Summary
Ikaros zinc finger transcription factors are important regulators of the gene programs underlying the development of hematopoietic cell lineages. The family consists of five members: Ikaros, Helios, Aiolos, Eos, and Pegasus, which engage in both homo- and heterotypic intrafamilial interactions to exert diverse functional effects. Pioneering studies focused on the role of these factors in early lymphoid development, as their absence resulted in severe defects in lymphocyte populations. More recent work has now begun to define nuanced, stage-specific roles for Ikaros family members in the differentiation and function of mature T, B, and innate lymphoid cell populations including natural killer (NK) cells. The precise transcriptional mechanisms by which these factors function, both independently and collaboratively, is an area of active investigation. However, several key themes appear to be emerging regarding the pathways influenced by Ikaros family members, including the end-to-end regulation of cytokine signaling. Here, we review roles for Ikaros factors in lymphoid cell development, differentiation, and function, including a discussion of the current understanding of the transcriptional mechanisms they employ, and considerations for the future study of this important transcription factor family.
Keywords: Ikaros transcription factors, T Cells, B Cells, Innate Lymphoid Cells, Transcriptional Regulation, Cytokine Signaling
History and Structure of Ikaros Zinc Finger Transcription Factors
The Ikaros zinc finger family of transcription factors has been studied since the early 1990’s, when the namesake and founding member of the family, Ikaros, was identified in a screen for transcriptional regulators of the Cd3d locus (1). Subsequently, 4 additional family members were characterized, which exhibit a high degree of amino acid conservation both across family members and between human and murine proteins. These include Aiolos, Helios, Eos, and Pegasus (2–6). To date, roles for all family members with the exception of Pegasus have been described in regulating hematopoietic cell populations (Fig. 1). Functions attributed to Ikaros family members were initially limited to roles in the early stages of lymphoid development, as the absence of functional Ikaros or Aiolos was found to result in severe disruption of T, B, and NK cell lineages (1, 7, 8). More recently, however, studies including conditional knockout of Ikaros factors in mature T cell populations have expanded understanding to include stage-specific transcriptional regulation mediated by this family in mature lymphoid populations. In this review, we focus on current understanding of the roles of these factors in immune cell development, differentiation, and function, and highlight both historical and emerging mechanisms by which these factors exert their functions across cell types.
Figure 1. Ikaros family member-dependent transitions and expression patterns in developmental pathways for B cell, T cell, and ILC lineages.

Ikaros zinc finger transcription factors exhibit distinct expression patterns and regulatory roles throughout lymphopoiesis. Expression of each factor is indicated by a colored circle in the nucleus of relevant cell populations. Speculated/likely expression patterns are denoted by faded circles and dashed outer lines. Where known, developmental transitions that require individual factors are indicated by inclusion of those factors at the indicated transition point.
The five members of the Ikaros family: Ikaros, Helios, Aiolos, Eos, and Pegasus (encoded by the genes IKZF1-IKZF5, respectively) have been studied in both human and murine settings. Full-length Ikaros family members exhibit a high degree of structural similarity, particularly in regions containing C2H2 Krüppel-like zinc finger (ZF) motifs, which facilitate family member DNA binding and protein-protein interactions (Fig. 2) (9–11). All family members contain 4 N-terminal ZFs (ZF1–4), with the exception of Pegasus, which has only 3. Structure-function analyses for Ikaros have determined that ZFs 1 and 4 specify its binding to target sites throughout the genome, while ZF2 and ZF3 stabilize protein-DNA interactions (12–15). Such interactions are dependent on the core DNA binding motif a/gGGAA, which is found in target gene regulatory regions in the form of both single sequences and tandem repeats, suggesting that multimeric binding of Ikaros family members may be dictated by the latter at specific loci (14). While this structure-function relationship has been characterized most thoroughly for Ikaros, the high degree of amino acid conservation across family members suggests that similar ZF motif functions and DNA binding patterns may be conserved across all Ikaros proteins.
Figure 2. Schematic depicting the structure of Ikaros zinc finger family members and both experimental murine Ikaros mutants.

(A) The basic structure of members of the Ikaros zinc finger family of transcription factors consists of 3–4 N-terminal C2H2 zinc finger (ZF) binding motifs, which mediate protein-DNA interactions, and 2 C-terminal ZFs required for Ikaros factor homo- and hetero-dimerization. The ‘x’ denotes the fourth N-terminal ZF that is present in all family members excepting Pegasus. (B) The founding family member, Ikaros, is encoded by 7 of its 8 exons (the first contains the 5’ UTR), with N-terminal ZFs located in exons 4–6, and the C-terminal protein-protein interaction ZFs in its final exon. Several mutant versions of the Ikaros protein have been generated to explore the role of Ikaros in immune cell populations. These include germline or conditional mutations, as noted. (C) A DNA binding mutant lacking most of exon 4 and all of exon 5. Given interactions between Ikaros proteins, this “dominant negative” (DN−/−) disrupts the activity of full-length Ikaros and its expression results in a number of defects in lymphoid populations as described. (D,E) Elimination of the majority of the C-terminal portion of Ikaros (“C−/−”) results in a destabilized protein and functionally “null” phenotype, which has been examined in the context of both germline mutation (D) and conditional mutation (E) under the control of the distal lck promoter, which restricts the mutation to single-positive (mature) thymocyte populations. The C-terminal mutation also results in lymphoid defects as noted.
Ikaros family members also contain a pair of C-terminal ZFs, which are highly conserved between all family members and are critical for their function. These domains are required for protein-protein interactions, including those with non-Ikaros factors, as well as homo- and hetero-dimerization between Ikaros family members. Functionally, such interactions result in the ability of Ikaros factors to regulate both the cellular localization and functions of other family members, depending upon cellular context and expression across immune cell subsets (16). Thus, this interdependence between factors is an important consideration in studies utilizing genetic knockouts or mutant forms of Ikaros family members to study their individual functions (16).
In addition to full-length proteins, many Ikaros factor genes also encode a number of splice variants which exhibit distinct cellular localization and functional characteristics (2, 14, 17–20). Such characteristics vary across isoforms based on the ZF domains they contain, and thus splice variants have been broadly divided into DNA-binding and non-DNA-binding “dominant negative” forms (13, 21). Due to the intrafamilial interactions between Ikaros family members, association of dominant negative isoforms with wildtype Ikaros proteins sequesters the latter from their DNA binding sites and ultimately alters their function. Although many Ikaros family member splice variants have been described in transformed cell lines, at least a subset of functionally distinct isoforms are expressed in healthy immune cell populations (22). Ultimately, however, the full repertoire of splice variants for each Ikaros family member, as well as their specific roles in transcriptional regulation, are areas ripe for further investigation.
Historical mechanisms of gene regulation by Ikaros family members
Despite distinct expression patterns and functions across immune cell populations, Ikaros family members exhibit a number of shared transcriptional mechanisms to exert their control over target gene loci. These include non-mutually exclusive mechanisms that may occur together or independently depending on cellular context and the presence of additional interacting factors, including other Ikaros family members (Fig. 3). Ikaros was originally identified via its interaction with the Cd3d locus, and subsequent studies determined that direct DNA binding is also a key feature of other Ikaros family members (Fig. 3A). Further early mechanistic work defined roles for Ikaros factors in altering the chromatin landscape of developing immune cells. It was demonstrated that Ikaros factors recruit co-factors including chromatin modifying enzymes/complexes, which result in subsequent alterations in both chromatin marks and structure at target gene loci (Fig. 3B–D) (11, 21, 23).
Figure 3. Established transcriptional mechanisms underlying Ikaros family member regulation of target gene expression.
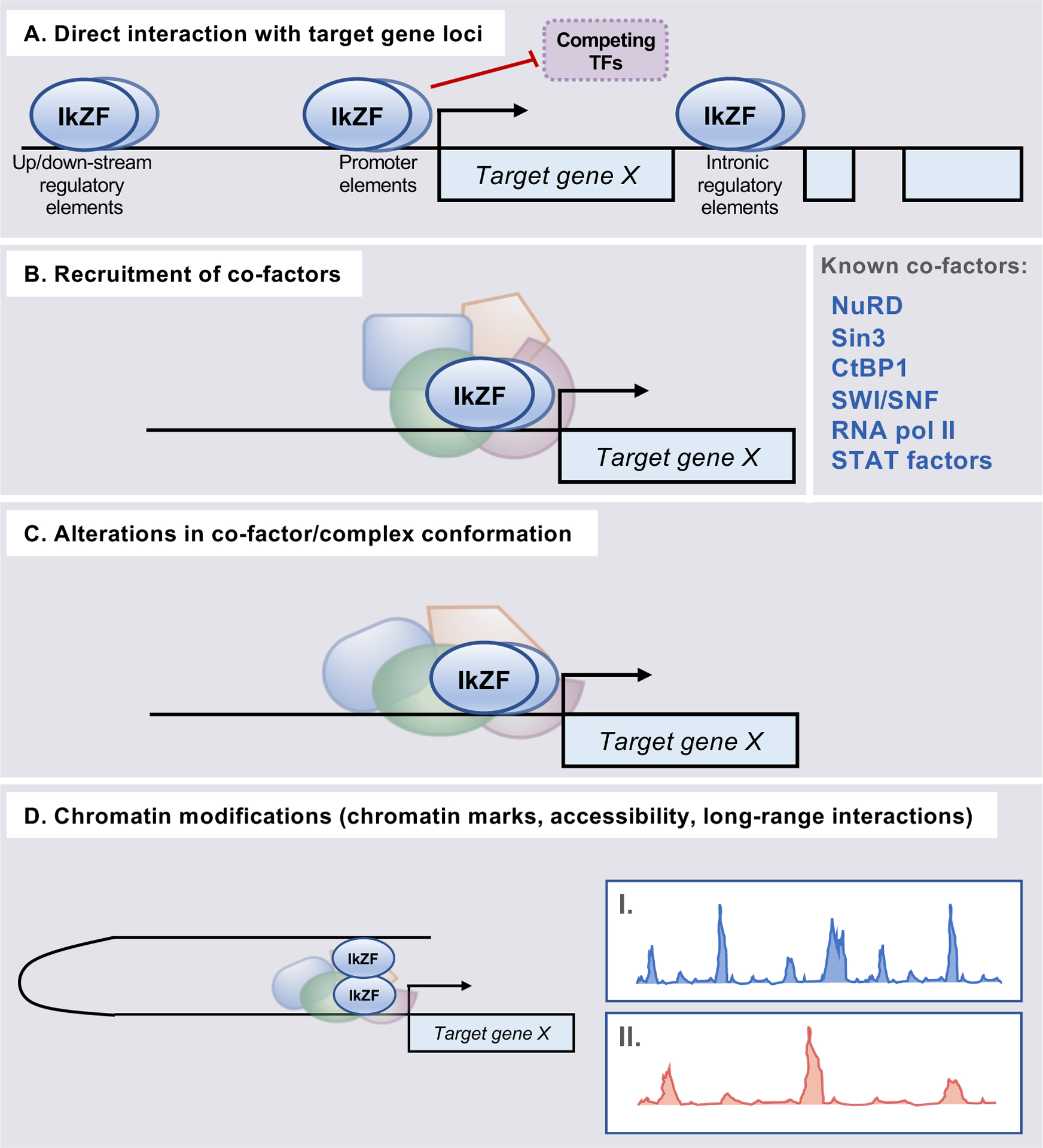
Ikaros family members have long been known to exert transcriptional control through a number of non-mutually exclusive DNA- and chromatin-dependent mechanisms. (A) All family members interact directly with DNA and associate with target gene loci. This allows for locus-specific control via both recruitment of co-factors to target gene regulatory regions, as noted, and by preventing association of competing transcriptional regulators. (B) Ikaros, Aiolos, Helios, and Eos have been found to interact with chromatin remodeling complexes/enzymes and/or additional cofactors to influence the accessibility and expression of target genes. (C) Association of the above co-factors with Ikaros family members has been suggested to result in alterations to co-factor conformation, which alters their function/binding. (D) The interactions described in A-C promote both local and long-range alterations to chromatin structure, including the deposition of chromatin marks (illustrated as example ChIP-seq peaks for chromatin mark enrichment in “I” and “II”). These include changes in regulatory region acetylation and methylation patterns. Ikaros factors have also been implicated in mediating long-range interactions (i.e. chromatin looping) between distal target gene regulatory elements.
