The local vitamin and nutrition store is popular in my neighborhood. Recently, I walked into this store and realized it is similar to the biotechnology companies in which many scientists purchase their amino acids, media, and cell-culture reagents. There were the usual supplements to treat every disease you can imagine, but by far the most popular items showcased were amino acids. The clerk told me that many people purchase amino acids and spike their smoothie drinks to boost muscle performance and enhancement. Of course, many of us prefer to eat eggs, lentils, fish, and meat as a source of protein, which is digested to generate the amino acids required for intracellular de novo protein synthesis. There are 20 amino acids. Animals have seven conditionally essential amino acids that can be synthesized and are usually not required in the diet. However, they are essential components of the diet for specific populations that cannot synthesize them in adequate amounts. There are four “nonessential” amino acids can be synthesized and, thus, are not required in diet. The remaining nine “essential” amino acids are obtained from diet. Most plants and bacteria, however, can synthesize all 20 amino acids. Most of us usually think of amino acids as important for protein synthesis, but they can also generate glucose, ATP, and fatty acids, and they are metabolic precursors for numerous biomolecules, including heme groups, nucleotide bases, and signaling molecules (catecholamines, neurotransmitters). In addition, they are essential for epigenetic modifications. This review covers (1) generation of amino acids, (2) degradation of amino acids, and (3) generation of catecholamines and heme by amino acids and their role in epigenetics (Fig. 1).
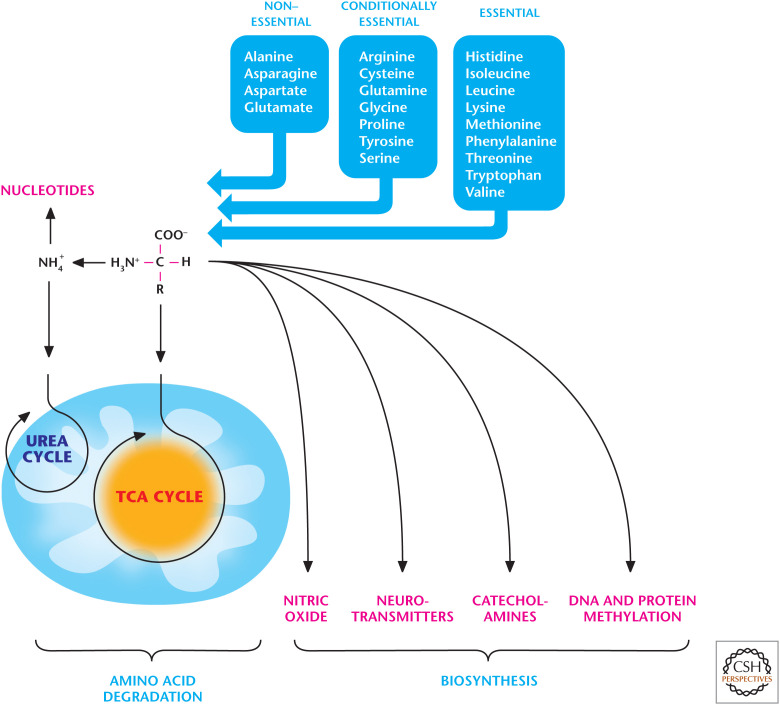
Figure 1.
An overview of amino acid metabolism. Amino acids are degraded to yield NH4+, which enters the urea cycle, and a carbon skeleton that can enter metabolic pathways to generate ATP, glucose, and fatty acids. Amino acids are also used to produce NO, neurotransmitters, and catecholamines. The amino acid methionine provides the methyl group for many DNA and histone methyltransferases to regulate epigenetics.
QUICK GUIDE TO AMINO ACIDS
There are 20 amino acids. Animals cannot synthesize nine amino acids, which are called “essential” amino acids; therefore, they are taken from diet (Fig. 1). The remaining 11 are “nonessential” or “conditionally essential” amino acids.
Glycolytic and TCA cycle intermediates generate the nonessential and conditionally essential amino acids.
Free amino acids produced from degradation of either cellular proteins or dietary proteins are deaminated to yield NH4+ and a carbon skeleton. NH4+ enters the urea cycle and the carbon skeleton can enter metabolic pathways to generate ATP, glucose, and fatty acids (Fig. 1).
Glutamate acts as both a nitrogen donor and acceptor and is the central amino acid for the movement of nitrogen among amino acids.
Tyrosine is the precursor to generation of norepinephrine, epinephrine, dopamine, and melanins.
Methionine provides the methyl group for many DNA and histone methyltransferases to regulate epigenetics.
Cysteine, glutamate, and glycine generate the antioxidant glutathione.
Nitric oxide synthases use arginine to generate nitric oxide (NO).
Glycine and glutamate can serve as neurotransmitters. Glutamate can generate another amino acid neurotransmitter, γ-aminobutyric acid (GABA), which does not participate in protein synthesis.
Tryptophan is used as a precursor for generation of another neurotransmitter, serotonin. Serotonin is used to generate melatonin.
METABOLIC PATHWAYS THAT PRODUCE NONESSENTIAL AMINO ACIDS
Animals lack the enzymes to generate the essential amino acids, thus, these amino acids must be obtained from the diet. The structures of the essential amino acids, generally, are more complex than the nonessential amino acids. Essential amino acids require a significantly greater number of enzymatic reactions for synthesis and are found in plants and lower organisms. Tyrosine and arginine are conditionally essential. Tyrosine is derived from the essential amino acid phenylalanine by the enzyme phenylalanine hydroxylase. Endogenous tyrosine production is dependent on dietary phenylalanine, and a significant portion of tyrosine is obtained from diet. A small amount of arginine can be generated from argininosuccinate in the urea cycle (see the section The Urea Cycle Is Necessary for Amino Acid Degradation).
Animals can synthesize the conditionally essential and nonessential amino acids using glycolytic and TCA cycle intermediates. The glycolytic intermediate 3-phosphoglycerate generates serine and glycine. These reactions are outlined in Figure 2. In the initial step, 3-phosphoglycerate is oxidized by 3-phosphoglycerate dehydrogenase to generate 3-phosphohydroxypyruvate by coupling this reaction to the reduction of NAD+ to NADH. Subsequently, 3-phosphohydroxypyruvate undergoes a glutamate-linked transamination reaction catalyzed by phosphoserine aminotransferase 1 to form 3-phosphoserine, which is then hydrolyzed by phosphoserine phosphatase to yield serine. The enzyme serine hydroxytmethyltransferase converts serine into glycine, coupling this reaction to the conversion of tetrahydrofolate (THF) to N5,N10-methylene THF. Serine is also required for cysteine production. Cystathionine β synthase converts homocysteine and serine into cystathionine, which cystathionine γ-lyase converts to cysteine (see Fig. 11).
