Abstract
Embryogenesis in seed plants is the process during which a single cell develops into a mature multicellular embryo that encloses all the modules and primary patterns necessary to build the architecture of the new plant after germination. This process involves a series of cell divisions and coordinated cell fate determinations resulting in the formation of an embryonic pattern with a shoot-root axis and cotyledon(s). The phytohormone auxin profoundly controls pattern formation during embryogenesis. Auxin functions in the embryo through its maxima/minima distribution, which acts as an instructive signal for tissue specification and organ initiation. In this review, we describe how disruptions of auxin biosynthesis, transport, and response severely affect embryo development. Also, the mechanism of auxin action in the development of the shoot-root axis and the three-tissue system is discussed with recent findings. Biological tools that can be implemented to study the auxin function during embryo development are presented, as they may be of interest to the reader.
In seed plants, embryogenesis is the process of embryo development that begins with the fertilization of an ovule (de Vries and Weijers 2017). This process will determine the overall architecture of the new plant after germination. The coordinated growth of embryonic tissues governs the final shape and size of the seed that ultimately influence the overall quality and quantity of seed production. In recent years, multiomics approaches, including genetics, epigenetics, transcriptomics, and metabolomics are being used to advance our understanding on embryogenesis mostly in the model plant Arabidopsis (Robert et al. 2013; Palovaara et al. 2017; Gao et al. 2019; Han et al. 2019; Radoeva et al. 2019a; Zhou et al. 2019).
Like other developmental processes, embryo development involves the concerted action of several genetic and environmental factors, where the phytohormone auxin plays a prominent role (Mroue et al. 2018). Auxin allows proper polarity and patterning of the developing embryo through its distribution, which is maintained by a local auxin production together with well-coordinated transport (Friml et al. 2003; Cheng et al. 2007; Stepanova et al. 2008; Robert et al. 2013, 2015b; Locascio et al. 2014). The auxin signaling pathway interprets these auxin maxima into proper developmental programs (Hardtke and Berleth 1998; Hamann et al. 2002; Schlereth et al. 2010).
This review aims to offer a comprehensive overview of auxin actions during embryo development. We will highlight recent findings that together define the role of auxin in proper embryo patterning, including some of the latest tools to study this role.
PLANT EMBRYOGENESIS
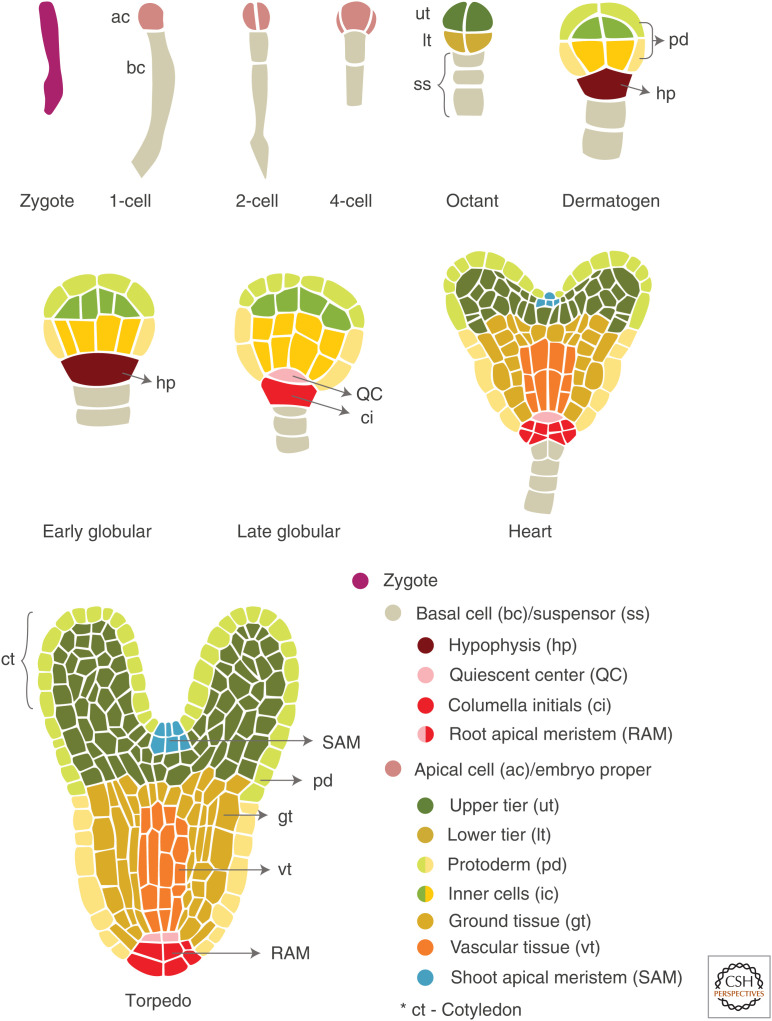
During plant embryogenesis, the formation of an axial–radial pattern ultimately determines the developmental fates of the embryo (Capron et al. 2009). In Arabidopsis, it starts with the asymmetrical zygotic division that generates a small apical cell with dense cytoplasm and a large vacuolated basal cell (Fig. 1). After two rounds of longitudinal divisions and one round of transverse division, the apical cell converts into an octant-stage embryo (eight-celled). The basal cell divides anticlinally. It produces six to nine suspensor cells that help in connecting the embryo to the vascular system of the plant for nutritional purposes (Capron et al. 2009). The eight-celled embryo has two domains, an upper tier and a lower tier (Fig. 1). The upper tier domain of the embryo proper, corresponding to the apical domain, gives rise to the shoot apical meristem (SAM) and the cotyledons (embryonic leaves). The lower tier will generate the embryonic root and hypocotyl (protoderm, ground, and vascular tissues). At the globular stage, the upper suspensor cell, the hypophysis, divides asymmetrically to contribute to the formation of the root apical meristem (RAM).
Figure 1.
Schematic overview of Arabidopsis embryo morphogenesis. Embryo development is shown from the zygote to the torpedo stage. See the text for details.
Radial patterning begins when periclinal divisions give rise to the protoderm, an embryonic epidermis, resulting in a 16-celled embryo (Fig. 1). Divisions of the inner cells, aligned with the apicobasal axis, will form the vascular tissues surrounded by ground tissue cells that will generate endodermis and cortical cell types. The development of cotyledons marks the end of the globular stage, thus giving the embryo a bilateral symmetry and a heart-shaped appearance. Further, the development of cotyledons and cell elongation along the embryo apicobasal axis give rise to a torpedo-shaped embryo (Fig. 1). Accumulation of seed storage reserves such as proteins, fats, lipids, and carbohydrates marks the beginning of the seed-filling phase (Baud et al. 2008). Finally, in the maturation stage, the seed becomes tolerant to desiccation by losing water and enters into the state of dormancy.