Direct interaction of Ikaros family members with DNA depends upon the presence of N-terminal ZFs, as mutations in these regions have been found to abrogate both factor DNA binding and subsequent regulation of target gene expression (14). Curiously, mutations in the C-terminal protein interaction domain of Ikaros has also been found to disrupt its high-affinity DNA binding and ability to regulate transcription, suggesting that protein-DNA interactions may also be guided by homo- and hetero-dimerization of Ikaros family members (24, 25). Such protein-DNA interactions have been found within promoter, enhancer, and super-enhancer elements, and function not only as a platform for the recruitment of co-factors to regulate target genes, but also via competitive inhibition of other transcriptional regulators (Fig. 3A) (26). To date, however, the full scope of how Ikaros factor homo- and hetero-dimerization, or multimerization, may affect such competition, and detailed understanding regarding multi-Ikaros factor binding at individual gene loci, is incomplete.
Enrichment of Ikaros family members at specific target genes has also been associated with the recruitment of a number of chromatin modifying enzymes and remodeling complexes (Fig. 3B). Perhaps the most well-characterized interactions are those between Ikaros family members and the ATPase Mi2b within the histone deacetylate (HDAC)-containing nucleosome-remodeling and deacetylase (NuRD) complex (16, 21, 27). NuRD/Ikaros factor enrichment patterns at target gene loci vary dramatically throughout lymphocyte development, implicating this mechanism in the regulation of a broad range of genes in lymphoid populations (16). Mechanistically, Ikaros and Aiolos associate with and recruit the NuRD complex to target gene loci, leading to hypoacetylation of histones at target gene promoters and reduced gene expression. Curiously, Aiolos and Ikaros appear to function both collaboratively and independently to recruit the NuRD complex, as loss of Ikaros alone was found to reduce, but not eliminate, observed NuRD enrichment (16). It has also been suggested that association of Ikaros and Aiolos with the NuRD complex results in conformational changes that impact its function, which highlights the possibility that Ikaros factors may exert similar control via their interaction with other transcriptional complexes (Fig. 3C) (16, 28). Finally, histone deacetylation appears to be a conserved mechanism regulated by Ikaros family members, as Ikaros has been found to interact with members of the Sin3 family of HDAC-associating transcription factors (13). Ultimately, interactions between Ikaros and Aiolos with NuRD or Sin3 result in regulatory region hypoacetylation and gene repression.
Aside from HDAC-containing complexes, Ikaros has also been found to repress gene expression through recruitment of the polycomb repressive complex PRC2 in both double-negative thymocytes and during B cell development (23, 29). PRC2 mediates tri-methylation of H3 lysine 27. Thus, loss of Ikaros results in reduced PRC2 association and the repressive chromatin mark H3K27me3 at target gene regulatory regions (23). Another HDAC-independent role for Ikaros factors in remodeling chromatin structure is through their interaction with C-terminal binding protein 1 (CtBP1), which functions as a co-repressor in conjunction with Ikaros and Eos in different lymphocyte populations (13, 30). Both Ikaros and Eos associate with CtBP1 to affect promoter methylation and the repression of target gene expression. Curiously, despite homology between Ikaros, Eos, and Aiolos, Aiolos has not been found to associate with CtBP1, suggesting that differential functions of these factors may be due to either their distinct expression patterns or preferential interactions of individual family members with chromatin modifying enzymes (13).
Ikaros family member interactions with chromatin remodeling enzymes are not limited to repressive complexes, as Ikaros and Aiolos have also been found to associate with the activating ATP-dependent chromatin remodeling Switch/Sucrose Non-Fermentable (SWI/SNF) complex component BRG1 (13, 21, 31–33). Finally, in addition to local epigenetic changes at target gene loci, Ikaros has been implicated in mediating long-range interactions/chromosome looping between distal regulatory elements (34–36). Additionally, both Ikaros and Helios have been found to associate with centromeric heterochromatin, suggesting that they may be involved in mediating the repression of target genes by directing them to heterochromatin-containing foci (2, 37). It has been suggested that the Ikaros family member Aiolos may also contribute to this silencing mechanism (38). While the extent to which Ikaros factors may control chromatin restructuring is currently limited, similarities between Ikaros family members, as well as heterodimerization between individual Ikaros proteins, suggests that this mechanism may impact large-scale chromatin looping/remodeling in a broad range of cell types (35, 36). Collectively, these studies implicate Ikaros family members in both positive and negative regulation of gene targets through their interaction with chromatin remodeling complexes and both local and long-range alterations to chromatin structure.
Regulation of lymphocyte populations by Ikaros family members
Early lymphoid cell development
Ikaros family members have long been known to regulate key processes throughout early immune cell development (Fig. 1). Ikaros is expressed throughout hematopoeisis, and has been found to localize to pericentromeric heterochromatin in hematopoietic cell populations to regulate key genes involved in their development (12, 39). Of all of its family members, Ikaros has been studied most extensively in lymphocyte populations (1, 7, 22, 40). To elucidate its role in lymphopoeisis, early work involved the generation of a number of transgenic mouse models in which Ikaros had been functionally altered (Fig. 2). These included mutations to specific ZF domains in both the N- and C-terminus of the Ikaros protein, which serve to disrupt its function or stability. Pioneering studies utilizing such mutants established that they caused dramatic defects in lymphoid development and proliferation in vivo (7, 25).
First, deletion of N-terminal DNA binding ZFs resulted in the formation of a dominant negative form of Ikaros, which is incapable of interacting with DNA, but can still form homo- and hetero-dimers between Ikaros family members (7). Mice homozygous for the dominant negative mutation exhibited dramatic defects in lymphocyte development, including a complete lack of T, B, and natural killer cells (7). Subsequent work utilizing mice heterozygous for this mutation found that these animals displayed normal patterns of thymic development up to one month of age (likely due to the presence of wildtype Ikaros). However, they began to lose discrete double- and single-positive thymocyte populations shortly thereafter. This stage was followed by general lymphoproliferation and ultimately resulted in rapid development of leukemia and lymphoma, identifying wildtype Ikaros as a potent tumor suppressor in lymphoid populations (41).
In addition to N-terminal mutations, early work also assessed the functional consequences of altering the Ikaros C-terminal protein-protein interaction domain. Deletion of the C-terminal region of Ikaros resulted in a “null” version of the protein, which was unstable at the protein level and ultimately non-functional (Fig. 2D) (26). This mutation resulted in an absence of T and B cell precursors throughout murine fetal development, a continued absence of B cells postnatally, and a reduced and aberrant number of T lymphocytes (26). Ultimately, these early studies established that both DNA binding and protein-protein interactions are required for Ikaros to drive normal development of lymphocyte populations.
While the above studies were key in establishing Ikaros as a driver of lymphopoeisis, its precise role in individual developmental stages was unclear. Subsequent work utilizing the above mutants established that disruption of either the N- or C-terminal ZFs of Ikaros results in decreased long-term repopulation activity of the hematopoietic stem cell (HSC) population (Fig. 1A) (42). While differentiation of the lympho-myeloid primed progenitor (LMPP) population did not require Ikaros, it is responsible for activating a lymphoid gene signature while repressing stem cell gene programs at the this stage (43). Thus, subsequent generation of common lymphoid progenitors (CLP) does not occur in the absence of Ikaros (44, 45). Following lymphoid lineage specification, Ikaros continues to play a critical role in the development of both T and B cell populations (Figs. 1B, C) (4, 20).
Early thymocyte development
To date, understanding of the roles of Ikaros family members in early thymocyte development has been limited to Ikaros and Aiolos, although the role of Aiolos is less well-characterized. While Ikaros is expressed throughout thymocyte development, Aiolos is not strongly upregulated until the T cell precursor stage (4). Studies utilizing the dominant negative Ikaros mutant have implicated Ikaros in the regulation of both pre-T cell receptor (TCR) and TCR signaling, which ultimately disrupts thymocyte differentiation across double-negative and double-positive stages (Fig. 1C). As described above, initial work using mice heterozygous for the dominant negative mutation established that these mice exhibited augmented thymocyte proliferation in response to TCR stimulation at the double-positive stage (Fig. 2C) (41). Further, double-negative thymocytes in these animals were capable of differentiating into double-positive cells in the absence of the pre-TCR complex. These augmented double-positive cells were also able to transition into single-positive populations without an appropriate TCR-coreceptor combination, resulting in cells that never fully matured and failed to reach the periphery (46). Finally, Ikaros and Aiolos have been found to regulate CD8α expression in thymocyte populations (33). As such, loss of functional forms of Ikaros or Aiolos results in increased generation of immature CD4+ T cell populations in vivo. Mechanistically, Ikaros was shown to interact with and directly regulate the CD8α gene locus (33). Thus, Ikaros appears to provide multi-stage regulation of T cell development, though the full repertoire of Ikaros gene targets, and the roles of additional Ikaros family members, including Aiolos, in these populations, are unclear.
