Abstract
Objectives
The aim of the study was to evaluate completeness and timeliness of the rapidly developed surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in England using patient-level data.
Study design
This is an observational study wherein public health surveillance systems are evaluated.
Methods
Data were collected in the Public Health England's Second-Generation Surveillance System through routine laboratory reporting processes, as well as via enhanced testing in collaboration with commercial partners. Three periods were chosen to present developments in disease surveillance around the first pandemic wave in England. Completeness of valid entries for key demographic and epidemiological fields was summarised. Timeliness was assessed using recorded date intervals: from sample collection to the laboratory reporting a positive result, the positive result being received by the national surveillance system and the data being available for epidemiological analysis.
Results
In each period, demographic variables were more than 95% complete and enhanced ethnicity more than 85%, allowing a rich understanding of the general characteristics of COVID-19 cases in England. The proportion of cases completing all reporting stages of the national system within 3 days of when the specimen was taken increased from 69.1% in period 1 to 76.6% in period 3. In period 3, the median number of days to complete all reporting stages decreased to 2, from 3 in previous periods. Analysis of each reporting stage offers suggestive evidence that timeliness of the system has improved as reporting has become established over time.
Conclusions
Timely processing of data for epidemiological use was consistent and rapid once received by the national system. Delays in timeliness were most likely to occur in the first stage of the reporting process, before laboratory input to the surveillance platform. Existing national surveillance mechanisms enhanced during the response have succeeded in providing rapid collection and reporting of case data to facilitate epidemiological monitoring and analysis and guide public health policy and strategy.
Keywords: COVID-19, SARS-CoV-2, Surveillance, Evaluation, Epidemiology, Infectious diseases, Public health, Respiratory, SARS
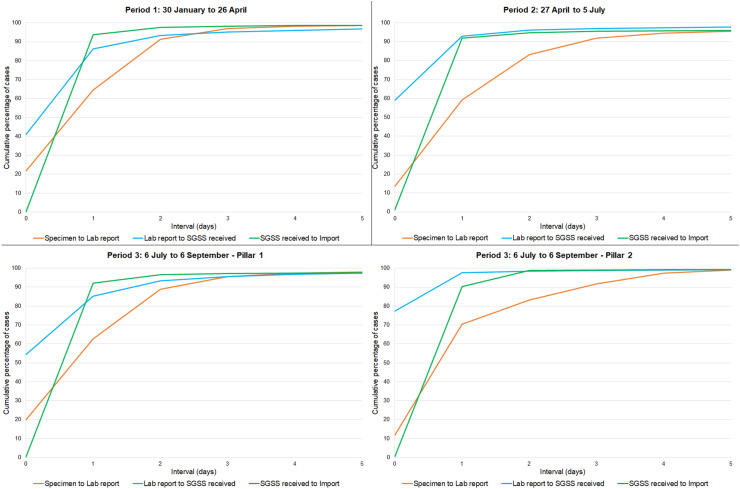
Surveillance of the novel coronavirus disease, COVID-19, was escalated in England in early 2020, with initial cases reported in January 2020.1 Rapid detection of new incident cases was a key priority, and initial processes were built into existing laboratory reporting systems including the Second-Generation Surveillance System (SGSS) and Respiratory DataMart.2 Urgent need to improve case ascertainment, as well as to alleviate testing capacity challenges, resulted in the UK government's deployment of a strategy to scale up testing for COVID-19 in April 2020.3 This policy referred to testing ‘pillars’, with three pillars that contributed to detection of cases with current infection: pillar 1, aiming to strengthens established testing pathways, such as National Health Service (NHS) and Public Health England (PHE) laboratories; pillar 2, initiating testing capacity through commercial partners, and pillar 4, swab testing for surveillance studies. This expansion of testing aimed to provide more rapid results to improve data collection to better understand the epidemiological characteristics of infection and to support key workers' ability to return to work with reduced risk. Based on key priorities of data completeness and timeliness, we evaluated the rapidly developed and expanded laboratory surveillance system for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) around the first pandemic wave in England (see Fig. 1 ).
Fig. 1.
Time interval between reporting stages, by period. SGSS, Second-Generation Surveillance System.
