ABSTRACT
Groucho-related genes (GRGs) are transcriptional co-repressors that are crucial for many developmental processes. Several essential pancreatic transcription factors are capable of interacting with GRGs; however, the in vivo role of GRG-mediated transcriptional repression in pancreas development is still not well understood. In this study, we used complex mouse genetics and transcriptomic analyses to determine that GRG3 is essential for β cell development, and in the absence of Grg3 there is compensatory upregulation of Grg4. Grg3/4 double mutant mice have severe dysregulation of the pancreas gene program with ectopic expression of canonical liver genes and Foxa1, a master regulator of the liver program. Neurod1, an essential β cell transcription factor and predicted target of Foxa1, becomes downregulated in Grg3/4 mutants, resulting in reduced β cell proliferation, hyperglycemia, and early lethality. These findings uncover novel functions of GRG-mediated repression during pancreas development.
KEY WORDS: Pancreas development, β cells, Groucho/TLE co-repressor, Foxa1, Mouse
Summary: Pancreatic β cells require Groucho co-repressors during development to repress a master regulator of the liver genetic program, Foxa1, and to allow Neurod1-mediated β cell expansion.
INTRODUCTION
The pancreas is a multifunctional organ comprising exocrine and endocrine tissue. The exocrine tissue represents the majority of the organ and secretes digestive enzymes into the duodenum. The adult endocrine cell compartment is primarily composed of insulin-secreting β cells, glucagon-secreting α cells, somatostatin-secreting δ cells and pancreatic polypeptide-secreting PP cells that are clustered into discrete cellular bundles called the islets of Langerhans. Together, the islet endocrine cells secrete hormones to regulate glucose homeostasis. Dysfunction in this regulation results in diabetes mellitus, an increasingly prevalent disease worldwide characterized by hyperglycemia due to the inability to adequately respond to or secrete endogenous insulin (Cernea and Dobreanu, 2013; American Diabetes, 2020). Understanding the processes underlying normal pancreas development can inform mechanisms of dysfunction in diabetes and provide insights into developing β cell replacement therapies.
Pancreas organogenesis in the developing mouse begins between embryonic days (e) 8.5-9.0 when pancreatic duodenal homeobox (PDX1)+ cells are specified from the foregut endoderm (Jorgensen et al., 2007). All functional cells of the adult pancreas are derived from a PDX1+ multipotent progenitor population. As pancreas development progresses, a distinct neurogenin 3 (NEUROG3)+ endocrine progenitor population arises, from which all pancreatic endocrine cells will be specified. Following endocrine specification, the maturation, expansion and organization of islet cells occurs throughout the rest of development and postnatally, resulting in the functional adult endocrine pancreas (Pan and Wright, 2011).
Development of the pancreas is controlled by a complex network of transcription factors that regulate gene expression with cellular and temporal specificity (Dassaye et al., 2016). Each of these factors has a unique repertoire of target genes that they can activate or repress. Often, the regulatory function of transcription factors relies on interactions with co-activators or co-repressors. One family of such co-repressors are encoded by the Groucho-related [GRG; also known as transducer-like enhancer of split (TLE)] genes. groucho was originally discovered in Drosophila and is conserved in mammals, which have four full-length GRG genes (Paroush, 1994; Buscarlet and Stifani, 2007; Turki-Judeh and Courey, 2012; Agarwal et al., 2015). There are also two truncated family members – GRG5 (the mouse ortholog of human AES; also known as TLE5) and GRG6 (TLE6) – that are thought to inhibit the function of full-length GRG proteins (Marcal et al., 2005; Zhang et al., 2008). GRG proteins are unable to bind DNA directly; instead, they are recruited to the genome by interactions with specific peptide motifs found in an array of transcription factors. The repressive function of GRGs is due in part to their ability to recruit histone deacetylases (HDACs) and the Polycomb repressive complex (PRC), which condense chromatin and prevent transcription of target genes (Chen et al., 1999; Papizan et al., 2011; Patel et al., 2012; Turki-Judeh and Courey, 2012).
GRGs are involved in many aspects of mammalian development and cellular functions, with each GRG having a unique tissue expression profile. Previous studies have identified crucial roles for GRGs in neurodevelopment, osteogenesis, hematopoiesis, adipogenesis and kidney development (Muhr et al., 2001; Cai et al., 2003; Villanueva et al., 2011; Wheat et al., 2014). Additionally, a study of embryonic pancreas explants from Grg3 (Tle3) null mice demonstrated that endocrine cells failed to delaminate from the trunk epithelium and displayed defective differentiation ex vivo (Metzger et al., 2012). In an in vitro β cell model, GRG3 repressed expression of Arx and glucagon to promote monohormonal β cell identity (Metzger et al., 2014). Together, these studies suggest there is a crucial role for GRG3 during pancreas development; however, the analyses were limited by short time survival of the explants and caveats associated with β cell lines.
There are several known interacting partners of GRGs that have been characterized in different tissue contexts, including members of the HES, FOXA, NKX and PAX families of transcription factors, which also represent essential factors that regulate pancreas development (Eberhard et al., 2000; Muhr et al., 2001; Yao et al., 2001; Milili et al., 2002; Iype et al., 2004; Linderson et al., 2004; Sekiya and Zaret, 2007; Jangal et al., 2014). For example, HES1 promotes differentiation of the non-endocrine pancreas progenitor population, and FOXA1 and FOXA2 are expressed in the foregut endoderm from which the pancreas and liver are derived (Ang et al., 1993; Monaghan et al., 1993; Jensen et al., 2000; Kaestner, 2010). Whereas Foxa1 transcript is primarily associated with the liver lineage and cannot be detected in early stage pancreatic tissue (Krentz et al., 2018; Li et al., 2018), Foxa2 remains expressed in the pancreas lineage and FOXA2 induces early expression of PDX1 (Gao et al., 2008). Later in development, FOXA2 promotes endocrine differentiation and β cell function (Gao et al., 2010). Furthermore, PAX4 and PAX6 regulate the β/δ and β/α cell fates, respectively, and NKX2-2 and NKX6-1 are necessary for endocrine cell differentiation and maintenance of β cell identity (Sander et al., 1997; Sosa-Pineda et al., 1997; St-Onge et al., 1997; Sussel et al., 1998; Collombat et al., 2003, 2005; Henseleit et al., 2005; Taylor et al., 2013; Gutierrez et al., 2017). In the context of pancreas development, we have previously shown that the interaction between NKX2-2 and GRG3 is necessary to prevent β-to-α cell reprogramming by repressing the α cell master regulatory gene, Arx (Papizan et al., 2011).
To characterize the role of GRGs in pancreas development more comprehensively, we determined that Grg2 (Tle2) and Grg3 were the two family members predominantly expressed during pancreas development, which corresponds with previously published expression data of the developing pancreatic epithelium (Hoffman et al., 2008). Surprisingly, mice with global deletion of Grg2 had no discernable phenotype. Furthermore, pancreas-specific deletion of Grg3 or a combination of Grg2 and Grg3 had surprisingly mild phenotypes. Pancreas-specific deletion of Grg3 in vivo also did not exhibit many of the previously reported phenotypes seen in pancreatic explants and cell culture models (Metzger et al., 2012, 2014). Remarkably, expression analysis revealed that in the absence of Grg3, Grg4 (Tle4), the family member that is normally not expressed in the pancreas, is highly upregulated. Combined pancreatic deletion of Grg3 and Grg4 resulted in an exacerbated pancreas developmental phenotype and had significantly reduced numbers of β cells embryonically and perinatally, demonstrating that Grg4 upregulation can partially compensate for loss of Grg3. At the molecular level, we determined that GRG activity was essential for repressing Foxa1 expression. In the absence of Grg3 and Grg4, Foxa1 becomes erroneously expressed, resulting in downregulation of Neurod1 and causing defects in β cell expansion, a phenotype similar to that seen in mice lacking Neurod1 in the pancreas and β cells (Naya et al., 1997; Romer et al., 2019). Taken together, these findings provide new insights into the necessity of GRG transcriptional co-factors in pancreas organogenesis.
RESULTS
Grg3 is highly expressed in the embryonic pancreas and adult islets
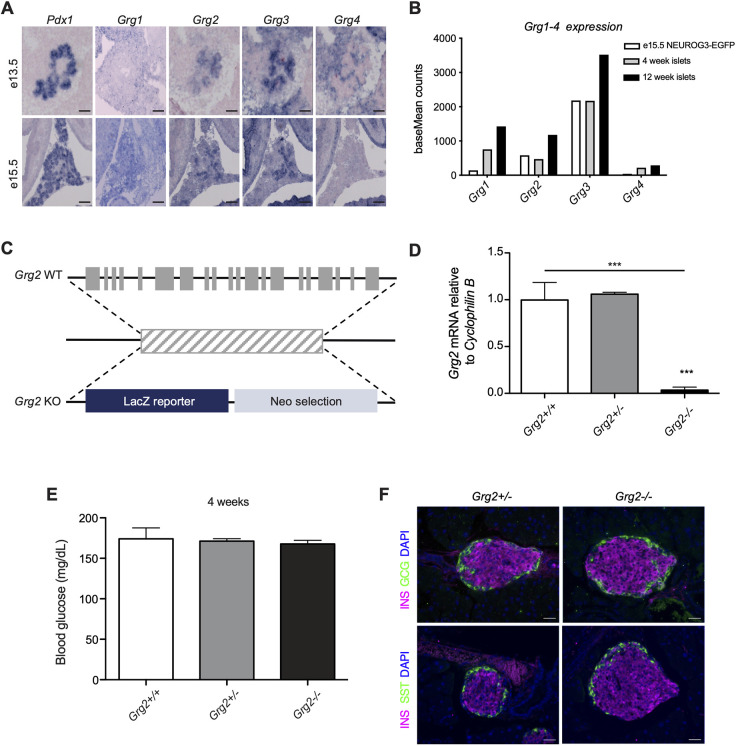
Each member of the family of mammalian GRGs has a unique tissue-expression profile (Agarwal et al., 2015). To determine which GRGs are expressed in the pancreas in the absence of appropriate antibodies specific for each of the GRG proteins, we performed RNA in situ hybridization on embryonic tissue from wild-type mice, using probes for Grg1-4 as well as Pdx1 to delineate the developing pancreas (Fig. 1A). At both e13.5 and e15.5, epithelial Grg1 expression could not be detected above background signal; however, Grg2 and Grg3 expression extensively overlapped with Pdx1 (Fig. 1A). Grg4 expression could be detected in the pancreatic mesenchyme at e13.5 but was no longer present at e15.5 (Fig. 1A). Because expression of the different GRG family members differed from previously published data (Hoffman et al., 2008), we also analyzed existing RNA-sequencing (RNA-seq) data for GRG gene expression. In sorted NEUROG3+ (NEUROG3-EGFP) endocrine progenitors from e15.5 pancreata, we confirmed expression of Grg2 and Grg3 in the pancreatic endocrine progenitor compartment, with Grg3 being the most highly expressed Groucho family member (Fig. 1B, white bars). RNA-seq data from islets isolated from 4- and 12-week-old mice show that Grg3 levels remained high in the endocrine population through adulthood (Fig. 1B), whereas Grg1 and Grg2 were both upregulated in adult islets (Gutierrez et al., 2017; Romer et al., 2019). Several published single-cell RNA-seq datasets also confirm these expression patterns (Scavuzzo et al., 2018; Tabula Muris Consortium, 2018). Based on these results, we focused our analyses of embryonic pancreas-specific GRG function on Grg2 and Grg3.
