SUMMARY
The development of single-cell RNA sequencing (scRNA-seq) has allowed high-resolution analysis of cell-type diversity and transcriptional networks controlling cell-fate specification. To identify the transcriptional networks governing human retinal development, we performed scRNA-seq analysis on 16 time points from developing retina as well as four early stages of retinal organoid differentiation. We identified evolutionarily conserved patterns of gene expression during retinal progenitor maturation and specification of all seven major retinal cell types. Furthermore, we identified gene-expression differences between developing macula and periphery and between distinct populations of horizontal cells. We also identified species-specific patterns of gene expression during human and mouse retinal development. Finally, we identified an unexpected role for ATOH7 expression in regulation of photoreceptor specification during late retinogenesis. These results provide a roadmap to future studies of human retinal development and may help guide the design of cell-based therapies for treating retinal dystrophies.
In Brief
Lu et al. performed scRNA-seq on the developing human retina and retinal organoids. Using comprehensive analyses, they deduced mechanisms regulating human retinal cell-fate specification and foveagenesis and contrasted these with murine retinal development. The data are a valuable resource for understanding human retinal development and disease.
INTRODUCTION
The vertebrate retina is an accessible system for studying central nervous system (CNS) development. The retina develops from a polarized layer of neuroepithelial cells that gives rise to six major classes of neurons and one class of glia in temporally distinct, but often overlapping, intervals during development. Certain cell types, such as retinal ganglion cells (RGCs), horizontal cells, cone photoreceptors, and GABAergic amacrine cells, are born relatively early, while glia, bipolar cells, glycinergic amacrine, and most rod photoreceptors are born relatively late (Cepko, 2014; La Vail et al., 1991; Voinescu et al., 2009; Wong and Rapaport, 2009; Young, 1985). The birth order of these cell types is evolutionarily conserved and regulated by largely intrinsic mechanisms (Gomes et al., 2011; He et al., 2012).
Despite high evolutionary conservation among many aspects of retinal development, there are important species-specific differences. Species differ in the number of subtypes of horizontal cells, with mice having only one subtype, but macaques and chicks have two or three (Boije et al., 2016). The relative ratio of rods to cones, and of photoreceptors to inner retinal cell types, also varies (Peichl, 2005). For instance, in mice the distribution of major cell types does not vary across the retina (Jeon et al., 1998) whereas humans and other primates have a fovea in the central retina that is specialized for high-acuity vision and enriched in cones and retinal ganglion cells (Collin, 1999). The gene regulatory networks that control how these species-specific differences arise are poorly understood.
An understanding of mechanisms controlling human retinal development is particularly important for treatment of human-specific diseases such as retinoblastoma and macular degeneration. Retinoblastoma is caused by inactivation of the RB1 tumor suppressor gene. The retina is exquisitely sensitive to RB1 loss in humans, whereas it is highly resistant in other species, with murine models of retinoblastoma requiring the combined loss of Rb1 and other tumor suppressors (Chen et al., 2004; Dannenberg et al., 2004; MacPherson et al., 2004; Sangwan et al., 2012; Zhang et al., 2004). Whereas the murine disease originates from inner retinal neurons, human retinoblastoma arises from cone precursors (Xu et al., 2009, 2014). These phenotypic differences likely reflect differences in gene regulatory networks in human and murine cones (Xu et al., 2009). A deeper understanding of human and murine cone gene-regulatory networks could generate new insights into tumorigenesis (Ajioka et al., 2007; Bremner and Sage, 2014; Chen et al., 2004; Xu et al., 2014).
Macular degeneration, which in its age-related (AMD) form affects up to 25% of the US population aged over 80 years (Jager et al., 2008), results in central vision loss from the death of photoreceptors in the foveal region of the macula (Curcio et al., 2005). The relevance of animal models for AMD remains unclear since the macula is specific to primates, and most common animal models lack regions of high-acuity vision. Although both bulk RNA-seq analysis and small-scale single-cell RNA-seq (scRNA-seq) studies (Hoshino et al., 2017; Hu et al., 2019) have been used to profile gene-expression changes during human retinal neurogenesis, these data have not shed light on human-specific mechanisms that regulate retinal development, particularly with respect to cone photoreceptor specification and foveal patterning.
Single-cell RNA sequencing technologies provide a powerful tool to comprehensively classify cell types of the central nervous system and the gene regulatory networks that control their development (Fan et al., 2018; Farrell et al., 2018; Liu et al., 2017; Tasic et al., 2018; Wagner et al., 2018; Zeisel et al., 2018; Zhong et al., 2018). Studies in both macaques and humans have examined the diversity of cellular subtypes within the mature retina (Cowan et al., 2019; Liang et al., 2019; Lukowski et al., 2019; Menon et al., 2019; Peng et al., 2019; Voigt et al., 2019). Furthermore, recent studies in mice using scRNA-seq have identified changes in gene expression during multiple aspects of retinal development (Buenaventura et al., 2019; Clark et al., 2019; Lo Giudice et al., 2019). This has led to the identification of genes, such as the NFI family of transcription factors, which directly regulate retinal neurogenesis and cell-fate specification. These large datasets also have the potential to identify both evolutionarily conserved and species-specific gene regulatory networks controlling human retinal development.
In this study, we employ a similar approach to generate a comprehensive scRNA-seq profile of human retinal development. We profile 16 stages of human retinal development and four different maturation stages of retinal organoids, ranging from early neurogenesis through adulthood, analyzing 118,555 retinal cells in total. Comparing human and mouse, we observe broadly similar changes in the gene expression profiles of both retinal progenitor cells (RPCs) and most postmitotic retinal cell types, but we also observe species-specific differences. These include the expression of transcription factors that control the specification of cone photoreceptors and horizontal interneurons and gene regulatory networks that pattern the macula. Most notably, we find that the neurogenic bHLH factor ATOH7, which is expressed in early-stage neurogenic RPCs, and regulates the formation of early-born cells, is also expressed in late-stage neurogenic RPCs and regulates human photoreceptor (rod versus cone) specification. This underscores the importance of obtaining gene-expression data directly from primary human cells, illustrates the limitations of existing animal models for studying human disease, and provides insight into therapeutic approaches for treating retinal diseases.
RESULTS
Construction and Analysis of Human Retinal scRNA-Seq Libraries
To comprehensively profile gene-expression changes across retinal development, we performed scRNA-seq on dissociated retinas from both human retinal organoids and primary tissues. To profile very early stages of retinal development, we profiled human retinal organoids, generated at 24, 30, 42, and 59 days in vitro (Eldred et al., 2018; STAR Methods). These time points correspond to early stages of retinogenesis for which we were unable to procure primary tissue samples and include stages at which the first RGCs are generated. Strikingly, a majority of the cells from organoids profiled between 24 and 42 days in culture (13,552/19,861; 68.2%; Table S1) were annotated as non-eye-field cells, both by expression of ventral telencephalic and hypothalamic markers (FOXG1, NKX2–1, DLX5, ARX, and LHX6; Shimogori et al., 2010), and lack of detectable expression of eye-field specification markers (VSX2, PAX6, RAX, and LHX2). In addition, we generated single-cell transcriptional profiles from whole developing retinas obtained at 9, 11, 12, 13, 14, 15, 16, 17, 18, 19, 22, 24, and 27 gestational weeks (GW), macular and peripheral samples from 20 GW and 8 days postnatal (PND), and whole retina from a healthy 86-year-old donor (Figures 1A, 1B, S1A, S1D, and S1E). Biological and technical replicates were performed at GW24 and GW19, respectively (Figures S1A and S1E). Replicate samples and samples from similar ages (i.e., GW9 and GW11) display high concordance in average gene expression across samples (Figure 1D). Samples were profiled to a mean depth of 3,472.61 unique molecular identifiers (UMIs; SD = 1,862.78) and 1,502.14 genes (SD = 589.41) per cell (Figures S1A–S1C). These samples were then integrated using Monocle 2.99.3 and UMAP dimension reduction on high variance genes to obtain a 3D embedding of retinal development (Figures 1B, 1C, S1D–S1G; Videos S1 and S2; Table S2) (Becht et al., 2018; Qiu et al., 2017). Major cell types were annotated using known cell-type marker genes (Blackshaw et al., 2004; Clark et al., 2019; Macosko et al., 2015; Siegert et al., 2012) expressed within the 126 clusters of cells in the dimension reduction space (Figures S1G and S1H).
Figure 1. Single-Cell RNA-Seq Profiling of the Developing Human Retina.
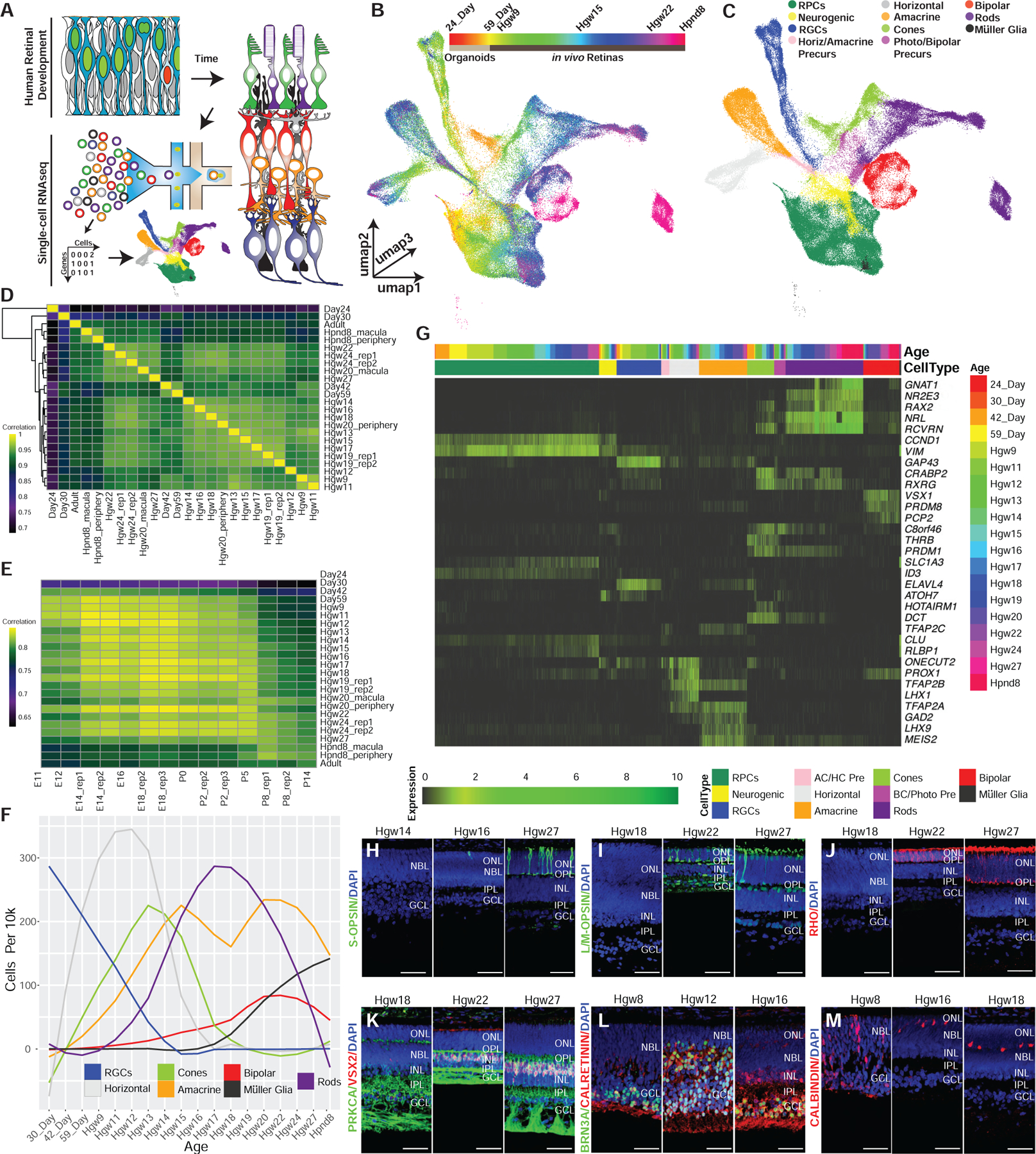
(A) Schematic of experimental design.
(B and C) 3D UMAP embedding of the retina dataset, with individual cells colored by (B) age and (C) annotated cell types.
(D and E) Spearman correlation between the transcriptomes of (D) human samples, or (E) across human and mouse retinal samples.
(F) Normalized specification windows of retinal cell types.
(G) Heatmap showing relative expression of transcripts with high specificity to individual cell types, ordered by cell type and developmental age (top annotation bars).
(H–M) Immunohistochemistry on primary human retinal tissue validating the dynamic expression of cell-type markers, including (H) S-OPSIN (short wavelength cones); (I) L/M-OPSIN (long/medium wavelength cones); (J) Rho (rods); (K) PRKCA and VSX2 (bipolar cells); (L) BRN3A (RGCs) and calretinin (horizontal, amacrine, and RGC cells.); and (M) calbindin (Horizontal cells). Nuclei are counterstained with DAPI. Scale bar, 50 mm. Abbreviations: Hgw, human gestational weeks; Hpnd, human postnatal day; RPCs, retinal progenitor cells; RGCs, retinal ganglion cells; AC/HC Pre, amacrine cell-horizontal cell precursors; BC/Photo Pre, bipolar cell-photoreceptor cell precursors; NBL, neuroblast layer; GCL, ganglion cell layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer.
Although these data were obtained from three different sources (Figure S1A), involving different dissociation techniques (see STAR Methods), this nonetheless yielded an integrated cell distribution and a set of developmental trajectories that broadly resemble those seen in mouse (Clark et al., 2019). Two major classes of RPCs are observed, which match the primary and neurogenic subtypes in mouse (Clark et al., 2019). Differentiating Müller glia form a continuous developmental trajectory that emerges from primary RPCs, while all retinal neurons emerge from the neurogenic fraction. As in the mouse, three major neuronal trajectories are observed: RGCs; amacrine and horizontal interneurons; and rods, cones, and bipolar cells (Figures 1C and S1F; Video S2). RPCs and RGCs from organoids integrate into this distribution, comprising the earliest developmental ages (Figures 1B, S1D, and S1E). Unlike in mice, we observed a distinct trajectory of human horizontal cells (Figures 1C and S1F), likely due to the increased abundance of captured horizontal cells within this dataset compared with mouse (5.8% in human versus 1.5% in mouse; Table S1; Clark et al., 2019).
We next examined the temporal windows of cell-fate specification of each major retinal cell type (Figures 1F, S1I, and S1J). Analysis of the proportions of cell types captured at each age indicated that RGCs are the first cell type specified, detected in the earliest retinal organoid samples (24 and 30 days in vitro; Figures S1I and S1J). The emergence of RGCs was closely followed by cones (30 and 42 days in vitro) and horizontal cells (day 59 in vitro). RGCs, cones, and horizontal cells were all present within the earliest (GW 9) in vivo samples. We observed the emergence of amacrine cells, rods, and bipolar cells shortly thereafter, with significant numbers of each cell type detected at GW12, GW12, and GW14–15, respectively. Müller glia were the last to emerge and were not present in significant numbers until GW19–20. To identify specification windows for individual cell types, we examined the cell type proportions by age within proximal clusters to neurogenic or precursor cell populations (see STAR Methods; Figure 1F). The cell specification windows for human retina largely reflect those observed in mice (Clark et al., 2019; Voinescu et al., 2009).
We next identified selective markers of each major progenitor and postmitotic cell type using feature ranking within annotated cell types with genesorteR (Ibrahim and Kramann, 2019), revealing known and unique markers of specific cell types (Figure 1G). While most show identical cell-type specificity in both mice and humans, discrete patterns of cell-specific expression were evident in some cases. For instance, in mice, the lipid-binding protein Clu is selectively expressed in Müller glia (Blackshaw et al., 2004). In humans, CLU is strongly expressed in both primary RPCs and Müller glia. Aditionally, the retinoid-binding protein CRABP2 is detected exclusively in early-stage primary RPCs in mice (Clark et al., 2019); however, in humans it is also strongly expressed in cone precursors. Other examples of cone-enriched genes in humans that show different patterns of expression in mouse include DCT, which is restricted to the retinal pigmented epithelium in mouse, and HOTAIRM1, which is not detectably expressed in mouse retina (Figure 1G). Other differences include human-specific genes, such as RAX2, which is expressed in both cone and rod photoreceptors. Immunohistochemical analysis demonstrates that protein expression patterns of known marker genes also broadly reflect cell-specific and temporally dynamic patterns of transcript expression detected by scRNA-seq (Figures 1H–1M, S1K–S1Q).
Early and Late-Stage Human RPCs Show Distinct Gene Expression Profiles
Previous scRNA-seq analysis of mouse retinal development identified clear transcriptional signatures of early and late-stage primary RPCs (Clark et al., 2019), reflecting differential expression of genes that control proliferation, neurogenesis, and cell-fate determination. To determine the extent of evolutionary conservation in temporal patterning of RPCs, we conducted pseudotemporal analysis of gene-expression changes across primary RPCs and Müller glia. Interestingly, organoid-derived RPCs displayed gene-expression patterns akin to early-stage RPCs from primary tissue (Figures 2A–2D and S3G). Pseudotemporal ordering of RPCs identified bimodal densities of RPCs across pseudotime, followed by Müller glia (Figure S2A), reflecting the sequential developmental ages of input tissue (Figures 2A–2C and S2B). This mirrored the transcriptional changes observed in mice, although the transition between early and late-stage RPCs occurred more gradually. In mice, the transition from early and late-stage RPCs occurs rapidly between E16 and E18 (Clark et al., 2019), but in humans this process occurs between 11 and 15 GW (Figure S2B) and likely reflects the differences in the timing of human retina development between central and peripheral regions (Diaz-Araya and Provis, 1992; van Driel et al., 1990). The fraction of human RPCs in G1 is increased relative to mouse (p < 0.0001; chi-squared test; Figure S2C), which may reflect the longer time course of retinal neurogenesis in humans relative to mice (Centanin and Wittbrodt, 2014).