Early B cell development
As early studies utilizing the Ikaros N- and C-terminal mutants described above resulted in a complete absence of B cells in vivo, the development of additional models was necessary to further investigate the role of Ikaros factors in B cell development (7, 26). Use of a murine model in which a β-galactosidase reporter was inserted into the second exon of Ikaros resulted in a low level of Ikaros expression (47). In this model, fetal B cells were completely absent, but the B cell compartment developed postnatally from a dramatically reduced number of precursors in the bone marrow. B cells in these mice exhibited marked defects, notably from pro- to pre-B cell stages, supporting a role for Ikaros in the regulation of the pre-B cell compartment (Fig. 1B) (47). To this end, a number of studies determined that disruption of Ikaros signaling prevents normal pre-B cell development and disrupts pre-BCR cell signaling (48–50).
Mechanistically, Ikaros and Aiolos have both been identified as key, stage-specific regulators of pre-BCR formation (38, 49, 51, 52). Specifically, both Ikaros and Aiolos have been found to regulate Igll1, the gene encoding the pre-BCR component λ5, at specific stages of pre-B cell development. Ultimately, this mechanism allows for downregulation of pre-BCR and progression of the B cell lineage to light chain rearrangement (38). Further, use of a conditional Ikaros mutant with an inactive DNA binding domain specifically in early pre-B cells established that expression of this mutant results in an arrest of B cell differentiation at the large pre-B stage (52). Several studies have also established, in addition to the regulation of pre-BCR expression, that Ikaros influences both V(D)J recombination and cell cycle progression (49, 51). Of note, one such study found that the induction of Ikaros expression in cycling pre-B cells was sufficient to redirect this population to express a resting pre-B cell gene program (51). Finally, Ikaros was found to play a role in antagonizing the IL-7-dependent regulation of key genes involved in the progression of B cell development (49).
In addition to the Igll1 locus, Ikaros and Aiolos have been shown to regulate B cell developmental processes by influencing a number of additional gene targets. Specifically, Ikaros supports recombination events by activating Rag expression, modulating accessibility to the VH gene, and regulating and compaction of the Igh locus (48). Further, both Ikaros and Aiolos inhibit pre-B-cell differentiation by directly suppressing c-Myc expression, which is necessary for pre-B cells to progress from this stage of differentiation in normal development (53). Collectively, the available literature indicate that Ikaros and Aiolos play important roles in regulating normal B cell lineage progression by modulating a number of key processes.
Early innate lymphoid cell (ILC) development
ILCs are non-T, non-B lymphocytes that function as sentinels of the immune system and are among the body’s initial responders and protectors against invading pathogenic viruses, bacteria, and parasites (54). ILCs also perform unique and vital functions in other processes including pregnancy, formation and repair of lymphoid tissues, wound healing, mucosal homeostasis, and cancer immune surveillance (54–59). The ILC family includes cytotoxic NK cells and non-cytotoxic “helper” ILC1s, ILC2s, and ILC3s, which are typically regarded as the innate counterparts to cytotoxic CD8+ T cells and to helper CD4+ TH1, TH2, and TH17 T cell subsets, respectively. Although Ikaros family members are required for the development of lymphoid progenitor cells and therefore for the ultimate generation of all ILCs, the specific roles of Ikaros family members in early ILC development are still being characterized (60). Pioneering work regarding the role of Ikaros in immune cell populations established that the absence of functional Ikaros protein resulted in a complete lack of NK cells, as well as T and B cell populations (26, 45, 60). It was also demonstrated that Ikaros-deficient mice lacked peripheral lymphoid tissues, which may be attributed to the loss of lymphoid tissue inducer cells (LTis, a subset of ILC3s) in the absence of Ikaros (15). Ikaros is also expressed in all other ILC subsets, however, it may function as a repressor of certain ILC differentiation pathways, perhaps at more downstream developmental stages (61). For instance, conditional deletion of Ikaros exon 4 in Rorγt-expressing cells resulted in the expansion and hyperactivity of IL-22-producing ILC3s due to disinhibition of the aryl hydrocarbon receptor (AHR) pathway, which is required for IL-22-producing ILC3s (62–64). This suggests that Ikaros may play diverse roles in the regulation of ILC gene programs.
Regarding roles for other Ikaros family members, recent work has demonstrated that in addition to Ikaros, Aiolos and Helios are relatively highly expressed in NK cells/ILC1s, and ILC3s, respectively (61, 65–67). Functionally, Aiolos-knockout mice have been shown to exhibit dysregulated NK cell maturation, while liver ILC1s were reportedly unaltered (68). As with Ikaros, the precise gene targets of Aiolos and its role in individual NK cell developmental stages remains poorly characterized. Interestingly, Helios is also expressed in murine NK cells at early stages in their development, but is subsequently downregulated. This suggests that Helios may participate in the regulation of NK cell development, perhaps at initial stages (69). Ultimately, the study of Ikaros family members in ILC populations is in its infancy, and much remains to be explored with regard to both pathways targeted by Ikaros factors and the mechanisms they employ to support later stages of ILC development.
Collectively, Ikaros family members have been identified as key regulators of lymphopoeisis, as their absence or altered function result in loss or disrupted lymphocyte populations. However, much remains to be explored regarding specific gene targets regulated by Ikaros factors throughout early lymphocyte development, as well as the molecular mechanisms they employ to exert such functions.
Regulation of mature T cell populations by Ikaros factors
T cell populations are divided into two distinct subtypes with a shared developmental pathway that diverges during T cell maturation in the thymus (Fig. 1C) (70, 71). CD8+ “cytotoxic” T cells recognize cognate antigens presented via MHCI, which allows them to identify and eliminate infected self cells through the production of pro-inflammatory cytokines and effector molecules including granzymes and perforin. Complementing CD8+ T cell function, CD4+ T cells recognize cognate antigen presented via MHCII by professional antigen presenting cells, including dendritic cells and macrophages. Functionally, effector CD4+ T cells coordinate pathogen-specific immune responses through both cell-cell interactions and the production of infection-specific effector cytokines (72). These functions ultimately orchestrate responses by both innate and adaptive immune cell populations, including CD8+ T cells, to eliminate pathogens, infected self-cells, and certain cancers. In response to concurrent antigenic stimulation and infection-specific cytokine environments, both CD4+ and CD8+ T cells are able to differentiate into a number of subsets that perform distinct effector functions.
The differentiation and function of mature T cell populations is dependent upon both environmental and autocrine cytokine signals, which specify the expression of subset-specific transcriptional programs (72–74). Such programs include the expression of key, “lineage-defining” transcription factors, which make up a transcriptional network that ultimately dictates T cell differentiation and function. Until relatively recently, much of the understanding concerning the role of Ikaros family members in T cell populations was limited to early thymocyte development. However, more recent studies have shed light on the role of these factors in regulating the differentiation of individual T cell populations and their effector functions. This includes regulation of the expression of both key transcriptional regulators and cytokine signaling pathways to influence mature T cell populations. Roles for each Ikaros family member in T cell subsets are highly context-dependent, likely due to 1) dependence of Ikaros factor expression on subset-specific environmental signals, 2) alterations in the expression of different function-regulating cofactors (including other Ikaros family members), and 3) the specific Ikaros family member isoforms expressed in each T cell setting. Broadly speaking, however, Ikaros family member functions in T cell subsets appear to regulate the expression of T cell gene programs by modulating expression of either 1) cytokines responsible for directing the specification and function of individual T cell subsets and/or 2) key transcriptional regulators involved in promoting or repressing T cell fates. Our group recently detailed current understanding of subset-specific roles for Ikaros family members in individual CD4+ T helper cell populations (75). Here, we have attempted to summarize understanding of broad roles of these factors in mature T cell contexts, and highlight important emergent themes in Ikaros family mediated gene regulation.
Ikaros
As with early lymphocyte populations, Ikaros has been the most extensively studied in mature T cell populations. Curiously, it appears to play broad and differential roles across individual T helper cell subsets, in which it is widely expressed. However, many of the studies described here have relied on experimental/animal models with germline mutations in Ikaros (Fig. 2). As Ikaros influences lymphocyte development well before the single-positive thymocyte stage, utilizing such studies to interpret its role in mature T cell populations represents a limitation with much of the current literature.
In mature T cells, Ikaros-mediated effects begin as early as T cell activation, during which it appears to shape the chromatin landscape to regulate both T cell responsiveness and proliferation in response to TCR engagement (76). As in thymocyte development, Ikaros also differentially influences the differentiation and function of T helper cell populations via not only direct induction of cytokines and key transcription factors to promote T helper cell fate, but also repression of such target genes to suppress gene programs associated with alternative T helper cell gene programs. For instance, Ikaros has been found to support T helper 2 (TH2) cell differentiation by simultaneously inducing the expression of the pro-TH2 cytokine interleukin 4 (IL-4) and repressing the expression of key T helper 1 (TH1) genes, including those encoding the TH1 lineage-defining transcription factor T-bet (Tbx21) and the TH1 effector cytokine interferon gamma (IFN-γ; Ifng), through the direct interaction with their regulatory regions in TH2 populations (77–79). Curiously, while Ikaros is also expressed in TH1 populations, it is not found at the Tbx21 promoter in this context, suggesting that differential mechanisms and/or interacting partners alter Ikaros localization to known target genes between individual T helper cell subsets (78).
In further support of a role for Ikaros in regulating the expression of key T cell transcription factors, Ikaros has been shown to positively regulate the expression of the T follicular helper (TFH) cell lineage-defining transcription factor Bcl-6, through direct interaction with its promoter (24). As Bcl-6 is an important regulator of both effector (TFH) and memory T cell populations, as well as B cells, this suggests that Ikaros may also play an as-yet-undescribed role beyond the TFH cell context. Additionally, Ikaros has been implicated in the positive regulation of T helper 17 (TH17) cell differentiation and function. Specifically, Ikaros was found to support TH17 differentiation by promoting the expression of a number of pro-TH17 genes, including those encoding AHR (Ahr), the TH17 lineage-defining factor ROR-γ (Rorc), and TH17 effector cytokines IL-17a (Il17a) and IL-22 (Il22), while repressing Tbx21 and Foxp3 (80). Collectively, these findings suggest that Ikaros plays broad and diverse roles in the regulation of effector CD4+ T cell subsets.
In order to more precisely define the role of Ikaros specifically in mature T cell populations, a recent study utilized mice in which Ikaros had been knocked out under the control of the distal lck promoter (81). Loss of Ikaros in mature T cells alone resulted in increased Ifng expression in TH2 cells, but did not ultimately impact T-bet expression in this context (81). This suggests that while Ikaros exhibits stage-specific functions in mature T cell populations, previous associations between Ikaros and transcription factor expression could possibly be linked to developmental effects in thymocytes. Finally, use of the above model also determined that while the absence of Ikaros in mature populations does not appear to ultimately disrupt TH1 or TH2 differentiation, Ikaros null CD4+ T cells were unable to differentiate into iTreg populations in vitro, possibly due to disrupted pro-inflammatory cytokine expression (81).