PROPER EMBRYO DEVELOPMENT REQUIRES AUXIN BIOSYNTHETIC GENES
Auxin biosynthesis in plants is extremely complex, involving multiple pathways for de novo auxin production. Indole-3-acetic acid (IAA) is the predominant natural auxin in higher plants, which is mainly synthesized via tryptophan-dependent biosynthetic pathways, named after their first-formed intermediate metabolite (Mashiguchi et al. 2011; Ljung 2013; Zhao 2014). One of these pathways, the indole-3-pyruvate (IPyA) pathway, involves two steps, first the conversion of tryptophan to IPyA by the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) family, followed by the oxidative decarboxylation of IPyA by the YUCCA (YUC) family to produce IAA. The importance of TAA1 and YUC genes in the auxin biosynthesis pathway has been demonstrated by analyzing their mutant phenotypes. Loss-of-function mutants of TAA1 and TAA-RELATED 1 (TAR1) genes and multiple yuc mutants exhibit severe auxin-deficient phenotypes such as impaired embryogenesis, defects in vascular and floral development, and complete sterility, demonstrating the predominant developmental role of the TAA1-YUC pathway in auxin biosynthesis (Cheng et al. 2007; Stepanova et al. 2008; Robert et al. 2015a; Brumos et al. 2018).
In Arabidopsis, five TAA and 11 YUC genes have been identified (Cao et al. 2019). YUC1, 4, 10, and 11 display an overall similar expression pattern in developing embryos with concentrated expression in the apical region of the globular embryo and later in cotyledons (Cheng et al. 2007). The yuc1 yuc4 yuc10 yuc11 quadruple mutants exhibited impaired embryo development, where the hypophysis failed to develop, and only one cotyledon was observed in most mutant embryos (Cheng et al. 2007). Similarly, GFP-TAA1 activity was observed in the apical parts of the embryo, and later in root meristematic regions. Subsequent analysis of their mutant phenotypes (taa1 tar1 tar2) corroborated their role in proper embryo development as the triple mutant exhibited developmental defects in the embryo similar to the quadruple yuc mutant phenotypes (Stepanova et al. 2008).
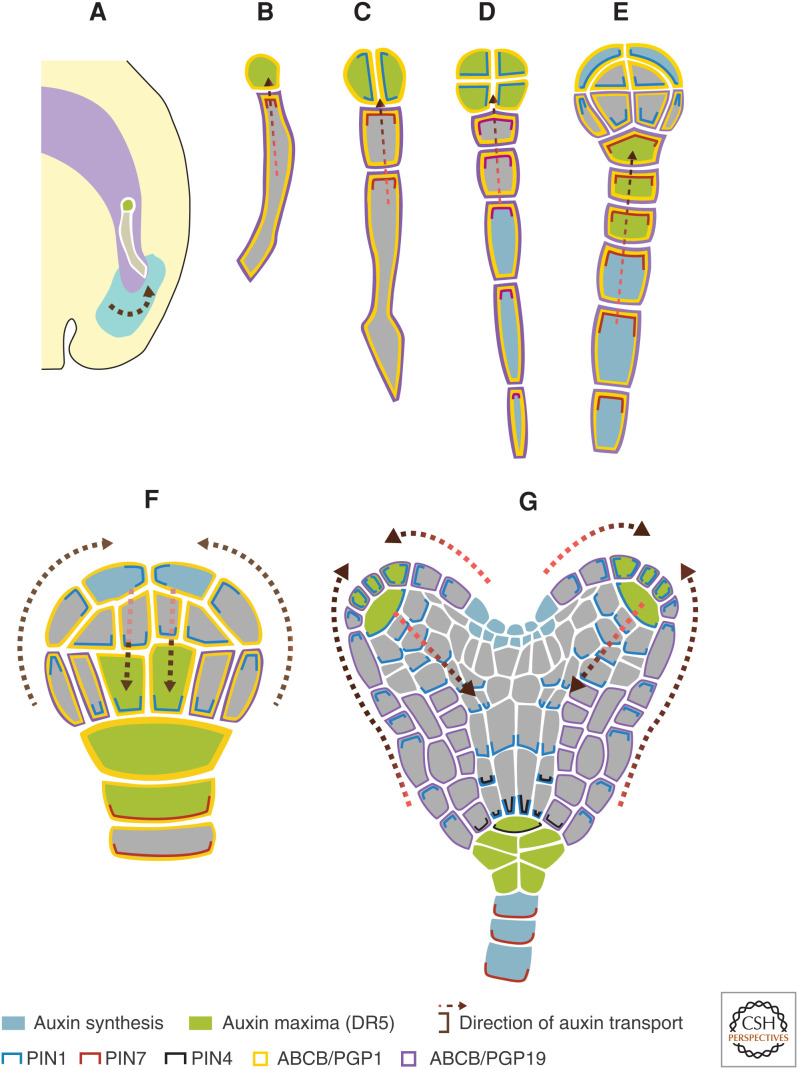
YUC3, 4, and 9 are expressed in the suspensor of the (pre-)globular embryo, indicating their involvement in local auxin biosynthesis in the basal region (Fig. 2D,E; Robert et al. 2013). However, the respective mutants were phenotypically affected in the apical region, suggesting a non-cell-autonomous action of auxin biosynthesis in the development of the apical part of the embryo. Noteworthy, transport played an essential role as auxin is transported from the site of synthesis (suspensor) to the apical embryonic region by PIN7 auxin carriers (Fig. 2; Friml et al. 2003; Robert et al. 2013). Likewise, the onset of auxin biosynthesis by TAA1, YUC1, and YUC4 was observed in the apical region of the globular embryo (Robert et al. 2013). From there, auxin flow, mediated by PIN1 proteins, led to the formation of auxin maxima in the basal region, which is required for the specification of the future root pole (Fig. 2; Friml et al. 2003; Robert et al. 2013). In a recent study, it was shown that the auxin biosynthesis machinery involving TAA1/TAR and YUC genes acts in the seed integuments to produce auxin. This maternally produced auxin contributes to early embryo development (Fig. 2; Robert et al. 2018).
Figure 2.
An illustration depicting auxin synthesis (in blue, contributed by TAA1 and YUC proteins), auxin maxima (in green), and transport (shown PIN1 [blue lines], PIN4 [black lines], PIN7 [red lines], ABCB/PGP1 [yellow lines], and ABCB/PGP19 [purple lines]) during Arabidopsis embryo development. (A) During the early development of an embryo, maternal tissue supplies auxin to the developing embryo, depicted by the arrow. (B–G) One-cell, two-cell, eight-cell, 16-cell, 32-cell, and heart-stage embryos, respectively. Dashed arrows depict the direction of auxin transported, extrapolated by the cellular localization of the auxin efflux carriers. Auxin influx carriers are not indicated.
