, we used BTH101 cells co-transformed with vectors containing no inserts. For confirmation of positive interactions, the cells expressing a T18 and a T25 tagged protein were grown over-night at 30°C in LB with appropriate antibiotics. The overnight cultures were diluted with 0.9 % NaCl and dilutions were spotted on LB agar plates with appropriate antibiotics, 0.5 mM IPTG, and 40 µg ml−1 X-gal and incubated 40 - 48 hours at 30°C.
For β-galactosidase assays, cells were grown overnight at 30°C in LB with appropriate antibiotics and 0.5 mM IPTG and β-galactosidase activities were measured as described by Miller (Miller J. S., 1972).
Protein expression and co-purification
TraVN, TraVF, and TraWN were overproduced in E. coli BL21(DE3) while TrbCF was overproduced in both E. coli BL21(DE3) and E. coli Origami2(DE3). For TraVN and TraVF expression, the N-terminal lipobox was replaced with a pelB signal sequence for periplasmic expression while a his-tags were added in the C-terminal end. The entire reading frame of TraWN was expressed while E. coli TrbCF was expressed without the signal sequence since the attempt to express TrbCF with the signal sequence gave a low level of expression. Cells of E. coli BL21(DE3) carrying either pET22bTraVN, pET22bTraVF, pCOLATraWN, or pCOLATrbCF constructs or E. coli Origami2(DE3) carrying pCOLATrbCF were grown in LB at 37°C to an OD600 of approx. 0.4. Overproduction was induced by the addition of IPTG to a final conc. of 1 mM and incubation was continued for 3 h at 37°C. The cells were harvested by centrifugation and the cell pellets were frozen (−20). For the TraVN /TraWN and TraVF/TraWN co-purifications the pellets were resuspended in buffer I (25 mM NaH2PO4, 150 mM NaCl, 5 mM imidazole. 5 mM MgCl2 pH 8) for the TraVF/TrbCF and TraVN/TrbCF co-purifications the pellets were resuspended in buffer II (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole. pH 8 ). The resuspended pellets were incubated 30 min. with 1 mg/ml lysozyme on ice followed by sonication. After sonication, the cells were centrifugated for at 7000 g 40 min. at 4°C. For TraWN and TrbCF the supernatants were used for the co-purification experiments. For TraVN and TraVF pellets were resuspended in buffer I or buffer II and frozen at −20°C. After one round of freezing and thawing partial binding of TraVN and TraVF to nickel resin was observed (Fig. S4) and samples treated this way were used for co-purification and control experiments. For control experiments, cells carrying expression plasmids were replaced with cells carrying pCOLADuet-1 or pET22b. For resin binding samples were applied to 0.5 ml of washed and equilibrated Ni-NTA agarose beads (QIAGEN, Hilden, Germany) and incubated overnight at 4°C, with mixing. The beads were washed with 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole. pH 8. Subsequently, bound proteins were stepwise eluted with 1 ml 25 mM NaH2PO4, 150 mM NaCl, 125 mM imidazole. pH 8 and 1 ml 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole (Fig. S4). Equal amounts of the eluted sample from co-purification and control experiments were analyzed by SDS-PAGE.
Supplementary Material
Acknowledgements
This work was supported by EPSRC Grant Nos EP/L001489/2, EP/J004111/2, EP/N031926/1 and a Royal Academy of Engineering Chair in Emerging Technology to N.K and NIH grant R01AI047958 to J.P.D. We thank Scot P. Ouellette for providing plasmids pUTM18C and pSTM25. NK thanks S. Heeb for bringing N. gonorrhoeae’s type IV secretion system to his attention many years ago.
