Abstract
Background
Organoids are a reliable model used in the study of human brain development and under pathological conditions. However, current methods for brain organoid culture generate tissues that range from 0.5 to 2 mm of size, which need to be constantly agitated to allow proper oxygenation. The culture conditions are, therefore, not suitable for whole-brain organoid live imaging, required to study developmental processes and disease progression within physiologically relevant time frames (i.e. days, weeks, months).
Results
Here we designed 3D-printed microplate inserts adaptable to standard 24 multi-well plates, which allow the growth of multiple organoids in pre-defined and fixed XYZ coordinates. This innovation facilitates high-resolution imaging of whole-cerebral organoids, allowing precise assessment of organoid growth and morphology, as well as cell tracking within the organoids, over long periods. We applied this technology to track neocortex development through neuronal progenitors in brain organoids, as well as the movement of patient-derived glioblastoma stem cells within healthy brain organoids.
Conclusions
This new bioengineering platform constitutes a significant advance that permits long term detailed analysis of whole-brain organoids using multimodal inverted fluorescence microscopy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42490-021-00049-5.
Keywords: Brain organoids, Live-imaging, Fluorescence microscopy, Glioblastoma
Background
Brain organoids [1] have emerged as promising models to better understand brain development and the mechanisms that contribute to the pathology of different brain diseases including Alzheimer’s Disease, Parkinson’s Disease and brain cancer [2–4]. Cerebral organoids are generated from human embryonic (hESC) or induced pluripotent (iPSC) stem cells, which, after directed differentiation, lead to the emergence of different cell populations that self-organise to form tissue structures resembling the developing (foetal) human cerebral cortex [1, 5–7]. Since the original publication of this method [1], various protocols have been reported, including those suitable for the growth of brain region-specific organoids [1, 7–12]. It is now possible to induce systematic differentiation of stem cells into dorsal and ventral cortices [1, 13], hindbrain, midbrain [1, 7–12], cerebellum [14] and subcortical structures such as the hypothalamus [11] and hippocampus [15]. Furthermore, neurons within brain organoids produce electrical activity [12, 16] and can support functional neuromuscular junctions [17]. These results indicate that brain organoids provide an important in vitro platform technology with which to mimic the cellular composition as well as the cytoarchitecture and function of brain tissue [1, 18].
Brain organoids can also be generated from patient-derived tissue samples, which – in many cases – reproduce critical features of the brain disease [11, 19–22], constituting a significant technological advantage for the study of these diseases. For example, this technology has revealed how impairment of neural progenitor cells (NPCs) found in Zika virus infection at various stages of human embryogenesis contribute to microcephaly [11] as well as used to recapitulate different Alzheimer’s Disease pathologies, such as amyloid aggregation, Tau hyper-phosphorylation and endosome abnormalities [23]. Patient-derived brain organoids could also be used to produce patient-specific tissue for tissue regeneration after insults such as tumour biopsies, neurodegenerative disease and neurological trauma [24, 25].
Brain organoids generated from patient-derived tissue have also attracted significant interest in the clinical setting, particularly in brain cancer, including glioblastoma, and thus constitute a powerful pre-clinical model for the development of personalised treatment strategies [3]. In this regard, glioblastoma-brain organoid models reproduce key characteristics of the brain cancer microenvironment found in glioblastoma patients, including hypoxic gradients [20], pathological features [26], cellular diversity and gene expression [22]. Furthermore, glioblastoma-brain organoid models can be used to assess patient response to current standard of care treatments in addition to novel immunotherapies [22, 26, 27]. Patient-derived brain organoids have the potential to predict patient prognosis rapidly, assess treatment response and improve diagnosis. Taken together, increasing the clinical management of aggressive cancers such as glioblastoma [28] and avoiding the ‘trial and error’ approaches of current clinical practice, which can be costly, ineffective and present limited benefits to patients.
However, a significant limitation of brain organoid technology that hinders its application to personalised medicine is the lack of a methodology that allows temporal (e.g. days, weeks, months) live high-resolution imaging of multiple organoids in a multi-well plate. Such a method would allow evaluation of treatments using patient-derived organoids, while concomitantly determining the effect of these treatments on various disease parameters. This is particularly important in brain cancer, where current approaches generally focus on cancer cell proliferation and neglect fundamental malignant processes such as invasion in which cancer cells interact with both the cellular and non-cellular microenvironment [3, 28, 29]. In this report, we describe the use of 3D-printed microplate inserts that solve this problem, by enabling the culture of multiple brain organoids in specified XYZ coordinates, thus facilitating high-resolution imaging using fluorescence microscopy.
Results
3D-printed microplate inserts to culture multiple brain organoids
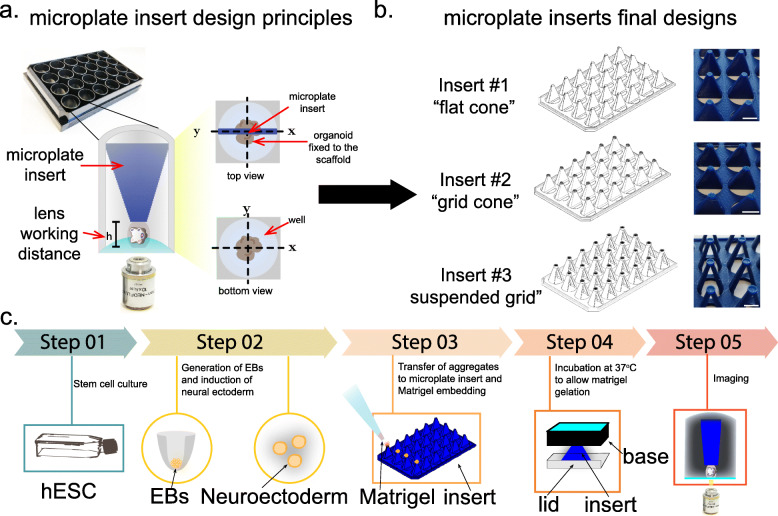
To facilitate the culture of multiple brain organoids in specified XYZ positions within a microplate, we designed a series of 3D-printed microplate inserts for standard 24-well microplates (Fig. 1a) that permit:
the culture of multiple brain organoids (30–60 day-old) using glass bottom microplates suitable for imaging applications.
- the confinement of the brain organoid to the axial (Z) position to facilitate whole-organoid live imaging by:
-
i.maintaining the brain organoid within the working distance of the objective (2.5 mm).
-
ii.minimising the effect (contact, pressure) that the presence of the inserts may exert on brain organoids, as this confinement is compatible with > 30 day-old organoid dimensions (~ 1 mm average diameter, [30]).
-
i.
the fixation of the XY position of the organoid within individual wells to facilitate automated imaging acquisition and post-acquisition analysis as the organoid develops.
Fig. 1.
Microplate inserts for brain organoid culture. a Design principle for microplate inserts for brain organoid culture in fixed XYZ positions compatible with high-resolution imaging. b Final design of the three types of microplate inserts. Scale bar = 10 mm. c Pipeline for growth of cerebral organoids using microplate inserts
We anticipate that by controlling these three parameters, the engineered microplate inserts will be ideal for high-resolution, whole-organoid live imaging, and be suitable for automated microscopy that is required for relatively fast and systematic readouts like those employed in drug screening.
As a result of our design requirements, we were able to produce three different microplate inserts that satisfy the stated design principles (Fig. 1b). For simplicity, we named these microplate inserts as flat-cone (insert #1), grid-cone (insert #2), and suspended-grid (insert #3). A shared design element of each insert is the ability to secure the organoid tissue close to the glass bottom of the microplate within the objective working distance (Fig. 1a).
A fine mesh (“grid”) present on the tip of inserts #2 and #3, is a unique feature that minimises the potential interaction surface that might occur between the organoid and the insert. In the case of insert #3, a hollow space present on the side opposite to the grid aids with media exchange and oxygenation of the organoids [30].