Figure 2.
The glycolytic intermediate 3-phosphoglycerate generates serine and glycine. 3-Phosphoglycerate dehydrogenase (PHGDH) oxidizes 3-phosphoglycerate to generate 3-phosphohydroxypyruvate, which is converted into 3-phosphoserine by phosphoserine aminotransferase 1 (PSAT1). Subsequently, phosphoserine phosphatase (PSPH) converts 3-phosphoserine into serine. The enzyme serine hydroxytmethyltransferase (SHMT1) converts serine into glycine by coupling this reaction to the conversion of THF to N5,N10-methylenetetrahydrofolate (5,10-CH2-MTHF).
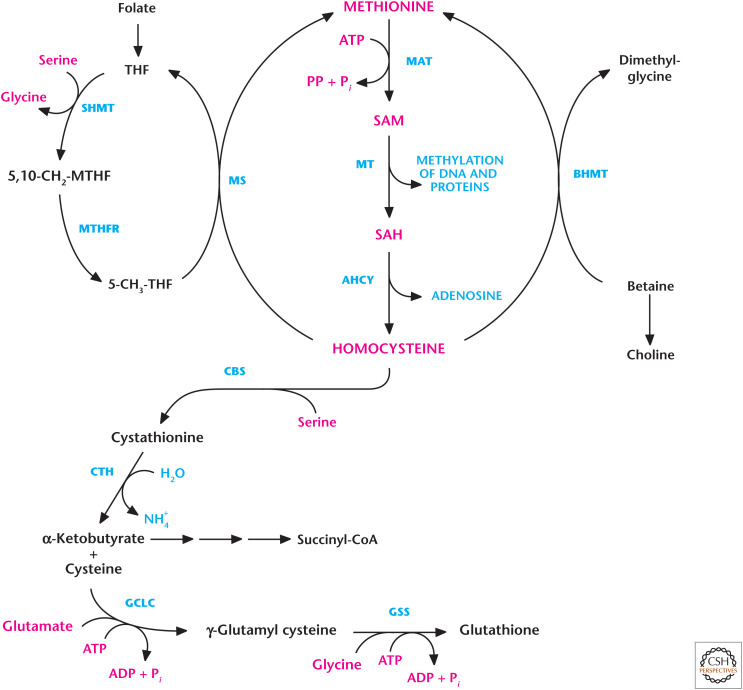
Figure 11.
Methionine metabolism is used for methylation and cysteine production. Methionine adenosyltransferase (MAT) generates S-adenosylmethionine (SAM) using methionine and ATP as substrates. Methyltransferases (MTs) use SAM for methylation. The demethylation of SAM results in the formation of S-adenosylhomocysteine (SAH), which is converted into homocysteine and adenosine by adenosylhomocyteinase (AHCY). Methionine synthase can convert homocysteine back to methionine, a reaction that requires 5-methyl-tetrahydrofolate (5-MTHF) as the methyl donor. The resulting tetrahydrofolate (THF) is converted to 5,10-methylenetetrahydrofolate (5,10-CH2-THF) by serine hydroxymethyl transferase (SHMT). 5,10-CH2-THF is then reduced to 5-MTHF by methylenetetrahydrofolate reductase (MTHFR). Betaine-homocysteine S-methyltransferase (BHMT) can also generate methionine from homocysteine by coupling betaine conversion to dimethylglycine. Homocysteine generates cysteine by a series of reactions known as trans-sulfuration. Serine condenses with homocysteine to generate cystathionine by cystathionine synthase (CBS). Subsequently, cystathionine is cleaved by cystathionine γ-lyase (CTH) to produce cysteine and α-ketobutyrate. Glutamate-cysteine ligase (GCLC) uses glutamate and cysteine as substrates to generate γ-glutamyl cysteine, which, in combination with glycine, is converted into glutathione by glutathione synthetase (GSS).
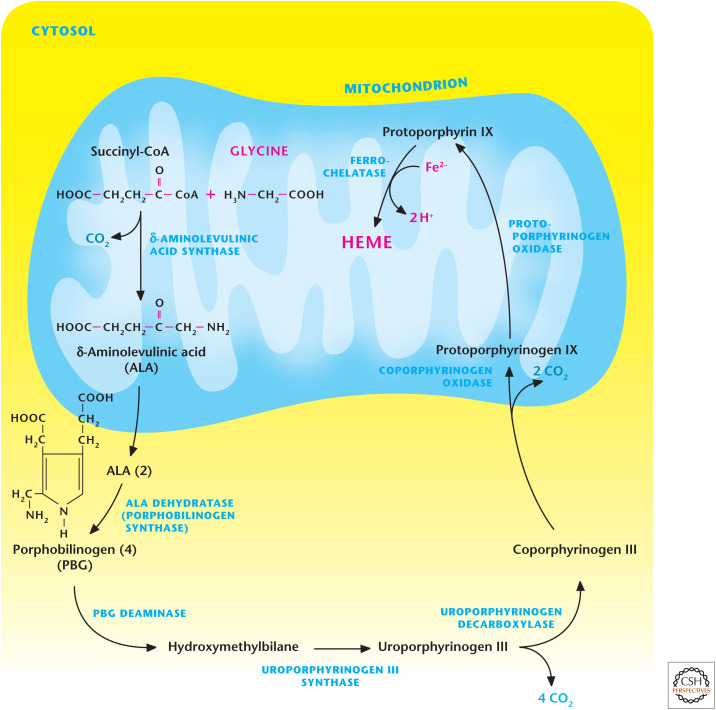
An important role of glycine is that it is a metabolic precursor to heme, as shown in Figure 3. The initial step catalyzed by δ-aminolevulinic acid synthase combines glycine with the TCA cycle intermediate succinyl-CoA to generate δ-aminolevulinic acid in the mitochondrial matrix. Next, δ-aminolevulinic acid is exported to the cytosol, where two molecules condense to generate porphobilinogen. A series of reactions occur in the cytosol and mitochondria, culminating in the final step in which ferrochelatase incorporates Fe2+ into the heme ring.
Figure 3.
Glycine is a precursor in heme synthesis. The initial step of heme synthesis is catalyzed by δ-aminolevulinic acid synthase, which combines glycine with the TCA cycle intermediate succinyl-CoA to generate δ-aminolevulinic acid in the mitochondrial matrix. Next, δ-aminolevulinic undergoes a series of reactions, which occur in the cytosol and mitochondria, to generate the heme ring.