To this end, in addition to the regulation of TH1, TH2, and TH17 effector cytokines, Ikaros has also been directly implicated in the repression of IL-2 production in anergic CD4+ T cells, where it has been found to directly associate with the Il2 locus and recruit repressive histone deacetylases (82–84). While further work is necessary to define the role of Ikaros in regulating IL-2 expression in effector CD4+ T cell populations beyond anergic cells, a similar relationship appears to be at play in CD8+ T cell populations. Specifically, Ikaros has been found to negatively regulate the autocrine expression of IL-2 in CD8+ T cells, though whether it directly associates with the Il2 locus in this population is unclear (85). While more work is needed to determine the precise role of Ikaros in both CD4+ and CD8+ populations, given important roles for IL-2 signaling in directing T cell responses, these aspects of Ikaros-mediated regulation may have broad implications across mature T cell populations (86).
Further, corollary data using germline Ikaros-knockout animals suggest that Ikaros may also function to regulate the expression of IL-10, IL-17, IL-21, and IL-22, while loss of Ikaros exclusively in mature T helper cells resulted in increased expression of IL-2, IFN-γ, TNF-α and GM-CSF when these cells were stimulated in vitro (79–81, 87). Supporting a similar role for Ikaros in CD8+ T cells, a recent study in CAR T cell populations found that treatment of these cells with the immunomodulatory drug lenalidomide, which is known to semi-selectively degrade Ikaros and Aiolos proteins, resulted in increased proliferation and function of these cells. This included both increased cytotoxicity and augmented expression of IL-2, IFN-γ, TNF-α and GM-CSF, similar to the phenotype observed in T helper cell populations upon loss of Ikaros (88). Finally, Ikaros may also play a role in regulating responsiveness to cytokine signals, including IL-2, as overexpression of the dominant negative form of Ikaros resulted in increased expression of the IL-2 receptor alpha subunit (CD25) in IL-12 treated CD8+ cells (89). Thus, this relationship will be interesting to explore in future studies across immune cell populations.
Collectively, the above studies support a role for Ikaros in varied, differential regulation of T helper cell subsets, with Ikaros supporting the differentiation of TH2, TH17, TFH, and regulatory T cell types, while suppressing expression of the TH1 gene program (75). This regulation appears to be exerted, at least in part, via the conserved regulation of effector cytokine expression and possibly that of cytokine receptors. However, the mechanisms employed by Ikaros to regulate the above target gene programs remain incompletely characterized. Thus, a key area of future study will be to determine 1) subset-specific co-factors that may drive this association and/or Ikaros function at target gene regulatory regions, and 2) mechanisms by which Ikaros is differentially targeted to specific gene loci in different T helper cell populations.
Aiolos
While comparatively less is known regarding roles for Aiolos in mature T cell populations, currently available data suggest that it functions similarly to Ikaros in terms of supporting T helper cell gene programs, including positive associations with TH17, TFH, and TREG populations, and a negative association with the TH1 cell type (75). Mechanistically, Aiolos also appears to share target genes with Ikaros in these contexts. Specifically, both Ikaros and Aiolos have been implicated in the direct regulation of Il2 expression. As IL-2 is a key, differential regulator of a number of T helper cell gene programs, this mechanism has the potential to broadly impact T helper subset differentiation and function (86, 90). To this end, Aiolos has been found to directly repress IL-2 in Th17 cells to support their differentiation and effector function (91). Whether Aiolos-mediated support of TH17 differentiation is limited to IL-2 regulation is unclear, as the same study demonstrated that Aiolos expression correlated with that of the TH17 effector cytokine genes Il17a and Il17f, as well as Il21, and the production of IL-17 in vitro (91). Similar to findings in TH17 cells, one study suggested that Aiolos may also regulate the expression of IL-2 in human regulatory T cells (92). However, whether this relationship is due to direct interaction of Aiolos with the IL-2 promoter is as yet unknown.
In addition to its direct regulation of IL-2 production, Aiolos has recently been linked to the positive regulation of the anti-inflammatory cytokine IL-10 in human CD4+ T cell populations (93, 94). Given that IL-10 is produced by a number of T helper cell subsets, it will be interesting to determine whether Aiolos may similarly regulate its expression in additional T helper cell contexts. Finally, as discussed above, recent work in CD8+ CAR T cell populations indicated that Aiolos may positively regulate their cytotoxicity and pro-inflammatory cytokine production, as lenalidomide treatment caused increases in both. However, whether this effect is due to loss of Aiolos, Ikaros, or another lenalidomide-sensitive factor, is yet to be determined (88). Regardless, the above data are supportive of a role for Aiolos, like Ikaros, in the differential regulation of pro- and anti-inflammatory cytokine production in a number of individual T helper cell subsets.
Much like Ikaros, in addition to cytokine regulation, Aiolos has also been implicated in the regulation of transcription factor expression. This includes support of the T follicular helper (TFH) gene program through its direct induction of Bcl-6 expression. Like Ikaros, Aiolos has been shown to bind to the Bcl6 promoter, where it cooperates with STAT3 to directly induce Bcl-6 expression in vitro (24). While Aiolos expression is elevated in TFH populations in vivo, the full extent of its regulatory relationship to the TFH gene program, and TFH cell function, is an area of active investigation in our laboratory. To this end, as with Ikaros, limited data are available regarding Aiolos enrichment throughout the genome in mature T helper cell populations, and much remains to be learned regarding both Aiolos-specific target genes and the transcriptional mechanisms underlying Aiolos function at these loci.
Finally, as Ikaros and Aiolos have been shown to physically interact and cooperate to regulate gene expression, it is possible that overlapping functions observed for these two factors in T cell populations may be due to their collaborative activities observed in these and other lymphocyte populations. While the full extent of this relationship is still being elucidated across mature T cell subsets, it is an important consideration when examining phenotypes in experimental systems in which one of these factors has been functionally compromised. To this end, similar to Ikaros, the role of Aiolos has been explored predominantly in the context of germline knockouts or mutants, as conditional disruption of this protein has never been explored. Thus, generation of tools to allow for more precise control of Aiolos manipulation will further endeavors to explore its function in mature T cells and beyond.
Eos
Available literature demonstrates that Eos is highly expressed in regulatory T cell (TREG) populations, and plays a prominent role in their maintenance and function (30, 95–97). Loss of Eos function has been implicated in both reduced TREG suppressive capabilities and, in some cases, a gain of pro-inflammatory effector characteristics in this population (30, 95, 96). Ultimately, this phenotype has been associated with the spontaneous generation of autoimmune disease in mice lacking Eos expression (96, 98). One key function of Eos in supporting TREG populations appears to be its role as a co-repressor in conjunction with the Treg lineage-defining factor Foxp3 (30, 97). Eos and Foxp3 cooperate to repress a number of gene targets to support the TREG gene program, including Il2 (30, 95, 97). However, the full repertoire of mechanisms by which Eos supports Treg differentiation and function are still being determined.
Curiously, despite structural similarities between Eos, Ikaros, and Aiolos, and a degree of overlapping gene targets (including Il2), broadly speaking, Eos appears to function in opposition to Ikaros and Aiolos in the context of mature T cell populations. Early studies found that CD4+ T cells lacking Eos in a mixed bone marrow chimera model of EAE exhibited reduced expression of the TH1 effector cytokine IFN-γ (which is thought to be suppressed by both Ikaros and Aiolos), suggesting that Eos may function to promote its expression (98). Furthermore, studies from the same group, as well as our lab, found that Eos expression positively correlated with that of TH1-associated genes, including IL-2 and CD25, in conventional T cells (24, 98). Of note, while Eos cooperates with Foxp3 to repress IL-2 production in TREG populations, Eos expression was found to correlate with an increase in IL-2 expression in the absence of Foxp3 in conventional T cells (98). As discussed previously, this supports the notion that subset-specific roles for Ikaros family members are regulated, in part, by the presence and absence of target gene- and cell type-specific co-factors.
Beyond TH1 populations, Eos was found to repress TH17 development (which is supported by both Ikaros and Aiolos), as loss of Eos in the above EAE model resulted in increased IL-17 production by conventional CD4+ T cell populations, and Eos knockdown resulted in increased TH17 cell numbers (98, 99). Similarly, TREGS lacking Eos produce IL-17, and loss of Eos correlates with increased TH17 cells in the CNS (98). Further, conditional knockout of Eos in TREGS leads to the development of severe Experimental Autoimmune Encephalomyletis (EAE) and a loss of TREG suppressive function in vivo (96). Intriguingly, mice with a global deficiency of Eos or deficiency in T cell populations alone developed autoimmune disease at a much older age, supporting a role for Eos not only in TREG populations but potentially other pro-inflammatory effector populations (96). To this end, at least one study has established that Eos is also upregulated in CD8+ T cells upon stimulation, suggesting that it may play a regulatory role in this population as well (98). Ultimately, the available literature support a dynamic role for Eos in the regulaton of T helper cell gene programs, though the precise mechanisms by which it functions, and its role across CD4+ and CD8+ T cell populations is an area open for exploration.
Helios
Helios is expressed in a number of mature T cell populations, however, its role in the generation of individual T cell subsets is still being defined. To date, roles for Helios in mature T cell populations have been described predominantly in the context of TREG populations. Helios was first shown to be a marker for distinguishing stable TREG cells generated in the thymus, and subsequent work has established that Helios expression correlates with TREG suppressive function (100–104). Further work demonstrated that Helios is required for the stable inhibitory activity of both CD4+ TREG cell populations (105). Recent work has shown that Helios plays a role in enhancing differentiation of human CD4+ naïve T cells into regulatory T cells, as ablation of Helios in fetal cells impaired their differentation into iTREG cells upon TCR stimulation. This model further indicated, similar to phenotypes observed with Eos, that loss of Helios impaired immunosuppressive gene expression in favor of a pro-inflammatory phenotype. While these data suggest that Helios may positively regulate TREG populations in humans, its role in murine TREG populations is less clear. While some work has suggested that Helios is required for murine TREG cell suppressive function, work utilizing homologous recombination to inactivate the Helios gene found no impairment of the differentiation or effector function of regulatory T cells (106). Mechanistically, similar to Eos, Helios has been found to cooperate with Foxp3 to support TREG cell differentiation, suggesting that all of these factors may function together to drive the TREG phenotype and/or provide redundancy to support critical immune tolerance (30, 97, 103, 107). Finally, while the expression of Helios has been shown to increase in murine TH2 and TFH cell populations in vivo, its loss does not appear to impact these populations. Thus, it is possible that another Ikaros family member may compensate upon loss of Helios in these cell types (108).
