Abstract
Introduction
Hepatocellular carcinoma (HCC) is a leading cause of cancer related mortality in the world and it has limited treatment options. Understanding the molecular drivers of HCC is important to develop novel biomarkers and therapeutics.
Purpose of Review
HCC arises in a complex background of chronic hepatitis, fibrosis and liver regeneration which lead to genomic changes. Here, we summarize studies that have expanded our understanding of the molecular landscape of HCC.
Recent Findings
Recent technological advances in next generation sequencing (NGS) have elucidated specific genetic and molecular programs involved in hepatocarcinogenesis. We summarize the major somatic mutations and epigenetic changes have been identified in NGS-based studies. We also describe promising molecular therapies and immunotherapies which target specific genetic and epigenetic molecular events.
Summary
The genomic landscape of HCC is incredibly complex and heterogeneous. Promising new developments are helping us decipher the molecular drivers of HCC and leading to new therapies.
Keywords: liver cancer, HCC, carcinogenesis, mutation, genomics, therapeutics
INTRODUCTION
The global incidence of hepatocellular carcinoma (HCC), the most common primary liver malignancy and the 6th most common cancer worldwide, is expected to significantly increase over the next 10 years [1,2]. Unfortunately, HCC survival still remains dismal, with 5-year survival rates of 32.6%, 10.8%, and 2.4% for localized, regional, and distant stages of disease respectively [3]. Hepatitis B (HBV) and hepatitis C (HCV) are the most common risk factors for HCC, and the incidence of HCC has historically mirrored the incidence of these infectious diseases [4]. With the advent of the HBV vaccine and HCV antiviral therapy, there is hope that the burden of hepatitis-related HCCs will decrease. However, viral hepatitis is still expected to drive increased incidences of HCC over the next 10 years [2,5]. Furthermore, alcoholic liver disease (ALD), obesity, and non-alcoholic fatty liver disease (NAFLD) remain important risk factors for HCC, and these etiologies are actually increasing in incidence [6,7].
Even though risk factors for HCC vary from region to region, the mechanisms of hepatocellular carcinogenesis mostly converge on the processes of chronic liver inflammation and regeneration. Chronic liver injury secondary to either virus-induced inflammation, alcohol-induced hepatocellular damage, or lipotoxicity-induced oxidative stress lead to a vicious cycle of regeneration and fibrosis that increases the risk of genomic instability and hepatocarcinogenesis [8,9]. These pathogenic mechanisms underscore why 80–90% of HCC arise in a cirrhotic liver [10]. However, HCC can also arise in the non-cirrhotic liver, especially in patients with HBV or NAFLD [11,12]. Thus, the genomic landscape in which hepatocarcinogenesis occurs is incredibly complicated.
Currently, the only definitive therapeutic cure for HCC is liver transplantation (LT), and even LT is associated with recurrence rates of 10–15% [13]. ●For unresectable tumors, few therapies exist. The oral multi-kinase inhibitor sorafenib has been the therapeutic workhorse for unresectable HCCs ever since it was approved by the Food and Drug Administration (FDA) in 2007. However, its therapeutic efficacy has been greatly limited by rapid drug resistance and toxicities [14,15]. ●Despite the recent approval of new first line therapies like atezolizumab/bevacizumab or Lenvatinib and second line therapies like regorafenib, nivolumab, and cabozantinib, there still remains a pressing need for effective therapeutics that can significantly improve long term survival [16–18].
Genomic Landscape of HCC
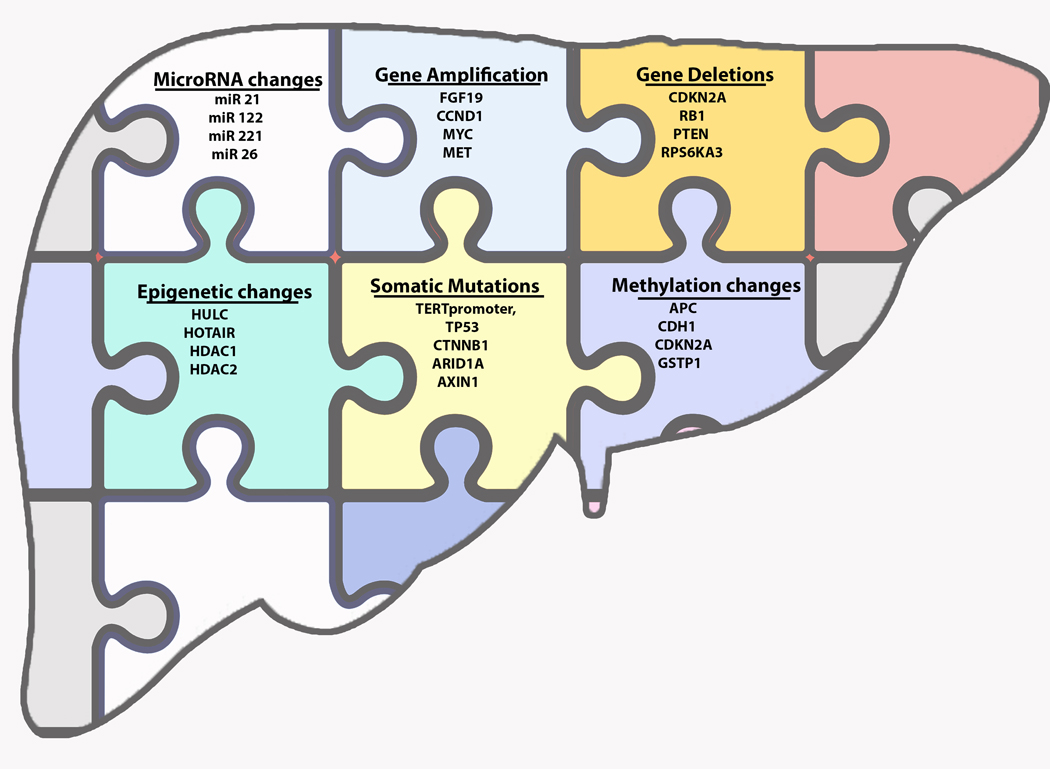
The dearth of therapeutic options for HCC continues to propel research into the mechanisms of hepatocarcinogenesis. Over the last five years, significant progress has been made in the identification of somatic mutations, copy number variations (CNVs), and epigenetic modifications that drive hepatocarcinogenesis and contribute to disease outcomes (Table 1). Improvements in genome wide screening and high-throughput genomics have led to the identification of new gene signatures and proteomic targets that can help to diagnose and prognosticate HCC. Recent developments in single-cell RNA-sequencing (scRNA-Seq) are opening a window into the pathophysiology of tumor heterogeneity. This review summarizes the genomic landscape of HCC (Figure 1), and identifies studies that have recently expanded our understanding of hepatocarcinogenesis in a meaningful way.
