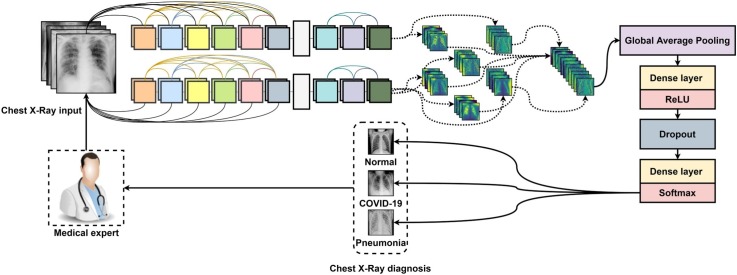
Graphical abstract
Abbreviations: AP, Average Pooling; AUC, Area Under the Curve; BS, Batch Size; BN, Batch Normalization; CAD, Computer-Aided Diagnosis; CCE, Categorical Cross-Entropy; CV, Computer Vision; CNN, Convolutional Neural Networks; CT, Computer Tomography; CXR, Chest X-Rays; DCNN, Deep Convolutional Neural Networks; DR, Dropout Rate; DL, Deep Learning; GAP, Global Average Pooling; GRAD-CAM, Gradient-Weighted Class Activation Maps; JPG, Joint Photographic Group; LR, Learning Rate; MP, Max-Pooling; PEPX, Projection-Expansion-Projection-Extension; rRT-PCR, real-time Reverse-Transcription Polymerase Chain Reaction; P-R, Precision-Recall; ROC, Receiver Operating Characteristic; ReLU, Rectified Linear Unit; SGD, Stochastic Gradient Descent; WHO, World Health Organization
Keywords: Chest x-rays, Computer-aided diagnosis, Covid-19, Deep learning, Densely connected neural networks
Abstract
Due to the unforeseen turn of events, our world has undergone another global pandemic from a highly contagious novel coronavirus named COVID-19. The novel virus inflames the lungs similarly to Pneumonia, making it challenging to diagnose. Currently, the common standard to diagnose the virus's presence from an individual is using a molecular real-time Reverse-Transcription Polymerase Chain Reaction (rRT-PCR) test from fluids acquired through nasal swabs. Such a test is difficult to acquire in most underdeveloped countries with a few experts that can perform the test. As a substitute, the widely available Chest X-Ray (CXR) became an alternative to rule out the virus. However, such a method does not come easy as the virus still possesses unknown characteristics that even experienced radiologists and other medical experts find difficult to diagnose through CXRs. Several studies have recently used computer-aided methods to automate and improve such diagnosis of CXRs through Artificial Intelligence (AI) based on computer vision and Deep Convolutional Neural Networks (DCNN), which some require heavy processing costs and other tedious methods to produce. Therefore, this work proposed the Fused-DenseNet-Tiny, a lightweight DCNN model based on a densely connected neural network (DenseNet) truncated and concatenated. The model trained to learn CXR features based on transfer learning, partial layer freezing, and feature fusion. Upon evaluation, the proposed model achieved a remarkable 97.99 % accuracy, with only 1.2 million parameters and a shorter end-to-end structure. It has also shown better performance than some existing studies and other massive state-of-the-art models that diagnosed COVID-19 from CXRs.
1. Introduction
In an unexpected turn of events, our world is once again experiencing another historical struggle from a novel coronavirus, namely SARS-CoV-2. As stated in a study, the origin of the virus started in Wuhan, China, around December 2019. Shortly after, the coronavirus became known as COVID-19 and had spread across nearby countries within a short period, and soon after, around March 2020, the World Health Organization (WHO) declared a global pandemic [1].
According to recent studies, COVID-19 can cause inflammation to the lungs that radiate common symptoms of high fever, dry cough, and fatigue. In some cases, patients may also experience the loss of taste, sore throat, and even skin rashes. However, due to the virus penetrating the lungs more than the other organs, people with weaker immune systems suffer from severe complications like shortness of breath, chest aches, and decreased mobility [2].
With the use of a diagnostic procedure, the real-time Reverse Transcription Polymerase Chain Reaction (rRT-PRC) can detect signs of COVID-19 from specimens acquired from nasopharyngeal or oropharyngeal swabs [3]. However, to perform such a test, the process requires a well-trained medical practitioner equipped with specialized equipment. Even today, some countries still lack the said expertise and diagnostic equipment, specifically in underdeveloped countries, and it continues to worsen as the spread of the virus progresses [4].
In such cases, due to the lack of the said testing equipment and personnel, like most common lung diseases, potential COVID-19 patients can also receive screenings through a lesser alternative like Chest X-Rays (CXR), compared to an rRT-PRC or a Computer Tomography (CT) scan. In some well-developed countries, having experts and the said technologies helps them cope with diagnosing COVID-19 in various methods. However, even with the X-rays' availability, the results still require high proficiency and experience to identify signs of COVID-19 infections accurately. Even for some expert radiologists, producing an accurate diagnosis specifically for COVID-19, is still complicated. The lack of such tends to cause slow, inaccurate, and higher examination costs [5,6].
Recently, technology progressed at an unimaginable rate. With the help of Computer-Aided Diagnosis (CAD), Deep Learning (DL), and Computer Vision (CV) methods, several studies produced ways to reduce such difficulty in the medical field. A study initiated by Wang et al. [7] proposed a solution using the mentioned methods to develop a Convolutional Neural Network (CNN) called the COVID-Net, explicitly designed to identify COVID-19 infected CXRs automatically from a curated dataset of various CXRs. Their proposed COVID-Net consisted of a lightweight residual pattern called Projection → Expansion → Projection → extension (PEPX). Their PEPX module utilizes 1 × 1 convolutional filters to address the image's lower dimensions at the first phase. With their added expansion method, the images expand differently from their original input form. A 3 × 3 depth-wise convolution contributes further by extracting additional image characteristics with less learning complexity and computational strain. The following projection phase then reverts the enlarged image to its previous dimension and expands the entire feature set's depth. With a test data of 300 images composed of three CXR classes, namely, Normal, COVID-19, and Pneumonia, their proposed COVID-Net achieved an overall accuracy of 93.3 % in detecting COVID-19 from CXRs. Their study also yielded better performance than VGG19 with 83.0 % and ResNet50 with 90.6 %.
Unlike COVID-Net, Al-Falluji resolved the same problem using a more extensive pre-trained Deep Convolutional Neural Network (DCNN) model, ResNet-18 [8]. Their proposed model had modifications to suit the given problem by adding a noise filter, a Global Average Pooling (GAP) layer, and compression layers. For an improved learning process, they used transfer learning to acquire readily available features to improve the model's image recognition and train it in a short period of only 30 epochs [9]. Such a method also improved their model's overall performance compared to the standard handcrafted feature extraction approach. Upon evaluation, their proposed modified ResNet-18 attained an accuracy of 96.73 % from the classification of non-infected, COVID-19, and Pneumonia CXRs.
Rather than relying on a single DCNN for the task of detecting COVID-19 from CXRs, Chowdhury et al. had the initiative to use an ensemble method [10]. Their study chose a recent DCNN model named EfficientNet as their base model. With transfer learning and fine-tuning, they managed to train several EfficientNet models to recognize several CXRs, including COVID-19 cases. Their trained EfficientNet models then classified samples of CXRs and had their prediction scores averaged as an ensemble. As stated in their results, their proposed ensembled EfficinetNet with a coefficient of three achieved 97 % accuracy with a low overhead demand than most state-of-the-art DCNNs that performed the same task.