AUXIN TRANSPORT IN GOVERNING EMBRYO DEVELOPMENT
Auxin is transported by translocation through the phloem, chemiosmotic gradient (passive membrane diffusion), and directional (polar) transport by influx and efflux carriers located in the plasma membrane. The current understanding of auxin transport during embryo development has been acquired by studying auxin reporter lines (such as DR5) and mutant phenotypes (Friml et al. 2003; Furutani et al. 2004; Blilou et al. 2005; Mravec et al. 2008; Ugartechea-Chirino et al. 2010; Robert et al. 2015b; Swarup and Bhosale 2019).
Active auxin transport by cellular influx is mediated in the embryo by three importer proteins, AUX1 (AUXIN RESISTANT 1), LAX1 (LIKE AUX1), and LAX2, belonging to the AUX/LAX family, part of the amino acid permease superfamily (Mironova et al. 2017; Swarup and Bhosale 2019). AUX1 expression begins from the 32-cell stage in the inner cells and later restricted to the provascular cells. LAX2 follows a similar expression pattern, in addition to the uppermost suspensor cell and hypophysis. In contrast, LAX1 is expressed in the apical cell, then restricted to the upper tier and later to the cotyledon tips (Robert et al. 2015b). These influx proteins were reported to be important for the formation and organization of shoot and root apex (Ugartechea-Chirino et al. 2010; Robert et al. 2015b).
Auxin efflux can only be achieved by active transport via efflux carriers. It is mediated by PIN (PIN-FORMED) and ABCB/PGP (B subfamily of ATP binding cassette transporters/P-glycoproteins) carriers (Fig. 2). Four PIN proteins (PIN1, 3, 4, and 7) are active during embryo development. Their redundant function is demonstrated by an absence or the presence of subtle embryonic phenotypes in any single pin mutants. However, a quadruple mutant (pin1 pin3 pin4 pin7) shows severe patterning defects stating the importance of PINs (Friml et al. 2003; Blilou et al. 2005). Nevertheless, the mutant embryos remain viable and can develop into seedlings (Verna et al. 2019).
PIN7 is restricted to the suspensor cells. Its expression is detectable after the zygotic division. It localizes to the apical membrane of the basal cell (Fig. 2B), driving the auxin from the integuments into the apical cell (Fig. 2A; Robert et al. 2018). Auxin accumulation in the apical cell is crucial for its specification into an embryo proper. The pin7 mutant embryos are filamentous without proper apical cell establishment (Friml et al. 2003). At the 32-cell stage, PIN7 switches to the basal membranes of the suspensor cells, supposedly transporting auxin back to the seed integuments (Fig. 2F). Although PIN7 plays a critical role in transporting auxin between the seed tissues and the embryo, its regulation is poorly understood. Recently, Xiong et al. (2019) reported the importance of JANUS, a spliceosome subunit, in the expression regulation of PIN7 in the basal cell. PIN1 expresses in the apical cell lineage. Before the 32-cell stage, PIN1 shows no apparent polarity and is assumed to mediate a uniform auxin distribution within the cells of the embryo (Fig. 2B–E). However, coinciding with the PIN7 polarity switch at the 32-cell stage, PIN1 polarizes to the basal membranes of the inner cells (Fig. 2F). This polarity switch drives auxin into the upper suspensor cells, a critical step for the proper root pole formation. Also, PIN1 plays a key role in positioning and partitioning of cotyledon primordia and boundaries. PIN3 and PIN4 expression starts at the late heart stage in columella precursors and hypophysis, respectively (Fig. 2G; Friml et al. 2003).
ABCB/PGP1 and 19 contribute to the auxin efflux. ABCB1 localizes apolarly to the suspensor and all embryonic cells until the mid-globular stage, whereas ABCB/PGP19 expression is restricted to suspensor lineage (Fig. 2B–F). Later, the ABCB/PGP19 expression extends to the lower tier in the octant and 16-cell stages. However, at the late-globular and mid-heart stages, ABCB/PGP19 overlaps with PIN1 in protoderm cells and expresses in the ground tissue cells (Fig. 2G). In contrast to pin multiple mutants, pgp1 pgp19 embryos are not morphogenetically defective. However, these transporters exert synergistic effects. Indeed, the pin1 pgp1 pgp19 triple mutant displays stronger defects in the development of cotyledons than pin1, pgp1, and pgp19 alone (Mravec et al. 2008).
AUXIN PERCEPTION AND RESPONSE IN MEDIATING EMBRYO DEVELOPMENT
The signaling mechanisms of auxin action are of central importance in modulating different developmental aspects and responses to environmental cues (Taylor-Teeples et al. 2016; Mroue et al. 2018). Auxin acts as a “molecular glue” that promotes interaction between an F-box protein as auxin receptor, TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB), and the members of Aux/IAA transcriptional repressors (Leyser 2018). This auxin-mediated binding between TIR1/AFB and Aux/IAA promotes the ubiquitination-dependent degradation of Aux/IAA proteins, which in turn releases the transcription factors (TFs) belonging to the AUXIN RESPONSE FACTOR (ARF) family (Lavy and Estelle 2016; Leyser 2018). These TFs bind to auxin response elements (AuxREs) present over the promoter sequences of different target genes and regulate their expression (Mironova et al. 2014).
The TIR1/AFB F-box proteins are the substrate recognition component of SCF (Skp1, Cullin, and F-box protein) ubiquitin-ligase complexes (Salehin et al. 2015; Leyser 2018). Six AFB genes, including TIR1 and AFB1-5, have been identified in Arabidopsis (Dharmasiri et al. 2005a; Prigge et al. 2020). Genetic and molecular analysis revealed their involvement in mediating auxin response throughout plant development, including embryogenesis (Dharmasiri et al. 2005b; Parry et al. 2009; Prigge et al. 2016, 2020). Mutant phenotypes of all six TIR1/AFB genes were analyzed individually and in 63 mutant combinations (Dharmasiri et al. 2005b; Prigge et al. 2020). Embryogenesis was not affected in any of the single mutants, probably due to the functional redundancy. In contrast, the mutant combinations displayed a wide range of developmental defects during the root, shoot, gametophyte, and embryo development. Out of those mutants, the triple (tir1 afb2 afb3), quadruple (tir1 afb2 afb3 afb5 and tir1 afb1 afb2 afb3), and sextuple (tir1 afb1 afb2 afb3 afb4 afb5) mutants exhibited severe embryonic phenotypes such as abnormal cell division in the upper and lower tiers of the embryo, lack of cotyledons, aberrant hypophysis development, and overproliferating suspensors (Dharmasiri et al. 2005b; Prigge et al. 2020). Also, one-quarter of the sextuple mutant embryos were either lethal or aborted. Besides, defects in radial patterning events were also observed in the tir1 afb2 afb3 afb5 mutant. These studies demonstrate the roles of the TIR/AFB subunit of SCFTIR1/AFB complexes during embryo development.