Fig. 1.
Grg3 is highly expressed in the embryonic pancreas and adult islets. (A) Grg2 and Grg3 expression overlaps with pancreatic Pdx1 expression at e13.5 and e15.5. Grg4 is expressed in the mesenchyme surrounding the pancreas at e13.5. Scale bars: 100 μm. (B) baseMean counts from previously published RNA-seq experiments. Grg3 is highly expressed in sorted NEUROG3+ endocrine progenitor cells at e15.5 and remains expressed in isolated adult islets at 4 weeks and 12 weeks. (C) Schematic of Grg2 knockout (KO) allele. (D) RT-qPCR validation of Grg2 knockout showing loss of transcript in P0 Grg2−/− homozygous pancreata. n=4-7 for all genotypes. ***P≤0.0001; two-tailed Student's t-test. (E) Ad libitum blood glucose measurements of 4-week-old Grg2−/− mice. n=4-6 for all genotypes. (F) Immunofluorescence analysis of 6-week-old Grg2 heterozygous and homozygous mutants showing no visible islet phenotype. Scale bars: 25 μm.
Pancreas-specific loss of Grg3 leads to hyperglycemia and upregulation of Grg4
Because genetic disruption of Grg2 has not been previously performed in mice, we generated mice carrying a null mutation of Grg2 using a Grg2/Tle2tm1(KOMP)Vlcg embryonic stem cell line (Fig. 1C,D). Surprisingly, given the severe phenotypes associated with global knockout of Grg3 (embryonic lethality due to placental defects) or Grg4 (lethality around 4 weeks of age due to hematopoietic and bone development defects), the Grg2 homozygous null mice lived into adulthood with no apparent defects (Villanueva et al., 2011; Gasperowicz et al., 2013; Wheat et al., 2014). These mice were euglycemic and did not appear to have any overt pancreatic islet phenotypes (Fig. 1E,F).
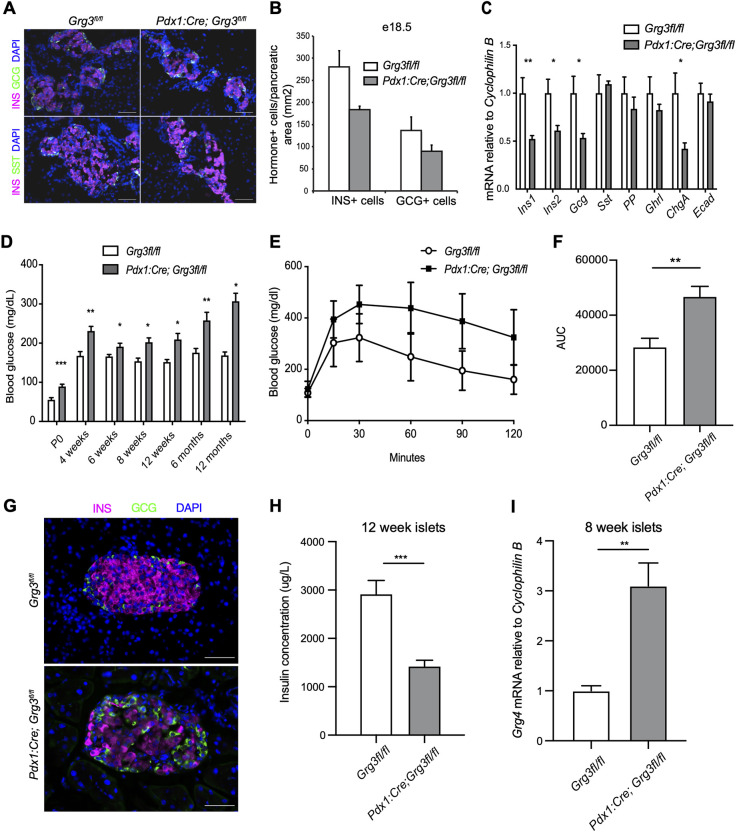
Owing to the embryonic lethality associated with a global deletion of Grg3 (Gasperowicz et al., 2013), we generated pancreas-specific Grg3 mutants using Pdx1:Cre to drive recombination of a floxed Grg3 allele (Pdx1:Cre; Grg3fl/fl) (Hingorani et al., 2003; Villanueva et al., 2011) (Fig. S1A,B). Previous analysis of embryonic pancreas explants derived from Grg3−/− mice suggested that Grg3 was essential for the appropriate differentiation of all pancreatic endocrine cells (Metzger et al., 2012). Based on these previous findings, we anticipated that the Pdx1:Cre; Grg3fl/fl mice would be neonatal lethal, and therefore assessed the phenotype of Pdx1:Cre; Grg3fl/fl embryos just prior to birth. Surprisingly, at e18.5, the Pdx1:Cre; Grg3fl/fl islets appeared remarkably normal, exhibiting only a trend in decreased β cell numbers (Fig. 2A,B). Consistent with the immunofluorescence data, there was a small, but significant, decrease in Ins1, Ins2 and Gcg mRNA expression in e18.5 pancreata (Fig. 2C). There were no apparent defects in the number of non-β endocrine cells or levels of the other endocrine hormones at this stage (Fig. 2A-C), although we were able to detect a small number of insulin (INS) and somatostatin (SST) co-expressing cells in adult islets (Fig. S1C).
Fig. 2.
Pancreas-specific loss of Grg3 results in hyperglycemia, impaired glucose tolerance and upregulation of Grg4. (A) Immunofluorescence analysis of islet hormones insulin (INS) and glucagon (GCG) in e18.5 pancreata from control and Pdx1:Cre; Grg3fl/fl embryos. Scale bars: 50 μm. (B) Quantification of hormone+ cells from the experiment shown in A. n=3 for each genotype. (C) RT-qPCR analysis of islet hormones, chromogranin A (ChgA) and E-cadherin (Ecad) in e18.5 whole pancreata. n=3-4 for each genotype. *P≤0.05, **P≤0.01; Student's two-tailed t-test. (D) Conditional loss of Grg3 in the pancreas results in elevated ad libitum blood glucose levels at all time points between P0 and 12 months. n=3-10 for each time point. *P≤0.05, **P≤0.01, ***P≤0.001; two-tailed Student's t-test. (E) Pdx1:Cre; Grg3fl/fl mice are glucose intolerant at 6 weeks of age. n=13-19 for each genotype. (F) Area under the curve (AUC) calculations for the data shown in E. **P≤0.01; two-tailed Student's t-test. (G) Immunofluorescence staining of INS and GCG of 6-week-old islets show fewer INS+ β cells in Pdx1:Cre; Grg3fl/fl islet. Scale bars: 50 μm. (H) Intracellular insulin content of isolated islets from 12-week-old mice as measured with an insulin ELISA assay. n=3 for each genotype. ***P≤0.001; two-tailed Student's t-test. (I) RT-qPCR analysis of Grg4 mRNA expression in isolated islets from control and Pdx1:Cre; Grg3fl/fl 8-week-old animals. n=3.**P≤0.01; Student's two-tailed t-test.
Previous work suggested that the defect in endocrine cell differentiation was due to defects in the suppression of E-cadherin (cadherin 1) (Metzger et al., 2012). However, consistent with our observation that there were no general defects in the formation of the non-β endocrine cell lineages at e16.5, nor any observable delamination defects, we could not detect increased expression of E-cadherin mRNA in the Pdx1:Cre; Grg3fl/fl mutants (Fig. 2C, Fig. S1D).
In light of the subtle defects observed in the Pdx1:Cre; Grg3fl/fl embryos, we next assessed the phenotype of postnatal mutant animals. Pdx1:Cre; Grg3fl/fl mice were born at normal Mendelian ratios; however, they were hyperglycemic compared with control littermates at all time points tested (Fig. 2D). At 6 weeks of age, Pdx1:Cre; Grg3fl/fl mice displayed impaired glucose clearance (Fig. 2E,F). Consistent with this, immunofluorescent analysis of Pdx1:Cre; Grg3fl/fl islets isolated from 6-week-old mice revealed a discernable decrease in the number of β cells, with no apparent effect on the other endocrine cell types (Fig. 2G). Consistent with the decreased number of β cells, lower levels of Ins1 and Ins2 genes, and impaired glucose clearance, islets isolated from 12-week-old Pdx1:Cre; Grg3fl/fl mice also had decreased intracellular insulin content (Fig. 2H).