In another study by Singh et al. [11], they proposed a modified structure of the Xception model comprised of six convolution layers and twelve depth-wise convolutions. Their proposed model trained for 100 epochs using specialized hardware, augmentation techniques, and hyper-parameter tuning. Their combined methods achieved a 95.80 % accuracy that outperformed other studies like the DarkCovidNet [12] with 87.02 %, DeTraC-ResNet18 [13] with 95.12 %, and a Hierarchical EfficientNetB3 [14] with 93.51 %.
Table 1 presents a summary of the discussed recent studies that performed the detection of COVID-19 infected CXRs. As discussed, the recent studies yielded exceptional performances towards the diagnosis of COVID-19 from CXRs using various methods and techniques. However, other methods can still induce improvements to reduce the computational cost and optimization efforts further while attaining better accuracy. Therefore, this work proposes a lightweight model using various techniques like network truncation, partial layer freezing, and feature fusion of a pre-trained DCNN model to diagnose COVID-19 infected CXRs.
Table 1.
Summary of studies that diagnosed COVID-19 chest x-rays with DCNNs.
| Model | Accuracy (%) | Classes | Type |
|---|---|---|---|
| COVID-Net [7] | 93.30 | Normal, COVID-19, Pneumonia | CXR |
| Modified ResNet-18 [8] | 96.37 | Normal, COVID-19, Pneumonia | CXR |
| ECOVNet-EfficientNetB3 base [10] | 97.00 | Normal, COVID-19, Pneumonia | CXR |
| Modified Xception [11] | 95.70 | Normal, COVID-19, Pneumonia | CXR |
| DarkCovidNet [12] | 87.02 | Normal, COVID-19, Pneumonia | CXR |
| DeTraC-ResNet18 [13] | 95.12 | Normal, COVID-19, SARS | CXR |
| Hierarchical EfficientNetB3 [14] | 93.51 | Normal, COVID-19, Pneumonia | CXR |
Primarily, this work strongly contributes based on the following:
-
•
This work truncated a pre-trained DCNN, the DenseNet121 model. The said method further reduced its network size and complexity without sacrificing a significant fraction of its performance. With that said, the truncated DenseNet attained a faster training time due to a shorter end-to-end structure while maintaining a rich extraction of relevant features.
-
•
Compared to other studies, this work fused a mirror image of a truncated DenseNet model, where the other half had its entire network fully re-trained from a curated CXR dataset with COVID-19 cases and ImageNet. On the other hand, the other half had its upper layers partially frozen to generate a different feature batch. Both models' distinct features are then fused right before the network's end, handled by a proposed set of layers equipped with regularization layers to prevent overfitting problems. This method provided a broad spectrum of diverse features compared to a traditionally trained DCNN model.
-
•
Unlike most existing works that performed the detection of COVID-19 from CXRs, this proposed method achieved remarkable results. It even outperformed other state-of-the-art models and studies without the need for heavy optimization, data augmentation, specialized hardware, long training epochs, ensemble methods, and an expensive computing cost. With that said, the proposed method can become easily deployable, reproducible, upgradeable, and most importantly, usable in future cases.
2. Materials and methods
2.1. Dataset collection and preparation
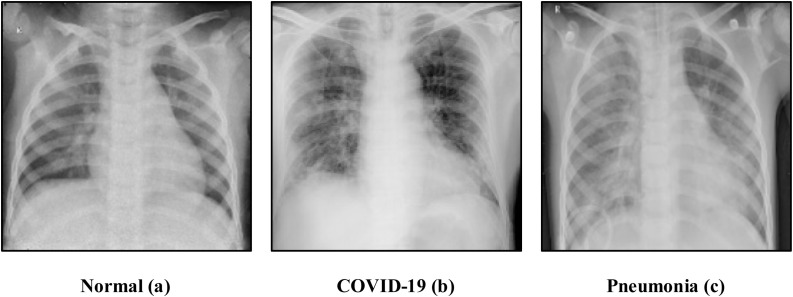
Due to the difficulty of acquiring CXRs, more or so samples with COVID-19, this work relied on a readily available curated dataset from Sait et al. [15]. Fig. 1 illustrates samples from the sourced dataset containing 3270 Normal (a), 1281 COVID-19 infected (b), and 4657 Pneumonia (c) CXR images.
Fig. 1.
Sample images of normal (a), covid-19 infected (b), and (c) pneumonia chest x-rays.
The images had no fixed dimensions as all came from various reliable sources. With that said, this work had all images resized and normalized into a 299 × 299 dimension, maintained to its Joint Photographic Group (JPG) format to prevent large consumptions of memory and to add better stability during both training and validation phases [16].
In Table 2 , the entire collection of 9208 CXR images had proper distributions to train and validate the proposed model, in which 80 % of the entire dataset became the train data and the remaining 20 % as the validation data selected stochastically to prevent the case of bias [17]. It is worth mentioning that this work relied only based on the presented number of CXR samples to showcase the proposed model's performance towards an unbalanced dataset without relying on data augmentation.
Table 2.
Specification of the curated dataset, with Normal, COVID-19, and Pneumonia chest x-rays.
| Class label | Train (80 %) | Validation (20 %) | Total (100 %) |
|---|---|---|---|
| Normal | 2616 | 654 | 3270 |
| COVID-19 | 1025 | 256 | 1281 |
| Pneumonia (Bacterial and Viral) | 3726 | 931 | 4657 |
| Total | 7367 | 1841 | 9208 |
2.2. Proposed model and construction
This section focuses on the proposed model's developmental process for the automated diagnosis of COVID-19 and the other mentioned CXRs. The following includes the DenseNet model's background that served as the base structure and concept, added with this work's proposed method that decreased the model's parameter size that still sufficiently generated a robust feature pool.
2.2.1. DenseNet
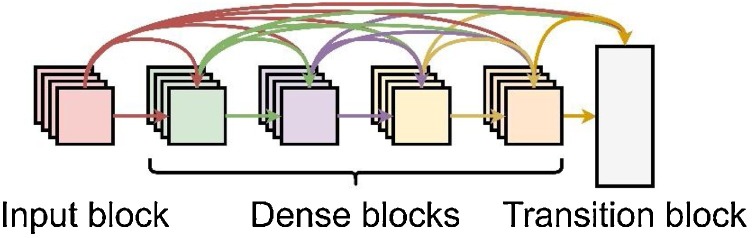
For the proposed model, this work selected to use a densely connected neural network, the DenseNet model, illustrated in Fig. 2 . The purpose of having a DenseNet model delivers a flawless propagation of features throughout the entire network without the saturation of performance, even with a more extensive depth. DenseNet also solves the parameter inflation through concatenation rather than the addition of layers [18].
Fig. 2.
A simplified visual concept of the DenseNet model [18].
