Orphan G protein-coupled receptors (GPCRs) are largely intractable therapeutic targets, owing to the lack of chemical tools for exploring their pharmacology. The discovery of such tools, however, is hampered by a number of unknowns, such as effector coupling and appropriate positive controls. In our 2017 Nature Chemical Biology paper1, we developed a computational chemical tool discovery approach called GPCR Contact-Informed Neighboring Pocket (GPCR-CoINPocket). This method predicted pharmacological similarity of GPCRs in a ligand- and structure-independent manner, to enable the discovery of off-target activities of known compounds at orphan GPCRs and hence the identification of so-called surrogate ligands. Our orphan GPCR target for prospective surrogate ligand discovery efforts was GPR37L1, a brain-specific receptor linked to cerebellar development2 and seizures3. We had previously demonstrated that GPR37L1 constitutively coupled to Gαs and generated ligand-independent increases in intracellular cAMP4. Thus, the inverse agonist activities of computationally predicted surrogates were tested in the cAMP response element luciferase (CRE-luc) reporter gene assay in human embryonic kidney (HEK293) cells expressing either vector control or what we thought was untagged GPR37L1 in pcDNA3.1. However, we recently discovered that the GPR37L1 construct used in both studies was incorrect: instead of pcDNA3.1, it carried the receptor inserted backwards into a yeast p426GPD vector (hereafter referred to as p426-r37L1). We reported the issue to the editors of Sci Signal and Nat Chem Biol, which eventually led to the retraction of both papers4 and1. Here, we correct the cloning error and describe our subsequent unsuccessful efforts to re-test the computationally predicted GPR37L1 ligands.
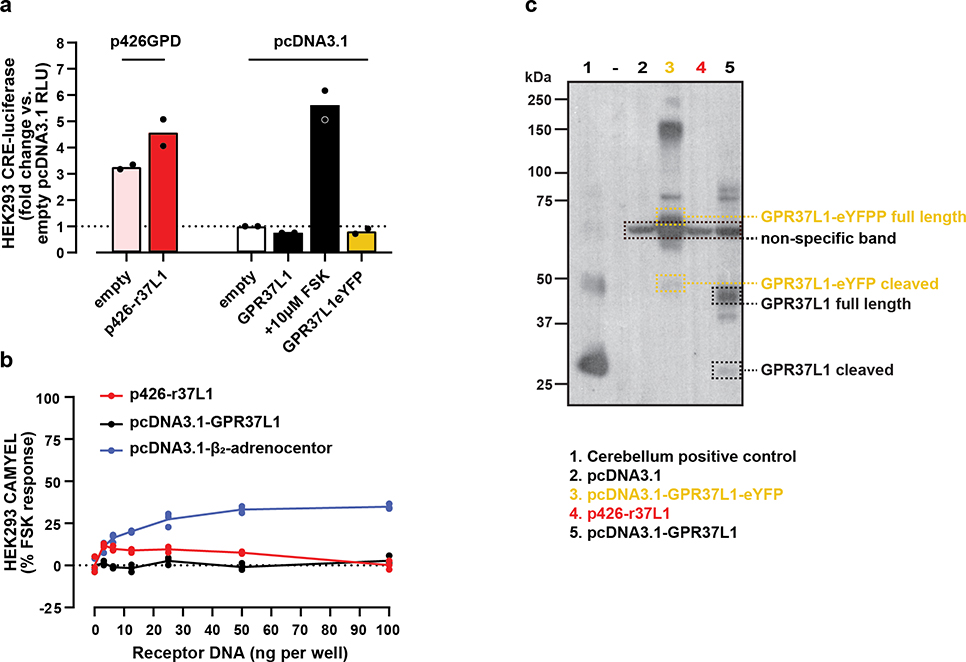
In the CRE-luc assay, p426-r37L1 had considerably elevated transcriptional reporter activity when compared to pcDNA3.1, which had led to the interpretation that GPR37L1 constitutively activated Gαs and promoted cAMP production; however this elevated signal is also evident for empty p426GPD vector (Fig1a). In contrast, neither the corrected wild type GPR37L1 construct, nor GPR37L1 C-terminally tagged with enhanced yellow fluorescent protein (GPR37L1-eYFP), were different from baseline. DNA titration of p426-r37L1 in an upstream orthogonal, real-time cAMP assay with the BRET-based CAMYEL biosensor5, produced results that were no different from pcDNA3.1 vector alone, while titration with the positive control β2-adrenoceptor resulted in a concentration-dependent signal increase (Fig1b). The titration with pcDNA3.1-GPR37L1 also produced no increase in CAMYEL signal, in agreement with the CRE-luc assay and consistent with lack of constitutive Gαs-directed activity. Receptor expression by the corrected construct was confirmed by Western blotting (Fig1c). Together these results suggest that the aberrant signal generated by p426-r37L1 was due to interference caused by the p426-GPD vector and specific to the CRE-luc assay, and that the real GPR37L1 is not constitutively active towards Gαs.