Table 1:
Overview of Major Genomic Landscape and Clinicopathologic Correlations in HCC
| Type of Genomic Change | Class | Main Gene Target | References | Clinicopathologic Associations | Therapeutic Relevance |
|---|---|---|---|---|---|
| Somatic Mutations | Tumor Suppressors | TP53 (p63, p73, p14ARF, mdm2, mdm4, ATM) |
Teramoto et al, 1994 Chan et al, 2004 Villanueva et al, 2011 Chittmittrapap et al 2013 Zhan et al, 2013 Zhan et al, 2014 Torrecilla et al, 2017 Long et al, 2019 Li et al, 2020 |
Clinical: HBV-HCC (32) Histopathology: increased stem cell markers (30), high Edmonson gade (29, 32), decreased in areas of cholestasis and tumor infiltrating lymphocytes (32), increased in areas of necrosis (32) Survival: high recurrence rates, poor OS (30–33) and DFS (31,32), downregulated immune responses (39) |
Resistance: doxirubicin (34–35)/paclitaxel (34) (in vitro) Therapeutics: p53 MVA vaccine + pembrolizumab (Phase I, 40), recombinant adenovirus p53 (Phase II, 41, 43, 44) |
| ARID1A ARID1B |
Schulze et al, 2015 Nahon et al, 2017 Hu et al, 2018 Qu et al, 2019 Moore et al, 2019 Li et al, 2020 Peng et al, 2020 |
Clinical: ALD-HCC (45), HBV-HCC (45–47), tobacco use (45) Histopathology: hepatic steatosis (48–49), larger more aggressive tumors (51–52), increased angiogenesis (53), poor OS (51, 52), mismatch repair (52) |
-- | ||
| Telomerases | TERT promoter |
Nault et al, 2013 Nault et al, 2016 Pezzuto et al, 2016 Torrecilla et al, 2017 Yu et al 2017 Pezutto et al 2017 Kawai-Kitahata et al, 2017 Li et al, 2018 Cheng et al, 2019 Li et al, 2020 |
Clinical: Older age, African or European ancestry, HCV-HCC, HBV-HCC (61–63) Histopathology: low/high grade dyspastic nodules (56–57) Survival: shorter DFS (60, 64–65), late intrahepatic recurrence (60) |
Therapeutics: INO-1400 or INO-1401 ± INO-9012 (Phase I, 66) | |
| Growth Factor Pathways |
CTNNB1 (Wnt/β-catenin) (APC, AXIN1, AXIN2, PRC1, AKIP1, TXNDC12, DICER1) |
Zulehner et al, 2010 Schulze et al, 2015 Torrecilla et al, 2017 Ataide et al, 2017 Li et al, 2020 |
Clinical: ALD-HCC (46) Histopathology: micro- and macrovascular invasion, increased pathologic grade, increased tumor size, and multifocal disease (73–74) Survival: HCC recurrence (73–74) |
Therapeutics: DKK1 antibody (Phase I/II, 79) | |
| Signal Transduction | RPS6KA3 (MAP/ERK) | TCGA, 2017 Calderaro et al, 2017 Doycheva et al, 2019 |
Histopathology: poor differentiation, macrovascular invasion, high proliferation, and chromosomal instability, multifocal disease (80–82) | -- | |
| STAT3 (JAK/STAT) | Kan et al, 2013 | Histopathology: promotes proliferation, angiogenesis, metstasis, Warburg effect, inhibition of apoptosis | Therapeutics: STAT inhibitors: OPB-111077 (Phase I, 97), OPB-31121 (Phase I, 98), Napabucasin (Phase I, 99), and AZD9150 (Phase 1b, 100) | ||
| Other | KEAP1 | Zheng et al, 2019 | -- | Resistance: acquired resistance to sorafenib, lenvatinib, and regorafenib (83) | |
| HNF1A |
Reznik et al, 2004 Rebouissou et al, 2007 Zeng et al, 2011 Hetchman et al, 2019 |
Clinical: negative viral status, female sex, no cirrhosis (84–86) Histopathology: reduced expression associated with proliferation, migration, and invasion in HCC cells (87) forced expresioon leads to differentiation into mature hepatocytes (87–88) |
-- | ||
| CNVs | Gene Amplification | MYC |
Chan et al, 2004 Poon et al, 2006 Schlaeger et al, 2008 Pedica et al, 2013 Ao et al, 2016 Yu et al, 2017 |
Clinical: viral hepatitis, ALD (105–106) Histopathology: large undifferentiated tumors (107–110) Survival: poor DFS, metatastasis, HCC recurrence (107–110) |
|
| E2F1/E2F3 | Kent et al, 2017 | Histopathology: advanced HCC (111) | -- | ||
| MMP9 | Yu et al, 2020 | Clinical: AFP level (112) Histopathology: tumor size, differentiation, invasion, stage (112) |
-- | ||
| UBE2Q1 | Zhang et al, 2018 | Survival: Poor OS and DFS (114) | -- | ||
| EXT1 | Dong et al, 2018 | Clinical: AFP level (115) Survival: Poor DFS (115) |
-- | ||
| WNK2 | Zhou et al, 2019 | Histopathology: increased tumor growth, macrophage infiltration (116) Survival: early tumor recurrence, metastasis (116) |
-- | ||
| FGF19 | Kaibori et al, 2016 | Histopathophysiology: agressive, undifferentiated tumors (in vivo) (Sawey et al, 2012), increased proliferation/invasion (in vitro) (Miura et al, 2012) Survival: poor overall and disease free survival (Miura et al, 2012) |
Resistance: complete response after sorafenib treatment | ||
| Gene Deletions | CDKN2A |
Kawai-Kitahata et al, 2016 Cancer Genome Atlas Research Network et al, 2017 |
Histopathology: Cell cycle arrest in G1/G2 Survival: Poor Prognosis, advanced aggressive cancer |
-- | |
| RB1 |
Kawai-Kitahata et al, 2016 Cancer Genome Atlas Research Network et al, 2017 Sun et al, 2019 |
Clinicals: HBV/HCV HCCs (113) | -- | ||
| Epigenetic Modifications | DNA Methylation | APC |
Um et al, 2011 Shen et al, 2013 |
-- | Treatment: 1st-Line: azacytadine and decitabine, 2nd Line: guadecitabine and zebularine (Phase I/II, 126–127) |
| GSTP1 | Shen et al, 2013 | Clinicals: HBV | |||
| RASSF1a | Shen et al, 2013 | Clinicals: Aflatoxin B1 | |||
| p16 | Um et al, 2011 | Clinicals: Aflatoxin B1 | |||
| COX2 | Um et al, 2011 | Histopathophysiology: Dysplastic nodules (123) | |||
| CDH1 | Wu et al, 2019 | Survival: Poor OS(124) | |||
| miRNAs | mIR21 | Xu et al, 2013 | Histopathology: HCC proliferation and migration (129–134) | -- | |
| mIR221/222 | Pineau et al, 2010 | Treatment: Miravirsin (Phase II, 137–138) | |||
| mIR224 | Lin et al, 2016 | -- | |||
| mIR-26 | Yang et al, 2014 | Histopathology: Suppression of HCC proliferation and angiogenesis (129–134) | -- | ||
| mIR-122 | Coulouarn et al, 2009 | -- | |||
| mIR-199 | Guo et al, 2015 | -- | |||
| mIR 200a | Chen et al, 2017 | Histopathology: inhibit cell growth, migration and invasion (135, 136) | -- | ||
| lncRNAs | HULC | Li et al, 2016 | Histopathology: proliferation, EMT, angiogenesis (140) | -- | |
| HOTAIR | Fujisaka et al, 2017 | Histopathology: no impact on cell proliferation, migration (141), actively recruited macrophages | Resistance: chemoresistance (140,142) | ||
| HDAC | HDAC3 |
Lu et al, 2018 Ji et al, 2018 |
Histopathology: HCC proliferation (143–144) Survival: HCC recurrence, reduced DFS (143) |
Treatment: Panabinostat, Belinostat, Resminostat, CUDC-101 (Phase I/II, 148–150) | |
| HDAC1/2 |
Ler et al, 2015 Yang et al, 2019 |
Survival: predict mortality (145, 146) | Resistance: regulation doxorubicin sensitivity (145) |
AFP-alpha-fetoprotein; ALD-alcoholic liver disease; HBV-hepatitis B virus; HCC-hepatocellular carcinoma; HCV-hepatitis C virus; HDAC-histone deacetylases; OS-overall survival; DFS-disease-free survival, lncRNAs-long non-coding RNAs; miRNAs-microRNAs
Figure 1: Major Genetic Alterations in Human Hepatocellular carcinoma (HCC).
Somatic Mutations
The normal aging liver is thought to acquire 30–40 somatic mutations per year, either induced by genotoxic stress or random mutations arising from DNA replication [19,20]. Hepatic stem cells and differentiated hepatocytes have both been shown to acquire these mutations, with a recent study revealing that mature hepatocytes in the normal liver have twice the rate of somatic mutations as hepatic stem cells [21].● Still, compared to other tissues heavily dependent on stem-cell regeneration, the overall mutational burden and malignant potential of the normal adult liver remains low [19,20]. In the setting of chronic liver disease and inflammation, however, hepatocytes are susceptible to additional proliferation-induced mutagenesis by way of mitochondrial damage and oxidative/endoplasmic reticulum stress [22]. Another mechanism of mutagenesis is via genomic viral integrations as seen in HBV-HCCs, which typically have the highest rates of somatic mutation [23]. HCV, on the other hand, typically promotes HCC carcinogenesis through double strand breaks that result in missense mutations. Lastly, somatic mutations in NAFLD, ALD, and toxin-driven HCCs are typically caused by direct DNA damage through chronic inflammation and reactive oxygen species.
Clinically significant driver mutations in HCC have been shown to involve these major pathways: tumor suppressor genes (TP53, ARID 1/2, RB1, TSC1/2), telomerases (TERT/TERC), the WNT-beta catenin pathway (CTNNB1, AXINI, AXIN2), PI3K/Akt/MTOR pathways, MYC pathway, JAK/STAT pathways (JAK1, IL6R, IL6ST), oxidative stress pathways (KEAP1, NFE2L2), RAS/RAF/MAP kinases (RPS6KA3), and the MET pathway. We will now discuss the mutations in these major pathways in further detail.
Tumor Suppressor Genes
As the “guardian of the genome”, the TP53 tumor suppressor gene is responsible for the regulation of cellular processes like cell death and angiogenesis. It is the most frequently altered gene in human cancer, with the International Agency for Research on Cancer (IARC) reporting over 29,000 TP53 mutations in human cancers [24]. TP53 is altered or inactivated in 30–50% of all HCCs- 80% of aflatoxin B1-HCCs, 45% of HBV-HCCs, and 13% of HCV-HCCs [24–26]. Notable gain of function (GOF) mutations in HCC include TP53 V157F and TP53 R249S, the latter being associated with aflatoxin and hepatitis B exposure [24,27,28].
Clinically, mutations in the TP53 family (including p63 and p73), activator p14ARF and inhibitors MDM2 and MDM4, are associated with higher expression of stem-cell like markers, high Edmonson grade, high rates of recurrence, lower disease-free survival, and therapy resistance [29–32]. TP53 may also downregulate the immune response [33],●● making these HCCs a potential target for immunotherapy [34]. ● Therapies targeting TP53 are aimed at supplementing wild-type p53 or blocking its interaction with cytoplasmic partners. Palbociclib, an oral cyclin-dependent kinase 4/6 inhibitor, has been shown to inhibit p53 DNA-damage partner Ataxia telangiectasia mutated (ATM) and to increase radiosensitivity of HCC cell lines, with the potential implication that gain-of-function p53 expression can be suppressed [35]. ● Furthermore, recombinant adenovirus p53, when combined with transarterial chemoembolization (TACE), has been shown to increase overall survival (OS) and disease-free survival (DFS) in patients with HCC [36].