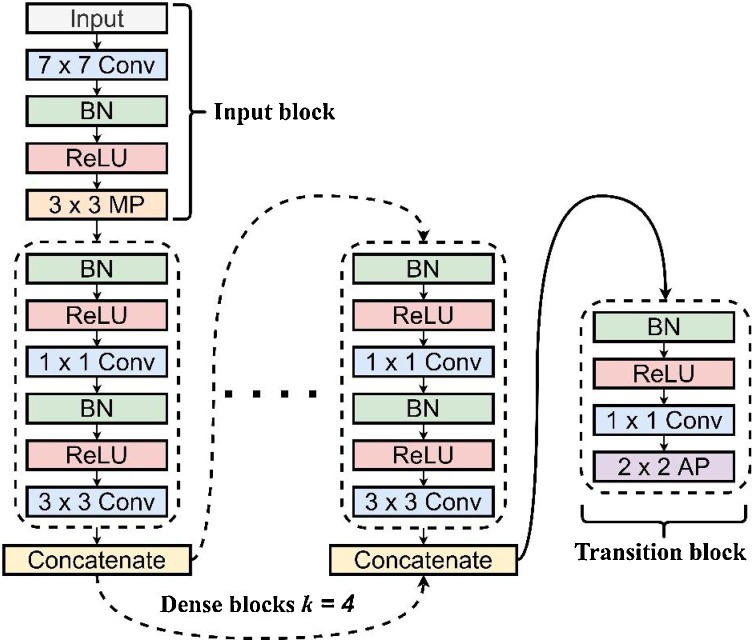
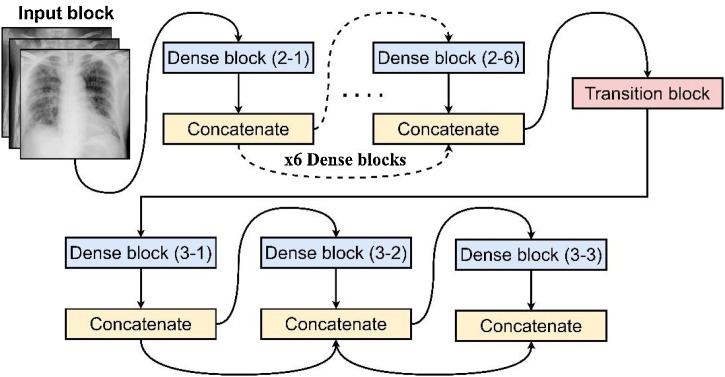
Fig. 3 illustrates an in-depth view of a 5-layer DenseNet with a k = 4 number of dense blocks. As presented, the DenseNet model begins with an input block (layer 1) composed of a 7 × 7 Convolutional (Conv) layer → Batch Normalization (BN) → Rectified Linear Unit Activation (ReLU) → 3 × 3 Max-Pooling (MP) layer. Subsequently, a set of dense blocks follows with a BN → ReLU → 1 × 1 Conv layer pattern followed by another BN → ReLU → 3 × 3 Conv. Unlike residual networks and other deep models consisting of massive parameters via feature summation, the DenseNet model uses a dense block with a k growth rate concatenated to all network layers. This technique became an effective transmission of feature inputs from the preceding layers towards the other succeeding layers end-to-end [18]. Such a design generates a rich gradient quality even at greater depths while sustaining a low parameter count, making it an ideal choice for this task.
Fig. 3.
A DenseNet model that presents the internal specifications of its dense block and transition layer.
Like most DCNN models, the DenseNet model requires a downsampling layer to prevent exhaustion of resources during feature extraction [18], where it utilizes a transition layer that downsamples the feature maps using a 1 × 1 Conv and a 2 × 2 Average Pooling (AP) with strides of 1 and 2, respectively.
2.2.2. Truncation method
Though the DenseNet model already has significantly fewer parameters than most DCNNs, the proposed method aims to reduce the parameters further without affecting too much of its performance, considering that even the smallest member of the DenseNet family, the DenseNet121, still has about 7 million parameters [18]. Since the goal of the original DenseNet revolves around massive datasets like the ImageNet with more than 14 million images and 1000 classes, training and reproducing this model can become tedious due to the lack of adequate computing resources. Also, with the limited dataset for this task, using the entire model's structure only adds complexity and consumes immense resources. Therefore, through a proposed truncation of its entire structure, most of its layers were removed, reducing the number of parameters and further shortened the end-to-end flow of features.
Fig. 4 presents the proposed truncated DenseNet-Tiny with only six dense blocks followed by a transition layer connected to another set of three dense blocks left. The dedicated output layers will then connect when the model had fused. The proposed design significantly decreased the parameter size and feature depth of the DenseNet121 model by 93 %. From the initial 8 million parameters, DenseNet121, the truncated DenseNet-Tiny had reduced its parameters to only about half a million.
Fig. 4.
The structure of the proposed truncated DenseNet-Tiny.
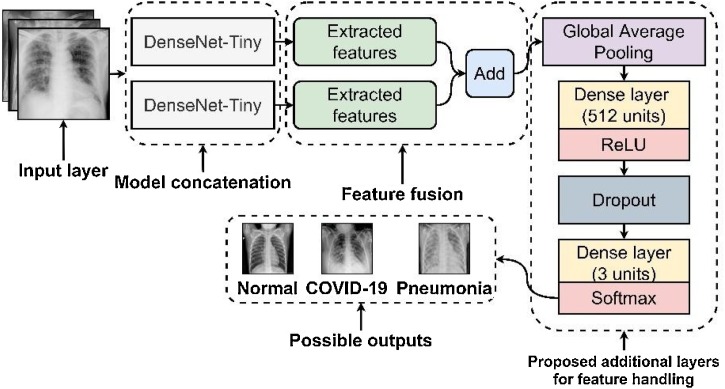
2.2.3. Model concatenation and feature fusion
Due to the reduced network size of the truncated DenseNet-Tiny, it resulted in a smaller parameter size. In this case, the relatively small parameter size emanated consequences that affected the DenseNet-Tiny's performance in recognizing the three classes of CXRs effectively [19]. However, putting additional depth of layers can only make the truncation method pointless and distort the extraction of features.
Therefore, an additional proposed method referred to as model concatenation [20] and feature fusion [21] provided insights that solved this problem. In Fig. 5 , a mirror image of the DenseNet-Tiny with the exact specifications combined to form the Fused-DenseNet-Tiny pipeline. Through model concatenation and feature fusion, the Fused-DenseNet-Tiny became wider instead of longer, maintaining the desired rapid end-to-end extraction of features during training and validation. Also, to add better support in handling the robust features produced by the fused model, this work added a new set of layers consisting of a GAP [22], Dense with 512 units with a ReLU activation, and Dropout [23] connected to another Dense layer with three units activated by a Softmax classifier [24]. The added layers aim to induce an improved performance and regularization that prevents overfitting issues [25], potentially allowing the model to perform better even with the provided unseen validation dataset and other future real-world CXR images.
Fig. 5.
The blueprint of the proposed Fused-DenseNet-Tiny.
Such a practical and less tedious solution provides an advantage of improving the feature production without designing or using another model that can re-initiate complexity in the network.