Despite these inconsistencies, current work does support a consistent role for Helios and Ikaros family members across mature T cell populations. Similar to other family members, Helios appears to regulate both transcription factor and cytokine expression. As with Eos, Helios suppresses Il2 in TREG populations by binding directly to the IL2 promoter and inducing repressive epigenetic modifications, including histone deacetylation (107). Additionally, loss of Helios results in decreased binding of Foxp3 to the IL2 promoter and TREG suppressive function (107). Similarities between this effect and those observed upon loss of Eos suggest that these factors may cooperate (or provide redundant regulation) at the Il2 locus. Beyond regulation of IL-2, Helios also appears to regulate the pro-inflammatory cytokines IFN-γ, IL-17, and TNFα, as its loss in TREG cells results in their increased expression (105). These data are corroborated by the observation that Helios– Foxp3+ human TREG cells secrete IL-17, unlike their Helios+ counterparts (109). Curiously, such expression patterns are similar to those observed in the absence of Ikaros, once again suggesting potential cooperation/redundancy between these two factors. Finally, similar to Ikaros, Helios may also regulate cytokine responsiveness, as overexpression of a dominant form of Helios in CD8+ T cell populations stimulated with IL-12 resulted in increased expression of CD25 (89). Like Eos, Helios is upregulated in CD8+ T cells upon activation, and has been detected in certain CD8+ populations during chronic HIV infection in vivo, suggesting it may play a role in regulating these populations (110, 111). Ultimately, however, more work will be required to more precisely define the role of Helios in both effector and regulatory T cell types.
Ikaros factor regulation of mature B cell populations
Mature B cells are critical mediators of the humoral response, through their role in the formation of germinal centers (GCs) to promote selection and the generation of high-affinity antibody producing B cells and long-lived plasma cell populations. The activation of mature B cells is initially reliant upon engagement of the B cell receptor (BCR), which can occur via T-cell-dependent or independent mechanisms. Following their activation, B cells form GCs in collaboration with TFH cells and follicular dendritic cells. Within the GC, B cells undergo rapid proliferation, differentiation, somatic hypermutation, and selection. This process ultimately results in differentiation of plasma cells and the production of high-affinity antibodies. Some B cells will also enter the long-term memory B-cell population, which are poised to differentiate into long-lived, high-affinity plasma cells upon stimulation with cognate antigen. Broadly speaking, the roles of Ikaros family members in the regulation of mature B cells can be divided into effects on B cell activation and antibody production. To date, these functions are limited to Ikaros and Aiolos, which are highly expressed in the B-cell committed lineage.
Roles for Ikaros factors in B cell activation
Ikaros and Aiolos both support the proper function of mature B cell populations, in part by regulating thresholds for B cell activation. Ikaros has been found to play a critical role in limiting the response of naïve splenic B cells to BCR signals, as Ikaros-deficient follicular B cells display increased proliferation and early entry into the cell cycle following stimulation (112). Further, this study determined that naïve Ikaros-deficient cells display constitutive phosphorylation of B cell signaling kinases, including p38 and ERK1/2, supporting a role for Ikaros in limiting signaling even before BCR stimulation (112). Additionally, Ikaros has been implicated in preventing autoimmune responses and controlling both anergy and Toll-like receptor (TLR) signaling in B cells (113). Specifically, B cells from mice with mature B cell-specific deletion of Ikaros exhibited a failure to induce anergy after chronic exposure to self-antigen. These findings correlated with RNA-seq data demonstrating that Ikaros target genes include a large part of the anergy expression program (113). Finally, stimulation of Ikaros-deficient B cells with common TLR antagonists resulted in increased activation and proliferation, as well as a failure to upregulate feedback inhibitors of the pro-inflammatory MyD88-NF-κB pathway, further supporting a role for Ikaros in preventing inappropriate inflammatory responses by restraining TLR signaling (113).
Similar to Ikaros, Aiolos has also been shown to play a role in mediating activation and proliferative B cell responses to stimulation. Early work utilizing transgenic mice containing an “Aiolos-null” mutation determined that, in the absence of functional Aiolos, peripheral B cells display increased surface expression of activation markers, increased BCR signaling strength, and undergo BCR-mediated proliferation even under limiting amounts of stimulation (8, 114). Further, these mice displayed abberant T-cell-dependent B cell responses, including germinal center formation and elevated serum levels of IgG and IgE in the absence of immunization. This response was accompanied by a lack of marginal zone (MZ) and peritoneal B cells and increased production of autoreactive antibodies (8). Mechanistically, Aiolos has been established as a negative regulator of Bruton’s tyrosine kinase (Btk), which represses MZ B cell formation (114). Thus, as with Ikaros, Aiolos appears to limit potential autoimmunity by serving as a strong negative regulator of B cell activation and antibody production in resting B cell populations.
Roles for Ikaros factors in antibody generation
In addition to its effects on B cell activation, Ikaros has also been implicated in the regulation of class switching recombination (CSR). First, Ikaros deficiency results in increased switching to IgG2b and IgG2a at the expense of other isotypes, regardless of stimulation conditions (36). Mechanistically, Ikaros has been shown to regulate a number of genes involved in CSR, including direct interaction with the Igh locus and isotype gene promoters and maintenance of a repressive chromatin state at γ2b and γ2a genes. Further, Ikaros controls transcriptional competition between S regions (36).
Further supporting a role for Ikaros in regulating antibody responses, it has been shown to interact with the transcription factor interferon regulatory factor 4 (IRF4) to promote both germinal center reactions and plasma cell differentiation (115). Mechanistically, Ikaros was found to be essential for IRF4 binding to Zinc finger IRF-composite elements (ZICE). Binding of IRF4/Ikaros heterodimeric complexes to ZICE resulted in the repression of key genes that prevent differentiation to the plasma cell state. Interestingly, this work also established that if the ZICE regulatory element was proximal to an Ets motif, Ikaros/IRF4/PU.1 complexes could function as transcriptional activators, illustrating a mechanism whereby Ikaros can function as both transcriptional activator and repressor in a context-dependent fashion (115).
In the same study, the authors also demonstrated that Aiolos is capable of similarly interacting with IRF4 (115). However, Aiolos did not appear to be essential for IRF4-ZICE interactions. As with NuRD interactions, these findings also demonstrate that while Aiolos and Ikaros may exhibit conserved co-factor interactions, the functional outcomes of such interactions can be distinct. Finally, a more definitive role for Aiolos in plasma cell differentation came from an earlier study in which the authors demonstrated that Aiolos-null mice produce far fewer high affinity antibody producing cells following immunization when compared to WT controls (116). Collectively, the above studies illustrate the importance of Ikaros and Aiolos to the regulation of humoral immunity, via both the activation of mature B cell populations, as well as their transition into antibody-secreting plasma cells.
Ikaros factor regulation of mature ILC and NK cell populations
Mature ILCs are enriched throughout the body at anatomical barrier sites and are poised to rapidly produce cytokines and chemokines that orchestrate immune responses to infection. ILCs interact with—and share many functional and phenotypic features with—T cells. However, unlike T cell populations, which depend upon TCR rearrangement and activation via TCR engagement with cognate antigen, ILCs instead rely on germline-encoded receptors, including pattern-recognition receptors, to detect endogenous and foreign antigenic signals. Mature ILCs consist of three groups that exhibit specialized functions in host immunity and are distinguished by their expression of subset-specific transcription factors and cytokines (54). Group 1 ILCs (NK cells and ILC1s) protect against viruses and other intracellular pathogens, express T-bet, and produce IFN-γ (117–119). NK cells express cytolytic granules (e.g. perforin), as well as the transcription factor Eomesodermin, and are distinct among ILCs in their capacity to directly kill infected or malignant cells (117). Group 2 ILCs (ILC2s) protect against parasites, express Rorα and Gata3, and produce IL-4, IL-5, and IL-13 (120). Group 3 ILCs (ILC3s and LTis) protect against pathogenic bacteria and facilitate lymphoid tissue formation, express Ahr and Rorγt, and produce IL-17 and/or IL-22 (121, 122).
The specific roles of Ikaros family members in mature NK cell and ILC functions are not yet clearly defined, though as mentioned earlier, accumulating data in humans and in mice indicate specific expression patterns of Ikaros family members within ILC and NK cell subsets: all ILCs express Ikaros, Group 1 ILCs (NK cells and ILC1s) exhibit relatively higher—if not exclusive—expression of Aiolos, and ILC3s display increased expression of Helios relative to other ILC populations (61, 66, 67, 69). Moroever, among human NK cell subsets, Aiolos transcript was reportedly higher in the more mature and cytotoxic CD56dim NK cell subset compared to the less mature CD56bright NK cell subset (123).
Regarding functions for individual Ikaros factors in the above populations, recent genetic deletion studies in mice demonstrated that Aiolos is not required for NK cell or ILC1 differentiation or survival, but its absence results in the functional dysregulation of NK cell populations (68). Further, Aiolos-deficient mice exhibited NK cells that were hyperreactive to RMAS and B16 tumor cells in vivo and also showed increased homeostatic and IL-15-induced proliferation. However, some of the above changes may have been due to Aiolos-dependent effects on other immune cell populations, as analysis of AIolos-deficient NK cells in a mixed bone marrow chimera study exhibited reduced expression of the NK effector genes Ifng and Gzmb (68). Thus, these cells were functionally impaired in their ability to control MCMV infection in vivo.
Mechanistically, a comparative analysis of the transcriptomes of wildtype versus Aiolos-deficient NK cells found that approximately two-thirds of the differentially expressed genes were upregulated in the absence of Aiolos, suggesting that it has a major suppressive role in this population (68). These findings are in line with some human data demonstrating that lenalidomide treatment, which facilitates ubiquitin-mediated Ikaros and Aiolos degradation, promotes NK cell functional activity, although this drug was also shown to activate NK cells (and T cells) through an alternative mechanism dependent upon Zap-70 (124). Thus, further work will be necessary to determine the potential role of lenalidomide-dependent degradation of Ikaros and Aiolos on NK cell function. Finally, it is also interesting to note that small nucleotide polymorphisms were detected within enhancer regions of both the Ikaros (IKZF1) and Aiolos (IKZF3) genes in patients with inflammatory bowel disease (IBD), in which Group 1 ILCs are dysregulated and often expanded (66, 67, 125). Thus, it will be useful in future investigations to determine the impacts of these polymorphisms on Ikaros and Aiolos expression and function in this clinical setting.