The tumor suppressors ARID1A and ARID1B are components of SWI/SNF complexes that allow DNA repair machinery to access chromatin. Loss-of-function mutations in these genes have historically been associated with alcoholic liver disease and HBV infection [37,38], and recent studies suggest ARID1 may contribute to the development of hepatic steatosis [39,40]. ●ARID mutations typically occur in the later stages of HCC development, and result in larger, more aggressive tumors [41–43] with high tumor mutational burdens and increased angiogenesis, both of which potentially make them susceptible to immunotherapy and anti-angiogenic therapies [44,45]. ●●
Telomerases
Telomeres are repetitive nucleotide sequences that provide a docking location for the DNA polymerase complex during replication, and also protect chromosomes from deterioration or fusion. When telomeres shorten beyond a critical length after successive rounds of DNA replication, the telomerase complex, composed of a core catalytic telomerase reverse transcriptase (TERT) and RNA template telomerase RNA component (TERC) is activated to lengthen telomeres and restore the liver’s regenerative capacity. The TERT promoter mutation is one of the most common genetic alterations in HCC, with an overall frequency of 30–60% [46]. Its presence in low-grade and high dysplastic nodules reveal its role in the early stages of hepatocarcinogenesis [47,48], which is to drive malignant transformation by selecting for those HCC precursors that escape apoptosis with indefinite telomerase activity. Ironically, loss of function of telomerase gene variants also predispose hepatocytes to malignant transformation by impairing hepatocyte response to chronic injury and accelerating cirrhosis [49].
Clinically, TERT promoter mutations are associated with shorter DFS and late intrahepatic recurrence after surgical resection [50]. They are also more frequent in patients with older age, African or European ancestry, and HCV-HCC [51–53]. Other clinically meaningful TERT gene alterations include HBV viral integrations at the TERT gene promoter locus and TERT gene amplifications [50], both of which are associated with decreased OS [54,55]. ● The therapeutic promises of TERT mutations are yet to be realized. There are currently no approved therapies targeting TERT mutations, although a Phase I clinical trial with an immunotherapeutic agent against hTERT in solid tumors is currently in progress [56]. TERT mutations in circulating DNA may also be a novel way of screening for patients at high risk patients for HCC [57,58]. ●
Wnt/β-catenin pathway
The Wnt/β-catenin pathway, which in normal tissues is critical for embryonic body axis patterning, cell migration and cell fate specification, is commonly exploited in hepatocarcinogenesis. CTNNB1, which encodes β-catenin, a multifunctional protein that links the intracellular actin cytoskeleton to adherens junctions and also serves as a key nuclear effector of canonical Wnt signaling [59], is mutated in 20–40% of all HCCs [60,61]. Missense mutations in CTNNB1 result in higher nuclear and cytoplasmic β-catenin expression in HCCs compared to normal liver, para-carcinoma tissue, and cirrhotic liver [62]. Nuclear expression in HCCs has been associated with more aggressive histopathologic features, such as micro- and macrovascular invasion, increased pathologic grade, increased tumor size, multifocal disease, and tumor recurrence [52,63]. Furthermore, mutations in any of the proteins responsible for the activation or destruction of β-catenin can lead to aberrant nuclear accumulation. For example, loss-of-function mutations in the APC, AXIN1 and AXIN2 genes result in the sustained activation of the Wnt pathway by disruption of the multiprotein destruction complex that tags β-catenin for degradation. Other recently identified members of the Wnt/β-catenin pathway upregulated in HCC include protein regulator of cytokinesis 1 (PRC1), AKIP1, and thioredoxin protein TXNDC12 [64–66].● Germline mutations in the microRNA processing gene DICER1 have also been associated with CTNNB1 mutations in familial HCC, although the mechanistic relationship remains unclear [67]. So far, Wnt pathway proteins and genes have not proved to be druggable targets. An antibody that currently targets DKK1, a protein regulator of the Wnt pathway, is currently in phase I clinical trials for HCCs [68]. ●●
Other Notable Mutations
Several other notable pathways have been implicated in hepatocarcinogenesis. Mutations in RPS6KA3, a MAP/ERK pathway kinase that was recently shown to be mutated in 4–10% of HCCs, were associated with poor differentiation, macrovascular invasion, high proliferation, and chromosomal instability [69–71].●● Genome-wide screening revealed mutations in KEAP1, a master regulator and ubiquitinator of antioxidant gene NFR2, to be the top cause of acquired resistance to sorafenib, lenvatinib, and regorafenib in HCC cell lines [72]. ●HNF1A is a liver-enriched transcription factor (TF) that regulates cellular homeostasis and metabolism. Inactivated or mutated HNF1A has been found in HCCs in patients with negative viral status, female sex, and no cirrhosis [73–75]. A specific point mutation (c.A1532 > T/p.Q511L) causes reduced expression, proliferation, migration, and invasion in HCC cells, while forced expression induces differentiation of these cells into mature hepatocytes [76]. Additionally, the combinatorial transduction of TFs NF4A, HNF1A and FOXA3 was shown to suppress cellular proliferation of HCC cells [77]. ●Lastly, the transmembrane receptor Janus kinases (JAKs) and the signal transducers STATs are commonly deregulated in HCC [78]. STAT3 mutations promote a number of cancer hallmarks, such as proliferation, angiogenesis and metastasis [79–83]. ●●Several small molecule STAT inhibitors, including Stattic, OPB-111077, OPB-31121, Napabucasin, and AZD9150 are currently in preclinical or Phase I clinical trials for HCC [84–88].●
Thus, recent NGS based studies have allowed a comprehensive understanding of the somatic mutation landscape of HCC. Although none of the major mutations are directly druggable at present, there are several promising candidates in the pipeline.
Copy Number Alterations
Somatic copy number alterations (SCNAs) result from the gain or loss of individual genes, or more commonly, entire chromosomal arms. The molecular consequence of SCNAs is the potential activation of oncogenes and loss of tumor suppressors, both of which drive carcinogenesis. Multiple studies have shown that copy number gains in chromosomes 1q and 8q, and losses in 8p and 17p, are the most frequent chromosomal arm level alterations in HCC [69,89,90]. Apart from the arm-level changes, gene-level changes are also important to identify. The well-known driver oncogenes CCND1, FGF19, MYC, MET, VEGFA, MCL1, and TERT, were recently shown to be significantly amplified in HCCs [69]. ●●Amplification of MYC, a transcription factor known to regulate all of the programs that are hallmarks of cancer, is thought to be an early genomic event in liver carcinogenesis. It has been found in both chronic liver disease, and in 70% of viral and alcohol-related HCCs [91]. MYC amplification at 8q24.1 has been repeatedly associated with large undifferentiated liver tumors, poor prognosis, metastasis and HCC recurrence [92–94]. Other well-known CNV amplifications include RB-regulated transcription factors E2F1 and E2F3. Amplification of these genes resulted in spontaneous HCC in murine models, and queries of the Cancer Genome Atlas (TCGA) datasets revealed a significant increase in the E2F family gene dosage in tumors of patients with advanced HCC [95]. ●Furthermore, copy number increases in matrix metalloproteinase-9 (MMP9), which promotes tumor metastasis via the breakdown of the extracellular matrix, were also shown to be associated with key clinicopathological features of HCC such as alpha-fetoprotein (AFP) level, tumor size, differentiation, invasion, and stage [96]. ●The selective presence of MMP9 CNVs in tumor tissue over normal tissue makes it a potentially promising diagnostic biomarker for HCC. On the other hand, analysis of HCC tumors has also revealed significant gene deletions in CDKN2A and tumor suppressors like ERRFI1, NCOR1, and RB1 [97], the latter of which is a common mechanism for the development of HBV and HCV HCCs [98]. ●
Other recently identified clinically significant CNVs include UBE2Q1, EXT1, WNK2, and JAGGED1. UBE2Q1 is an E2 ubiquitin-conjugating enzyme thought to promote carcinogenesis via the β-catenin/EGFR-PI3K-Akt-mTOR signaling pathway. Copy number gains in this gene are associated with poorer OS and DS [99]. ●EXT1, which encodes an endoplasmic reticulum glycosyltransferase, has previously been shown to prognosticate breast cancer, cholangiocarcinoma, and acute lymphoblastic leukemia (ALL). EXT1 mRNA was recently shown to be correlated with serum AFP, and its upregulation was found to be associated with worse DFS [100]. ●Analysis of 736 primary HCC samples revealed copy number loss of WNK2, a potential tumor suppressor, to be associated with early tumor recurrence, macrophage infiltration, tumor growth, and metastasis, likely via ERK1/2 signaling activation [101]. ●Amplifications of JAGGED1, which encodes a NOTCH pathway ligand, were also recently shown to be associated with poor OS and early HCC recurrence. Lastly, copy number mutations can also have positive therapeutic consequences. Increased FGF19 copy numbers were associated with a complete response after sorafenib treatment [102].