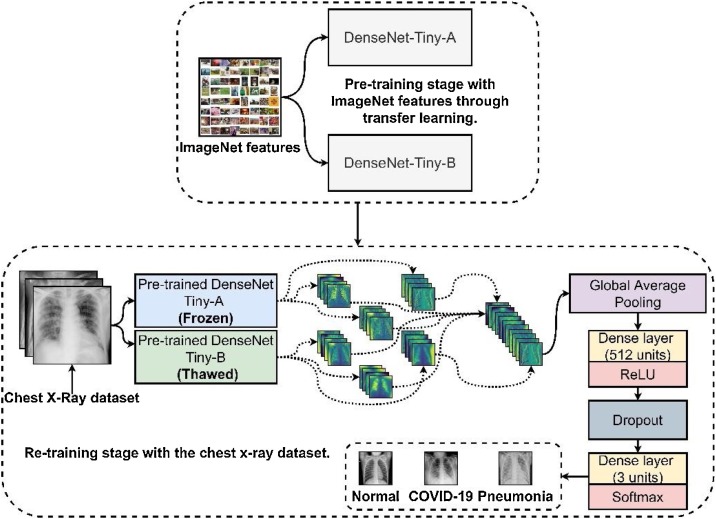
2.2.4. Transfer learning, fine-tuning, and partial layer freezing
Due to the scarcity of the desired CXR images for training, this work employed both transfer learning [9] and fine-tuning [26] to supply additional features for both DenseNet-Tinys. However, transferring the same set of features from ImageNet for both pipelines may only produce redundant features. Therefore, this work employed a fine-tuning technique that partially froze specific layers in one of the mirrored DenseNet-Tiny's network to produce independent features [27].
Fig. 6 illustrates that after the transfer learning process, instead of allowing both DenseNet-Tinys to re-train and extract features from the CXR dataset entirely, the other half had its extraction layers frozen. Unlike other studies, this work had a mirrored version fused, dubbed as the DenseNet-Tiny-B, where all its layers thawed and re-trained simultaneously to received newly generated features, contrasting to its other half, the DenseNet-Tiny-A. With that said, the DenseNet-Tiny-B produced a distinct set of shared features using the ImageNet and the CXR dataset, while the other DenseNet-Tiny-A preserved most of the ImageNet features on its upper layers and only had its lower dense layers trained with the CXR dataset.
Fig. 6.
The transfer learning and partial layer freezing framework to train the Fused-DenseNet-Tiny.
With the two similar models having a different set of features fused, the proposed model's feature pool became robust even with fewer parameters involved.
2.3. Model compilation and training
Before the actual training process began, the model had its hyper-parameters and a loss function selected. Hyper-parameters represent the tunable parts of a DL model that can affect most of its learning procedure, which cannot be tweaked during training [28]. Together, an added loss function calculates and reduces errors during both the train and validation phases. For an efficient result, an appropriate selection of hyper-parameters and loss function is imperative. It is worth mentioning that, unlike other studies, this work did not perform any stringent optimization techniques to tune its hyper-parameters. Therefore, providing less assistance to train the model to prove its ease of reproducibility and adaptive ability towards an imported dataset.
2.3.1. Hyper-parameters
The tuned hyper-parameters of the model consisted of the Learning Rate (LR), Batch Size (BS), optimizer, Dropout Rate (DR), and epochs. In selection, the following values provided in Table 3 produced the best results during the experiments. A BS of 16 gave the model a fast yet non-exhausting training process combined with an Adam optimizer [29]. The selected optimizer recently had tremendous success and started to become a de facto algorithm to optimize most DCNN models that involved medical images [30]. Also, Adam attains a faster convergence with less memory consumption compared to the likes of a Stochastic Gradient Descent (SGD) [31] and RMSProp [32], making it a fitting candidate for this work. The LR had a value of 0.0001 that worked well with the rest of the selected hyper-parameters. Included, the DR of 0.5 provided an adequate regularization that reduced the size of dense units at random during learning that prevented the model from overfitting due to the robust flow of features [33].
Table 3.
Selected hyper-parameters to train the Fused-DenseNet-Tiny.
| Hyper-Parameter | Value |
|---|---|
| LR | 0.0001 |
| BS | 16 |
| Optimizer | Adam |
| DR | 0.5 |
| Epochs | 25 |
2.3.2. Loss function
Selecting an appropriate loss function helps the model identify errors efficiently by calculating the dissimilarities between a predicted and a ground truth class [34].
The proposed model with three CXR classes went with a Categorical Cross-Entropy (CCE) loss function that worked effectively with the Softmax classifier [35], considering it already falls under a multi-class task.
In Eq. (1), M represents the three classes, Normal, COVID-19, and Pneumonia. Each class had its loss calculated separately upon each observation o from the ground truth label y that identified if its prediction p of sample c is correct [36].
| (1) |
3. Experimental results and discussion
In DL, proper evaluation using the appropriate metrics can determine how well a model performs according to a specific task. In this section, the Fused-DenseNet-Tiny undergoes a series of evaluations to validate its overall performance in diagnosing the three CXRs: Normal, COVID-19, and Pneumonia. As mentioned, this work used 20 % of the dataset or 1841 images for validation.
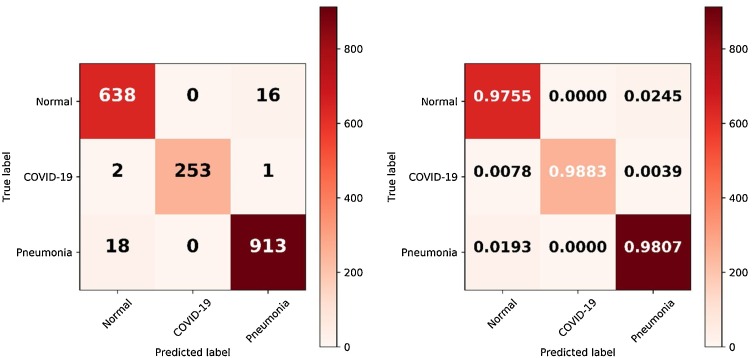
3.1. Confusion matrix
For a visual interpretation of how well the Fused-DenseNet-Tiny classified the validation samples individually, this work used a Confusion Matrix [37]. As illustrated in Fig. 7 , both confusion matrices consist of a True label or the ground truth, representing each image sample's labels, while the Predicted label represents the diagnosed samples. In which the CM uses a True Positive (TP), True Negative (TN), False Positive (FP), and a False Negative (FN) to identify whether the model had a correct or incorrect diagnosis of specific CXR. A TP indicates that the model correctly diagnosed a COVID-19 or Pneumonia CXR. In contrast, a TN implies that the model diagnosed a Normal CXR as Normal. On the other hand, the FP means that the model diagnosed a Normal CXR as any of the two infections, while FN diagnosed an infected CXR as Normal.
Fig. 7.
The classification results of the Fused-DenseNet-Tiny visualized with a confusion matrix.
Upon evaluation, the Fused-DenseNet-Tiny had the most difficulty diagnosing Normal CXRs with 16 FP CXRs, having 2.45 % of its entire set of samples diagnosed as Pneumonia. In terms of COVID-19, the model had a remarkable performance with only two FN and one FP, achieving 98.83 % correct diagnoses from all the COVID-19 CXRs. Simultaneously, the proposed model had 18 FN of Pneumonia CXRs being Normal and having the model diagnose the entire 98.07 % of the Pneumonia CXRs correctly.
3.2. Sensitivity and specificity
From a medical standpoint, a sensitivity test serves as an essential metric to indicate the correctness of identifying a diseased person, while a specificity test pertains the other way around of identifying a person without a disease [38]. In DL, such a concept delivers a similar objective [39].
