ABSTRACT
To gain a deeper understanding of pancreatic β-cell development, we used iterative weighted gene correlation network analysis to calculate a gene co-expression network (GCN) from 11 temporally and genetically defined murine cell populations. The GCN, which contained 91 distinct modules, was then used to gain three new biological insights. First, we found that the clustered protocadherin genes are differentially expressed during pancreas development. Pcdhγ genes are preferentially expressed in pancreatic endoderm, Pcdhβ genes in nascent islets, and Pcdhα genes in mature β-cells. Second, after extracting sub-networks of transcriptional regulators for each developmental stage, we identified 81 zinc finger protein (ZFP) genes that are preferentially expressed during endocrine specification and β-cell maturation. Third, we used the GCN to select three ZFPs for further analysis by CRISPR mutagenesis of mice. Zfp800 null mice exhibited early postnatal lethality, and at E18.5 their pancreata exhibited a reduced number of pancreatic endocrine cells, alterations in exocrine cell morphology, and marked changes in expression of genes involved in protein translation, hormone secretion and developmental pathways in the pancreas. Together, our results suggest that developmentally oriented GCNs have utility for gaining new insights into gene regulation during organogenesis.
KEY WORDS: WGCNA, Gene networks, Pancreas, Protocadherin, CRISPR, Zfp800, β-cells
Summary: Calculation of a gene co-expression network for pancreatic β-cell development reveals crucial roles for the clustered protocadherin genes and Zfp800 in pancreas development.
INTRODUCTION
Pancreatic β-cells are crucially important for metabolic regulation and abnormalities in their function and/or survival play a central role in the pathophysiology of both type 1 and type 2 diabetes mellitus. β-Cells and other pancreatic cells arise by a stepwise differentiation process that begins during early embryological development and ends during early postnatal life (Pan and Wright, 2011). Over this time, cells advance through multiple successive developmental stages, including formation of the definitive endoderm, specification of pre-pancreatic epithelium; branching of the pancreatic epithelium and lineage segregation through proliferation and differentiation of pancreatic multipotent progenitor cells; endocrine cell priming and differentiation; and specification and postnatal maturation of β-cells (Bastidas-Ponce et al., 2017; Larsen and Grapin-Botton, 2017).
The identity of both nascent and adult pancreatic cells is determined by cell type-specific gene regulatory networks (GRNs) that consist of transcription factors (TFs) and cis-regulatory elements, non-coding RNAs and chromatin-modifying enzymes (Conrad et al., 2014; Kaestner et al., 2003). GRNs encompassing several dozen well-defined TFs have been derived from the manual curation of published literature (Arda et al., 2013). These GRNs are useful conceptual models that concisely summarize our understanding of how specific TFs act during pancreas organogenesis to affect changes in both cell identify and function. However, owing to the massive number of regulatory factors and elements, as well as the wide variety and non-linearity of mechanisms that affect their activity, the modeling of gene regulation at a whole-transcriptome scale remains a prohibitively daunting task.
An alternative approach applicable to large transcriptomic datasets is to calculate a gene co-expression network (GCN) (Oldham et al., 2008; Zhang and Horvath, 2005). By revealing modules of co-expressed genes, GCNs have utility for forming hypotheses about the roles of specific genes in cellular processes (Wang et al., 2017). In a developmental context, GCNs enable sets of genes that exhibit similar expression patterns during development to be identified. Furthermore, network topology, including the actual position of the genes/nodes within the network and node centrality (measures of node connectivity) may be useful for identifying genes that are important at certain development stages or in specific cellular functions (Azuaje, 2014).
Cell adhesions play important roles in tissue development and morphogenesis. Molecules in the cadherin gene families contribute to cell recognition and sorting, boundary formation and maintenance, cell movements, and the induction and maintenance of structural and functional cell and tissue polarity (Halbleib and Nelson, 2006). A particularly important category of cadherin molecules are the clustered protocadherins. The protocadherin family consists of over 70 genes, 50 of which are located in three tightly linked gene clusters (Pcdha, Pcdhb and Pcdhg) and is expressed in both neuronal and non-neuronal tissues (Chen and Maniatis, 2013). Protocadherins provide crucial molecular diversity for cell-cell interactions that underlie neuronal differentiation and migration, axon outgrowth, dendrite arborization, and synaptogenesis (Peek et al., 2017). Little is known about the expression or regulation of protocadherin genes during pancreatic organogenesis.
Similarly, zinc finger proteins (ZFPs) play many important roles in regulating gene expression during development. ZFPs containing C2H2 zinc finger motifs are widespread in nature and represent nearly 3% of human proteins (Klug, 2010). The majority of C2H2 ZFPs are DNA-binding proteins (Najafabadi et al., 2015). However, ZFPs may not only regulate transcription, but also serve as chromatin maintenance factors, and can bind RNA and other molecules (Vilas et al., 2018). KRAB-ZFPs, which are characterized by an N-terminal Krüppel-associated box (KRAB) domain and a C-terminal array of C2H2 zinc fingers, are likely important for silencing of transposable elements (TEs) in the genome (Ecco et al., 2017) and KRAB-ZFP-mediated transcriptional repression is linked to the regulation of cell proliferation, differentiation and apoptosis (Lupo et al., 2013). The binding of KRAB-ZFPs to TEs has also been shown to play a role in the tissue-specific regulation of expression of neighboring genes (Imbeault et al., 2017). Other less common domains that occur within C2H2-containing ZFPs include DNA-binding homeodomains, and protein-protein interacting SCAN and BTB domains (Fedotova et al., 2017). INSM1, a C2H2-containing ZFP, plays a well-defined role in pancreas development (Osipovich et al., 2014); however, little is known about the contribution of most ZFPs to pancreas development.
Here, we used iterative WGCNA (Greenfest-Allen et al., 2017 preprint), an extension of standard weighted gene correlation network analysis (WGCNA), to calculate a lineage-directed GCN that reflects mouse β-cell development. The network is based on the RNA sequencing of 11 temporally and genetically defined murine cell populations, and organizes 7894 of the 15,949 genes expressed over nearly a 75-day time frame into 91 highly coherent modules and 11 meta-modules that provide a useful overview of the organization of the β-cell developmental transcriptome. We demonstrate the utility of this GCN by using it to discover the stage-specific expression of different clusters of protocadherin genes during pancreas development, to extract and analyze sub-networks of transcriptional regulators important for endocrine differentiation and β-cell function, and to aid our selection of three ZFPs for further analysis using CRISPR-induced global gene knockouts in mice. These studies illustrate how deriving a GCN may provide new insights into gene expression and function during development. An online resource that contains these data (https://markmagnuson.github.io/BetaCell-GCN/genebrowser) enables investigators to identify expression patterns of genes of interest during pancreas development.
RESULTS
RNA sequencing of genetically and temporally defined pancreatic cell populations
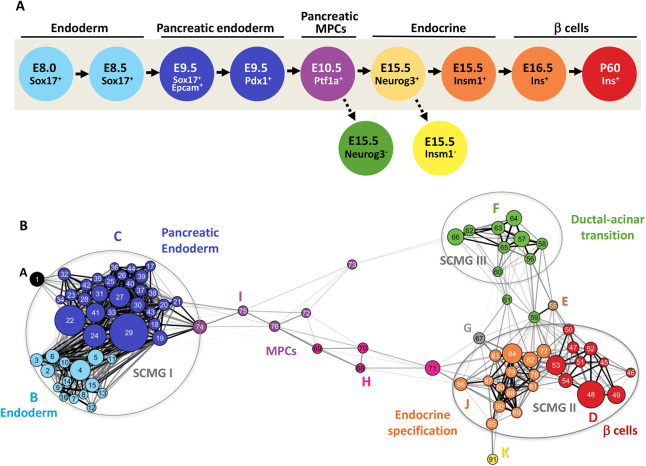
Genetically driven fluorescent reporter genes and fluorescence-activated cell sorting (FACS) were used to purify cells from 11 different stages of pancreas/endocrine/β-cell development and to generate 33 different RNA-Seq datasets (Fig. 1A). These cell populations covered a developmental time frame from embryonic day (E) 8.0 to postnatal day (P) 60 (Fig. S1), or nearly 75 days. Nine of the 11 different cell populations consisted of normal development stages including endoderm (defined by Sox17GFPCre expression at E8.0 and E8.5), ventral foregut endoderm (marked by the combination of Sox17GFPCre expression and EPCAM-positive immunoreactivity at E9.5), foregut endoderm (marked by Pdx1CFP expression at E9.5), pancreatic multipotent progenitor cells (marked by Ptf1aYFP expression at E10.5), endocrine progenitor cells (marked by Neurog3GFP or Insm1GFPCre at E15.5), and both immature and mature β-cells [identified by expression of a mouse Ins1 (MIP)-GFP transgene at both E16.5 and P60]. Additionally, two abnormal cell populations were included to identify genes for which expression was dependent on either Neurog3 (Neurog3GFP/GFP at E15.5) or Insm1 (Insm1GFPCre/GFPCre at E15.5) (Figs S1 and S2, Table S1). In total, the RNA-Seq datasets revealed 15,949 different genes that were expressed in at least one cell population (Table S2).
Fig. 1.
Developmentally oriented gene co-expression network for β-cells. (A) A developmental scheme outlining 11 populations representing different stages of β-cell development profiled by RNA-Seq. Nine murine cell populations along the β-cell lineage and two additional mutant populations were profiled. Each profiled cell population has been shown in genetically altered mice to reflect a defined progenitor cell population that precedes the formation of mature β-cells, the populations are: gut tube endoderm (Sox17+/− at E8.5), posterior foregut endoderm (Pdx1+/− at E9.5), pancreatic multipotent progenitor cells (MPCs) (Ptf1a+/− at E10.5), endocrine progenitor cells (Neurog3+/− and Insm1+/− at E15.5), nascent β-cells (Ins+ at E16.5), and adult β-cells (Ins+ at P60). Also profiled were two mutant conditions for endocrine progenitor cells (Neurog3−/− and Insm1−/− at E15.5). (B) A meta-network view of GCN constructed by applying iterative WGCNA analysis on the obtained developmental RNA-Seq profiles. Each node in the meta-network represents a module of highly co-expressed genes; module numbers are indicated inside the node; node sizes are proportional to the number of genes in the module. Node color represents meta-module memberships and is coordinated with colors of developmental stages (as in A) it represents. The meta-network is defined by correlations between module eigengenes and partitions modules into three distinct strongly connected module groups (SCMGs). Edge lengths are inversely proportional to the topological overlap between modules; two modules with a high degree of topological overlap are strongly connected to the same group of modules. The strongest connections (topological overlap>0.45) are further highlighted by heavy black lines. A high-resolution version of this image is provided in Fig. S4, as is an interactive Cytoscape file for the entire network as well as individual meta-modules via https://markmagnuson.github.io/BetaCell-GCN/.