We employed a Raise 3D Pro printer for printing the final microplates (Fig. 1b; see also Methods) and cultured brain organoids following standard protocols developed by Lancaster and Knoblich with small modifications [1, 30] (Fig. 1c). For Step 1 of this procedure, we used hESC H9 [31], H9-GFP [32] or a mixture of both in a ratio 100 H9: 1 H9-GFP (“chimera organoids”) to generate embryoid bodies (EBs) in low-attachment U-shaped 96 well plates (Fig. 1c). In Step 2, EBs were matured to form neuroectodermal aggregates. In Step 3, we placed the microplate inserts facing upwards over a multi-well plate lid, which permits the transfer and positioning of the aggregates on the tip of the inserts. In Step 4, a droplet of Matrigel was added on top of each organoid and then the plate base (without the lid and facing downwards) was placed on top of the insert. Following Matrigel gelation by incubation at 37 °C, the multi-well plate containing the inserts was returned to the normal orientation before the addition of culture medium for further organoid culture, differentiation and imaging (Step 5, Fig. 1c).
Microplate inserts allow long-term monitoring and individual assessment of brain organoid growth using automated fluorescence microscopy
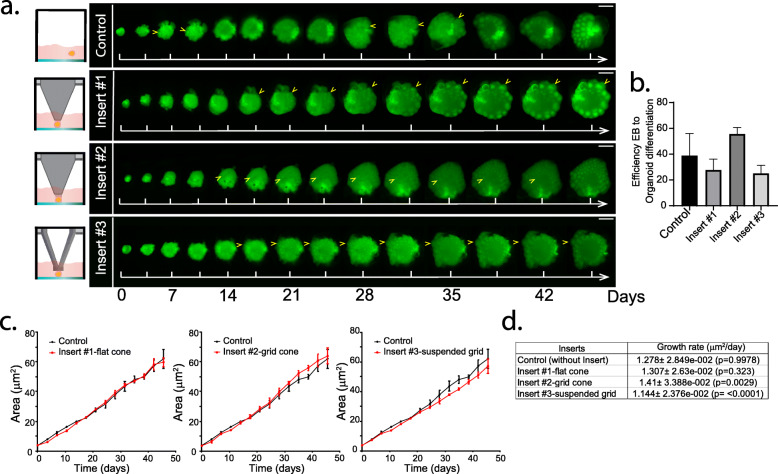
In order to test the advantages of using microplate inserts for culture and imaging of mature brain organoids, we performed long term imaging of individual brain organoids starting immediately after Matrigel embedding (Day 0) for 50 continuous days, which has not previously been reported. For this, organoids grown in multi-well plates with and without inserts were imaged every 3.5 days (84 h) for 50 days using wide field automated fluorescence microscopy (2x objective, InCell Analyzer 2200, Cytiva). Figure 2a shows still images from different time points taken for organoids grown with and without microplate inserts. A key advantage of using microplate inserts is that the organoids are grown in a fixed position, preserving their orientation and permitting the tracking of individual features within the organoid (yellow arrows) in contrast to the control organoids grown without inserts (Fig. 2a and Supplementary Movie 1).
Fig. 2.
Time course of live brain organoid imaging using microplate inserts. a Images were taken every ~ 3.5 days using an InCell Analyzer microscope using a 2.1x objective. Control refers to organoids grown without microplate inserts. Organoids grown with inserts #1 to #3 maintain their orientation and produce characteristic cortical structures (yellow arrows). Panels were made using cropped images (same size for all time points and conditions) placed on a black background. Image’s intensity levels were contracted (same histogram contraction applied to panels from all time points across all conditions) from their original 16-bit range to 8-bit RGB range for Figure preparation in Adobe Illustrator. Scale Bar = 1 mm. b Percentage of EBs that successfully matured to brain organoids under different conditions was determined by fluorescence microscopy at day 50. Successfully grown organoids were identified by an increase in size over time and identification of characteristic cortical structures. Data are means ± SEM for three independent experiments. c Organoid 2D projected area (Area) vs time plots for brain organoid cultures in the presence or absence of microplate inserts. Data points correspond to mean values for three independent experiments (n = 3), and for each experiment at least 3 organoids per condition were analysed. d Results of linear regression analysis (Area = initial organoid size + Growth rate * Days) of plots in c. The table shows the p values for the comparison of organoid growth rates between the control and microplate insert groups. Non-Significant (n.s) and Significant (*, p < 0.05)
We then assessed the effect of the different microplate inserts on organoid formation. For this, we determined the efficiency of EB to organoid differentiation by counting the proportion of organoids that successfully formed and exhibited characteristic cortical structures (yellow arrows Fig. 2a) by day 50. The different microplate inserts were compared to the control condition without insert. Figure 2b shows the results of 3 independent experiments with 24 organoids (i.e. one multi-well plate) per experiment. The differentiation efficiency was ~ 40% in all experimental and control conditions. We observed less variability (measured as standard error of mean) for conditions using microplate inserts compared to the control conditions without inserts. Moreover, cultures using microplate insert #2 showed a trend (although non-significant) of yielding a larger proportion of organoids when compared to all the other conditions (with or without insert, Fig. 2b). Together, these results suggest that the use of microplate inserts and the restriction of organoid movement within the well, did not alter the capacity of EBs to differentiate to organoids.
We then evaluated the effect of microplate inserts on brain organoid growth rates. For this we analysed the time series of acquired images (Fig. 2a) by automatically segmenting organoids based on their fluorescence intensity and measuring the organoid z-projected area (Scripts for image segmentation are provided in the Supplementary Information). For successfully grown organoids (Fig. 2b), the organoid projected area was plotted as a function of time (Fig. 2c) and organoid growth rates and organoid initial projected area determined by linear regression (Fig. 2d and Supplementary Table 1).
Firstly, we confirmed that the organoid projected area at time 0 (i.e. immediately after embedding in Matrigel) was similar either in the presence or absence of microplate inserts (Fig. 2c, Supplementary Table 1). This suggested that the presence of the insert does not exert pressure on the organoids which might significantly alter their shape and projected area [33, 34]. This also reduced the potential for differences in organoid growth rates arising from different initial EB size.
Analysis of growth rates at later time points revealed that cultures grown with microplate insert #1 grew at rates comparable to control conditions without an insert (Fig. 2c and d). Organoids grown using microplate insert #2 exhibited a significantly higher growth rate, compared to the controls after 30 days while organoids grown using microplate insert #3 resulted in a significantly slower growth rate (Fig. 2c, d). Of note, although we found slight differences in organoid growth rates in different conditions, the size of the organoids was not significantly different between inserts and controls at any time point of our analysis. For example, at day 50, calculations using data from our linear regression analysis of growth rates, determined organoid area to be 66.3 ± 2.2, 66.7 ± 2.0, 71.1 ± 2.6 and 59.8 ± 1.8 μm2 for control (no insert), insert 1, insert 2 and insert 3, respectively. These values correspond to 0.6 ± 6%, 7 ± 7% and − 10 ± 6% differences in size between organoids grown using inserts 1–3 compared to organoids grown without inserts, respectively. One-way ANOVA statistical analysis of the variation in size was determined to be non-significant indicating that the presence of the inserts did not lead to altered 2D organoid areas as would have been expected by an increase in pressure or a highly forced confinement of the organoids due to the presence of the microplate inserts. Importantly, we performed staining for cleaved-caspase 3 in organoid sections to assess apoptotic cell death. Although we found cleaved caspase 3 positive cells principally in the periphery of cortical structures, the distribution of these apoptotic cells was not dramatically different between organoids using the different microplate inserts (Supplementary Figure 1).
Altogether, these results indicate that the presence of the microplate inserts, which facilitate long-term fluorescence microscopy imaging of live brain organoids, do not interfere with organoid growth when compared to the reference methods of organoid production [1, 30, 33].