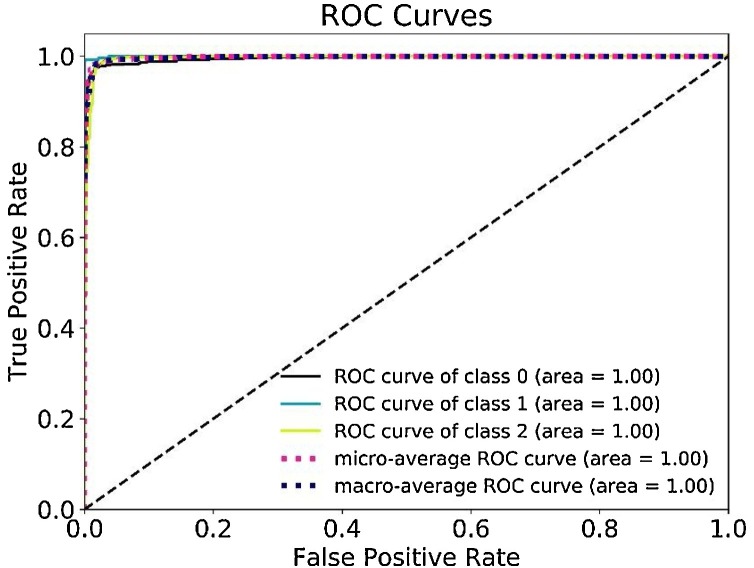
Illustrated in Fig. 8 , using a Receiver Operating Characteristic (ROC) curve and its Area Under the Curve (AUC), this work identified the performance of the Fused-DenseNet-Tiny based on sensitivity and specificity test in a varying threshold. ROCs with a larger AUC indicate better performance, whereas an AUC < 0.5 implies that the model merely conducted guesses and has no discrimination capability to diagnose the CXRs, making it unreliable [40].
Fig. 8.
The Receiver Operating Characteristic and its Area Under the Curve.
As evaluated, the proposed model achieved a remarkable performance in sensitivity and specificity with a uniformed 1.00 AUC for all three classes of CXRs.
3.3. Precision and recall
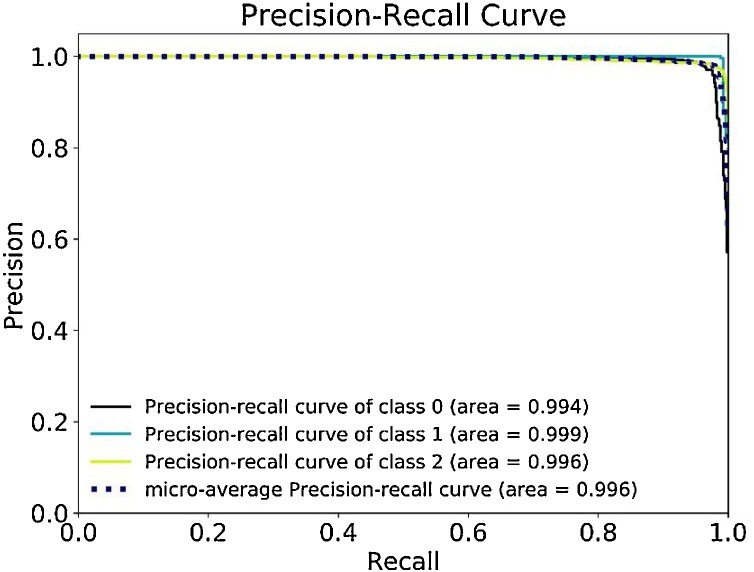
In situations like this work with unbalanced dataset samples, a Precision-Recall (P-R) curve becomes a critical evaluation tool to determine FP and FN rates yielded through a discriminating threshold. In similar terms with the ROC, the P-R curve also uses an AUC for evaluation, whereas a smaller AUC indicates that the model is more prone to false predictions [41].
From the given illustration in Fig. 9 , the diagnosis for the Normal CXR had a 0.994 AUC, while the COVID-19 had 0.999, Pneumonia with 0.996, rendering a micro-average of 0.996 AUC. Even with the slight fluctuations from the unbalanced distribution of validation samples, the model still efficiently handled the diagnostic process as seen on the P–R curve.
Fig. 9.
The Precision-Recall curve and its Area Under the Curve.
3.4. Saliency maps
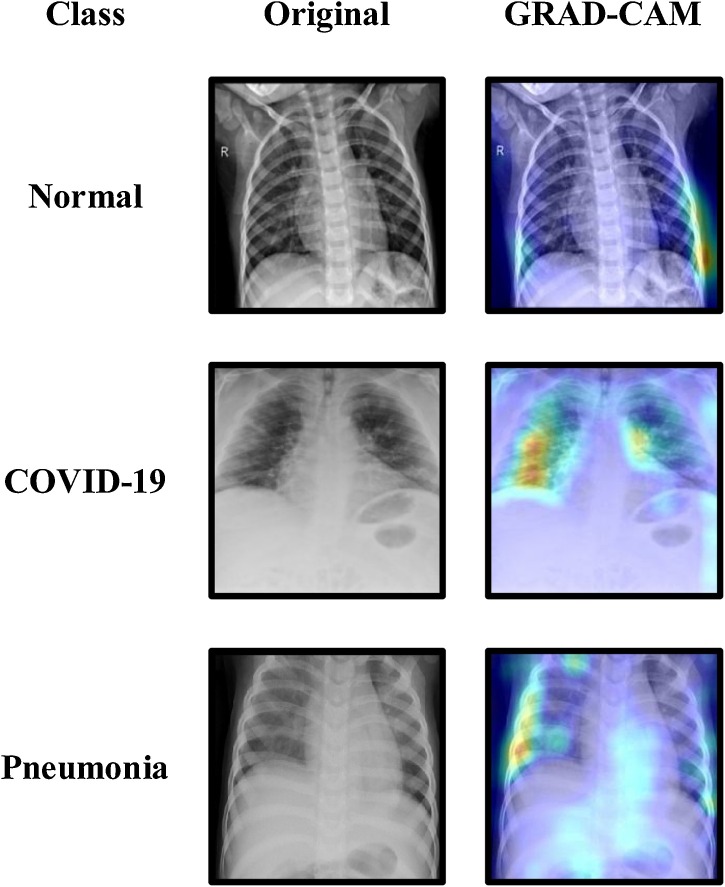
For added transparency and visualization, this work used the Gradient-Weighted Class Activation Maps (GRAD-CAM) algorithm that functions by capturing a specific class's vital features from the last Conv layer of a CNN model to localize its important regions [42].
From the given samples in Fig. 10 , using the GRAD-CAM algorithm with the Fused-DenseNet-Tiny had its salient areas identified. The highlighted patched areas indicated the most salient features that the model believes to be important for diagnosing each CXR accordingly. The proposed model also shows that it did not rely on random guesses during its diagnosis. Though not precisely what some medical experts might expect, the GRAD-CAM still has its limitations, as it cannot entirely capture the desired areas without flaws [43]. Nonetheless, having an algorithm to visualize what CNNs like the Fused-DenseNet-Tiny sees emanates better trust and reliability. It is worth mentioning that the GRAD-CAM did not affect any of the proposed models' workability as it only served as a visualization tool for this work.
Fig. 10.
Gradient-Weighted Class Activation Maps of the Fused-DenseNet-Tiny.