Generation of a GCN for pancreatic β-cell development
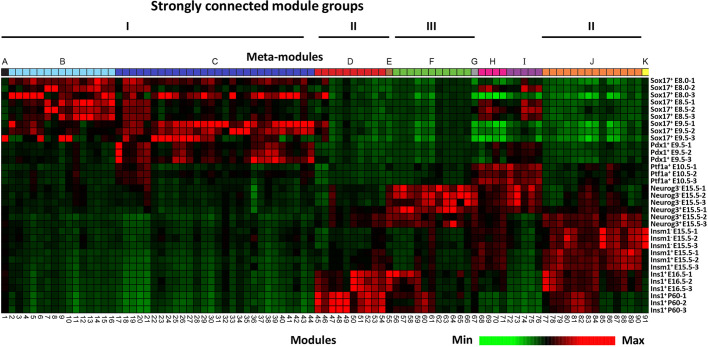
Next, we utilized iterative WGCNA (Fig. S3A-C) to calculate a GCN from the 33 RNA-Seq datasets (Greenfest-Allen et al., 2017 preprint). The use of iterative WGCNA improved the clustering of genes into modules compared with standard WGCNA (Fig. S3D), with 49% (n=7894) of the expressed genes being partitioned into 91 modules that exhibited distinctive patterns of expression across the pancreatic/endocrine/β-cell developmental transcriptome (Fig. 2). Although most modules contain genes that are expressed in multiple cell populations, 295 genes were identified that were expressed in only a single cell population (Fig. S4, Table S2).
Fig. 2.
Meta-network defined by module eigengenes highlights a shift in transcriptome dynamic prior to endocrine specification. A heatmap-based clustering of the eigengenes of the 91 modules comprising the meta-network illustrated in Fig. 1B reveals a distinct shift in expression pattern just prior to endocrine specification. Modules (columns) are organized according to meta-modules denoted as colored blocks at the top (as in Fig. 1B) and clustered based on correlation between eigengenes within each meta-module. Samples (rows) are clustered by correlation in eigengene-standardized expression across the modules.
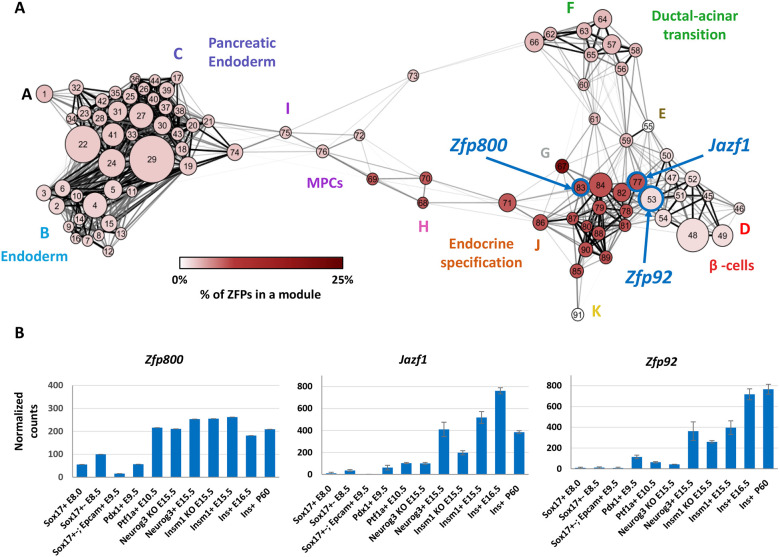
To group and visualize the relationships between co-expression modules further, we also performed a community-detection analysis that identified 11 groups of modules, hereafter termed meta-modules. The higher level of organization provided by grouping of modules (Figs 1B and 2, Table 1), makes the network easier to interpret by associating the meta-modules with specific developmental stages. Most of the meta-modules were further segregated into three discrete sub-nets defined by a core of highly interconnected modules that we refer to as strongly connected module groups (SCMGs). The two largest SCMGs (I and II), which together contain 80% (n=6332) of the genes that were successfully classified, reflect a major shift in the transcriptome that occurs when pancreatic endoderm differentiates into pre-endocrine and endocrine cells. The majority of expressed genes not contained in SCMG I and II are present either in several intermediate modules representing multipotent progenitor cells, or in SCMG III, branched out meta-modules that reflect the re-specification of cellular fate in the absence of Neurog3.
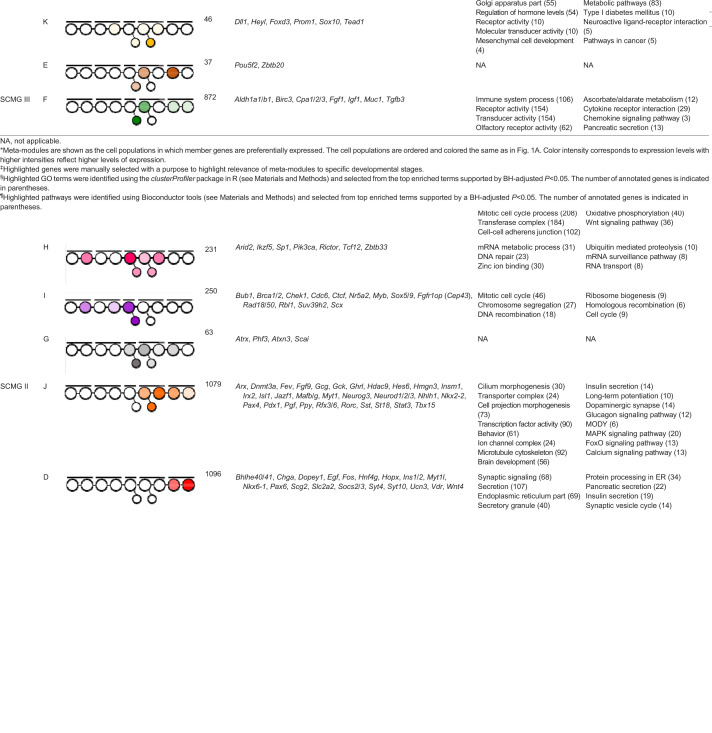
Table 1.
Meta-module highlights
Description of GCN for pancreatic β-cell development
SCMG I is a very densely connected grouping of modules that correspond to early stages of endoderm and/or pancreatic specification (Figs 1B and 2, Table 1). Meta-module B contains genes that are preferentially expressed in definitive endoderm and includes TFs known to govern the patterning of early gut endoderm (Cdx4, Hoxa5, Gata2, Eomes) and genes involved in several morphogenic signaling pathways [e.g. WNT (Wnt5a/b, Wnt6), TGF/Bmp (Tgfb1i1, Bmp2, Bmp5) and FGF (Fgf10, Fgfr1)]. Meta-module C contains genes expressed at higher levels in E9.5 foregut endoderm cells, including both ventral foregut endoderm, as marked by Sox17 and EPCAM expression, and Pdx1-expressing posterior foregut endoderm. TFs important for the specification of pancreas and the establishment of different endocrine lineages (Gata4, Foxa1, Onecut1) are found in this meta-module (Zaret et al., 2008). Interestingly, meta-module C contains more than a dozen cadherin-expressing genes that may be important for determining the morphology of the pancreatic epithelium, including the γ cluster protocadherins, which are essential for neuronal development (Chen et al., 2012). Meta-module C also contains several well-known endoderm markers (Sox17, Cldn6) (Anderson et al., 2008); genes involved in cell division (Cdk2, Aurka), RNA transcription and processing (Nono, Hnrnpc) and carbohydrate catabolism (Hk2, Ldha); and TFs involved in regulation of metabolic processes (e.g. Cebpa, Pprc1).
The transition from unspecified endoderm to specified endocrine cells, or from SCMG I to SCMG II, is spanned by meta-modules I and H. Meta-module I contains genes with low expression in early endoderm (E8.5), higher expression in pancreatic multipotent progenitor cells (MPCs; Ptf1a+ at E10.5) and Neurog3−/− cell populations, followed by very low expression in later endocrine stages. This meta-module includes Sox9, a key transcription factor involved in maintaining MPCs, as well as Nr5a2, a nuclear receptor controlling both MPC formation and acinar cell differentiation (Hale et al., 2014). Meta-module I is also enriched for genes involved in cell cycle regulation (Bub1, Chek1, Cdc6, Myb, Rbl1), chromatin regulation (Ctcf, Suv39h2) and DNA damage repair (Rad50, Brca1). Meta-module H contains genes initially co-expressed with Ptf1a at E10.5 and at low levels in endocrine progenitor cells, but not in P60 mature β-cells. Similar to the transitional meta-module I, meta-module H is enriched for genes involved in chromatin maintenance, mRNA processing and degradation. This meta-module is also enriched for ZFPs (e.g. Zbtb33, Ikzf5, Sp1) and several transcriptional co-regulators (Arid2, Tcf12). The expression of transcriptional and post-transcriptional DNA and RNA regulators at this stage may be important for the transition from multipotent progenitor cells towards endocrine cell transcriptome.
SCMG II encompasses two large meta-modules (J and D) that, together, contain 24 modules of genes expressed during endocrine cell differentiation and β-cell specification and maturation, respectively. Meta-module J contains genes expressed during endocrine differentiation, and the individual modules within it exhibit widely variable expression patterns. For example, genes in module 85 are highly expressed in Insm1−/− cells (arrested endocrine progenitors) and include the key endocrine and β-cell fate regulators Neurog3 and Pax4. In contrast, the genes in module 88 are preferentially expressed in Neurog3+ and Insm1+ endocrine progenitors and include the pro-endocrine TFs Myt1, Rfx3 and Fev. Genes in module 79, including Insm1, Neurod1 and St18 (Myt3), are expressed in endocrine progenitors and then maintain lower expression levels in nascent β-cells. Module 77 contains genes with peak expression in Ins1+ cells at E16.5, including the pro-β-cell TFs Mafb and Mlxipl (ChREPB). Overall, meta-module J contains many putative transcriptional regulators, including 67 ZFPs, most of which are uncharacterized, but may play a role in regulating the differentiation of endocrine cells. SCMG II also contains meta-module D, which groups modules of genes for which expression is highest in nascent and/or mature β-cells. This meta-module includes TFs Nkx6-1, Pax6 and Tshz1, all of which have been shown to be important for maintenance of adult β-cell identity (Perelis et al., 2016; Raum et al., 2015; Swisa et al., 2017; Taylor et al., 2013), as well as circadian rhythm regulators Clock, Nr1d1, Bhlhe40 and Bhlhe41, underlying their importance for normal β-cell physiology (Perelis et al., 2016). Modules 48 and 49 contain genes that exhibit highest expression levels at P60. These modules contain Ins1 and Ins2, genes involved in protein synthesis and secretion (Syt4, Scg2, Chga), the glucose transporter Slc2a2, and Ucn3, a marker for β-cell maturation, and several TFs important for the function of β-cells (Vdr, Hnf4g and Hopx). Interestingly, all α cluster protocadherin genes reside in module 48 of meta-module D, whereas the β cluster is in meta-module J (modules 79, 81, 90) and the γ cluster is in meta-module C (module 36). This suggests that different families of protocadherin genes, each of which are known to be essential for cell adhesion, are crucial for establishing epithelial cell surface diversity in a stage-specific manner during β-cell development.