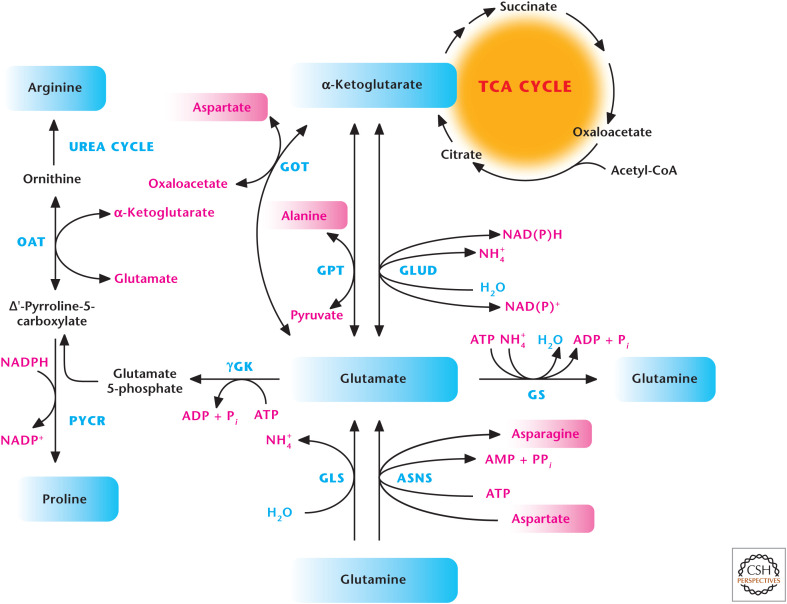
The major TCA cycle intermediate α-ketoglutarate is at the heart of generating multiple amino acids. In particular, the interconversion of glutamate to α-ketoglutarate allows the generation of multiple amino acids, including alanine, aspartate, and arginine. Glutamate undergoes two transamination reactions, which generate alanine and aspartate, both in the cytosol and mitochondrial matrix. Aspartate aminotransferase uses glutamate and oxaloacetate to generate aspartate and α-ketoglutarate, and alanine aminotransferase uses glutamate and pyruvate to generate alanine and α-ketoglutarate (Fig. 4). Ornithine aminotransferase generates ornithine by coupling to glutamate conversion into α-ketoglutarate. Subsequently, ornithine can generate arginine through the urea cycle (see Fig. 7). Glutamate can be generated from α-ketoglutarate and glutamine. Glutamate dehydrogenase converts α-ketoglutarate and NH4+ to glutamate by coupling NADPH to NADP+. Glutamine synthetase uses ATP and NH4+ to generate glutamine from glutamate. Conversely, glutaminase converts glutamine into glutamate with the release of ammonium (NH4+). Glutamine can also be converted into glutamate and asparagine by asparagine synthetase, a reaction that uses aspartate and ATP as substrates (Fig. 4). Finally, γ-glutamyl kinase uses ATP to convert glutamate into glutamate 5-phosphate, which is then converted into δ-pyrroline-5-carboxylate and proline by pyrroline-5-carboxylate reductase (Fig. 4).
Figure 4.
Glutamate generates multiple amino acids. Aspartate aminotransferase (GOT) uses glutamate and oxaloacetate to produce aspartate and α-ketoglutarate, and alanine aminotransferase (GPT) uses glutamate and pyruvate to produce alanine and α-ketoglutarate. Ornithine aminotransferase (OAT) generates ornithine, which can generate arginine through the urea cycle. Glutamate dehydrogenase (GLUD) converts α-ketoglutarate and NH4+ to glutamate by coupling NADPH to NADP+. Glutamine synthetase (GS) uses ATP and NH4+ to generate glutamine from glutamate. Conversely, glutaminase (GLS) converts glutamine into glutamate with the release of ammonium (NH4+). Asparagine synthetase (ASNS) uses aspartate, glutamine, and ATP to generate asparagine. γ-Glutamyl kinase (γGK) uses ATP to convert glutamate into glutamate 5-phosphate, which is subsequently converted into δ-pyrroline-5-carboxylate and proline by pyrroline-5-carboxylate reductase (PYCR).
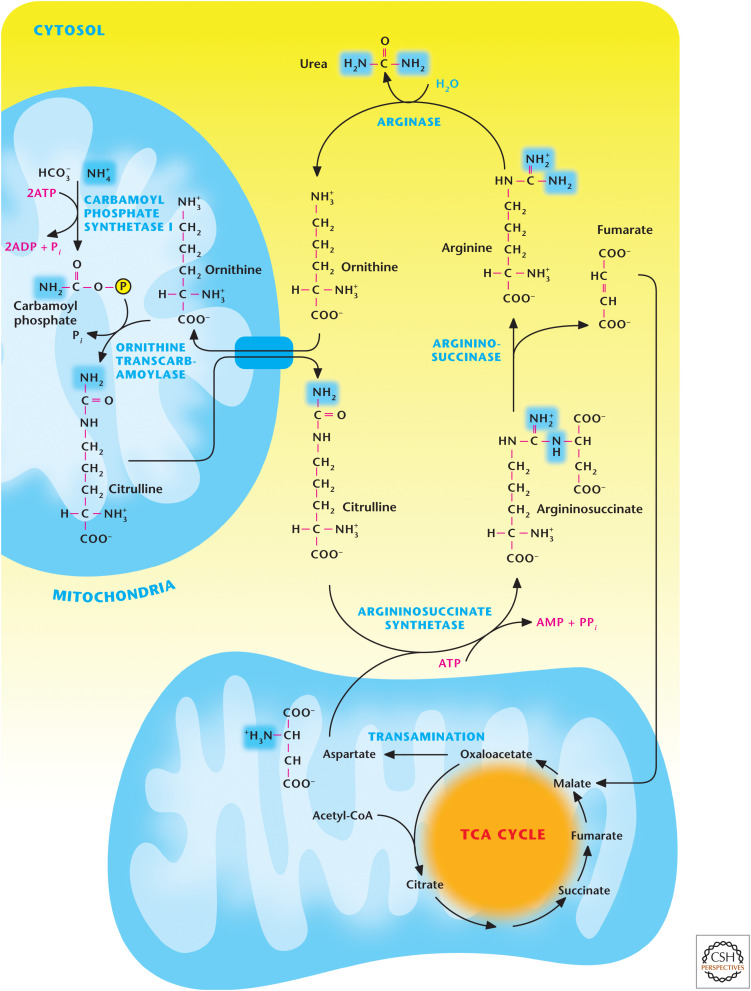
Figure 7.