In addition to the above phenotypes, Aiolos-deficient NK cells also exhibited relatively increased expression of Helios (Ikzf2) (68). Increased Helios expression (and concomitant ILC3 expansion) was also demonstrated in human IBD patients that were treated with lenalidomide (61). This suggests that the upregulation of Helios may be compensatory, or that Ikaros family members regulate the expression of one another in ILC populations, either directly or indirectly. It is also notable that increased expression of Helios was observed in murine NK cells bearing a W32R mutation in the Ncr1 gene that abolishes cell surface expression of NKp46 (i.e. Noé mice) (69). The NK cells in these mice were hyperreactive to a number of stimuli in vitro and in vivo, and were capable of providing increased protection against MCMV and influenza infection. However, it is not yet clear if increased Helios expression was the cause of this hyperreactivity. Finally, further supporting a role for Helios in modulating ILC function, recent clinical data in humans in the setting of liver transplantation showed decreased NK cell functionality in association with lower expression of Helios transcript (126). Together, these correlative data suggest that Helios may promote NK cell functions, though the mechanism is as yet unknown. Indeed, further investigation is warranted to determine the various roles of Ikaros family members in NK cell and ILC function.
Emerging Roles for Ikaros family members in lymphoid cell regulation
Ikaros family members appear to exert their effects at target gene loci through a number of conserved mechanisms, the full range of which are still being determined. However, the identities of Ikaros factor target gene loci suggest that there are conserved pathways through which these factors may functionally regulate immune cell populations. Of particular interest are emerging roles for Ikaros family members in the end-to-end regulation of cytokine signaling pathways. While this work has been described predominantly in the context of mature T cell populations, cytokine signaling is a critical driver of development, differentiation, and function across T, B, and ILC populations.
Regulation of pro- and anti-inflammatory cytokine expression
Given distinct phenotypes positively and negatively regulated by individual Ikaros family members, it is interesting to note that Ikaros, Aiolos, Eos, and Helios all appear to play roles in the regulation of cytokine expression. These include both anti-inflammatory (e.g. IL-10) and pro-inflammatory (e.g. IFN-γ, IL-17a/f, TNF-α, GM-CSF) cytokines, which exert a broad range of effects on immune cell populations. Curiously, the expression of these cytokines appears to be regulated by different Ikaros family members across mature T helper cell subsets, suggesting that subset-specific Ikaros factor functions drive the expression of distinct effector programs. As much of the data linking Ikaros family members to the above cytokines is corollary, it will be of interest to define both independent and potential cooperative roles for Ikaros factors in the direct regulation of their target gene loci.
Beyond the above cytokines, Ikaros, Helios, Aiolos, and Eos have all been linked to the direct regulation of the immunomodulatory cytokine IL-2 in various cellular contexts (30, 85, 91). While much of the above work has defined roles for Ikaros family members in repressing IL-2 expression, recent work, including a study from our laboratory, suggests that Eos may differentially regulate the Il2 locus depending on cellular context. Specifically, Eos represses IL-2 production in TREG cells, but does not appear to exhibit this function in effector T helper cell populations (30, 98). The conserved role for Ikaros factors in regulating IL-2 in multiple T cell populations suggests several possibilities: first, that regulation of IL-2 signaling is dependent upon multiple, interacting Ikaros family members within a single cellular context, or, alternatively, that the importance of IL-2 regulation makes it a key target for redundant Ikaros factor functions across the immune system. Ultimately, determining the independent and/or cooperative functions of Ikaros family members, as well as co-factors recruited by Ikaros factors at the regulatory regions of such gene loci will provide much needed insight into the roles of these factors in regulating individual immune cell programs.
Ikaros factor-dependent regulation of cytokine-driven feed-forward mechanisms
While the regulation of cytokine expression is critical for the effector functions of individual immune populations, such signals are also responsible for driving the development and differentiation of early and mature lymphocyte populations, respectively. Thus, individual lymphoid populations require the expression of distinct, and often opposing, cytokine-dependent transcriptional programs. One cytokine known to differentially regulate such programs is IL-2, which positively or negatively effects the formation and function of individual mature T cell subsets. Specifically, IL-2 promotes the differentiation of TH1, TH2, and TREG cell populations, while inhibiting the development of TH17 and TFH subsets. Curiously, Ikaros factor regulatory axes also appear to exist between these subsets. Specifically, Aiolos supports TFH and TH17 cells, while Eos has been implicated in the positive regulation of both TH1 and TREG populations. Supporting a role for the cytokine dependence of such regulatory axes, work from our laboratory also indicates that the expression of Aiolos and Eos are differentially regulated by signals from IL-2. Specifically, expression of Aiolos is repressed, while Eos expression is activated, by IL-2 signals (24, 75). Thus, given the role for Ikaros family members in modulating cytokine expression, it is possible that cytokine-dependent feed-forward mechanisms exist to support both the expression of Ikaros family members and their subsequent regulation of subset-specific gene programs. Unlike Aiolos and Eos, Ikaros uniquely exhibits regulatory roles across individual T helper cell subsets. This, along with recent work describing possible pioneering activity of Ikaros (127), suggests that it may broadly support mature T cell populations, while its interaction(s) with specific “lineage-supporting” family members ultimately specify T cell fate and function.
In addition to the regulation of cytokine expression, Ikaros factors may also play roles in modulating lymphoid cell responsiveness to cytokine signals by influencing the expression of cytokine receptors or receptor subunits. For example, in addition to the regulation of Bcl-6, our laboratory determined that Aiolos cooperates with signal transducer and activator of transcription 3 (STAT3) to promote the expression of the gene encoding IL-6 receptor alpha (Il6ra) in CD4+ T cell populations (24). Similarly, Eos has been positively linked to the expression of IL-2Rα (CD25; Il2ra) in conventional CD4+ T, as its loss results in diminished CD25 expression in these populations (98). Further implicating IL-2 signaling as a key process regulated by Ikaros factors, Ikaros and Helios have also been implicated in regulating Il2ra in CD8+ T cell populations. However, unlike Eos, Ikaros and Helios appear to negatively regulate CD25 expression. To date, whether Ikaros factors directly bind to and activate or repress cytokine receptor loci is unclear.
Regulation of STAT factor activity by Ikaros family members
As cytokine signaling itself is known to regulate receptor expression both positively and negatively, it will be of interest to determine whether Ikaros family members govern receptor expression through direct transcriptional regulation of cytokine receptor gene loci or an alternative, indirect mechanism. To this end, recent data from our laboratory support the exciting possibility of end-to-end roles for Ikaros family members in regulating cytokine signaling pathways. Specifically, our findings suggest that Ikaros family members also interact with, and may regulate the activity of, STAT factors—key signaling molecules downstream of cytokine receptors with roles across immune cell populations (128, 129). As discussed above, our initial findings in CD4+ T cells indicated that Aiolos interacts and cooperates with STAT3 to induce the expression of Bcl-6 and IL-6Ra (24). We also observed co-enrichment of Ikaros with Aiolos/STAT3 at the Bcl6 locus, suggesting that it may also cooperate with STAT3 to regulate target gene expression. The precise role of Aiolos, Ikaros, and STAT3 in mechanistically regulating Bcl-6 expression, as well as additional TFH genes, is an area that we are actively investigating.
Ikaros/STAT factor interactions are not limited to Aiolos/STAT3, however. Our laboratory has identified a second complex composed of Eos and STAT5 in TH1 cells (unpublished data). As discussed above, relatively recent findings support opposing roles for Aiolos and Eos in mature T cell populations, as Aiolos supports, while Eos represses, TFH and TH17 cell identity and function (75). Conversely, Eos supports, while Aiolos antagonizes, TH1 cell differentiation. Our previous data indicated that Eos was highly expressed upon IL-2 treatment under TH1-polarizing conditions (24). As IL-2 is known to differentially regulate the above T helper cell subsets, we hypothesized that Eos may function to promote the IL-2 signaling pathway to support TH1 cell differentiation. Indeed, we found that Eos physically interacts with STAT5 to support both STAT5 activation and its association with TH1 target genes, including IL2ra. Thus, STAT/Ikaros family member interactions appear to be conserved in both TFH and TH1 cell populations.
Given homology between individual members of the Ikaros and STAT factor families, and dependence of individual lymphoid subsets upon specific members of these families, the above relationships support potential roles for Ikaros/STAT factor regulatory complexes across T, B, and ILC populations. Additionally, homo- and hetero-dimerization have been observed for both Ikaros factors and STAT family members. Should the above interactions be conserved across factors and immune populations, these interactions would allow for complex, stage- and cell-type-specific interplay between individual family members and would expand their gene-specific regulatory activities exponentially. Ultimately, these findings, in conjunction with current understanding of Ikaros family member regulation of cytokine and cytokine receptor expression, support exciting roles for Ikaros factors in the end-to-end regulation of cytokine signaling, which may have implications across immune cell populations.
Much of the work detailing the role of Ikaros family members in regulating cytokine pathways has been performed in the context of mature effector T cell populations. However, the importance of cytokine signaling is by no means limited to this population, as it is responsible for mediating early lymphoid development, B cell maturation, plasma cell generation, the formation of ILC populations, and the formation of immunological memory. As such, the mechanisms described herein have the potential to regulate key developmental and effector processes in cell types across the immune system.
Future Perspectives
Key aspects of Ikaros factor biology which are responsible for their function and localization have long been understood, including the presence of distinct splice variants, the dependence of factor function on homo- and hetero-dimerization between family members, and the varied expression of Ikaros factors throughout the stages of lymphopoiesis. These functional characteristics/relationships exert profound effects on Ikaros factor regulation of target genes and, ultimately, the differentiation and function of individual immune cell populations. However, to date, their roles in discrete immune cell populations and at specific target gene loci are incompletely understood.
First, current findings do not preclude the function of both chromatin-dependent and -independent mechanisms for Ikaros proteins. These may include the interaction of Ikaros family members with STAT proteins prior to DNA binding, as well as co-enrichment of these factors at individual target gene loci. To this end, our recent findings suggest that Eos interacts with STAT5 to promote its activation and subsequent interaction with target genes (unpublished data). Whether this interaction occurs in the cytosol as well as the nucleus is unclear. However, previous work has established that certain isoforms of both Ikaros and Aiolos preferentially localize to the nucleus vs. cytosol, suggesting differential functions for these factors based on their cellular location (130). Ultimately, it will be of interest to further explore Ikaros factor localization and interacting factors in both the nucleus and cytosol, via co-immunoprecipitation and/or microscopy analyses.
Next, most studies regarding Ikaros family member regulation of lymphocyte populations have focused upon the analysis of a single family member. However, it is well established that these factors are co-expressed across immune cell populations, that they interact, and that they exhibit both overlapping and distinct phenotypic effects. Thus, future studies that analyze co-enrichment of these factors and association of heterodimers with transcriptional complexes will provide necessary insight into the precise roles of these factors in the regulation of hematopoietic cell regulation.