SCMG III is a group of modules that includes most of meta-module F. These modules delineate genes that become upregulated in the absence of Neurog3, which are likely indicative of fate re-specification in the absence of this key transcription factor. Meta-module F contains ductal and acinar-specific genes, including the ductal marker Muc1 and the acinar markers Cpa1 and Aldh1a1. This finding is consistent with the failure of Neurog3-deficient trunk progenitor cells to delaminate and form endocrine cells, instead being redirected towards ductal and acinar cell lineages (Wang et al., 2010). On account of this, we observe the divergent clustering of genes expressed in Neurog3−/− and Insm1−/− cell populations, two populations of cells that do not normally occur during pancreas development. Inclusion of these cell populations in our study resulted in two distinct meta-modules (F and K, respectively) the gene content of which reflects the effects that knocking out Neurog3 or Insm1 has on β-cell development. Meta-module F is fully contained in its own SCMG (III), consistent with the loss of Neurog3 blocking the formation of endocrine cells and causing a re-specification of cellular fate (Wang et al., 2010). In contrast, meta-module K is closely associated with meta-module J in SCMG II, consistent with the less-consequential effect of knocking out Insm1 compared with Neurog3 (Osipovich et al., 2014) as endocrine-like cells continue to be formed in mice lacking Insm1. The classifications of all identified genes within the GCN and their developmental expression patterns are available at https://markmagnuson.github.io/BetaCell-GCN/genebrowser.
Distribution of regulatory genes in the network
We next explored the distribution of different categories of regulatory genes within the modules of the GCN and found 563 TF-encoding genes, 1904 signaling pathway genes and 151 lincRNAs (long non-coding intergenic RNAs) expressed in the developmental lineages leading to a mature β-cell (Tables S2 and S3). Interestingly, although TFs are found in 93% (85/91) of the detected modules, a large proportion of them reside in just 11 modules within meta-modules B, H, G and J (Table S3). Similarly, signaling pathway genes are also very widely distributed across the GCN but only nine modules are enriched in these genes, with almost half of them being in early endodermal meta-module B. Like TFs and signaling pathway genes, lincRNAs are also non-randomly distributed, as they are present in 58% of the modules but enriched in only ten modules. Five of the lincRNA-enriched modules are in meta-module F, and, notably, three modules are in adult β cell-associated meta-module D (modules 48, 50 and 52) and in module 77 (meta-module J), which comprises genes active in nascent β cells.
In addition to regulatory genes, we examined the distribution of genes for which expression changes are associated with glucose intolerance and type 2 diabetes (T2D) in human islets (Fadista et al., 2014) and found 1557 mouse orthologs of the aforementioned human genes, 726 of which were present in our β-cell network and were non-randomly distributed. Perhaps not surprisingly, most of the modules containing genes important in glucose homeostasis and metabolism reside in meta-module D (modules 48, 52, 53, 54) and J (modules 78, 79, 82, 84). A full listing of enriched modules for each regulatory gene set and T2D genes is provided in Table S3.
Sub-families of clustered protocadherin genes are preferentially expressed at different stages during β-cell development
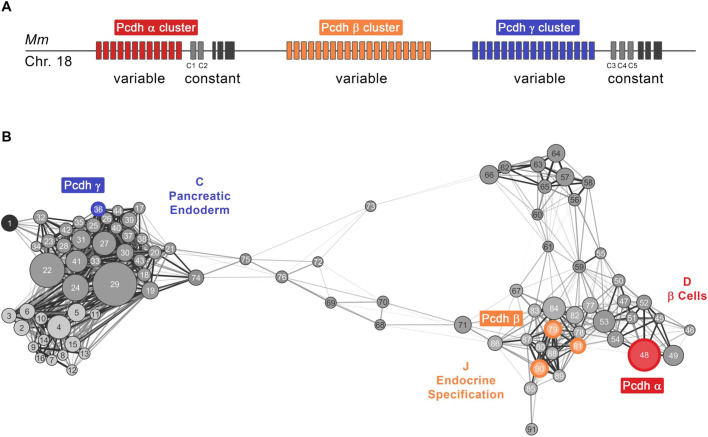
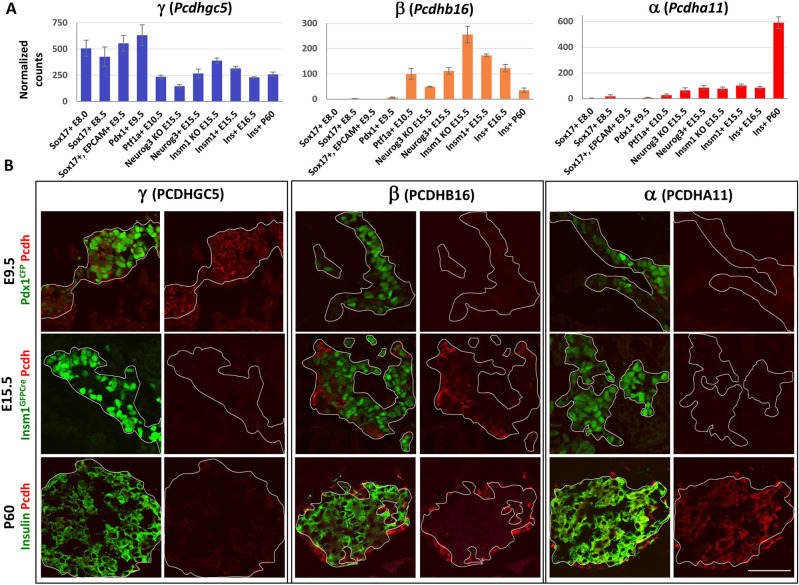
As previously noted, we observed the developmental stage-dependent expression of the clustered protocadherins (Pcdh) genes, which are present in the mouse genome as three closely linked sequential gene clusters designated α (Pcdha), β (Pcdhb) and γ (Pcdhg) (Fig. 3A). The selective transcription of individual Pcdh isoforms is achieved by promoter choice followed by alternative pre-mRNA cis-splicing, which generates a wide diversity of Pcdh isoforms within single cells (Chen and Maniatis, 2013). In the network, Pcdhγ genes are preferentially expressed during pancreatic endoderm formation (meta-module C, module 36), Pcdhβ genes during early endocrine differentiation (meta-module J, modules 78, 81 and 90) and Pcdhα genes in mature β-cells (meta-module D, module 48) (Fig. 3B, Table S2).
Fig. 3.
Distribution of clustered protocadherins in β-cell development GCN. (A) Schematic of genomic DNA locus (mouse chromosome 18) containing clustered protocadherins (Pcdh) genes organized in three closely linked gene clusters designated α, β and γ. Variable exons of the Pcdhα cluster sub-family are colored in red, Pcdhβ in orange and Pcdhγ in blue. Homologous Pcdhα and Pcdhγ C family variable exons are in light gray and constant exons are shown in black. (B) Different cluster sub-families of protocadherins are found in modules (square nodes) falling into meta-modules representing different developmental parts of the network. A meta-network view of GCN highlights that the module containing Pcdhα genes (red) is in meta-module D (mature β-cells), modules with Pcdhβ genes (orange) are in meta-module J (endocrine specification), and a module with Pcdhγ genes (blue) is in meta-module C (pancreatic endoderm).
To validate the developmental stage-dependent expression of clustered Pcdh genes, we co-stained foregut endoderm (marked by Pdx1CFP reporter allele expression at E9.5), endocrine progenitor cells (marked by InsmGFPCre reporter allele expression at E15.5) and mature β-cells (marked by insulin at P60) with antibodies against proteins expressed from Pcdh genes representing different clusters: Pcdhgc5 (cluster γ), Pcdhb16 (cluster β) and Pchda11 (cluster α) (Fig. 4A). These experiments revealed that PCDHGC5 is preferentially expressed at E9.5 in Pdx1-expressing foregut endoderm, whereas its expression is diminished or absent in endocrine progenitor cells and mature β-cells. Moreover, PCDHGC5 colocalizes with cells co-stained with anti-PDX1 antibodies at E9.5 (Fig. 4B; Fig. S5A). In contrast, PCDHB16 is preferentially expressed in endocrine progenitors at E15.5, whereas PCHDA11 is preferentially expressed in mature islet cells at P60 (Fig. 4B). Immunostaining confirmed that PCDHB16 is expressed in sub-populations of INSM1- and NEUROG3-expressing cells at E15.5. Co-staining with anti-hormone antibodies also indicated that PCDHB16 is co-expressed in glucagon- and pancreatic polypeptide-expressing endocrine cells but not in insulin- or somatostatin-expressing endocrine cells at E15.5 (Figs S5B and S6A). Furthermore, at P60, PCDHB16 expression is restricted to the glucagon-expressing α-cells and pancreatic polypeptide-expressing PP cells (Fig. S6A). At the same time point, PCHDA11-positive immunostaining is enriched in insulin-positive β-cells, with only occasional α and PP cells demonstrating co-expression with PCHDA11 (Fig. S6B). Thus, immunostaining confirms that Pcdhγ cluster genes are expressed preferentially during early pancreas specification, β cluster genes are expressed preferentially during endocrine specification, and α cluster genes are expressed in mature β-cells. These findings illustrate the GCN has utility in revealing the developmental stage-specific expression of groups of genes.
Fig. 4.
Differential expression of clustered protocadherins during β-cell development. (A) Expression levels of representative clustered protocadherins show that the cluster γ protocadherin Pcdhgc5 is preferentially expressed at earlier stages of pancreas development (E8.0 Sox17+-E9.5 Pdx1+), whereas cluster β protocadherin Pcdhb16 is preferentially expressed in E15.5 endocrine cell populations, and the cluster α protocadherin Pcdha11 is preferentially expressed in mature β-cells at P60. (B) Immunofluorescence staining confirms preferential expression of PCDHGC5 protein in Pdx1.CFP-marked pancreatic progenitor cells at E9.5, PCDHB16 expression in Insm1.GFP-marked endocrine progenitor cells at E15.5, and preferential expression of PCDH11 in insulin-positive adult β-cells at P60. Scale bar: 50 µm.
Sub-networks of transcriptional regulators involved in endocrine differentiation and β-cell function
The analysis of topological features of sub-networks has potential utility in prediction of functionally critical or important genes that often exhibit high centrality within GCNs (Carlson et al., 2006). In particular, the degree of centrality, or the number of connections associated with a gene, has been associated with biologically important candidate genes in several systems (Azuaje et al., 2013, 2011). For this reason, we extracted sub-networks for each of the 11 meta-modules, and for transcriptional regulators in each of the meta-modules and performed network topology analysis. All of the extracted sub-networks are viewable at https://markmagnuson.github.io/BetaCell-GCN/ where gene connectivities and degrees of centrality can be readily determined. As an example, a sub-network of transcriptional regulators from meta-module J is shown in Fig. S7. Within this sub-network Neurod1, Insm1 and Isl1, three TFs known to have important functions in endocrine cell development, all exhibit high centrality, as do several other important regulatory factors. For instance, Ncoa6, which has the highest value for degree centrality among transcriptional regulators in meta-module J, encodes a transcriptional co-regulator that is broadly involved in regulation of β-cell identity (Scoville et al., 2015). Similarly, Nsd1, a histone methyltransferase with high centrality, is associated with hyperinsulinemic hypoglycemia (Nakamura et al., 2015). Inspection of a sub-network of transcriptional regulators from meta-module D (Fig. S8) also revealed several TFs known to play a role in β-cell development and mature function (Nkx6-1, Pax6, Clock). In this sub-network, Esr1 has the highest centrality, consistent with this gene being important for the formation of β-cells during development and after injury (Yuchi et al., 2015).