Footnotes
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article. In addition, we have created version-controlled cell repositories (cellrepo) as recommended in (Tellechea-Luzardo et al., 2020) to facilitate reproduction and derivative work from this paper. These repositories focuse on the TraV proteins and describes the plasmids containing them. The cellrepo for our data is available here:
References
- Alvarez-Martinez CE, and Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73: 775–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arutyunov D, Arenson B, Manchak J, and Frost LS (2010) F Plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J Bacteriol 192: 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arutyunov D, and Frost LS (2013) F conjugation: back to the beginning. Plasmid 70: 18–32. [DOI] [PubMed] [Google Scholar]
- Battesti A, and Bouveret E (2012) The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods San Diego Calif 58: 325–334. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Widdick D, Palmer T, and Brunak S (2005) Prediction of twin-arginine signal peptides. BMC Bioinformatics 6: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan MM, Heilers JH, van der Does C. and Dillard JP. (2017) Secretion of Chromosomal DNA by the Neisseria gonorrhoeae Type IV Secretion System. Curr. Top Microbiol. Immunol 413: 323–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Durand R, Rivard N, Anquetil C, Barrette C, Humbert M, and Burrus V (2017) Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLOS Genet 13: e1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casu B, Smart J, Hancock MA, Smith M, Sygusch J, and Baron C (2016) Structural analysis and inhibition of TraE from the pKM101 type IV secretion system. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran Darbari V, and Waksman G (2015) Structural biology of bacterial type IV secretion systems. Annu Rev Biochem 84: 603–629. [DOI] [PubMed] [Google Scholar]
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, and Waksman G (2009) Structure of the outer membrane complex of a type IV secretion system. Nature 462: 1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ (2016) The Mosaic Type IV Secretion Systems. EcoSal Plus 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, and Edwards DH (2008) Control of the cell elongation–division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol 68: 1029–1046. [DOI] [PubMed] [Google Scholar]
- Das A, Anderson LB, and Xie YH (1997) Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol 179: 3404–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, and Xie YH (2000) The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol 182: 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP, and Seifert HS (2001) A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol 41: 263–277. [DOI] [PubMed] [Google Scholar]
- Ding Z, Zhao Z, Jakubowski SJ, Krishnamohan A, Margolin W, and Christie PJ (2002) A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J Bacteriol 184: 5572–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran TJ, Loh SM, Firth N, and Skurray RA (1994) Molecular analysis of the F plasmid traVR region: traV encodes a lipoprotein. J Bacteriol 176: 4182–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton TC, Holland SJ, Frost LS, and Hazes B (2005) F-like type IV secretion systems encode proteins with thioredoxin folds that are putative DsbC homologues. J Bacteriol 187: 8267–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth N and Skurray R (1992) Characterization of the F plasmid bifunctional conjugation gene, traG. Mol. Gen. Genet 232 (1): 145–153. [DOI] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, and Waksman G (2009) Structure of a type IV secretion system core complex. Science 323: 266–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama JA, Zilhão R, and Dionisio F (2017) Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: Plasmids promote the immigration of other plasmids but repress co-colonizing plasmids. Plasmid 93: 6–16. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Phan IQH, Scheib H, Subramanian S, Edwards TE, Lehman SS, et al. (2015) Structural insight into how bacteria prevent interference between multiple divergent Type IV Secretion Systems. mBio 6: e01867–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour MW, Lawley TD, Rooker MM, Newnham PJ, and Taylor DE (2001) Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol Microbiol 42: 705–715. [DOI] [PubMed] [Google Scholar]
- Goessweiner-Mohr N, Grumet L, Arends K, Pavkov-Keller T, Gruber CC, Gruber K, et al. (2013) The 2.5 Å structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem 288: 2018–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JE, Costa TRD, Patel RS, Gonzalez-Rivera C, Sarkar MK, Orlova EV, et al. (2017) Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation: Chimeric T4SSs reveal compositional flexibility of outer membrane subassemblies. Mol Microbiol 105: 273–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Neron B, Abby SS, Garcillan-Barcia MP, la Cruz F. d., and Rocha EPC (2014) Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 42: 5715–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, and Dillard JP (2005) Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol 55: 1704–1721. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Schwartz KJ, and Dillard JP (2001) Insertion-duplication mutagenesis of Neisseria: Use in characterization of DNA transfer genes in the Gonococcal Genetic Island. J Bacteriol 183: 4718–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer CJ, and Hall RM (2015) The A to Z of A/C plasmids. Plasmid 80: 63–82. [DOI] [PubMed] [Google Scholar]
- Harris RL, Hombs V, and Silverman PM (2001) Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol Microbiol 42: 757–766. [DOI] [PubMed] [Google Scholar]