Human brain organoids grown using microplate inserts maintain human cerebral cortical organisation
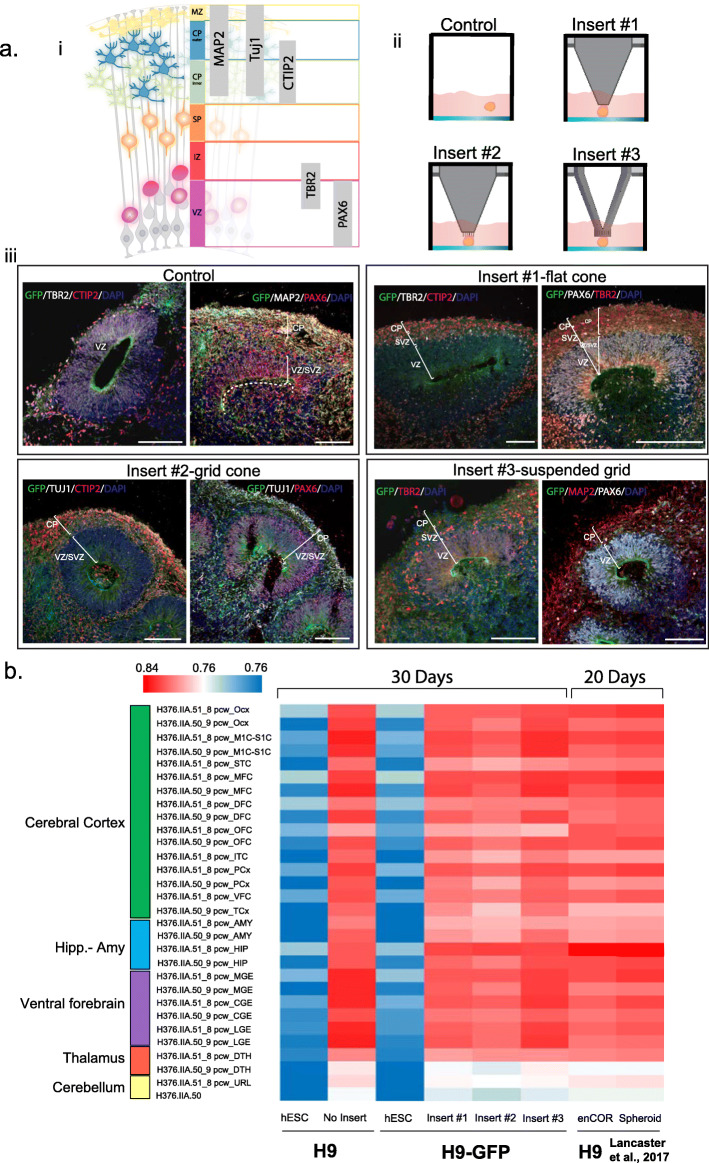
We then used immunofluorescence to analyse in more detail the cytoarchitecture (Fig. 3a, i) of 30 day-old brain organoids grown in the presence or absence of microplate inserts (Fig. 3a, ii). For this, brain organoids were embedded and frozen in OCT, sectioned and immune-labelled with different markers (Fig. 3a, iii). Staining with neural progenitor marker (Pax6) revealed a typical ventricular region within organoids, which was comparable between all conditions analysed (Fig. 3a, iii, control and inserts #1-#3). Tbr2-positive intermediate progenitors cells (IPCs), which are neuronal progenitors that divide away from the ventricular surface [34], were present in the proliferative region of the cerebral organoids grown in different conditions, an observation that is in agreement with previously published reports for brain organoids [1, 7]. Furthermore, staining for CTIP2, MAP2 and TUJ1 confirmed that brain organoids grown using microplate inserts contain neurons that have properly migrated within the preplate and which should therefore subsequently mature in a cortical plate structure [35]. Thus, cortical zones in brain organoids generated using microplate inserts display typical progenitor zone organisation as previously shown for brain organoids grown using standard methods [1, 30, 33].
Fig. 3.
Brain organoids grown using microplate inserts show cerebral cortex identity. a i. Schematic showing the distribution of neuronal differentiation markers in the brain cortex. ii. Schematic representation of brain organoid cultures using the different microplate inserts. Control refers to the condition without insert. iii. Representative sections of cerebral organoids grown using the microplate inserts. Each tissue contains neural progenitors (PAX6+), intermediate progenitors (TBR2+), and neurons (CTIP2+, Tuj1+, MAP+), with no significant differences observed in the cytoarchitecture of the cerebral cortex of the brain organoids grown under the different experimental conditions (i.e. with or without microplate inserts). Panels were made with cropped images (same size for all panels shown). Image’s intensity levels were contracted at the same extent for all individual channels, across all panels shown to increase visibility and maximize compatibility between original 16-bit range -microscope image output- and 8-bit range (RGB) for Figure preparation in Adobe Illustrator. Scale Bar = 100 μm b Heatmap of Spearman correlation analysis of gene expression between H9 and H9-GFP hESC, brain organoids (grown with and without microplate inserts), previously published organoid RNA-seq data [33] and BrainSpan database https://www.brainspan.org/static/download.html. Hipp-Amy: Hippocampus-Amygdala
To further validate these results, we performed RNA sequencing (RNA-seq) experiments to determine the gene expression profile of 30 day-old organoids grown using microplate inserts for comparison with gene expression signatures reported for different brain regions early in development [36, 37] and with brain organoids grown using reference methods [1]. To achieve this, we prepared single cell suspensions of 30 day-old brain organoids derived from H9 and H9-GFP cells and performed RNA-seq. Gene expression profiles from brain organoids cultured using microplate inserts and data previously reported [33] were then compared to those derived from different brain regions (cerebral cortex, hippocampus, amygdala, ventral forebrain, thalamus and cerebellum) collected from the BrainSpan database (https://www.brainspan.org/static/download.html [36, 37]). Comparison of gene expression patterns for markers from different brain regions was performed by calculating Spearman correlation coefficients between gene expression values derived from the organoids and those reported in the BrainSpan data base [33]. As a control, we also included non-differentiated hESCs. In general, we found gene expression profiles of H9-GFP organoids (inserts #1–3), but not h9-GFP hESC, correlated well with the profiles from different brain regions, particularly the cerebral cortex, hippocampus/amygdala and ventral forebrain. These patterns agree with data obtained for organoids grown using standard protocols (20 day-old enCOR, Lancaster dataset, Fig. 3b). We also observed that the RNA expression profiles of 30 day-old organoids grown in our experimental conditions correlate less with thalamus and cerebellum, similar to the findings of Lancaster et al. using 20 day-old enCOR organoids [33]. Comparing gene expression, we observed a high correlation between conditions using microplate inserts #3, no insert H9 (Control), and 20-day-old enCOR (Fig. 3b). These results show that the expression profiles of organoids using microplate inserts are comparable to those generated using standard protocols [1, 30, 33].
Use of microplate inserts to track cerebral cortex development process and monitor tumour growth and invasion within brain organoids
After determining that microplate inserts allow the proper growth and development of brain organoids, we wanted to test whether this technology enables whole organoid live imaging and the tracking of individual cells within the tissue using high-resolution microscopy. For this we focused on two biological processes that are important in brain development and disease: (i) neuronal progenitor behaviour including interkinetic nuclear migration (INM) and cell division orientation, which are key processes that occur during neocortex expansion [38, 39] and (ii) glioma stem cell migration, invasion and proliferation, central steps in glioblastoma development and progression [29].