Characterization of ZFPs in meta-module J and D
Because ZFPs are well-established regulators of cell fate, we also determined the proportion of ZFP-coding genes in each module and found that meta-modules H and J contained many ZFPs, suggesting their importance in the formation of pancreatic endocrine cells (Fig. 5A). Moreover, meta-module D also contains several ZFPs that may specifically regulate β-cell function. Overall, 81 C2H2 class ZFPs were associated with endocrine specification and β-cell maturation and are summarized in Table S4. In meta-module J, the ZFPs also exhibited high centrality, suggesting a role in regulating transcription of many other genes (Fig. S7). For example, Casz1 and Zhx1, two ZFPs known to interact with transcriptional repressive complexes (Kim et al., 2007; Liu et al., 2015), both have high network connectivity. Other less well-understood ZFPs, such as Zfp318, Zbtb6 and Zfp800, also show high levels of network connectivity. Interestingly, several ZFPs in meta-module J, including Zfp174, Zfp445 and Jazf1, are associated with T2D (Fadista et al., 2014). Similarly, meta-module D contains ZFPs such as Zfp9, Zfp941 and Zfp92 that may be involved in mature β-cell function (Fig. S8). Identification of multiple ZFPs with potential transcription factor activity that may be involved in endocrine specification and β-cell function provides a second example of GCN utility.
Fig. 5.
Distribution of zinc finger proteins (ZFPs) in β-cell development GCN. (A) A meta-network view of GCN shows module nodes colored according to the proportion of ZFP-coding genes in the modules. Genes coding for ZFPs are enriched in meta-module J (endocrine specification). The C2H2-type ZFPs within each developmental module were identified via GO analysis. Quantified enrichments are represented as a percentage of C2H2 ZPF coding genes per total number of genes within a given module. Thick blue outlines indicate modules that contain the three ZFPs selected for further analysis: Zfp800, Jazf1 and Zfp92. (B) Expression levels of Zfp800, Jazf1 and Zfp92 in profiled developmental lineages show that Zfp800 is preferentially expressed in pancreatic multipotent and endocrine progenitor cells, Jazf1 in endocrine progenitors and nascent β-cells, and Zfp92 in nascent and mature β-cells.
To explore further the role of ZFPs in pancreas development and function, we selected three specific ZFP-containing genes, Jazf1, Zfp92 and Zfp800, for further study. Our selections were based on their differing temporal expression patterns during β-cell development, network degree centrality measures (Zfp800 is highly connected), regulation by Neurog3 (Jazf1 and Zfp92 are strongly regulated by Neurog3), and possible association with T2D (Jazf1). During development, Zfp800 is preferentially expressed in pancreatic MPCs and endocrine progenitor populations, Jazf1 is upregulated in nascent β-cells, and Zfp92 is highly expressed in mature β-cells (Fig. 5B). RT-qPCR of adult tissues showed that Zfp800 is broadly expressed with the highest levels observed in testis and pancreas tissues. Jazf1 is highly expressed in testis, pancreatic islet, adipose and brain tissues, and Zfp92 is predominantly expressed in islets (Fig. S9A). Using scRNA-Seq data from the Tabula Muris Consortium (2018), we found that Zfp800 is highly expressed in β-cells, endocrine cells, and other pancreatic cell types (Fig. S9B), Jazf1 is expressed in α- and β-cells and to a lesser degree in other endocrine cells, and Zfp92 is predominantly expressed in β-cells and in δ-cells.
Zfp800 knockout mice display abnormalities in pancreas development
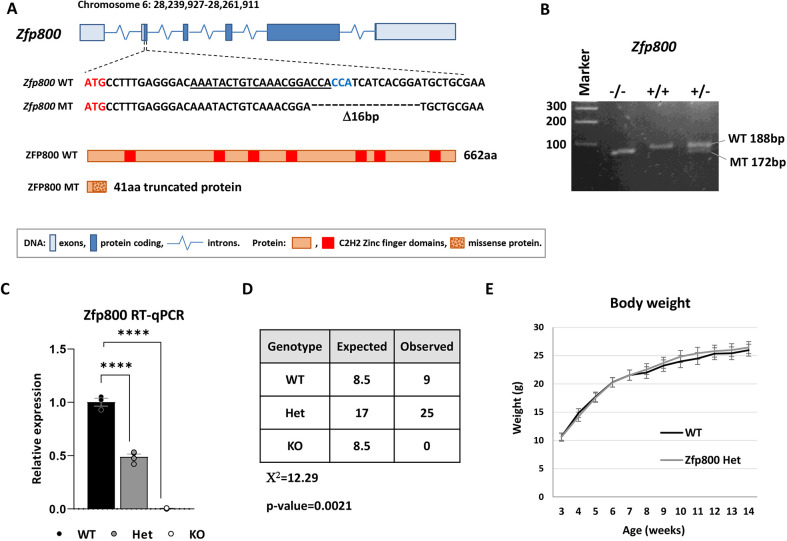
To determine whether one or more of these ZFPs might be important for the development or function of the pancreatic endocrine cells, we performed CRISPR/Cas9 mutagenesis to generate global knockout alleles for Zfp800, Jazf1 and Zfp92. Injection of gene-specific guide RNAs into the Cas9-expressing mouse embryos yielded founder animals bearing frameshift deletions in the first protein-coding exons of each ZFP. For instance, the 16 bp frameshift deletion in the second exon of Zfp800 gene is located 32 bp downstream of the start codon and results in translation of truncated peptide with a missense protein sequence (Fig. 6A,B). Similarly, an 8 bp deletion in the first exon of Jazf1 and a 7 bp deletion in the third exon of Zfp92 are also predicted to cause early frameshifts that would impair function of these ZFPs or expression of their mRNAs (Fig. S10). Consistent with this, Zfp800 mRNA was absent in Zfp800 null mice, probably due to a nonsense-mediated decay (Fig. 6C).
Fig. 6.
Zfp800 knockout mice are postnatally lethal. (A) Schematic of the Zfp800 gene and the location and sequence of the CRISPR/Cas9-generated 16 bp deletion. The guide crRNA sequence that caused the mutation is underlined and the PAM sequence is in a blue font. The deletion of 16 bp leads to a frameshift that abrogates normal ZFP800 662-aa protein translation and leads to translation of a truncated 41-aa peptide with 31 missense amino acids. (B) Representative image of a PCR genotyping gel for Zfp800 wild type (WT), heterozygous (Het) and knockout (KO). (C) RT-qPCR analysis of Zfp800 mRNA expression in pancreatic RNA samples collected at E18.5. n=3, error bars represent s.e.m.; ****P≤0.0001 (one-way ANOVA). (D) χ2 analysis of progenies from Zfp800 Het×Het crossings genotyped at weaning (P21). No Zfp800 knockout animals survived past weaning demonstrating postnatal lethality with complete penetrance. (E) Zfp800 Het and WT animals have similar weights at different ages. n=8 (4 males and 4 females) for each genotype.
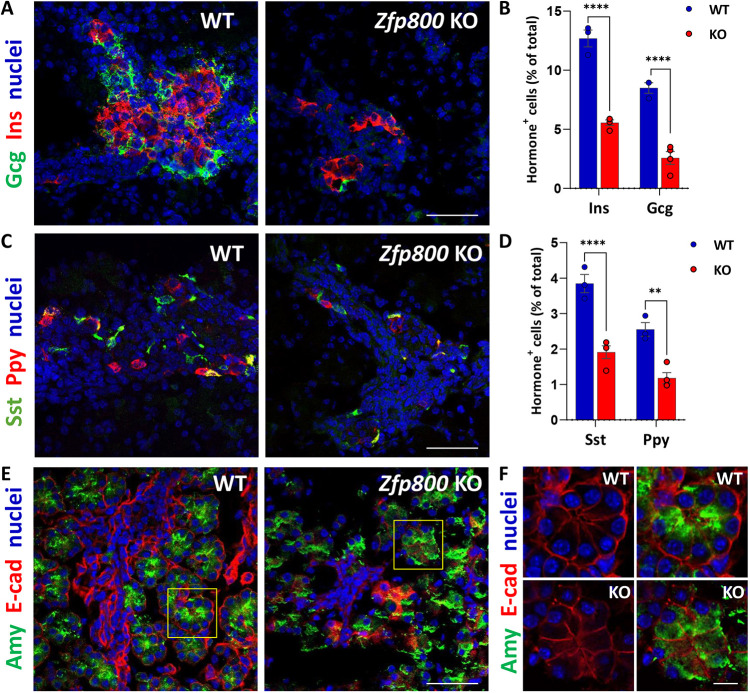
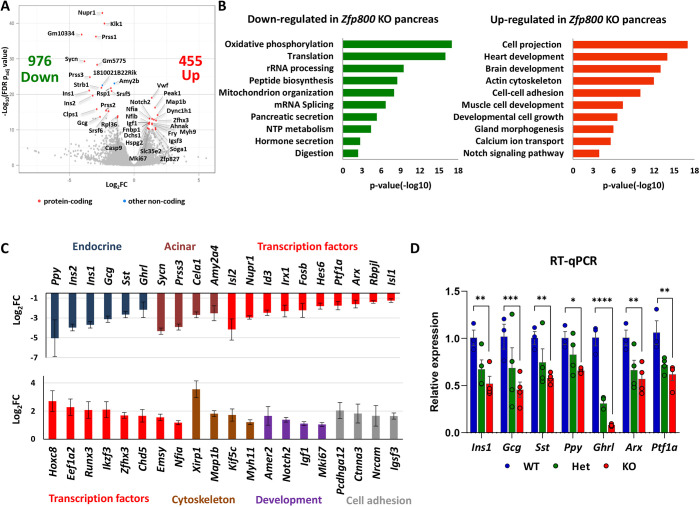
As we established lines from the founder animals, we discovered that although we could efficiently produce Jazf1 and Zfp92 null animals, mice that lacked Zfp800 did not survive past a few days of birth (Fig. 6D). Because Jazf1 and Zfp92 homozygous knockout mice were viable, fertile and did not demonstrate any gross abnormalities, for the purpose of this study we focused on further characterization of Zfp800 knockout embryos. Heterozygous Zfp800 animals were found to survive into adulthood, have normal weight and are fertile (Fig. 6E). To determine whether abnormalities in the pancreas might contribute to the premature demise of Zfp800 null pups, we isolated knockout and wild-type pancreata from E18.5 embryos and analyzed tissue morphology and gene expression by RNA-Seq. Immunostaining revealed reduced numbers of insulin, glucagon, somatostatin and pancreatic polypeptide-positive cells in the knockout embryos, indicating an impairment in endocrine cell development (Fig. 7A-D). In addition, staining for amylase and the acinar cell membrane marker E-cadherin (cadherin 1), revealed defects in acinar cell polarity and organization. Interestingly, in the knockout embryos amylase was no longer concentrated at the apical region of the cell (Fig. 7E,F) and the typical rosette organization of acini was impaired. RNA-Seq of knockout versus wild-type embryos revealed a large number of dysregulated genes, with 976 genes being down- and 455 upregulated (log2FC<−1 or >1, Padj<0.05) in Zfp800 knockouts (Fig. 8A; Table S5).