Figure 2. Pseudotime Analysis Reveals Genes Differentially Expressed across Primary and Neurogenic RPCs.
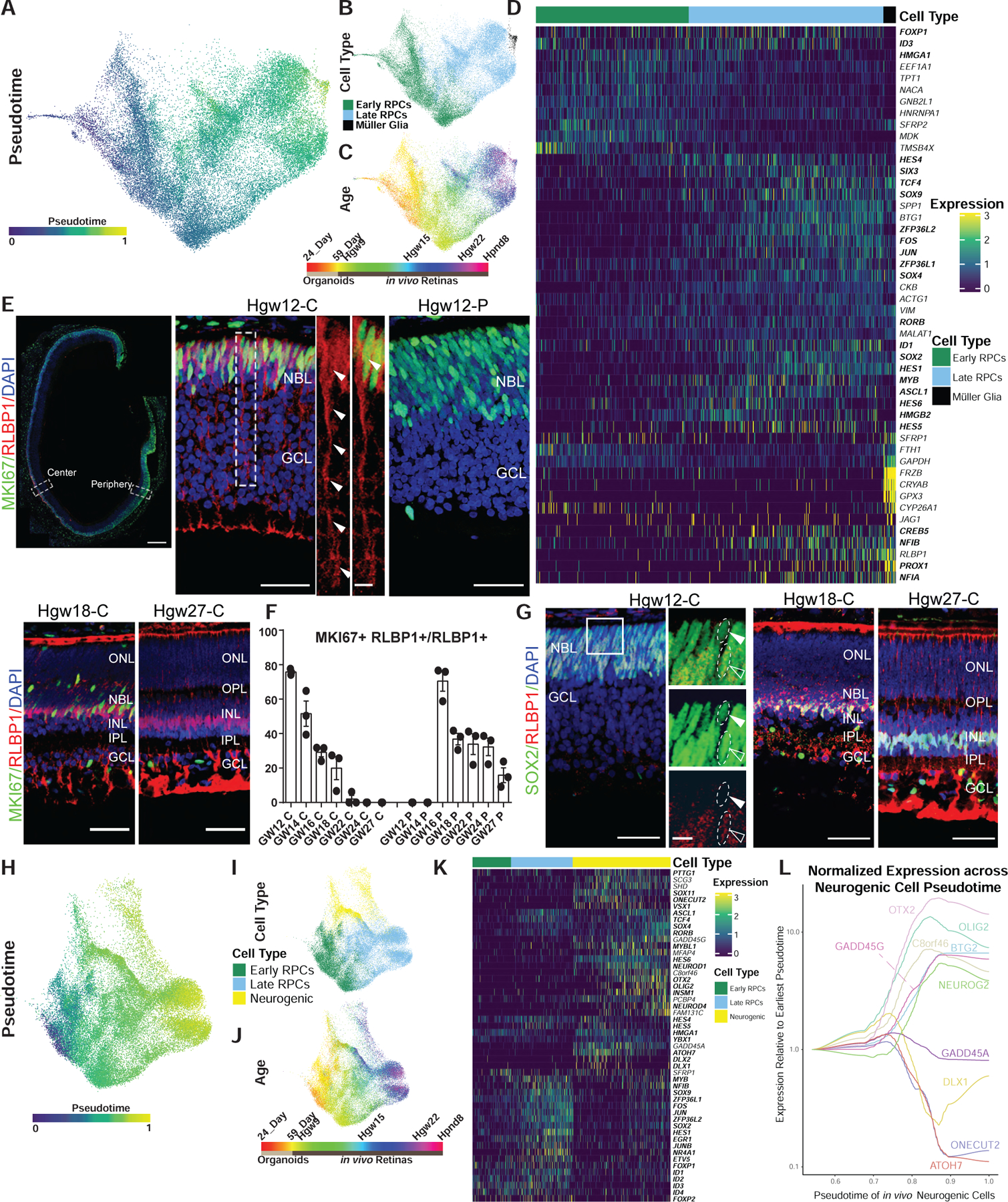
(A–C) UMAP embedding of the developmental trajectories of primary RPCs and Müller glia with cells colored by (A) pseudotime, (B) cell type, and (C) developmental age.
(D) Heatmap of differentially expressed transcripts along pseudotime from primary RPC to Müller glia. Cells are ordered by cell type and pseudotime with transcription factors listed in bold.
(E) Immunohistochemistry detection for RLBP1 and MKI67 in GW12 (top panels), GW18-Central (bottom left), and GW27-Central retina (bottom right) with magnified views of GW12 central (top center) and peripheral (top right) regions. Nuclei are counterstained with DAPI. GW12 scale bar: 300 μm(left), 50 μm(center and middle), 10 μm (center side panels); GW18-C and GW27-C scale bar, 50 μm.
(F) Bar chart showing the proportion of actively proliferating (MKI67+) cells among the RLBP1+ population, as detected by immunohistochemistry, in the central and peripheral regions across the developing human retina (GW12, GW14, GW16, GW18, GW22, GW24, and GW27). Data are mean ± SEM.
(G) Immunohistochemistry of SOX2 and RLBP1 in central regions of developing human retinas at GW12, 18, and 27. High-magnification images are of the boxed region in the GW12 image. Open arrowheads indicate co-localization, with closed arrowheads indicating failure to detect RLBP1 expression. Nuclei are counterstained with DAPI. GW12 scale bar, 50 μm (left), 10 μm (right); GW18-C and GW27-C scale bar, 50 μm.
(H–J) UMAP embedding of primary and neurogenic RPCs with cells colored by (H) pseudotime, (I) cell type, and (J) developmental age.
(K) Heatmap of differential transcript expression along pseudotime from primary RPC to neurogenic RPC. Cells are ordered by cell type and pseudotime with transcription factors listed in bold.
(L) Graph showing the normalized and smoothed expression of early and late neurogenic genes across the neurogenic trajectory relative to expression at the earliest point of neurogenic cell pseudotime. Abbreviations: Hgw, human gestational weeks; GW, gestational weeks; Hpnd, human postnatal day; NBL, neuroblast layer; GCL, ganglion cell layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; C, central retina; P, peripheral retina.
We observed similar expression patterns of markers for early and late-stage primary RPCs in humans as in mice, including SFRP2 and NFIA, respectively (Figures 2D and S2I). Likewise, many of the genes upregulated in Müller glia begin their expression in late-stage primary RPCs (i.e., RLBP1 and NFIB) (Figure 2D). Unlike the Müller glia-specific expression pattern seen in mice (Blackshaw et al., 2004; Reichenbach and Bringmann, 2013), both CLU and VIM are expressed in primary RPCs at all stages of neurogenesis (Figure 1G). Known inhibitors of the WNT pathway (SFRP1/2 and FRZB) are expressed first in early RPCs and then again at later stages in Müller glia (Figure 2D). Immunohistochemistry revealed that expression of RLBP1 is observed in KI67+ mitotic late-stage RPCs in the central retina by 12 GW and displays a more developmentally delayed peripheral retinal expression by 16 GW (Figures 2E, 2F, and S2D). The loss of MKI67 and RLBP1 co-localization coincided with the emergence of co-localization of SOX2 and RLBP1 in Müller glia (Figures 2G, S2F, and S2G). Similar trends of MKI67/RLBP1 and RLBP1/SOX2 co-expression were observed within the scRNA-seq data (Figures S2E and S2H). Notably, the GW20 macula sample displayed very few MKI67/RLBP1 cells but large numbers of SOX2/RLBP1 positive cells, consistent with an advanced differentiation of the macular region compared with the rest of the retina (Figures S2E and S2H).
Neurogenic RPCs are observed in all developmental samples except the day 24 organoids and PND8 samples (Figures 2H–2J; Table S1), and pseudotemporal ordering of neurogenic RPCs reflected transcriptional signatures of age-matched primary RPCs. Neurogenic RPCs likewise showed broadly similar temporal expression patterns in humans as in mice (Figures 2I–2K and S2J). As in mice, a subset of human genes show enriched expression in either early or late-stage neurogenic RPCs (Figures 2K and S2J); however, pseudotime analyses in humans did not reveal a clear signature of early and late-stage neurogenic RPCs, as is observed in mice (Figures S2K–S2L) (Clark et al., 2019). Transcripts that are enriched in early-stage neurogenic RPCs compared with late-stage neurogenic RPCs in both humans and mice include DLX1/2, ONECUT2, and ATOH7 (Figures 2K, 2L, and S2J). Conversely, ASCL1, OTX2, and SOX4 display increased expression levels in late-stage neurogenic RPCs, compared with early-stage neurogenic cells (Figures 2K, 2L, and S2J). A handful of genes, however, showed substantial species-specific differences in expression. Several genes that are expressed across neurogenic RPC development in mice—such as OLIG2, NEUROG2, and BTG2—display increased expression in late-stage human neurogenic RPCs (Figures 2L and S2J). Moreover, C8orf46 (3110035E14Rik; Vexin), which is selectively expressed in early-stage neurogenic RPCs in mice (Clark et al., 2019; Moore et al., 2018), has higher expression in late stage, compared with early-stage, neurogenic RPCs in humans (Figure 2L). As in primary RPCs, a greater fraction of human neurogenic RPCs are in G1 than in mice (p < 0.0001; chi-square test), and the fraction of cells in G1 increases steadily over the course of neurogenesis (Figure S2M).
Identification of Differentiation Trajectories for Each Major Human Retinal Cell Type
To identify gene regulatory networks that control retinal cell specification and differentiation, we conducted independent pseudotime analyses along developmental trajectories from RPCs to each of the major retinal cell types (Figures 3 and S3). We observed many transcription factors that show highly enriched expression at various stages of differentiation of each cell type in humans; observations largely recapitulated in mice. Both human and mouse retinal ganglion cells express the well-characterized transcript markers POU4F1, POU4F2, POU6F2, ISL1, NHLH2, RXRG, EBF1, and EBF3 (Clark et al., 2019; Rheaume et al., 2018; Tran et al., 2019) (Figures 3A–3D); however, human RGCs also express MYC (Figure 3D). Human horizontal cells express TFAP2A/B, ONECUT1/2, LHX1, and ESRRB, consistent with expression in mice (Figures 3E–3H; S3H; (Boije et al., 2016; Clark et al., 2019), and show considerable overlap in transcription-factor expression with amacrine cells (Figures 3I–3L and S3H). However, differentiating human starburst amacrine cells (SACs) selectively express NEUROG3, which is not detected in mouse retina in any cell type. Human SACs also express the transcription factors SOX2, ISL1, and FEZF1, which are also selectively expressed in mouse SACs (Figures S3A–S3C and S3H; Balasubramanian and Gan, 2014; Clark et al., 2019; Peng et al., 2020).
Figure 3. Pseudotime Analyses Identify Transcription Factor Networks Controlling Human Retinal Cell Fate Specification.
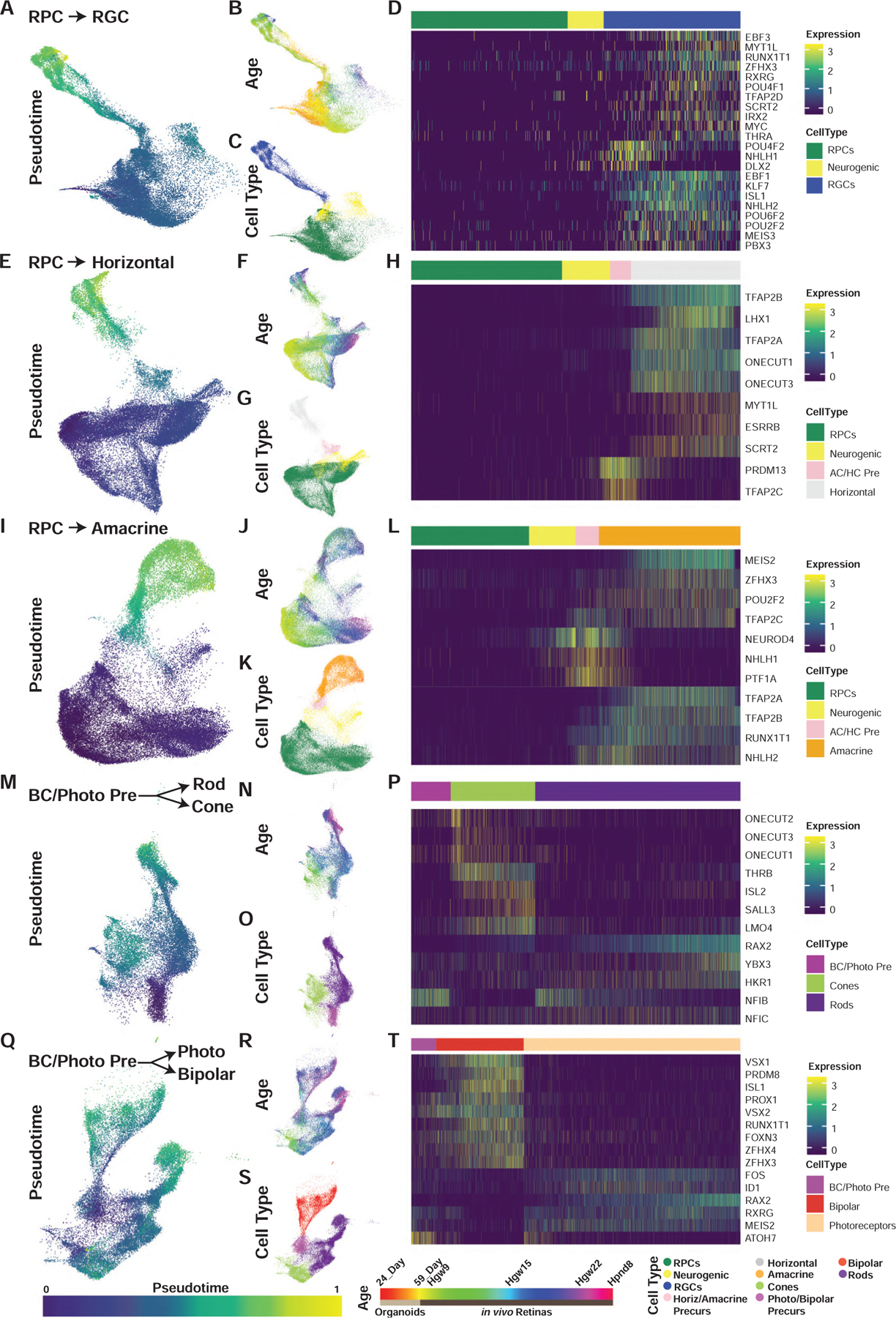
(A–T) UMAP embeddings of cellular inputs for pseudotime analyses of (A–D) retinal ganglion cells, (E–H) horizontal cells, (I–L) amacrine cells, (M–P) rods-cones, and (Q–T) photoreceptor-bipolar cells. UMAP plots are colored by (A, E, I, M, and Q) cellular pseudotime values, (B, F, J, N, and R) age, and (C, G, K, O, and S) cell type. UMAP embeddings shown are subsets of initial dimension reductions. (A–C) contain only RPCs that contribute to the RGC trajectory while (E–T) include all RPCs. (D, H, L, P, and T) heatmap showing relative expression of differentially expressed transcription factors across pseudotime, highlighting transcription factors with enriched expression in endpoint cell types. Abbreviations: Hgw, human gestational weeks; Hpnd, human postnatal day; RPC, retinal progenitor cells; RGC, retinal ganglion cells; Photo, photoreceptors; BC/Photo Pre, bipolar cell/photoreceptor precursors.
Some of the more pronounced differences in transcription-factor expression between human and mouse are observed in differentiating photoreceptors. In the mouse, ISL2 is strongly expressed in retinal ganglion cells, but only weakly expressed in cone photoreceptors (Triplett et al., 2014). In both humans and chick, however (Edqvist et al., 2006; Triplett et al., 2014), ISL2 is strongly expressed in cones, along with its cofactor LMO4 (Figures 3M–3P). The Kruppel class zinc finger transcription factor, HKR1, which is selectively expressed in developing human rods, is entirely absent from the mouse genome. Strikingly, ATOH7 is also expressed in human photoreceptor-bipolar precursors and immature cones even late in development (Figures 1G and 3Q–3T), in sharp contrast to the mouse and also every other species examined to date, where it is preferentially expressed in early-stage neurogenic RPCs and newly differentiated, early-born retinal neurons (Brown et al., 1998; Kanekar et al., 1997; Kay et al., 2001). These findings imply the existence of species-specific differences in the transcriptional regulatory networks controlling rod and cone photoreceptor specification.
We also observed an organoid-specific cone trajectory in our dimension reductions (Figures 1B and 1C). While this trajectory likely represents batch effects of in vitro differentiation versus in vivo development, this motivated us to assess the similarities and differences between cone specification and differentiation between these conditions. To do this, we performed pseudotemporal analysis on organoids versus in vivo cone development without batch-effect correction, identifying some unexpected differences in differentiating cones between retinal organoids and primary tissue (Figures S3D–S3F). While organoid-derived cones express substantially lower levels of cone precursor-enriched transcription factors, such as CRX, they also express neurogenic factors typically associated with nonphotoreceptor cell types including LHX9, NHLH1, and SOX11 (Figures S3F and S3H). Subsets of organoid-derived cones, however, do express high levels of other cone photoreceptor-enriched genes. These include THRB, ISL2, LMO4, RXRG, SALL3, and DCT (Figure S3F). Comparisons of average gene expression of putative organoid-derived cones to cell types annotated from primary retinal tissue revealed that organoid-derived cones most closely match transcriptional signatures of developing cones and photoreceptor precursor cells (Figure S3G).
While we identified many transcripts and transcription factors as differentially expressed across the pseudotemporal analyses of individual cell trajectories (Figures 3 and S3), it should be noted that many of the genes are not specific to any one cell trajectory (Figure S3H). This suggests that many transcription factors are reused during the specification of multiple cell types.