Overview of the urea cycle. The initial step for urea synthesis is catalyzed by the mitochondrial matrix enzyme carbamoyl phosphate synthetase I to incorporate the first nitrogen molecule. Subsequently, ornithine transcarbamoylase uses carbamoyl phosphate and ornithine as substrates to generate citrulline in the mitochondrial matrix. Next, citrulline is exported to the cytosol and converted into argininosuccinate by argininosuccinate synthetase. This reaction results in the incorporation of a second nitrogen atom from aspartate. Next, the enzyme argininosuccinase cleaves argininosuccinate to produce fumarate and arginine. Finally, the enzyme arginase converts arginine to urea and ornithine to complete the urea cycle.
THE UREA CYCLE IS NECESSARY FOR AMINO ACID DEGRADATION
Free amino acids are generated from degradation of either cellular proteins or dietary proteins. Free amino acids cannot be simply stored; thus, they are recycled for protein synthesis or deaminated to yield NH4+ and a carbon skeleton. The NH4+ is toxic to cells and removed either by synthesis of nitrogen-containing compounds, such as nucleotides or amino acids, or excreted in the form of urea. The carbon skeletons of amino acids feed into metabolic pathways to generate ATP, glucose, and fatty acids (Fig. 5).
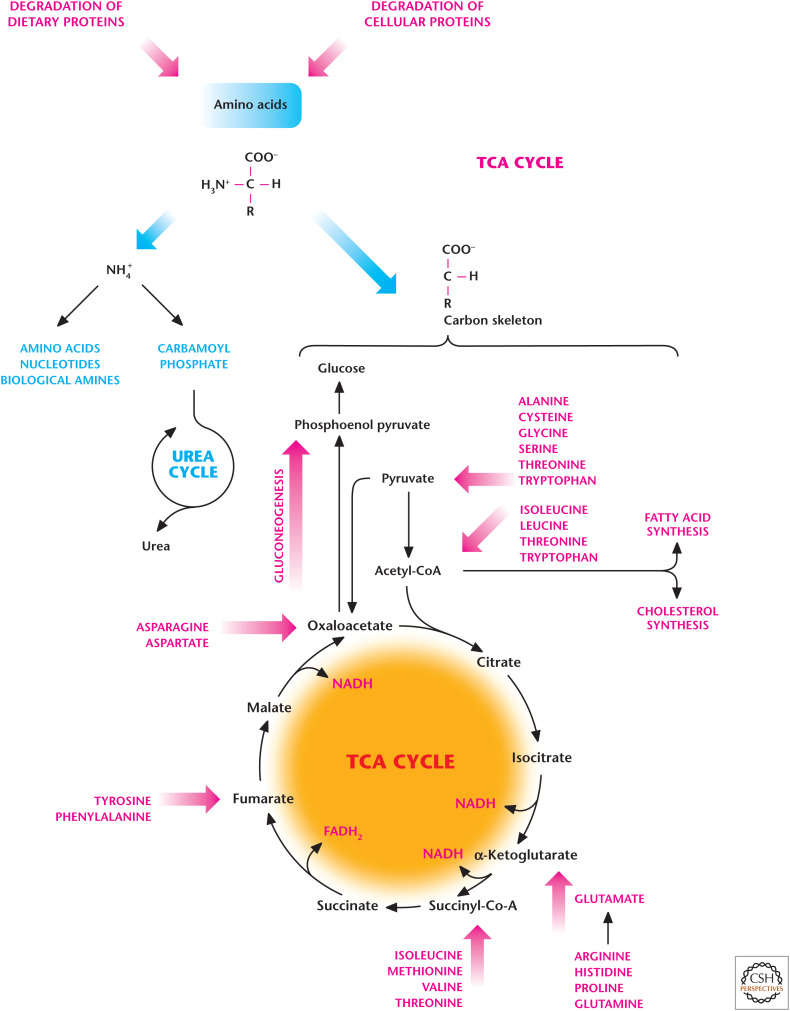
Figure 5.
Amino acid degradation produces ammonium (NH4+) and a carbon skeleton. The NH4+ is removed either by synthesis of nitrogen-containing compounds, such as nucleotides, or excreted in the form of urea. The carbon skeletons of amino acids can be converted into TCA cycle intermediates, which are used either to generate ATP through oxidative phosphorylation or provide the precursors for fatty acid synthesis and gluconeogenesis.
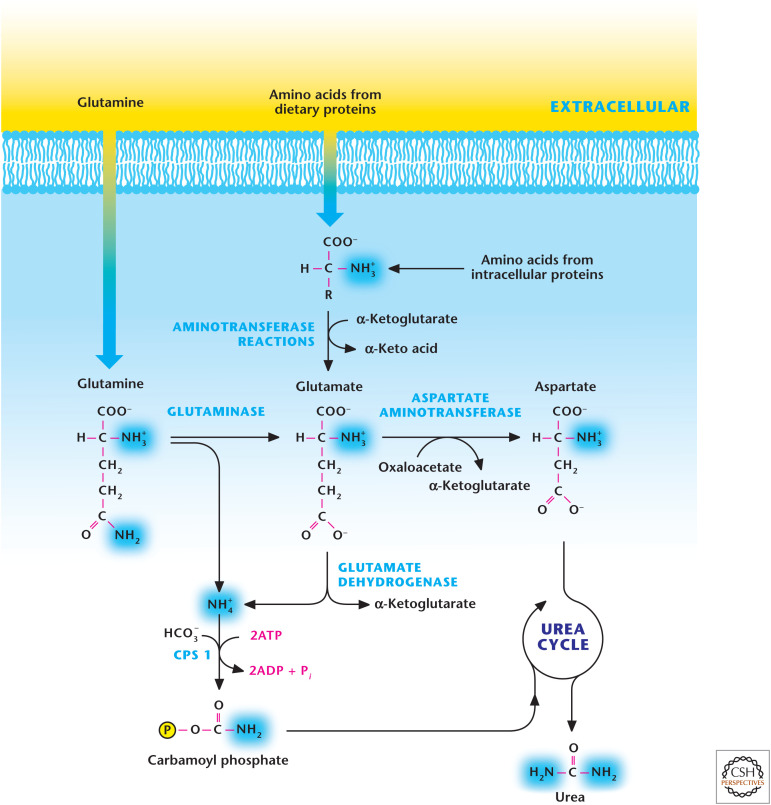
The carbon skeleton of amino acids can be converted into TCA cycle intermediates that can be used either to generate ATP through oxidative phosphorylation or provide the precursors for fatty acid synthesis and gluconeogenesis (Fig. 5). Amino acids normally account for 10%–15% of ATP generated. However, under high-protein diets or conditions of starvation in which the muscle protein is degraded, a significant portion of ATP generated is from amino acids. Conversely, if cells have abundant ATP, then excess free amino acids can be converted into TCA cycle intermediates, which can then generate glucose and fatty acids. The excess nitrogen (NH4+) due to amino acid degradation can be removed by urea, which is synthesized in the liver by the urea cycle and exported to the kidneys, where it is concentrated and enters the bladder for excretion in the urine. Hans Krebs and a medical student who worked in his laboratory, Kurt Henseleit, discovered the urea cycle in 1932 (see Box 1). Urea has two nitrogen and one carbon molecule. The two substrates of the urea cycle are carbamoyl phosphate and aspartate. Glutamine and glutamate contribute one of the nitrogen molecules, and the oxidative metabolism of the TCA cycle contributes HCO3− for carbamoyl phosphate synthesis, which takes place in the mitochondrial matrix. Aspartate contributes the second source of nitrogen molecules to complete the generation of urea. The two primary carriers of nitrogen molecules in the blood are glutamine and alanine, which are absorbed by the liver to participate in the urea cycle (Fig. 6). The excess ammonia generated by tissues reacts with glutamate to produce glutamine by the enzymatic action of glutamine synthetase. The glutamine generated from tissues is released into the bloodstream and absorbed in the liver, where it can generate NH4+ and glutamate by the enzyme glutaminase. Alanine is generated by muscle tissues, which catabolize amino acids as a source of energy. In the muscle tissue, excess ammonia is transferred to pyruvate by alanine aminotransferase to produce alanine and released into the bloodstream. The liver absorbs the alanine, where alanine-aminotransferase converts it to glutamate and pyruvate. The other sources of glutamate in the liver are amino acids generated from dietary intake and muscle and intracellular protein degradation. These amino acids can generate glutamate by α-ketoglutarate-dependent aminotransferase enzymes in the liver. Subsequently, glutamate dehydrogenase in the mitochondrial matrix can convert glutamate into α-ketoglutarate, liberating NH4+ (Fig. 6). Aspartate aminotransferase also converts glutamate to aspartate, which feeds into the urea cycle to provide the second source of nitrogen molecules.
BOX 1.
HANS KREBS AND THE UREA CYCLE
In his 1953 Nobel lecture, Hans Krebs pointed out that his elucidation of the urea cycle in 1932 provided insights that helped him dissect the TCA, or citric acid or Krebs, cycle in 1937. Perhaps more importantly, his publication of the urea cycle made him instantly famous and was instrumental in enabling Krebs, a Jew, to escape Nazi Germany.