Overall, the observed Pdx1:Cre; Grg3fl/fl phenotype was not as severe as expected based on the previous knockout studies and the known functional interactions of GRG3 with NKX2-2 and other essential islet transcription factors (Buscarlet and Stifani, 2007; Jennings and Ish-Horowicz, 2008; Papizan et al., 2011; Metzger et al., 2012; Agarwal et al., 2015). The embryonic expression pattern of Grg2 (Fig. 1A,B) suggested a possible functional redundancy between Grg2 and Grg3 that might compensate for the lack of Grg3 expression in the pancreas. To test this, we intercrossed the Grg2−/− and Pdx1:Cre; Grg3fl/fl mice to generate Grg2/3 double-mutant mice; however, there was no worsening of the Grg3 single mutant phenotype in the Grg2/3 double-mutant mice (Fig. S2). Although Grg1 and Grg4 are not normally expressed in the developing endocrine pancreas, it was also possible that one or both of these family members became upregulated in response to deletion of Grg3; indeed, we observed a 3-fold increase in Grg4 expression in isolated Pdx1:Cre; Grg3fl/fl islets (Fig. 2I), whereas Grg1 was not significantly altered. This discovery led us to hypothesize that Grg4 upregulation was compensating for the loss of Grg3 and preventing a more severe pancreas phenotype.
Grg3/4 double mutants are extremely hyperglycemic with a reduction in β cells
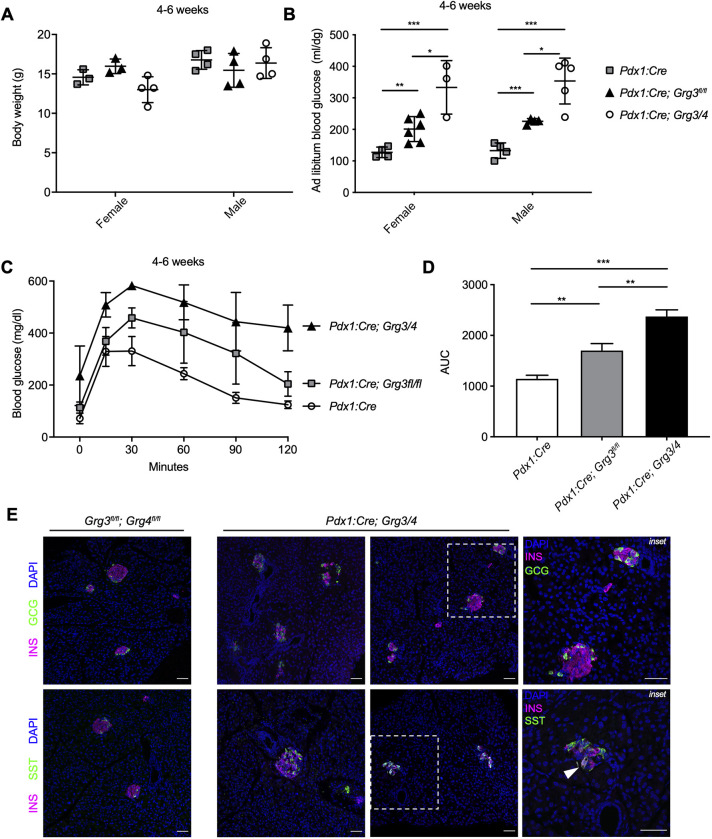
To determine whether the upregulation of Grg4 functionally compensates for the loss of Grg3 in the Pdx1:Cre; Grg3fl/fl mutants, we next generated Grg3; Grg4 double knockout mice. Because the Grg4 null mutation results in lethality around 4 weeks of age owing to bone and hematopoiesis defects, we obtained a floxed allele of Grg4 and generated pancreas-specific Grg3/Grg4 mutant mice (Pdx1:Cre; Grg3/4) (Wheat et al., 2014). Unlike the single Pdx1:Cre; Grg3fl/fl mutants, we observed fewer than expected double knockout mice at weaning (12% versus 25%, χ2 test, 10 litters, n=96, P=0.0193), suggesting that simultaneous deletion of Grg3 and Grg4 caused early postnatal lethality and confirming that upregulation of Grg4 could partially compensate for lack of Grg3. The small number of double mutant mice that did survive to weaning, likely as a result of inefficient deletion of both Grg3 and Grg4 alleles, had normal body weight compared with Pdx1:Cre controls and Pdx1:Cre; Grg3fl/fl single mutants (Fig. 3A). However, double Grg3/4 mutants displayed more severe hyperglycemia and glucose intolerance phenotypes compared with Pdx1:Cre; Grg3fl/fl mice (Fig. 3B-D). Along with having disorganized islet morphology, we also observed a small number of INS and SST co-expressing cells, similar to observations in Pdx1:Cre; Grg3fl/fl mice (Fig. 3E). INS+/GCG+ cells were not observed in either the single or double knockout animals.
Fig. 3.
Pdx1:Cre; Grg3/4 mutants that survive to weaning age are extremely hyperglycemic with fewer β cells and bihormonal INS+/SST+ cells. (A) Body weight measurements of control and Pdx1:Cre; Grg3/4 young adults show no difference between genotypes. n=3-4 for each genotype. (B) Ad libitum blood glucose measurements of 4- to 6-week-old mice showing hyperglycemia in Pdx1:Cre; Grg3/4 double mutants compared with both Pdx1:Cre control and Pdx1:Cre; Grg3fl/fl animals. n=3-6 for each genotype.*P≤0.05; **P≤0.01; ***P≤0.0001; two-tailed Student's t-test. (C) Glucose tolerance tests of 4- to 6-week-old Pdx1:Cre control mice and Pdx1:Cre;Grg3 and Pdx1:Cre;Grg3/4 mutants. n=3-5 for each genotype. (D) AUC of the data shown in C showing increased AUC of Grg3/4 double mutants compared with Pdx1:Cre control and Grg3 single mutants. **P≤0.01, ***P≤0.001; two-tailed Student's t-test. (E) Immunofluorescence staining of islets from 4-week-old control and Pdx1:Cre; Grg3/4 mutant mice showing a visible decrease in INS+ β cells and disorganized islet morphology. Right-hand panels are magnifications of the boxed areas showing examples of INS+SST+ bihormonal cells in Pdx1:Cre; Grg3/4 mutant. Arrowhead indicates INS+SST+ bihormonal cell. Scale bars: 50 μm.
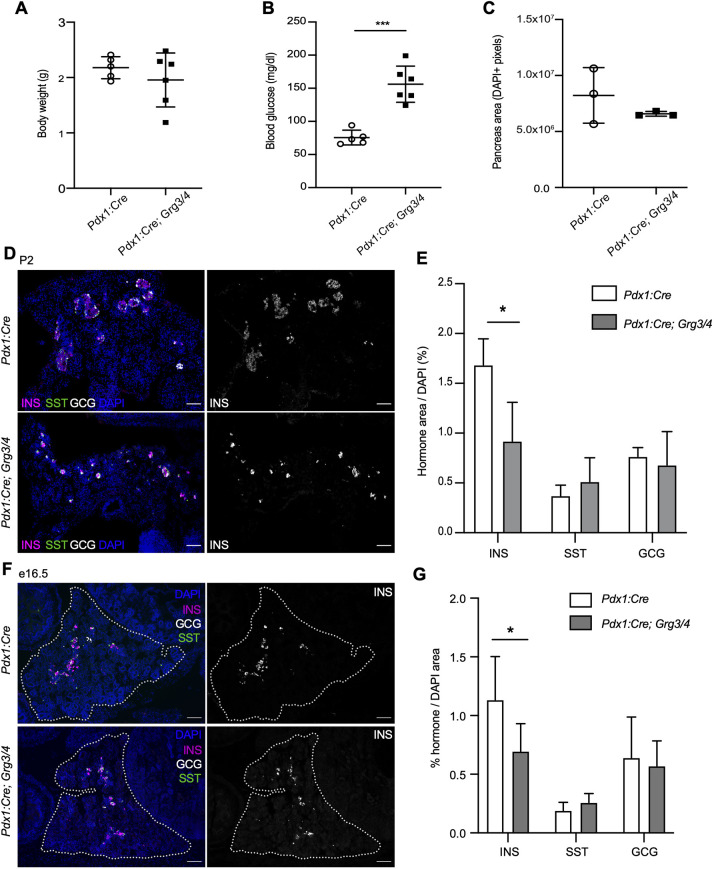
Because of the loss of most Pdx1:Cre; Grg3/4 double knockout mice by weaning, we wanted to assess whether mice lacking both Grg3 and Grg4 alleles survived through gestation. We analyzed mice at postnatal day (P) 2 and found that Pdx1:Cre; Grg3/4 double mutants were born at normal Mendelian ratios. Mutants showed no difference in body weight or general pancreas area, but already displayed elevated blood glucose levels as neonates (Fig. 4A-C), suggesting a possible defect in β cell development. Immunofluorescence analysis of islet hormone expression identified a significant reduction in the number of INS+ cells, without apparent alterations in SST+ or GCG+ cells, indicating that the loss of Grg3 and Grg4 only affected development of the β cell lineage, although to a more severe extent than loss of Grg3 alone (Fig. 4D,E).
Fig. 4.
Loss of Grg3 and Grg4 leads to perinatal hyperglycemia and a decrease in β cell area at P2 and e16.5. (A) Pancreas-specific knockout of Grg3 and Grg4 (Pdx1:Cre; Grg3/4) does not result in body weight changes in P2 mice. n=6. (B) Pdx1:Cre; Grg3/4 mice have elevated blood glucose levels at P2 compared with Cre-only controls. n=6. ***P≤0.0001; two-tailed Student's t-test. (C) Pancreas area as measured by DAPI immunofluorescence is unchanged in Pdx1:Cre; Grg3/4 mice at P2. n=3. Representative images are shown in D. (D) Immunofluorescence staining of P2 pancreata for INS, SST and GCG. Scale bars: 100 μm. (E) Quantification of hormone area relative to pancreas area (DAPI). Pdx1:Cre; Grg3/4 mice have decreased INS+ area with no changes in SST or GCG area. n=3-4 for each genotype. *P≤0.05; two-tailed Student's t-test. (F) Immunofluorescence staining of e16.5 pancreata for INS, SST and GCG. Dotted line delineates the pancreas. Scale bars: 50 μm. (G) Quantification of hormone+ area relative to pancreas (DAPI) area as shown in F. n=6 for each genotype. *P≤0.05; two-tailed Student's t-test.