Central versus Peripheral Differences in Retinal Transcript Expression
To identify regional differences in gene expression associated with the development of the macula compared to the peripheral retina, we examined differential gene expression between macula and peripheral cells from the GW20 and PND8 retinal samples using the “fit_models” function from Monocle3. We observed broad transcriptional differences between the regions within individual cell types (Figures S4A–S4H); however, analyses are partially confounded by low sample numbers and differences in capture efficiency for different cell types that are inherent to the datasets (Figure S4I). Furthermore, since macular retina is developmentally advanced relative to peripheral retina (Diaz-Araya and Provis, 1992; van Driel et al., 1990; La Vail et al., 1991), we examined the correlation of differentially expressed transcripts with pseudotime trajectories across each individual cell type from the full dataset as part of analyses associated with Figure 3 (Figures S4J–S4Q). The correlation with pseudotime helps to distinguish genuine enrichment in cells in macular retina from differences in developmental maturation of cells between macular and peripheral retina (Diaz-Araya and Provis, 1992; van Driel et al., 1990; La Vail et al., 1991). The pseudotime analyses suggest many of the observed regional differences are correlated (both positively and negatively) with cell-type differentiation and maturation, and hence are unlikely to reflect bona fide differences in gene expression signatures between macula and periphery in the mature retina. However, other transcripts such as the cone macula-enriched transcript DST display little correlation with pseudotime and may reflect true regional differences.
We next performed differential expression analysis of macular and peripheral RPCs from the GW20 datasets in order to identify potential candidate genes involved in macular development. To minimize the effect of regional differences in developmental timing (Diaz-Araya and Provis, 1992; van Driel et al., 1990; La Vail et al., 1991), we identified multiple genes that were differentially expressed between macular and peripheral RPCs within the GW20 datasets that showed low correlation with the RPC pseudotime performed in analyses associated with Figure 2 (correlation close to 0; Figure S4Q). These include CYP26A1, DIO2, CDKN1A, ANXA2, and FRZB (Figures 4A and S4Q). Of particular interest is CYP26A1, which encodes an enzyme that degrades retinoic acid, marks the developing rod-free zone in the chicken (da Silva and Cepko, 2017), and is selectively expressed in RPCs and Müller glia of the developing and mature primate fovea (Cowan et al., 2019; Peng et al., 2019).
Figure 4. Identification of Macular RPC Transcripts for Regional Specification of the Developing Human Retina.
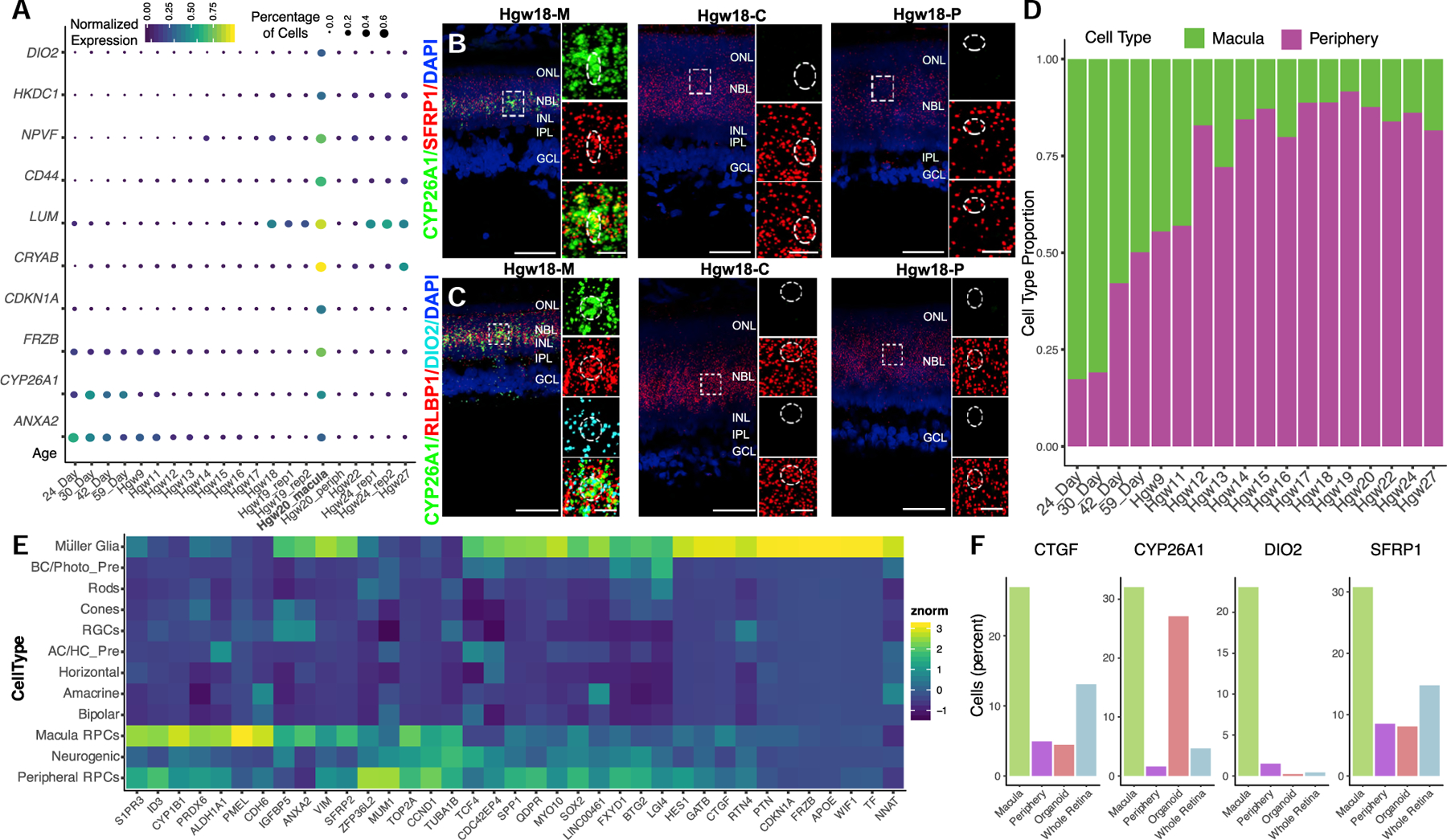
(A–C) (A) Dot plot of differentially expressed genes between macular and peripheral retina RPCs and their relative expression and percentage of expressing cells in RPCs in each sample. The bolded Hgw20_rep1 sample highlights a macular sample containing significant numbers of RPCs. (B and C) RNAscope detecting (B) CYP26A1 and SFRP1 and (C) CYP26A1, RLBP1, and DIO2 transcripts in macular, central, and peripheral Hgw18 retina samples with high-magnification images of boxed regions. Nuclei are counterstained with DAPI. Scale bar, 50 μm and 10 μm (magnified views).
(D) Proportion of macular and peripheral RPCs as classified by CYP26A1, CDKN1A, DIO2, ANXA2, or FRZB expression at each age.
(E) Heatmap showing cell-type expression enrichment of differentially expressed transcripts between the inferred macular and peripheral RPCs.
(F) Bar plots showing proportion of cells expressing macular RPC enriched genes within each sample type. Abbreviations: Hgw, human gestational weeks; M, macular retina; C, central retina; P, peripheral retina; NBL, neuroblast layer; GCL, ganglion cell layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; RPCs, retinal progenitor cells; AC/HC Pre, amacrine cell/horizontal cell precursors; BC/Photo Pre, bipolar cell/photoreceptor precursors; RGCs, retinal ganglion cells.
We then validated enrichment of CYP26A1 and DIO2 mRNA, and co-expression of SFRP1 or RLBP1 within macula cells of the GW18 retina (Figures 4B and 4C). This allowed us to extrapolate the expression of any of the five listed marker genes to the rest of the RPCs within the dataset to identify additional potential macular RPCs within whole-retina dissociations (Figure 4D). Subsequent differential tests were performed on the inferred macular and peripheral primary RPCs, identifying additional potential candidate regulators of macula development. Many of these candidate regulators of regionalization from these differential expression tests display enriched expression within the inferred macula RPCs or Müller glial cells when analyzing expression across the entire dataset (Figure 4E). One interesting candidate regulator of macula development that displays enriched expression within our macular RPC population is CTGF (Figure 4F). CTGF is a downstream target of the Hippo-signaling pathway. While CTGF displays enriched expression within Müller glia compared to RPCs (Figure 4E), recent studies have identified a FGF15 (19 in humans)/FGFR4-mediated pathway for Hippo-pathway activation (Ji et al., 2019). Since FGF19 expression marks early RPCs, activation of Hippo signaling and degradation of retinoic acid may function concordantly to confer macular specialization. Based on these initial results, additional studies are required to integrate the role of the identified candidate genes in both the temporal and spatial gene expression within the developing macula.
Specification and Differentiation of Two Human Horizontal Cell Subtypes
While mouse retinas contain a single subtype of horizontal cell, primate retinas contain at least two distinct subtypes—H1 cells, which receive input from both rods and cones, and H2 cells, which receive input from cones. Recent scRNA-seq analysis of macaque retina was able to distinguish these two horizontal cell subtypes by the presence or absence of CALB1 expression (Peng et al., 2019). Our analysis of human horizontal cell precursors also clearly identifies two subtypes of differentiating horizontal cells, distinguished by differential expression of the LIM homeodomain transcription factors LHX1 and ISL1 (Figures 5A–5C and S5F). Using smfISH, we confirm that LHX1+ and ISL1+ horizontal cells are distinct populations (Figures 5D–5H). Both horizontal cell populations express ONECUT1–3 (Figures S5A–S5C, S5G, and S5H). The presence of ISL1+ horizontal cells in humans is similar to the chicken, where two populations of Isl1+ horizontal cells with distinct morphology are observed (Suga et al., 2009). The ISL1+ horizontal cells are also CALB1+, likely corresponding to the identified CALB1+/ISL1+ H2 horizontal cells of macaque (Figure S5D) (Peng et al., 2019). However, since all horizontal cells are both Lhx1+ and Calb1+ in mice (Boije et al., 2016; Clark et al., 2019; Lo Giudice et al., 2019), this represents a major species-specific difference in gene expression. Differential expression and pseudotemporal analyses identify additional markers of these horizontal subtypes, including PCDH9, C1QL1, and SOSTDC1 for LHX1+ cells and FAM135A and SYNPR for ISL1+ horizontal cells (Figures 5I, S5E, and S5I). The two populations of horizontal cells also show different expression of cell-surface receptors. LHX1+ cells specifically express the dopamine receptor DRD2, while no other dopamine receptor genes are expressed at detectable levels in either horizontal cell population (Figure 5I). LHX1+ H1 cells also express MEGF11, which has been shown to regulate horizontal cell mosaicism in mice (Kay et al., 2012), suggesting that an alternative mechanism is likely used to form ISL1+ H2 horizontal cell mosaics (Figure 5I).
Figure 5. Identification and Differentiation of Two Horizontal Cell Subtypes within the Developing Human Retina.
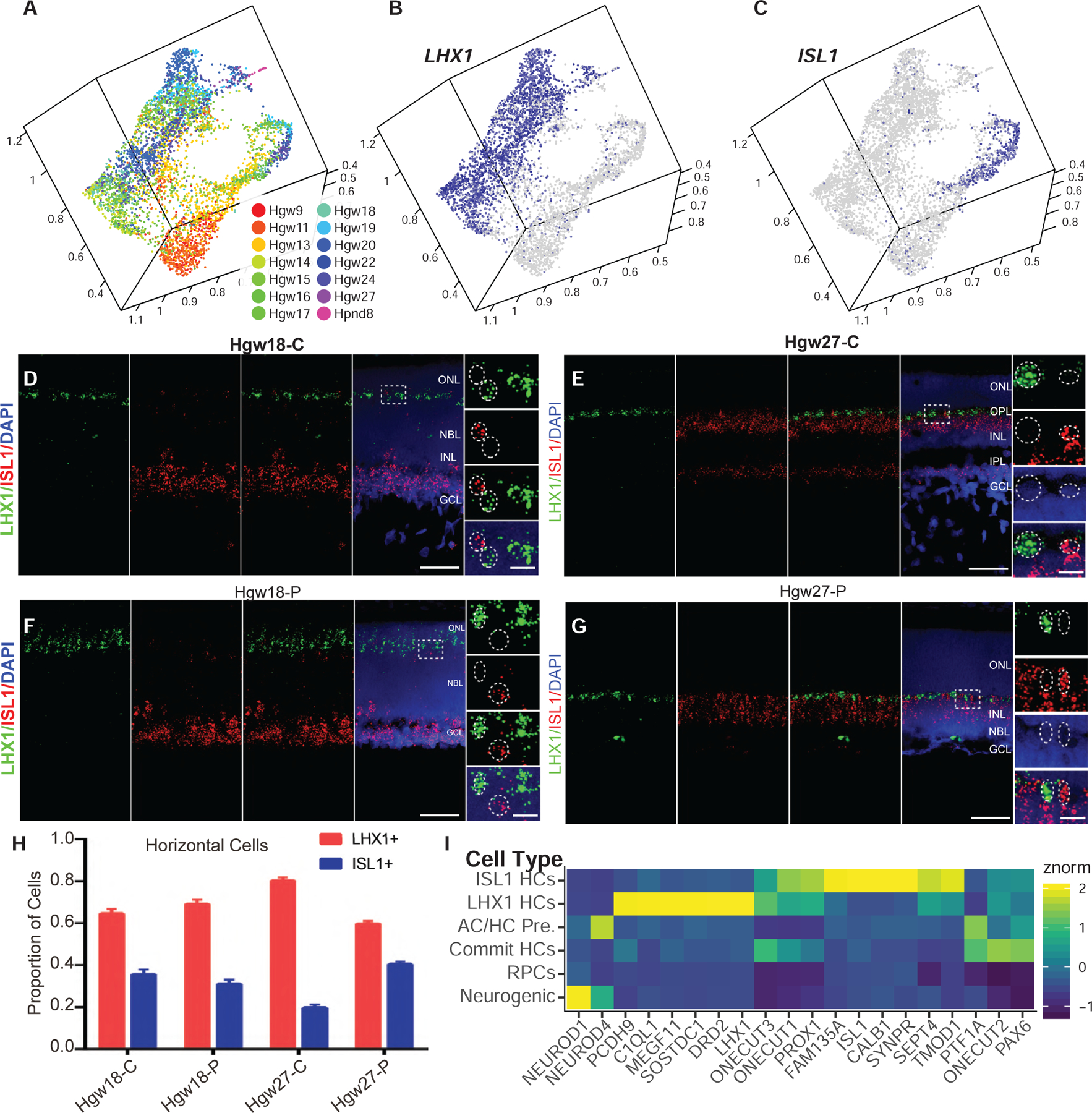
(A–C) UMAP embedding of horizontal cells, colored by (A) age or (B–C) relative expression of (B) LHX1 and (C) ISL1.
(D–G) RNAScope detecting expression of LHX1 and ISL1 transcripts in central (D and E) and peripheral (F and G) Hgw18 and Hgw27 human retina, with higher-magnification views of the boxed regions. Nuclei are counterstained with DAPI. Scale bar, 50 μm and 10 μm (magnified views).
(H) Quantification of the proportions of each horizontal cell subtype in central and peripheral retina at ages Hgw18 and 27 from fluorescent in situ hybridization experiments. Data are mean ± SEM.
(I) Heatmap showing relative cell type expression of horizontal cell commitment, differentiation, and subtype specification genes. Abbreviations: Hgw, human gestational weeks; Hpnd, human postnatal day; C, central retina; P, peripheral retina; NBL, neuroblast layer; GCL, ganglion cell layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; AC/HC Pre., amacrine cell/horizontal cell precursors; Commit HCs, committed horizontal cells; ISL1 HCs, ISL1-positive horizontal cells; LHX1 HCs, LHX1-positive horizontal cells.
Identification of Convergent and Divergent Gene-Expression Patterns in Mouse and Human Retina Using scCoGAPS and projectR
The recently developed nonnegative matrix factorization technique, scCoGAPS, allows unsupervised identification of patterns of gene set usage (both co-expression or lack of detected expression) that represent common features across cells (Sherman et al., 2019; Stein-O’Brien et al., 2019). As the input matrix is based on gene expression, scCoGAPs inherently identifies a continuum of gene usage across all cells and is not limited to finite, user-defined parameters, such as cell type designation or developmental age. These “patterns” of gene set use reflect the contribution of each gene set in defining shared features, previously shown to be associated with biological processes (Fertig et al., 2016). Through use of scCoGAPS and projectR, a transfer-learning approach, we can then quickly assess shared or divergent features across datasets, identifying both evolutionarily conserved and species-specific sets of co-regulated genes (Sherman et al., 2019; Stein-O’Brien et al., 2019). These tools were used to assess the congruence of gene expression signatures across both the human and mouse retinal developmental scRNA-seq datasets in an unbiased manner, highlighting species-specific features of gene expression governing retinal development.
To identify patterns of gene expression across the human retinal development dataset, we used 3,113 genes displaying high variance across the dataset (Table S2; see STAR Methods). We identified 97 patterns within our human scRNA-seq data that correlate with specific features of the dataset. Through projectR (see STAR Methods), we then projected our published mouse retinal development scRNA-seq dataset, excluding all nonneuroretinal cells, onto these 97 human retinal patterns (Figures 6A–6G, S6A–S6D, and S6I; Clark et al., 2019; Stein-O’Brien et al., 2019). By analyzing correlations of pattern features (pattern weights) and cell-type annotation of each individual cell in both human and mouse datasets, we are able to identify patterns that are maximally correlated with an individual cell type in an individual species and compare the congruence of cell-type association between the two species (Figures 6A, S6A–S6D, and S6I). In general, patterns highly correlated with individual cell types showed broadly similar cellular expression patterns in both species. For example, Pattern 67 strongly correlates with amacrine cells in both human and mouse datasets (Figure 6A). Examining the genes strongly influencing Pattern 67, we ascertain that many known regulators of amacrine cell specification drive this pattern including RBFOX2, PAX6, and MEIS2 (Bumsted-O’Brien et al., 2007; Gu et al., 2018; Hitchcock et al., 1996; Zaghloul and Moody, 2007). Pattern 67 also highlights unstudied genes within amacrine cells, including the amacrine-cell-enriched transcription factor, ZNF385D, which has been implicated in GWAS studies of reading disability (Eicher et al., 2013).