Data on laboratory-confirmed cases of SARS-CoV-2 infection are legally required to be submitted to PHE by the operators of diagnostic laboratories; submitted laboratory data are managed within the SGSS. Three periods were chosen to present developments in the surveillance system's timeliness and completeness. Cases were assigned to a period by laboratory report date, which has 100% completeness and validity. The periods were January 30–April 26 (the set-up period; ends with the week with the highest number of cases reported), April 27–July 5 (the peak, including some of the highest testing demand and rapid escalation of new systems; ends at the low point of reported cases after the peak) and July 6–September 6 (beginning of the post–first-wave period, as defined in national surveillance reporting).4
Some criteria of this analysis were assessed based on the reporting pillar. It is important to note that pillar testing stratification mainly defines the reporting pathways and may not always represent homogenous populations. Pillar 1 includes testing of patients in hospitals (through routine diagnostic investigations or due to COVID-19 symptoms), as well as testing of healthcare and social care workers. Pillar 2 testing broadly represents community testing in the wider population, including mildly symptomatic cases and testing from mobile units. Both pillar 1 and pillar 2 contain some outbreak investigations and care home testing, wherein reporting is based on whether the testing is carried out by a PHE/NHS (pillar 1) or commercially contracted (pillar 2) laboratory. Pillar 4 swab tests can be reported into either pillar 1 or pillar 2 depending on the diagnostic laboratory contracted for the study, and pillar 4 results are not consistently distinguishable within the surveillance system. While pillar 1 is built upon existing laboratory reporting pathways with established data flows, pillar 2 and 4 required new processes to be created for both data collection and submission. This can lead to differences in both timeliness and completeness of reported data fields by the reporting pillar.
Key demographic and epidemiological fields were reviewed for completeness, and the percentage of records containing valid entries was summarised for each period. These fields included surname, forename, sex, date of birth, NHS number, residential postcode and ethnicity as well as epidemiological measures such as the date of symptom onset, hospital-acquired infection, travel exposure and symptom status indicators.
All data apart from the residential postcode and ethnicity fields are unmodified from the SGSS. Data recorded explicitly as ‘Unknown’ or as a default value (i.e., 01/01/1900) were classified as missing. Data on ethnicity were obtained from the NHS Digital Hospital Episode Statistics (HES) and Secondary Uses Service (SUS) databases.5 , 6 Ethnicity assignment follows the same process as HES-Office for National Statistics mortality linkage, whereby personal identifiers (NHS number, sex, age and postcode) from HES and SUS are linked to people testing positive for COVID-19 in an iterative manner as per eight predefined matching criteria.7 Where there are differing ethnicities for the same personally identifiable information, priority is given based on (a) a valid ethnicity (i.e., not including ‘Unknown’ or ‘Prefer not to say’), (b) the most recent date and (c) higher ranked data sets. The data sets are ranked, starting with the highest, as follows: SUS live feed, HES Admitted Patient Care, Outpatient HES and HES Accident and Emergency. Where this linkage did not result in a valid ethnicity for cases reported through pillar 2, the self-reported pillar 2 ethnicity was used. Postcodes that were indicated as being populated with laboratory or GP default information were considered missing for the purpose of assigning patient residential information.
Timeliness was assessed using four key date fields to construct three intervals: (a) from specimen date to laboratory report date, which is the time between the sample being collected and the laboratory reporting positive results to its systems; (b) from laboratory report date to SGSS receipt date, indicating the time taken from the positive result being available to the result being received by the national surveillance system; and (c) from SGSS receipt date to import date, the time between receipt in the SGSS and the data being imported so that it can be used by epidemiologists, statisticians and modellers. Some of these intervals occur on the same day; for instance, intervals 2 and 3 could occur on the same day. The timeliness analysis included only case records from April 14 for pillar 1 and from May 26 for pillar 2 owing to limitations on available date fields before then, and the end of the analysis period was September 6, 2020.
There were 303,082 cases that met the inclusion criteria for this analysis: 125,779 cases in period 1, 120,403 in period 2 and 56,900 in period 3. Completeness of these data is described in Table 1. Demographic variables, including name, sex, postcode and date of birth, were more than 95% complete in each period, and ethnicity was more than 85% complete owing to the enhancement process. This is a detailed demographic data set allowing a rich understanding of the general characteristics of COVID-19 cases in England, as demonstrated in its use informing the evidence base and in the wider public health literature across various mediums. Examples include daily dissemination of data to local and national public health to inform policy decision-making (including local public health restrictions),8 , 9 modelling to provide forecasting and tracking of the pandemic in real time,10, 11, 12 routine surveillance reporting of official statistics4 , 13 and peer-reviewed research.14 , 15
The least complete demographic field was the NHS number, an identifier linked to a patient's electronic health record. This field is routinely enhanced in the SGSS through matching to the Demographic Batch Service.16 Low completeness is likely due to matching requiring a high level of precision that is not always available for self-reported information (such as through pillar 2). Completeness for this field decreased across the study period from 92.9% to 80.8%. While part of this decrease reflects an increasing proportion of national COVID-19 cases being reported through the pillar 2 reporting pathway, pillar 1 completeness also decreased from approximately 94% in periods 1 and 2 to 80.9% in period 3.