To determine whether the decrease in β cell numbers was due to a defect in the formation of β cells during embryogenesis, we quantified the islet hormone-positive areas of e16.5 Pdx1:Cre; Grg3/4 double knockout embryos. We found that although INS+ β cells were specified by this stage, the overall INS+ area was significantly smaller in the double mutant embryos, with no apparent changes in GCG- or SST-expressing cells (Fig. 4F,G). This did not appear to be associated with a decrease in the endocrine progenitor pool as we did not observe reduced numbers of NEUROG3-expressing endocrine progenitor cells (Fig. S3), consistent with previous reports that Grg3 is expressed downstream of Neurog3 (Metzger et al., 2012).
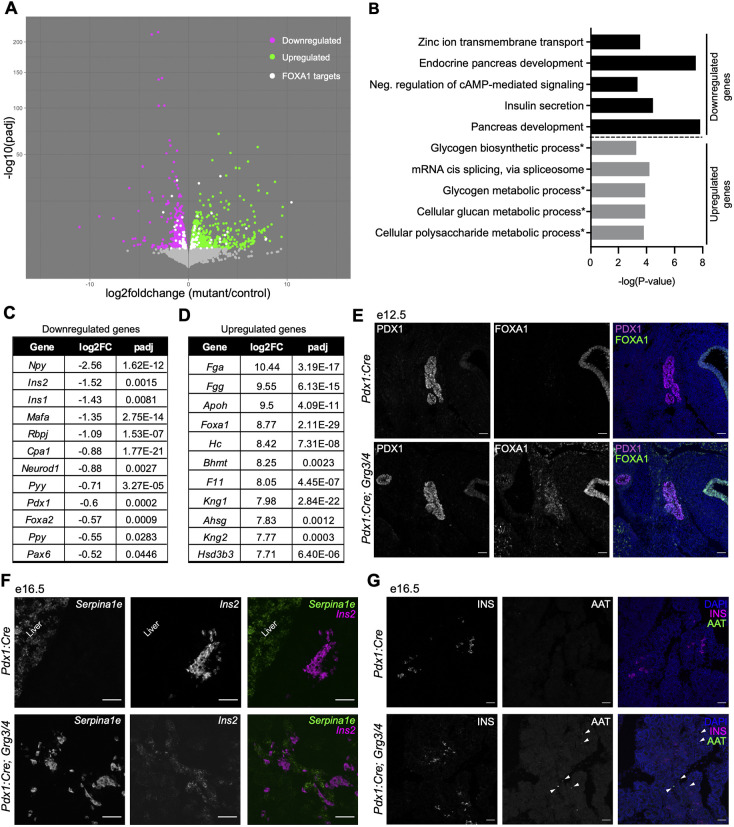
Pancreas genes are downregulated in Grg3/4 mutant pancreata with ectopic expression of liver genes and the master liver regulator Foxa1
Our data revealed that β cells were being specified from the endocrine progenitor population, but there was decreased β cell mass at e16.5 and P2. Although the reduction in β cell numbers was significant, it was not sufficient to explain the resulting in perinatal hyperglycemia (Fig. 4). This led us to hypothesize that the remaining β cells were also dysfunctional due to developmental and/or maturation defects. To understand what was occurring in Pdx1:Cre; Grg3/4 mutant pancreata after β cell formation but prior to birth (to avoid the many changes associated with the transition to milk-feeding), we performed transcriptomic analyses on e18.5 pancreata from Pdx1:Cre; Grg3fl/fl, Pdx1:Cre; Grg3/4 and Pdx1:Cre control embryos. DESeq2 analysis between the single mutant and control pancreata identified 1181 significantly differentially expressed genes, whereas double mutant and control pancreata had 1973 significant differentially expressed genes with 1007 upregulated and 966 downregulated genes (Fig. S4, Fig. 5A). Similar gene expression changes were observed in both the single and double mutant datasets, although the levels of gene expression changes were more severe in the Pdx1:Cre; Grg3/4 double-mutant mice (Fig. S4).
Fig. 5.
Pancreas genes are downregulated with ectopic expression of FOXA1 and liver genes in Grg3/4 mutants. (A) Volcano plot showing differentially expressed genes in e18.5 pancreata from Pdx1:Cre controls versus Pdx1:Cre; Grg3/4 mutants using DESeq2 analysis. Downregulated genes are indicated by magenta dots and upregulated genes are indicated by green dots. White dots represent predicted Foxa1 target genes that are differentially expressed in the dataset. Padj cutoff=0.05. n=3 for each genotype. (B) Gene ontology (GO) term analysis of up- and downregulated genes. Asterisks indicate pathways associated with liver metabolism. (C) Select list of downregulated genes related to pancreas development and hormone expression. (D) Top 10 upregulated genes sorted by fold change. (E) Immunofluorescence staining of e12.5 pancreata showing overlap of PDX1 and FOXA1 protein in Pdx1:Cre; Grg3/4 mutants. Scale bars: 50 μm. (F) Validation of the upregulated liver gene Serpina1e using RNAscope in e16.5 Pdx1:Cre; Grg3/4 pancreas. Scale bars: 50 μm. (G) Immunofluorescence staining in e16.5 mice of alpha1 antitrypsin (AAT, encoded by Serpina1a-e genes) shows that upregulation of mRNA does not correlate with protein expression levels. White arrowheads indicate AAT cells in Pdx1:Cre; Grg3/4 pancreas. Scale bars: 50 μm.
Using PANTHER gene ontology (GO) software on the list of significantly downregulated genes, we found multiple pancreatic processes to be perturbed in the Pdx1:Cre; Grg3/4 mutant pancreata (Fig. 5B, top section). Insulin secretion, zinc ion transmembrane transport and cAMP-mediated signaling, both essential components involved in insulin secretion, were among the top 5 GO terms identified. Genes encoding multiple islet hormones were also downregulated, including Ins1, Ins2, Npy, Pyy and Ppy (Fig. 5C), whereas Gcg and Sst were unchanged, consistent with our immunofluorescence analysis (Fig. 4D-G). In addition to GO terms associated with islet function, downregulated genes were also significantly associated with pancreas and endocrine pancreas development (Fig. 5B, top section), including Foxa2, Pdx1, Neurod1 and Pax6 (Fig. 5C). These gene expression changes could be explained by the decreased insulin area observed in e16.5 and P2 mutant pancreata (Fig. 4D-G), but could also be attributed to alterations in β cell function. Although the acinar and ductal tissues in Pdx1:Cre; Grg3/4 mutants appeared normal, a small subset of acinar and ductal genes were downregulated, suggesting that loss of Grg3/4 could have subtle effects on exocrine functions (Fig. S5).
GRGs act primarily as transcriptional co-repressors, suggesting that these downregulated genes are likely an indirect consequence of defective upstream GRG-mediated gene repression. To identify genes that were more likely to be directly linked to loss of GRG function, we focused on genes that were significantly upregulated in Pdx1:Cre; Grg3/4 mutant pancreata. Surprisingly, GO analysis of these genes revealed enrichment of metabolic processes normally found in the liver, including glycogen biosynthesis and metabolism, glucan metabolism, and polysaccharide metabolism (Fig. 5B, bottom section). Consistent with this, most of the genes with the highest fold changes were canonical liver genes (Fig. 5D), including the transcription factor Foxa1, which acts with Foxa2 as major regulators of hepatocyte differentiation (Lee et al., 2005).
Foxa1 is expressed in the early foregut endoderm from where the pancreas and liver are both derived. During normal development, Foxa1 expression diminishes in the pancreas and becomes restricted to the liver lineage (Li et al., 2018; Scavuzzo et al., 2018). In e18.5 Pdx1:Cre; Grg3/4 mutants, Foxa1 RNA expression was increased 8.7-fold (Fig. 5D). To determine whether ectopic FOXA1 could also be detected at the protein level, we used immunofluorescence to assess FOXA1 protein expression at e12.5, P2 and 6 weeks of age. Consistent with increased expression at the RNA level, FOXA1 protein could be detected in the pancreas of Pdx1:Cre; Grg3/4 mutants at all ages tested (Fig. 5E, Fig. S6). In addition to FOXA1 protein expression, predicted Foxa1 target genes were also significantly dysregulated in the Pdx1:Cre; Grg3/4 mutant pancreata. Of the significantly differentially expressed genes, we identified 102 predicted Foxa1 targets, of which 53 were upregulated and 49 were downregulated, in agreement with Foxa1 being both an activator and repressor of gene expression (Fig. 5A, white dots; Table S1).
In addition to confirming the ectopic expression of FOXA1, we also sought to confirm expression of the liver genes identified by RNA-seq. Although we were able to detect robust expression of a candidate liver gene Serpina1e mRNA (Fig. 5F), expression of SERPINA1A-E protein (also known as alpha1 antitryspin, AAT), could only be detected in very few cells in the Pdx1:Cre; Grg3/4 mutant pancreata at e16.5 (Fig. 5G). This suggests that although liver genes were upregulated, they could not be translated. Dysregulation of the liver gene expression program did not influence pancreas patterning or overall pancreas morphology (Fig. S4B).