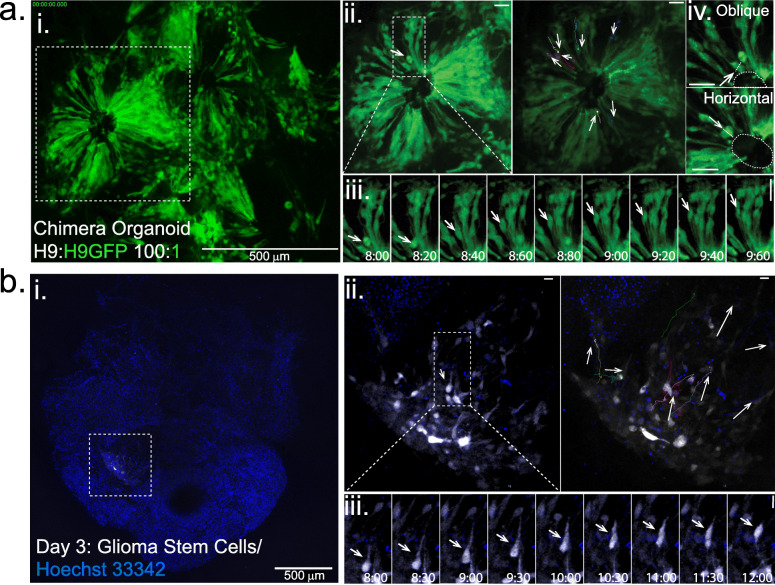
For the analysis of INM, and to increase the contrast of individual cells, we prepared “chimera organoids”, which result from the differentiation of EBs produced using a mixture of H9 and H9-GFP in a 100:1 ratio (Fig. 4a, i). After 30 days’ growth using microplate insert #3, these organoid chimeras showed a visible mosaic of cells expressing GFP, which correspond to neuronal progenitors and neurons at different stages of differentiation (Fig. 4a, i-ii). At this point in differentiation, we performed confocal laser scanning for live cell imaging to analyse cell movement within the organoid (Fig. 4b, iii, Supplementary Movie 2). Analysis of spindle orientation of dividing cells throughout the movie revealed that the majority of cells dividing at the apical surface predominately exhibited horizontal and oblique spindle orientation (Fig. 4a, iv), as is expected from the division of neuronal progenitors [38]. Thus, live-imaging of intact whole cerebral organoids using the microplate inserts captures key characteristics of neuronal progenitor cells’ behaviour. Note that it was not possible to generate comparable data for organoids grown without inserts, due to an inability to keep the organoids in focus and track the same area over time, thus highlighting the importance of our new approach.
Fig. 4.
Live brain organoid imaging. a Neuronal progenitor behaviour in brain organoids (see also Supplementary Movie 2). i. Whole brain organoid projection. ii. Magnified view of a brain organoid cortical area. iii. Neuronal movement tracking in a brain organoid region and still images showing INM. iv. Examples of neuronal progenitor oblique and horizontal cell division. Scale bars are 500 μm (i) and 50 μm (ii-iv). b Visualisation of GSC tumour growth and invasion within healthy brain organoids using confocal microscopy (see also Supplementary Movie 3). i. Time-lapse (h) of cerebral organoids injected with patient-derived glioma stem cells (PD-GSC). ii. Magnified view of region highlighted in i (left) with individual cell trajectories (right). iii. Still images showing GSC movement within the brain organoid. In a and b, Panels were made with cropped images (same size for all time points and conditions). Image’s intensity levels were contracted (same extent for all time points across all conditions but different across different channels) from their original 16-bit range to 8-bit RGB for Figure preparation in Adobe Illustrator
We then analysed glioma stem cell behaviour within brain organoids using live fluorescence microscopy imaging (Fig. 4b, i, Supplementary Movie 3). For this, we used patient-derived glioma stem cells (PD-GSC), which we fluorescently labelled using DNA-encoded fluorescent reporter RFP using lentivirus transduction [40, 41]. PD-GSC stably expressing RFP were injected directly into H9-GFP derived 30 day-old brain organoids grown using microplate insert #2. For injection, the microplate inserts with attached organoids were taken out of the microplate and inverted to allow injection of PD-GSC cells on top of the organoid. After injection, the microplate insert was returned to the microplate and cultured overnight before initiating live imaging to monitor the growth and invasion of the injected tumour cells into the surrounding healthy brain organoid (Fig. 4b, ii). Using live imaging, we were able to track individual cancer cells invading the surrounding healthy brain organoid, a process that showed some degree of collective behaviour (Fig. 4b, iii, Supplementary Movie 3). Thus, this assay permitted the assessment of PD-GSC behaviour within a physiologically relevant tissue context, which could have applications for the pre-clinical evaluation of potential anti-cancer therapies. Such assays have the potential to identify effective therapies that are efficient at targeting tumour heterogeneity and resistance to therapy [42–47], tumour invasion [26, 48], as well as drug-toxicity to the surrounding brain tissue [3].
Discussion
Previously, different approaches have been taken to develop multi-well plate based microscopy assays for neural spheroids [49, 50] and brain organoids [51] and such approaches have been applied to brain cancer [48, 52]. Although useful, these methods have several disadvantages. Firstly, most of them are optimised for the culture of neurospheres [50], which are 3D aggregates of neurons without a brain tissue cytoarchitecture or very immature organoids (i.e. < 10 day-old [48]), that lack key features present in mature cerebral organoids (> 30 days old) such as the presence of distinct cortical brain regions [7, 53], electrical waves [12, 16] and presence of individual neuronal cell populations [54]. Unfortunately, 3D culture protocols, as well as brain organoids grown on chips [51], are not suitable for high-resolution imaging of > 30 day-old cultured organoids and are incapable of tracking individual cells within the organoid, due to their non-adherent nature and the agitation conditions required for culturing [11].
In addition, the design of these microplate inserts contrasts with previous efforts [55, 56] where human brain organoids were cultured confined to a semi 2D-dimensional compartment (150 μm thickness). Although such a sub-millimetre confinement [55–57] facilitates high-resolution imaging, this approach lacks utility for mature organoids which are large in size, thus reducing their pre-clinical applicability [22].
In this report, we present the development of new 3D-printed microplate inserts that allow the growth of brain organoids in standard multi-well plates and permit the scale-up of cerebral organoid culture. Development of 3D-microplate inserts allowed the direct observation of individual cells in whole live organoids to assess their growth, migration and proliferation within a three-dimensional and more physiologically relevant microenvironment. Moreover, immunofluorescence, RNAseq, and analysis of organoid growth revealed that organoids grown using microplate inserts are comparable to those generated using standard protocols [1, 30, 33].
Although cerebral organoids grown using the methods described here did not produce a significant number of astrocytes and oligodendrocytes [7, 58] extending the culture for longer time periods using the microplate inserts and the addition of PDGF-AA and IGF-1 from days 50–60 and T3 from days 60–70 [58] would produce more complex organoids which could then be analysed via whole-organoid live high resolution microscopy.
The ability to perform long-term high-resolution imaging of mature brain organoids, makes this technology more amenable to high-content analyses used for screening cellular responses to therapies, an important process for personalised phenotypic screening for brain cancer and other diseases.
We expect our whole organoid imaging method, along with deep imaging methods such as multiphoton, light sheet and single plane illumination microscopy [59–63] that enable single cell resolution deep into tissues, will significantly improve our understanding of the spatiotemporal relationships between different cell types in development and during disease progression.
Conclusions
This new bioengineering platform based on the use of 3D printed micro-plate inserts that fit standard tissue culture microplates constitutes a significant advance in the culture and analysis of mature brain organoids by enabling long term detailed analysis using high-resolution multimodal inverted fluorescence microscopy to better understand spatiotemporal relationship between cells in the organoid.
Methods
3D printing
3D drawings of microplate inserts were performed in AUTOCAD (Autodesk) and sliced for 3D printing using IdeaMaker (Raise3D) software. 3D printing was performed using a Pro2 Dual Extruder 3D Printer (Raise3D) using polylactic acid (PLA) filaments (Raise3D).
Culture and maintenance of hESC
H9 [31] and H9 Cre-LoxP (H9-GFP, [32] were obtained from WiCell https://www.wicell.org/ and grown in feeder-independent conditions as described in [30]. Briefly, cells were cultured in growth factor reduced Matrigel (Corning, cat#356230)-coated T-25 flasks at 2.25 μg Matrigel/flask using mTeSR1 medium (Stem Cell Technologies, cat# 85870). ROCK inhibitor (Y-27632, Stem Cell Technologies, cat#72304) was used for cells when thawing and splitting only. After cells reached 60–70% confluence (~3 days of culture), cells were detached using EDTA 0.5 mM (Invitrogen, cat#15575020) in sterile D-PBS without calcium and magnesium (Gibco, cat#14200166) and plated again on fresh Matrigel-coated flasks. For the experiments shown here, cells were kept for no longer than ten passages.