Figure 6. Cross-Species Comparisons of Gene Usage Using scCoGAPS and ProjectR Reveals Conserved and Divergent Pattern Usage across Human and Mouse Retinal Cell Types.
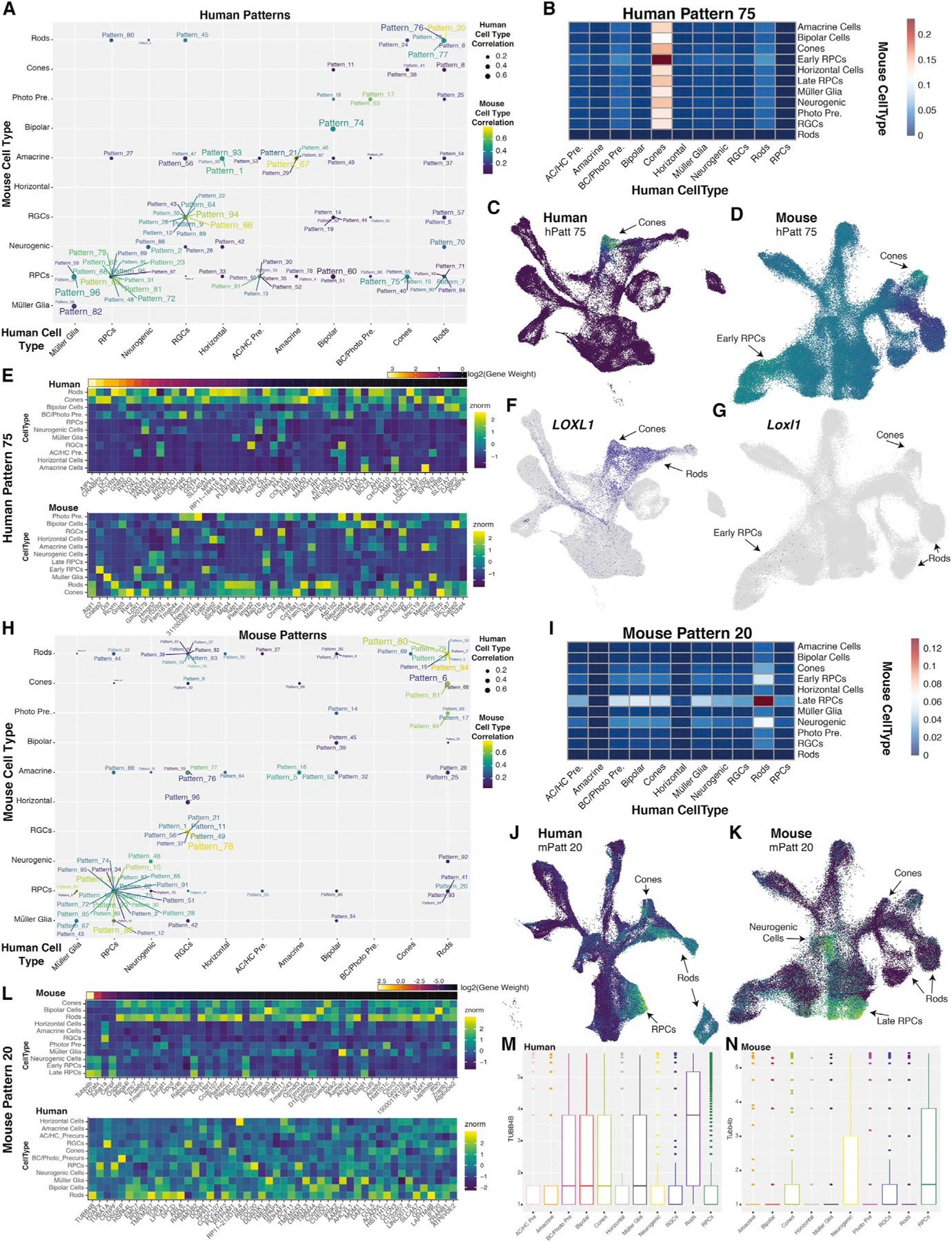
(A) Plot indicating maximally correlated retinal cell types in mouse (y axis) and human (x axis) of human patterns. Correlation values are indicated through size of the point for human cell types, and color of the dot and label for mouse cell types.
(B) Heatmap indicating the intersection of correlations of Human Pattern 75 for human (x axis) and mouse (y axis) cell types.
(C and D) UMAP embeddings of (C) human and (D) mouse scRNA-seq datasets, with cells colored by Human Pattern 75 pattern weights.
(E) Heatmap of relative gene expression within (top) human or (bottom) mouse cell types for the top 50 weighted genes of Human Pattern 75. Genes are ordered by relative gene weights.
(F and G) UMAP embeddings displaying LOXL1/Loxl1 expression within (F) human and (G) mouse retinal scRNA-seq datasets.
(H) Plot of maximally correlated retinal cell types in mouse (y axis) and human (x axis) of mouse patterns. Correlation values are indicated through size of the point for human cell types, and color of the dot and label for mouse cell types.
(I) Heatmap indicating the intersection of correlations of Mouse Pattern 20 for human (x axis) and mouse (y axis) cell types.
(J and K) UMAP embeddings of (J) human and (K) mouse scRNA-seq datasets, with cells colored by Mouse Pattern 20 pattern weights.
(L) Heatmap of relative gene expression within (top) mouse or (bottom) human cell types for the top 50 weighted genes of Mouse Pattern 20 and their orthologs in human (bottom). Genes are ordered by relative gene weights.
(M and N) Boxplots displaying the log2(expression + 1) of TUBB4B in each (M) human and (N) mouse retinal cell type. Abbreviations: AC/HC Pre., amacrine cell/horizontal cell precursors; RGCs, retinal ganglion cells; RPCs, retinal progenitor cells; BC/Photo Pre, bipolar cell/photoreceptor precursors; Photo Pre, photoreceptor precursors; hPatt, human pattern; mPatt, mouse pattern.
However, several patterns displayed discordance among human and mouse cell types with which they maximally correlated, particularly patterns correlated with human cone development (Figure 6A). One example of divergent patterns between mouse and human is Human Pattern 75, which marks human cones but highlights both early RPCs and cones in mice (Figures 6B–6D, S6A, S6C, and S6I). Examination of the top marker genes of Human Pattern 75 indicate that the divergence between the species is driven by transcripts that include CRABP2, DCT, and LOXL1 (Figure 6E). Specifically, human LOXL1 is expressed in developing neurons, with high expression in cones and developing rods (Figure 6F). In contrast, mouse Loxl1 shows little expression within the developing retina, being detected in only a few early--stage RPCs (Figure 6G). A similar expression enrichment within human cones and mouse RPCs is observed for CRABP2/Crabp2 (Figure 6E).
We also identified 97 gene patterns across 3164 highly variable genes within the neuroretinal cells from the mouse scRNA-seq dataset (Table S2) and repeated the reverse comparison process (Clark et al., 2019; Stein-O’Brien et al., 2019). Of note, 753 of the > 3,000 input genes for scCoGAPS were shared across both species-specific analyses. A similar overall picture was seen within the mouse scCoGAPS results; many patterns displaying high correlation with individual cell types highlight the same cell type in projections to the human dataset. Again, however, we observed instances of patterns with high correlations with incongruent cell types across the species (Figures 6H and S6E–S6H, and S6J). Notable among these is Pattern 20, which in mice is enriched in late-stage RPCs but in humans is enriched in rods, cones, and late-stage RPCs (Figures 6H–6K, S6E, S6G, and S6J). Tubb4b, a marker for Pattern 20, shows differences in cellular expression levels in late-stage RPCs, rods, and cones in humans and mice (Figures 6L–6N). Other examples include mouse Rod patterns 13, 22, 35, and 59, which fail to project well into the human dataset (Figures S6E and S6J). These unbiased cross-species analyses highlight both similarities and disparities in gene usage across mouse and human retinal development, in particular in the control of photoreceptor specification and differentiation and can be used to identify instances where mouse models may not recapitulate human disease.
ATOH7 Controls the Relative Ratio of Human Rod and Cone Photoreceptors
Multiple lines of evidence from this study suggest that the gene regulatory networks controlling photoreceptor development differ substantially between mice and humans. We identified ATOH7 expression in late-stage neurogenic RPCs, amacrine-horizontal cell precursors, bipolar-photoreceptor precursors and immature photoreceptors, in addition to its expression in early-stage neurogenic RPCs, which has been observed in other species (Figures 7A, S7A, and S7B; Brown et al., 1998; Kanekar et al., 1997; Kay et al., 2001; Matter-Sadzinski et al., 2001). ATOH7 expression in immature photoreceptors was confirmed using immunostaining for ATOH7 and the early photoreceptor-enriched markers OTX2 and CRX. At both early and late stages of photoreceptor development, we observe a fraction of both OTX2-(Figures 7B, 7C, and S7C) and CRX-positive cells (Figures 7D, 7E, and S7D) that also express ATOH7. This co-localization was temporally dynamic and showed differences between central and peripheral retina. Nearly 10% of all OTX2+ cells in the central retina were also ATOH7-positive at GW10, but at later stages very little co-localization was observed (Figure 7C). In contrast, at stages examined between GW10 and GW20, 12%–20% of all OTX2-positive cells in peripheral retina were also ATOH7-positive (Figure 7C). Co-localization of ATOH7 and CRX also showed a large difference between central and peripheral retina, with very little co-localization observed in the central retina. More than 30% of CRX-positive cells in peripheral retina also displayed detectable expression of ATOH7 at GW14, though the fraction of co-expressing cells declined rapidly thereafter (Figures 7D and 7E). Similar trends of OTX2 and CRX transcript co-expression with ATOH7 was confirmed within the scRNA-seq dataset (Figures S7K–S7L).
Figure 7. Knockdown of ATOH7 Promotes Differentiation of Rod Photoreceptor at the Expense of Late-Born Cones.
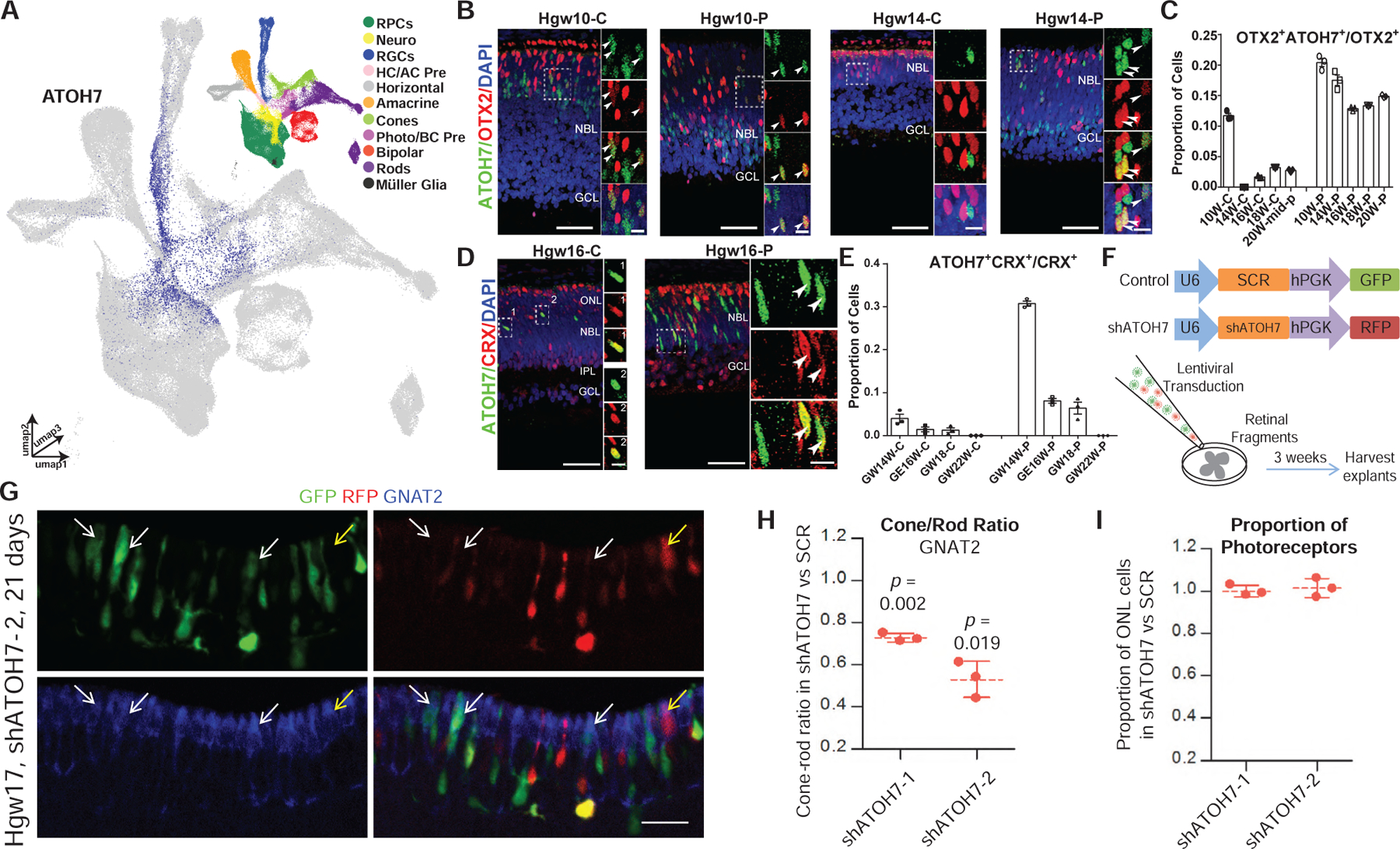
(A) UMAP embedding of the retinal scRNA-seq dataset, colored by relative expression of ATOH7 and cell type annotation (top right).
(B) Immunostaining for ATOH7 and OTX2 in central and peripheral Hgw10 and Hgw14 retinas, with magnified views of boxed regions. Scale bar, 50 μm and 10 μm (magnified views)
(C) Quantification of OTX2+ cells that are also ATOH7+ in central and peripheral retina at various ages. Data are presented as means ± SEM.
(D) Immunostaining for ATOH7 and CRX in the central and peripheral Hgw16 retina, with zoomed in views of boxed regions. Hgw16-C scale bar, 50 (left), 5 μm (right); Hgw16-P scale bar: 50 (left), 10 μm (right). Arrowheads indicate co-localization of markers and nuclei are counterstained with DAPI in panels B+D.
(E) Quantification of CRX+ cells that are also ATOH7+ in central and peripheral retina at various ages. Data are presented as means ± SEM.
(F) Schematic diagram of ATOH7 knockdown experiment.
(G) Representative image from human retinal explants co-transduced with shSCR (GFP) and shATOH7–2 (RFP) lentiviruses and stained with cone marker GNAT2 (blue). White arrows, GNAT2+ cells expressing shSCR; yellow arrow, GNAT2+ cells expressing shATOH7. Scale bars, 20 μm.
(H) Ratio of cones/rods in shATOH7 versus shSCR cells. Data are presented as means ± SD with p-values indicating results of a paired t-test.
(I) Ratio of the ONL proportions in shATOH versus shSCR cells. Data are presented as means ± SD. Abbreviations: RPCs, retinal progenitor cells; Neuro, neurogenic cells; RGCs, retinal ganglion cells; HC/AC Pre, horizontal cell-amacrine cell precursors; Photo/BC Pre, photoreceptor-bipolar cell precursors; Hgw, human gestational weeks; GW, gestational weeks; C, central retina; P, peripheral retina; NBL, neuroblast layer; GCL, ganglion cell layer; ONL, outer nuclear layer; INL, inner nuclear layer; IPL, inner plexiform layer; C, central retina; P, peripheral retina.
This implies that ATOH7 may promote the specification and differentiation of cones in human peripheral retina. To directly test whether ATOH7 regulates human photoreceptor genesis, we co-transduced GW17–19 explants with lentiviral vectors that express control or ATOH7 shRNAs together with GFP or RFP, respectively (Figure 7F). Quantifying control and test shRNAs transduced cells in the same explant avoids the confounding effects of positional variation in developmental stage. We confirmed efficient ATOH7 depletion by shATOH7 vectors relative to a scrambled (shSCR) control (Figures S7E and S7F). Explants were assessed 21–23 days after co-transduction. Cone and rod proportions were quantified in control and test cells using positive and negative labeling in the outer nuclear layer (ONL) for one of two cone markers (RXRγ or GNAT2) or a rod marker (NRL). The cone-rod ratio in shATOH7 cells relative to shSCR cells was then calculated. Irrespective of which of the three staining methods was used, depleting ATOH7 with either of two separate shRNAs reduced the relative fraction of cones, while increasing the fraction of rods (Figures 7G, 7H, and S7G–S7J), without affecting overall photoreceptor numbers (Figure 7I). Together, these data indicate that ATOH7 promotes cone genesis in the developing human retina.
DISCUSSION
Our transcriptomic analysis, using both embryonic stem cell-derived retinal organoids and primary tissue at single-cell resolution, encompasses nearly the full time course of human retinal development. While bulk RNA-seq of the developing human retina and scRNA-seq of human retinal organoids are already available, they have important limitations (Collin et al., 2019; Hoshino et al., 2017; Hu et al., 2019; Mellough et al., 2019). Bulk RNA-seq cannot resolve cell-type-specific changes in gene expression. Furthermore, the extent to which retinal organoids fully recapitulate in vivo development at the individual cell level is still not entirely clear (Cowan et al., 2019). The combinatorial use of scRNA-seq and human developing retinal tissue in our dataset overcomes some of these limitations. Combined, the dataset offers a valuable resource for identifying the gene-regulatory networks that occur in human retinal development. Using this dataset in parallel with data generated from mice (Buenaventura et al., 2019; Clark et al., 2019; Lo Giudice et al., 2019) can highlight similarities and discrepancies across evolution and help determine whether specific mouse models are actually relevant to human retinal disease. Furthermore, these findings open new avenues of research with respect to spatial patterning, cell-fate specification, and the function of individual neuronal subtypes of the human retina. However, it is recognized that, due to sequence depth limitations associated with scRNA-seq technique and the availability of primary human retinal tissue, our dataset does not detect all differentially expressed transcripts or accurately capture genetic differences associated with race, sex, health status, or age.