Key epidemiological surveillance variables reported by laboratories were mainly incomplete. Availability of the date of symptom onset decreased from 2.2% to 0.2% from period 1 to 3, as the proportion of cases detected through pillar 2 increased, with almost entirely incomplete data for this field, after its inclusion in pillar 2 data collection in May. The asymptomatic indicator has shown the greatest completeness improvement, increasing from 1.4% to 88.5% across the analysis periods. This is almost entirely due to improvements in completeness for pillar 2 testing, in which this became a mandatory variable in late June 2020. Other indicators, such as travel exposure and hospital-acquired infection status, were generally unavailable through pillar 1 and not collected through pillar 2.
Analysing the three key date intervals in the system reporting process shows that most timeliness variance between the three periods occurs in the first 3 days from when the specimen is collected. (Fig. 1) The interval between the specimen date and laboratory report date—which incorporates the time taken for specimens to arrive, be tested and be processed within laboratories—was the longest interval in each period. This interval was completed within 3 days for 90% of cases in each period. The timeliness of the second reporting stage, from laboratory report to SGSS receipt date, improved significantly over time, completing within 1 day from 41% to 74.5% of reports between periods 1 and 3. The final reporting stage, from SGSS receipt to import date, occurred within 1 day for 90% of cases in all periods, demonstrating that processing for epidemiological use was consistent and rapid once data were received by the national system.
The two primary COVID-19 case reporting pathways (i.e., pillars) show distinct patterns in reporting by interval. The first interval, from the specimen date to the laboratory report date, is typically shorter for those within the pillar 1 system, with 95% processed within 3 days, whereas it takes up to 4 days to see that level of completeness for pillar 2. Conversely, reporting from the laboratory to the SGSS is quicker through pillar 2, with more than three-quarters of cases received by the SGSS on the same day as the laboratory report (77.3%, compared with 54.4% of cases from pillar 1 laboratories).
Combining the three reporting stages describes the overall timeliness of case data being reported through the surveillance system from the date a patient is tested. The largest improvements in timely reporting occurred between days 1 and 3. The proportion of cases completing all reporting stages within 2 days increased from 27.2% in period 1 to 53% in period 3 and within 3 days increased from 69.1% to 76.6% over the same time. The proportion completing within 4 days was relatively stable in each period (from 84.4% to 86.8%). In period 3, the median number of days to complete all reporting stages decreased to 2, from 3 in previous periods.
Analysis of each reporting stage of the new surveillance system offers suggestive evidence that the timeliness of the system has improved as COVID-19 reporting has become established over time. Delays in timeliness are most likely to occur in the first stage of the reporting process, before laboratory input to the surveillance platform. Efforts to consistently improve system-wide timeliness, in each reporting pillar, should be directed to strengthening this first reporting stage.
Data-driven insights to inform decision-making for the pandemic response rely on timely and complete data on laboratory-confirmed cases. The SGSS is the principal data source used by stakeholders for these purposes, but relies on data being reported by diagnostic laboratories with sufficient information to rapidly inform the epidemiology. The limited collection and reporting of key information by laboratories, such as the date of symptom onset, hospitalisation and travel exposure, prevents the identification of detailed risk factors for transmission and severity of infection. Increase in lack of patient NHS number submitted by diagnostic laboratories imposes a burden on secondary mechanisms such as deterministic and probabilistic data linkages and poses a hurdle to facilitating broader health informatics linkages going forward.
The COVID-19 pandemic has changed the landscape of public health surveillance in England. Existing surveillance mechanisms that have been enhanced during the response, such as the SGSS, have succeeded in providing rapid collection and reporting of case data to facilitate epidemiological monitoring and analysis and guide the public health policy and strategy. Larger-scale health service or diagnostic laboratory reporting improvements, as well as an emphasis on high-quality data collection, may be required to address the remaining limitations. The surveillance and health information structures that have been developed, and will continue to be refined, will allow public health services to better characterise the pandemic to the benefit of healthcare professionals and the public, with potential learning and application for the surveillance of other infectious diseases in the future.
Author statements
Ethical approval
Ethical approval was not required. The authors were already able to access the anonymised data set and it is not possible to identify individuals from the information provided.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Competing interests
All co-authors are employed by Public Health England, who run the surveillance system described. The authors have no competing interests to declare.