Krebs was born in Germany in 1900 and was initially destined to become an otolaryngologist like his father. But after receiving his M.D. from the University of Hamburg in 1925, he took a job in the laboratory of Otto Warburg at the Kaiser Wilhelm Institute for Biology at Berlin-Dahlem. In Warburg's laboratory, Krebs learned the techniques of manometry for measuring oxygen consumption in tissue slices and spectrophotometry. Warburg's manometer could be used to study respiration in vitro in tissue slices only 10-layers-of-cells thick. He also learned how to formulate the experimental approaches to solving research problems. Krebs later said that “Warburg had taught me the alphabet of scientific research.” Why he left Warburg's laboratory has had varying explanations. Apparently, Warburg did not keep scientists who were developing a reputation and becoming more senior, although Krebs has said that Warburg did not think he would be successful in a research career. Also, Krebs and Warburg disagreed about how to investigate respiration within cells. Warburg was interested in comparing normal and cancer cells, whereas Krebs wanted to investigate the steps of specific metabolic reactions, an approach that Warburg regarded as a distraction from his research goals. At any rate, Krebs left Warburg's laboratory in 1930 for the University of Freiburg, where he returned to medical practice, but also did laboratory research.
Krebs chose the synthesis of urea in the liver as his first project “because it appeared to be one of the simplest biosynthetic processes and one that occurs at a high rate.” Using a liver tissue slice assay, Krebs and his research associate, Kurt Henseleit, showed that ornithine acted as a catalyst, as did citrulline and arginine, in the production of urea from ammonium and carbon dioxide. These results were published in a series of three papers that gave Krebs immediate recognition. His key insight was realizing that this pathway was a cycle and not a linear metabolic pathway. This mental preparation and his familiarity now with amino acid metabolism were key elements in his Nobel Prize-winning dissection of the TCA cycle in 1937. Remarkably, Krebs determined, with Hans Kornberg, a third metabolic cycle, the glyoxylate cycle, the glyoxylate bypass of the TCA cycle, in 1957.
Figure 6.
Glutamine and glutamate generate two nitrogen molecules for the urea cycle. Urea contains two nitrogen molecules. Glutaminase generates NH4+ and glutamate. Subsequently, glutamate can be converted into α-ketoglutarate to liberate another NH4+ by glutamate dehydrogenase. Carbamoyl phosphate synthetase (CPS1) uses NH4+, HCO3−, and ATP as substrates to generate carbamoyl phosphate, which provides the first source of nitrogen molecules for urea generation. Amino acids, such as alanine, can also be converted into glutamate through aminotransferases. Next, the aspartate aminotransferase converts glutamate into aspartate, which feeds into the urea cycle to provide the second source of nitrogen molecules for urea production.
The urea cycle contains five enzymatic reactions (Fig. 7). The two initial steps occur in the mitochondria and the other three reactions take place in the cytosol. The initial step for urea synthesis is catalyzed by the enzyme carbamoyl phosphate synthetase I, which produces carbamoyl phosphate from NH4+ and HCO3−, and requires two ATP molecules. Next, carbamoyl phosphate combines with ornithine to generate citrulline in the mitochondrial matrix, a reaction catalyzed by the enzyme ornithine transcarbamoylase. Citrulline is exported to the cytosol, where it is converted into argininosuccinate by argininosuccinate synthetase. This reaction results in the incorporation of a second nitrogen atom from aspartate. In the next reaction, the enzyme argininosuccinase cleaves argininosuccinate to produce fumarate and arginine. Finally, the enzyme arginase converts arginine to urea and ornithine to complete the urea cycle. Note that ornithine is recycled in a fashion similar to oxaloacetate in the TCA cycle. The other molecule that is recycled here is aspartate. The fumarate generated in the urea cycle in the cytosol is converted by cytosolic fumarase to malate, which is transported into the mitochondrial matrix by the malate-aspartate shuttle. Subsequently, malate is converted into oxaloacetate by malate dehydrogenase, generating the reducing equivalent NADH, which can generate ATP through oxidative phosphorylation. Thus, the ATP cost of the urea cycle is offset by the generation of this NADH molecule. The oxaloacaetate generated can be used as a substrate by aspartate aminotransferase to generate aspartate, which is transported back into the cytosol by the malate-aspartate shuttle to be used by the urea cycle. Hence, the urea and TCA cycles are metabolically linked through common metabolic intermediates.