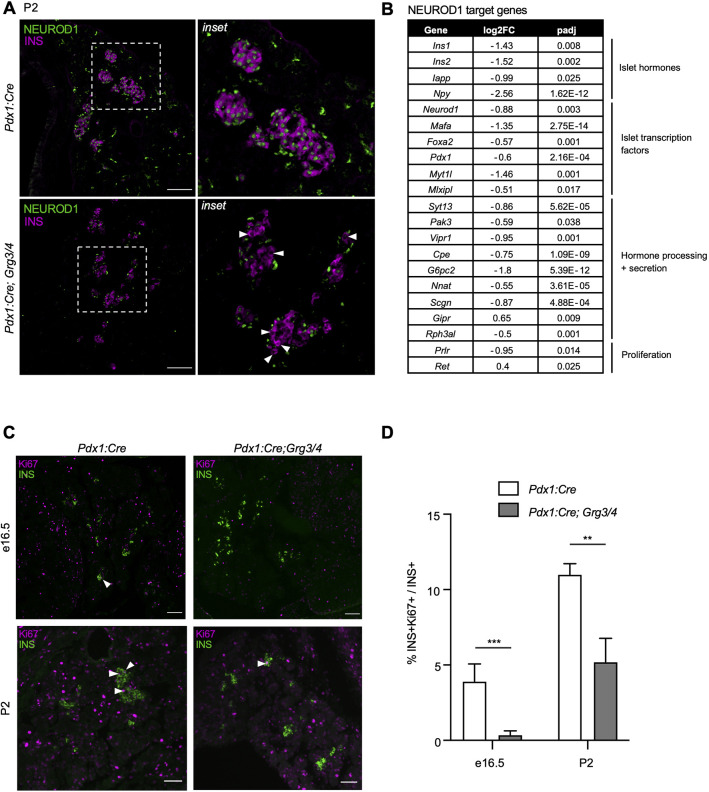
Neurod1 expression is lost in Grg3/4 mutant β cells with a reduction in β cell proliferation
One of the predicted Foxa1 targets that was downregulated in Pdx1:Cre; Grg3/4 mutant pancreata was Neurod1. NEUROD1 is a transcription factor required for β cell proliferation, survival and maturation (Naya et al., 1997; Gu et al., 2010; Romer et al., 2019). Interestingly, Pdx1:Cre; Grg3/4 mutant β cells lacked expression of NEUROD1 (Fig. 6A, white arrowheads), whereas all β cells in control pancreata co-stained for insulin and NEUROD1. INS−/NEUROD1+ cells likely represent non- β endocrine cells as previously described (Chao et al., 2007). Additionally, we found that many of the direct Neurod1 target genes were significantly downregulated in Pdx1:Cre; Grg3/4 mutant pancreata (Romer et al., 2019) (Fig. 6B). Neurod1−/− mice fail to form pancreatic islets and have a significant reduction in β cell number perinatally, and β cell-specific loss of Neurod1 results in the failure of β cell expansion by proliferation (Naya et al., 1997; Gu et al., 2010; Mastracci et al., 2013a; Romer et al., 2019). To determine whether Pdx1:Cre; Grg3/4 mutants phenocopy Neurod1−/− mice, we assessed β cell proliferation and apoptosis. In both e16.5 and P2 pancreata, the percentage of proliferating Ki67+ β cells was significantly decreased in Pdx1:Cre; Grg3/4 mutants compared with control pancreata (Fig. 6C,D), whereas, by TUNEL staining, we observed no β cell apoptosis in either control or mutant pancreata (data not shown). These findings suggest that the decrease in β cells in the Pdx1:Cre; Grg3/4 mutants was primarily due to defects in β cell proliferation, consistent with previously described Neurod1 mutant phenotypes (Romer et al., 2019).
Fig. 6.
Grg3/4 mutants have a loss of NEUROD1, target gene expression, and proliferation of β cells. (A) Immunofluorescence staining for INS and NEUROD1 in P2 pancreata. White arrowheads indicate INS+ β cells without NEUROD1. INS−/NEUROD1+ cells likely represent non-β endocrine cells. Right-hand panels are magnifications of the boxed areas on the left. Scale bars: 100 μm. (B) List of differentially expressed Neurod1 target genes in e18.5 pancreata. (C) Immunofluorescence analysis of proliferating β cells using the proliferative marker Ki67 in e16.5 and P2 mice. White arrowheads indicate INS+Ki67+ cells. Scale bars: 50 μm. (D) Percentage of Ki67+ cells in INS+ cells as shown in C. n=3-6 for each genotype. **P≤0.01; ***P≤0.001; two-tailed Student's t-test.
DISCUSSION
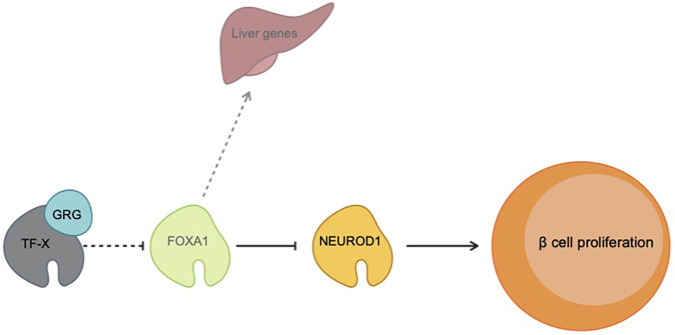
The transcription factor networks controlling pancreas development and function have been extensively characterized. Although it is known that many of these transcription factors rely on interactions with co-factors to allow cell-specific gene expression, only a small number of studies have explored their respective functions in vivo (McKenna et al., 2015; Spaeth et al., 2016, 2019; Yang et al., 2020). The Groucho family of co-repressors has been documented in many developmental systems to play integral roles in promoting cell fate decisions via transcriptional repression (Agarwal et al., 2015). In the present study, we used phenotypic and transcriptomic analyses to characterize GRG function in the developing murine pancreas. We found that pancreas-specific loss of Grg3 resulted in ectopic expression of Grg4 as a compensatory mechanism preventing a more severe phenotype in Grg3 single mutant animals. When both Grg3 and Grg4 were deleted from pancreatic progenitors, the hepatic transcription factor FOXA1 is no longer repressed in the pancreas lineage, resulting in ectopic expression of liver genes in embryonic pancreata (Fig. 7). We further show that β cells from Pdx1:Cre; Grg3/4 mutants lack NEUROD1 and fail to proliferate normally (Fig. 7). Together, these results uncover novel gene regulatory roles of GRGs in pancreas development and improve our understanding of co-factor-mediated transcriptional repression.
Fig. 7.
Model of GRG function in pancreas development. During normal pancreas development, GRG3/4 interacts with transcription factors (TF-X) to either directly or indirectly repress the master liver regulator Foxa1 in the pancreas lineage. Loss of GRG3/4-mediated repression results in ectopic FOXA1 protein, which both erroneously activates the liver program in the pancreas and represses expression of the β cell transcription factor gene Neurod1. Decreased NEUROD1 levels in β cells leads to a loss of β proliferation, early lethality, and hyperglycemia.
Until recently, there have been very few studies exploring the role of transcriptional repression in pancreas development. Two prior publications that explored GRG-mediated repression in the pancreas were limited by the early embryonic lethality of the Grg3 null mice (Metzger et al., 2012, 2014). These studies provided evidence of the importance of Grg3 the pancreas, but lacked the precision afforded by the pancreas-specific deletion of Grg3 using a Cre-Lox system. Although the use of cultured e12.5 pancreatic bud explants from Grg3−/− embryos allowed the investigators to assess later stages of pancreas development, the caveats associated with ectopic explant techniques may have affected their data interpretation, as we were not able to replicate their findings that endocrine cells failed to delaminate from the trunk epithelium due to a loss of E-cadherin repression, resulting in severe endocrine differentiation defects (Metzger et al., 2012). It is also possible that their pancreatic phenotypes were affected by pancreas non-autonomous defects as Grg3−/− embryos are significantly smaller and have defects in many different tissues that could have indirect effects on pancreas development (Gasperowicz et al., 2013).
Previous work from our lab demonstrated that the GRG3-interacting domain of NKX2-2 (Nkx2.2TN) is required to repress the α cell master regulator Arx in β cells. Loss of this interaction resulted in demethylation of the Arx promoter, significant β-to-α reprogramming, and lethality due to severe hyperglycemia (Papizan et al., 2011). Interestingly, perturbing this interaction by removing GRG3/4 from the pancreas did not phenocopy the Nkx2.2TN mutation – we did not observe upregulation of Arx nor an increase in α cells embryonically or postnatally. This suggests that the NKX2-2 TN domain has additional functions beyond its interaction with GRGs or that GRG-mediated repression during pancreas development extends beyond interactions with NKX2-2 to counteract the Nkx2.2TN phenotype. It was also surprising that deletion of Grg3 and combined deletion of Grg3/4 did not display a more severe phenotype. NKX6-1, another essential β cell transcription factor that results in a >90% reduction of β cells when deleted (Sander et al., 2000; Henseleit et al., 2005; Taylor et al., 2013), interacts with GRG4 in the central nervous system and promotes proper neural tube patterning (Muhr et al., 2001). Therefore, NKX6-1 would be expected to at least partially rely on GRG interactions in the pancreas to exert its function. Additional pancreas transcription factors that contain GRG interaction domains are PAX4/6, FOXA1/2, HES1, SOX9 and ARX (Buscarlet and Stifani, 2007; Jennings and Ish-Horowicz, 2008; Agarwal et al., 2015). Although these candidates have not yet been confirmed to interact with GRGs in the pancreas, we expected that loss of GRG function in the pancreas would produce an extremely severe phenotype that at least partially reflected the combined phenotypes of these essential pancreatic transcriptional regulators. Because mice deleted for Grg3/Grg4 in the pancreas do not phenocopy mutations in interacting transcription factors, either these essential pancreatic transcriptional regulators do not rely on GRG interactions to exert their functions or there is inherent redundancy in the system to compensate for the absence of GRG functions. This redundancy would likely operate through non-GRG dependent pathways as GRG2 deletion had no effect on pancreas development, either alone or in combination with deletion of Grg3, and GRG1 expression could not be detected in embryonic pancreas of either wild-type or any of the Grg mutant animals.
One novel phenotype we did discover in the Pdx1:Cre; Grg3/4 mutants was that FOXA1, a master regulator of the hepatic program, was no longer repressed in the pancreas throughout development and postnatally. Previous studies have suggested that FOXA1 acts redundantly in FOXA2-deficient pancreata; however, recent single-cell RNA-seq data suggest that Foxa1 is expressed at extremely low to undetectable levels in the developing pancreas (Krentz et al., 2018) and loss of FOXA1 alone does not affect pancreas function (Gao et al., 2010). Overexpression of Foxa1 in the pancreas has not been previously reported and the direct consequences on gene expression were difficult to dissect given the large number of dysregulated genes in Pdx1:Cre; Grg3/4 mutants. Interestingly, a number of liver genes were also ectopically expressed in the pancreas, which is consistent with previous studies that demonstrated that Grg3 expression was necessary to suppress the liver program in the developing foregut endoderm (Santisteban et al., 2010). However, we were unable to detect upregulation of the corresponding liver proteins. As the general translational machinery should be intact in the mutant islets, the disconnect between RNA and protein expression could reflect the absence of liver-specific proteins that would be needed to stabilize and/or facilitate the translation of these cell-specific transcripts. With the advent of single-cell genomic and proteomic technologies, there is a growing appreciation for a disconnect between mRNA and protein levels in several cellular contexts (Liu et al., 2016). Future studies that explore tissue-specific translational control could provide important information on novel mechanisms that regulate pancreas versus liver lineage programs.