3.5. Classification performance
For an overall identification of the Fused-DenseNet-Tiny's performance in diagnosing the three distinct CXRs, the use of the following metrics evaluated its critical points. The following includes the accuracy, precision, recall, and f1-score, calculated based on the following equations below [44].
| (2) |
| (3) |
| (4) |
| (5) |
Table 4 presents the overall performance of the Fused-DenseNet-Tiny in diagnosing 1841 samples of Normal, COVID-19, and Pneumonia CXRs. Upon evaluation, the Fused-DenseNet-Tiny achieved a reliable performance of 99.84 % accuracy with COVID-19, 98.10 % with Pneumonia, and 98.04 % with Normal CXRs. In terms of precision, recall, and f1-score, the Fused-DenseNet-Tiny model performed considerably well even with the unbalanced data and had no perceptible signs of bias or class superiority issues as reflected from the given and previous results.
Table 4.
Performance of the trained Fused-DenseNet-Tiny in diagnosing the chest x-rays.
| Classes | Accuracy (%) | Precision | Recall | F1-score | Sample size |
|---|---|---|---|---|---|
| Normal | 98.04 | 0.98 | 0.97 | 0.97 | 654 |
| COVID-19 | 99.84 | 0.99 | 1.00 | 0.99 | 256 |
| Pneumonia | 98.10 | 0.98 | 0.98 | 0.98 | 931 |
3.6. Discussion
The Fused-DenseNet-Tiny had proven to perform efficiently, even with fewer parameters and less depth based on the evaluated results. However, to further showcase its contribution and improvements, this work compared its overall accuracy with other state-of-the-art models and studies that performed a similar diagnosis of COVID-19 in CXRs.
From the presented results in Table 5 , the proposed Fused-DenseNet-Tiny achieved the highest accuracy of 97.99 % without the employment of hyper-parameter optimization, data augmentation, lengthy training epochs, ensembling, and any specialized hardware. It is worth mentioning that the proposed Fused-DenseNet-Tiny only performed under a standard GTX 1070 GPU (released 2016), an i5 4th generation CPU, and 16GB of RAM.
Table 5.
Performance comparison of the proposed Fused-DenseNet-Tiny with other studies.
| Model | Accuracy (%) | Classes | Type |
|---|---|---|---|
| Fused-DenseNet-Tiny (this work) | 97.99 | Normal, COVID-19, Pneumonia | CXR |
| COVID-Net [7] | 93.30 | Normal, COVID-19, Pneumonia | CXR |
| Modified ResNet-18 [8] | 96.37 | Normal, COVID-19, Pneumonia | CXR |
| ECOVNet-EfficientNetB3 base [10] | 97.00 | Normal, COVID-19, Pneumonia | CXR |
| Modified Xception [11] | 95.70 | Normal, COVID-19, Pneumonia | CXR |
| DarkCovidNet [12] | 87.02 | Normal, COVID-19, Pneumonia | CXR |
| DeTraC-ResNet18 [13] | 95.12 | Normal, COVID-19, SARS | CXR |
| Hierarchical EfficientNetB3 [14] | 93.51 | Normal, COVID-19, Pneumonia | CXR |
Furthermore, this work also trained several state-of-the-art DCNNs with the same dataset. Table 6 shows that the proposed Fused-DenseNet-Tiny did not attain the highest overall performance rating due to its lesser and more straightforward feature learning process. However, compared with the significantly larger DenseNet121 that attained the highest accuracy of 98.48 %, the Fused-DenseNet-Tiny only falls 0.49 % less accurate with 97.99 %.
Table 6.
Comparison of performance with other state-of-the-art models.
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| DenseNet121 [18] | 98.48 | 98.71 | 98.59 | 98.48 |
| EfficientNetB0 [45] | 98.21 | 98.59 | 98.18 | 98.39 |
| Fused-DenseNet-Tiny (this work) | 97.99 | 98.38 | 98.15 | 98.26 |
| InceptionV3 [46] | 97.99 | 98.31 | 98.23 | 98.26 |
| ResNet152V2 [47] | 97.88 | 98.25 | 98.09 | 98.17 |
| Xception [48] | 97.61 | 97.92 | 97.83 | 97.87 |
| MobileNetV2 [49] | 97.12 | 97.46 | 97.75 | 97.58 |
| VGG16 [50] | 96.58 | 97.06 | 96.94 | 96.97 |
| InceptionResNetV2 [51] | 96.14 | 94.48 | 96.90 | 95.59 |
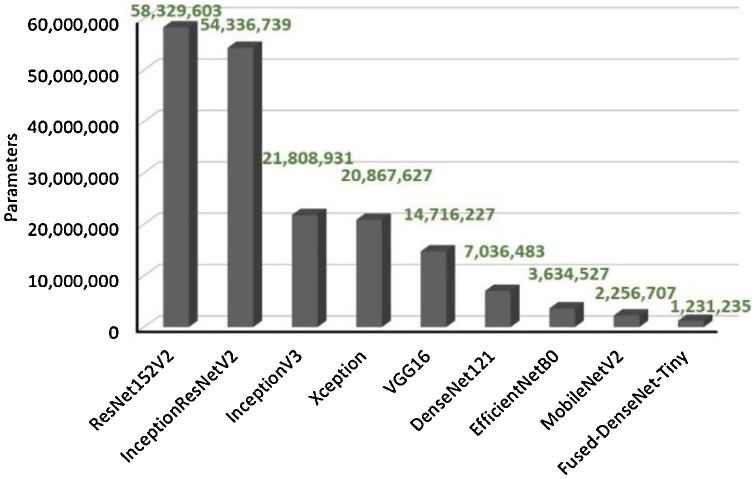
Even though the Fused-DenseNet-Tiny fell short in attaining the highest accuracy in this work, in terms of the parameter size, the Fused-DenseNet-Tiny had shown an advantage over the rest of the presented state-of-the-art models having only around 1.2 million, as shown in Fig. 11 . Convincingly, the Fused-DenseNet-Tiny, even with a slightly minimal performance difference compared to InceptionV3, EfficientNetB0, and DenseNet121, still garnered a remarkable performance to size ratio.
Fig. 11.
The parameter sizes of the Fused-DenseNet-Tiny and other state-of-the-art models.
As presented by the calculated results throughout this article, the proposed method to re-structure and train a state-of-the-art DCNN model like the DenseNet can considerably maintain most of its performance and save a large disk capacity and computing cost simultaneously. Such a solution can become easily deployable in smaller or lower-performing machines, including mobile devices, giving developing countries and locations that cannot confidently perform tests or diagnose CXRs infected by COVID-19 an opportunity to emanate better performance. Medical experts and the likes can gain more confidence in conducting their diagnosis and not require costly equipment through the proposed DL-based solution's assistance [52].
It is essential to know that even with the increase of training data, the model's weight size would not inflate compared to re-structuring and adding layers, making it conveniently scalable in most situations. Adding layers or depth also does not mean an increase in performance [53,54]. The Fused-DenseNet-Tiny and its small parameter size and dense structure can train faster even with more data than larger state-of-the-art models that train with lesser data, thus yielding immediate improvements from additional reliable data in the future.