Generation of human brain organoids
We used the protocol by Lancaster and Knoblich [30] with some modifications for the use of microplate inserts. Briefly, 70–80% confluent hESC in T-25 flasks were incubated with Accutase (Sigma-Aldrich, cat# A6964) to produce a single-cell suspension in DMEM/F12 medium (Gibco, cat# 11330057) supplemented with Knockout Serum Replacement (Gibco, cat# 10828028), bFGF (4 ng/ml, Preprotech, cat# AF-100-18B) and ROCK inhibitor (100 μM). Cells (18,000 cells per well) were then seeded in a low attachment 96-well plate (Corning, cat#CLS7007), to form EBs. For chimera organoids 18,000 cells of a mixture 100 (H9):1(H9-GFP) was used.
EBs with smooth edges [30] were then transferred to a low-attachment 24-well plate (Corning, cat# CLS3473) to induce the formation of the neuroectoderm by feeding with DMEM/F12 supplemented with N2 (Gibco, cat# 17502048) and 1 μg/μl of Heparin (Sigma-Aldrich, cat#H3149). The neuroepithelial aggregates were fed every day for a week and then processed for embedding into regular Matrigel (Corning, cat#354234) droplets in the presence or absence of microplate inserts.
For conditions using microplate inserts, first, microplate inserts were sterilised by immersion in 80% ethanol followed by 30 min exposure to UV light within a biosafety cabinet. After this, EBs were added on top of the insert tips, by placing the microplate inserts –with the insert tips facing upwards– on a lid of a 24-well glass-bottom plate (CellVis, Cat#P24–1.5H-N). The neuroepithelial aggregates were then placed on the top of the micro-plate inserts using 1000 μL wide-bore pipette tips (Thermo Scientific, cat#2079G), and covered with 20 μL of regular Matrigel. Then, and in an inverted position, the base of the microplate was placed on top of the microplate inserts. To keep the neuroepithelial aggregates in place within the wells of the microplate, matrigel gelation was induced for 20 min at 37 °C in this orientation. After gelation, the whole assembly was inverted, and 400 μL neural induction media was added [30].
For conditions not using microplate inserts (Control), embedding and Matrigel gelation was performed as described in [30] and then Matrigel domes containing neuroepithelial aggregates were transferred to a 24-glass-bottom microplate (CellVis, Cat#P24–1.5H-N). Two days after Matrigel embedding, all cultures were fed with Improved Differentiation Medium without vitamin A [30], supplemented with 3 μM CHIR99021 (Sigma-Aldrich, cat#SML1046). Finally, organoid cultures were cultured in an incubator with constant agitation using an orbital shaker (Thermo Scientific, Cat#88881102).
Cerebral organoid analysis and immunofluorescence staining
Organoids were fixed in 4% paraformaldehyde for 30 min at room temperature, washed with Dulbecco’s phosphate-buffered saline (D-PBS) and dehydrated with sucrose (Sigma-Aldrich, cat#84097) 30% over-night at 4 °C. Individual tissues were frozen by embedding in a solution of 10% sucrose and 7.5% gelatine (Sigma-Aldrich, cat#G1890) and submerged in 2-methylbutane (Sigma-Aldrich, cat#270342) at − 50 °C. The cerebral organoids were cut with using cryostat (Leica, CM1850) generating 20 μm serial sections. After cutting, sections were kept at − 80 °C for long-term storage. For immunostaining, tissues sections were washed with DPBS to remove the remnants of gelatine, then permeabilised with a solution of 0.2% Triton X-100 (Sigma-Aldrich, cat#T9284) for 30 min at room temperature, and blocked with 5% BSA (Sigma-Aldrich, cat#A7030) for 1 h at room temperature. A mix containing different primary antibodies was prepared in blocking buffer (5% BSA in D-PBS). The antibody mixture (100 μL) was added to each glass slide and then covered with Parafilm and incubated overnight at 4 °C. Tissue sections were then washed with D-PBS and incubated with a secondary mix (prepared in 1% BSA) for 1 h at room temperature. Coverslips were mounted on the glass slides containing the tissue sections using ProLong™ Diamond Antifade Mountant (Molecular Probes, cat#P36961) for 1 h at room temperature and imaged on a Leica SP8 STED microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Antibodies
Primary antibodies were Pax6 (Mouse, BD Pharmigen, cat#561462, dilution 1:100), Pax6 (Rabbit, Abcam, cat#ab195045, dilution 1:300), Sox2 (Rabbit, Merck, cat#AB5603, dilution 1:200), TBR1 (Rabbit, Abcam, cat#ab31940, dilution 1:100), TBR2 (Rabbit, Abcam, cat#ab23345, dilution 1:100), TUBB3 (Mouse, BioLegend, cat#801201, dilution 1:300), MAP 2 (Mouse, Merck, cat#MAB3418, dilution 1:50), CTIP2 (Rat, Abcam, cat#ab18465, dilution 1:100) and Cleaved Caspase-3 (Asp175) Antibody (Cat# 9661, Cell Signaling Technology, dilution 1:100). Secondary antibodies were species-specific antibodies conjugated with AlexaFluor 488, 546, 594 or 647 (Invitrogen).
Immunofluorescence imaging
All confocal images of organoid sections were taken using a TCS SP8 STED 3X microscope (Leica) using a HC PL APO CS2 20x/0.75 DRY objective lens at 0.1 μm/pixel. A white light laser (50% power) tuned at 488, 568 and 647 nm laser line using tuneable acoustic-optical beam splitter was used for excitation of GFP, Alexa 568 and Alexa 647 (5–20% intensity was used for excitation). A diode 405 laser was used for Hoechst 33342 (nuclei), with a 405 nm laser line (2% laser transmission was used for excitation). Emission was collected in the range 415–445 nm (Hoescht33342); 500–530 nm (GFP), 580–620 nm (Alexa 568) and 660-700 nm (Alexa 647) using spectral detectors (Leica) and Hybrid sensors (HyD, gain: 100, offset: 0). Z slices (600 nm optical section, 1 AU) were captured, but single optical slices at the plane of ventricular zones were shown (Fig. 3). Images were acquired and processed using Leica LAS X software equipped with Lighting module (Leica).
PD-GSC culture
Freshly resected glioblastoma tumour tissue was obtained from the hospital operating theatre by the South Australian Neurological Tumour Bank and further processed to generate patient-derived glioma stem cells. For this, small pieces of the resected tumour tissue, or a slurry of tissue fragments generated by cavitron ultrasonic surgical aspirator (CUSA), were added to a GentleMACS C-tube (cat#130–093-237). Tissue dissociation was performed using the Miltenyi Human Tumour Dissociation kit (cat# 130–095-929), by adding enzymes to the tissue according to manufacturer’s instructions, placing the C-tube on the GentleMACS dissociator and running the recommended program. The tissue mixture was filtered with a 70 μm cell strainer and the resulting suspension centrifuged (5 min at 250 x g). The cell pellet was washed twice with DMEM (Gibco, cat#11995065) and an aliquot of cells was resuspended in StemPro NSC serum-free medium kit (Life Technologies; cat#A10509–01) supplemented with Glutamax (Gibco, cat#35050061) and transferred to a T-25 Matrigel-coated flasks (coating performed at 1/100 dilution in D-PBS). We used Matrigel-coated flasks for ongoing culture, and Accutase for passaging. Passaging was performed when cells reached 80–100% confluence.
Lentivirus infection of PD-GSC was performed for expression of RFP in PD-GSC. For lentivirus preparation, HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) with lentiviral plasmid CMV-RFP-T2A-Luciferase (System Biosciences) and packaging plasmids pLP1, pLP2 and pVSVG (Invitrogen), according to the manufacturer’s instructions. Viral supernatant was harvested after 96 h and added to primary PD-GSC cells with 2 μg/ml polybrene (Sigma) in StemPro NSC serum-free media (Life Technologies). Successful stable transduction of PD-GSC was confirmed by fluorescence microscopy.