Because the consensus sequence and targets of FOXA1 have been identified in other biological contexts, we were able to predict additional genes that were differentially regulated by FOXA1 in this system. From these analyses, we identified Neurod1 as a potential FOXA1 target that was significantly downregulated in Pdx1:Cre; Grg3/4 mutants. The phenotype identified here is strikingly similar to that of Neurod1−/− mice: reduced β cell proliferation resulting in failed expansion of the β cell population, leading to severe hyperglycemia and early lethality (Naya et al., 1997; Itkin-Ansari et al., 2005; Mastracci et al., 2013a; Romer et al., 2019). Although we can attribute much of the pancreas phenotype of Pdx1:Cre; Grg3/4 mutant mice to the dysregulation of Foxa1 and Neurod1, there were almost 2000 genes perturbed in mutant pancreata, suggesting that additional pathways are upregulated when GRG function is disrupted. These results provide strong evidence for the necessity of GRG-mediated transcriptional repression, both direct and indirect, in pancreas development. Future studies will focus on identification of the full cohort of GRG-interacting factors in the pancreas, the genes that are regulated by these complexes, and molecular mechanism underlying GRG-mediated gene repression in this system.
Transcriptional repression during organogenesis is widely recognized as a crucial component of proper tissue development (Gary and Levin, 1996; Meehan, 2003; Golson and Kaestner, 2017; Jambhekar et al., 2019). Loss of repression leads to aberrant gene expression and perturbations in cell-specific genetic programs. Although many studies have previously focused on the mechanisms activating tissue-specific gene expression, our understanding of the factors involved in inducing and maintaining repression of other genetic programs is still evolving. Ultimately, further insight into the mechanisms that promote cell- and tissue-specific gene programs while concurrently repressing other cell and tissue identities could enable us to improve therapeutic options for diabetic patients.
MATERIALS AND METHODS
Animal models and maintenance
Animals were maintained under protocol 00045 as approved by the University of Colorado Denver Institutional Animal Care and Use Committee (IACUC). All mice were maintained on a mixed C57BL6 genetic background. Animals were group-housed by sex with up to five siblings per cage with ad libitum food and water. Cages were held at 22°C, changed once every 2 weeks, and regularly monitored for virus and parasite infection. Euthanasia was performed by CO2 inhalation with cervical dislocation as a secondary method following asphyxiation. For timed pregnancies, presence of a vaginal plug in the morning was defined as e0.5. All mice and embryos were genotyped with primers listed in Table S2 using PCR with GoTaq DNA Polymerase Mastermix (Promega). The generation of Pdx1:Cre, Grg3flox, and Grg4flox mice have been previously described (Hingorani et al. Tuveson, 2003; Villanueva et al., 2011; Wheat et al., 2014). Pdx1:Cre mice are available at Jackson Laboratories: B6.FVB-Tg(Pdx1-cre)6Tuv/J (cat. #RRID:IMSR_JAX:014647).
Generation of Grg2 mice
We obtained Grg2/TLE2 knockout embryonic stem cells (Grg2/Tle2tm1(KOMP)Vlcg) through the Mouse Mouse Resource and Research Center (RRID:MMRRC_050067-UCD; http://velocigene.com/komp/detail/12600). The Grg2 null embryonic stem cells were injected into the blastocysts of F1 hybrid (C57BL/6×129) mice at the Columbia University Transgenic core to generate three independent lines of mice carrying a null mutation that eliminated Grg2 expression. The mice were genotyped using primers for the Grg2 wild-type alleles oRS31 (TLE2WT-R): 5′-GGGATTCTAGGATTCTAGGCAGGGC-5′ and oRS32 (TLE2WT-F): 3′-TTGAGGCATGGTCTTGCTTTGTAGC-3′; and the mutant alleles oRS28 (NeoF): 5′-GCAGCCTCTGTTCCACATACACTTCA-3′ and oRS30 (TLE2-R): 5′-AGAGCCAGGAAGATGGTTCAGTTGG-3′. These mice were subsequently bred and maintained on a C57BL6 genetic background. Complete deletion of Grg2 was verified by qRT-PCR (Fig. 1D).
RNA in situ hybridization
All GRG probes were generated from full-length cDNAs. Sense and antisense probes were labeled with the DIG RNA labeling mix (Roche Applied Science). RNA in situ hybridization was performed as previously described (Mastracci et al., 2013b).
Blood glucose
Ad libitum blood glucose measurements were taken using tail vein blood samples. Glucose concentrations were determined using a Contour 7151H glucose meter with Contour 7097C test strips.
Glucose tolerance tests
Mice were fasted overnight for 12 h followed by an intraperitoneal injection with glucose (2 mg/g body weight). Tail vein blood samples were collected at 0, 15, 30, 60 and 120 min after injection. Glucose concentrations were determined using a Contour 7151H glucose meter with Contour 7097C test strips.
Insulin content
Islets (20 per well) were cultured in Kreb's buffer with a glucose concentration of 2.8 mM for 1 h in a 12-well plate. Islets were then disrupted using a homogenizer in 50 μl of lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1% IGEPAL) and cellular insulin content was extracted by acid ethanol and measured by ultrasensitive insulin ELISA (CrystalChem).
Tissue preparation and immunofluorescence
Timed pregnant females were sacrificed and embryos were removed and placed directly into ice-cold PBS (pH 7.4). For e12.5-e16.5 embryos, heads were removed and remaining embryos were placed in 4% paraformaldehyde (PFA) for 4 h at 4°C. For P2 animals, abdominal organs were dissected and placed in 4% PFA for 4 h at 4°C. Adult whole pancreata were dissected and placed in 4% PFA for 4 h at 4°C. Tissue was cryo-preserved in 30% sucrose-PBS solution for overnight at 4°C followed by embedding and freezing in Optimum Cutting Temperature (O.C.T.) on dry ice. Tissue was stored at −80°C until 10 μm cryo-sections were collected for analyses using a Microm HM 525 cryostat.
For immunofluorescence analysis, sections were washed in PBS and 0.01% Triton X-100 (PBS-T) and blocked with 2% normal donkey serum (NDS) for 30 min at room temperature (RT). Primary antibodies were diluted in 2% NDS and incubated on sections overnight at 4°C in a humidified chamber. Sections were washed with PBS-T and incubated for 2-3 h at RT with secondary antibodies diluted in 2% NDS. Sections were washed with PBS-T and incubated with DAPI (Fisher Scientific, D1306) diluted 1:1000 in PBS-T for 15 min at RT. Sections were washed with PBS-T and mounted with VECTASHIELD Hardset Antifade Mounting Medium (Vector Laboratories, H-1400). All primary and secondary antibodies can be found in Table S3.
High-magnification images, including all images showing bihormonal cells, were taken using a Zeiss Confocal LSM800 microscope and processed with Zen, ImageJ and Adobe Photoshop software. Lower magnification images for quantification purposes were taken using a Leica DM5500B microscope.
RNAscope
RNAscope was performed as described in ACD Biosciences RNAscope Fluorescent Multiplex Kit Quick Guide (https://acdbio.com/documents/product-documents). Samples were fixed and sectioned as described above. The samples were then post-fixed at 4°C overnight in 4% PFA, followed by dehydration by ethanol and drying. Slides were treated with hydrogen peroxide for and antigen retrieval by boiling in Target Retrieval. Slides were washed with deionised (DI) water and subjected to mild protease digestion (Protease III) for 20 min in a humified chamber, washed again in DI water and incubated with probes purchased from ACD Biosciences [RNAscope Probe-Mm-Ins2-C1 (414661), -Serpina1e-C2 (88277)] for 2 h at 40°C. Slides were washed with Wash Buffer and placed in 5× SSC overnight. Slides were again washed and incubated in AMP1, -2 and -3 solutions for 30 min, 30 min,and 15 min, respectively, at 40°C, with washes in between each incubation. To develop signal, we used Opal Dyes 570 and 620 (RNAscope Multiplex Fluorescent Detection Kit v2) diluted in Multiplex TSA buffer. Each channel was developed sequentially by incubating samples in HRP-C1/2 for 15 min at 40°C, washed, and then incubated in the pre-diluted Opal Dye for 30 min at 40°C. Slides were then washed and incubated in HRP Blocker for 15 min at 40°C. This solution was removed, and slides were again washed followed by a 10 min incubation at RT with DAPI. Slides were mounted using Prolong Gold Mounting Media (Invitrogen) and imaged at 40× on a Zeiss LSM800 confocal microscope.
Morphometric analysis
Quantification of hormone and pancreas area was performed using ImageJ software and thresholding of individual color channels. Pancreas area was outlined and DAPI+ pixels were quantified. Hormone area was calculated as the hormone+ area divided by the DAPI+ area. For Ki67 quantification, all insulin+ β cells and Ki67/insulin double-positive cells were counted and percentage proliferation was calculated by dividing Ki67/insulin+ cell counts by total insulin+ cell counts. A total of five slides and ten slides, evenly spaced throughout the entire pancreas, were quantified per sample for e16.5 and P2 time points, respectively.
RNA extraction and RT-qPCR
Whole pancreata were collected from embryos and flash frozen in Buffer RLT (Qiagen). Total RNA extractions were performed using the RNeasy Mini Kit (Qiagen) and eluted in 30 μl RNase-free water. cDNA was generated with 500 ng of RNA as template using the iScript cDNA Synthesis Kit (Bio-Rad). The resulting cDNA was diluted to 1 ng/μl and 4 μl were used in each qPCR reaction using the SsoAdvanced Universal Probes Supermix (Bio-Rad) with Taqman probes (Table S4). Reactions were run on the Bio-Rad CFX96 Real Time PCR Detection System. Expression levels were normalized to cyclophilin B (also known as peptidylprolyl isomerase B) or Actb and quantified using the 2−ΔΔCT. Control samples were set to one for each gene to determine expression changes in mutants.