4. Conclusion
Due to the relentless spread of COVID-19 infections, mass testing became an essential aspect of most people's lives today. However, the gold standard testing procedure like rRT-PCR requires specialized testing equipment and a trained medical practitioner. Even with a lesser substitute like CXRs, a rapid and less expensive method to rule out the infections of COVID-19 still induces complexity in most underdeveloped countries. Hence, people started to automate such a difficult task through DL. This work served as an additional contribution with its lightweight yet efficient design that requires less effort to reproduce and does not require expensive equipment to help diagnose COVID-19 infections from CXRs.
As a result, the proposed model yielded a slight performance improvement but a massive decrease in computing cost and parameter size over other state-of-the-art models and existing studies. However, even with its lightweight design, a specific caveat still shows that the Fused-DenseNet-Tiny cannot outperform its larger counterpart due to its reduced extraction capabilities. Though not far in terms of performance, this work hypothesizes that hyper-parameter optimization, and the potential addition of more data may yield additional improvements that can alleviate such a problem in the future.
Nonetheless, even with minimal shortcomings, this work still concludes that the fusion of a mirrored truncated DenseNet, with its equivalent partially trained with ImageNet features and the other with shared weights from the CXR and ImageNet, the proposed model still performed efficiently even with less computing cost, data, and dependence to other sophisticated optimization methods towards the diagnosis of COVID-19 from CXRs.
Furthermore, the model can still induce further improvements upon applying the mentioned methods above, besides adding more data.
Authorship statement
I, Francis Jesmar P. Montalbo as the sole author of this paper, dedicated myself and time to the following.
-
•
Conceptualizing the study.
-
•
Acquiring, handling, and analyzing data.
-
•
Performing experiments, testing, and evaluating results.
-
•
Illustrating figures.
-
•
Drafting, revising, and finalizing the manuscript.
With this document, I certify and guarantee that this material has not been or will not be submitted or published in any other journal. This document was solely submitted to the Biomedical Signal Processing and Control journal with an ISSN of 1746–8094.
Ethical procedure
-
•
This research article meets all applicable standards with regards to the ethics of experimentation and research integrity, and the following is being certified/declared true.
-
•
As a researcher in my field of specialization, the paper has been submitted with full responsibility, following the due ethical procedure, and there is no duplicate publication, fraud, plagiarism, or concerns about animal or human experimentation.
Ethical approval
This work did not involve humans, animals, and other living specimens during experiments.
Code and data availability
The author provides the links to the source code and dataset used in this work → https://github.com/francismontalbo/fused-densenet-tiny
Dataset and code availability
For ease of evaluation, this work provides both the dataset source and code required to simulate the proposed model. https://github.com/francismontalbo/fused-densenet-tiny.
Acknowledgments
The author acknowledges Batangas State University for supporting this work. Without its help and support, this work would not have become possible.
Acknowledgments
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91(October):264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; Treasure Island, FL, USA: 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) [updated 2020 Apr 6]https://www.ncbi.nlm.nih.gov/books/NBK554776/ in StatPearls [Internet]Jan. [Online]. Available: [PubMed] [Google Scholar]
- 3.Li M., et al. Coronavirus disease (covid-19): spectrum of CT findings and temporal progression of the disease. Academic Radiol. 2020;27(5):603–608. doi: 10.1016/j.acra.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giri A., Rana D. Charting the challenges behind the testing of covid-19 in developing countries: nepal as a case study. J. Biosaf. Health Educ. 2020;2:53–56. doi: 10.1016/j.bsheal.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durrani M., Haq I., Kalsoom U., Yousaf A. Chest X-rays findings in COVID 19 patients at a University Teaching Hospital - A descriptive study. Pak. J. Med. Sci. 2020;36(19–4) doi: 10.12669/pjms.36.covid19-s4.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Costa Y.M.G. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020;(May):105532. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Lin Z., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R.A. Al-Falluji, Z.D. Katheeth, B. Alathari, Automatic Detection of COVID-19 Using Chest X-Ray Images and Modified ResNet18-Based Convolution Neural Networks, Computers, Materials, & Continua, pp. 1301-1313, doi: 10.32604/cmc.2020.013232.
- 9.Weiss K., Khoshgoftaar T.M., Wang D. A survey of transfer learning. J. Big Data. 2016;3(1):1–40. doi: 10.1186/s40537-016-0043-6. [DOI] [Google Scholar]
- 10.Garg A., Salehi S., La Rocca M., Garner R., Duncan D. 2020. Efficient and Visualizable Convolutional Neural Networks for COVID-19 Classification Using Chest CT.https://arxiv.org/abs/2012.11860 arXiv preprint arXiv:2012.11860 Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh K.K., Siddhartha M., Singh A. Diagnosis of coronavirus disease (covid-19) from chest x-ray images using modified xceptionnet. Romanian J. Inform. Sci. Technol. 2020;23:S91–105. https://romjist.ro/abstract-657.html Available: [Google Scholar]
- 12.Ozturk T., et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbas A., Abdelsamea M., Gaber M. Classification of covid19 in chest x-ray images using detrac deep convolutional neural network. medRxiv. 2020 doi: 10.1101/2020.03.30.20047456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luz E., Silva P.L., Silva R., Silva L., Moreira G., Menotti D. 2020. Towards an Effective and Efficient Deep Learning Model for COVID-19 Patterns Detection in X-ray Images.http://arxiv.org/abs/2004.05717 arXiv preprint arXiv:2004.05717 Available: [Google Scholar]
- 15.U. Sait, K.V. Gokul Lal, S.P. Prajapati, R. Bhaumik, T. Kumar, S. Sanjana, K. Bhalla, Curated Dataset for COVID-19 Posterior-Anterior Chest Radiography Images (X-Rays). Mendeley Data, V1, doi: 10.17632/9xkhgts2s6.1.