RNA sequencing
Single cell suspensions from organoids were generated by Accutase treatment followed by mechanical dissociation and passage through 70 μm cell strainer. Total RNA was isolated from one organoid per condition using TRIzol Reagent (Invitrogen, cat#15596026) according to the manufacturer’s instructions. Isolated RNA was further purified with Monarch RNA Cleanup Kit (NEB, cat#2040 L). RNA quality was validated in Agilent 2100 Bioanalyzer. All samples had RNA Integrity Number (RIN) > 8. RNA quantity was validated in Qubit 4 Fluorometer (Invitrogen). RNA-seq libraries were generated with KAPA standard RNA-seq HyperPrep kit (Kapa Biosystems) using 300 ng total RNA. Library quality was validated with the Agilent 2100 Bioanalyzer. PolyA+ enriched RNA-seq libraries from 16 human samples were multiplexed and sequenced on two separate runs using the Illumina NextSeq 500 platform and the stranded single end protocol with a read length of 75 bp. Sequenced reads derived from both runs were merged for each sample before further processing. Raw data, averaging 70.5 million reads per sample were analysed and quality checked using the FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were mapped against the human reference genome (hg19) using the STAR spliced alignment algorithm [64] (version 2.5.3a with default parameters and --chimSegmentMin 20, −-quantMode GeneCounts) returning an average unique alignment rate of 90%. The gene counts were TMM normalized using R (version 3.2.3) and edgeR [65] (version 3.3).
For comparison of our RNA-seq gene expression data to the Allen Human Brain Atlas transcriptome data set, we downloaded the gene expression values from FPKM (https://www.brainspan.org/static/download.html). We analysed the data provided for 8 and 9 post-conception weeks (pcw), filtered by the genes expressed in our samples, and then matched through gene symbol. In addition, we extended our correlation analysis with previously published RNA-seq data [33] from 20 and 60 day-old human brain organoids (Spheroid) and engineered cerebral organoids (enCOR), (GEO: GSE80538). Gene expression was compared between conditions using Spearman correlation analysis (MATLAB, see Supplementary Information).
Live organoid imaging
For low resolution imaging, cerebral organoids grown on multi-well plates with different microplate inserts were imaged using the InCell Analyser 2200 imaging system (Cytiva) equipped with a 2X objective, 0.1 NA (Cytiva), a 5.5Mp scientific-grade 16-bit CMOS camera (Cytiva) and In Cell Analyzer Acquisition Software v4.5 28–9630-76UM AE (Cytiva). Images (2048 × 2048 pixels at 3 μm/pixel) were taken every 3.5 days, using 0.1 s exposure for each channel. The differential interference contrast (DIC) imaging modality was used to take brightfield images, while GFP signal was imaged using FITC excitation (475/28 nm) and emission filters (511.5/23 nm) and appropriate dichroic mirrors (QUAD1, Cytiva). Images from the entire plate were analysed using a custom-made Image J script included in the Supplementary Information. This script imports all images of the plate at all different time points into one stack and then uses GFP fluorescence for segmentation of the organoid, which is then used to measure GFP mean fluorescence and organoid size for different wells at each time point. After running the script, the results table was exported to Excel (Microsoft) and then to PRISM (GraphPad) for regression analysis and graphical representation. Data points correspond to mean values for three independent experiments (n = 3), with at least 3 organoids per condition analysed from each experiment.
Confocal images of whole organoid chimeras alone or injected with RFP-expressing PD-GSCs were taken using a TCS SP8 STED 3X microscope (Leica) using a NF 488 (Leica, for GFP) or SMD1 NF 405/470 notch filter (Leica, for RFP/Hoechst) and LAS X (Leica, version 3.5.5.19976) acquisition software under controlled environmental conditions (5% CO2, 37 °C).
For long term imaging, cerebral organoid chimeras grown in multi-well plates on microplate insert #3 were imaged with the TCS SP8 STED 3X microscope (Leica) under controlled environmental conditions (5% CO2, 37 °C). Confocal images of GFP expressing cells were taken every 20 min for 20.3 h with an HC PL APO CS2 20x/0.75 DRY objective lens (1024 × 1024 pixels at 0.568 μm/pixel). A 488 nm (20%) laser line transmission of a white light laser (50% power) was used for excitation. A Hybrid detector (HyD, gain: 100, offset: 0) was used for detection, with a gated range of 500–598 nm. A total of 141 Z slices were captured, each 0.685 μm apart. Of these, slices 66–100 were selected for maximum Z projection (LAS X) and further analysis, with the remaining slices excluded to improve visualisation of individual cellular movements.
To investigate RFP-expressing PD-GSCs within cerebral organoids and analyse the process of tumour cell invasion, channels were imaged sequentially, with a white light laser (50% power) used to excite RFP (PD-GSCs) using a 594 nm laser line (20% intensity), while a diode 405 laser was used for Hoechst 33342 (nuclei), with a 405 nm laser line (2% intensity). Hybrid detectors (HyDs, gain: 100, offset: 0) were used to collect emission within the ranges of 417–546 nm for Hoescht 33342 and 605–690 nm for RFP. Time lapse imaging was performed by acquisition every 30 mins for 17.5 h at 20X magnification and 0.75 zoom with an HC PL APO CS2 20x/0.75 DRY (Leica) objective lens (2048 × 2048 pixels at 0.379 μm/pixel). 63 Z slices 2.409 μm apart (total distance 150 μm) were taken for each time point and maximum Z projections were generated using the Leica LAS X image processing software (Leica).
To generate images of whole organoids, images were taken with an HC PL APO CS2 10x/0.40 DRY (Leica) objective lens using the LAS X Tile Scan acquisition mode. Four tiles (each 2048 × 2048 pixels at 0.757 μm/pixel) completed the total field of view, with 52 Z slices 5 μm apart (total distance 255 μm) taken for each tile. The LAS X software was used to generate maximum Z projections from the raw images for each tile and channel, and the tif files were exported. The single channel images were merged in FIJI, with no manipulation. The resulting 4 tiles were stitched into a single image (3536 × 3540 pixels, 0.757 μm /pixel) using the Image J plugin developed by Preibisch and collaborators [66], with the following settings distinct from default [Tile overlap: 27.7%; Fusion method: Intensity of random input tile; Compute overlap: active; Subpixel accuracy: active; Grid size: 2 × 2] for the most accurate alignment while minimising duplication of nuclei in the overlap region.
For figure preparation, all images were worked on their original resolutions and only altered their size for the purpose of preparation of figures to adjust to a resolution of 300 ppi in Adobe Illustrator.
PD-GSC microinjection in brain organoids
For microinjection of RFP expressing PD-GSC into 30 day-old hESC-derived brain organoids, we used a Microinjection Syringe Pump/SMARTouch Controller (UMP3T-1, World Precision Instruments, USA) equipped with a glass 25 μL syringe and pulled pipette. For this, a suspension of cells was made in DPBS containing Brilliant Blue colorant. We used a 25 μL glass syringe (Hamilton) with a glass pulled pipette and a 30 μm tip (Fivephoton Biochemicals, USA). The cell concentration per injection was 9 × 103 cells/μL (2 × 0.25 μL per organoid) and injections were performed at 3 nL/sec.
Supplementary Information
Additional file 1: Supplementary Figure 1 (related to Fig. 2). Representative sections of cerebral organoids grown on the micro-well inserts and stained with Hoechst 33342 and against cleaved caspase 3. Cleaved caspase 3 is observed in the periphery of cortical structures within organoids, without significant differences across the different experimental conditions (i.e. with or without microplate inserts). Panels were made with cropped images (same size for all time points and conditions). Image’s intensity levels were contracted (same extent for each of the channels across all conditions) from their original 16-bit range to 8-bit RGB for Figure preparation in Adobe Illustrator. Scale Bar = 100 μm.
Additional file 2: Supplementary Movie 1 (related to Fig. 2). Time-course imaging of brain organoids using different microplate inserts. Images were taken every 3.5 days for 50 days.
Additional file 3: Supplementary Movie 2 (related to Fig. 4a). Time course fluorescence microscopy imaging of chimera organoids over 20 h (images taken every 20 min time interval), showing neuronal progenitor cell behaviour.
Additional file 4: Supplementary Movie 3 (related to Fig. 4d-e). Time course fluorescence microscopy imaging of PD-GSC (grey) injected into hESC-derived brain organoids. At 48 h post-injection, Hoechst 33342 was added to the culture media for a 20 min incubation. Following a wash and media change, images were acquired every 30 min for 17.5 h.