Epigenetic modifications
Epigenetic changes, which can occur via DNA methylation, histone modification, chromatin remodeling and non-coding RNAs, alter the way that genetic code is expressed, rather than directly affecting the nucleotide sequence. Dysregulated DNA methylation has been shown to be an important early event in the pathogenesis of HCC. Studies have noted greater global hypomethylation in HCC tumor tissue, particularly CpG dinucleotides within CpG islands, compared to adjacent tissue, with anywhere from 500 to 684 CpG sites being significantly hypermethylated in matched HCC and normal adjacent tissue comparisons [103,104]. A 2012 study suggested that these hypermethylated genes may be good early biomarkers for HCC, and five randomly selected genes (CDKL2, STEAP4, HIST1H3G, CDKN2A and ZNF154) from the top 18 hypermethylated genes in their study were detectable in the plasma of 63% of patients [103]. A more recent study identified 6 hypermethylated genes (NEBL, three FAM55C sites, GALNT3, and DSE) from 375 HCC samples that, when used as biomarkers for HCC, achieved a 98% specificity for HCC [105]. ●Other individual genes historically found to be hypermethylated in HCC include APC (81.7%), GTP1 (33.3%), RASSF1a (66.7%), p16 (48.3%), COX2 (35.0%), and Cadherin-1 (CDH1)(33.3%)[103,106]. A meta-analysis of 12 relevant HCC studies covering 981 patients showed that CDH1 hypermethylation was significantly higher in HCC tissues compared to normal liver, and was correlated with worse OS [107]. ●
Epigenetic changes in HCC can potentially be targeted using small molecule inhibitors of DNA methytransferases (DNMTs), which have historically been used in the treatment of myelodysplastic syndrome. Several first generation DNMTs like azacytadine and decitabine have been shown to reduce tumor formation by inducing hepatic cell differentiation and increasing cell sensitivity to sorafenib in preclinical studies, with decitabine Phase I/II clinical trials revealing acceptable safety and toxicity [108,109]. ●●Second generation DNMTs like guadecitabine and zebularine were created to improve upon the short half-lives of first generation DNMTs, and are also being tested in Phase I/II clinical trials [110].
Non-coding RNAs, which include micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs), constitute a well-studied class of epigenetic regulators in HCC. Upregulated expression of mIR 21, mIR 221/222, and mIR 224 have been associated with increased HCC proliferation and migration, while decreased expression of mIR-26, mIR 122, and mIR-199 have been shown to suppress HCC proliferation and angiogenesis [111].●● The downregulated expression of mIR 200a was also recently shown to inhibit cell growth, migration and invasion [112].● These miRNAs may prove to be promising therapeutic targets. Anti-mIR 221 is currently in pre-clinical trials, and treatment with Miravirsin, a mIR 122 inhibitor that has completed Phase IIa trials, resulted in a substantial and prolonged decrease in plasma mIR 122 in patients with HCV [113,114]. Notable upregulated lncRNAs include HULC and HOTAIR [115,116]. HULC is thought to promote HCC proliferation and carcinogenesis indirectly by activating the CREB transcription factor, and is also associated with the epithelial-to-mesenchymal transition and angiogenesis [115]. HOTAIR, on the other hand, is thought to maintain the tumor microenvironment via CCL2 expression [116] ● and its loss was shown to sensitize HCC cells lines to chemotherapy [115,117].
Histones are protein octamers that help to condense DNA. Modifications to the histone tails that protrude from the DNA/histone nucleosome structure play an important role in the regulation of gene transcription and expression. The placement and removal of acetyl groups from these histone tails by histone acetyltransferases (HATs) and histone deacetylases (HDACs) are often dysregulated in HCC. Even though increased expression of histone deacetylase 3 (HDAC3) has repeatedly been shown to promote HCC proliferation and predict HCC recurrence, deficiency of HDAC3 was recently shown to promote HCC carcinogenesis in a murine model via a defect in the H3K9ac/H3K9me3 transition [118,119]. ● Upregulated HDACs 1 and 2 have also been shown to predict mortality in patients with HCC, and they may regulate doxorubicin sensitivity in HCC cell lines [120]. Although they have been used in the treatment of hematological malignancies, HDAC inhibitors have unknown efficacy in HCC. Panabinostat is currently in the preclinical phase of investigation, whereas Belinostat, Resminostat, and CUDC-101 are currently in Phase I/II clinical trials [121–124].
Gene Signatures
Transcriptomic studies in HCC have helped identify gene signatures, or clusters of differentially expressed genes, that can diagnose and prognosticate HCC, and also predict therapeutic response. Historically, gene signatures in HCC have focused on hepatocyte proliferation gene clusters and the EPCAM-positive hepatic cancer stem cell (hCSC) gene clusters. The proliferation cluster, which is expressed across a broad spectrum of human malignancies, includes the A- and B- type cyclins that control the cell cycle at G1/S and/or G2/M transition (CCNA2, CCNB2), cell division cycle proteins (CDC2, CDC7, CDC14, CDC20), heterohexamer DNA helicase minichromosome maintenance protein complex (MCM3–7), proliferating cell nuclear antigen (PCNA), and DNA topoisomerase 2 α (TOP2A), among others [125,126]. HCCs that express this gene signature, which largely mirrors a c-MYC- regulated gene signature, are associated with poorer OS, and were more likely to also have decreased expression of liver-specific genes that promoted hepatocyte dedifferentiation.
The hCSC gene cluster, on the other hand, was more likely to have increased expression of cell adhesion molecule EpCAM, epithelial marker CK19, and AFP, and has clinically been associated with chemotherapeutic resistance [127,128]. A recent study determined that EpCAM-regulated intramembrane proteolysis helps to drive the hCSC signature, and can potentially be targeted for inhibition in HBV-HCCs [129]. The molecular consequence of an hCSC gene signature is de-differentiation of tissues and loss of epithelial morphology, both of which promote malignant tumor behaviors and predict a worse prognosis for patients with HCC [130]. ●●Moreover, hCSC signatures may also be correlated with chemokine networks thought to create a hospitable inflammatory niche for tumor progression and metastasis [131]. A recent study identified an eight gene signature (TK1, CTTN, CEP72, TRIP13, FTH1, FLAD1, CHRM2, AMBP) in HBV-HCC tumors similar to that found in a previous study that is controlled by transcription factor OCT4, which is abundantly expressed in pluripotent stem cells [132].
Several clinically meaningful gene signatures have recently been identified. Through the application of two different algorithms to screen for differentially expressed genes in paired and unpaired HCC, Zhang et al identified a 14 gene signature in the cell-cycle-related gene cluster (BIRC5, BUB1B, CDC45, DTL, GINS2, KIF23, KIF2C, MAD2L1, MCM4, OIP5, PLK4, PTTG1, and ZWINT) that predicts poor OS and HCC recurrence [133]. ●●Another study revealed evidence of network reprogramming in carbon metabolism and cancer pathway genes through the identification of a 22-carbon metabolism gene signature that predicts poorer OS and DFS [134]. Li et al identified a DNA repair-related prognostic signature of seven genes (ADA, FEN1, POLR2G, SAC3D1, UPF3B, SF3A3, and SEC61A1) that, when used to stratify patients into high-risk and low-risk groups, predicts survival in HCC [135].●● None of the gene signatures have yet been validated or approved for clinical use, but significant progress is being made towards this goal.
Proteomics
Genetic, epigenetic, and post-translational dysregulation in HCC ultimately results in changes in protein expression levels and protein-protein interactions. Early proteomic studies in HCC, while useful in identifying potential protein targets related to early HCC diagnosis, were limited by smaller sample sizes, lack of validation, and absence of additional functional characterization [136]. These limitations have largely been addressed by advances in high-throughput protein analysis techniques and have resulted in detailed proteomic maps of HCC. Recently, a proteomic and phospho-proteomic comparison between 110 paired HBV-HCC and non-tumor tissues revealed enrichment of cell cycle, integrin, PDGF signaling, MAPK, TNF, and MET pathways, as well as hyperphosphorylation of the p38, RHO, myosin, RB1, and IL1 pathways [137].●● Metabolic reprogramming was found to be a key feature of HBV-HCC l on paired tumor and adjacent non-tumor liver tissues of 316 patients [138]. ●●Forty-two proteins known to play a role in amino acid metabolism and oxidoreductase activity were dysregulated in HCC. Importantly, the study found that with the exception of a few key metabolic enzymes (SOAT1, SOAT2, GLS, GLUD2), most proteins in liver-specific pathways, including gluconeogenesis, detoxification, and ureagenesis-ammonia, were significantly decreased in tumors.