Figure 1: The corrected GPR37L1 construct does not have the apparent Gαs constitutive activity seen with p426-r37L1.
(a–b) Examination of the effects of p426-r37L1 on reporter signal in (a) CRE-luc and (b) CAMYEL assays. HEK293 cells were transiently co-transfected with the CRE-luc reporter or the CAMYEL BRET biosensor together with incorrect or corrected GPR37L1 construct or β2-adrenoceptor, as indicated. ‘Empty’ refers to empty plasmid DNA. For CRE-luc assay, data represent n=2 biological replicates each performed in triplicate on separate days; each data point is the average of the technical replicates for the corresponding biological replicate, and the bar height indicates the mean of biological replicates. For CAMYEL assay, data represent n=1 with technical replicates shown as dots. RLU, relative light units; FSK, forskolin. (c) Western blot for transient HEK293 cellular expression of r37L1, corrected GPR37L1 or controls, as indicated. Cerebellum from a male C57BL/6J mouse was used as a positive control. Dashed lane contained protein ladder only. Image is n=1. All raw data is available in the accompanying Source Data file.
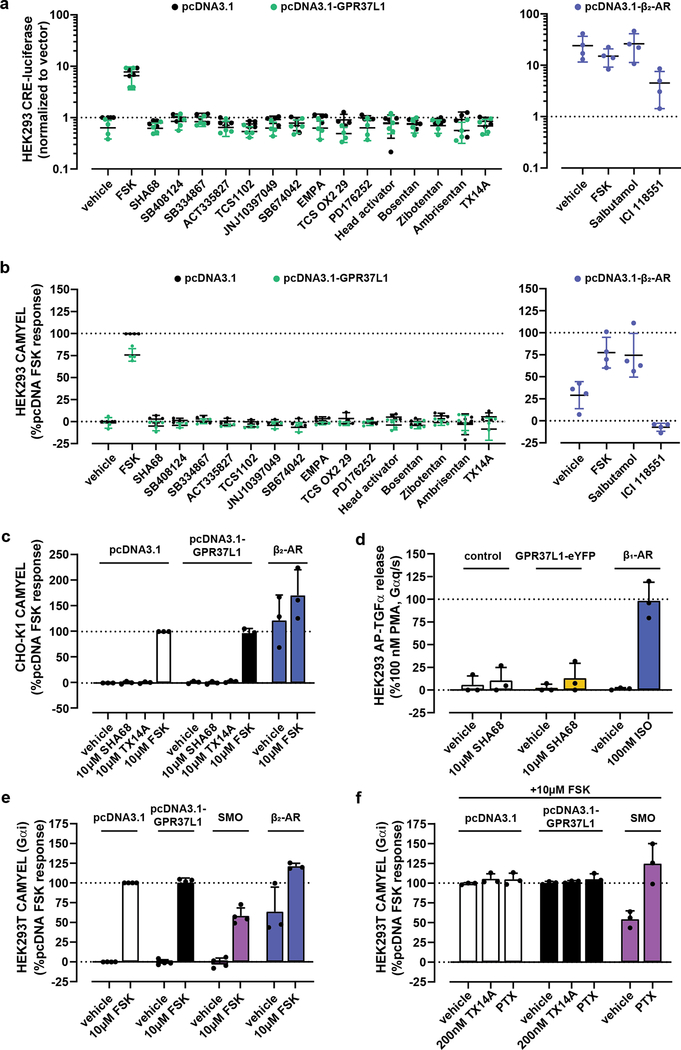
Next, we focused on the computationally predicted compounds that were reported in1 to be inverse agonists of Gαs-directed constitutive activity of GPR37L1. Because the corrected construct did not generate such activity (Fig1a–c), their inverse agonism could not be confirmed. The compounds were not Gαs-directed agonists of GPR37L1 (Fig2a–b) and their antagonism could not be tested for the lack of a control agonist; the previously reported GPR37L1 agonist prosaptide TX14A6,7 had no activity towards Gαs (Fig2a–c). These findings held true in two cellular contexts, HEK293 (Fig2a–b) and CHO cells (Fig2c), and in two Gαs-cAMP pathway assays, CRE-luc (Fig2a) and CAMYEL (Fig2b–c). A positive control, the constitutively active Gαs-coupled β2 adrenoceptor, behaved as expected. The “lead” compound from1, SHA68, was also tested in the receptor-proximal AP-TGFα sheddase assay8 in FlpIN TREx HEK293 cells inducibly expressing GPR37L1-eYFP and co-transfected with Gαq/s (Fig2d); this chimeric G protein directs Gαs-coupled receptor signaling through the Gαq/TGFα pathway. Gαs-directed agonism was apparent for the positive control, isoproterenol, in β1 adrenoceptor-expressing cells, but not for SHA68 at GPR37L1 (Fig2d). Altogether, this suggests that the compounds reported in1 do not activate Gαs-mediated signaling by GPR37L1, whereas their ability to inhibit such signaling could not be tested.