THE ROLE OF AMINO ACIDS BEYOND BIOSYNTHESIS OF MACROMOLECULES AND BIOENERGETICS
An important feature of amino acids, sometimes overlooked, is their central role in the production of catecholamines and neurotransmitters, epigenetics, and modulation of reactive oxygen and nitrogen species. The following sections discuss amino acids that are centrally involved in the production of these important molecules. It is also important to note that certain amino acids can activate signaling pathways. Notably, leucine is a potent activator of mTOR, a powerful kinase that promotes anabolism, as discussed in Chandel (2020).
AMINO ACIDS ARE NECESSARY FOR NEUROTRANSMISSION
Initially, amino acids were not considered as neurotransmitters because they are produced ubiquitously and required for protein synthesis. However, today it is well recognized that the amino acids glutamate and glycine can cause excitatory and inhibitory neurotransmission in the nervous system, respectively. Amino acids are also precursors for neurotransmitters. For example, the amino acid neurotransmitter GABA uses glutamate as a substrate and is produced by an enzyme (glutamic acid decarboxylase; GAD). It is located in neurons and not involved in protein synthesis, unlike glycine and glutamine. Amino acids used for synaptic transmission are compartmentalized into synaptic vesicles, which are released by calcium-dependent exocytosis at presynaptic neurons. Thus, the amino acids used for protein synthesis and neurotransmission are in distinct compartments.
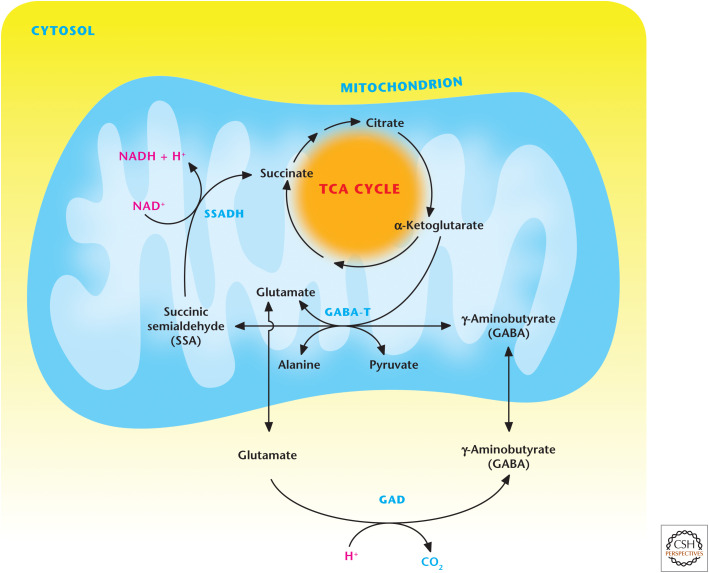
Receptors for glutamate and GABA neurotransmitters are located on postsynaptic neurons can either directly open an ion channel (ionotropic) or couple to the G-protein coupled receptor to elicit their neurotransmission. GABA levels in the brain are produced and conserved by the GABA shunt (Fig. 8). The first step in the GABA shunt is generation of glutamate from the Krebs cycle or glutamine. Subsequently, GAD catalyzes the decarboxylation of glutamate to form GABA. GABA and α-ketoglutarate are metabolized by 4-aminobutyrate aminotransferase (GABA-T) to generate succinic semialdehyde and glutamate. Succinic semialdehyde can be oxidized by succinic semialdehyde dehydrogenase (SSADH) to produce succinate, which can then reenter the TCA cycle, completing the loop. Interestingly, recent studies indicate that lipopolysaccharide-stimulated macrophages also use the GABA shunt to generate succinate, which activates the transcription factor HIF-1 to enhance the production of the cytokine interleukin-1β for innate immunity.
Figure 8.
The GABA shunt produces and conserves GABA levels. The GABA shunt is initiated by GAD, catalyzing the decarboxylation of glutamate to form GABA. Subsequently, GABA and α-ketoglutarate are metabolized by GABA-T to generate succinic semialdehyde and glutamate. Succinic semialdehyde can be oxidized by SSADH succinate and can then reenter the Krebs cycle.
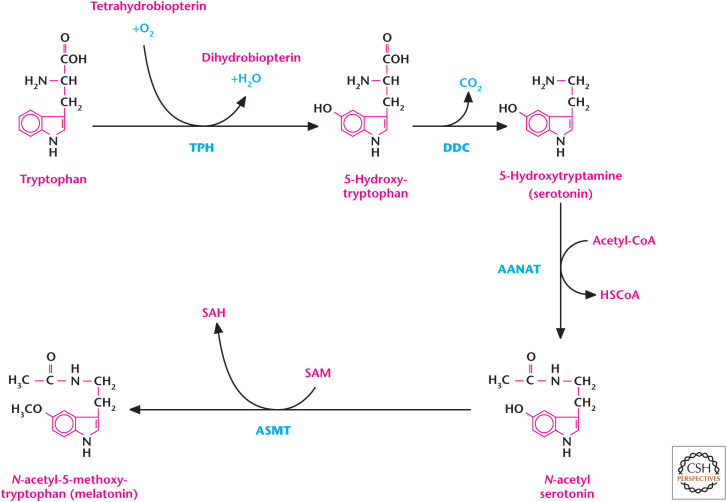
The amino acid tryptophan is used as a precursor for generation of the neurotransmitter serotonin. Increased levels of serotonin are associated with feelings of happiness, whereas low-serotonin levels are linked to depression. The initial rate-limiting step in synthesis of serotonin is the conversion of l-tryptophan to 5-hydroxytryptophan by the enzyme l-tryptophan hydroxylase (Fig. 9). The subsequent metabolic step is the decarboxylation of 5-hydroxytryptophan to generate 5-hydroxytryptamine (i.e., serotonin) by the enzyme aromatic amino acid decarboxylase. Interestingly, the generation of the sleep hormone melatonin is downstream from serotonin synthesis (Fig. 9). Serotonin N-acetyltransferase (arylalkylamine N-acetyltransferase) converts serotonin and acetyl-CoA into N-acetyl-serotonin, which becomes melatonin by acetylserotonin O-methyltransferase.