Many other proteomic studies are focused on identifying protein targets for either diagnostic or therapeutic purposes. Using an absolute quantitation-based multidimensional liquid chromatography-tandem mass spectrometry technique, Liu et al identified 27 differentially abundant proteins, mostly in the ERK1/2 and nuclear factor-κ beta (NF-KB) pathways, in the serum of patients post-radical resection that were associated with HCC early recurrence [139]. ●PGK1, a glycolysis enzyme that has been detected in the serum of patients with a broad spectrum of malignancies [140], was specifically identified as an independent predictor of HCC recurrence and OS. Zhao et al used a high throughout urinary proteome analysis platform to compare the urine of 74 HCC and 82 high-risk patients with HBV-HCC to identify seven features that distinguish HCC from the high risk control population in a non-invasive fashion [141]. ●●Thus, large scale proteomics are adding to our understanding of the functional pathways activated in HCCs and are identifying promising diagnostic and predictive biomarkers for HCC.
Conclusions
HCC is a heterogeneous disease with such a complex genomic landscape. Understanding the molecular drivers of HCC carcinogenesis is essential both to identify biomarkers and to develop molecular targeted therapies. Promising new developments in this field are enabling us to develop therapies that can target the various drivers of HCC and evolve personalized therapeutic strategies. Future genomic studies promise to advance our understanding of this malignancy in meaningful ways and will hopefully ultimately lead to improvement in clinical outcomes for patients with HCC.
Acronyms
- 2D LC-MS/MS
multidimensional liquid chromatography-tandem mass spectrometry
- AFP
alpha-fetoprotein
- ALD
alcoholic liver disease
- ALL
acute lymphoblastic leukemia
- ATM
Ataxia telangiectasia mutated
- CDH1
Cadherin-1
- CNV
copy number variations
- DFS
disease-free survival
- DNMTs
DNA methytransferases
- FDA
Food and Drug Administration
- GOF
gain of function
- HATs
histone acetyltransferases
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- hCSC
hepatic cancer stem cell
- HCV
hepatitis C virus
- HDACs
histone deacetylases
- HDAC3
histone deacetylase 3
- JAKs
Janus kinases
- lncRNAs
long non-coding RNAs
- LT
Liver Transplantation
- MAT1A
methionine adenosyltransferase 1
- MMP9
matrix metalloproteinase-9
- micro RNAs
micro RNAs
- NAFLD
nonalcoholic fatty liver disease
- NGS
next generation sequencing
- NF-KB
nuclear factor-κ beta
- OS
overall survival
- PLEC1
plectin 1
- PRC1
protein regulator of cytokinesis 1
- SCNAs
somatic copy number alterations
- scRNA-Seq
single-cell RNA-sequencing
- TACE
transarterial chemoembolization
- TCGA
the Cancer Genome Atlas
- TERC
telomerase RNA component
- TERT
telomerase reverse transcriptase
- TF
transcription factor
- TOP2A
topoisomerase 2 α
- JAKs
transmembrane receptor Janus kinases
Footnotes
Compliance with Ethics Guidelines
Nia Adeniji and Renumathy Dhanasekaran declare that they have no conflict of interest
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
No Conflict of interest
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Valery PC, Laversanne M, Clark PJ, Petrick JL, McGlynn KA, Bray F. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology. 2018;67: 600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seegobin K, Majeed U, Ritter A, Wylie N, Starr JS, Jones JC, et al. Factors affecting time to treatment in hepatocellular cancer. Journal of Clinical Oncology. 2020. pp. e19088–e19088. doi: 10.1200/jco.2020.38.15_suppl.e19088 [DOI] [Google Scholar]
- 4.Huang Y-T, Jen C-L, Yang H-I, Lee M-H, Su J, Lu S-N, et al. Lifetime Risk and Sex Difference of Hepatocellular Carcinoma Among Patients With Chronic Hepatitis B and C. Journal of Clinical Oncology. 2011. pp. 3643–3650. doi: 10.1200/jco.2011.36.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumert TF, Jühling F, Ono A, Hoshida Y. Hepatitis C-related hepatocellular carcinoma in the era of new generation antivirals. BMC Med. 2017;15: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marengo A, Rosso C, Bugianesi E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu Rev Med. 2016;67: 103–117. [DOI] [PubMed] [Google Scholar]
- 7.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl: S2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park EJ, Lee JH, Yu G-Y, He G, Ali SR, Holzer RG, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zámbó V, Simon-Szabó L, Szelényi P, Kereszturi É, Bánhegyi G, Csala M. Lipotoxicity in the liver. World Journal of Hepatology. 2013. p. 550. doi: 10.4254/wjh.v5.i10.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe J, Ghouri Y, Mian I. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. Journal of Carcinogenesis. 2017. p. 1. doi: 10.4103/jcar.jcar_9_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Do AL, Wong CR, Nguyen LH, Nguyen VG, Trinh H, Nguyen MH. Hepatocellular carcinoma incidence in noncirrhotic patients with chronic hepatitis B and patients with cirrhosis of all etiologies. J Clin Gastroenterol. 2014;48: 644–649. [DOI] [PubMed] [Google Scholar]
- 12.Tobari M, Hashimoto E, Taniai M, Kodama K, Kogiso T, Tokushige K, et al. The characteristics and risk factors of hepatocellular carcinoma in nonalcoholic fatty liver disease without cirrhosis. J Gastroenterol Hepatol. 2020;35: 862–869. [DOI] [PubMed] [Google Scholar]
- 13.Menahem B, Lubrano J, Duvoux C, Mulliri A, Alves A, Costentin C, et al. Liver transplantation versus liver resection for hepatocellular carcinoma in intention to treat: An attempt to perform an ideal meta-analysis. Liver Transpl. 2017;23: 836–844. [DOI] [PubMed] [Google Scholar]
- 14.Tang W, Chen Z, Zhang W, Cheng Y, Zhang B, Wu F, et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther. 2020;5: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marisi G, Cucchetti A, Ulivi P, Canale M, Cabibbo G, Solaini L, et al. Ten years of sorafenib in hepatocellular carcinoma: Are there any predictive and/or prognostic markers? World Journal of Gastroenterology. 2018. pp. 4152–4163. doi: 10.3748/wjg.v24.i36.4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamoorthy SK, Relias V, Sebastian S, Jayaraman V, Saif MW. Management of regorafenib-related toxicities: a review. Therap Adv Gastroenterol. 2015;8: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Liu K, Chen M, Sun J-Y, McCaughan GW, Lu X-J, et al. Immunotherapy for hepatocellular carcinoma: recent advances and future perspectives. Ther Adv Med Oncol. 2019;11: 1758835919862692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Personeni N, Pressiani T, Rimassa L. Lenvatinib for the treatment of unresectable hepatocellular carcinoma: evidence to date. J Hepatocell Carcinoma. 2019;6: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538: 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunner SF, Roberts ND, Wylie LA, Moore L, Aitken SJ, Davies SE, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazhnik K, Sun S, Alani O, Kinkhabwala M, Wolkoff AW, Maslov AY, et al. Single-cell analysis reveals different age-related somatic mutation profiles between stem and differentiated cells in human liver. Sci Adv. 2020;6: eaax2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L-X, Ling Y, Wang H-Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2018;2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B-L, Ji X, Yu L-X, Gao Y, Xiao C-H, Liu J, et al. Somatic mutation profiling of liver and biliary cancer by targeted next generation sequencing. Oncol Lett. 2018;16: 6003–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37: 865–876. [DOI] [PubMed] [Google Scholar]
- 25.Teramoto Tadahisa, Satonaka Kazuhiro, Kitazawa Sohei, Fujimori Takahiro, Hayashi Kozaburo and Maeda Sakan. p53 Gene Abnormalities Are Closely Related to Hepatoviral Infections and Occur at a Late Stage of Hepatocarcinogenesis. Cancer Res. 1994;54. Available: https://cancerres.aacrjournals.org/content/54/1/231.short [PubMed] [Google Scholar]
- 26.Zhang W, He H, Zang M, Wu Q, Zhao H, Lu L-L, et al. Genetic Features of Aflatoxin-Associated Hepatocellular Carcinoma. Gastroenterology. 2017;153: 249–262.e2. [DOI] [PubMed] [Google Scholar]
- 27.Chittmittrapap S, Chieochansin T, Chaiteerakij R, Treeprasertsuk S, Klaikaew N, Tangkijvanich P, et al. Prevalence of aflatoxin induced p53 mutation at codon 249 (R249s) in hepatocellular carcinoma patients with and without hepatitis B surface antigen (HBsAg). Asian Pac J Cancer Prev. 2013;14: 7675–7679. [DOI] [PubMed] [Google Scholar]
- 28.Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol. 2011;55: 724–725. [DOI] [PubMed] [Google Scholar]
- 29.Kancherla V, Abdullazade S, Matter MS, Lanzafame M, Quagliata L, Roma G, et al. Genomic Analysis Revealed New Oncogenic Signatures in TP53-Mutant Hepatocellular Carcinoma. Frontiers in Genetics. 2018. doi: 10.3389/fgene.2018.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z, Guo D, Wang Q, Liu P, Xiao Y, Wu P, et al. Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics. 2019;9: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z-Z, Bao L-L, Zhao K, Xu Q, Zhu J-Y, Zhu K-X, et al. Copy Number Aberrations of Multiple Genes Identified as Prognostic Markers for Extrahepatic Metastasis-free Survival of Patients with Hepatocellular Carcinoma. Curr Med Sci. 2019;39: 759–765. [DOI] [PubMed] [Google Scholar]