Fig. 7.
Zfp800 knockout mice have defects in pancreatic endocrine and acinar tissue development. (A-D) Immunofluorescence staining (A,C) and quantification (B,D) of E18.5 pancreatic sections from Zfp800 knockout (KO) and wild-type (WT) mice for pancreatic endocrine hormones insulin (Ins, red) and glucagon (Gcg, green) (A,B), and somatostatin (Sst, green) and pancreatic polypeptide (Ppy, red) expressing cells (C,D) show a reduction in the number of hormone-positive cells in the Zfp800 KO. n=3, error bars represent s.e.m.; ****P≤0.0001; **P≤0.01 (one-way ANOVA). (E,F) Immunofluorescence staining with the acinar cell enzyme amylase (Amy, green) and cell membrane marker E-cadherin (E-Cad, red) of E18.5 pancreatic sections from KO and WT mice demonstrates defects in acinar tissue formation in the absence of Zfp800. Amylase is localized near the apical regions of the cells in the center of the acinus in WT tissues, whereas KO cells are disorganized, with amylase staining throughout. Yellow squares show areas that are enlarged in F. Cell nuclei are stained with To-Pro-3 (blue). Scale bars: 100 µm (A,C,E); 10 µm (F).
Fig. 8.
RNA-Seq analysis of pancreatic tissues from Zfp800 knockout mice. (A) Volcano plot showing differentially expressed genes (log2FC over P-value) in Zfp800 KO versus WT pancreata from E18.5 embryos. The top ten differentially expressed genes (based on Padj value) are indicated by names and total numbers of downregulated (Down) and upregulated (Up) genes are provided (log2FC<−1 or >1, Padj<0.05). (B) Functional enrichment analysis of downregulated and upregulated genes. Select top enriched pathways are shown. (C) Differential expression of select top downregulated (top graph) and upregulated (bottom graph) genes. Colors indicate gene functional associations. Log2FC: log2 fold change Zfp800 KO versus WT (Padj<0.05), error bars represent s.e.m. (Log2FC) determined by DEseq. (D) RT-qPCR analysis of mRNA expression for select pancreatic genes in pancreatic RNA samples collected at E18.5. Zfp800 wild-type (WT), heterozygous (Het) and knockout (KO) samples. n=3-4, error bars represent s.e.m. ****P≤0.0001; ***P≤0.001; **P≤0.01; *P≤0.05 (two-way ANOVA).
Functional enrichment analysis showed that downregulated genes are involved in oxidative phosphorylation, translation, rRNA processing, and pancreatic and hormone secretion, whereas upregulated genes are involved in cell projection morphogenesis, actin cytoskeleton, and heart, brain and muscle development (Fig. 8B; Table S5). Among the top downregulated genes are pancreatic hormones (Ins1, Ins2, Gcg), acinar genes (Prss3, Cela1) and TFs important for pancreas development (Id3, Arx, Ptf1a, Isl1) (Fig. 8C). At the same time, among the top upregulated genes were genes coding for other developmental TFs (Hoxc8, Runx3, Nfia), cytoskeletal components (Xirp1, Map1b) and developmental signaling proteins (Amer2, Notch2). The decrease in expression of pancreatic hormones and TFs in Zfp800 knockout pancreata was confirmed by RT-qPCR (Fig. 8D). Combined, our data show that Zfp800 is required for pancreas development and its disruption leads to multiple alterations in both tissue histology and gene expression. Although the function of Zfp800 in pancreas and other tissues remains to be analyzed further, these findings provide a third example of the utility of our GCN.
DISCUSSION
A robust, developmentally based GCN
Using 11 different FACS-purified cell populations, we calculated a GCN that reflects multiple steps in the developmental lineage resulting in pancreatic β-cells. By utilizing iterative WGCNA, an extension of standard WGCNA, and by using an eigengene connectivity threshold of 0.85 (Greenfest-Allen et al., 2017 preprint), we classified more genes into modules and obtained more modules than prior studies (Benitez et al., 2014; Juhl et al., 2008; Scearce et al., 2002; White et al., 2008) (Fig. S3). Furthermore, grouping of the modules into meta-modules based on their overall eigengene similarity, and identification of three SCMGs based on high topological overlap, enabled similarities in gene expression to be viewed at high, medium and low levels of resolutions, respectively. Indeed, the 11 meta-module networks we describe associate closely related modules with specific developmental stages.
Predictive value of the GCN
The collection of modules, meta-modules and SCMGs we describe provides a great deal of unexplored information about both pancreatic and β-cell development. By grouping genes into a multi-level framework, patterns and associations can be identified that enable new insights into β-cell development. First, our exploration of the network revealed that genes necessary for TFGβ and Notch signaling, and cell-cell adhesion involving cadherins, are preferentially expressed in modules associated with the early cell populations, but not those associated with endocrine cells, reflecting a transition from cellular morphogenesis to that of hormone synthesis and secretion (Table 1). Second, we found that transcriptional regulators, signaling proteins and lincRNAs are non-randomly distributed among the developmentally delineated gene-modules. Third, by extracting 11 different sub-networks for each meta-module from the GCN, we were able to identify many unstudied transcriptional regulators from the two meta-modules, J and D, most closely associated with endocrine differentiation and β-cell function.
Stage-specific expression of the clustered protocadherin genes
To demonstrate the general utility of our GCN, we first used it to identify changes in the temporal expression pattern of clustered protocadherin genes. We found that Pcdhγ genes are preferentially expressed during pancreas specification (meta-module C), Pcdhβ genes are preferentially expressed during early endocrine differentiation (meta-module J), and Pcdhα genes are preferentially expressed in mature β-cells (meta-module D). These differences are consistent with developmental stage-specific activation of Pdch cluster-specific promoters, and suggest that structural differences within the many different protocadherin isoforms may dictate their divergent functions in pancreatic endoderm, which express cluster γ, endocrine progenitor cells, which express cluster β, and mature β-cells, which express cluster α. Consistent with the early developmental function of cluster γ protocadherins, mice deficient in Pcdhg cluster genes die as neonates (Wang et al., 2002), whereas those deficient in Pcdha cluster genes are viable but have abnormalities in specific neuron functions (Hasegawa et al., 2008; Katori et al., 2009). Moreover, the importance of α cluster protocadherins for mature β-cell function is suggested by the identification of a T2D-associated quantitative trait locus (eQTL) in a promoter region that likely drives expression of several Pcdha genes (Fadista et al., 2014). Given that adhesive and repulsive cell-cell communications are likely to be important for endocrine cell migration, islet cell aggregation, and three-dimensional organization and function of mature islets, our findings point to the need for more in-depth analysis of how different protocadherin genes affect pancreatic morphogenesis.
Pro-endocrine and β-cell-associated meta-modules contain many putative transcriptional regulators
To make it easy for others to explore potential gene-gene associations, we extracted sub-networks for all genes in each meta-module, as well as sub-networks of transcriptional regulators for each meta-module. As a second example of GCN utility, we used two of these sub-networks to explore transcriptional regulators in meta-modules J and D, which contain genes preferentially expressed during endocrine differentiation and mature β-cells. Although these sub-networks contain many established pro-endocrine and β-cell-specific TFs, they also revealed many uncharacterized ZFPs. We identified and described 81 ZFPs in meta-modules J and D that could play yet-to-be-defined roles in endocrine specification and β-cell maturation (Table S4). Similar analysis of the meta-modules associated with definitive endoderm or pancreatic endoderm specification could also yield new targets for understanding endoderm differentiation.
Exploring the function of three ZFPs in pancreas development
Finally, to demonstrate the functional importance of several of the ZFPs in meta-modules J and D in pancreas and/or endocrine development, we selected Zfp800, Jazf1 and Zfp92 for a more detailed analysis. These selections were based on their location in the network, regulation by Neurog3, and possible association with T2D. Zfp800, Jazf1 and Zfp92 each contain classical C2H2 zinc finger DNA-binding domains, with Zfp92 also containing a repressive KRAB domain. In adult tissues, Zfp800 is broadly expressed but has high levels of expression in the pancreas, and both Jazf1 and Zfp92 are highly expressed in pancreatic islets, with Jazf1 also being expressed in other tissues at lower levels. According to our developmental GCN, Zfp800 begins to be expressed at E10.5, peaks at E15.5 in endocrine cells, and is maintained in adult β-cells. Jazf1 and Zfp92 become highly expressed in E15.5 endocrine cells, and peak in nascent β-cells at E16.5 and adult β-cells, respectively. These variations may provide clues to the function of each protein. For instance, Zfp800 may be required during early development, whereas Jazf1 and Zfp92 may be more important for later stages of development and cellular function of adult β-cells. Moreover, a high degree centrality measure for Zfp800 suggests that this gene may play a more important functional role (Azuaje, 2014).
In support of this network-based prediction, we found that Zfp800 knockout animals exhibited early postnatal lethality whereas Jazf1 and Zpf92 knockouts survive into adulthood. Although the deletion of Jazf1 and Zfp92 did not cause any profound developmental phenotypes, these proteins may contribute to the function of adult pancreatic endocrine cells, or in other tissues. Indeed, recent studies of β-cell specific Jazf1 knockout mice have revealed a role for Jazf1 in regulating ribosomal protein synthesis and protection against endoplasmic reticulum stress-induced apoptosis (Kobiita et al., 2020). Interestingly, a role for Jazf1 was only revealed when the knockout allele was analyzed in the context of the highly diabetogenic Akita mutation.
Zfp800 is crucial for pancreas development
Our observations of early postnatal lethality of mice lacking Zfp800, a decrease in the number of pancreatic endocrine cells, and disturbances in pancreatic exocrine cell morphology, indicate that Zfp800 has a crucial, albeit undefined, role in the development and/or function of the pancreas, and possibly other tissues as well. Although expression of many pancreatic genes was decreased in the absence of Zfp800, the downregulation of Ptf1a is noteworthy because Ptf1a regulates acinar cell maturation as well as pancreatic MPCs at earlier developmental time points (Kawaguchi et al., 2002). As Zfp800 is expressed in Ptf1a-expressing MPCs that develop into endocrine, ductal and acinar cell types, a decrease in Ptf1a expression in the MPC lineage in Zfp800 knockouts could be a factor that affects later development of all pancreatic lineages (Fukuda et al., 2008). Similarly, alterations seen in the expression of both Arx and Isl1 are likely to affect the development of pancreatic endocrine cells by altering the production of specific endocrine cell types (Bastidas-Ponce et al., 2017).