Figure 9.
Tryptophan produces the neurotransmitter serotonin. The initial rate-limiting step in synthesis of serotonin is the conversion of l-tryptophan to 5-hydroxytryptophan by the enzyme l-tryptophan hydroxylase (TPH). Subsequently, aromatic l-amino acid decarboxylase (DDC) generates serotonin from 5-hydroxytryptophan. Serotonin can also generate melatonin. Arylalkylamine N-acetyltransferase (AANAT) adds an acetyl group to serotonin, resulting in the production of N-acetyl serotonin. Acetylserotonin O-methyltransferase (ASMT) adds a methyl group from S-adenosylmethionine (SAM) to N-acetyl serotonin, resulting in the formation of S-adenosylhomocysteine (SAH) and melatonin.
TYROSINE METABOLISM GENERATES NEUROTRANSMITTERS, CATECHOLAMINES, AND MELANINS
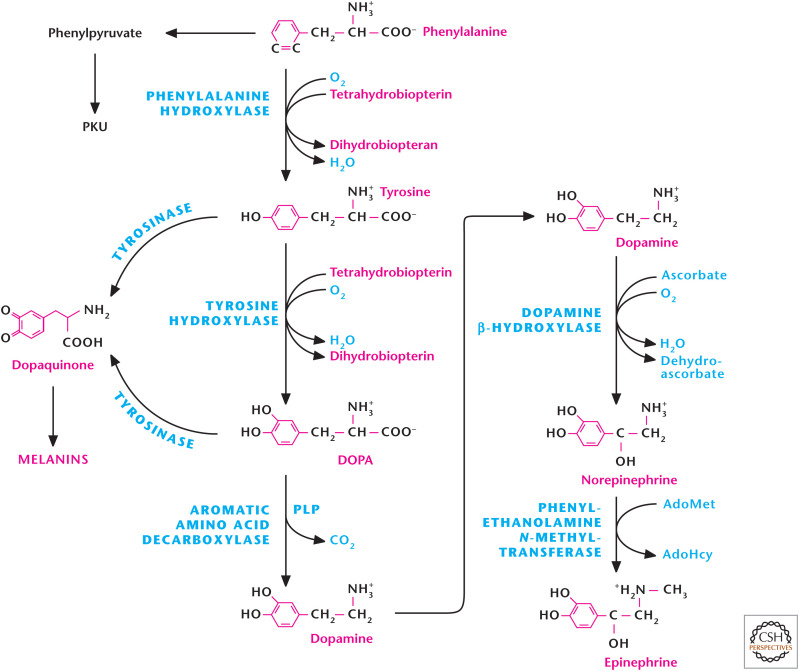
Tyrosine is the precursor for the catecholamines norepinephrine, epinephrine, and dopamine (Fig. 10). Tyrosine hydroxylase converts tyrosine to generate dihydroxyphenylalanine (l-DOPA), a metabolic precursor to dopamine and dopaquinone. Tyrosine hydroxylase requires the enzyme cofactor tetrahydrobiopterin. l-DOPA is converted to dopamine by the enzyme aromatic amino acid decarboxylase. Dopamine is a potent neurotransmitter that is required for numerous brain functions. Notably, patients with Parkinson's disease, a debilitating condition marked by motor impairment and tremors, have few normal dopamine-producing cells in the midbrain area called the substantia nigra. Thus, l-DOPA is often prescribed to patients with Parkinson's disease to elevate their dopamine levels. There is also accumulating evidence that too-high levels of dopamine are observed in schizophrenia. The antipsychotic drugs for treatment work, in part, by limiting dopamine levels. Dopamine is also a precursor to epinephrine and norepinephrine, produced in the adrenal medulla. Catecholamines are associated with the fight-or-flight response and prepare the body to deal with environmental stress. The effects of catecholamines are associated with the sympathetic nervous system and increase blood pressure, heart rate, and blood glucose levels. Tyrosine is also a precursor to the production of the melanins, eumelanins, and pheomelanins by melanocytes to provide skin and hair pigmentation (Fig. 10). Eumelanins provide dark pigments, such as brown or black, and pheomelanins give rise to red or yellow pigmentation. The ratio of eumelanins and phomelanins in melanocytes dictates skin and hair pigmentation. People lacking the gene tyrosinase cannot generate pigments; thus, they exhibit alibinism.
Figure 10.
Tyrosine metabolism produces neurotransmitters, catecholamines, and melanins. Tyrosine hydroxylase converts tyrosine to generate dihydroxyphenylalanine (DOPA), which is converted to dopamine by the enzyme aromatic amino acid carboxylase. Dopamine generates norepinephrine through dopamine β-hydroxylase. Phenylethanolamine N-methyltransferase converts norepinephrine to epinephrine. Tyrosine is also a precursor for melanins through tyrosine or DOPA oxidation to dopaquinone by the enzyme tyrosinase. Phenylalanine hydroxylase produces tyrosine from phenylalanine. Genetic defects in the phenylalanine hydroxylase gene result in the metabolic disease phenylketonuria (PKU). PKU patients show high levels of phenylalanine that generate toxic metabolites, such as phenylpyruvate, resulting in neurological and developmental problems.
Given the multiple roles of tyrosine metabolism, the maintenance of tyrosine levels within cells is vital. Tyrosine can be obtained from the diet or generated from phenylalanine. The enzyme phenylalanine hydroxylase converts dietary phenylalanine to tyrosine (Fig. 10). Genetic defects in the phenylalanine hydroxylase gene result in the metabolic disease phenylketonuria (PKU), in which phenylalanine accumulates significantly in the blood (Fig. 10). PKU patients show neurological and developmental problems because of the high levels of phenylalanine, which generate toxic metabolites, such as phenylpyruvate, phenylacetate, and phenyllactate. PKU disease is autosomal recessive and one of the more common metabolic genetic disorders. PKU patients are diagnosed shortly after birth as a result of a simple and routine blood test and must be on lifelong strict phenylalanine-limited diet to limit the buildup of phenylalanine and so prevent neurological and developmental complications.