Answering the questions above will require the generation of more precise tools, including conditional knockouts/transgenic lines to assess the function of individual Ikaros factors in specific immune cell populations. The use of such tools in conjunction with Next-Generation Sequencing approaches will offer powerful methods to define individual and cooperative functions of Ikaros family members, as well as the co-factors required for these activities. First, the use of Cut&Run or chromatin immunoprecipitation (ChIP)-seq assays of lymphocyte populations from wildtype versus Ikaros factor transgenic mouse lines would provide critical information regarding whole-genome enrichment patterns for Ikaros family members in individual lymphocyte populations. This analysis would allow for both the assessment of Ikaros family member abundance at target gene loci, and the identification of binding sites throughout the genome. Further, such analyses could be utilized to define the role of homo- and hetero-dimerization of Ikaros factors at individual regulatory regions, as well as the presence or absence of individual factors within target gene loci upon loss of another family member. Finally, Cut&Run/ChIP-seq may be leveraged to analyze co-enrichment of Ikaros factors with additional transcriptional regulators including STAT factors, as well as define binding site sequences at such loci. The above enrichment patterns may also be assessed alongside RNA-sequencing data to define functional gene targets regulated by Ikaros factors. Expanding upon standard RNA sequencing, the use of single-cell (sc)RNA-seq would offer the opportunity to define expression patterns for each Ikaros family member throughout lymphocyte differentiation and across distinct mature lymphocyte populations.
As Ikaros family members have been implicated in the remodeling of chromatin structure, it will also be of interest to utilize ATAC-seq in conjunction with transgenic mouse models to evaluate alterations to chromatin accessibility in the presence and absence of functional Ikaros proteins. As above, these data could be also assessed alongside enrichment and transcript expression patterns, to determine how Ikaros factor enrichment affects chromatin availability and subsequent changes in gene expression. Finally, the use of Hi-C analyses to evaluate chromatin looping events mediated by Ikaros factors will be useful in conjunction with binding pattern assessments described above to define precise roles for Ikaros family members in promoting interactions between distal regulatory elements within and across target gene loci. Ultimately, a layered approach combining many or all of the above analyses would provide a comprehensive understanding of the roles of Ikaros family members in lymphoid populations. Such understanding will offer critical information for hypothesis generation and the analysis of Ikaros factor roles both independently, cooperatively, and in conjunction with additional transcriptional mediators including STAT factors. Given the emerging importance of Ikaros family members in regulating cytokine and cytokine receptor signaling, this work will provide developmental understanding and powerful insight for the therapeutic manipulation of cytokine signaling pathways, which are some of the most readily targeted in current clinical approaches to treat human disease.
Acknowledgements
This work was supported by grants from The National Institutes of Health (CA199447 and CA208353 to A.G.F. and AI134972 to K.J.O). K.A.R. was supported in part by a Nell Mondy and Hartley Corporation graduate fellowship from the Graduate Women in Science (GWIS) foundation. A.G.F. and K.J.O. were also supported by funds from The Ohio State University College of Medicine and The Ohio State University Comprehensive Cancer Center.
Footnotes
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest
The authors declare no competing interests.
References
- 1.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258(5083):808–12. [DOI] [PubMed] [Google Scholar]
- 2.Hahm K, Cobb BS, McCarty AS, et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12(6):782–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley CM, Ikeda T, Koipally J, et al. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. 1998;8(9):508–15. [DOI] [PubMed] [Google Scholar]
- 4.Morgan B, Sun L, Avitahl N, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16(8):2004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honma Y, Kiyosawa H, Mori T, et al. Eos: a novel member of the Ikaros gene family expressed predominantly in the developing nervous system. FEBS Lett. 1999;447(1):76–80. [DOI] [PubMed] [Google Scholar]
- 6.Perdomo J, Holmes M, Chong B, Crossley M. Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J Biol Chem. 2000;275(49):38347–54. [DOI] [PubMed] [Google Scholar]
- 7.Georgopoulos K, Bigby M, Wang JH, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79(1):143–56. [DOI] [PubMed] [Google Scholar]
- 8.Wang JH, Avitahl N, Cariappa A, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9(4):543–53. [DOI] [PubMed] [Google Scholar]
- 9.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48(9–10):1272–8. [DOI] [PubMed] [Google Scholar]
- 10.Payne MA. Zinc finger structure-function in Ikaros Marvin A Payne. World J Biol Chem. 2011;2(6):161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida T, Georgopoulos K. Ikaros fingers on lymphocyte differentiation. Int J Hematol. 2014;100(3):220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14(17):2146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koipally J, Heller EJ, Seavitt JR, Georgopoulos K. Unconventional potentiation of gene expression by Ikaros. J Biol Chem. 2002;277(15):13007–15. [DOI] [PubMed] [Google Scholar]
- 14.Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14(12):8292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schjerven H, McLaughlin J, Arenzana TL, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14(10):1073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Jackson AF, Naito T, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2011;13(1):86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14(11):7111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Perez-Casellas LA, Savic A, Song C, Dovat S. Ikaros isoforms: The saga continues. World J Biol Chem. 2011;2(6):140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liippo J, Nera KP, Veistinen E, et al. Both normal and leukemic B lymphocytes express multiple isoforms of the human Aiolos gene. Eur J Immunol. 2001;31(12):3469–74. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell JL, Seng A, Yankee TM. Expression and splicing of Ikaros family members in murine and human thymocytes. Mol Immunol. 2017;87:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Sif S, Jones B, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345–55. [DOI] [PubMed] [Google Scholar]
- 22.Georgopoulos K The making of a lymphocyte: the choice among disparate cell fates and the IKAROS enigma. Genes Dev. 2017;31(5):439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oravecz A, Apostolov A, Polak K, et al. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat Commun. 2015;6:8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Read KA, Powell MD, Baker CE, et al. Integrated STAT3 and Ikaros Zinc Finger Transcription Factor Activities Regulate Bcl-6 Expression in CD4(+) Th Cells. J Immunol. 2017;199(7):2377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15(19):5358–69. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JH, Nichogiannopoulou A, Wu L, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5(6):537–49. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida T, Hu Y, Zhang Z, et al. Chromatin restriction by the nucleosome remodeler Mi-2beta and functional interplay with lineage-specific transcription regulators control B-cell differentiation. Genes Dev. 2019;33(13–14):763–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oestreich KJ, Weinmann AS. Ikaros changes the face of NuRD remodeling. Nat Immunol. 2011;13(1):16–8. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y, Yoshida T, Georgopoulos K. Transcriptional circuits in B cell transformation. Curr Opin Hematol. 2017;24(4):345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan F, Yu H, Dang EV, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325(5944):1142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgopoulos K Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2(3):162–74. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-del Arco P, Koipally J, Georgopoulos K. Ikaros SUMOylation: switching out of repression. Mol Cell Biol. 2005;25(7):2688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harker N, Naito T, Cortes M, et al. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10(6):1403–15. [DOI] [PubMed] [Google Scholar]
- 34.Bottardi S, Mavoungou L, Milot E. IKAROS: a multifunctional regulator of the polymerase II transcription cycle. Trends Genet. 2015;31(9):500–8. [DOI] [PubMed] [Google Scholar]
- 35.Keys JR, Tallack MR, Zhan Y, et al. A mechanism for Ikaros regulation of human globin gene switching. Br J Haematol. 2008;141(3):398–406. [DOI] [PubMed] [Google Scholar]
- 36.Sellars M, Reina-San-Martin B, Kastner P, Chan S. Ikaros controls isotype selection during immunoglobulin class switch recombination. J Exp Med. 2009;206(5):1073–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–54. [DOI] [PubMed] [Google Scholar]
- 38.Thompson EC, Cobb BS, Sabbattini P, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26(3):335–44. [DOI] [PubMed] [Google Scholar]
- 39.Gurel Z, Ronni T, Ho S, et al. Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J Biol Chem. 2008;283(13):8291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heizmann B, Kastner P, Chan S. The Ikaros family in lymphocyte development. Curr Opin Immunol. 2018;51:14–23. [DOI] [PubMed] [Google Scholar]
- 41.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83(2):289–99. [DOI] [PubMed] [Google Scholar]
- 42.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190(9):1201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30(4):493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allman D, Sambandam A, Kim S, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4(2):168–74. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7(4):382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190(8):1039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32(3):720–30. [DOI] [PubMed] [Google Scholar]
- 48.Reynaud D, Demarco IA, Reddy KL, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9(8):927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heizmann B, Kastner P, Chan S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med. 2013;210(13):2823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwickert TA, Tagoh H, Gultekin S, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15(3):283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreiros-Vidal I, Carroll T, Taylor B, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121(10):1769–82. [DOI] [PubMed] [Google Scholar]
- 52.Joshi I, Yoshida T, Jena N, et al. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol. 2014;15(3):294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30(17):4149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vivier E, Artis D, Colonna M, et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174(5):1054–66. [DOI] [PubMed] [Google Scholar]
- 55.Rak GD, Osborne LC, Siracusa MC, et al. IL-33-Dependent Group 2 Innate Lymphoid Cells Promote Cutaneous Wound Healing. J Invest Dermatol. 2016;136(2):487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vacca P, Vitale C, Munari E, Cassatella MA, Mingari MC, Moretta L. Human Innate Lymphoid Cells: Their Functional and Cellular Interactions in Decidua. Front Immunol. 2018;9:1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diefenbach A, Gnafakis S, Shomrat O. Innate Lymphoid Cell-Epithelial Cell Modules Sustain Intestinal Homeostasis. Immunity. 2020;52(3):452–63. [DOI] [PubMed] [Google Scholar]
- 58.Onder L, Ludewig B. A Fresh View on Lymph Node Organogenesis. Trends Immunol. 2018;39(10):775–87. [DOI] [PubMed] [Google Scholar]
- 59.Kim MY, Kim KS, McConnell F, Lane P. Lymphoid tissue inducer cells: architects of CD4 immune responses in mice and men. Clinical and experimental immunology. 2009;157(1):20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida T, Ng SY, Georgopoulos K. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol. 2010;22(2):154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazzurana L, Forkel M, Rao A, et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3-ILC1/NK cell transdifferentiation. Eur J Immunol. 2019;49(9):1344–55. [DOI] [PubMed] [Google Scholar]