Data statement
The data analysed during this study are not publicly available owing to a need to protect the individual's anonymity. These data are confidential, but fully anonymised data may be available from the corresponding author on reasonable request. Aggregated and anonymised output from the data set described is publicly available at https://coronavirus.data.gov.uk/details/cases.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.puhe.2021.03.012.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Holden B, Quinney A., Padfield S., Morton W., Coles S., Manley P. COVID-19: public health management of the first two confirmed cases identified in the UK. Epidemiol Infect. 2020 doi: 10.1017/S0950268820001922. 28 August. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Public Health England . 2020. Sources of COVID-19 surveillance systems.https://www.gov.uk/government/publications/national-covid-19-surveillance-reports/sources-of-covid-19-systems 2 October 2020. Available at: [Google Scholar]
- 3.Department of Health & Social Care . 2020. Coronavirus (COVID-19): scaling up testing programmes.https://www.gov.uk/government/publications/coronavirus-covid-19-scaling-up-testing-programmes 4 April 2020. Available at: [Google Scholar]
- 4.Public Health England . 2020. National flu and COVID-19 surveillance weekly reports 2020.https://www.gov.uk/government/statistics/national-flu-and-covid-19-surveillance-reports 8 October 2020. Available at: [Google Scholar]
- 5.NHS Digital . 2020. Hospital Episode statistics (HES)https://digital.nhs.uk/data-and-information/data-tools-and-services/data-services/hospital-episode-statistics 19 November 2020. Available at: [Google Scholar]
- 6.NHS Digital . 2020. Secondary uses service (SUS)https://digital.nhs.uk/services/secondary-uses-service-sus 14 December 2020. Available at: [Google Scholar]
- 7.Health and Social Care Information Centre . 2015. A guide to linked mortality data from hospital Episode statistics and the office for national statistics.https://digital.nhs.uk/binaries/content/assets/legacy/pdf/r/q/hes-ons_linked_mortality_data_guide.pdf June 2015. Available at: [Google Scholar]
- 8.Department of Health & Social Care . 2020. COVID-19 contain framework: a guide for local decision-makers.https://www.gov.uk/government/publications/containing-and-managing-local-coronavirus-covid-19-outbreaks/covid-19-contain-framework-a-guide-for-local-decision-makers#data 29 December 2020. Available at: [Google Scholar]
- 9.Department of Health & Social Care . 2020. Coronavirus (COVID-19): full list of local restriction tiers by area.https://www.gov.uk/guidance/full-list-of-local-restriction-tiers-by-area 26 November 2020. Available at: [Google Scholar]
- 10.Davies N., Barnard R., Jarvis C., Russell T., Semple M., Jit M. Association of tiered restrictions and a second lockdown with COVID-19 deaths and hospital admissions in England: a modelling study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30984-1. 24 December. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MRC Biostatistics Unit COVID-19 Working Group . 2021. Nowcasting and forecasting of the COVID-19 pandemic.https://www.mrc-bsu.cam.ac.uk/tackling-covid-19/nowcasting-and-forecasting-of-covid-19/ 12 February 2021. Available at: [Google Scholar]
- 12.Scientific Advisory Group for Emergencies . 2020. Scientific evidence supporting the government response to coronavirus (COVID-19): evidence considered by the Scientific Advisory Group for Emergencies (SAGE)https://www.gov.uk/government/collections/scientific-evidence-supporting-the-government-response-to-coronavirus-covid-19#modelling-inputs 26 June 2020. Available at: [Google Scholar]
- 13.Public Health England . 2021. Statistics at PHE.https://www.gov.uk/government/statistics/national-flu-and-covid-19-surveillance-reports Available at: [Google Scholar]
- 14.Mensah A., Sinnathamby M., Zaidi A., Coughlan L., Simmons R., Ismail S. SARS-CoV-2 infections in children following the full re-opening of schools and the impact of national lockdown: prospective, national observational cohort surveillance, July-December 2020. England. Journal of Infection. 2021 doi: 10.1016/j.jinf.2021.02.022. 25 February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harman K., Allen H., Kall M., Dabrera G. Interpretation of COVID-19 case fatality risk measures in England. J Epidemiol Commu Health. 2021 doi: 10.1136/jech-2020-216140. 29 January. [DOI] [PubMed] [Google Scholar]
- 16.NHS Digital . 2020. Access data on the personal demographics service - demographics Batch service (DBS)https://digital.nhs.uk/services/demographics/access-data-on-the-personal-demographics-service 7 December 2020. Available at: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.