Zfp800-deficient pancreata exhibit reduced expression of genes involved in metabolic energy production, ribosomal protein synthesis and pancreatic secretion, and increased expression of cytoskeletal genes, and genes associated with muscle and brain development. Zfp800 clearly has effects within the pancreas, but based on its expression pattern it might also contribute to the development and/or function of other cell types. Interestingly, little is known about the role of ZNF800 in humans although several nearby single nucleotide polymorphisms (rs17866443, rs77084830, rs28947805 and rs74788757) are associated with T2D (Mahajan et al., 2018). Moreover, PDX1, NKX2-2 and NKX6-1 have been reported to bind near the ZFP800 promoter in human pancreatic progenitor cells (Cebola et al., 2015; Mularoni et al., 2017).
Conclusion
We expect the GCN we have calculated to be useful for further understanding the developmental transitions that occur along the developmental lineage from endoderm to mature pancreatic β-cells. We also expect that the many ZFPs that we have found to be expressed during the formation of pancreatic endocrine cells will be of both developmental and functional importance.
MATERIALS AND METHODS
Mouse strains
Sox17tm2(EGFP/cre)Mgn (Sox17GFPCre), Pdx1tm2Mgn (Pdx1CFP), Ptf1atm1.1Mgn (Ptf1aYFP), Neurog3tm1(EGFP)Khk (Neurog3GFP), Insm1tm1.1Mgn (Insm1GFPCre) and Tg(Ins1-EGFP)1Hara (MIP-GFP) mice (Mus musculus) have been previously described (Burlison et al., 2008; Choi et al., 2012; Hara et al., 2003; Lee et al., 2002; Osipovich et al., 2014; Potter et al., 2012). Embryos were isolated from timed matings where the presence of vaginal plug at noon was considered as E0.5. All animals were housed at Vanderbilt University and the protocols used to isolate cells were approved by the Vanderbilt Institutional Animal Care and Use Committee.
FACS isolation
Sox17GFPCre, Pdx1CFP, Ptf1aYFP and Insm1GFPCre dissected embryonic tissues were prepared as previously described (Choi et al., 2012). Dissected tissues from Neurog3GFP and MIP-GFP embryos were dissociated using trypsin, filtered, centrifuged at 180 g for 5 min, and cells were suspended in PBS containing 10% fetal bovine serum with DAPI or propidium iodide to exclude dead cells. For the postnatal MIP-GFP samples, pancreata were perfused, dissociated, and islets were hand-selected. Cell populations were analyzed and isolated by FACS using an Aria II or III (BD Biosciences) to purities of at least 95%. GFP and YFP were excited with a 488 nm laser and emission detected with a 502 nm long pass and 530/30 band pass filter. CFP was excited using 445 nm laser and emission detected with a 470 nm long pass and 510/80 band pass filter. Diagrams of the mouse alleles and representative FACS isolation profiles are provided in Figs S1 and S2.
RNA isolation, amplification, library construction, and sequencing for the GCN
For each developmental cell population, transcriptome profiling was performed on amplified RNA from three independent biological replicates. Cells were sorted either into PBS containing 10% fetal bovine serum, centrifuged at 180 g for 5 min and suspended in TRIzol or into TRIzol LS (Invitrogen). Total RNA was isolated, DNase-treated (Ambion), column-purified (Zymo Research) and assessed using a Bioanalyzer (Agilent Technologies), and RNAs with an RNA integrity number >7 were processed for RNA-Seq. The Ovation RNA-Seq system, version 1 (NuGEN), was used to amplify 10 ng of total RNA. cDNA was sheared using the Covaris S2 and multiplexed libraries were constructed using the TruSeq DNA sample prep kits A&B (Illumina). Libraries were hybridized using the TruSeq SR cluster kit, version 3, and the TruSeq SBS kit, version 3. Single-end tags of at least 100 nucleotides were obtained using a HiSeq 2000 (Illumina). RNA amplification, library preparation and sequencing were performed at Vanderbilt Technologies for Advanced Genomics (VANTAGE) core. Feature extraction, deconvolution, conversion of BCL to FastQ was accomplished using HCS 1.4.8 or HCS 1.5.15.1, RTA 1.12.4.2 or 1.13.48, and CASAVA 1.8.0a19 or 1.8.2. Reads were mapped to the mm10 genome with the Spliced Transcripts Alignment to a Reference (STAR) v.2.3.0 software package. More than 3×107 reads were obtained for each sample, ∼70% of which were uniquely mapped to the reference genome (Table S1).
RNA-Seq for Zfp800 knockout mice
Bulk RNA-Seq of the E18.5 embryonic pancreas samples was performed by using unamplified RNA. Libraries were prepared and sequenced by Novogene Corporation using a NovaSeq6000 (Illumina) platform. Raw sequencing reads were processed using Trim Galore! (0.5.0) and FastQC (0.11.8) was used to assess quality before and after the removal of adapter sequences and pairs that were either <20 bp or that had Phred scores <20. The Spliced Transcripts Alignment to a Reference (STAR 2.6.0) (Dobin et al., 2013) was used to align the short reads against the mm10 (GRCm38) mouse genome reference and used GENCODE comprehensive gene annotations (Release M17). The average uniquely mapped reads count per sample was 22 M (75% of total) Differential gene expression analysis utilized DESeq2 (Dobin et al., 2013) and clusterProfiler (Yu et al., 2012). Genes exhibiting <20 normalized read counts in three or more samples or genes not expressed in all samples were removed.
Normalization and filtering
Expression was quantified as normalized counts using the PORT pipeline (available at https://github.com/itmat/Normalization). PORT extends the idea of resampling discussed by Li and Tibshirani (2013) to not only correct for different read depths, but also other factors such as exon/intron/intergenic balance and different success in ribosomal depletion. We combined UCSC known genes and RefSeq genes obtained from UCSC Genome Bioinformatics as our mm10 gene models. Genes exhibiting <10 normalized read counts in two or more replicates of a cell population were considered not expressed in that population; genes not expressed in all cell populations were removed from subsequent analyses.
Co-expression network
We applied iterative WGCNA (Greenfest-Allen et al., 2017 preprint) using the algorithm available via GitHub (https://github.com/cstoeckert/iterativeWGCNA) and an eigengene connectivity (kME) value of 0.85. Iterative WGCNA was used to construct a meta-network and estimate topological overlap among the modules based on correlations between detected module eigengenes. The Spin-glass community detection algorithm, as implemented in igraph library (Csardi and Nepusz, 2006), was then used to group modules with a large degree of topological overlap into meta-modules using R, version 3.3.2 (R Core Team, 2015). This method uses simulated annealing to maximize the ratio of intra-community to inter-community connectivity to better segregate highly connected and potential overlapping sub-networks. Hierarchical clustering (correlation-based) of modules within meta-modules was performed on their eigengenes. The module meta-network was visualized using Cytoscape (Shannon et al., 2003).
Functional annotation and pathway analysis
Enriched pathways were identified using the package KEGGprofile (Zhao and Shyr, 2015) in R. Enriched gene functions were identified using gene ontology (GO) annotations (Ashburner et al., 2000) obtained from the Bioconductor genome-wide mouse annotation database org.Mm.eg.db (version 3.2.3) and the R package clusterProfiler (Yu et al., 2012). The complete set of expressed genes in the β-cell developmental transcriptome was used as the reference set in all enrichment analyses. The enrichment for C2H2-type ZFPs within each module were identified via GO analyses using DAVID v6.8 (Huang et al., 2009). All genes from each module were queried using default parameters in DAVID, and identified C2H2-type genes were counted. Quantified enrichments were represented as a percentage of the total number of genes within a given module and the network enrichment visualization was produced using Cytoscape.
Regulatory gene enrichment analysis
TFs were identified as genes annotated directly with either the GO term ‘sequence-specific DNA binding transcription factor activity’ (GO:0003700) or one of its children. Genes involved in signaling pathways identified as those annotated by REACTOME (Croft et al., 2014; Milacic et al., 2012) as associated with signal transduction pathways. lincRNAs were identified from ENSEMBL exon annotations using the UCSC gene table browser. Enrichment of each gene set within individual modules was evaluated using the Fisher's exact test; non-random distributions of the regulatory gene sets across the network was assessed with a χ2 test. All statistical analyses were performed with the base R package, version 3.3.2.
Expression specificity analysis
Cell population-specific gene expression was assessed using the H and Q measures described by Schug et al. (2005). H is based on Shannon-entropy and measures overall specificity: a gene with a low H value shows a highly non-uniform expression across the conditions considered. Q combines H with the relative expression level of a gene in a given condition: a low value of Q indicates specificity of that gene expression in that condition. Here, H and Q were computed at the exon level and exons filtered for H<1.5 (Fig. S3). The first quartile of Q-values was used as an initial threshold to generate a list of condition-specific exons that was adjusted downward as necessary to refine the gene list for cell populations of interest.
Sub-network analysis and availability
Network topology analysis of sub-networks was performed using the NetworkAnalyzer plugin (Doncheva et al., 2012) for Cytoscape. The transcriptional regulators within meta-modules J and D sub-network analysis were compiled manually based on GO annotation (GOTERM_BP_DIRECT, GO:0006355) using the DAVID functional annotation resource (Huang et al., 2009). The sub-network files for all meta-modules are available to download as Cytoscape-compatible JSON files and as interactive web pages using the CytoscapeJS (Franz et al., 2016) at https://markmagnuson.github.io/BetaCell-GCN/.
Immunohistochemistry
Immunofluorescence staining and imaging of frozen sections was performed as previously described (Choi et al., 2012). Co-staining with anti-protocadherin antibodies was carried out on pancreatic tissues isolated from Pdx1CFP/+ mice at E9.5, Insm1GFP/+ mice at E15.5 and wild-type mice at P60. Zfp800 knockout and litter matched wild-type pancreatic tissues were isolated at E18.5. Primary antibodies and dilutions used were as follows: rabbit anti-PCDHG5 (MBS9204539, MyBioSource), 1:200; rabbit anti-PCDHB16 (MBS2525954, MyBioSource), 1:200; rabbit anti-PCDHA11 (orb1299, Biorbyt), 1:500; chicken anti-GFP/CFP (A10262, Invitrogen), 1:500; guinea pig anti-PDX-1 (a gift from Chris Wright, Vanderbilt University, Nashville, TN, USA), 1:1000; guinea pig anti-INS (PA1-26938, Invitrogen), 1:1000; rabbit anti-GCG (4030-01, Linco Research), 1:1000; sheep anti-SST (13-2366, American Research Products), 1:1000; rabbit anti-GHRL (19-031-30, Phoenix Pharmaceuticals), 1:1000, guinea pig anti-PPY (4041-01, Linco Research), 1:1000; goat anti-NEUROG3 (AB2774, BCBC Antibody Core), 1:500; mouse anti-INSM1 (AB2154, BCBC Antibody Core), 1:500; goat anti-amylase (sc-12821, Santa Cruz Biotechnology), 1:1000; mouse anti-E-cadherin (13-1700, Invitrogen), 1:500. Secondary antibodies (Invitrogen, 1:1000) were as follows: goat anti-chicken IgG Alexa 488 (A-11039) or 555 (A-21437) conjugated; goat anti-guinea pig IgG Alexa 555 (A-21435) or 647 (A-21450) conjugated; donkey anti-rabbit IgG Alexa 555 (A-31572) or 680 (A-10043) conjugated; donkey anti-sheep IgG Alexa 555 (A-21099) conjugated. Nuclei were stained with TO-PRO-3 dye (T3605, Invitrogen, 1:1000). Images were acquired using an LSM Meta 510 confocal microscope with either 20× or 40× objectives.