Additional file 5: Supplementary Table 1 (Related to Fig. 2). Linear regression results for measurements of organoid growth rates from brain organoids grown using different microplate inserts.
Additional file 6: Supplementary Table 2 (Related to Fig. 3). RNA-seq results from brain organoids grown using different microplate inserts.
Additional file 7: Supplementary Information. This contains information related to 1) Image J script for image segmentation and quantification of different organoid parameters and 2) Matlab script for Spearman correlation coefficient analysis between RNA expression profiles.
Acknowledgments
The authors are grateful to Dr. Madeline Lancaster (MRC Laboratory, UK) for her initial support and advice on this project and providing protocols for brain organoid culture. We thank Dr. Dario Arrua (Future Industries Institute, University of South Australia) and Dr. Cedric Bardy (South Australian Health and Medical Research Institute –SAHMRI-) for encouraging discussions, the SA Brain Neurological Tumour Bank for providing access to patient-derived tissue samples and our laboratory colleagues for their continuous support and fellowship. RNA-seq and imaging experiments were performed at the Australian Cancer Research Foundation (ACRF) Cancer Genomics and Cancer Discovery Accelerator facilities, established with the generous support of the Australian Cancer Research Foundation and Cancer Council SA: Beat Cancer Project.
Abbreviations
- AD
Alzheimer disease
- BSA
Bovine serum albumin
- D-PBS
Dulbecco’s phosphate-buffered saline
- EBs
Embryoid bodies
- ESC
Embryonic stem cells
- enCOR
Engineered cerebral organoids
- GBM
Glioblastoma multiforme
- GSC
Glioma stem cells
- hESC
Human embryonic stem cells
- iPSC
Induced pluripotent stem cells
- NPCs
Neuronal progenitor cells
- PD-GSC
Patient-derived glioma stem cells
- RNA-seq
Bulk RNA sequencing
Authors’ contributions
M.O.M and G.A.G. conceived the project with inputs from C.S-H, V.T., S.P., M.P.B and L.M.E. M.O.M. performed most of the experiments with support from S.L.P., K.G.S., S.L., E.P. and K.S. G.A.G., C.S-H and M.O.M. contributed to microplate insert design. C.S-H and G.A.G. performed 3D printing. G.C. and J.T. contributed to RNA-seq experiments and analysis. M.O.M., K.G.S., S.L.P. and G.A.G. contributed to organoid imaging. M.O.M. and G.A.G. wrote scripts for RNAseq and image analysis. M.N.T. contribute to lentivirus work and preparation of RFP-labelled glioma stem cells. R.J.O., S.M.P., M.N.T. and L.M.E. contributed to bio-specimen collections and preparation of patient-derived glioma stem cells. E.C.F.Y. and K.S. contributed to cleaved caspase immunostaining. All authors contributed to manuscript editing. M.O.M., R.J.O. and G.A.G analysed the data and wrote the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by grants from the National Health and Medical Research Council of Australia [grant numbers 1067405, 1123816 to G.A.G. and 1156693 to S.M.P.]; the Cure Brain Cancer Foundation [to G.A.G., S.M.P., R.J.O., S.P., V.T., and M.P.B.]; the University of South Australia and the MAWA Foundation [to M.O.M. and G.A.G]; the Neurosurgical Research Foundation [to R.J.O., S.P., L.M.E., M.P.B., M.O.M., S.M.P. and G.A.G.]; the Cancer Council SA Beat Cancer Project [to M.P.B. and G.A.G.]; a Hospital Research Package funded by the Beat Cancer Project and Health Services Charitable Gifts Board [to MPB]; the Fay Fuller Foundation (to S.M.P.); the Australian Research Council [FT160100366 to G.A.G.] and a Premier’s Research and Industry Fund grant provided by the South Australian Government Department for Innovation and Skills to V.T. and G.C. S.L.P was supported by a UniSA Research Training Program Scholarship. The funders had no role in study design, data collection and analysis, interpretation of data, decision to publish, or preparation of the manuscript.
Availability of data and materials
Computational scripts for image segmentation and RNA-seq analysis are provided in the Supplementary Information. Other data generated and analysed during this study are included in this published article [and its supplementary information files], except for microscopy image datasets, which are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Brain tumour tissue was obtained from the South Australian Neurological Tumour Bank (SANTB) from patients undergoing resection of their brain tumour at Flinders Medical Centre in Adelaide, South Australia, Australia. Written informed consent was obtained from study participants and consent forms and ethical approval to enable the collection of brain tumour tissues was approved by the Southern Adelaide Clinical Human Research Ethics Committee (#286.10). SANTB specimens were de-identified prior to use in this study and all specimens were linked with non-identifiable, comprehensive clinical information. In vitro and in vivo culture of cells derived from tumour tissues for this project was approved by the Royal Adelaide Hospital and Southern Adelaide Clinical Human Research Ethics Committees (HREC/17/RAH/358 and HREC/18/SAC/16).
Consent for publication
Images presented in the manuscript correspond to de-identified patient-derived stem cells and there are no details on individuals reported within the manuscript.
Competing interests
M.O.M., C.S-H and G.A.G have filed a patent on microplate inserts. Other authors have declared no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mariana Oksdath Mansilla, Email: mariana.om8@gmail.com.
Guillermo A. Gomez, Email: Guillermo.Gomez@unisa.edu.au
References
- 1.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benito-Kwiecinski S, Lancaster MA. Brain organoids: human neurodevelopment in a dish. Cold Spring Harb Perspect Biol. 2019. [DOI] [PMC free article] [PubMed]
- 3.Gomez GA, Oksdath M, Brown MP, Ebert LM. New approaches to model glioblastoma in vitro using brain organoids: implications for precision oncology. Transl Cancer Res. 2019. [DOI] [PMC free article] [PubMed]
- 4.Marton RM, Pasca SP. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2019. [DOI] [PubMed]
- 5.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112(51):15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17(12):3369–3384. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, Chung K, Knoblich JA. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36(10):1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545(7652):54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Goke J, Tan ZY, Saw TY, Tan CP, Lokman H, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19(2):248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, Jarazo J, Walter J, Bruggemann I, Boussaad I, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Reports. 2017;8(5):1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545(7652):48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21(3):383–398 e387. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10(4):537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019. [DOI] [PMC free article] [PubMed]
- 17.Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22(4):669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oksdath M, Perrin SL, Bardy C, Hilder EF, DeForest CA, Arrua RD, Gomez GA. Synthetic scaffolds to control the biochemical, mechanical, and geometrical environment of stem cell-derived brain organoids. APL Bioeng. 2018;2(4). [DOI] [PMC free article] [PubMed]
- 19.Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18(10):573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, Rich JN. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilieva M, Fex Svenningsen A, Thorsen M, Michel TM. Psychiatry in a dish: stem cells and brain organoids modeling autism spectrum disorders. Biol Psychiatry. 2018;83(7):558–568. doi: 10.1016/j.biopsych.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180(1):188–204 e122. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS One. 2016;11(9):e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hebert JM, Vijg J. Cell replacement to reverse brain aging: challenges, pitfalls, and opportunities. Trends Neurosci. 2018;41(5):267–279. doi: 10.1016/j.tins.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig PE, Thankam FG, Patil AA, Chamczuk AJ, Agrawal DK. Brain injury and neural stem cells. Neural Regen Res. 2018;13(1):7–18. doi: 10.4103/1673-5374.224361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y, et al. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26(12):3203–3211 e3205. doi: 10.1016/j.celrep.2019.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi HG, Jeong YH, Kim Y, Choi YJ, Moon HE, Park SH, Kang KS, Bae M, Jang J, Youn H, et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng. 2019;3(7):509–519. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- 28.Aldape K, Brindle KM, Chesler L, Chopra R, Gajjar A, Gilbert MR, Gottardo N, Gutmann DH, Hargrave D, Holland EC, et al. Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16(8):509–520. doi: 10.1038/s41571-019-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrin SL, Samuel MS, Koszyca B, Brown MP, Ebert LM, Oksdath M, Gomez GA. Glioblastoma heterogeneity and the tumour microenvironment: implications for preclinical research and development of new treatments. Biochem Soc Trans. 2019;47(2):625–638. doi: 10.1042/BST20180444. [DOI] [PubMed] [Google Scholar]