This analysis identified some major differences between the gene-expression patterns of immature cones analyzed in human retinal organoids and primary tissue (Figures S3D–S3F). We hypothesize that this difference results from the abrupt changes in culturing conditions associated with the addition or subtraction of growth factors, signaling-pathway inhibitors, or oxygen levels during the human retinal organoid-culturing process. One of these treatments, in particular, is the addition of the gamma-secretase and Notch-pathway inhibitor DAPT from days 29–45 of organoid culture to assist with photoreceptor specification (Jadhav et al., 2006; Wahlin et al., 2017; Yaron et al., 2006). Interestingly, 92.7% (1,591 of 1,716) of organoid-derived cones captured in our dataset were derived from organoids within this treatment window (i.e., 30 or 42 days in culture organoid samples). This suggests that while the treatment of organoids with DAPT does stimulate photoreceptor specification, the differentiation trajectory of these organoid-derived cones differs markedly from that seen in vivo, even though more mature organoid-derived cones and native cones eventually show broadly similar gene-expression profiles (Cowan et al., 2019). Additional time points and further direct comparisons of both in vivo and retinal organoid development will further resolve the degree of developmental conservation across primary and cultured tissue (Sridhar et al., 2020).
Human retinal neurogenesis occurs over a much longer interval than in a mouse. While many of the identified cell-type-specific markers and differentially expressed genes across the developmental trajectory of each major retinal cell type are conserved in mice, there are still some major differences. For example, Clu is specific to Müller glia in mice, but is expressed in both RPCs and Müller glia in humans. Interestingly, a previous study has shown that Clu+ revival stem cells in the intestine are multipotent, capable of giving rise to the major cell types of the intestine and transiently expanding in a YAP1-dependent manner (Ayyaz et al., 2019). Since Clu is strongly expressed in both human RPC and Müller glia, it is possible that manipulating the YAP1 pathway will induce the regenerative pathway of human Müller glia. Consistent with this, two recent papers have implicated Hippo-pathway signaling in regulating Müller glial quiescence, and YAP overexpression was sufficient to induce Müller glia to reenter the cell cycle (Hamon et al., 2019; Rueda et al., 2019). A number of genes, including several transcription factors, such as HES4 and HKR, show highly cell-type-specific expression in human retina, but are absent from the mouse genome altogether. This raises the question of whether these genes are used in regulating human-specific aspects of retinal development or have simply taken over evolutionarily conserved functions carried out by related genes in mice.
The presence of the fovea is perhaps the most obvious anatomical difference between the retinas of primates and other mammals. We found differentially expressed genes between peripheral and macular RPCs, allowing us to postulate the molecular underpinnings of cone-rich foveal formation. One of the genes examined, the retinoic acid-degrading enzyme CYP26A1 is conserved in chicken, and is necessary and sufficient for creating a rod-free zone (da Silva and Cepko, 2017). This suggests that conserved mechanisms inhibiting the retinoic-acid-signaling pathway are involved in the formation of a high-acuity, cone-rich, rod-free zone. Other genes that are strong candidates for controlling foveal development and enriched in macular RPCs include DIO2, which promotes differentiation of L/M cones at the expense of S-cones (Eldred et al., 2018), the Wnt-pathway inhibitors SFRP2 and FRZB, and secreted antiproliferative factors such as CTGF and PTN. Wnt signaling acts during early stages of retinal development to inhibit central retinal identity (Cho and Cepko, 2006; Liu et al., 2007), and its presence here suggests that it may play an analogous role in control of foveagenesis. Antiproliferative factors such as CTGF and PTN may mediate the early cessation of proliferation seen in the foveal region (Diaz-Araya and Provis, 1992; van Driel et al., 1990), although the fact that CTGF expression is also enriched in foveal Müller glia of adult macaque suggests that it may also have other functions related to controlling mature glial function or morphology (Peng et al., 2019). The coordinated action of these signaling pathways in RPCs may provide the initial signals that pattern the fovea. Although macular-enriched genes that are not positively correlated with pseudotime are identified as enriched in multiple neuronal cell types, including several that are enriched in both developing photoreceptors of the human macula and adult macaque fovea (Peng et al., 2019), their function is less clear and awaits direct functional analysis. Furthermore, additional histochemical analyses across the full course of macula development are required to confirm the differential expression results derived from the relatively few age-matched samples analyzed from both central and peripheral retina, and to minimize the possibility that these results reflect regional differences in cell maturation.
We observed two subtypes of horizontal cells in the human retina, consistent with those classified in previous studies of macaque retina (Peng et al., 2019). Although morphological studies of adult human retina have identified three distinct subtypes of horizontal cells (Kolb et al., 1994), we only observed clear transcriptional evidence for two. In humans, these two subtypes express different combinations of neurotransmitter receptor genes. Most prominently, LHX1+ horizontal cells specifically express the dopaminergic receptor DRD2, while no other dopaminergic receptor genes are expressed in either horizontal cell subtypes. While dopaminergic amacrine cells are thought to be the only retinal cells that produce dopamine, we detected weak expression of the dopamine transporter SLC6A3 in both rods and bipolar cells. The expression of DRD2 on LHX1+ horizontal cells and potential dopamine transporter transcript expression in rods and bipolar cells, therefore, implies that LHX1+ horizontal cells may selectively respond to a dopamine signal within the rod spherule.
We also observed that ISL1+/CALB1+, unlike LHX1+, horizontal cells do not express detectable levels of MEGF11 expression. MEGF11 regulates homotypic repulsion of horizontal cells in mice (Kay et al., 2012). Since retinal cell subtypes are frequently spaced evenly across the retina, but randomly positioned with respect to other subtypes (Rockhill et al., 2000), it will be interesting to identify the determinants of ISL1+ horizontal cell retinal mosaic spacing independent of detectable MEGF11 and define these mechanisms of horizontal cell homotypic repulsion in the developing human retina. Other differences include the presence of separate populations of LHX1+ and CALB1+ horizontal cells in primate retinas (Peng et al., 2019), whereas the genes are co-localized in the single type of mouse horizontal cell. Since chickens and primates also possess distinct populations of Isl1+ and Lhx1+ horizontal cells, with two morphologically distinct populations of Isl1+ cells observed in chickens (Suga et al., 2009), we hypothesize that nocturnal rodents consolidated these two subtypes into one cell type, which then subsumed many functions of both. Further examination of the number of cellular subtypes within the retina across evolutionarily divergent species will address whether the number of horizontal cell subtypes directly correlates with the number of cone subtypes.
To examine gene regulatory networks in an unbiased manner and without a priori knowledge of gene interactions, we used scCoGAPS pattern identification and projectR latent-transfer learning. These tools provide a robust, transferable way for comparing gene-expression networks across species (Sherman et al., 2019; Stein-O’Brien et al., 2019). These techniques identified patterns of gene usage that were both shared and divergent across species. Furthermore, these analyses highlighted genes, such as LOXL1, implicated in pseudoexfoliation syndrome (Thorleifsson et al., 2007), to have distinct expression patterns across human and mouse retinal development. This not only implies that humans harboring mutations in LOXL1 may present with a retinal pathology during development, but also highlights some of the unexpected pitfalls of modeling human diseases in mice. It is not clear how these evolutionarily divergent expression patterns arise. It has been hypothesized that coordinated changes in regulatory domains of co-expressed genes drive human-specific cortical expansion (Reilly et al., 2015). This is further highlighted by large transcriptional differences in gene expression in developing cortical cell types within the primate lineage despite little sequence divergence in homologous protein coding genes (Khrameeva et al., 2019). Hence, it is possible that the divergent gene-expression patterns observed in human and mouse retinal development mostly correspond to changes in the use and/or sequence of cis-regulatory elements.
Finally, we examined the role of ATOH7 within late stages of retinal neurogenesis. Patients with mutations within the ATOH7 coding or regulatory sequences exhibit persistent fetal vasculature (PFV) and/or nonsyndromic congenital retinal nonattachment (NCRNA), respectively (Ghiasvand et al., 2011; Prasov et al., 2012). Both diseases, however, are associated with defects in RGC development. Here, we show ATOH7 is also involved in the specification of late-born cones, suggesting alterations in the rod/cone ratio may also be observed in patients with either PHPV or NCRNA. Atoh7 mutant mice generate additional cones at the expense of RGCs (Brown et al., 2001), although atoh7 zebrafish mutant retinas exhibit no change in cone development (Kay et al., 2001). Together, these data suggest that ATOH7 may have different mechanisms of function in retinal cell fate specification across evolution. Furthermore, our results highlight a fundamental difference between the mechanisms by which human and mouse cones are specified. The exquisite sensitivity or resistance of RB1-deficient human or murine cones to tumorigenesis, respectively, reflects the selectively high expression of oncogenes such as MDM2 and MYCN in human cone precursors (Xu et al., 2009). Therefore, understanding species-specific differences in gene expression within developing cones may provide insight into differential susceptibility to RB1 loss.
The rapid improvement of techniques for the culturing of human ES- and iPS-derived retinal organoid preparations makes it feasible to directly study the functional role of candidate extrinsic and intrinsic regulators of human retinal development identified in this study. While functional studies using human retinal organoids have thus far largely focused on analysis of evolutionarily conserved gene-regulatory networks (Eldred et al., 2018), they provide an accessible and tractable system for studying aspects of retinal development that cannot be readily modeled in mice. The combinatorial application in organoid preparations of growth and differentiation factors identified in our study has the potential to identify molecular signals that are sufficient to induce foveagenesis. Coupled with the use of CRISPR-mediated knockout approaches, this will allow direct functional analysis of human retinal genes that lack mouse orthologs or show markedly different expression patterns in developing retina.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and fulfilled by the Lead Contact, Brian Clark (brian.s.clark@wustl.edu).
Materials Availability
The authors report no new materials were generated in conjunction with this study
Data and Code Availability
Single-cell RNA-sequencing files are available through GEO with the following accession numbers: GEO: GSE116106, GEO: GSE122970, GEO: GSE138002. Processed, finalized matrices are available through GEO accession number GEO: GSE138002. Visualization and analysis of processed data (dimension reduction, gene expression, heatmaps) is available at https://proteinpaint.stjude.org/?genome=hg38&singlecell=files/hg38/NEI.AGI.retina/singlecell/Human_Retina_Development.json. Details of analysis and input parameters is available within the STAR Methods (below) and in Table S3. Additional details and inquiries should be addressed to the Lead Contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Details of developmental stage, sex and ethnicity (where known) are provided in Figure S1A.
Ethics Statements
Canadian Samples
Retinas were obtained from the Morgentaler Clinic in Toronto with approval from the Research Ethics Board (REB, REB13–0132-E) of Mount Sinai Hospital in Toronto, Canada. All donors read the consent form approved by the REB before surgical procedures, and voluntarily donated developing eye samples. Informed consent did not include recordings of ethnicity.
Chinese Samples
The human tissue collection and research analysis was approved by the Reproductive Study Ethics Committee of Beijing Anzhen Hospital and The First Affiliated Hospital of University of Science and Technology of China. The informed consent forms were designed under ISSCR guidelines for human developing tissue donation and were in strict observance of the legal and institutional ethical regulations for elective pregnancy termination specimens at Beijing Anzhen Hospital and The First Affiliated Hospital of University of Science and Technology of China. All protocols were in compliance with the ‘Interim Measures for the Administration of Human Genetic Resources’ administered by the Ministry of Science and Technology of China.
US Sample
The use of human globes obtained from Tissue Banks was approved by the Johns Hopkins Institutional Review Board. A human globe from an 86 year old Caucasian female who died of a myocardial infarction and had no known ocular disease other than cataracts, was obtained from the Alabama Eye Bank (Birmingham, AL).
Human Pluripotent Cell Lines
Human retinal organoids were prepared from induced pluripotent stems cells (iPSCs) derived from the IMR-90 cell line obtained from ATCC (RRID:CVCL_C437).
METHODS DETAILS
Preparation of Retinal Organoids
Retinal organoids were prepared from induced pluripotent stem cells (iPSCs) derived from the IMR-90 (ATCC) cell line (Wahlin et al., 2017). On Day 0, iPSCs were plated at 3,000 cells/well in 96-well U-bottom plates for forced aggregation into embryoid bodies, cultured in mTeSR1 medium (Stem Cell technologies) + 5μM blebbistatin in hypoxic conditions (5%O2/10%CO2). Cells are then transferred to normoxia (20%O2/5%CO2) after 24 hours and medium is gradually (Days 1–7) changed to BE6.2 (10ml E6 stock (97mg insulin 53.5mg holo-transferrin, 230mg L-ascorbic acid, 5μl 14mg.ml sodium selenite, to 100ml with H2O - of note, no NaHCO3 was used in E6 stocks), 5.0ml B27 (without vitamin A), 2.5ml Glutamax (100X), 2.5ml NEAA (100X), 2.5ml Pyruvate (100x) 1.0ml NaCL (21.9g/.1L) and DMEM to 250ml) + 1%(v/v) matrigel supplemented with 3uM Wnt inhibitor, IWR-1. Medium exchange on Days 8 and 9 in culture removes the WNT inhibitor IWR-1 and matrigel. On Day 10, organoids are washed with HBSS and transferred to 10cm dishes and fed with BE6.2 medium + 100nM Smoothened agonist SAG. Optic vesicles are manually excised from organoids between Days 10 and 14. From Days 12–18 medium is changed every other day with LTR (125ml F12, 50ml FBS, 10ml B27, 5ml Glutamax (100X), 5ml NEAA (100X), 5ml pyruvate (100X), 500 μl taurine (1000X - 1M stock), DMEM to 500ml) medium + 100nM SAG. On Day 18, culture medium is changed to LTR medium without SAG. To promote retinal differentiation and maturation, medium is changed to LTR medium supplemented with 500nM all-trans retinoic acid (ATRA). From Days 28–42, LTR medium is supplemented with both 500nM ATRA and 10μM DAPT.
Retinal Dissociation for single cell sequencing
Canadian Samples
The gestational age was estimated by a combination of clinic intakes, ultrasound, crown-rump, and foot length measurements where possible (FitzSimmons et al., 1994; Shepard, 1975). Eye samples collected were held on ice for up to 6 hours in retina culture medium, containing IMDM with 10% FBS and 1X Antibiotic-Antimycotic (Life Technologies, Cat#15240062). Developing eyes at gestational ages week 9 to week 19 were removed from donor embryos, sterilized in 70% ethanol for 3 seconds, rinsed twice in cold phosphate-buffered saline (PBS) (Wisent Bioproducts, Cat#311–010-CL), and transferred to IMDM for retinal dissection. Retinas were dissociated with Papain Dissociation System (Worthington Biochemical, Cat# LK003150). Briefly, retinas were incubated in papain solution for about 15 minutes at 37°C and 5% CO2, with gentle pipetting every 5 minutes. With dissociation to ~20-cell clusters, cells were suspended in 10 volumes of sterile phosphate buffered saline (PBS), pelleted by centrifugation at 300 x g for 10 minutes, and rewashed with PBS, followed by digestion with 0.05% trypsin/EDTA (Wisent Bioproducts, Cat# 325–042-EL) with gentle pipetting to produce single cell suspension, washed with 10 volumes of retina culture medium, and resuspended in proper volumes of the culture medium for single cell sequencing procedure. This entire process took 35–45 minutes. Cell viability (>90%) was confirmed by negative staining with trypan blue (Life Technologies, Cat#15250061). Single cell libraries were then prepared using the 10x Single Cell 3’ v2 Reagent Kits according to the manufacturer’s instructions and sequenced on an Illumina NextSeq500 using recommended sequencing parameters (Read 1 – 26bp; Read 2 – 98bp; i7 Index - 8bp; i5 Index - 0).
Chinese Samples
Gestational age was measured in weeks from the first day of the woman’s last menstrual cycle to the sample collecting date. The eyeballs were removed first from donor embryos and immediately stored in ice-cold artificial cerebrospinal fluid (ACSF) containing 125.0 mM NaCl, 26.0 mM NaHCO3, 2.5 mM KCl, 2.0 mM CaCl2, 1.0 mM MgCl2, 1.25 mM NaH2PO4; pH 7.4, bubbled with 95% O2/5% CO2. Tissues were then transferred to the laboratory on ice within 30 minutes. Retinas were then immediately removed in the ACSF on ice immediately, a process which takes about 10 ~ 15 minutes. The retina macula samples (2mm) were defined by lack of vasculature. Whole retinal samples were gently separated into small pieces and then centrifuged at 200g for 2min. The supernatant was removed and 500ul digestion buffer (2mg/ml collagenase IV (Gibco), 10 U/μl DNase I (NEB), and 1mg/ml papain (Sigma) in PBS) was added. The tissue was then rotated and incubated at 37°C on a thermo cycler with 300g for 15–20 min. Samples were triturated every 5 min to digest the tissue sample into single cell suspensions. Finally, inactivation of enzymatic digestion was induced through the addition of an equivalent volume of 10% fetal bovine serum (Gibco) in PBS. Cells were pelleted and resuspended in 0.04% BSA/PBS and stained with 7-amino-actinomycin D (7-AAD) for 10 min on ice in order to check for cell viability. 7-AAD-negative cells were collected by FACS and resuspended in 0.04% BSA/PBS. Single cell libraries were then prepared using the 10x Single Cell 3’ v2 Reagent Kit according to the manufacturer’s instructions and sequenced on an Illumina HiSeq4000 with 150bp paired-end reads.