METHIONINE METABOLISM IS NECESSARY FOR EPIGENETIC REGULATION AND CYSTEINE PRODUCTION
Epigenetics is a powerful mechanism by which genes are modulated without altering the underlying DNA code. The histone proteins that wrap DNA can undergo modifications that make the DNA either accessible or inaccessible to proteins that either activate or repress genes. One modification is methylation catalyzed by histone methyltransferase enzymes that add methyl groups to specific residues on different histones. There are also histone demethylases that remove the methyl groups. Methionine provides the methyl group for many histone methyltransferases, as well as other type of methyltransferases, including those involved in the conversion of norepinephrine to epinephrine. These methyltransferases use S-adenosylmethionine (SAM). SAM is generated by condensation of ATP and methionine catalyzed by methionine adenosyltransferase. The methyl group (CH3) is attached to the methionine sulfur atom in SAM. During the generation of SAM, all the ATP phosphates are lost so that only the adenosine component is attached to methionine (Fig. 11).
SAM is converted into S-adenosylhomocysteine (SAH) upon transferring its methyl group to DNA or proteins. SAH is then cleaved by adenosylhomocyteinase to yield homocysteine and adenosine. Methionine synthase can convert homocysteine back to methionine, a reaction that requires 5-MTHF as the methyl donor. The resulting THF can undergo a series of reactions, known as the folate cycle, to generate 5-MTHF (Fig. 11). Dietary folate provides THF. Betaine-homocysteine S-methyltransferase can also generate methionine by using homocysteine and betaine as substrates. Homocysteine can generate cysteine by a series of reactions known as trans-sulfuration (Fig. 11). Homocysteine condenses with serine to produce cystathionine, which is subsequently cleaved by cystathionine γ-lyase (also called cystathionase) to produce α-ketobutyrate and cysteine. α-Ketobutyrate is converted to propionyl-CoA and then, via a three-step process, to the TCA cycle intermediate succinyl-CoA. The cysteine can undergo a series of reactions with glutamate and glycine to generate the cellular antioxidant glutathione (GSH) (Fig. 11). The rate-limiting enzyme of GSH synthesis is the generation of γ-glutamyl cysteine by glutamate-cysteine ligase. It is important to note that cysteine found in the blood is typically cystine, an amino acid formed by the oxidation of two cysteine molecules covalently linked by a disulfide bond. Cystine can be transported into cells and readily converted into cysteine by nonenzymatic reduction and provide another source of cysteine for GSH production. The transporter proteins SLC7A11 or xCT transport cystine in exchange for glutamate.
ARGININE METABOLISM PRODUCES NO
NO has emerged as an important molecule in the past three decades. In 1998, the Nobel Prize in Medicine or Physiology was awarded to Robert F. Furchgott, Louis J. Ignarro, and Ferid Murad “for their discoveries concerning nitric oxide as a signalling molecule in the cardiovascular system.” Robert Furchgott showed that acetylcholine relaxation of smooth muscle cells lining the blood vessels was dependent on an intact endothelium (Fig. 12). This laid the foundation for the discovery of endothelium-derived relaxing factor (EDRF). Louis Ignarro reported that EDRF relaxed blood vessels and determined that EDRF was identical to NO. Ferid Murad went on to show that NO increased cyclic guanosine monophosphate (cGMP) levels, causing relaxation of muscle. A fourth scientist overlooked by the Nobel Committee was Salvador Moncada, who also showed EDRF was NO and that NO is biosynthesized from the terminal guanidine nitrogen atom of l-arginine. There are three nitric oxide synthases (NOSs)—inducible, endothelial, and neuronal NOS—that use l-arginine and NADPH to generate NO and the product citrulline (Fig. 12). NO stimulation of soluble guanylate cyclase generates cGMP, which is normally broken down by phosphodiesterase type 5 (PDE5). The blockbuster drug Viagra used to treat erectile dysfunction (impotence) has the active ingredient sildenafil, which is a PDE5 inhibitor. Viagra prevents the breakdown of cGMP, which sustains blood vessel relaxation, improving blood flow to the penis and maintaining an erection. In addition to NO activation of soluble guanylate cyclase, NO can modulate cellular respiration by competing for the oxygen binding of cytochrome c oxidase in the mitochondrial electron transport chain. Furthermore, NO can cause protein posttranslational modification by incorporating the NO moiety into thiol groups to form S-nitrosothiol (SNO) (Fig. 12). The SNO modification results in a change in protein activity that is analogous to protein phosphorylation. This is a reversible reaction in which thioredoxin proteins can reverse protein SNO modification. This modification has been observed in all phylogenetic kingdoms and is thought to be an ancient signal transduction mechanism.
Figure 12.
Arginine generates NO. NOs use l-arginine and NADPH to generate NO and the product citrulline. NO stimulation of soluble guanylate cyclase (GC) generates cGMP, which is normally broken down by phosphodiesterase type 5 (PDE5) to guanosine monophosphate (GMP). Viagra is a PDE5 inhibitor. NO can also cause protein posttranslational modifications by incorporating the NO moiety into thiol groups to form S-nitrosothiol (SNO). This modified thiol group can return to its reduced form through the actions of thioredoxin (TRX)/thioredoxin reductase (TrxR) or glutathione (GSH)/S-nitrosoglutathione reductase (GSNOR).
Footnotes
From the recent volume Navigating Metabolism by Navdeep S. Chandel
Additional Perspectives on Metabolism available at www.cshperspectives.org
SUGGESTED READING
*This reference is also in this collection.
- Altman LA. 1981. Sir Hans Krebs, Winner of Nobel for Research on Food Cycles, Dies. The New York Times, Obituary, December 9, 1981. [Google Scholar]
- *.Chandel NS. 2020. Signaling and metabolism. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Stamler JS. 2012. Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem 287: 4411–4418. doi:10.1074/jbc.R111.285742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H. 2000. Krebs and his trinity of cycles. Nat Rev Mol Cell Biol 1: 225–228. doi:10.1038/35043073 [DOI] [PubMed] [Google Scholar]
- Krebs HA. 1964. The citric acid cycle. Nobel lecture, December 11, 1953. In Nobel Lectures, Physiology or Medicine 1942–1962. The Nobel Foundation, Elsevier, Amsterdam. [Google Scholar]
- Krebs HA. 1973. The discovery of the ornithine cycle of urea synthesis. Biochem Education 1: 19–23. [Google Scholar]
- Locasale JW. 2013. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13: 572–583. doi:10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. 2012. Metabolic regulation of epigenetics. Cell Metab 16: 9–17. doi:10.1016/j.cmet.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada S, Higgs EA. 2006. The discovery of nitric oxide and its role in vascular biology. Br J Pharmacol 147: Suppl. 1: S193–S201. doi:10.1038/sj.bjp.0706458 [DOI] [PMC free article] [PubMed] [Google Scholar]