In Arabidopsis, 15 out of 29 Aux/IAA short-lived repressors are expressed in embryo and suspensor (Weijers and Jürgens 2005; Ploense et al. 2009; Rademacher et al. 2012). Only a few have been functionally studied in combination with their ARF counterparts. ARFs, downstream components of the auxin response pathway, act either as activators or repressors to regulate the expression of their target genes. In Arabidopsis, out of 23 ARF genes, 12 ARFs exhibited diverse expression patterns throughout embryogenesis, suggesting their potential role in different developmental stages of Arabidopsis embryo (detailed in Rademacher et al. 2011). However, until now, strong embryonic phenotypes in Arabidopsis have only been observed for mutations in the ARF5/MONOPTEROS (MP) gene and its repressor IAA12/BODENLOSS (BDL) (Berleth and Jurgens 1993; Schlereth et al. 2010; Rademacher et al. 2012). Also, ARF9 and ARF13, and their common repressor IAA10, regulate hypophysis specification and suspensor cell identity in Arabidopsis embryo (Rademacher et al. 2012).
AUXIN IN SUSPENSOR-DERIVED EMBRYOGENESIS
Derived from the basal cell of the two-celled embryo, suspensor cells remain extraembryonic except for the uppermost suspensor cell. This uppermost suspensor cell becomes the hypophysis in dermatogen embryos and ultimately participates to the embryonic root development. The other suspensor cells become quiescent after the suspensor is fully developed by the early globular stage (Mansfield and Briarty 1991). Later, the suspensor degenerates via programmed cell death and, therefore, does not contribute to the mature seed (Peng and Sun 2018). In Arabidopsis, despite their quiescence, suspensor cells can develop secondary embryos after impairment of the proembryo through mutations, treatments that damage or kill embryonic cells (Schwartz et al. 1994; Vernon and Meinke 1994), cell ablation (Weijers et al. 2003), or laser disruption (Gooh et al. 2015; Liu et al. 2015). This suggests that suspensor cells have an innate capability to generate embryos, and this activity is suppressed by an embryonic signal. Rademacher et al. (2012) demonstrated that a cell-autonomous auxin response (mediated by IAA10 and ARF9) is required to maintain the extraembryonic identity of suspensor cells (Fig. 3B). When this local auxin response is inhibited, suspensor cells express embryonic marker genes and acquire embryonic fate, resulting in the formation of a second embryo and eventually twin-like seedlings. This auxin-mediated, suspensor-derived embryogenesis is a complex process, which involves several genes associated with auxin homeostasis and response (Radoeva et al. 2019b). Among them, the prominent ones are bHLH genes (bHLH49, 60, 63, and 100) that mediate the auxin response and control suspensor proliferation and/or embryonic fate conversion. In particular, bHLH49 is a direct mediator of the auxin response that controls the developmental potential of the suspensor (Radoeva et al. 2019b). Reprogramming of suspensor cells toward embryonic identity is triggered by defined regulators. This process is marked by the activation of cell divisions that generate cells in which reprogramming occurs, followed by loss of suspensor identity and activation of embryonic fate in newly formed cell clusters (Radoeva et al. 2020).
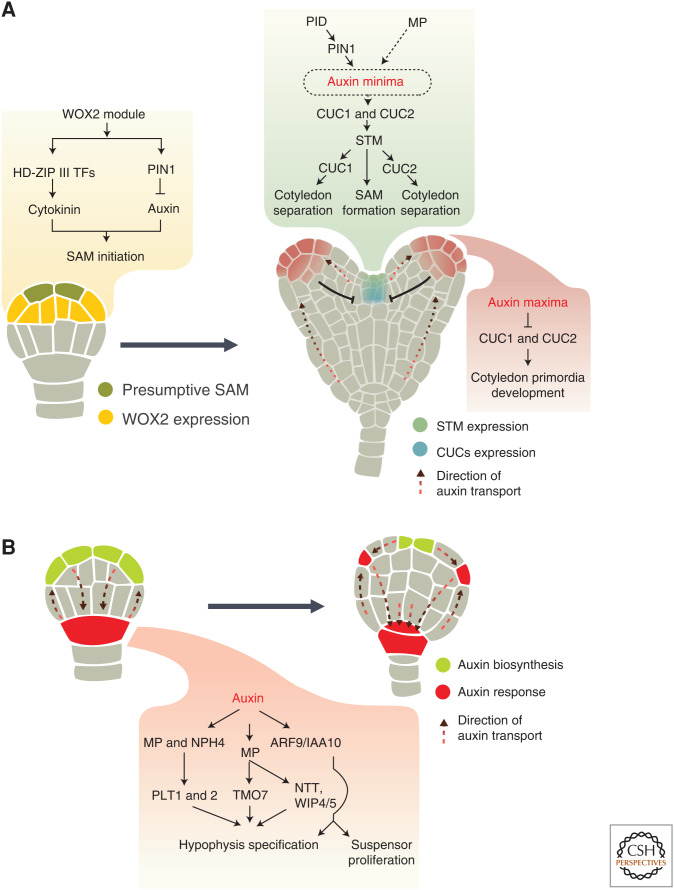
Figure 3.
A model depicting initiation of shoot apical meristem (SAM) and root apical meristem (RAM) during embryogenesis. (A) At the globular stage, the WOX2 module acts in SAM initiation by promoting the expression of HD-ZIP III transcription factors (TFs) and PIN1 in the apical region, which in turn enhances cytokinin response and represses auxin response, respectively. At the early heart stage, the expression of CUC1 and CUC2 is regulated by the activity of PIN1, PINOID (PID), and MP, most probably by creating an auxin gradient with a maximum in the cotyledon primordia and minimum in the center of the embryonic apex. CUC genes are preferentially expressed in the region with low auxin levels (i.e., between the cotyledon primordia where they promote cotyledon separation). CUC proteins are also required to activate STM expression. STM promotes SAM formation and also contributes to cotyledon separation by regulating CUC1 and CUC2 expression. PIN1 and PID-mediated auxin maxima suppress CUC genes expression in cotyledon primordia that eventually allows primordia to develop and also prevents (T-shaped arrow) the expression of CUC genes from expanding. (B) PIN-mediated basal auxin accumulation promotes MP and NPH4 activity, which is required for hypophysis specification. MP, through its targets (PLT1, PLT2, TMO7), triggers the asymmetric division of hypophysis. The ARF9/IAA10 module is required for hypophysis specification and to control suspensor cell proliferation. The dashed black circle and arrow in the green inset in A indicate signaling steps that need further experimental validation. The direction of auxin transport is represented by a dashed line.