US Samples
Retinal organoids were processed as described in Clark et al. (2019). Briefly, multiple organoids were pooled and placed in 200μl of cold HBSS per organoid, with an equivalent amount of Papain dissociation solution (for 1ml - 100μl freshly prepared 50mM L-Cysteine (Sigma), 100μl 10mM EDTA, 10μl 60mM 2-mercaptoethanol (Sigma), and Papain added to 1mg/ml (Worthington); to 1ml with reagent-grade water). Organoids in dissociation solution were then placed at 37°C for 10 minutes, with slight trituration every 1–2 minutes. Enzymatic digestion was halted with addition of 600μl Neurobasal Medium supplemented with 10% FBS for every 400μl of HBSS/dissociation solution. μml/organoid DNaseI (RNase free Recombinant DNaseI; Roche) was added and incubated 5–10 minutes at 37°C. The dissociation solution was then gently triturated using a P1000 pipette tip and cells were pelleted through centrifugation for 5 minutes at 300RCF. Supernatant was carefully aspirated off the cell pellet, followed by resuspension of the cellular pellet in 1–5ml Neurobasal media with 1% FBS. Cellular aggregates were removed by straining cells through a 50μm filter. Single-cell libraries were then prepared using the 10x Single Cell 3’ v2 Reagent Kits according to the manufacturer’s instructions and sequenced on an Illumina NextSeq500 using recommended sequencing parameters (Read 1 – 26bp; Read 2 – 98bp; i7 Index - 8bp; i5 Index - 0).
Human Adult Retinal Sample
The eye globe was removed within 3.3 hours after death, immediately preserved on ice for overnight shipping to Hopkins, and dissected no later than 16 hours postmortem. To dissect the neural retina, the anterior segment was first removed by incising the scleral behind the limbus, to remove the anterior parts, lens and vitreous body. The neural retina was then peeled off from the eyecup, and retinal cells were dissociated using Papain Dissociation System (Worthington Biochemical, Lakewood, NJ) following the manufacturer’s instructions. Dissociated cells were resuspended in ice-cold PBS, 0.04% BSA and 0.5 U/μl of RNAse inhibitors. Cells were then filtered through a 50μm filter and processed for single-cell RNA-sequencing consistent with the retinal organoids.
Library Preprocessing
Resulting sequencing outputs were processed through the CellRanger2 mkfastq and count pipelines using default parameters. Transcript reads were quantified using the 10x Genomics Human reference index (refdata-cellranger-GRCh38–3.0.0). Cells were then given unique, sample-specific cell identifiers to prevent duplicate cell names in the aggregate dataset from re-use of barcodes across samples.
Aggregation of Datasets, Initial Processing and Cell Type Assignment
Human Retinal Organoids
Resulting count matrix files from Cellranger alignments/counting of retinal organoids were imported and aggregated in Monocle2 R/Bioconductor (Qiu et al., 2017). First, cells with > 40,000 Total_mRNAs were removed as outliers. We then identified genes with high biological coefficient of variation by first normalizing sequencing depth across all organoid-derived cells using the Waddington-OT transformation to transcript copies per 10,000 (CPT) (Schiebinger et al., 2019) and then using a generalized additive model (MGCV R package; (Wood et al., 2015)) fit to the log2 mean CPT versus a cubic spline fit to the log2 coefficient of variation across all genes with detectable expression in >10 cells. Transcripts that displayed >1.1 residual to the fit were chosen as ‘high-variance’ genes. Dimension reduction was then performed on the resulting 2441 high variance genes (Table S1) using the first 24 principal components as input into largeVis dimension reduction (Tang et al., 2016). Cell type annotation of Retina/RPE versus non-Retina was performed using eye-field marker genes including RAX, PAX6, and VSX2 while excluding markers of ventral telencephalon and hypothalamus, including NKX2.1, DLX5 and DLX6. The resulting 11,758 Retinal/RPE cells of the original 25,461 organoid cells were then used for aggregation with the in vivo samples.
Human Retinas
All 20 samples of primary retinal tissue were manually aggregated from the individual Cellranger matrix files, resulting in 113,999 individual cells. Initial processing proceeded as above with the following differences: 1) Cells with >10,000 Total_mRNAs were removed, 2) A residual cutoff for determination of high variance genes was set at 1.15, resulting in 2719 highly variable genes that were used for input into dimension reduction, 3) the first 17 dimensions were used as input to PCA, and 4) dimension reduction was performed using UMAP within the ‘reduceDimension’ function of Monocle3alpha, with the following parameters: max_components = 3, metric= ‘canberra’, min_dist = 0.34, n_neighbors = 50, residualModelFormulaStr = “~Total_mRNAs + sample”, random_state = 123456L. The resulting 3-dimensional structure was clustered using the ‘clusterCells’ function of Monocle3alpha with the following parameters: use_pca = FALSE, k=15, res=4.0e-4, method = ‘louvain’, gaussian = TRUE, louvain_inter=5, set.seed(123456). Preliminary cell type annotation was performed based on cluster markers from the ‘find_cluster_markers’ function and examination of expression of known marker genes within the individual clusters. Non-retinal derived cells (microglia, vasculature, etc.) and annotated doublets (identified through incompatible gene expression) were removed from the cell dataset at this time, resulting in an accumulated cell total of 107,013 in vivo retinal cells.
Final Aggregation
Cell datasets from the organoids and in vivo retinas were merged to create a single cell dataset. We again removed cells with >10,000 Total_mRNAs, thus reducing the final dataset to 11,542 organoid-derived cells and 107,013 human retina-derived cells (118,555 total cells). The same exact dimension reduction parameters were used for the total aggregate as the in vivo retinas, using the high variance genes from the in vivo developing retina dataset. Clustering on the 3-dimensional UMAP was performed using ‘clusterCells’ function of Monocle3alpha using the following parameters: use_pca = FALSE, k=15, res=1.0e-3, method = ‘louvain’, gaussian = TRUE, louvain_inter=5, set.seed(123456). Determination of corresponding cell type of clusters was performed based on marker gene expression within clusters (See Figures S1G and S1H). Cell cycle phase of primary and neurogenic RPCs was determined using the ‘CellCycleScoring’ function of Seurat.
Correlations of Sample/Cell Type Relatedness
Related to Figures 1D, 1E, and S3G-
Relatedness of Human Samples (Figure 1D)
Individual sample expression matrices were normalized to the total number of UMIs per cell. Average gene expression was then determined across the entirety of the sample. Sample correlation was then performed using a pairwise spearman correlation with correlation values plotted within the heatmap. Plotting of the heatmap and clustering to generate the dendrogram are performed using the ‘pheatmap’ package.
Human and Mouse Sample Relatedness (Figure 1E)
As above, average gene expression was determined across each sample, normalized for read depth within individual cells. The normalized expression matrix was then reduced to the intersection of Mouse genes and Human orthologs, as determined by biomaRt. Human gene names were then referenced as their corresponding Mouse ortholog to put both species into one expression matrix. If no ortholog was detected for either species, the gene was removed from the expression matrix. If a gene mapped to multiple homologous genes, the gene expression of both orthologs was used. This resulted in an expression matrix of 18,492 unique mouse genes, with 22,211 total gene comparisons. Human/mouse sample correlations were performed using pairwise spearman correlations, with sample gene expression correlations plotted in the heatmap in Figure 1E. Samples are ordered by developmental age for both species.
Human Retinal Organoid and In Vivo Cell Type Relatedness (Figure S3G)
Same analysis design as for the relatedness of human samples (above), except that matrices were subset by cell type and sample_type (organoid versus in vivo) samples. Sample correlation was performed using pairwise spearman correlation.
Cell Type Specification Windows
Related to Figure 1F - Specification windows were determined by analyzing the proportions of definitively called cell types within clusters proximal to RPC, Neurogenic, or Precursor cell populations. We reasoned that these clusters contained the earliest definitively called cell types within the specification and differentiation process. The dataset was subset down to clusters 96, 30, 26, 119, 54, 27, and 120 for RGCs, horizontal cells, amacrine cells, Müller glia, cones, rods, and bipolar cells, respectively. We then generated a data frame of sample age by cell types within the subset data and normalized each age column to the total number of cells captured at that age. We then multiplied by 10,000 to give an estimated value of the number of newly specified cells expected to be captured at each individual age when 10,000 cells are captured. Values across the developmental time course were then smoothed to give a distribution using the ‘stat_smooth’ function in R for local polynomial regression fitting
Immunohistochemistry of Retinal Tissue
Human retina tissue samples were fixed overnight in 4% paraformaldehyde. The fixed retinas were dehydrated in 20% and 30% sucrose in PBS at 4°C and embedded in optimal cutting temperature medium (Thermo Scientific). Thin 20–40μm cryosections were collected on superfrost slides (VWR) using a Leica CM3050S cryostat. For immunohistochemistry, antibodies against the following proteins were used at the following dilutions: Goat anti-SOX2 (1:250, Santa Cruz), Rabbit anti-Mki67 (1:200, Millipore), Mouse Anti-Brn3a (1:500, Millipore), Mouse anti-RLBP1 (1:500, Abcam), Mouse anti-Rod-OPSIN (1:1000, Sigma), Rabbit anti-S-OPSIN (1:500, Millipore), Rabbit anti-L/M-OPSIN (1:500, Millipore), Mouse anti-Calbindin (1:500, Abcam), Rabbit anti RRKCA (1:500,Abcam), Sheep anti VSX2 (1:400, Exalpha Biologicals) ,Rabbit anti MCM2(1:200 Cell Signaling), Sheep anti ONECUT2 (1:500 R and D Systems), Mouse anti LHX1(1:200 DSHB), Rabbit anti ISL1(1:200 Abcam), Goat anti OTX2 (1:200 R and D Systems), Mouse anti CRX(1:200 Abnova Corporation), Rabbit anti ATOH7(1:200 Novus) and Goat anti-Calretinin (1:500, Millipore). Primary antibodies were diluted in blocking buffer containing 10% donkey serum, 0.2% Triton X-100 and 0.2% gelatin in PBS at pH 7.4. Alexa Fluor 488, Alexa Fluor 594 or Alexa Fluor 647 fluorophore-conjugated secondary antibodies (1:500) (Life Technologies) were used as appropriate. Cell nuclei were stained with DAPI (1:10000). Images were collected using an Olympus FV1000 confocal microscope.
RNAscope
RNAscope® detection was performed in strict accordance with the ACD RNAscope® protocol (Wang et al., 2012). Briefly, retina sections were dehydrated in sequential incubations with ethanol, followed by 30 min Protease III treatment and washing in ddH2O. Appropriate combinations of hybridization probes were incubated for 2 hours at 40°C, followed by fluorescence labeling, DAPI counterstaining, and mounted with Prolong Gold mounting medium.
Diffusion Pseudotime & Trajectory Analysis
We exported the cell dataset matrices used in Monocle to create an AnnData object for pseudotime inference in Scanpy v1.4 (Wolf et al., 2018). The dataset was subset to relevant cell types based on the desired analysis, with cells from samples displaying significant batch effects (Hgw12) or discontinuous trajectories in UMAP dimension reductions – likely resulting from gaps in sampling ages (Hpnd8 and Adult) – removed from the analyses. In the case of RGC analysis, RPC louvain clusters moving away from the RGC trajectory and expressing late RPC-enriched genes (i.e. HES1 and BTG1) in the re-visualized UMAP embedding were further removed.
Preprocessing
For all trajectories, except neuro- and gliogenesis, raw expression matrices were used after filtering out genes in which < 1 count was observed (< 3 total counts in the case of Rod/Cone and in vivo/ex vivo cone analyses). Counts were normalized on a per cell basis to transcripts per 10,000 transcripts in all trajectories except for in the bipolar cell/photoreceptor trajectory where counts per cell were normalized to the median counts per cell. Matrices subsequently underwent natural log transformation. For the rod/cone and horizontal cell trajectories, matrices were scaled to unit variance and zero mean. For the bipolar cell/photoreceptor trajectory, data was scaled before and after each batch correction step. For the photoreceptor trajectory, only cells with mitochondrial fraction < 0.05 were retained following gene filtering step. For the cone trajectory, only cells with mitochondrial fraction < 0.2 were retained before genes were filtered.
Dimension Reduction and Pseudotime Assignment
PCA was calculated using high variance genes used as input. High variance genes are listed in Table S2 for each cell type analysis. A neighborhood graph was computed and used to find a representative diffusion map of the data. Diffusion Pseudotime reconstructed the trajectory from least mature cells to final cell fate(s) (Haghverdi et al., 2016; Wolf et al., 2018). Batch effect corrections, if necessary, were conducted prior to determination of highly variable genes, unless the BBKNN test was used (Polański et al., 2020). The datasets/cell types and parameters used for batch effect corrections, dimension reduction and diffusion pseudotime value assignment within Scanpy are listed in Table S3.
Differential Gene Expression Analysis
Corresponding pseudotime values from scanpy were assigned to cells within the Monocle cell dataset. Differential genes across pseudotime were assessed using the ‘differentialGeneTest’ function in Monocle, requiring that all differential transcripts being expressed in >= 10 cells. The following parameters were used for the differential gene test:
if one terminal cell fate, fullModelFormulaStr= “~sm.ns(Pseudotime,df=3)”
if multiple terminal cell fates, fullModelFormulaStr = “~sm.ns(Pseudotime,df=3)*Branch” and reducedModelFormulaStr = “~sm.ns(Pseudotime,df=3)”.
If needed, known batch effects were regressed through inclusion in the ‘reducedModelFormulaStr’ input parameters within the Monocle ‘differentialGeneTest’ function (Qiu et al., 2017; Trapnell et al., 2014). The full and reduced models used for running the ‘differentialGeneTest’ analyses on each trajectory can be found in Table S3.
Regression of Cell Cycle
For analyzing the trajectories from RPCs to Müller Glial or Neurogenic Cells, cell cycle was regressed from the expression matrix by first using Seurat v3.0 to assign Cell Cycle Scores and Phase (Butler et al., 2018). A Seurat object was created from the subset of cells from the original dataset on which the analyses were to be performed (See Table S3). The Seurat object was then normalized and scaled. The ‘CellCycleScoring’ function assigned cell cycle scores and phases to each cell using the following parameters: ‘CellCycleScoring(object=glia, s.features=s.genes, g2m.features=g2m.genes, set.ident=TRUE)’. s.genes and g2m.genes contain annotated S and G2 cell cycle genes, respectively, as included within Seurat (Tirosh et al., 2016). The mouse gene names were converted to the human homologs through uppercasing the gene names. Resulting cell cycle scores and phases were assigned to corresponding cells within the Monocle cell dataset. Cell Cycle Scores were regressed within the ‘preprocessCDS’ function via the “residualModelFormulaStr” (Qiu et al., 2017; Trapnell et al., 2014). Differential gene expression across pseudotime analysis was then conducted on the regressed expression matrix as described above.
Annotation of Retinal Progenitor Cell Subtypes
Examination of the density of RPCs across pseudotime identified two clear density peaks. We annotated early versus late RPCs based on positioning across the pseudotime axis in relation to the trough between the peaks (dotted line - Figure S2A; pseudotime value of 0.4609), with early RPCs corresponding to pseudotime values ≤ 0.4609 and late RPCs corresponding to pseudotime values >.04609.
UMAP Embedding of Amacrine/Starburst Amacrine Cell Trajectory –
Related to Figures S3A and S3B
In Scanpy, an AnnData object including Amacrine Cells, RPCs, Neurogenic Cells, AC/HC precursors at all ages except Adult, PND8 and GW12 was created. To preprocess the data, genes < 1 count were filtered out, counts were normalized to reads per 10,000 transcripts, and log transformed. Highly variable genes were annotated using the default settings under flavor ‘Seurat’. UMAP was performed with the following parameters: neighbors=30, 9 principal components, distance=0.75, metric = ‘canberra’, and random_state = 123456. 15 diffusion components were calculated and used as input to recompute distances and connectivities of neighbors. Louvain clustering was then performed at a resolution of 1. Clusters expressing SOX2 and CHAT were re-annotated as Starburst Amacrine Cell.
scCoGAPS Pattern Discovery
Following Stein O’Brien and Clark et al. 2019, we used scCoGAPS, a non-negative matrix factorization algorithm, to factor the expression matrix (genes x cells) into two separate matrices of lower dimensions: Pattern and Amplitude matrices. In the Amplitude matrix (gene x pattern), each value represents the weight of a gene that contributes to each pattern. In the Pattern matrix (cell x pattern), each value represents the weight of each pattern in each cell. In order to determine the relatedness of patterns with features of the dataset, we calculated the correlation of each pattern with global features assigned to each cell, including features such as cell type assignment or read depth.
Human Patterns
scCoGAPs analysis was performed on all retinal cells excluding the adult retinal samples. The large expression matrix size (33,694 genes by 118,555 cells) was subset down to a set of 3113 (Table S2) highly variable genes that excluded both mitochondrial and ribosomal protein coding genes. This down-sampling of genes allowed both faster implementation of scCoGAPS and was implemented to reduce the number of patterns that highlight sample batch effects of cellular stress or read depth. The log2 transformation of the expression matrix was used as input into the scCoGAPS function from CoGAPS v3.5 (Sherman et al., 2019; Stein-O’Brien et al., 2019). The parameters were default singleCell parameters, except nPatterns=100, nIterations=500, sparseOptimization = True, seed =830, nSets=27 (~3960 cells/set) with annotation weights set to sample underrepresented cell types more.
Mouse Patterns
We used scCoGAPS from CoGAPS v3.5 to reanalyze the mouse retinal development dataset from Clark et al. (2019), but limited the dataset to include only the retinal cells unlike (Clark et al., 2019) where all cells were used. We used 3164 high variance genes across the neuroretinal mouse dataset, again excluding mitochondrial and ribosomal genes and used nSets = 25 (4414 cells/set). Otherwise, all other scCoGAPS parameters remained the same as human pattern discovery.
For each dataset, the input parameters to scCoGAPS for nPatterns was determined empirically. In previous analysis of the mouse retinal dataset (Clark et al., 2019; Stein-O’Brien et al., 2019), we identified 80 consensus patterns, however, we reasoned that additional patterns would further stratify the dataset as we observed some previous patterns in which 2 biological processes were highlighted. This suggested to us that additional patterns would dimensionalize the data further. Therefore, we used 100 Patterns as input to scCoGAPS. As scCoGAPS features a ‘patternMatch’ function, identifying similar patterns and merging them into one pattern while removing spurious patterns, our analysis resulted in 97 consensus mouse patterns that reflected known biology better than previous iterations. For the human dataset, we first tested 120 patterns, however, many of the patterns highlighted few cells or corresponded solely to batch effects, suggesting an over-dimensionalization of the data. Subsequent re-analyses obtained stable results when 100 patterns were used as input. The final human scCoGAPS dataset stabilized at 97 patterns.