- 30.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 32.Du ZW, Hu BY, Ayala M, Sauer B, Zhang SC. Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells. 2009;27(5):1032–1041. doi: 10.1002/stem.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35(7):659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25(1):247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanes DH, Reh TA, Harris WA. Development of the nervous system. 3. Burlington: Academic Press; 2012. [Google Scholar]
- 36.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Curr Opin Neurobiol. 2012;22(5):737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67(6):906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 40.Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 2017;14(1):53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- 41.Chertkova A, Mastop M, Postma M, van Bommel N, van der Niet S, Batenburg K, Joosen L, Gadella TWJ, Okada Y, Goedhart J. bioRxiv. 2017. Robust and bright genetically encoded fluorescent markers for highlighting structures and compartments in mammalian cells. [Google Scholar]
- 42.Darmanis S, Sloan SA, Croote D, Mignardi M, Chernikova S, Samghababi P, Zhang Y, Neff N, Kowarsky M, Caneda C, et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, Watchmaker PB, Yagnik G, Di Lullo E, Malatesta M, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18(1):234. doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller S, Liu SJ, Di Lullo E, Malatesta M, Pollen AA, Nowakowski TJ, Kohanbash G, Aghi M, Kriegstein AR, Lim DA, et al. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol. 2016;12(11):889. doi: 10.15252/msb.20166969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849 e821. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan J, Levitin HM, Frattini V, Bush EC, Boyett DM, Samanamud J, Ceccarelli M, Dovas A, Zanazzi G, Canoll P, et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018;10(1):57. doi: 10.1186/s13073-018-0567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krieger TG, Tirier SM, Park J, Jechow K, Eisemann T, Peterziel H, Angel P, Eils R, Conrad C. Modeling glioblastoma invasion using human brain organoids and single-cell transcriptomics. Neuro-Oncology. 2020. [DOI] [PMC free article] [PubMed]
- 49.Durens M, Nestor J, Williams M, Herold K, Niescier RF, Lunden JW, Phillips AW, Lin YC, Dykxhoorn DM, Nestor MW. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J Neurosci Methods. 2020;335:108627. doi: 10.1016/j.jneumeth.2020.108627. [DOI] [PubMed] [Google Scholar]
- 50.Madoux F, Tanner A, Vessels M, Willetts L, Hou S, Scampavia L, Spicer TP. A 1536-well 3D viability assay to assess the cytotoxic effect of drugs on spheroids. SLAS Discov. 2017;22(5):516–524. doi: 10.1177/2472555216686308. [DOI] [PubMed] [Google Scholar]
- 51.Yin F, Zhu Y, Wang Y, Qin J. Engineering brain organoids to probe impaired neurogenesis induced by cadmium. ACS Biomater Sci Eng. 2018;4(5):1908–1915. doi: 10.1021/acsbiomaterials.8b00160. [DOI] [PubMed] [Google Scholar]
- 52.da Silva B, Mathew RK, Polson ES, Williams J, Wurdak H. Spontaneous glioblastoma spheroid infiltration of early-stage cerebral organoids models brain tumor invasion. SLAS Discov. 2018;23(8):862–868. doi: 10.1177/2472555218764623. [DOI] [PubMed] [Google Scholar]
- 53.Kelava I, Lancaster MA. Stem cell models of human brain development. Cell Stem Cell. 2016;18(6):736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570(7762):523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat Phys. 2018;14(5):515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karzbrun E, Tshuva RY, Reiner O. An on-chip method for long-term growth and real-time imaging of brain organoids. Curr Protoc Cell Biol. 2018;81(1):e62. doi: 10.1002/cpcb.62. [DOI] [PubMed] [Google Scholar]
- 57.Lee SY, Doh I, Nam DH, Lee DW. 3D cell-based high-content screening (HCS) using a micropillar and microwell chip platform. Anal Chem. 2018;90(14):8354–8361. doi: 10.1021/acs.analchem.7b05328. [DOI] [PubMed] [Google Scholar]
- 58.Madhavan M, Nevin ZS, Shick HE, Garrison E, Clarkson-Paredes C, Karl M, Clayton BLL, Factor DC, Allan KC, Barbar L, et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat Methods. 2018;15(9):700–706. doi: 10.1038/s41592-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruns T, Bauer M, Bruns S, Meyer H, Kubin D, Schneckenburger H. Miniaturized modules for light sheet microscopy with low chromatic aberration. J Microsc. 2016;264(3):261–267. doi: 10.1111/jmi.12439. [DOI] [PubMed] [Google Scholar]
- 60.Galland R, Grenci G, Aravind A, Viasnoff V, Studer V, Sibarita JB. 3D high- and super-resolution imaging using single-objective SPIM. Nat Methods. 2015;12(7):641–644. doi: 10.1038/nmeth.3402. [DOI] [PubMed] [Google Scholar]
- 61.Maioli V, Chennell G, Sparks H, Lana T, Kumar S, Carling D, Sardini A, Dunsby C. Time-lapse 3-D measurements of a glucose biosensor in multicellular spheroids by light sheet fluorescence microscopy in commercial 96-well plates. Sci Rep. 2016;6(1):37777. doi: 10.1038/srep37777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponjavic A, Ye Y, Laue E, Lee SF, Klenerman D. Sensitive light-sheet microscopy in multiwell plates using an AFM cantilever. Biomed Opt Express. 2018;9(12):5863–5880. doi: 10.1364/BOE.9.005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rakotoson I, Delhomme B, Djian P, Deeg A, Brunstein M, Seebacher C, Uhl R, Ricard C, Oheim M. Fast 3-D imaging of brain organoids with a new single-objective planar-illumination two-photon microscope. Front Neuroanat. 2019;13:77. doi: 10.3389/fnana.2019.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25(11):1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1 (related to Fig. 2). Representative sections of cerebral organoids grown on the micro-well inserts and stained with Hoechst 33342 and against cleaved caspase 3. Cleaved caspase 3 is observed in the periphery of cortical structures within organoids, without significant differences across the different experimental conditions (i.e. with or without microplate inserts). Panels were made with cropped images (same size for all time points and conditions). Image’s intensity levels were contracted (same extent for each of the channels across all conditions) from their original 16-bit range to 8-bit RGB for Figure preparation in Adobe Illustrator. Scale Bar = 100 μm.
Additional file 2: Supplementary Movie 1 (related to Fig. 2). Time-course imaging of brain organoids using different microplate inserts. Images were taken every 3.5 days for 50 days.
Additional file 3: Supplementary Movie 2 (related to Fig. 4a). Time course fluorescence microscopy imaging of chimera organoids over 20 h (images taken every 20 min time interval), showing neuronal progenitor cell behaviour.
Additional file 4: Supplementary Movie 3 (related to Fig. 4d-e). Time course fluorescence microscopy imaging of PD-GSC (grey) injected into hESC-derived brain organoids. At 48 h post-injection, Hoechst 33342 was added to the culture media for a 20 min incubation. Following a wash and media change, images were acquired every 30 min for 17.5 h.
Additional file 5: Supplementary Table 1 (Related to Fig. 2). Linear regression results for measurements of organoid growth rates from brain organoids grown using different microplate inserts.
Additional file 6: Supplementary Table 2 (Related to Fig. 3). RNA-seq results from brain organoids grown using different microplate inserts.
Additional file 7: Supplementary Information. This contains information related to 1) Image J script for image segmentation and quantification of different organoid parameters and 2) Matlab script for Spearman correlation coefficient analysis between RNA expression profiles.
Data Availability Statement
Computational scripts for image segmentation and RNA-seq analysis are provided in the Supplementary Information. Other data generated and analysed during this study are included in this published article [and its supplementary information files], except for microscopy image datasets, which are available from the corresponding author on reasonable request.