- 32.Zhan P, Ji Y-N. Prognostic significance of TP53 expression for patients with hepatocellular carcinoma: a meta-analysis. Hepatobiliary Surg Nutr. 2014;3: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42: 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MultiVir, Inc. Safety and Efficacy of p53 Gene Therapy Combined With Immune Checkpoint Inhibitors in Solid Tumors. 2018. Available: https://clinicaltrials.gov/ct2/show/NCT03544723?term=p53&cond=Hepatocellular+Carcinoma&draw=2&rank=7
- 35.Huang C-Y, Hsieh F-S, Wang C-Y, Chen L-J, Chang S-S, Tsai M-H, et al. Palbociclib enhances radiosensitivity of hepatocellular carcinoma and cholangiocarcinoma via inhibiting ataxia telangiectasia–mutated kinase–mediated DNA damage response. European Journal of Cancer. 2018. pp. 10–22. doi: 10.1016/j.ejca.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Yan J, Wang F. Effects of TACE combined with precise RT on p53 gene expression and prognosis of HCC patients. Oncol Lett. 2018;16: 5733–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahon P, Nault J-C. Constitutional and functional genetics of human alcohol-related hepatocellular carcinoma. Liver Int. 2017;37: 1591–1601. [DOI] [PubMed] [Google Scholar]
- 38.Sung W-K, Zheng H, Li S, Chen R, Liu X, Li Y, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44: 765–769. [DOI] [PubMed] [Google Scholar]
- 39.Qu Y-L, Deng C-H, Luo Q, Shang X-Y, Wu J-X, Shi Y, et al. Arid1a regulates insulin sensitivity and lipid metabolism. EBioMedicine. 2019;42: 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore A, Wu L, Chuang J-C, Sun X, Luo X, Gopal P, et al. Arid1a Loss Drives Nonalcoholic Steatohepatitis in Mice Through Epigenetic Dysregulation of Hepatic Lipogenesis and Fatty Acid Oxidation. Hepatology. 2019;69: 1931–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasileiou G, Ekici AB, Uebe S, Zweier C, Hoyer J, Engels H, et al. Chromatin-Remodeling-Factor ARID1B Represses Wnt/β-Catenin Signaling. Am J Hum Genet. 2015;97: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, Li L, Rao X, Wen Z, Ding X, Wang X, et al. Implications of driver genes associated with a high tumor mutation burden identified using next-generation sequencing on immunotherapy in hepatocellular carcinoma. Oncology Letters. 2020. doi: 10.3892/ol.2020.11372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng Y, Gao B, Zhou Z, Chen T, Xie W, Huang M, et al. Hepatocellular carcinoma with ARID1A mutation is associated with higher TMB and poor survival. Journal of Clinical Oncology. 2020. pp. e16667–e16667. doi: 10.1200/jco.2020.38.15_suppl.e16667 [DOI] [Google Scholar]
- 44.Hu C, Li W, Tian F, Jiang K, Liu X, Cen J, et al. Arid1a regulates response to anti-angiogenic therapy in advanced hepatocellular carcinoma. J Hepatol. 2018;68: 465–475. [DOI] [PubMed] [Google Scholar]
- 45.Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51: 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nault J-C, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40: 9–14. [DOI] [PubMed] [Google Scholar]
- 47.Torrecilla S, Sia D, Harrington AN, Zhang Z, Cabellos L, Cornella H, et al. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol. 2017;67: 1222–1231. [DOI] [PubMed] [Google Scholar]
- 48.Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4: 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donaires FS, Scatena NF, Alves-Paiva RM, Podlevsky JD, Logeswaran D, Santana BA, et al. Telomere biology and telomerase mutations in cirrhotic patients with hepatocellular carcinoma. PLOS ONE. 2017. p. e0183287. doi: 10.1371/journal.pone.0183287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu JI, Choi C, Ha SY, Park C-K, Kang SY, Joh J-W, et al. Clinical importance of TERT overexpression in hepatocellular carcinoma treated with curative surgical resection in HBV endemic area. Sci Rep. 2017;7: 12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pezzuto F, Buonaguro L, Buonaguro FM, Tornesello ML. Frequency and geographic distribution of TERT promoter mutations in primary hepatocellular carcinoma. Infect Agent Cancer. 2017;12: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zulehner G, Mikula M, Schneller D, van Zijl F, Huber H, Sieghart W, et al. Nuclear beta-catenin induces an early liver progenitor phenotype in hepatocellular carcinoma and promotes tumor recurrence. Am J Pathol. 2010;176: 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pezzuto F, Izzo F, Buonaguro L, Annunziata C, Tatangelo F, Botti G, et al. Tumor specific mutations in TERT promoter and CTNNB1 gene in hepatitis B and hepatitis C related hepatocellular carcinoma. Oncotarget. 2016;7: 54253–54262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Y, Huang M, Xie W, Gao C, Cai S, Ji J, et al. Chromosome 8q24 amplification predicts prognosis for patients with advanced hepatocellular carcinoma (HCC). Journal of Clinical Oncology. 2019. pp. e15654–e15654. doi: 10.1200/jco.2019.37.15_suppl.e15654 [DOI] [Google Scholar]
- 55.Li X, Xu W, Kang W, Wong SH, Wang M, Zhou Y, et al. Genomic analysis of liver cancer unveils novel driver genes and distinct prognostic features. Theranostics. 2018;8: 1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pharmaceuticals I. hTERT Immunotherapy Alone or in Combination With IL-12 DNA Followed by Electroporation in Adults With Solid Tumors at High Risk of Relapse (TRT-001). 2016. Available: https://clinicaltrials.gov/ct2/show/NCT02960594
- 57.Trung NT, Hoan NX, Trung PQ, Binh MT, Van Tong H, Toan NL, et al. Clinical significance of combined circulating TERT promoter mutations and miR-122 expression for screening HBV-related hepatocellular carcinoma. Sci Rep. 2020;10: 8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiao J, Watt GP, Stevenson HL, Calderone TL, Fisher-Hoch SP, Ye Y, et al. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: Prevalence and risk factors. Hepatology Communications. 2018. pp. 718–731. doi: 10.1002/hep4.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31: 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waisberg J. Wnt−/−β-catenin pathway signaling in human hepatocellular carcinoma. World Journal of Hepatology. 2015. p. 2631. doi: 10.4254/wjh.v7.i26.2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khalaf AM, Fuentes D, Morshid AI, Burke MR, Kaseb AO, Hassan M, et al. Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. Journal of Hepatocellular Carcinoma. 2018. pp. 61–73. doi: 10.2147/jhc.s156701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li P, Cao Y, Li Y, Zhou L, Liu X, Geng M. Expression of Wnt-5a and β-catenin in primary hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7: 3190–3195. [PMC free article] [PubMed] [Google Scholar]
- 63.Ataide EC, Perales SR, Silva MG, Filho FC, Sparapani AC, Latuf Filho PF, et al. Immunoexpression of Heat Shock Protein 70, Glypican 3, Glutamine Synthetase, and Beta-Catenin in Hepatocellular Carcinoma After Liver Transplantation: Association Between Positive Glypican 3 and Beta-Catenin With the Presence of Larger Nodules. Transplant Proc. 2017;49: 858–862. [DOI] [PubMed] [Google Scholar]
- 64.Yuan K, Xie K, Lan T, Xu L, Chen X, Li X, et al. TXNDC12 promotes EMT and metastasis of hepatocellular carcinoma cells via activation of β-catenin. Cell Death Differ. 2020;27: 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cui Y, Wu X, Lin C, Zhang X, Ye L, Ren L, et al. AKIP1 promotes early recurrence of hepatocellular carcinoma through activating the Wnt/β-catenin/CBP signaling pathway. Oncogene. 2019;38: 5516–5529. [DOI] [PubMed] [Google Scholar]
- 66.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65: 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caruso S, Calderaro J, Letouzé E, Nault J-C, Couchy G, Boulai A, et al. Germline and somatic DICER1 mutations in familial and sporadic liver tumors. J Hepatol. 2017;66: 734–742. [DOI] [PubMed] [Google Scholar]
- 68.Johannes Gutenberg University Mainz, Leap Therapeutics, Inc. DKN-01 Inhibition in Advanced Liver Cancer. 2018. Available: https://clinicaltrials.gov/ct2/show/NCT03645980?term=Wnt&cond=Hepatocellular+Carcinoma&draw=2&rank=1
- 69.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu, Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169: 1327–1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc J-F, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67: 727–738. [DOI] [PubMed] [Google Scholar]