RNA sequencing
RNA was extracted as described and RNA integrity number (RIN) was determined using the Eukaryote Total RNA Nano Kit for the Agilent 2100 Bioanalyzer. Libraries were prepared from samples with RIN values >8.0 using the Universal Plus mRNA-Seq with NuQuant kit (NuGen), then subjected to high-throughput RNA sequencing using the NovaSEQ 6000 for paired end sequencing (2×150) from PolyA selected total RNA by the University of Colorado Cancer Center Genomics and Microarray Core. Reads were quality checked using fastQC and adapters trimmed using trim-galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Reads were aligned to the mm10 genome using HISAT2 and mapped to genes using HTSeq with the Ensemble genome: Mus_musculus.GRCm38.95.gtf (Anders et al., 2015; Kim et al., 2015). Differential gene expression was determined using DESeq2 in R (Love et al., 2014). Gene ontology enrichment analysis was performed using Panther (http://pantherdb.org/). Differentially expressed Foxa1 known and predicted target genes were assessed using Harmonizome with the TRANSFAC Curated Transcription Factor Targets database (http://amp.pharm.mssm.edu/Harmonizome/).
Western blotting
Nuclear protein lysates were prepared from excised P0 pancreata using the Nuclear Extract Kit (Active Motif). Approximately 20 µg of each sample was loaded onto a 10% Bis-Tris polyacrylamide gel (Invitrogen). Proteins were transferred to PVDF membrane and the membrane blocked in 5% milk for 30 min, incubated with rabbit anti-TLE3 (1:100, sc-9124, Santa Cruz Biotechnology) and rabbit anti-GAPDH (1:1000, ab9485, Abcam) overnight at 4°C, washed, incubated with anti-rabbit-HRP (1:10,000, 32260, Zymed) for 1 h at RT, washed, and developed with the Western Lightning chemiluminescence kit (GE Biosciences).
Quantification and statistical analysis
Graphs and statistical analyses were generated using GraphPad Prism 8. All values are shown as mean±s.d. P-values were calculated using two-tailed Student's t-test. Each n represents an individual animal and n values for all experiments are listed in the figure legends. For the RNA-sequencing experiments, Padjusted ≤0.05 was used as the cutoff for differentially expressed genes (defined by the Benjamini–Hochberg procedure for multiple hypothesis testing).
Supplementary Material
Acknowledgements
We thank members of the Sussel lab for helpful feedback on this project and especially Michelle Guney, Nicole Moss and David Lorberbaum for critical reading of the manuscript. We also thank Dr Peter Tontonoz (University of California Los Angeles) and Dr David Sweetser (Massachusetts General Hospital) for generously providing us with the Grg3 and Grg4 floxed mice, respectively. We are grateful for technical assistance for many specialized techniques and analyses presented here: RNA sequencing was conducted by the University of Colorado Denver Microarray and Genomics core and advice to A.T. for the RNA sequencing computational analysis was provided by the CU Denver RNA Biosciences Initiative.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.T., R.A.S., L.S.; Methodology: A.T., R.A.S., L.S.; Validation: A.T.; Formal analysis: A.T., R.A.S., D.G., A.P., A.N., L.S.; Investigation: A.T., R.A.S., D.G., A.P., A.N., L.S.; Resources: L.S.; Data curation: A.T.; Writing - original draft: A.T.; Writing - review & editing: A.T., R.A.S., L.S.; Supervision: L.S.; Project administration: L.S.; Funding acquisition: L.S.
Funding
Support for the project was provided by the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK082590 and P30 DK116073 to L.S., F31 DK122634 to A.T., F31 DK107028 to R.A.S.). Deposited in PMC for release after 12 months.
Data availability
Raw RNA-seq fastq files have been deposited in Gene Expression Omnibus under accession numbers GSE149889 (e15.5 sorted NEUROG3-EGFP cells) and GSE149891 (e18.5 Pdx1:Cre; Grg3 and Pdx1:Cre; Grg3/4 pancreata).
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.192401.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.192401.reviewer-comments.pdf
References
- Agarwal, M., Kumar, P. and Mathew, S. J. (2015). The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 67, 472-481. 10.1002/iub.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Diabetes Association. (2020). 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care 43, S14-S31. 10.2337/dc20-S002 [DOI] [PubMed] [Google Scholar]
- Anders, S., Pyl, P. T. and Huber, W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, S. L., Wierda, A., Wong, D., Stevens, K. A., Cascio, S., Rossant, J. and Zaret, K. S. (1993). The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development 119, 1301-1315. [DOI] [PubMed] [Google Scholar]
- Buscarlet, M. and Stifani, S. (2007). The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 17, 353-361. 10.1016/j.tcb.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Cai, Y., Brophy, P. D., Levitan, I., Stifani, S. and Dressler, G. R. (2003). Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 22, 5522-5529. 10.1093/emboj/cdg536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernea, S. and Dobreanu, M. (2013). Diabetes and beta cell function: from mechanisms to evaluation and clinical implications. Biochem. Med. 23, 266-280. 10.11613/BM.2013.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, C. S., Loomis, Z. L., Lee, J. E. and Sussel, L. (2007). Genetic identification of a novel NeuroD1 function in the early differentiation of islet α, PP and ε cells. Dev. Biol. 312, 523-532. 10.1016/j.ydbio.2007.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G., Fernandez, J., Mische, S. and Courey, A. J. (1999). A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13, 2218-2230. 10.1101/gad.13.17.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat, P., Mansouri, A., Hecksher-Sørensen, J., Serup, P., Krull, J., Gradwohl, G. and Gruss, P. (2003). Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 17, 2591-2603. 10.1101/gad.269003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat, P., Hecksher-Sorensen, J., Broccoli, V., Krull, J., Ponte, I., Mundiger, T., Smith, J., Gruss, P., Serup, P. and Mansouri, A. (2005). The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the α- and β-cell lineages in the mouse endocrine pancreas. Development 132, 2969-2980. 10.1242/dev.01870 [DOI] [PubMed] [Google Scholar]
- Dassaye, R., Naidoo, S. and Cerf, M. E. (2016). Transcription factor regulation of pancreatic organogenesis, differentiation and maturation. Islets 8, 13-34. 10.1080/19382014.2015.1075687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, D., Jiménez, G., Heavey, B. and Busslinger, M. (2000). Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19, 2292-2303. 10.1093/emboj/19.10.2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, N., LeLay, J., Vatamaniuk, M. Z., Rieck, S., Friedman, J. R. and Kaestner, K. H. (2008). Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 22, 3435-3448. 10.1101/gad.1752608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, N., Le Lay, J., Qin, W., Doliba, N., Schug, J., Fox, A. J., Smirnova, O., Matschinsky, F. M. and Kaestner, K. H. (2010). Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature β-cell. Mol. Endocrinol. 24, 1594-1604. 10.1210/me.2009-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, S. and Levin, M. (1996). Transcriptional repression in development. Curr. Opin. Cell Biol. 8, 358-364. 10.1016/S0955-0674(96)80010-X [DOI] [PubMed] [Google Scholar]
- Gasperowicz, M., Surmann-Schmitt, C., Hamada, Y., Otto, F. and Cross, J. C. (2013). The transcriptional co-repressor TLE3 regulates development of trophoblast giant cells lining maternal blood spaces in the mouse placenta. Dev. Biol. 382, 1-14. 10.1016/j.ydbio.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Golson, M. L. and Kaestner, K. H. (2017). Epigenetics in formation, function, and failure of the endocrine pancreas. Mol. Metab. 6, 1066-1076. 10.1016/j.molmet.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, C., Stein, G. H., Pan, N., Goebbels, S., Hörnberg, H., Nave, K.-A., Herrera, P., White, P., Kaestner, K. H., Sussel, L.et al. (2010). Pancreatic β cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 11, 298-310. 10.1016/j.cmet.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, G. D., Bender, A. S., Cirulli, V., Mastracci, T. L., Kelly, S. M., Tsirigos, A., Kaestner, K. H. and Sussel, L. (2017). Pancreatic β cell identity requires continual repression of non-β cell programs. J. Clin. Investig. 127, 244-259. 10.1172/JCI88017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henseleit, K. D., Nelson, S. B., Kuhlbrodt, K., Hennings, J. C., Ericson, J. and Sander, M. (2005). NKX6 transcription factor activity is required for α- and β-cell development in the pancreas. Development 132, 3139-3149. 10.1242/dev.01875 [DOI] [PubMed] [Google Scholar]
- Hingorani, S. R., Petricoin, E. F., Maitra, A., Rajapakse, V., King, C., Jacobetz, M. A., Ross, S., Conrads, T. P., Veenstra, T. D., Hitt, B. A.et al. (2003). Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437-450. 10.1016/S1535-6108(03)00309-X [DOI] [PubMed] [Google Scholar]
- Hoffman, B. G., Zavaglia, B., Beach, M. and Helgason, C. D. (2008). Expression of Groucho/TLE proteins during pancreas development. BMC Dev. Biol. 8, 81. 10.1186/1471-213X-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin-Ansari, P., Marcora, E., Geron, I., Tyrberg, B., Demeterco, C., Hao, E., Padilla, C., Ratineau, C., Leiter, A., Lee, J. E.et al. (2005). NeuroD1 in the endocrine pancreas: localization and dual function as an activator and repressor. Dev. Dyn. 233, 946-953. 10.1002/dvdy.20443 [DOI] [PubMed] [Google Scholar]
- Iype, T., Taylor, D. G., Ziesmann, S. M., Garmey, J. C., Watada, H. and Mirmira, R. G. (2004). The transcriptional repressor Nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic β-cell differentiation: evidence for feedback activation of the nkx6.1 gene by Nkx6.1. Mol. Endocrinol. 18, 1363-1375. 10.1210/me.2004-0006 [DOI] [PubMed] [Google Scholar]