AUXIN-MEDIATED EMBRYO PATTERNING
In plant embryo, body poles are anatomically termed as apex and base (Friml et al. 2006). During apicobasal patterning, SAM and cotyledons are originated from the apex (upper tier), whereas the base (lower tier) produces hypocotyl, radicle, and RAM. In contrast, the development and arrangement of a three-tissue system (protodermal, vascular, and ground tissue) occur during radial patterning (Capron et al. 2009). In this part, we will review the role of auxin in pattern formation during embryogenesis.
Auxin Control of Cell Division
Embryo patterning relies on coordinated formative cell divisions that produce different cell layers by orienting the division plane (Yoshida et al. 2014). Besides, embryo development heavily depends on differential cell specification, which is predicted by geometric asymmetry during cell division. Auxin overrides the default geometric division rule and forces the cell to select another division wall, therefore, controlling pattern formation during embryo morphogenesis (Yoshida et al. 2014). Auxin affects division plane orientation by influencing the orientation of cortical microtubule (MT) arrays (CMAs) (Chakrabortty et al. 2018). A link between CMA orientation and division plane orientation has been established (Wasteneys 2002; Ehrhardt and Shaw 2006). Auxin-mediated MT stability, the influence of the cell shape, and reduced edge catastrophe are the factors governing CMA orientation that predicts division plane orientation, leading to oriented cell division in the early embryos (one-cell to 16-cell stages) (Chakrabortty et al. 2018). Cell-polarization relative to body axes is also crucial for patterning events. This polarity information influences division orientation during multicellular development, which is mediated by polar, edge-localized SOSEKI proteins (Yoshida et al. 2019). SOSEKI1 (SOK1) has been identified in the transcriptome of the globular-stage embryo in which MP activity was locally inhibited (Möller et al. 2017). It was the second most down-regulated gene (Schlereth et al. 2010). It suggests that ARF5/MP transcriptionally regulates SOK1. SOK1 expression is localized to apical edges of lower-tier inner cells at globular-stage embryo, and in the corners of the hypophysis and vascular cells at the transition to heart stage. Misexpression of SOK1 leads to altered cell division orientations in all cell types in the embryo. However, the detailed cellular mechanism underlying SOK1 activity in division plane orientation is yet to be determined.
Initiation of Apicobasal Polarity: Emphasis on Auxin
Initial separation of the apical and basal cells after the first division of the zygote establishes a foundation for subsequent patterning events (Haecker et al. 2004; Laux et al. 2004; Capron et al. 2009). This crucial fate decision relies upon the differential expression of the WUSCHEL-related homeobox (WOX) TFs along the apicobasal axis (Haecker et al. 2004). WOX2, WOX8/STPL (STIMPY-LIKE; Wu et al. 2007), and WOX9 (STIMPY; Wu et al. 2005) are expressed in different regions of the eight-celled embryo. WOX2 is restricted to the apical region, whereas WOX8 is expressed in the suspensor cells and hypophysis. Conversely, Lie et al. (2012) observed a weak expression of WOX8 in the embryo proper. The expression of WOX9 is confined to the upper suspensor cell at four-cell and eight-cell stages, and the lower tier domain of the eight-celled embryo. The shift of WOX9 expression from the hypophysis to the lower tier is required for the specification of the lower tier domain identity (Haecker et al. 2004). These WOX9 expression dynamics are absent in arf5/mp and iaa12/bdl mutants, suggesting that the MP/BDL-mediated auxin response is required for the changes of WOX9 expression (Haecker et al. 2004).
WOX2 is the main regulator of embryonic shoot patterning in the cascade of WOX genes. The activity of WOX1, WOX3/PRS (PRESSED FLOWER), and WOX5 is only relevant when WOX2 activity is absent. Conversely, whereas expressed in the basal domain, both the basal and apical embryonic lineages in the wox8 wox9 double mutants are defective. Linking auxin to the WOX pathway, wox8 wox9 double mutants showed reduced PIN1 expression, suggesting that WOX8/WOX9 activity is required for normal PIN1 activity and localized auxin response maxima (Breuninger et al. 2008). Likewise, a genetic interaction between WOX2, WOX8, and ARF5/MP was observed during embryo patterning, with strongly enhanced defects in the wox2 wox8 mp triple mutants. WOX2, WOX8, and ARF5/MP act redundantly to regulate PIN1 expression in the cotyledons (Breuninger et al. 2008). Taken together, these observations suggest that WOX genes control embryo patterning in concert with auxin-mediated responses and may involve other auxin-independent factors.
Role of Auxin in Initiating SAM
The SAM development begins with the specification of stem cells between the presumptive cotyledons in the globular embryo (Fig. 1; Kaplan and Cooke 1997; Mayer et al. 1998). During apical patterning, a transition from radial to bilateral symmetry takes place with the development of the cotyledon primordia. The development of cotyledons relies upon two coordinated processes: outgrowth of the cotyledon primordia and development of the cotyledon boundaries (Lie et al. 2012). The well-proportioned separation of cotyledon primordia is obligatory for the establishment of SAM (Aida et al. 1997, 1999).
SAM development and cotyledon separation are regulated by a network of genes involving CUP-SHAPED COTYLEDON1 (CUC1), CUC2, CUC3, and SHOOT MERISTEMLESS (STM) (Barton and Poethig 1993; Aida et al. 1999; Takada et al. 2001; Vroemen et al. 2003). In agreement with their function, the three CUC genes have an overlapping expression domain in a central stripe of cells between the cotyledon primordia in early-heart-stage embryos (Fig. 3A). CUC1 and CUC2 act synergistically with STM to control SAM formation. CUC1 and CUC2 are required to activate STM expression, and STM in turn is necessary for CUC1 activity from early embryogenesis onward and spatial expression of CUC2 at the bending-cotyledon stage (Fig. 3A; Aida et al. 1999). Three essential factors of auxin transport and response pathway (i.e., PIN1, PINOID [PID]), and ARF5/MP, regulate CUC1 and CUC2 expression to confer cotyledon separation and establishment of the bilateral symmetry (Aida et al. 2002; Furutani et al. 2004). PID, a serine/threonine protein kinase, phosphorylates PIN1 proteins for their proper localization and activity (Huang et al. 2010; Zhang et al. 2010). To establish the cotyledon primordia, PIN1 and PID synergistically promote auxin accumulation in cotyledon primordia, which in turn represses the expression of CUC1 and CUC2 therein. Indeed, pin1 and pid mutant seedlings are both defective in the number of cotyledons (Furutani et al. 2004). Moreover, genetic and expression analysis indicated that PIN1 and ARF5/MP restrict CUC1 expression to the central embryonic apex and activate CUC2 expression in the cotyledon boundaries (Fig. 3A). It has been speculated that an auxin minimum in the presumptive SAM region due to the activity of PIN1 and MP may stimulate the expression of CUC genes between the cotyledon primordia (Fig. 3A). Furthermore, PIN1 promotes the establishment of bilateral symmetry in parallel to STM as pin1 stm1 double mutants have completely fused cotyledons (Aida et al. 2002).