- 62.Lee JS, Cella M, McDonald KG, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13(2):144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S, Heller JJ, Bostick JW, et al. Ikaros Inhibits Group 3 Innate Lymphoid Cell Development and Function by Suppressing the Aryl Hydrocarbon Receptor Pathway. Immunity. 2016;45(1):185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montaldo E, Teixeira-Alves LG, Glatzer T, et al. Human RORgammat(+)CD34(+) cells are lineage-specified progenitors of group 3 RORgammat(+) innate lymphoid cells. Immunity. 2014;41(6):988–1000. [DOI] [PubMed] [Google Scholar]
- 65.Bjorklund AK, Forkel M, Picelli S, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17(4):451–60. [DOI] [PubMed] [Google Scholar]
- 66.Collins PL, Cella M, Porter SI, et al. Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell. 2019;176(1–2):348–60 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuchs A, Vermi W, Lee JS, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38(4):769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holmes ML, Huntington ND, Thong RP, et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. 2014;33(22):2721–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Narni-Mancinelli E, Jaeger BN, Bernat C, et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335(6066):344–8. [DOI] [PubMed] [Google Scholar]
- 70.Schmitt TM, Zuniga-Pflucker JC. T-cell development, doing it in a dish. Immunol Rev. 2006;209:95–102. [DOI] [PubMed] [Google Scholar]
- 71.Kumar BV, Connors TJ, Farber DL. Human T Cell Development, Localization, and Function throughout Life. Immunity. 2018;48(2):202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu J T Helper Cell Differentiation, Heterogeneity, and Plasticity. Cold Spring Harb Perspect Biol. 2018;10(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol. 2012;24(3):297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weinmann AS. Roles for helper T cell lineage-specifying transcription factors in cellular specialization. Adv Immunol. 2014;124:171–206. [DOI] [PubMed] [Google Scholar]
- 75.Powell MD, Read KA, Sreekumar BK, Oestreich KJ. Ikaros Zinc Finger Transcription Factors: Regulators of Cytokine Signaling Pathways and CD4(+) T Helper Cell Differentiation. Front Immunol. 2019;10:1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity. 1999;10(3):333–43. [DOI] [PubMed] [Google Scholar]
- 77.Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol. 2009;182(2):741–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas RM, Chen C, Chunder N, et al. Ikaros silences T-bet expression and interferon-gamma production during T helper 2 differentiation. J Biol Chem. 2010;285(4):2545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Umetsu SE, Winandy S. Ikaros is a regulator of Il10 expression in CD4+ T cells. J Immunol. 2009;183(9):5518–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong LY, Hatfield JK, Brown MA. Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development. J Biol Chem. 2013;288(49):35170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lyon de Ana C, Arakcheeva K, Agnihotri P, Derosia N, Winandy S. Lack of Ikaros Deregulates Inflammatory Gene Programs in T Cells. J Immunol. 2019;202(4):1112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bandyopadhyay S, Dure M, Paroder M, Soto-Nieves N, Puga I, Macian F. Interleukin 2 gene transcription is regulated by Ikaros-induced changes in histone acetylation in anergic T cells. Blood. 2007;109(7):2878–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bandyopadhyay S, Montagna C, Macian F. Silencing of the Il2 gene transcription is regulated by epigenetic changes in anergic T cells. Eur J Immunol. 2012;42(9):2471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas RM, Chunder N, Chen C, Umetsu SE, Winandy S, Wells AD. Ikaros enforces the costimulatory requirement for IL2 gene expression and is required for anergy induction in CD4+ T lymphocytes. J Immunol. 2007;179(11):7305–15. [DOI] [PubMed] [Google Scholar]
- 85.O’Brien S, Thomas RM, Wertheim GB, Zhang F, Shen H, Wells AD. Ikaros imposes a barrier to CD8+ T cell differentiation by restricting autocrine IL-2 production. J Immunol. 2014;192(11):5118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones DM, Read KA, Oestreich KJ. Dynamic Roles for IL-2-STAT5 Signaling in Effector and Regulatory CD4(+) T Cell Populations. J Immunol. 2020;205(7):1721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heller JJ, Schjerven H, Li S, et al. Restriction of IL-22-producing T cell responses and differential regulation of regulatory T cell compartments by zinc finger transcription factor Ikaros. J Immunol. 2014;193(8):3934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Zhou G, Risu N, et al. Lenalidomide Enhances CAR-T Cell Activity Against Solid Tumor Cells. Cell Transplant. 2020;29:963689720920825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clambey ET, Collins B, Young MH, et al. The Ikaros transcription factor regulates responsiveness to IL-12 and expression of IL-2 receptor alpha in mature, activated CD8 T cells. PLoS One. 2013;8(2):e57435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23(5):598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quintana FJ, Jin H, Burns EJ, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13(8):770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gandhi R, Kumar D, Burns EJ, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol. 2010;11(9):846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Evans HG, Roostalu U, Walter GJ, et al. TNF-alpha blockade induces IL-10 expression in human CD4+ T cells. Nat Commun. 2014;5:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ridley ML, Fleskens V, Roberts CA, et al. IKZF3/Aiolos Is Associated with but Not Sufficient for the Expression of IL-10 by CD4(+) T Cells. J Immunol. 2020;204(11):2940–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bettini ML, Pan F, Bettini M, et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36(5):717–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gokhale AS, Gangaplara A, Lopez-Occasio M, Thornton AM, Shevach EM. Selective deletion of Eos (Ikzf4) in T-regulatory cells leads to loss of suppressive function and development of systemic autoimmunity. J Autoimmun. 2019:102300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma MD, Huang L, Choi JH, et al. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity. 2013;38(5):998–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rieder SA, Metidji A, Glass DD, et al. Eos Is Redundant for Regulatory T Cell Function but Plays an Important Role in IL-2 and Th17 Production by CD4+ Conventional T Cells. J Immunol. 2015;195(2):553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang HY, Barbi J, Wu CY, et al. MicroRNA-17 Modulates Regulatory T Cell Function by Targeting Co-regulators of the Foxp3 Transcription Factor. Immunity. 2016;45(1):83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Getnet D, Grosso JF, Goldberg MV, et al. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Mol Immunol. 2010;47(7–8):1595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shevach EM, Thornton AM. tTregs, pTregs, and iTregs: similarities and differences. Immunol Rev. 2014;259(1):88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sugita K, Hanakawa S, Honda T, et al. Generation of Helios reporter mice and an evaluation of the suppressive capacity of Helios(+) regulatory T cells in vitro. Exp Dermatol. 2015;24(7):554–6. [DOI] [PubMed] [Google Scholar]
- 103.Takatori H, Kawashima H, Matsuki A, et al. Helios Enhances Treg Cell Function in Cooperation With FoxP3. Arthritis Rheumatol. 2015;67(6):1491–502. [DOI] [PubMed] [Google Scholar]
- 104.Thornton AM, Korty PE, Tran DQ, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim HJ, Barnitz RA, Kreslavsky T, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350(6258):334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. 2009;183(4):2303–11. [DOI] [PubMed] [Google Scholar]
- 107.Baine I, Basu S, Ames R, Sellers RS, Macian F. Helios induces epigenetic silencing of IL2 gene expression in regulatory T cells. J Immunol. 2013;190(3):1008–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Serre K, Benezech C, Desanti G, et al. Helios is associated with CD4 T cells differentiating to T helper 2 and follicular helper T cells in vivo independently of Foxp3 expression. PLoS One. 2011;6(6):e20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raffin C, Pignon P, Celse C, Debien E, Valmori D, Ayyoub M. Human memory Helios- FOXP3+ regulatory T cells (Tregs) encompass induced Tregs that express Aiolos and respond to IL-1beta by downregulating their suppressor functions. J Immunol. 2013;191(9):4619–27. [DOI] [PubMed] [Google Scholar]
- 110.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLoS One. 2011;6(8):e24226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Naluyima P, Lal KG, Costanzo MC, et al. Terminal Effector CD8 T Cells Defined by an IKZF2(+)IL-7R(−) Transcriptional Signature Express FcgammaRIIIA, Expand in HIV Infection, and Mediate Potent HIV-Specific Antibody-Dependent Cellular Cytotoxicity. J Immunol. 2019;203(8):2210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heizmann B, Sellars M, Macias-Garcia A, Chan S, Kastner P. Ikaros limits follicular B cell activation by regulating B cell receptor signaling pathways. Biochem Biophys Res Commun. 2016;470(3):714–20. [DOI] [PubMed] [Google Scholar]
- 113.Schwickert TA, Tagoh H, Schindler K, Fischer M, Jaritz M, Busslinger M. Ikaros prevents autoimmunity by controlling anergy and Toll-like receptor signaling in B cells. Nat Immunol. 2019;20(11):1517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cariappa A, Tang M, Parng C, et al. The follicular versus marginal zone B lymphocyte cell fate decision is regulated by Aiolos, Btk, and CD21. Immunity. 2001;14(5):603–15. [DOI] [PubMed] [Google Scholar]
- 115.Ochiai K, Kondo H, Okamura Y, et al. Zinc finger-IRF composite elements bound by Ikaros/IRF4 complexes function as gene repression in plasma cell. Blood Adv. 2018;2(8):883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cortes M, Georgopoulos K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J Exp Med. 2004;199(2):209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity. 2017;47(5):820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fuchs A, Vermi W, Lee JS, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 2013;38(4):769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bernink JH, Peters CP, Munneke M, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14(3):221–9. [DOI] [PubMed] [Google Scholar]
- 120.Mjosberg JM, Trifari S, Crellin NK, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12(11):1055–62. [DOI] [PubMed] [Google Scholar]
- 121.Cupedo T, Crellin NK, Papazian N, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10(1):66–74. [DOI] [PubMed] [Google Scholar]
- 122.Cella M, Fuchs A, Vermi W, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457(7230):722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Allan DSJ, Cerdeira AS, Ranjan A, et al. Transcriptome analysis reveals similarities between human blood CD3(−) CD56(bright) cells and mouse CD127(+) innate lymphoid cells. Sci Rep. 2017;7(1):3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hideshima T, Ogiya D, Liu J, et al. Immunomodulatory drugs activate NK cells via both Zap-70 and cereblon-dependent pathways. Leukemia. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Forkel M, van Tol S, Hoog C, Michaelsson J, Almer S, Mjosberg J. Distinct Alterations in the Composition of Mucosal Innate Lymphoid Cells in Newly Diagnosed and Established Crohn’s Disease and Ulcerative Colitis. J Crohns Colitis. 2019;13(1):67–78. [DOI] [PubMed] [Google Scholar]
- 126.Jamil KM, Hydes TJ, Cheent KS, et al. STAT4-associated natural killer cell tolerance following liver transplantation. Gut. 2017;66(2):352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ding Y, Zhang B, Payne JL, et al. Ikaros tumor suppressor function includes induction of active enhancers and super-enhancers along with pioneering activity. Leukemia. 2019;33(11):2720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Villarino AV, Kanno Y, O’Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 2017;18(4):374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Caballero R, Setien F, Lopez-Serra L, et al. Combinatorial effects of splice variants modulate function of Aiolos. J Cell Sci. 2007;120(Pt 15):2619–30. [DOI] [PubMed] [Google Scholar]