Figure 2: The corrected GPR37L1 construct does not display constitutive or ligand-induced activity in any assays examined.
Examination of constitutive and ligand-induced cAMP elevation in HEK293 cells transfected with or without the corrected pcDNA3.1-GPR37L1, using (a) CRE-luc or (b) CAMYEL assays. β2-adrenoceptor +/− 10 μM salbutamol was used as a positive control for constitutive cAMP signaling. Final ligand concentrations were 10 μM. (a) n=4 biological replicates; (b) n=3 biological replicates except for controls, SHA68, head activator, bosentan, zibotentan, ambrisentan and TX14A, which were n=4. (c) CAMYEL assay performed as in (b) using CHO-K1 cells to control for cellular background. Data represents n=3 biological replicates. (d) Ligand-induced Gαs coupling was probed using the AP-TGFα-sheddase assay in stable FlpIN TREx HEK293 cells inducibly expressing GPR37L1-eYFP, transiently transfected with AP-TGFα and chimeric Gαq/s. β1 adrenoceptor stimulated with isoproterenol (ISO) was used as a positive control. Data represent n=3 biological replicates. (e–f) Constitutive suppression of FSK-stimulated cAMP was monitored using the CAMYEL BRET biosensor in HEK293T cells transiently co-transfected with CAMYEL and either the corrected GPR37L1 plasmid, Smoothened, or β2 adrenoceptor. Gαi3 was transiently co-transfected with GPR37L1 and Smoothened. Data represent n=4 biological replicates except for the β2 adrenoceptor (n=3). All data panels are presented as mean +/− standard deviation, where the bar height represents the average of all biological replicates each performed independently in three technical replicates. The averages of the technical triplicates for each biological replicate are shown as dots. All raw data is available in the accompanying Source Data file.
Because the inchoate GPR37L1 literature contains conflicting reports of Gαs vs Gαi coupling specificity of this receptor3,4,6,7,9,10, we turned our attention to the Gαi pathway. Unlike the positive control Smoothened11, GPR37L1 showed no Gαi-directed constitutive activity as indicated by the lack of constitutive, pertussis-toxin-sensitive suppression of forskolin-stimulated cAMP in the CAMYEL assay in HEK293T cells (Fig2e–f). No cAMP suppression was observed following cell pretreatment with prosaptide TX14A either (Fig2f). This lack of constitutive GPR37L1 activity or responses to the previously reported agonist precluded investigations of inverse agonism or antagonism of the compounds from1 in the Gαi pathway. Altogether, our data suggests that the pharmacological toolbox for GPR37L1 is once again empty.