Comparisons of Pattern Usage across Species
To examine the extent to which human or mouse patterns corresponded to similar biological processes in the opposite species, we used projectR to project the log2 of the scRNA-Seq expression matrix into the scCoGAPS pattern amplitude matrix (Stein-O’Brien et al., 2019). With the Amplitude matrix from source dataset (i.e. human) and the expression matrix from target dataset (i.e. mouse) as input, projectR estimates a new Pattern matrix defining the role each pattern plays in each cell of the target dataset. This allowed us to assess how each pattern defines features of the target dataset (i.e. cell types and processes.) Since human and mouse genes differ, the Ensembl database from BioMart was used to identify the corresponding homologs of the pattern genes in the source Amplitude matrix (Durinck et al., 2005, 2009). For certain genes, biomaRt retrieved no homologous genes. In these cases, the letter case of the gene short names was modified to match that of the target species. If these gene short names were still not in the expression matrix of the target species, they were removed. If a gene mapped to multiple homologous genes, the gene’s weights were reused for both target genes. If multiple genes map to the same homologous gene, their pattern weights were aggregated by the maximum pattern weights.
Construction of shATOH7 Lentiviral Vectors
To suppress ATOH7 expression in developing human retina, pLKO.1-TRC cloning vector (Moffat et al., 2006) was purchased from Addgene (Cat#10878). The puromycin selection marker in the vector was replaced with GFP-P2A-Puromycin or RFP-P2A-Puromycin ORF cassette with Gibson Assembly Master Mix (New England Biolabs, Cat#E2611L) to generate pLKO.1-GFP-P2A-Puro or pLKO.1-RFP-P2A-Puro cloning vector. Specific cloning details are available on request. The target sequence for scrambled shRNA is CCTAAGGTTAAGTCGCCCTCG available from Addgene (Cat#1864) (Sarbassov et al., 2005), and the sequences of the hairpin pair are as follows:SCR-F: CCGGCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTG;
SCR-R: AATTCAAAAACCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG.
The target sequence for shATOH7–1 is GTGAAGTTACAGTATCCATTA available from TRC library database (TRCN0000423538) (Moffat et al., 2006), and the sequences for the hairpin pairs are as follows:
shATOH7–1F: CCGGGTGAAGTTACAGTATCCATTACTCGAGTAATGGATACTGTAACTTCACTTTTTTGAAT;
shATOH7–1R: AATTCAAAAAGTGAAGTTACAGTATCCATTACTCGAGTAATGGATACTGTAACTTCAC.
The target sequence for shATOH7–2 is GATTCTCAGATTACCTTTATT designed with an online tool (http://www.broad.mit.edu/genome_bio/trc/rnai.html) (Moffat et al., 2006), and the sequences of the hairpin pair are as follows: shATOH7–2F:
CCGGGATTCTCAGATTACCTTTATTCTCGAGAATAAAGGTAATCTGAGAATCTTTTTG;
shATOH7–2R:
AATTCAAAAAGATTCTCAGATTACCTTTATTCTCGAGAATAAAGGTAATCTGAGAATC.
Both shRNAs are targeted to the 3′UTR of ATOH7 gene. The oligos of the hairpin pairs were purchased from Eurofins Genomics, annealed together, and cloned into AgeI and EcoRI cloning sites of pLKO.1-GFP-P2A-Puro or pLKO.1-RFP-P2A-Puro vector with Quick Ligation Kit (New England Biolabs, Cat#M2200S) (Moffat et al., 2006).
Production of shATOH7 lentivirus and virus validation
Lentivirus was produced with Lenti-X 293T packaging cells (Clontech, Cat#632180) transfected with viral vectors and packaging plasmids pMD2.G and psPAX2 (Addgene, Cat#12259 and Cat#12260) in DMEM medium (Wisent Bioproducts, Cat#319–005-CL) supplemented with 10% fetal bovine serum (FBS) (Wisent Bioproducts, Cat#080–150) (Sanjana et al., 2014). Briefly, for each virus, 5X 106 cells were seeded in a 10-cm plate the day before transfection. On the day of transfection, culture medium was removed and replaced with 5 ml of prewarmed fresh medium. A mixture of 3 μg viral vector, 2.25 μg pPAX2, 0.75 μg pMD2.G and 18 ul of Lipofectamine 2000 (Life Technologies, Cat#11668019) in 3.0 ml of Opti-MEM I Reduced-Serum Medium (1X) (Life Technologies, Cat#31985070) was added dropwise to the cells with gentle mixing by rocking the plate back and forth. The culture medium with transfection reagent was removed 12 hours later and replaced with 10 ml of warm fresh medium. Virus-containing supernatants were harvested 48 hours after transfection, filtered through a 0.45 μm low protein binding membrane (Sarstedt, Cat#83.1826), and concentrated with Lenti-X Concentrator (Clontech, Cat#631231) according to the inserted protocol. Virus pellets were dissolved in 100 μl of IMDM (Wisent Bioproducts, Cat#319–105-CL), aliquoted and stored at −80°C.
For validation, a developing human cone cell line was generated by transduction of human retina with shRB1 virus and selection for replicating cells. Identity was validated with multiple cone markers, then cells were transduced with shATOH7 lentivirus for 8 days. To assess ATOH7 knockdown efficiency, RNA and protein were extracted from the cells, and Real-Time qRT-PCR as well as Western blot (Chen et al., 2009) were carried to measure ATOH7 expression. Human β-ACTIN and TBP (TATA-Box Binding Protein) were used as qRT-PCR references, and β-ACTIN was used as a loading control for Western blot (Figure S7). The PCR primers are the following.
ATOH7-F: AGTACGAGACCCTGCAGATG
ATOH7-R: TGGAAGCCGAAGAGTCTCTG
β-ACTIN-F: AAAGCCACCCCACTTCTCTCTAA
β-ACTIN-R: ACCTCCCCTGTGTGGACTTG
TBP-F: ATGTTGAGTTGCAGGGTGTG
TBP-R: CAGCACGGTATGAGCAACTC
Transduction of Developing Human Retinal Explants with shATOH7 Lentivirus
Developing human retinas at gestational age from GW17 to GW19 were sterilely dissected in IMDM and cut radially. Tissue fragments were transferred on to cell-culture inserts (Millipore, Cat#PICM03050) with photoreceptor side down (Jin and Xiang, 2012). Inserts with retinal fragments were quickly put in 6-well plates with 1300 μl of prewarmed retina culture medium and incubated at 37°C with 5% CO2. Due to spatiotemporal difference in developing human retina development (Hoshino et al., 2017), each explant culture was co-transduced with both control and treatment viruses by adding freshly thawed virus mixture on top of retinal explants (Figure 1A). Explants were cultured for about 3 weeks, and the medium was changed twice per week, with half of the culture medium replaced with fresh medium.
Immunofluorescence Staining and Microscopy of Retinal Explants
After about 3 weeks of culture, the retinal explants were fixed with 4% paraformaldehyde overnight at 4°C, washed 3 times with cold PBS, dehydrated in 30% sucrose for 24 hours at 4°C, embedded in 30% sucrose/OCT compound (1:1), and stored at −80°C. The frozen tissue blocks were sectioned at 16 μm thickness, and air-dried at room temperature for 4 hours, stored at −20°C. For immunostaining, sections were rinsed in Tris-buffered saline with 0.1% Tween 20 (TBS-T), incubated in blocking buffer containing 2.5% bovine serum albumin (BSA) in TBS-T for 1 hour at room temperature, and labeled with primary antibodies diluted in blocking buffer overnight at 4°C. The following day, sections were washed 3 times with TBS-T, and incubated for 1 hour at room temperature with appropriate secondary antibodies conjugated to 488/568/647 fluorophores. Subsequently, sections were washed with TBS-T for 3 times, cover-slipped, mounted in VECTASHIELD Hardset Antifade Mounting Medium with DAPI (Vector Laboratories, Cat# H-1500) to counterstain nuclei, and imaged using Nikon Ti-E inverted confocal microscope. Images were processed using ImageJ (NIH). For each explant per staining, at least 10 images were processed. Cones were confirmed with RXRγ or GNAT2 staining, and rods with NRL staining. The ratios of percentages of cones versus rods in the control and treatment virus transduced photoreceptors were calculated. Antibodies were used at the following dilutions: ATOH7 – 1:700, GFP - 1:300 (rabbit polyclonal; Life Technologies), GNAT2 – 1:100, mCherry - 1:500, NRL - 1:00, RFP 1:300, RXRγ - 1:100.
QUANTIFICATION AND STATISTICAL ANALYSIS
Details of the statistical analyses used are present within the figure legends and Table S3. Statistical significance was determined using PRISM, R, or Monocle software (see Key Resources Table), unless otherwise noted.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-Sox2 | Santa Cruz | Cat# sc17320; RRID:AB_2286684 |
| Mouse monoclonal anti-RLBP1 | Abcam | Cat# ab15051; RRID:AB_2269474 |
| Mouse monoclonal anti-Opsin, Unconjugated, Clone RET-P1 | Sigma-Aldrich | Cat# O4886; RRID:AB_260838 |
| Rabbit Polyclonal anti-Ki-67 | Millipore | Cat# AB9260; RRID:AB_2142366 |
| Mouse monoclonal anti-Brn3a (POU-domain protein), Unconjugated | Millipore | Cat# MAB1585; RRID:AB_94166 |
| Mouse polyclonal anti-S-Opsin | Millipore | Cat# AB5407; RRID:AB_177457 |
| Mouse polyclonal anti-L/M-Opsin | Millipore | Cat# AB5405 RRID:AB_177456 |
| Mouse monoclonal anti-calbindin | Abcam | Cat# ab9481–500; RRID:AB_2811302 |
| Goat Polyclonal anti-Calretinin | Millipore | Cat# AB1550; RRID:AB_90764 |
| Sheep polyclonal anti-CHX10(VSX2) | Exalpha Biologicals | Cat# x1179p; RRID:AB_2314191 |
| PKC alpha antibody [Y124] | Abcam | Cat#ab32376; RRID:AB_777294 |
| MCM2 (D7G11) XP(tm) Rabbit mAb antibody | Cell Signaling Technology | Cat# 3619s; RRID:AB_2142137 |
| Human ONECUT2/OC-2 Affinity Purified Polyclonal Ab antibody | R&D Systems | Cat#AF6294; RRID:AB_10640365 |
| Mouse monoclonal anti-LHX1 | DSHB | Cat#4F2; RRID: AB_531784 |
| Human Otx2 Affinity Purified Polyclonal Ab antibody | R&D Systems | Cat# AF1979; RRID:AB_2157172 |
| Mouse Anti-Human CRX Monoclonal Antibody, Unconjugated, Clone 4G11 | Abnova Corporation | Cat# H00001406-M02; RRID:AB_606098 |
| Rabbit polyclonal anti-ATOH7 | Novus | Cat# NBP1–88639; RRID:AB_11034390 |
| Mouse monoclonal – β-ACTIN | Sigma-Aldrich | Cat# A5441; RRID:AB_476744 |
| Goat polyclonal anti-GFP | Abcam | Cat# Ab6673; RRID:AB_305643 |
| Rabbit polyclonal anti-GFP | Life Technologies | Cat# A11122; RRID:AB_221569 |
| Rabbit polyclonal anti-GNAT2 | LifeSpan BioSciences | Cat# LS-C321680; RRID:AB_2811301 |
| Chicken polyclonal anti-mCherry | Novus Biologicals | Cat# NBP2–25158; RRID:AB_2636881 |
| Goat polyclonal anti-NRL | R&D Systems | Cat# AF2945; RRID:AB_2155098 |
| Rabbit polyclonal anti-RFP | Abcam | Cat# Ab62341; RRID:AB_945213 |
| Mouse monoclonal anti-RXRγ | Santa Cruz Biotechnology | Cat# Sc-365252; RRID:AB_10850062 |
| Alexa Fluor 488 Conjugated Donkey anti-Mouse IgG(H+L) | Invitrogen | Cat# A-21202; RRID:AB_141607 |
| Alexa Fluor 488 Conjugated Donkey anti-Rabbit IgG(H+L) | Invitrogen | Cat# A-21206; RRID:AB_141708 |
| Alexa Fluor 594 Conjugated Donkey anti-Rabbit IgG(H+L) | Invitrogen | Cat# A21207; RRID:AB_141637 |
| Alexa Fluor 594 Conjugated Donkey anti-Mouse IgG(H+L) | Invitrogen | Cat# A-21203; RRID:AB_141633 |
| Alexa Fluor 594 Conjugated Donkey anti-Goat IgG(H+L) | Invitrogen | Cat# A-11058; RRID:AB_142540 |
| Alexa Fluor 647 Conjugated Donkey anti-Goat IgG(H+L) | Invitrogen | Cat# A21447; RRID:AB_10584487 |
| Alexa Fluor 647 Conjugated Donkey anti-Mouse IgG(H+L) | Invitrogen | Cat# A-31571; RRID:AB_162542 |
| Alexa Fluor 488 Conjugated Donkey anti-Goat IgG(H+L) | Invitrogen | Cat# A11055; RRID:AB_2534102 |
| Alexa Fluor 647 Conjugated Donkey anti-Rabbit IgG(H+L) | Invitrogen | Cat# A31573; RRID:AB_2536183 |
| Alexa Fluor 568 Conjugated Donkey anti-Rabbit IgG(H+L) | Invitrogen | Cat# A10042; RRID:AB_2534017 |
| Alexa Fluor 568 Conjugated Goat anti-Chicken | Molecular Probes | Cat# A11041; RRID:AB_2534098 |
| Biological Samples | ||
| Human retinal samples | Anzhen Hospital of Capital Medical University and The First Affiliated Hospital of University of Science and Technology of China | N/A, materials available on request from Dr. Xiaoqun Wang |
| Human retinal samples | Morgentaler Clinic, Mount Sinai Hospital, Toronto, Canada | |
| Human retinal sample | Alabama Eye Bank, Birmingham, AL | 86-year-old Female without retinal disease; Cause of death: mitral regurgitation Death to enucleation: 3.3 hr |
| Human retinal organoids | ATCC | Generated from iPSCs derived from IMR-90 cell line (RRID: CVCL_C437) |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RNAscope® Multiplex Fluorescent Reagent Kit v2 | ACD | Cat# 323100 |
| RNAscope® Probe- Hs-CYP26A1 | ACD | Cat#487741 |
| RNAscope® Probe- Hs-SFRP1-C2 | ACD | Cat#429381 |
| RNAscope® Probe- Hs-RLBP1-C2 | ACD | Cat#414221-C2 |
| RNAscope® Probe- Hs-DIO2-C3 | ACD | Cat#562211-C3 |
| RNAscope® Probe- Hs-LHX1 | ACD | Cat#493021 |
| RNAscope® Probe- Hs-ISL1-C2 | ACD | Cat#478591-C2 |
| 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) | Thermo Fisher | Cat# D1306; RRID: AB_2629482 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3′ Library and Gel Bead Kit v2 | 10x Genomics | Cat# PN-120237 |
| Chromium Single Cell 3′ Chip Kit v2 | 10x Genomics | Cat# PN-120236 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat# PN-120262 |
| Deposited Data | ||
| Single-cell RNA sequencing Visualization of processed data available at: https://proteinpaint.stjude.org/?genome=hg38&singlecell=files/hg38/NEI.AGI.retina/singlecell/Human_Retina_Development.json |
This paper | GEO: GSE118614 GEO: GSE116106 GEO: GSE122970 |
| Mouse retinal development single-cell RNA-sequencing | Clark et al., 2019 | GEO: GSE138002 |
| Software and Algorithms | ||
| GraphPad Prism 5.0/7 | GraphPad Software | http://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Adobe Photoshop CC2015 | Adobe Systems | http://www.adobe.com |
| Cell ranger 2.0.1 | 10x Genomics | http://10xgenomics.com |
| R version 3.6.0 | The R project | https://www.r-project.org/ |
| CoGAPS v3.5 | (Sherman et al., 2019; Stein-O’Brien et al., 2019) | https://doi.org/10.18129/B9.bioc.CoGAPS |
| projectR | (Stein-O’Brien et al., 2019) | https://doi.org/10.18129/B9.bioc.projectR |
| umap | (Becht et al., 2018) | https://github.com/lmcinnes/umap/archive/0.2.4.tar.gz. |
| scanpy | (Wolf et al., 2018) | https://scanpy.readthedocs.io/ |
| Monocle (multiple version; 2.6.1, 2.99.3) | Cole Trapnell | https://github.com/cole-trapnell-lab/monocle-release |
| Monocle3_0.1.0 | Cole Trapnell | https://cole-trapnell-lab.github.io/monocle3/ |
| Seurat | (Butler et al., 2018) | https://satijalab.org/seurat/ |
| largeVis | (Tang et al., 2016) | https://github.com/lferry007/LargeVis |
| BBKNN | (Polański et al., 2020) | https://bbknn.readthedocs.io/ |
| Combat | (Leek et al., 2012) | https://github.com/brentp/combat.py |
| BioMart | (Durinck et al., 2005, 2009) | https://doi.org/10.18129/B9.bioc.biomaRt |
| ComplexHeatmap | (Gu et al., 2016) | https://github.com/jokergoo/ComplexHeatmap |
| genesorteR | (Ibrahim and Kramann, 2019) | https://github.com/mahmoudibrahim/genesorteR |
| pheatmap_1.0.12 | Raivo Kolde | https://CRAN.R-project.org/package=pheatmap |
Supplementary Material
Highlights.