Image quantification
The number of endocrine hormone-positive cells per total number of pancreatic cells defined by nuclear staining was calculated by manual cell counting on stained step sections (50 µm apart). At least 4000 cells were counted for each E18.5 pancreatic sample, and tissues from at least three different animals of the same genotype were counted using ImageJ software. Data were expressed as the percentage of hormone-producing cells per total pancreatic cells as defined by nuclear staining.
RT-qPCR
RNA from adult (P60) mouse organs or E18.5 pancreata was isolated using the Maxwell 16 LEV simplyRNA Tissue Kit (Promega). Reverse transcription was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Two nanograms of cDNA were used in real-time qPCR with Power SYBR Green PCR master mix (Applied Biosystems) using a CFX96 Real Time PCR system (Bio-Rad). Primers are listed in Table S6. Relative expression in comparison with that of wild type was determined using the 2−ΔΔCt method, using Hprt expression for initial normalization.
CRISPR mutagenesis
Zfp800, Jazf1 and Zfp92 knockout mice were obtained by microinjecting target-specific guide RNA (gRNA) complexes into Cas9-expressing embryos obtained from the mating of homozygous B6J/129(Cg)-Gt(ROSA)26Sortm1.1(CAG−cas9*,-EGFP)Fezh/J mice (JAX 026179) with C57Bl/6N mice. For each target gene, a pool of three gRNA complexes (crRNA+tracrRNA) (Table S6) were microinjected into single-cell embryos at a final concentration of 20 µM. The guide RNA complexes were made by first diluting crRNA (IDT) and tracrRNA (IDT) to a 100 µM stock in IDT Duplex Buffer. A mixture of 2 µl of crRNA (100 µM), 2 µl of tracrRNA (100 µM) and 6 µl of water was then heated to 95°C for 5 min and then slowly cooled to 25°C to anneal. To make the final, pooled injection mixture, 1.5 µl (∼675 ng) of each complex was mixed with the injection buffer (1 mM Tris HCl, pH 7.5, 0.1 mM EDTA, filtered through 0.2 µm) to a final concentration of 20 µM, or approximately 10 ng/µl of each gRNA complex. Founder animals resulting from the gRNA complex microinjections were screened for INDEL mutations via T7-assays (NEB) and by determining heterodimer formation via PAGE-gel analyses. For screening by PAGE-gel, PCR products using primers flanking the targeted site were heated to 95°C for 5 min and then slowly cooled to room temperature to reanneal. PCR products were then run on a 10% TBE PAGE gel (Invitrogen). PCR products for mutant founders were then analyzed by Sanger sequencing (GENEWIZ). All microinjections and founder screenings were performed by the Vanderbilt Gene Editing Resource. Founder animals were then bred to C57Bl6/J mice for seven generations while the Cas9-expressing ROSA26 allele was segregated out. Pups were genotyped by PCR using primers surrounding the deletion sites (Table S6).
Acknowledgements
We thank Jody Peters, Susan Hipkens, Rama Gangula and Lily Kim for technical assistance, Travis Clark for performing the RNA amplification and sequencing, and Jennifer Skelton and Linda Gower for expert performance of pronuclear DNA microinjection and husbandry. All RNA-Seq data processing was performed at the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. The Vanderbilt Gene Editing Resource is funded by the National Institutes of Health (DK020593 and CA68485) and VANTAGE is supported by the National Institutes of Health (CA68485, EY08126 and RR030956).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.B.O., E.G.-A., G.G., C.J.S., M.A.M.; Methodology: A.B.O., E.G.-A., J.-P.C., E.M., L.P.C., E.C., C.J.S.; Software: E.G.-A., J.-P.C., E.M., C.J.S.; Validation: A.B.O., E.G.-A., J.-P.C., C.J.S.; Formal analysis: A.B.O., E.G.-A., J.-P.C., E.M., C.J.S., M.A.M.; Investigation: A.B.O., K.D.D., E.G.-A., E.M., L.P.C., E.C., A.G.C.; Resources: J.-P.C., E.C., G.G., C.J.S., M.A.M.; Data curation: A.B.O., K.D.D., E.G.-A., J.-P.C., E.M.; Writing - original draft: A.B.O., E.G.-A., H.W.C.; Writing - review & editing: A.B.O., C.J.S., M.A.M.; Visualization: A.B.O., E.G.-A., J.-P.C.; Supervision: C.J.S., M.A.M.; Project administration: M.A.M.; Funding acquisition: G.G., C.J.S., M.A.M.
Funding
This study was supported by funding from the National Institutes of Health (DK72473 and DK89523 to M.A.M.). Deposited in PMC for release after 12 months.
Data availability
RNA-Seq data and metadata are available in ArrayExpress (accession numbers E-MTAB-2816 and E-MTAB-9538). Zfp800, Jazf1 and Zfp92 knockout mice are available from the Vanderbilt Cryopreserved Mouse Repository.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.196964.supplemental
References
- Anderson, W. J., Zhou, Q., Alcalde, V., Kaneko, O. F., Blank, L. J., Sherwood, R. I., Guseh, J. S., Rajagopal, J. and Melton, D. A. (2008). Genetic targeting of the endoderm with claudin-6CreER. Dev. Dyn. 237, 504-512. 10.1002/dvdy.21437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda, H. E., Benitez, C. M. and Kim, S. K. (2013). Gene regulatory networks governing pancreas development. Dev. Cell 25, 5-13. 10.1016/j.devcel.2013.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T.et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuaje, F. J. (2014). Selecting biologically informative genes in co-expression networks with a centrality score. Biol. Direct 9, 12. 10.1186/1745-6150-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuaje, F. J., Rodius, S., Zhang, L., Devaux, Y. and Wagner, D. R. (2011). Information encoded in a network of inflammation proteins predicts clinical outcome after myocardial infarction. BMC Med Genomics 4, 59. 10.1186/1755-8794-4-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuaje, F., Zhang, L., Jeanty, C., Puhl, S.-L., Rodius, S. and Wagner, D. R. (2013). Analysis of a gene co-expression network establishes robust association between Col5a2 and ischemic heart disease. BMC Med Genomics 6, 13. 10.1186/1755-8794-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas-Ponce, A., Scheibner, K., Lickert, H. and Bakhti, M. (2017). Cellular and molecular mechanisms coordinating pancreas development. Development 144, 2873-2888. 10.1242/dev.140756 [DOI] [PubMed] [Google Scholar]
- Benitez, C. M., Qu, K., Sugiyama, T., Pauerstein, P. T., Liu, Y., Tsai, J., Gu, X., Ghodasara, A., Arda, H. E., Zhang, J.et al. (2014). An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. PLoS Genet. 10, e1004645. 10.1371/journal.pgen.1004645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlison, J. S., Long, Q., Fujitani, Y., Wright, C. V. E. and Magnuson, M. A. (2008). Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev. Biol. 316, 74-86. 10.1016/j.ydbio.2008.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M. R. J., Zhang, B., Fang, Z., Mischel, P. S., Horvath, S. and Nelson, S. F. (2006). Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics 7, 40. 10.1186/1471-2164-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebola, I., Rodríguez-Seguí, S. A., Cho, C. H.-H., Bessa, J., Rovira, M., Luengo, M., Chhatriwala, M., Berry, A., Ponsa-Cobas, J., Maestro, M. A.et al. (2015). TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 17, 615-626. 10.1038/ncb3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. V. and Maniatis, T. (2013). Clustered protocadherins. Development 140, 3297-3302. 10.1242/dev.090621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. V., Alvarez, F. J., Lefebvre, J. L., Friedman, B., Nwakeze, C., Geiman, E., Smith, C., Thu, C. A., Tapia, J. C., Tasic, B.et al. (2012). Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron 75, 402-409. 10.1016/j.neuron.2012.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, E., Kraus, M. R.-C., Lemaire, L. A., Yoshimoto, M., Vemula, S., Potter, L. A., Manduchi, E., Stoeckert, C. J., Jr, Grapin-Botton, A. and Magnuson, M. A. (2012). Dual lineage-specific expression of Sox17 during mouse embryogenesis. Stem Cells 30, 2297-2308. 10.1002/stem.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, E., Stein, R. and Hunter, C. S. (2014). Revealing transcription factors during human pancreatic β cell development. Trends Endocrinol. Metab. 25, 407-414. 10.1016/j.tem.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft, D., Mundo, A. F., Haw, R., Milacic, M., Weiser, J., Wu, G., Caudy, M., Garapati, P., Gillespie, M., Kamdar, M. R.et al. (2014). The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472-D477. 10.1093/nar/gkt1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi, G. and Nepusz, T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems 1695, 1-9. [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva, N. T., Assenov, Y., Domingues, F. S. and Albrecht, M. (2012). Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 7, 670-685. 10.1038/nprot.2012.004 [DOI] [PubMed] [Google Scholar]
- Ecco, G., Imbeault, M. and Trono, D. (2017). KRAB zinc finger proteins. Development 144, 2719-2729. 10.1242/dev.132605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista, J., Vikman, P., Laakso, E. O., Mollet, I. G., Esguerra, J. L., Taneera, J., Storm, P., Osmark, P., Ladenvall, C., Prasad, R. B.et al. (2014). Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. USA 111, 13924-13929. 10.1073/pnas.1402665111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedotova, A. A., Bonchuk, A. N., Mogila, V. A. and Georgiev, P. G. (2017). C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae 9, 47-58. 10.32607/20758251-2017-9-2-47-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz, M., Lopes, C. T., Huck, G., Dong, Y., Sumer, O. and Bader, G. D. (2016). Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics 32, 309-311. 10.1093/bioinformatics/btv557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, A., Kawaguchi, Y., Furuyama, K., Kodama, S., Horiguchi, M., Kuhara, T., Kawaguchi, M., Terao, M., Doi, R., Wright, C. V. E.et al. (2008). Reduction of Ptf1a gene dosage causes pancreatic hypoplasia and diabetes in mice. Diabetes 57, 2421-2431. 10.2337/db07-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfest-Allen, E., Cartailler, J.-P., Magnuson, M. A. and Stoeckert, C. J. (2017). iterativeWGCNA: iterative refinement to improve module detection from WGCNA co-expression networks. bioRxiv 234062. 10.1101/234062 [DOI] [Google Scholar]
- Halbleib, J. M. and Nelson, W. J. (2006). Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199-3214. 10.1101/gad.1486806 [DOI] [PubMed] [Google Scholar]
- Hale, M. A., Swift, G. H., Hoang, C. Q., Deering, T. G., Masui, T., Lee, Y.-K., Xue, J. and MacDonald, R. J. (2014). The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development 141, 3123-3133. 10.1242/dev.109405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, M., Wang, X., Kawamura, T., Bindokas, V. P., Dizon, R. F., Alcoser, S. Y., Magnuson, M. A. and Bell, G. I. (2003). Transgenic mice with green fluorescent protein-labeled pancreatic β-cells. Am. J. Physiol. Endocrinol. Metab. 284, E177-E183. 10.1152/ajpendo.00321.2002 [DOI] [PubMed] [Google Scholar]