GPCR-CoINPocket, the computational methodology developed in1, is unaffected by the GPR37L1 cloning error and remains a powerful tool for understanding pharmacological similarity of GPCRs solely from their sequences. CoINPocket-predicted GPCR-ligand pairings have been validated both retrospectively1 and prospectively (e.g. for GPR31 and HCA1 receptors12), and the approach was also successfully applied to an evolutionarily unrelated lysine methyltransferase family13. Furthermore, GPCR-CoINPocket has been used to guide GPCR homology modeling14 and to delineate common GPCR activation mechanisms15. However, confirming the CoINPocket ligand predictions for GPR37L1 was not possible in the absence of a validated GPR37L1 activity assay with appropriate controls. In other words, we were unable to experimentally test the hypothesised pairings. Indeed, a shortcoming in all functional GPR37L1 studies is that the limited armamentarium forces researchers to infer specificity from indirect and sometimes inconclusive data. For example, phenotypic or second messenger amplification assays with the read-out many steps removed from receptor activation are vulnerable to non-specific signal interference, as in our case for p426-r37L11,4, or to interference from the endogenous receptor context6,7. These challenges are common for orphan research, but the potential neuroprotective role of GPR37L1 and its links to seizures mandate that the pharmacology and physiology of GPR37L1 must continue to be explored.
Methods
Compounds and reagents
Tissue culture and Western blot reagents were purchased from Sigma-Aldrich (St. Louis, MO) or Life Technologies (Carlsbad, CA) unless otherwise indicated. Compound suppliers are listed in Ngo et al.1.
Plasmids
Full length untagged human GPR37L1 with the native signal peptide, in pcDNA3.1, was synthesized by Genscript. p426-r37L1 (incorrectly presumed to be pcDNA3.1-GPR37L1 in1) contained full-length WT GPR37L1 inserted backwards into a yeast p426GPD vector. pcDNA3.1-GPR37L1-eYFP was described in4 and human β2-AR in pcDNA3.1 was from cDNA Resource Centre (cat #AROB200000). For BRET-based cAMP reporter assays, pcDNA3.1-CAMYEL was used5. pGEN-SMO was a gift from Philip Beachy (Stanford University; Addgene plasmid # 37673), and modified with a 2xStop codon following the receptor C-terminus. Rat Gαi3 in pcDNA3.1+ was a gift from Pradipta Ghosh (UC San Diego). All plasmids used in TGFα shedding assays8, except GPR37L1, were a generous gift from Asuka Inoue (Tohoku University).
CRE-luciferase cAMP reporter assay
CRE-luciferase reporter gene assays were performed as previously described4 with minor modifications. Briefly, HEK293 cells were seeded onto transparent 96-well plates at a density of 20,000 cells/well in DMEM containing 10% FBS and 1% penicillin/streptomycin solution. The next day, CRE-reporter DNA (0.04 μg/well) (Promega) was co-transfected with either pcDNA3.1, corrected pcDNA3.1-GPR37L1, or the erroneous p426-r37L1 construct (0.04 μg/well), or pcDNA3.1-β2-adrenoreceptor (0.02 μg/well; positive control) using Lipofectamine LTX. The following day, compounds diluted in DMEM were added to the cells (10 μM final concentration). After approximately 21 hr, cells were washed once with PBS before lysis with 40 μl Passive Lysis Buffer (Promega). For ligand screening experiments, lysates were frozen and thawed the next day to allow uniform processing of samples. To measure luciferase activity, 4 μL of lysate was transferred to a white OptiPlate-384 in duplicate. Following injection of 10 μl Luciferase Assay Reagent II (Promega), luminescence was detected immediately using a PHERAstar FSX plate reader (BMG-Labtech).