WOX2 and WOX8 redundantly promote the establishment of cotyledon boundaries by regulating the symmetric expression of the three CUC genes during apical patterning (Lie et al. 2012). WOX2, along with its redundant paralogs (i.e., WOX1, WOX3, and WOX5) (collectively termed as the WOX2 module), is required for the initiation of the embryonic apical meristem (Fig. 3A; Zhang et al. 2017). The WOX2 module promotes the development of a three-layered shoot meristem by regulating the cell division pattern in the apical domain, including the presumptive SAM region. By promoting PIN1 expression, the WOX2 module prevents auxin accumulation in the SAM region. Furthermore, the WOX2 module is required to enhance the expression of HD-ZIP III genes (PHABULOSA, PHAVULOTA, and REVOLUTA) in the presumptive SAM region. These genes are known regulators of embryonic apical fate and are expressed in the central domain during shoot meristem formation (Smith and Long 2010). This suggests that HD-ZIP III TFs act downstream of the WOX2 module in the same genetic pathway to regulate stem cell initiation during embryonic apical patterning. In addition to the auxin pathway, the WOX2 module also balances the cytokinin pathway through the HD-ZIP III TFs, thus helping in the formation of embryonic shoot meristem (Fig. 3A; Zhang et al. 2017). Therefore, this module contributes to stem cell initiation and protects them from differentiation, thereby ensuring that the SAM region is correctly specified in the apical domain.
Role of Auxin in Embryonic Root Formation
The RAM harbors concentrically arrayed stem cells (initials of different tissue types) around a quiescent center (QC). The asymmetric division of the hypophysis marks the initiation of the RAM at the globular stage (Capron et al. 2009). This division generates an upper lens-shaped cell (progenitor of the QC) and a large basal cell (columella root cap progenitor). The QC functions in maintaining neighboring stem cells by keeping them in an undifferentiated state (van den Berg et al. 1997). The importance of the hypophysis specification in RAM initiation and the involvement of auxin signaling are reflected by the rootless phenotype of two well-studied mutants (i.e., mp and bdl) (Berleth and Jurgens 1993; Hamann et al. 1999, 2002; Weijers et al. 2006).
BDL and MP mRNAs are absent from the hypophysis until its division and confined to the central provascular cells (Hamann et al. 2002), suggesting that both proteins act non-cell-autonomously. Targets of the MP/BDL signaling pathway are AUX1, LAX2, and PIN1 auxin transporters (Schlereth et al. 2010; Robert et al. 2015b), acting in the embryonic inner cells to direct an auxin flow toward the hypophysis. The resulting accumulation of auxin in the hypophysis promotes the RAM specification. However, exogenous auxin treatment could not rescue the rootless phenotype of mp and bdl mutants, indicating the involvement of other MP/BDL-dependent signals in regulating hypophysis specification (Weijers et al. 2006).
Three direct targets of MP, named as TARGET OF MONOPTEROS (TMO) 3, TMO5, and TMO7, were identified in the embryo (Schlereth et al. 2010). TMO5 and TMO7 were found to be involved in MP-dependent embryonic root formation (Fig. 3B). TMO5 functions cell-autonomously as its expression was restricted to its transcriptional domain. In contrast, TMO7 moves from its site of synthesis to the future hypophysis, indicating that TMO7 acts as an intercellular MP-dependent signal in the hypophysis for RAM specification. Other direct targets of MP, such as NO TRANSMITTING TRACT (NTT) and its paralogues WIP DOMAIN PROTEIN 4 (WIP4) and WIP5, are expressed in the hypophysis and redundantly promotes its asymmetric division, which marks the RAM initiation (Fig. 3B; Crawford et al. 2015).
The PLETHORA (PLT) genes are another indispensable gene network regulating the RAM establishment (Fig. 3B). The AP2 domain-containing TFs PLT1 and PLT2 are redundantly fundamental to the development and specification of the root stem cell niche in the embryo (Aida et al. 2004). The expression of both genes is confined to the basal region of the embryo, from where hypocotyl and embryonic root originate. Owing to the lack of PLT1 and PLT2 activity, hypophyseal derivatives divide aberrantly and fail to express QC markers. PLT genes act downstream of the auxin response, as suggested by their elevated expression after prolonged exogenous auxin treatment. Their basal embryonic expression depends on the redundant function of MP/ARF5 and its homolog NON-PHOTOTROPIC HYPOCOTYL4 (NPH4)/ARF7 (Hardtke et al. 2004). Moreover, the ectopic activity of PLT1 and PLT2 induces basal features (i.e., hypocotyl and root stem cell niche) in the embryonic shoot region, even in the absence of auxin. Furthermore, whereas a PIN1- and PIN4-dependent auxin maximum promotes root pole specification (Friml et al. 2003), these same PIN proteins are required to restrict the PLT expression domain to the basal region of the embryo to initiate the root primordia formation (Blilou et al. 2005). Altogether, these observations strongly support the notion that PIN-mediated accumulation of auxin in the basal domain is required to initiate and establish RAM through the transcriptional regulation of PLT genes.
It has been recently demonstrated that PLT genes are required for vascular regeneration in response to wounding, by directly regulating CUC2 expression (Radhakrishnan et al. 2020). To promote vascular regeneration, both PLT and CUC2 modulate local auxin biosynthesis by regulating the YUC4 expression in a coherent feedforward manner. Although this mechanism of the PLT-CUC2 module was observed in leaf, similar mechanisms may exist in the embryo as well.