- 71.Doycheva I, Thuluvath PJ. Systemic Therapy for Advanced Hepatocellular Carcinoma: An Update of a Rapidly Evolving Field. J Clin Exp Hepatol. 2019;9: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng A, Chevalier N, Calderoni M, Dubuis G, Dormond O, Ziros PG, et al. CRISPR/Cas9 genome-wide screening identifies KEAP1 as a sorafenib, lenvatinib, and regorafenib sensitivity gene in hepatocellular carcinoma. Oncotarget. 2019;10: 7058–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hechtman JF, Abou-Alfa GK, Stadler ZK, Mandelker DL, Roehrl MHA, Zehir A, et al. Somatic HNF1A mutations in the malignant transformation of hepatocellular adenomas: a retrospective analysis of data from MSK-IMPACT and TCGA. Hum Pathol. 2019;83: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rebouissou S, Imbeaud S, Balabaud C, Boulanger V, Bertrand-Michel J, Tercé F, et al. HNF1alpha inactivation promotes lipogenesis in human hepatocellular adenoma independently of SREBP-1 and carbohydrate-response element-binding protein (ChREBP) activation. J Biol Chem. 2007;282: 14437–14446. [DOI] [PubMed] [Google Scholar]
- 75.Reznik Y, Dao T, Coutant R, Chiche L, Jeannot E, Clauin S, et al. Hepatocyte nuclear factor-1 alpha gene inactivation: cosegregation between liver adenomatosis and diabetes phenotypes in two maturity-onset diabetes of the young (MODY)3 families. J Clin Endocrinol Metab. 2004;89: 1476–1480. [DOI] [PubMed] [Google Scholar]
- 76.Zeng X, Lin Y, Yin C, Zhang X, Ning B-F, Zhang Q, et al. Recombinant adenovirus carrying the hepatocyte nuclear factor-1alpha gene inhibits hepatocellular carcinoma xenograft growth in mice. Hepatology. 2011;54: 2036–2047. [DOI] [PubMed] [Google Scholar]
- 77.Takashima Y, Horisawa K, Udono M, Ohkawa Y, Suzuki A. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci. 2018;109: 3543–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23: 1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin Z, Ma T, Lin Y, Lu X, Zhang C, Chen S, et al. IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J Cell Biochem. 2018;119: 9419–9432. [DOI] [PubMed] [Google Scholar]
- 80.Xiong S, Wang R, Chen Q, Luo J, Wang J, Zhao Z, et al. Cancer-associated fibroblasts promote stem cell-like properties of hepatocellular carcinoma cells through IL-6/STAT3/Notch signaling. Am J Cancer Res. 2018;8: 302–316. [PMC free article] [PubMed] [Google Scholar]
- 81.Bi Y-H, Han W-Q, Li R-F, Wang Y-J, Du Z-S, Wang X-J, et al. Signal transducer and activator of transcription 3 promotes the Warburg effect possibly by inducing pyruvate kinase M2 phosphorylation in liver precancerous lesions. World J Gastroenterol. 2019;25: 1936–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang L, Jian Z, Gao Y, Zhou P, Zhang G, Jiang B, et al. RPN2 promotes metastasis of hepatocellular carcinoma cell and inhibits autophagy via STAT3 and NF-κB pathways. Aging. 2019;11: 6674–6690. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Yang L, Zhang X-Y, Li K, Li A-P, Yang W-D, Yang R, et al. Protopanaxadiol inhibits epithelial-mesenchymal transition of hepatocellular carcinoma by targeting STAT3 pathway. Cell Death Dis. 2019;10: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu G, Zhu L, Wang Y, Shi Y, Gong A, Wu C. Stattic Enhances Radiosensitivity and Reduces Radio-Induced Migration and Invasion in HCC Cell Lines through an Apoptosis Pathway. Biomed Res Int. 2017;2017: 1832494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoo C, Kang J, Kim K-P, Lim HY, Kim JH, Lee MA, et al. Phase I dose-finding study of OPB-111077, a novel STAT3 inhibitor, in patients with advanced hepatocellular carcinoma. Journal of Clinical Oncology. 2018. pp. 4078–4078. doi: 10.1200/jco.2018.36.15_suppl.4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okusaka T, Ueno H, Ikeda M, Mitsunaga S, Ozaka M, Ishii H, et al. Phase 1 and pharmacological trial of OPB-31121, a signal transducer and activator of transcription-3 inhibitor, in patients with advanced hepatocellular carcinoma. Hepatol Res. 2015;45: 1283–1291. [DOI] [PubMed] [Google Scholar]
- 87.Shitara K, Yodo Y, Iino S. A Phase I Study of Napabucasin Plus Paclitaxel for Japanese Patients With Advanced/Recurrent Gastric Cancer. In Vivo. 2019;33: 933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reilley MJ, McCoon P, Cook C, Lyne P, Kurzrock R, Kim Y, et al. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial. Journal for ImmunoTherapy of Cancer. 2018. doi: 10.1186/s40425-018-0436-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ki Kim S, Ueda Y, Hatano E, Kakiuchi N, Takeda H, Goto T, et al. TERT promoter mutations and chromosome 8p loss are characteristic of nonalcoholic fatty liver disease-related hepatocellular carcinoma. Int J Cancer. 2016;139: 2512–2518. [DOI] [PubMed] [Google Scholar]
- 90.Zhou C, Zhang W, Chen W, Yin Y, Atyah M, Liu S, et al. Integrated Analysis of Copy Number Variations and Gene Expression Profiling in Hepatocellular carcinoma. Sci Rep. 2017;7: 10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47: 511–520. [DOI] [PubMed] [Google Scholar]
- 92.Ao L, Song X, Li X, Tong M, Guo Y, Li J, et al. An individualized prognostic signature and multi-omics distinction for early stage hepatocellular carcinoma patients with surgical resection. Oncotarget. 2016. pp. 24097–24110. doi: 10.18632/oncotarget.8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu M-C, Lee C-W, Lee Y-S, Lian J-H, Tsai C-L, Liu Y-P, et al. Prediction of early-stage hepatocellular carcinoma using OncoScan chromosomal copy number aberration data. World J Gastroenterol. 2017;23: 7818–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pedica F, Ruzzenente A, Bagante F, Capelli P, Cataldo I, Pedron S, et al. A re-emerging marker for prognosis in hepatocellular carcinoma: the add-value of fishing c-myc gene for early relapse. PLoS One. 2013;8: e68203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kent LN, Bae S, Tsai S-Y, Tang X, Srivastava A, Koivisto C, et al. Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J Clin Invest. 2017;127: 830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu X, Huang J, Wu S, Huang Y, Shan Y, Lu C. Copy number variations of MMP-9 are prognostic biomarkers for hepatocellular carcinoma. Translational Cancer Research. 2020. pp. 698–706. doi: 10.21037/tcr.2019.11.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kawai-Kitahata F, Asahina Y, Tanaka S, Kakinuma S, Murakawa M, Nitta S, et al. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J Gastroenterol. 2016;51: 473–486. [DOI] [PubMed] [Google Scholar]
- 98.Sun S, Li Y, Han S, Jia H, Li X, Li X. A comprehensive genome-wide profiling comparison between HBV and HCV infected hepatocellular carcinoma. BMC Med Genomics. 2019;12: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang B, Deng C, Wang L, Zhou F, Zhang S, Kang W, et al. Upregulation of UBE2Q1 via gene copy number gain in hepatocellular carcinoma promotes cancer progression through β-catenin-EGFR-PI3K-Akt-mTOR signaling pathway. Mol Carcinog. 2018;57: 201–215. [DOI] [PubMed] [Google Scholar]
- 100.Dong S, Wu Y, Yu S, Yang Y, Lu L, Fan S. Increased EXT1 gene copy number correlates with increased mRNA level predicts short disease-free survival in hepatocellular carcinoma without vascular invasion. Medicine. 2018;97: e12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou S-L, Zhou Z-J, Hu Z-Q, Song C-L, Luo Y-J, Luo C-B, et al. Genomic sequencing identifies WNK2 as a driver in hepatocellular carcinoma and a risk factor for early recurrence. Journal of Hepatology. 2019. pp. 1152–1163. doi: 10.1016/j.jhep.2019.07.014 [DOI] [PubMed] [Google Scholar]
- 102.Kaibori M, Sakai K, Ishizaki M, Matsushima H, De Velasco MA, Matsui K, et al. Increased FGF19 copy number is frequently detected in hepatocellular carcinoma with a complete response after sorafenib treatment. Oncotarget. 2016;7: 49091–49098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen J, Wang S, Zhang Y-J, Kappil M, Wu H-C, Kibriya MG, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55: 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gentilini D, Scala S, Gaudenzi G, Garagnani P, Capri M, Cescon M, et al. Epigenome-wide association study in hepatocellular carcinoma: Identification of stochastic epigenetic mutations through an innovative statistical approach. Oncotarget. 2017;8: 41890–41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng J, Wei D, Ji Y, Chen L, Yang L, Li G, et al. Integrative analysis of DNA methylation and gene expression reveals hepatocellular carcinoma-specific diagnostic biomarkers. Genome Med. 2018;10: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee S, Lee HJ, Kim J-H, Lee H-S, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163: 1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu X, Yao X, Cao Q, Wu Z, Wang Z, Liu F, et al. Clinicopathological and prognostic significance of CDH1 hypermethylation in hepatocellular carcinoma: a meta-analysis. Cancer Management and Research. 2019. pp. 857–864. doi: 10.2147/cmar.s179710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gailhouste L, Liew LC, Yasukawa K, Hatada I, Tanaka Y, Nakagama H, et al. Differentiation Therapy by Epigenetic Reconditioning Exerts Antitumor Effects on Liver Cancer Cells. Mol Ther. 2018;26: 1840–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu J, Liu Y, Meng L, Liu K, Ji B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol Rep. 2017;38: 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jueliger S, Lyons J, Cannito S, Pata I, Pata P, Shkolnaya M, et al. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics. 2016;11: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, et al. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9: 3557–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen S-Y, Ma D-N, Chen Q-D, Zhang J-J, Tian Y-R, Wang Z-C, et al. MicroRNA-200a inhibits cell growth and metastasis by targeting Foxa2 in hepatocellular carcinoma. J Cancer. 2017;8: 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van der Ree MH, van der Meer AJ, van Nuenen AC, de Bruijne J, Ottosen S, Janssen HL, et al. Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment Pharmacol Ther. 2016;43: 102–113. [DOI] [PubMed] [Google Scholar]