Real-time cAMP measurements with the CAMYEL Bioluminescence Resonance Energy Transfer (BRET) biosensor
For Fig1b and Fig2b, HEK293 cells were seeded onto white 96-well plates at 20,000 cells/well. The next day cells were transfected using Lipofectamine LTX according to manufacturer’s instructions. For the DNA titration experiment (Fig1b), CAMYEL (0.02 μg/well) and construct of interest (0–0.1 μg/well) were co-transfected. For the experiments in Fig2b, 0.02 μg CAMYEL and 0.02 μg of receptor DNA were co-transfected per well. The following morning, cells were washed with 200 μL/well Hank’s Balanced Salt Solution (HBSS) and then incubated in 85 μl HBSS at 37°C for 30 min. Luminescence was initiated by addition of a mixture of 2 μM coelenterazine-h (Promega) and 40 μM 3-isobutyl-1-methylxanthine (IBMX; final concentrations listed) in HBSS; kinetic readings were taken for 5 min baseline and then for 15 min after addition of ligands, all at 37°C, using the BRET1 filter set on a PHERAstar plate reader (BMG-Labtech). Data presented are the 15 min endpoint read. Experiments in Fig2c were performed similarly except CHO-K1 cells were maintained in Ham’s F12 media containing 10% FBS and 1% penicillin/streptomycin solution. For Fig2e–f, HEK293T cells were plated at 700,000 cells/well in a 6-well plate and transfected next day with 1.0 μg/well CAMYEL, 0.5 μg/well Gαi3 (for GPR37L1 and SMO only), and 1.0 μg/well of the indicated receptors using TransIT-X2 transfection reagent (Mirus Bio); the total transfected DNA was normalized to 3.0 μg/well for all wells using empty pcDNA3.1. The following day, cells were lifted by gentle pipetting and re-plated in a 96-well plate at 50,000 cells/well. On day 4, cells were serum-starved in assay buffer (1X HBSS, 20 mM HEPES, 0.05% BSA) for 1 hr and then IBMX and coelenterazine-h were added to each well to the final concentrations of 100 μM and 10 μM, respectively, for 10 min. Emission intensity was recorded for 5 min using 460 nm and 530 nm filters on the EnVision or Victor × Light plate reader (PerkinElmer). Next, 10 μM FSK was added to each well and emission intensities at 460 nm and 530 nm were continuously read for another 20 min. In Fig2f, 200 nM TX14A was added after 5 min, and continuously read for 10 mins before FSK addition. PTX (200 ng/μL) was added at least 3 h prior to serum starvation in assay buffer. Inverse BRET (1/BRET) was calculated as the window-averaged ratio of emission intensity at 460 nm and 530 nm at 5 min post FSK addition, normalized to FSK response from the pcDNA3.1-transfected cells in the same experiment.
AP-TGFα sheddase assay
AP-TGFα sheddase assays were performed as described previously8. Briefly, GPR37L1-eYFP expression was induced in stable FlpIN TREx HEK293 cells using 100 ng/μL doxycycline. Transient transfections of β1-adrenoceptor, AP-TGFα and the Gαq/s chimera were performed using Lipofectamine LTX. Cells were stimulated with 10 μL/well of either isoproterenol for β1-adrenoceptor conditions or SHA68 for GPR37L1-eYFP conditions to achieve final concentrations of 100 nM or 10 μM, respectively. Phorbol-12-myristate-13-acetate (PMA, 100 nM) served as a positive control. Absorbance at 405 nm was measured using a PHERAstar FSX plate reader (BMG-Labtech).