Role of Auxin for the Formation and Patterning of the Vascular and Ground Tissues
Vascular tissues are essential for transport and structural stability/support. In a recent report, Smit et al. (2020) used a panel of vascular markers to understand the specification and ontogeny of the vascular tissue identity during embryogenesis. Most of these markers are active in the inner cells of a 16-cell embryo, indicating that these cells are the first to contribute to the vasculature (Smit et al. 2020). Vascular markers such as SOK1 and GATA20 lost their activity in bdl embryos, where the MP/ARF5 activity is blocked. The auxin-mediated signaling through MP/ARF5 appears to be required for vascular tissue initiation, specification, and patterning in the early embryo. Indeed, in mp embryos, abnormal periclinal vascular divisions are observed (De Rybel et al. 2013). TMO5, one direct target of MP, forms a heterodimer with LONESOME HIGHWAY (LHW) (Ohashi-Ito and Bergmann 2007) to mediate an auxin response in vascular tissue initiation. This TMO5-LHW dimer is required to regulate vascular indeterminacy and to promote periclinal cell divisions during vasculature formation. Consistent with their function, both proteins colocalize in vascular precursor cells at the mid-globular embryo. At the heart stage, TMO5 accumulation is restricted to xylem initials, whereas LHW accumulates over a wide area with maximum accumulation in the root pole and cotyledon primordia (De Rybel et al. 2013).
An auxin-cytokinin hormonal interaction is critical for vascular tissue development. The TMO5-LHW dimer promotes local cytokinin biosynthesis by regulating the expression of LONELY GUY4 (LOG4), an enzyme involved in the final step of cytokinin biosynthesis (Kuroha et al. 2009; De Rybel et al. 2014). This auxin-dependent cytokinin production is crucial for patterning of the vascular tissue and vascular growth. Mathematical modeling for the activity of the “auxin-MP-TMO5/LHW-LOG4-cytokinin” module in both embryonic and postembryonic roots indicates that this network is required to create two distinct domains during vascular tissue patterning: xylem axis with high auxin response and surrounding procambium cells with high cytokinin response. This network also involves the activity of PIN proteins whose localization dynamics are regulated by auxin-cytokinin intricacies (De Rybel et al. 2014).
While some of the molecular mechanisms for the vascular tissue initiation have been uncovered, the ones for the ground tissue initiation are still mysterious. Möller et al. (2017) observed aberrant division in the first ground tissue cells in the absence of ARF5/MP-mediated auxin response. Embryonic markers for ground tissue initiation were identified by transcriptome profiling of the early globular embryos, in which MP activity was blocked in the first ground tissue cells. Among them, known regulators of postembryonic ground tissue patterning and maintenance, such as SHORT-ROOT (SHR) and its direct target genes, were also present (Helariutta et al. 2000; Nakajima et al. 2001; Levesque et al. 2006). However, the SHR network does not affect division in the first ground tissue cells, suggesting the involvement of distinct regulators in establishing the ground tissue in the early embryo (Möller et al. 2017). Nevertheless, transcriptional regulation of SHR by MP activity implies, so far, an unpredicted role for SHR in initial ground tissue specification, which needs to be elucidated.
Overall, these observations suggest that the apicobasal embryonic polarity is largely regulated by PIN-mediated auxin transport, from a source (local production) to a sink (local signaling). In addition, the activity of ARF5/MP in auxin-dependent regulation of embryonic patterning is intricated within a multiple of signaling cascades.
TOOLS TO STUDY AUXIN FUNCTION IN EMBRYOS
In recent years, a few noteworthy tools joined the auxin toolbox. Among them, we will present two tools (hormone-activated Cas9-based repressors [HACRs], pronounced “hackers” and cellular markers for embryogenesis) that may be used for studying embryo development (Fig. 4).
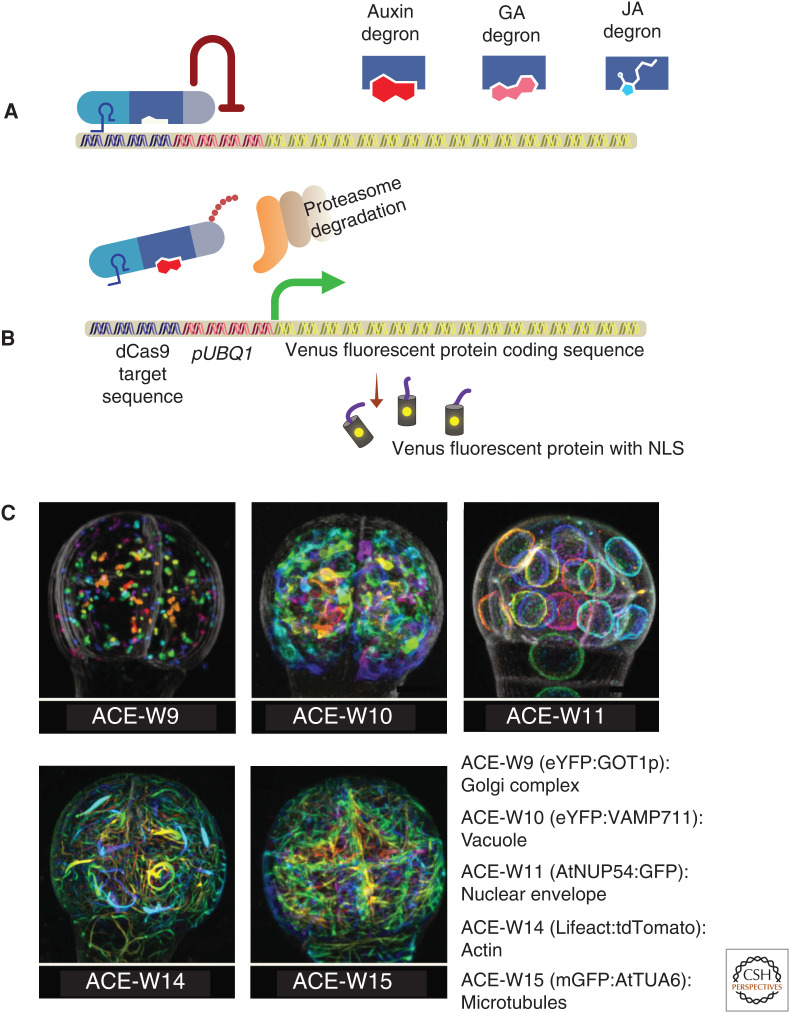
Figure 4.