- 16.Sabottke C., Spieler B. The Effect of Image Resolution on Deep Learning in Radiography. Radiology: Artificial Intelligence. 2020;2(1):e190015. doi: 10.1148/ryai.2019190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reitermanova Z. Data splitting. Proc. WDS's Contributed Papers Matfyzpress Prague, Czech Republic, vol. 10. 2010:31–36. https://www.mff.cuni.cz/veda/konference/wds/proc/pdf10/WDS10_105_i1_Reitermanova.pdf Available: [Google Scholar]
- 18.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Honolulu, HI; 2017. pp. 2261–2269. [DOI] [Google Scholar]
- 19.Gorban A., Mirkes E., Tyukin I. How deep should be the depth of convolutional neural networks: a backyard dog case study. Cognit. Comput. 2019;12(2):388–397. doi: 10.1007/s12559-019-09667-7. [DOI] [Google Scholar]
- 20.Noreen N., Palaniappan S., Qayyum A., Ahmad I., Imran M., Shoaib M. A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access. 2020;8:55135–55144. doi: 10.1109/ACCESS.2020.2978629. [DOI] [Google Scholar]
- 21.Cheng Y., Feng J., Jia K. A lung disease classification based on feature fusion convolutional neural network with X-ray image enhancement. 2018 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC); Honolulu, HI, USA; 2018. pp. 2032–2035. [DOI] [Google Scholar]
- 22.Lin M., Chen Q., Yan S. 2013. Network in network.https://arxiv.org/abs/1312.4400 arXiv preprint arXiv:1312.4400, Available: [Google Scholar]
- 23.Dahl G.E., Sainath T.N., Hinton G.E. Improving deep neural networks for LVCSR using rectified linear units and dropout. 2013 IEEE International Conference on Acoustics, Speech and Signal Processing; Vancouver, BC; 2013. pp. 8609–8613. [DOI] [Google Scholar]
- 24.Khan A.I., Shah J., Bhat M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Programs Biomed. 2020;196(November) doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S.-H. Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf. Fusion. 2021;67:208–229. doi: 10.1016/j.inffus.2020.10.004. 2020/10/09/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K., Kim J., Jeon E., Choi W., Kim N., Lee K. Evaluation of scalability and degree of fine-tuning of deep convolutional neural networks for COVID-19 screening on chest x-ray images using explainable deep-learning algorithm. J. Pers. Med. 2020;10(4):213. doi: 10.3390/jpm10040213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isikdogan L.F., Nayak B.V., Wu C.T., Moreira J.P., Rao S., Michael G. In: Vedaldi A., Bischof H., Brox T., Frahm J.M., editors. Vol 12372. Springer, Cham; 2020. “SemifreddoNets: Partially Frozen Neural Networks for Efficient Computer Vision Systems,” In proc. (Computer Vision – ECCV 2020. ECCV 2020. Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 28.Yu T., Zhu H. 2020. Hyper-Parameter Optimization: A Review of Algorithms and Applications.https://arxiv.org/abs/2003.05689 arXiv preprint arXiv:2003.05689, Available: [Google Scholar]
- 29.Kingma D.P., Ba J.L. Adam: a method for stochastic optimization. Proc. Int. Conf. Learn. Represent. 2015:1–41. https://hdl.handle.net/11245/1.505367 Available: [Google Scholar]
- 30.Kandel I., Castelli M., Popovič A. Comparative study of first order optimizers for image classification using convolutional neural networks on histopathology images. J. Imaging. 2020;6(9):92. doi: 10.3390/jimaging6090092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketkar N. Deep Learning With Python. Apress; Berkeley, CA, USA: 2017. “Stochastic gradient descent. [DOI] [Google Scholar]
- 32.Tieleman T., Hinton G. COURSERA: Neural Networks for Machine Learning. 2012. Lecture 6.5—RmsProp: divide the gradient by a running average of its recent magnitude.https://www.cs.toronto.edu/∼hinton/coursera/lecture6/lec6.pdf Available: [Google Scholar]
- 33.Srivastava N., et al. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(1):1929–1958. doi: 10.5555/2627435.2670313. [DOI] [Google Scholar]
- 34.Qu Z., Mei J., Liu L., Zhou D. Crack detection of concrete pavement with cross-entropy loss function and improved VGG16 network model. IEEE Access. 2020;8:54564–54573. doi: 10.1109/ACCESS.2020.2981561. [DOI] [Google Scholar]
- 35.Roelants P. Softmax classification with cross-entropy. Peterroelants.github.io. 2021 [Online] Available: https://peterroelants.github.io/posts/cross-entropy-softmax/ [Accessed: 28- Dec- 2020] [Google Scholar]
- 36.ML Cheatsheet, c2017. [Online]. Available: https://mlcheatsheet.readthedocs.io/en/latest/loss_functions.html. [Accessed: 15- Dec- 2020].
- 37.Ting K.M. Springer; Boston, MA, USA: 2017. Confusion Matrix; p. 260. [DOI] [Google Scholar]
- 38.Filleron T. Comparing sensitivity and specificity of medical imaging tests when verification bias is present: The concept of relative diagnostic accuracy. Eur. J. Radiol. 2018;98:32–35. doi: 10.1016/j.ejrad.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 39.Lee C.S., Baughman D.M., Lee A.Y. Deep learning is effective for classifying normal versus age-related macular degeneration optical coherence tomography images. Ophthalmol. Retina. 2017;124(8):1090–1095. doi: 10.1016/j.oret.2016.12.009. [DOI] [Google Scholar]
- 40.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J. Internal Med. 2013;4(2):627–635. https://pubmed.ncbi.nlm.nih.gov/24009950/ Available: [PMC free article] [PubMed] [Google Scholar]
- 41.Jeni L.A., Cohn J.F., De La Torre F. Facing imbalanced data—recommendations for the use of performance metrics. 2013 Humaine Association Conference on Affective Computing and Intelligent Interaction; Geneva; 2013. pp. 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. 2017 IEEE International Conference on Computer Vision (ICCV); Venice; 2017. pp. 618–626. [DOI] [Google Scholar]
- 43.Chattopadhay A., Sarkar A., Howlader P., Balasubramanian V.N. Grad-CAM++: generalized gradient-based visual explanations for deep convolutional networks. 2018 IEEE Winter Conference on Applications of Computer Vision (WACV); Lake Tahoe, NV; 2018. pp. 839–847. [DOI] [Google Scholar]
- 44.Hossin M., Sulaiman M.N. A review on evaluation metrics for data classification evaluations. Int. J. Data Mining Knowl. Manage. Process. 2015;5(2):1–11. doi: 10.5121/ijdkp.2015.5201. [DOI] [Google Scholar]
- 45.Tan M., Le Q. EfficientNet: rethinking model scaling for convolutional neural networks. Proc. 36th Int. Conf. Mach. Learn. 2019:6105–6114. http://proceedings.mlr.press/v97/tan19a.html Available: [Google Scholar]
- 46.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the inception architecture for computer vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, NV; 2016. pp. 2818–2826. [DOI] [Google Scholar]
- 47.He K., Zhang X., Ren S., Sun J. Vol. 9908. Springer, Cham; 2017. Identity mappings in deep residual networks. (ECCV, 2016, Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 48.Chollet F. Xception: deep learning with depthwise separable convolutions. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Honolulu, HI; 2017. pp. 1800–1807. [DOI] [Google Scholar]
- 49.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L. MobileNetV2: inverted residuals and linear bottlenecks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; Salt Lake City, UT; 2018. pp. 4510–4520. [DOI] [Google Scholar]
- 50.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. Proc. Int. Conf. Learn. Representations. 2015 https://arxiv.org/abs/1409.1556 Available: [Google Scholar]
- 51.Szegedy C., Ioffe S., Vanhoucke V., Alemi A. vol. 31. 2017. Inception-v4, inception-ResNet and the impact of residual connections on learning; p. 1.https://ojs.aaai.org/index.php/AAAI/article/view/11231 (Proceedings of the AAAI Conference on Artificial Intelligence). Available: [Google Scholar]
- 52.Sarkodie B.D., Osei-Poku K., Brakohiapa E. Diagnosing COVID-19 from chest X-ray in resource limited environment-case report. Int. Med. Case Rep. J. 2020;6(2):135. doi: 10.36648/2471-8041.6.2.135. [DOI] [Google Scholar]
- 53.Tran T.Lee, Kim J. Increasing neurons or deepening layers in forecasting maximum temperature time series? Atmosphere. 2020;11(10):1072. doi: 10.3390/atmos11101072. [DOI] [Google Scholar]
- 54.Google Developers; 2021. The Size and Quality of a Data Set. [Online]. Available: https://developers.google.com/machine-learning/data-prep/construct/collect/data-size-quality [Accessed: 20-Mar-2021] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The author provides the links to the source code and dataset used in this work → https://github.com/francismontalbo/fused-densenet-tiny