Western blot
HEK293 cells were seeded onto 6-well plates at a density of 400,000 cells/well. The next day, cells were transfected with pcDNA3.1, pcDNA3.1-GPR37L1-eYFP, p426-r37L1, or pcDNA3.1-GPR37L1 (0.4 μg/well) using Lipofectamine LTX. After 24 hr, cells were washed once with ice-cold PBS and lysed with ice-cold RIPA buffer containing cOmplete™ Protease Inhibitor Cocktail (Roche Diagnostics). For murine cerebellar samples, frozen tissue was homogenized in RIPA buffer containing cOmplete™ Protease Inhibitor Cocktail on ice using a POLYTRON homogenizer. The cell and tissue lysates were centrifuged at 18,000 g for 10 min and the resulting supernatant retained. Total protein concentrations were determined using a BCA assay. Loading samples were prepared by mixing cell lysates with NuPAGE LDS sample buffer containing 100 mM dithiothreitol, and heated for 15 min at 65°C. 10 μg of each sample was loaded on a NuPAGE 4–12% bis-tris polyacrylamide gel and separated over 1 hr at 187V. Proteins were transferred onto a polyvinylidene fluoride membrane (Merck Millipore), followed by blocking in Tris-buffered saline containing 0.1% Tween-20 (TBST) and 5% skim milk for 1 hr. Membranes were probed with goat anti-GPR37L1 primary antibody (1:1000, sc-164532, Santa Cruz Biotechnology) at 4°C overnight, followed by rabbit anti-goat horseradish peroxidase-conjugated secondary antibody (1:5,000, 61–1620, Invitrogen) for 1 hr. Chemiluminescence was detected with Clarity™ (Bio-Rad, USA) chemiluminescence substrate using X-ray film (Fujifilm).
Data analysis
Numerical data for Fig1a–b and Fig2 was analyzed and graphs were made in GraphPad Prism 8.4.3.
Data availability statement
Raw data for all figures is provided in the accompanying Source Data File.
Acknowledgements
We would like to thank Dr Robert M. Graham from the Victor Chang Cardiac Research Institute, Australia, for helpful discussions, Mr Theodore Nettleton, UNSW Sydney, for assistance with identifying the cloning error, and Dr Asuka Inoue, Tohoku University, for providing the AP-TGFα constructs and helpful advice. Funding - TN (NHMRC CJ Martin Early Career Fellowship 1145746), IK (NIH R01 AI118985 and R01 GM117424), NJS (National Heart Foundation Future Leader Fellowship 101153).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Ngo T et al. Orphan receptor ligand discovery by pickpocketing pharmacological neighbors. Nat Chem Biol 13, 235–242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Marazziti D et al. Precocious cerebellum development and improved motor functions in mice lacking the astrocyte cilium-, patched 1-associated Gpr37l1 receptor. Proc Natl Acad Sci USA 110, 16486–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giddens MM et al. GPR37L1 modulates seizure susceptibility: Evidence from mouse studies and analyses of a human GPR37L1 variant. Neurobiol Dis 106, 181–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman JL et al. Metalloprotease cleavage of the N terminus of the orphan G protein-coupled receptor GPR37L1 reduces its constitutive activity. Sci Signal 9, ra36 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Jiang LI et al. Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J Biol Chem 282, 10576–84 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer RC, Giddens MM, Schaefer SA & Hall RA GPR37 and GPR37L1 are receptors for the neuroprotective and glioprotective factors prosaptide and prosaposin. Proc Natl Acad Sci USA 110, 9529–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B et al. Glio- and neuro-protection by prosaposin is mediated by orphan G-protein coupled receptors GPR37L1 and GPR37. Glia 66, 2414–2426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue A et al. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods 9, 1021–9 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Zheng X, Asico LD, Ma X & Konkalmatt PR G protein-coupled receptor 37L1 regulates renal sodium transport and blood pressure. Am J Physiol Renal Physiol 316, F506–F516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster SR et al. Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Cell 179, 895–908 e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers BR, Neahring L, Zhang Y, Roberts KJ & Beachy PA Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci U S A 114, E11141–E11150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morita N et al. GPR31-dependent dendrite protrusion of intestinal CX3CR1(+) cells by bacterial metabolites. Nature 566, 110–114 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Rabal O, Castellar A & Oyarzabal J Novel pharmacological maps of protein lysine methyltransferases: key for target deorphanization. J Cheminform 10, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castleman PN, Sears CK, Cole JA, Baker DL & Parrill AL GPCR homology model template selection benchmarking: Global versus local similarity measures. J Mol Graph Model 86, 235–246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q et al. Common activation mechanism of class A GPCRs. Elife 8, e50279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data for all figures is provided in the accompanying Source Data File.