- Jambhekar, A., Dhall, A. and Shi, Y. (2019). Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625-641. 10.1038/s41580-019-0151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangal, M., Couture, J.-P., Bianco, S., Magnani, L., Mohammed, H. and Gévry, N. (2014). The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor α target genes. Nucleic Acids Res. 42, 11339-11348. 10.1093/nar/gku791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, B. H. and Ish-Horowicz, D. (2008). The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9, 205. 10.1186/gb-2008-9-1-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J., Pedersen, E. E., Galante, P., Hald, J., Heller, R. S., Ishibashi, M., Kageyama, R., Guillemot, F., Serup, P. and Madsen, O. D. (2000). Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36-44. 10.1038/71657 [DOI] [PubMed] [Google Scholar]
- Jørgensen, M. C., Ahnfelt-Rønne, J., Hald, J., Madsen, O. D., Serup, P. and Hecksher-Sørensen, J. (2007). An illustrated review of early pancreas development in the mouse. Endocr. Rev. 28, 685-705. 10.1210/er.2007-0016 [DOI] [PubMed] [Google Scholar]
- Kaestner, K. H. (2010). The FoxA factors in organogenesis and differentiation. Curr. Opin. Genet. Dev. 20, 527-532. 10.1016/j.gde.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Langmead, B. and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz, N. A. J., Lee, M. Y. Y., Xu, E. E., Sproul, S. L. J., Maslova, A., Sasaki, S. and Lynn, F. C. (2018). Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Rep. 11, 1551-1564. 10.1016/j.stemcr.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. S., Friedman, J. R., Fulmer, J. T. and Kaestner, K. H. (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944-947. 10.1038/nature03649 [DOI] [PubMed] [Google Scholar]
- Li, L.-C., Qiu, W.-L., Zhang, Y.-W., Xu, Z.-R., Xiao, Y.-N., Hou, C., Lamaoqiezhong, Yu, P., Cheng, X. and Xu, C.-R. (2018). Single-cell transcriptomic analyses reveal distinct dorsal/ventral pancreatic programs. EMBO Rep. 19, e46148. 10.15252/embr.201846148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderson, Y., Eberhard, D., Malin, S., Johansson, A., Busslinger, M. and Pettersson, S. (2004). Corecruitment of the Grg4 repressor by PU.1 is critical for Pax5-mediated repression of B-cell-specific genes. EMBO Rep. 5, 291-296. 10.1038/sj.embor.7400089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Beyer, A. and Aebersold, R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535-550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçal, N., Patel, H., Dong, Z., Belanger-Jasmin, S., Hoffman, B., Helgason, C. D., Dang, J. and Stifani, S. (2005). Antagonistic effects of Grg6 and Groucho/TLE on the transcription repression activity of brain factor 1/FoxG1 and cortical neuron differentiation. Mol. Cell. Biol. 25, 10916-10929. 10.1128/MCB.25.24.10916-10929.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci, T. L., Anderson, K. R., Papizan, J. B. and Sussel, L. (2013a). Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells in the pancreas. PLoS Genet. 9, e1003278. 10.1371/journal.pgen.1003278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci, T. L., Lin, C.-S. and Sussel, L. (2013b). Generation of mice encoding a conditional allele of Nkx2.2. Transgenic Res. 22, 965-972. 10.1007/s11248-013-9700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, B., Guo, M., Reynolds, A., Hara, M. and Stein, R. (2015). Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet β cells. Cell Rep. 10, 2032-2042. 10.1016/j.celrep.2015.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan, R. R. (2003). DNA methylation in animal development. Semin. Cell Dev. Biol. 14, 53-65. 10.1016/S1084-9521(02)00137-4 [DOI] [PubMed] [Google Scholar]
- Metzger, D. E., Gasperowicz, M., Otto, F., Cross, J. C., Gradwohl, G. and Zaret, K. S. (2012). The transcriptional co-repressor Grg3/Tle3 promotes pancreatic endocrine progenitor delamination and β-cell differentiation. Development 139, 1447-1456. 10.1242/dev.072892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger, D. E., Liu, C., Ziaie, A. S., Naji, A. and Zaret, K. S. (2014). Grg3/TLE3 and Grg1/TLE1 induce monohormonal pancreatic β-cells while repressing α-cell functions. Diabetes 63, 1804-1816. 10.2337/db13-0867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milili, M., Gauthier, L., Veran, J., Mattei, M.-G. and Schiff, C. (2002). A new Groucho TLE4 protein may regulate the repressive activity of Pax5 in human B lymphocytes. Immunology 106, 447-455. 10.1046/j.1365-2567.2002.01456.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan, A. P., Kaestner, K. H., Grau, E. and Schutz, G. (1993). Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119, 567-578. [DOI] [PubMed] [Google Scholar]
- Muhr, J., Andersson, E., Persson, M., Jessell, T. M. and Ericson, J. (2001). Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104, 861-873. 10.1016/S0092-8674(01)00283-5 [DOI] [PubMed] [Google Scholar]
- Naya, F. J., Huang, H.-P., Qiu, Y., Mutoh, H., DeMayo, F. J., Leiter, A. B. and Tsai, M.-J. (1997). Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11, 2323-2334. 10.1101/gad.11.18.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, F. C. and Wright, C. (2011). Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 240, 530-565. 10.1002/dvdy.22584 [DOI] [PubMed] [Google Scholar]
- Papizan, J. B., Singer, R. A., Tschen, S.-I., Dhawan, S., Friel, J. M., Hipkens, S. B., Magnuson, M. A., Bhushan, A. and Sussel, L. (2011). Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 25, 2291-2305. 10.1101/gad.173039.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush, Z. (1994). Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79, 805-815. 10.1016/0092-8674(94)90070-1 [DOI] [PubMed] [Google Scholar]
- Patel, S. R., Bhumbra, S. S., Paknikar, R. S. and Dressler, G. R. (2012). Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol. Cell 45, 185-195. 10.1016/j.molcel.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer, A. I., Singer, R. A., Sui, L., Egli, D. and Sussel, L. (2019). Murine perinatal β-cell proliferation and the differentiation of human stem cell-derived insulin-expressing cells require NEUROD1. Diabetes 68, 2259-2271. 10.2337/db19-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, M., Neubuser, A., Kalamaras, J., Ee, H. C., Martin, G. R. and German, M. S. (1997). Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 11, 1662-1673. 10.1101/gad.11.13.1662 [DOI] [PubMed] [Google Scholar]
- Sander, M., Sussel, L., Conners, J., Scheel, D., Kalamaras, J., Dela Cruz, F., Schwitzgebel, V., Hayes-Jordan, A. and German, M. (2000). Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 127, 5533-5540. [DOI] [PubMed] [Google Scholar]
- Santisteban, P., Recacha, P., Metzger, D. E. and Zaret, K. S. (2010). Dynamic expression of Groucho-related genes Grg1 and Grg3 in foregut endoderm and antagonism of differentiation. Dev. Dyn. 239, 980-986. 10.1002/dvdy.22217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavuzzo, M. A., Hill, M. C., Chmielowiec, J., Yang, D., Teaw, J., Sheng, K., Kong, Y., Bettini, M., Zong, C., Martin, J. F.et al. (2018). Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat. Commun. 9, 3356. 10.1038/s41467-018-05740-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya, T. and Zaret, K. S. (2007). Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell 28, 291-303. 10.1016/j.molcel.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda, B., Chowdhury, K., Torres, M., Oliver, G. and Gruss, P. (1997). The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature 386, 399-402. 10.1038/386399a0 [DOI] [PubMed] [Google Scholar]
- Spaeth, J. M., Walker, E. M. and Stein, R. (2016). Impact of Pdx1-associated chromatin modifiers on islet β-cells. Diabetes Obes. Metab. 18 Suppl. 1, 123-127. 10.1111/dom.12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth, J. M., Liu, J.-H., Peters, D., Guo, M., Osipovich, A. B., Mohammadi, F., Roy, N., Bhushan, A., Magnuson, M. A., Hebrok, M.et al. (2019). The Pdx1-Bound Swi/Snf chromatin remodeling complex regulates pancreatic progenitor cell proliferation and mature islet β-cell function. Diabetes 68, 1806-1818. 10.2337/db19-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge, L., Sosa-Pineda, B., Chowdhury, K., Mansouri, A. and Gruss, P. (1997). Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature 387, 406-409. 10.1038/387406a0 [DOI] [PubMed] [Google Scholar]
- Sussel, L., Kalamaras, J., Hartigan-O'Connor, D. J., Meneses, J. J., Pedersen, R. A., Rubenstein, J. L. R. and German, M. S. (1998). Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 125, 2213-2221. [DOI] [PubMed] [Google Scholar]
- Tabula muris consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, Principal investigators. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B. L., Liu, F.-F. and Sander, M. (2013). Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 4, 1262-1275. 10.1016/j.celrep.2013.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turki-Judeh, W. and Courey, A. J. (2012). Groucho: a corepressor with instructive roles in development. Curr. Top. Dev. Biol. 98, 65-96. 10.1016/B978-0-12-386499-4.00003-3 [DOI] [PubMed] [Google Scholar]
- Villanueva, C. J., Waki, H., Godio, C., Nielsen, R., Chou, W.-L., Vargas, L., Wroblewski, K., Schmedt, C., Chao, L. C., Boyadjian, R.et al. (2011). TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 13, 413-427. 10.1016/j.cmet.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat, J. C., Krause, D. S., Shin, T. H., Chen, X., Wang, J., Ding, D., Yamin, R. and Sweetser, D. A. (2014). The corepressor Tle4 is a novel regulator of murine hematopoiesis and bone development. PLoS ONE 9, e105557. 10.1371/journal.pone.0105557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Graff, S. M., Heiser, C. N., Ho, K.-H., Chen, B., Simmons, A. J., Southard-Smith, A. N., David, G., Jacobson, D. A., Kaverina, I.et al. (2020). Coregulator Sin3a promotes postnatal murine β-cell fitness by regulating genes in Ca2+ homeostasis, cell survival, vesicle biosynthesis, glucose metabolism, and stress response. Diabetes 69, 1219-1231. 10.2337/db19-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J., Lai, E. and Stifani, S. (2001). The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol. Cell. Biol. 21, 1962-1972. 10.1128/MCB.21.6.1962-1972.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Chen, H.-M., Jaramillo, E., Wang, L. and D'Mello, S. R. (2008). Histone deacetylase-related protein inhibits AES-mediated neuronal cell death by direct interaction. J. Neurosci. Res. 86, 2423-2431. 10.1002/jnr.21680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.