- 114.Moshiri F, Callegari E, D’Abundo L, Corrà F, Lupini L, Sabbioni S, et al. Inhibiting the oncogenic mir-221 by microRNA sponge: toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol Hepatol Bed Bench. 2014;7: 43–54. [PMC free article] [PubMed] [Google Scholar]
- 115.Li S-P, Xu H-X, Yu Y, He J-D, Wang Z, Xu Y-J, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7: 42431–42446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fujisaka Y, Iwata T, Tamai K, Nakamura M, Mochizuki M, Shibuya R, et al. Long non-coding RNA HOTAIR up-regulates chemokine (C-C motif) ligand 2 and promotes proliferation of macrophages and myeloid-derived suppressor cells in hepatocellular carcinoma cell lines. Oncology Letters. 2017. doi: 10.3892/ol.2017.7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang Z, Zhou L, Wu L-M, Lai M-C, Xie H-Y, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18: 1243–1250. [DOI] [PubMed] [Google Scholar]
- 118.Lu X-F, Cao X-Y, Zhu Y-J, Wu Z-R, Zhuang X, Shao M-Y, et al. Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transducer and activator of transcription 3 signaling pathway. Cell Death Dis. 2018;9: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ji H, Zhou Y, Zhuang X, Zhu Y, Wu Z, Lu Y, et al. HDAC3 Deficiency Promotes Liver Cancer through a Defect in H3K9ac/H3K9me3 Transition. Cancer Res. 2019;79: 3676–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Y, Zhang J, Wu T, Xu X, Cao G, Li H, et al. Histone deacetylase 2 regulates the doxorubicin (Dox) resistance of hepatocarcinoma cells and transcription of ABCB1. Life Sci. 2019;216: 200–206. [DOI] [PubMed] [Google Scholar]
- 121.Gahr S, Mayr C, Kiesslich T, Illig R, Neureiter D, Alinger B, et al. The pan-deacetylase inhibitor panobinostat affects angiogenesis in hepatocellular carcinoma models via modulation of CTGF expression. Int J Oncol. 2015;47: 963–970. [DOI] [PubMed] [Google Scholar]
- 122.Bitzer M, Horger M, Giannini EG, Ganten TM, Wörns MA, Siveke JT, et al. Resminostat plus sorafenib as second-line therapy of advanced hepatocellular carcinoma - The SHELTER study. J Hepatol. 2016;65: 280–288. [DOI] [PubMed] [Google Scholar]
- 123.Voi M, Fu S, Nemunaitis J, Bauman J, Bessudo A, Hamid O, et al. 590 Final Results of a Phase Ib Study of CUDC-101, a Multitargeted Inhibitor of EGFR, HER2, and HDAC, in Patients with Advanced Head and Neck, Gastric, Breast, Liver, and Non-small Cell Lung Cancer. European Journal of Cancer. 2012. p. 181. doi: 10.1016/s0959-8049(12)72387-0 [DOI] [Google Scholar]
- 124.Yeo W, Chung HC, Chan SL, Wang LZ, Lim R, Picus J, et al. Epigenetic Therapy Using Belinostat for Patients With Unresectable Hepatocellular Carcinoma: A Multicenter Phase I/II Study With Biomarker and Pharmacokinetic Analysis of Tumors From Patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. Journal of Clinical Oncology. 2012. pp. 3361–3367. doi: 10.1200/jco.2011.41.2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee J-S, Chu I-S, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40: 667–676. [DOI] [PubMed] [Google Scholar]
- 126.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36: 1090–1098. [DOI] [PubMed] [Google Scholar]
- 127.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang H-Y, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136: 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Y, Farmer RW, Yang Y, Martin RCG. Epithelial cell adhesion molecule in human hepatocellular carcinoma cell lines: a target of chemoresistence. BMC Cancer. 2016;16: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mani SKK, Zhang H, Diab A, Pascuzzi PE, Lefrançois L, Fares N, et al. EpCAM-regulated intramembrane proteolysis induces a cancer stem cell-like gene signature in hepatitis B virus-infected hepatocytes. J Hepatol. 2016;65: 888–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chao J, Zhao S, Sun H. Dedifferentiation of hepatocellular carcinoma: molecular mechanisms and therapeutic implications. Am J Transl Res. 2020;12: 2099–2109. [PMC free article] [PubMed] [Google Scholar]
- 131.Muramatsu S, Tanaka S, Mogushi K, Adikrisna R, Aihara A, Ban D, et al. Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology. 2013;58: 218–228. [DOI] [PubMed] [Google Scholar]
- 132.Ye C, Zhang X, Chen X, Cao Q, Zhang X, Zhou Y, et al. Multiple novel hepatocellular carcinoma signature genes are commonly controlled by the master pluripotency factor OCT4. Cell Oncol. 2020;43: 279–295. [DOI] [PubMed] [Google Scholar]
- 133.Zhang B-H, Yang J, Jiang L, Lyu T, Kong L-X, Tan Y-F, et al. Development and validation of a 14-gene signature for prognosis prediction in hepatocellular carcinoma. Genomics. 2020;112: 2763–2771. [DOI] [PubMed] [Google Scholar]
- 134.Zhang J, Baddoo M, Han C, Strong MJ, Cvitanovic J, Moroz K, et al. Gene network analysis reveals a novel 22-gene signature of carbon metabolism in hepatocellular carcinoma. Oncotarget. 2016;7: 49232–49245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li N, Zhao L, Guo C, Liu C, Liu Y. Identification of a novel DNA repair-related prognostic signature predicting survival of patients with hepatocellular carcinoma. Cancer Manag Res. 2019;11: 7473–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chan L-K, Ng IO-L. Proteomic profiling in liver cancer: another new page. Translational gastroenterology and hepatology. 2019. p. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567: 257–261. [DOI] [PubMed] [Google Scholar]
- 138.Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179: 561–577.e22. [DOI] [PubMed] [Google Scholar]
- 139.Liu H, Chen H, Wu X, Sun Y, Wang Y, Zeng Y, et al. The serum proteomics tracking of hepatocellular carcinoma early recurrence following radical resection. Cancer Manag Res. 2019;11: 2935–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He Y, Luo Y, Zhang D, Wang X, Zhang P, Li H, et al. PGK1-mediated cancer progression and drug resistance. Am J Cancer Res. 2019;9: 2280–2302. [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Y, Li Y, Liu W, Xing S, Wang D, Chen J, et al. Identification of noninvasive diagnostic biomarkers for hepatocellular carcinoma by urinary proteomics. J Proteomics. 2020;225: 103780. [DOI] [PubMed] [Google Scholar]
- 142.Chan K-T, Lung ML. Mutant p53 expression enhances drug resistance in a hepatocellular carcinoma cell line. Cancer Chemotherapy and Pharmacology. 2004. pp. 519–526. doi: 10.1007/s00280-004-0767-4 [DOI] [PubMed] [Google Scholar]
- 143.Zhan P, Ji Y-N, Yu L-K. TP53 mutation is associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a meta-analysis. Hepatobiliary Surg Nutr. 2013;2: 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Poon TCW, Wong N, Lai PBS, Rattray M, Johnson PJ, Sung JJY. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131: 1262–1270. [DOI] [PubMed] [Google Scholar]
- 146.Um T-H, Kim H, Oh B-K, Kim MS, Kim KS, Jung G, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011;54: 939–947. [DOI] [PubMed] [Google Scholar]
- 147.Shen J, Wang S, Zhang Y-J, Wu H-C, Kibriya MG, Jasmine F, et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8: 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107: 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin L, Lu B, Yu J, Liu W, Zhou A. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2016;40: 397–404. [DOI] [PubMed] [Google Scholar]
- 150.Yang X, Zhang X-F, Lu X, Jia H-L, Liang L, Dong Q-Z, et al. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factorcMet pathway. Hepatology. 2014;59: 1874–1885. [DOI] [PubMed] [Google Scholar]
- 151.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28: 3526–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62: 1132–1144. [DOI] [PubMed] [Google Scholar]
- 153.Ler SY, Leung CHOW, Khin LW, Lu G-D, Salto-Tellez M, Hartman M, et al. HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk regression model in a Southeast Asian population. Oncology Reports. 2015. pp. 2238–2250. doi: 10.3892/or.2015.4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miura S, Mitsuhashi N, Shimizu H, Kimura F, Yoshidome H, Otsuka M, et al. Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer. 2012. doi: 10.1186/1471-2407-12-56 [DOI] [PMC free article] [PubMed] [Google Scholar]