Identification of networks governing human retinal cell-type specification
Characterization of mechanisms controlling horizontal cell and foveal development
Analysis of conserved and divergent gene expression between human and mouse
ATOH7 loss during late neurogenesis inhibits specification of cone photoreceptors
ACKNOWLEDGMENTS
We thank J. Nathans, A. Kolodkin, R. Johnston Jr., S. Chen, P. Ruzycki, and W. Yap for comments on the manuscript. We thank the Transcriptomics and Deep Sequencing Core at Hopkins for assistance in sequencing and Dr. Jeff Wrana and Daniel Trcka for technical assistance with preparing single-cell libraries and both the Morgentaler Clinic in Toronto and the donors for access to developing retinal tissue. We also thank Xin Zhou, Michael Dyer, and Jian Wang (St. Jude Children’s Research Hospital) for help with hosting the scRNA-seq dataset. This work was supported by the Canadian Institute for Health Research (CIHR) grant #153128 (R.B.), the Krembil Foundation (R.B.), the Strategic Priority Research Program of the Chinese Academy of Sciences grants XDA16020601 and XDB32010100 (X.W.), National Basic Research Program of China grants 2019YFA0110100, 2017YFA0102601, and 2017YFA0103303 (X.W.), the National Natural Science Foundation of China (NSFC) grants 91732301, 31671072, 31771140, and 81891001 (X.W.), the Grants of Beijing Brain Initiative of Beijing Municipal Science & Technology Commission Z181100001518004 (X.W.), the National Science Fund for Distinguished Young Scholars of China grant 81925009 (T.X.), the National Natural Science Foundation of China grant 81790644 (T.X.), and by an unrestricted grant to the Department of Ophthalmology and Visual Sciences at Washington University from Research to Prevent Blindness (B.S.C.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2020.04.009.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ajioka I, Martins RAP, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, and Dyer MA (2007). Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 131, 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, and Gan L (2014). Development of retinal amacrine cells and their dendritic stratification. Curr. Ophthalmol. Rep 2, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 10.1038/nbt.4314. [DOI] [PubMed]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. (2004). Genomic analysis of mouse retinal development. PLoS Biol 2, E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boije H, Shirazi Fard S, Edqvist PH, and Hallböök F (2016). Horizontal cells, the odd ones out in the retina, give insights into development and disease. Front. Neuroanat 10, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner R, and Sage J (2014). Cancer: the origin of human retinoblastoma. Nature 514, 312–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, and Glaser T (1998). Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 125, 4821–4833. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, and Glaser T (2001). Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenaventura DF, Corseri A, and Emerson MM (2019). Identification of genes With enriched expression in early developing mouse cone photoreceptors. Invest. Ophthalmol. Vis. Sci 60, 2787–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumsted-O’Brien KM, Hendrickson A, Haverkamp S, Ashery-Padan R, and Schulte D (2007). Expression of the homeodomain transcription factor Meis2 in the embryonic and postnatal retina. J. Comp. Neurol 505, 58–72. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanin L, and Wittbrodt J (2014). Retinal neurogenesis. Development 141, 241–244. [DOI] [PubMed] [Google Scholar]
- Cepko C (2014). Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci 15, 615–627. [DOI] [PubMed] [Google Scholar]
- Chen D, Livne-Bar I, Vanderluit JL, Slack RS, Agochiya M, and Bremner R (2004). Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5, 539–551. [DOI] [PubMed] [Google Scholar]
- Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, and Bremner R (2009). Division and apoptosis of E2F-deficient retinal progenitors. Nature 462, 925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, and Cepko CL (2006). Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development 133, 3167–3177. [DOI] [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, et al. (2019). Single-cell RNA-Seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin J, Queen R, Zerti D, Dorgau B, Hussain R, Coxhead J, Cockell S, and Lako M (2019). Deconstructing retinal organoids: single cell RNA-Seq reveals the cellular components of human pluripotent stem cell-derived retina. Stem Cells 37, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin SP (1999). Behavioural ecology and retinal cell topography. In Adaptive Mechanisms in the Ecology of Vision, Archer SN, Djamgoz MBA, Loew ER, Partridge JC, and Vallerga S, eds. (Springer; ), pp. 509–535. [Google Scholar]
- Cowan CS, Renner M, Gross-Scherf B, Goldblum D, Munz M, Krol J, Szikra T, Papasaikas P, Cuttat R, Waldt A, et al. (2019). Cell types of the human retina and its organoids at single-cell resolution: developmental convergence, Transcriptomic identity, and disease map. SSRN Journal 10.2139/ssrn.3438371. [DOI]
- Curcio CA, Presley JB, Millican CL, and Medeiros NE (2005). Basal deposits and drusen in eyes with age-related maculopathy: evidence for solid lipid particles. Exp. Eye Res 80, 761–775. [DOI] [PubMed] [Google Scholar]
- da Silva S, and Cepko CL (2017). FGF8 expression and degradation of retinoic acid are required for patterning a high-acuity area in the retina. Dev. Cell 42, 68–81.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, Schuijff L, Dekker M, van der Valk M, and te Riele H (2004). Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev 18, 2952–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Araya C, and Provis JM (1992). Evidence of photoreceptor migration during early foveal development: a quantitative analysis of human fetal retinae. Vis. Neurosci 8, 505–514. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat. Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PHD, Myers SM, and Hallböök F (2006). Early identification of retinal subtypes in the developing, pre-laminated chick retina using the transcription factors Prox1, Lim1, Ap2alpha, Pax6, Isl1, Isl2, Lim3 and Chx10. Eur. J. Histochem 50, 147–154. [PubMed] [Google Scholar]
- Eicher JD, Powers NR, Miller LL, Akshoomoff N, Amaral DG, Bloss CS, Libiger O, Schork NJ, Darst BF, Casey BJ, et al. (2013). Genome-wide association study of shared components of reading disability and language impairment. Genes Brain Behav 12, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldred KC, Hadyniak SE, Hussey KA, Brenerman B, Zhang PW, Chamling X, Sluch VM, Welsbie DS, Hattar S, Taylor J, et al. (2018). Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362, eaau6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Dong J, Zhong S, Wei Y, Wu Q, Yan L, Yong J, Sun L, Wang X, Zhao Y, et al. (2018). Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res 28, 730–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, and Schier AF (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertig EJ, Ozawa H, Thakar M, Howard JD, Kagohara LT, Krigsfeld G, Ranaweera RS, Hughes RM, Perez J, Jones S, et al. (2016). CoGAPS matrix factorization algorithm identifies transcriptional changes in AP-2alpha target genes in feedback from therapeutic inhibition of the EGFR network. Oncotarget 7, 73845–73864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzSimmons J, Fantel A, and Shepard TH (1994). Growth parameters in mid-trimester fetal Turner syndrome. Early Hum. Dev 38, 121–129. [DOI] [PubMed] [Google Scholar]
- Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JA, Goldman D, and Glaser T (2011). Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci 14, 578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes FLAF, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, and Cayouette M (2011). Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 138, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Bok D, Yu F, Caprioli J, and Piri N (2018). Downregulation of splicing regulator RBFOX1 compromises visual depth perception. PLoS One 13, e0200417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Büttner M, Wolf FA, Buettner F, and Theis FJ (2016). Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848. [DOI] [PubMed] [Google Scholar]
- Hamon A, García-García D, Ail D, Bitard J, Chesneau A, Dalkara D, Locker M, Roger JE, and Perron M (2019). Linking YAP to Müller glia quiescence exit in the degenerative retina. Cell Rep 27, 1712–1725.e6. [DOI] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, and Harris WA (2012). How variable clones build an invariant retina. Neuron 75, 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock PF, Macdonald RE, VandeRyt JT, and Wilson SW (1996). Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J. Neurobiol 29, 399–413. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M, et al. (2017). Molecular anatomy of the developing human retina. Dev. Cell 43, 763–779.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wang X, Hu B, Mao Y, Chen Y, Yan L, Yong J, Dong J, Wei Y, Wang W, et al. (2019). Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol 17, e3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim MM, and Kramann R (2019). genesorteR: feature ranking in clustered single cell data. bioRxiv https://www.biorxiv.org/content/10.1101/676379v2.
- Jadhav AP, Mason HA, and Cepko CL (2006). Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913–923. [DOI] [PubMed] [Google Scholar]
- Jager RD, Mieler WF, and Miller JW (2008). Age-related macular degeneration. N. Engl. J. Med 358, 2606–2617. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, and Masland RH (1998). The major cell populations of the mouse retina. J. Neurosci 18, 8936–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Liu Q, Zhang S, Chen Q, Wang C, Zhang W, Xiao C, Li Y, Nian C, Li J, et al. (2019). FGF15 activates hippo signaling to suppress bile acid metabolism and liver tumorigenesis. Dev. Cell 48, 460–474.e9. [DOI] [PubMed] [Google Scholar]
- Jin K, and Xiang M (2012). In vitro explant culture and related protocols for the study of mouse retinal development retinal development. Methods Mol Biol 884, 155–165. [DOI] [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Jan YN, and Vetter ML (1997). Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron 19, 981–994. [DOI] [PubMed] [Google Scholar]
- Kay JN, Chu MW, and Sanes JR (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, and Baier H (2001). Retinal ganglion cell genesis requires Lakritz, a zebrafish atonal Homolog. Neuron 30, 725–736. [DOI] [PubMed] [Google Scholar]
- Khrameeva E, Kurochkin I, Han D, Guijarro P, Kanton S, Santel M, Qian Z, Rong S, Mazin P, Bulat M, et al. (2019). Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. bioRxiv https://www.biorxiv.org/content/10.1101/764936v1. [DOI] [PMC free article] [PubMed]
- Kolb H, Fernandez E, Schouten J, Ahnelt P, Linberg KA, and Fisher SK (1994). Are there three types of horizontal cell in the human retina? J. Comp. Neurol 343, 370–386. [DOI] [PubMed] [Google Scholar]
- La Vail MM, Rapaport DH, and Rakic P (1991). Cytogenesis in the monkey retina. J. Comp. Neurol 309, 86–114. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, and Storey JD (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Dharmat R, Owen L, Shakoor A, Li Y, Kim S, Vitale A, Kim I, Morgan D, Liang S, et al. (2019). Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun 10, 5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Xu S, Wang Y, Mazerolle C, Thurig S, Coles BLK, Ren JC, Taketo MM, van der Kooy D, and Wallace VA (2007). Ciliary margin trans-differentiation from neural retina is controlled by canonical Wnt signaling. Dev. Biol 308, 54–67. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu W, Yang L, Wu Q, Zhang H, Fang A, Li L, Xu X, Sun L, Zhang J, et al. (2017). The primate-specific gene TMEM14B marks outer radial glia cells and promotes cortical expansion and folding. Cell Stem Cell 21, 635–649.e8. [DOI] [PubMed] [Google Scholar]
- Lo Giudice Q, Leleu M, La Manno G, and Fabre PJ (2019). Single-cell transcriptional logic of cell-fate specification and axon guidance in early-born retinal neurons. Development 146, dev178103. [DOI] [PubMed] [Google Scholar]
- Lukowski SW, Lo CY, Sharov A, Nguyen QH, Fang L, Hung SSC, Zhu L, Zhang T, Nguyen T, Senabouth A, et al. (2019). Generation of human neural retina transcriptome atlas by single cell RNA sequencing. bioRxiv https://www.biorxiv.org/content/10.1101/425223v2.
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, and Jacks T (2004). Cell type-specific effects of Rb deletion in the murine retina. Genes Dev 18, 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter-Sadzinski L, Matter JM, Ong MT, Hernandez J, and Ballivet M (2001). Specification of neurotransmitter receptor identity in developing retina: the chick ATH5 promoter integrates the positive and negative effects of several bHLH proteins. Development 128, 217–231. [DOI] [PubMed] [Google Scholar]
- Mellough CB, Bauer R, Collin J, Dorgau B, Zerti D, Dolan DWP, Jones CM, Izuogu OG, Yu M, Hallam D, et al. (2019). An integrated transcriptional analysis of the developing human retina. Development 146, dev169474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M, Mohammadi S, Davila-Velderrain J, Goods BA, Cadwell TD, Xing Y, Stemmer-Rachamimov A, Shalek AK, Love JC, Kellis M, and Hafler BP (2019). Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun 10, 4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Moore KB, Logan MA, Aldiri I, Roberts JM, Steele M, and Vetter ML (2018). C8orf46 homolog encodes a novel protein Vexin that is required for neurogenesis in Xenopus laevis. Dev. Biol 437, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichl L (2005). Diversity of mammalian photoreceptor properties: adaptations to habitat and lifestyle? Anat. Rec. A Discov. Mol. Cell Evol. Biol 287, 1001–1012. [DOI] [PubMed] [Google Scholar]
- Peng Y-R, James RE, Yan W, Kay JN, Kolodkin AL, and Sanes JR (2020). Binary fate choice between closely related interneuronal types is determined by a Fezf1-dependent postmitotic transcriptional switch. Neuron 105, 464–474.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y-R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular classification and comparative Taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park JE (2020). BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L, Masud T, Khaliq S, Mehdi SQ, Abid A, Oliver ER, Silva ED, Lewanda A, Brodsky MC, Borchert M, et al. (2012). ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum. Mol. Genet 21, 3681–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A, and Bringmann A (2013). New functions of Müller cells. Glia 61, 651–678. [DOI] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, and Noonan JP (2015). Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill RL, Euler T, and Masland RH (2000). Spatial order within but not between types of retinal neurons. Proc. Natl. Acad. Sci. USA 97, 2303–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda EM, Hall BM, Hill MC, Swinton PG, Tong X, Martin JF, and Poché RA (2019). The hippo pathway blocks mammalian retinal Müller glial cell reprogramming. Cell Rep 27, 1637–1649.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan M, McCurdy SR, Livne-Bar I, Ahmad M, Wrana JL, Chen D, and Bremner R (2012). Established and new mouse models reveal E2F1 and Cdk2 dependency of retinoblastoma, and expose effective strategies to block tumor initiation. Oncogene 31, 5019–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, and Sabatini DM (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101. [DOI] [PubMed] [Google Scholar]
- Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. (2019). Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard TH (1975). Growth and development of human embryo and fetus. In Endocrine and Genetic Diseases of Childhood, Gardner LI 2nd, ed. (Philadelphia: W.B. Saunders: ), pp. 1–6. [Google Scholar]
- Sherman TD, Gao T, and Fertig EJ (2019). CoGAPS 3: Bayesian non-negative matrix factorization for single-cell analysis with asynchronous updates and sparse data structures. bioRxiv https://www.biorxiv.org/content/10.1101/699041v1. [DOI] [PMC free article] [PubMed]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, et al. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci 13, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, and Roska B (2012). Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci 15, 487–495. [DOI] [PubMed] [Google Scholar]
- Sridhar A, Hoshino A, Finkbeiner CR, Chitsazan A, Dai L, Haugan AK, Eschenbacher KM, Jackson DL, Trapnell C, Bermingham-McDonogh O, et al. (2020). Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep 30, 1644–1659.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien GL, Clark BS, Sherman T, Zibetti C, Hu Q, Sealfon R, Liu S, Qian J, Colantuoni C, Blackshaw S, et al. (2019). Decomposing cell identity for transfer learning across cellular measurements, platforms, tissues, and species. Cell Syst 8, 395–411.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga A, Taira M, and Nakagawa S (2009). LIM family transcription factors regulate the subtype-specific morphogenesis of retinal horizontal cells at post-migratory stages. Dev. Biol 330, 318–328. [DOI] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Jonasdottir A, Stefansdottir G, et al. (2007). Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science 317, 1397–1400. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, et al. (2019). Single-cell profiles of retinal neurons differing in resilience to injury reveal neuroprotective genes, 104, pp. 1039–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JW, Wei W, Gonzalez C, Sweeney NT, Huberman AD, Feller MB, and Feldheim DA (2014). Dendritic and axonal targeting patterns of a genetically-specified class of retinal ganglion cells that participate in image-forming circuits. Neural Dev 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel D, Provis JM, and Billson FA (1990). Early differentiation of ganglion, amacrine, bipolar, and Muller cells in the developing fovea of human retina. J. Comp. Neurol 291, 203–219. [DOI] [PubMed] [Google Scholar]
- Voigt AP, Whitmore SS, Flamme-Wiese MJ, Riker MJ, Wiley LA, Tucker BA, Stone EM, Mullins RF, and Scheetz TE (2019). Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res 184, 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, and Sanes JR (2009). Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J. Comp. Neurol 517, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, and Klein AM (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlin KJ, Maruotti JA, Sripathi SR, Ball J, Angueyra JM, Kim C, Grebe R, Li W, Jones BW, and Zack DJ (2017). Photoreceptor outer segment-like structures in long-term 3D retinas from human pluripotent stem cells. Sci. Rep 7, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, and Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LL, and Rapaport DH (2009). Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development 136, 1707–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SN, Goude Y, and Shaw S (2015). Generalized additive models for large data sets. J. R. Stat. Soc C 64, 139–155. [Google Scholar]
- Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar SC, Abramson DH, and Cobrinik D (2009). Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137, 1018–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Singh HP, Wang L, Qi DL, Poulos BK, Abramson DH, Jhanwar SC, and Cobrinik D (2014). Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature 514, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, and Ashery-Padan R (2006). Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133, 1367–1378. [DOI] [PubMed] [Google Scholar]
- Young RW (1985). Cell differentiation in the retina of the mouse. Anat. Rec 212, 199–205. [DOI] [PubMed] [Google Scholar]
- Zaghloul NA, and Moody SA (2007). Changes in Rx1 and Pax6 activity at eye field stages differentially alter the production of amacrine neurotransmitter subtypes in Xenopus. Mol. Vis 13, 86–95. [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schweers B, and Dyer MA (2004). The first knockout mouse model of retinoblastoma. Cell Cycle 3, 952–959. [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
- Tang J, Liu J, Zhang M, and Mei Q (2016). Visualizing large-scale and high-dimensional data. Proceedings of the 25th international conference on World Wide Web - WWW ‘16. 10.1145/2872427.2883041. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-sequencing files are available through GEO with the following accession numbers: GEO: GSE116106, GEO: GSE122970, GEO: GSE138002. Processed, finalized matrices are available through GEO accession number GEO: GSE138002. Visualization and analysis of processed data (dimension reduction, gene expression, heatmaps) is available at https://proteinpaint.stjude.org/?genome=hg38&singlecell=files/hg38/NEI.AGI.retina/singlecell/Human_Retina_Development.json. Details of analysis and input parameters is available within the STAR Methods (below) and in Table S3. Additional details and inquiries should be addressed to the Lead Contact.