- Harris RL, and Silverman PM (2004) Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol 186: 5480–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmis CW, and Schildbach JF (2013) Thioredoxin-like proteins in F and other plasmid systems. Plasmid 70: 168–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Khara P, and Christie PJ (2019) Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli. Proc Natl Acad Sci 116: 14222–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, and Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95: 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Ullmann A, and Ladant D (2001) Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J Mol Microbiol Biotechnol 3: 73–82. [PubMed] [Google Scholar]
- Karimova G, Dautin N, and Ladant D (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187: 2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DSJ, Peacock WLJ, Deacon WE, Brown L, and Pirkle DI (1963) Neisseria gonorrhoeae. virulence genetically linked to clonal variation. J Bacteriol 85: 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler PL, Chan YA, Hackett KT, Turner N, Hamilton HL, Cloud-Hansen KA, and Dillard JP (2013) Mating pair formation homologue TraG is a variable membrane protein essential for contact-independent type IV secretion of chromosomal DNA by Neisseria gonorrhoeae. J Bacteriol 195: 1666–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler PL, Hamilton HL, Cloud-Hansen K, and Dillard JP (2007) AtlA Functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol 189: 5421–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann G (2003) Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci CMLS 60: 2371–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T, Klimke W, Gubbins M, and Frost L (2003) F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224: 1–15. [DOI] [PubMed] [Google Scholar]
- Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, et al. (2014) Structure of a type IV secretion system. Nature 508: 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellechea-Luzardo J, Winterhalter C, Widera P, Kozyra J, de Lorenzo V and Krasnogor N (2020) Linking Engineered Cells to Their Digital Twins: A Version Control System for Strain Engineering. ACS Synthetic Biology 9: 536–545 [DOI] [PubMed] [Google Scholar]
- Miller JS (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press. NY. [Google Scholar]
- Oliveira LC, Souza DP, Oka GU, Lima F. da S., Oliveira RJ, Favaro DC, et al. (2016) VirB7 and VirB9 interactions are required for the assembly and antibacterial activity of a type IV secretion system. Struct Lond Engl 1993. [DOI] [PubMed] [Google Scholar]
- Ouellette SP, Gauliard E, Antosová Z, and Ladant D (2014) A Gateway ® -compatible bacterial adenylate cyclase-based two-hybrid system: A Gateway-compatible bacterial two-hybrid system. Environ Microbiol Rep 6: 259–267. [DOI] [PubMed] [Google Scholar]
- Pachulec E, Siewering K, Bender T, Heller E-M, Salgado-Pabon W, Schmoller SK, et al. (2014) Functional analysis of the Gonococcal Genetic Island of Neisseria gonorrhoeae. PLoS ONE 9: e109613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, and Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Ramsey ME, Bender T, Kilmowicz AK, Hackett KT, Yamamoto A, Jolicoeur A, Callaghan MM, Wasserman K.M. van der Does, C., and Dillard JP. (2015). Targeted mutagenesis of intergenic regins in the Neisseria gonorrhoeae gonococcal island reveals multiple regulatory mechanisms controlling type IV secretion. Mol. Microbiol 97(6): 1168–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Hackett KT, Bender T, Kotha C, van der Does C, and Dillard JP (2014) TraK and TraB are conserved outer membrane proteins of the Neisseria gonorrhoeae Type IV secretion system and are expressed at low levels in wild-type cells. J Bacteriol 196: 2954–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Hackett KT, Kotha C, and Dillard JP (2012) New complementation constructs for inducible and constitutive gene expression in Neisseria gonorrhoeae and Neisseria meningitidis. Appl Environ Microbiol 78: 3068–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi N, Cha SB, Shin SW, Jung BY, Lim S-K, and Yoo HS (2011) Plasmid typing and resistance profiling of Escherichia fergusonii and other Enterobacteriaceae isolates from South Korean farm animals. Appl Environ Microbiol 77: 3163–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele CW, Xian Y, Albrecht M, Faulstich M, Fraunholz M, Heinrichs E, Dittrich MT, Muller T, Reinhardt R and Rudel T (2014).Transcriptional landscape and essential genes of Neisseria gonorrhoeae.Nucleic Acids Res. 42: 10579–10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Pabón W, Jain S, Turner N, van der Does C, and Dillard JP (2007) A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol 66: 930–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Eisenlohr H, Domke N, and Baron C (1999) TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J Bacteriol 181: 5563–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shala-Lawrence A, Bragagnolo N, Nowroozi-Dayeni R, Kheyson S, and Audette GF (2018) The interaction of TraW and TrbC is required to facilitate conjugation in F-like plasmids. Biochem Biophys Res Commun 503(4): 2386–2392. [DOI] [PubMed] [Google Scholar]
- Silva C, Calva E, Calva JJ, Wiesner M, Fernandez-Mora M, Puente JL, and Vinuesa P (2015) Complete Genome Sequence of a Human-Invasive Salmonella enterica Serovar Typhimurium Strain of the Emerging Sequence Type 213 Harboring a Multidrug Resistance IncA/C Plasmid and a blaCMY-2-Carrying IncF Plasmid. Genome Announc 3: e01323-15–e01323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanesan D, Hancock MA, Villamil Giraldo AM, and Baron C (2010) Quantitative analysis of VirB8−VirB9−VirB10 interactions provides a dynamic model of type IV secretion system core complex assembly. Biochemistry (Mosc) 49: 4483–4493. [DOI] [PubMed] [Google Scholar]
- Tsirigos KD, Peters C, Shu N, Käll L, and Elofsson A (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43: W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