- Hasegawa, S., Hamada, S., Kumode, Y., Esumi, S., Katori, S., Fukuda, E., Uchiyama, Y., Hirabayashi, T., Mombaerts, P. and Yagi, T. (2008). The protocadherin-α family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell. Neurosci. 38, 66-79. 10.1016/j.mcn.2008.01.016 [DOI] [PubMed] [Google Scholar]
- Huang, D. W., Sherman, B. T. and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Imbeault, M., Helleboid, P.-Y. and Trono, D. (2017). KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550-554. 10.1038/nature21683 [DOI] [PubMed] [Google Scholar]
- Juhl, K., Sarkar, S. A., Wong, R., Jensen, J. and Hutton, J. C. (2008). Mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3-null mouse. Diabetes 57, 2755-2761. 10.2337/db07-1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner, K. H., Lee, C. S., Scearce, L. M., Brestelli, J. E., Arsenlis, A., Le, P. P., Lantz, K. A., Crabtree, J., Pizarro, A., Mazzarelli, J.et al. (2003). Transcriptional program of the endocrine pancreas in mice and humans. Diabetes 52, 1604-1610. 10.2337/diabetes.52.7.1604 [DOI] [PubMed] [Google Scholar]
- Katori, S., Hamada, S., Noguchi, Y., Fukuda, E., Yamamoto, T., Yamamoto, H., Hasegawa, S. and Yagi, T. (2009). Protocadherin-α family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. 29, 9137-9147. 10.1523/JNEUROSCI.5478-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, Y., Cooper, B., Gannon, M., Ray, M., MacDonald, R. J. and Wright, C. V. E. (2002). The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32, 128-134. 10.1038/ng959 [DOI] [PubMed] [Google Scholar]
- Kim, S.-H., Park, J., Choi, M.-C., Kim, H.-P., Park, J.-H., Jung, Y., Lee, J.-H., Oh, D.-Y., Im, S.-A., Bang, Y.-J.et al. (2007). Zinc-fingers and homeoboxes 1 (ZHX1) binds DNA methyltransferase (DNMT) 3B to enhance DNMT3B-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 355, 318-323. 10.1016/j.bbrc.2007.01.187 [DOI] [PubMed] [Google Scholar]
- Klug, A. (2010). The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys. 43, 1-21. 10.1017/S0033583510000089 [DOI] [PubMed] [Google Scholar]
- Kobiita, A., Godbersen, S., Araldi, E., Ghoshdastider, U., Schmid, M. W., Spinas, G., Moch, H. and Stoffel, M. (2020). The diabetes gene JAZF1 is essential for the homeostatic control of ribosome biogenesis and function in metabolic stress. Cell Rep. 32, 107846. 10.1016/j.celrep.2020.107846 [DOI] [PubMed] [Google Scholar]
- Larsen, H. L. and Grapin-Botton, A. (2017). The molecular and morphogenetic basis of pancreas organogenesis. Semin. Cell Dev. Biol. 66, 51-68. 10.1016/j.semcdb.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Lee, C. S., Perreault, N., Brestelli, J. E. and Kaestner, K. H. (2002). Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 16, 1488-1497. 10.1101/gad.985002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. and Tibshirani, R. (2013). Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat. Methods Med. Res. 22, 519-536. 10.1177/0962280211428386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Lam, N. and Thiele, C. J. (2015). Zinc finger transcription factor CASZ1 interacts with histones, DNA repair proteins and recruits NuRD complex to regulate gene transcription. Oncotarget 6, 27628-27640. 10.18632/oncotarget.4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo, A., Cesaro, E., Montano, G., Zurlo, D., Izzo, P. and Costanzo, P. (2013). KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr. Genomics 14, 268-278. 10.2174/13892029113149990002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, A., Taliun, D., Thurner, M., Robertson, N. R., Torres, J. M., Rayner, N. W., Payne, A. J., Steinthorsdottir, V., Scott, R. A., Grarup, N.et al. (2018). Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505-1513. 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milacic, M., Haw, R., Rothfels, K., Wu, G., Croft, D., Hermjakob, H., D'Eustachio, P. and Stein, L. (2012). Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 4, 1180-1211. 10.3390/cancers4041180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni, L., Ramos-Rodríguez, M. and Pasquali, L. (2017). The pancreatic islet regulome browser. Front. Genet. 8, 13. 10.3389/fgene.2017.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi, H. S., Mnaimneh, S., Schmitges, F. W., Garton, M., Lam, K. N., Yang, A., Albu, M., Weirauch, M. T., Radovani, E., Kim, P. M.et al. (2015). C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555-562. 10.1038/nbt.3128 [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Takagi, M., Yoshihashi, H., Miura, M., Narumi, S., Hasegawa, T., Miyake, Y. and Hasegawa, Y. (2015). A case with neonatal hyperinsulinemic hypoglycemia: It is a characteristic complication of Sotos syndrome. Am. J. Med. Genet. A 167, 1171-1174. 10.1002/ajmg.a.36996 [DOI] [PubMed] [Google Scholar]
- Oldham, M. C., Konopka, G., Iwamoto, K., Langfelder, P., Kato, T., Horvath, S. and Geschwind, D. H. (2008). Functional organization of the transcriptome in human brain. Nat. Neurosci. 11, 1271-1282. 10.1038/nn.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovich, A. B., Long, Q., Manduchi, E., Gangula, R., Hipkens, S. B., Schneider, J., Okubo, T., Stoeckert, C. J., Jr, Takada, S. and Magnuson, M. A. (2014). Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development 141, 2939-2949. 10.1242/dev.104810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, F. C. and Wright, C. (2011). Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 240, 530-565. 10.1002/dvdy.22584 [DOI] [PubMed] [Google Scholar]
- Peek, S. L., Mah, K. M. and Weiner, J. A. (2017). Regulation of neural circuit formation by protocadherins. Cell. Mol. Life Sci. 74, 4133-4157. 10.1007/s00018-017-2572-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelis, M., Ramsey, K. M., Marcheva, B. and Bass, J. (2016). Circadian transcription from beta cell function to diabetes pathophysiology. J. Biol. Rhythms 31, 323-336. 10.1177/0748730416656949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, L. A., Choi, E., Hipkens, S. B., Wright, C. V. E. and Magnuson, M. A. (2012). A recombinase-mediated cassette exchange-derived cyan fluorescent protein reporter allele for Pdx1. Genesis 50, 384-392. 10.1002/dvg.20804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2015). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Raum, J. C., Soleimanpour, S. A., Groff, D. N., Coré, N., Fasano, L., Garratt, A. N., Dai, C., Powers, A. C. and Stoffers, D. A. (2015). Tshz1 regulates pancreatic beta-cell maturation. Diabetes 64, 2905-2914. 10.2337/db14-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scearce, L. M., Brestelli, J. E., McWeeney, S. K., Lee, C. S., Mazzarelli, J., Pinney, D. F., Pizarro, A., Stoeckert, C. J., Jr., Clifton, S. W., Permutt, M. A.et al. (2002). Functional genomics of the endocrine pancreas: the pancreas clone set and PancChip, new resources for diabetes research. Diabetes 51, 1997-2004. 10.2337/diabetes.51.7.1997 [DOI] [PubMed] [Google Scholar]
- Schug, J., Schuller, W.-P., Kappen, C., Salbaum, J. M., Bucan, M. and Stoeckert, C. J., Jr. (2005). Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6, R33. 10.1186/gb-2005-6-4-r33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville, D. W., Cyphert, H. A., Liao, L., Xu, J., Reynolds, A., Guo, S. and Stein, R. (2015). MLL3 and MLL4 methyltransferases bind to the MAFA and MAFB transcription factors to regulate islet beta-cell function. Diabetes 64, 3772-3783. 10.2337/db15-0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B. and Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498-2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisa, A., Avrahami, D., Eden, N., Zhang, J., Feleke, E., Dahan, T., Cohen-Tayar, Y., Stolovich-Rain, M., Kaestner, K. H., Glaser, B.et al. (2017). PAX6 maintains β cell identity by repressing genes of alternative islet cell types. J. Clin. Invest. 127, 230-243. 10.1172/JCI88015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B. L., Liu, F.-F. and Sander, M. (2013). Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 4, 1262-1275. 10.1016/j.celrep.2013.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tabula Muris Consortium (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas, C. K., Emery, L. E., Denchi, E. L. and Miller, K. M. (2018). Caught with one's zinc fingers in the genome integrity cookie jar. Trends Genet. 34, 313-325. 10.1016/j.tig.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Weiner, J. A., Levi, S., Craig, A. M., Bradley, A. and Sanes, J. R. (2002). Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843-854. 10.1016/S0896-6273(02)01090-5 [DOI] [PubMed] [Google Scholar]
- Wang, S., Yan, J., Anderson, D. A., Xu, Y., Kanal, M. C., Cao, Z., Wright, C. V. E. and Gu, G. (2010). Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev. Biol. 339, 26-37. 10.1016/j.ydbio.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Bender, A., Wang, P., Karakose, E., Inabnet, W. B., Libutti, S. K., Arnold, A., Lambertini, L., Stang, M., Chen, H.et al. (2017). Insights into beta cell regeneration for diabetes via integration of molecular landscapes in human insulinomas. Nat. Commun. 8, 767. 10.1038/s41467-017-00992-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, P., May, C. L., Lamounier, R. N., Brestelli, J. E. and Kaestner, K. H. (2008). Defining pancreatic endocrine precursors and their descendants. Diabetes 57, 654-668. 10.2337/db07-1362 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang, L.-G., Han, Y. and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284-287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuchi, Y., Cai, Y., Legein, B., De Groef, S., Leuckx, G., Coppens, V., Van Overmeire, E., Staels, W., De Leu, N., Martens, G.et al. (2015). Estrogen receptor alpha regulates beta-cell formation during pancreas development and following injury. Diabetes 64, 3218-3228. 10.2337/db14-1798 [DOI] [PubMed] [Google Scholar]
- Zaret, K. S., Watts, J., Xu, J., Wandzioch, E., Smale, S. T. and Sekiya, T. (2008). Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb. Symp. Quant. Biol. 73, 119-126. 10.1101/sqb.2008.73.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17. 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- Zhao, S. and Shyr, Y. (2015). KEGGprofile: an annotation and visualization package for multi-types and multi-groups expression data in KEGG pathway. R package version 1.8.2. https://github.com/slzhao/KEGGprofile.