An illustration representing the mechanisms of HACRs (hormone-activated Cas9-based repressors) (A,B) and images representing a selection of the ACE-W lines (C). (A,B) The HACR system is designed to control the expression of NLS-Venus fluorescent proteins based on the concentration of auxin in the cell. In the absence of auxin (A), TOPLESS (TPL) represses the expression of NLS-Venus. The presence of auxin (B) in the cell targets the HACRs to proteasome-mediated degradation, thus derepressing the NLS-Venus expression. The HACR consists of gRNAs (light blue) targeting the UBQ1 promoter, an auxin-sensitive degron (dark blue), and a repressor domain from TPL (gray). Degrons can be switched to monitor GA or JA levels. (C) Representative images of Arabidopsis 32-cell-stage embryos expressing ACE markers labeling Golgi complex (ACE-W9), vacuole (ACE-W10), nuclear envelope (ACE-W11), actin cytoskeleton (ACE-W14), and microtubules (ACE-W15). (Panel C from Liao and Weijers 2018; reprinted, with permission, from John Wiley © 2018.)
HACRs
HACRs can be used as hormone reporters (auxin, gibberellins, and jasmonates) as well as degron-controlled genetic switches. HACRs are CRISPR-based genetic switches (Khakhar et al. 2018). HACRs are modular proteins designed to temper the expression of a target gene. It contains three domains (i.e., dCas9 [for the recognition of target sequence]), a degron, and a transcriptional regulator (to modulate the target gene expression). Khakhar et al. (2018) generated HACRs with degrons that respond to auxin, gibberellins, and jasmonates (Fig. 4A).
The auxin HACR functions as a radiometric auxin sensor similar to R2D2 (Liao et al. 2015). This HACR contains gRNAs targeting the UBQ1 promoter, an auxin-sensitive degron (from IAA17), and a repressor domain from TOPLESS (TPL) (Fig. 4A). When introduced in plants expressing a radiometric readout (pUBQ1:NLS-Venus; pUBQ1m:NLS-tdTomato, in which pUBQ1m contains a mutated gRNA target site), this HACR displayed a similar expression pattern as in R2D2, indicative of tissular auxin distribution (Fig. 4A,B).
HACRs have a great potential for rewiring the signaling pathways. It includes establishing new feedback loops, targeting genes of one hormone pathway, and using degrons of another hormone. This level of granular control would be challenging to achieve with any other technologies. Applied to embryo development as a readout for patterning programs, HACRs would be a powerful tool to study cell-specific processes and the function of TFs and other effectors.
Arabidopsis Cellular Markers for Embryogenesis
To study the cellular organization and organellar dynamics of early embryo development, Liao and Weijers (2018) developed a series of fluorescent marker lines named “Arabidopsis cellular markers for embryogenesis” (ACE). The ACE expression is under the control of two different promoters, a meristematic pRPS5A (ACE-R) and an embryo-specific pWOX2 (ACE-W). The ubiquitous expression of pRPS5A allows the imaging of the markers throughout embryogenesis. The WOX2 promoter, on the other hand, has a limited window of expression until the early globular stage, with the advantage to be embryo-specific. These subcellular reporters label plasma membrane, connectivity between the cells (plasmodesmata), cytoskeleton (actin and MTs), various organelles (nuclear pore complex for nuclear labeling, vacuole), and components of endomembrane systems (Fig. 4C). Also, the series includes polarly localized proteins to mark apical, basal, central, and peripheral domains of the plasma membrane. These markers are useful to study the polarity of cells and the dynamics of organelles in wild-type and defective mutant embryos in various auxin-related processes.
CONCLUDING REMARKS
Our understanding of auxin function in embryo development has increased substantially in the past decade. The data available so far indicates that auxin acts through its instructive maxima/minima distribution and regulates cell specification and organ initiation during embryo patterning. Such auxin distribution is established and sustained by a structured interplay between local auxin biosynthesis and polar auxin transport. However, many questions remained to be answered regarding auxin synthesis and transport during embryo development. For example, we are far away from knowing the mechanisms behind the transcriptional regulation of auxin biosynthetic genes during embryo development. Yet, another challenge would be to identify the factors coordinating auxin biosynthesis and transport in this complex tissue. MP/ARF5 has been identified as a critical player of auxin-dependent responses in the embryo. However, many distinct combinations of AUX/IAA and ARFs may exist for individual cellular auxin responses. Systematic analysis of these components and identification of their target genes in the embryo will shed new light on the mechanisms by which auxin exerts its function during embryo development. Further, to reach a new level of understanding, biological tools to monitor auxin synthesis, signaling, and responses, notably in developing embryos, are newly available, among them, HACR. Combined with state-of-the-art imaging techniques, like light-sheet microscopy, such tools would enable breakthroughs in embryonic auxin biology, as we have recently seen for male germline differentiation (Valuchova et al. 2020).
ACKNOWLEDGMENTS
We apologize to those colleagues whose work was not included here due to space constraints. This work was supported by the European Regional Development Fund-Project “Centre for Experimental Plant Biology” (No. CZ.02.1.01/0.0/0.0/16_019/0000738). The authors declare no conflict of interest. The funders had no role in the writing of the manuscript or in the decision to publish the results.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. 1997. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857. 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. 1999. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563. [DOI] [PubMed] [Google Scholar]
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M. 2002. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129: 3965. [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. 1993. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823. [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L. 2008. Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arabidopsis Book 6: e0113. 10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Jurgens G. 1993. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575. [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. 2008. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876. 10.1016/j.devcel.2008.03.008 [DOI] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN. 2018. Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47: 306–318.e5. 10.1016/j.devcel.2018.09.022 [DOI] [PubMed] [Google Scholar]
- Cao X, Yang H, Shang C, Ma S, Liu L, Cheng J. 2019. The roles of auxin biosynthesis YUCCA gene family in plants. Int J Mol Sci 20: 6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A, Chatfield S, Provart N, Berleth T. 2009. Embryogenesis: pattern formation from a single cell. Arabidopsis Book 7: e0126. 10.1199/tab.0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortty B, Willemsen V, de Zeeuw T, Liao CY, Weijers D, Mulder B, Scheres B. 2018. A plausible microtubule-based mechanism for cell division orientation in plant embryogenesis. Curr Biol 28: 3031–3043.e2. 10.1016/j.cub.2018.07.025 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2007. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439. 10.1105/tpc.107.053009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford BCW, Sewell J, Golembeski G, Roshan C, Long JA, Yanofsky MF. 2015. Genetic control of distal stem cell fate within root and embryonic meristems. Science 347: 655–659. 10.1126/science.aaa0196 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D. 2013. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24: 426–437. 10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda AS, Wendrich JR, Smit ME, Novák O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, et al. 2014. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345: 1255215. 10.1126/science.1255215 [DOI] [PubMed] [Google Scholar]
- de Vries SC, Weijers D. 2017. Plant embryogenesis. Curr Biol 27: R870–R873. 10.1016/j.cub.2017.05.026 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005a. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. 2005b. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119. 10.1016/j.devcel.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Shaw SL. 2006. Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol 57: 859–875. 10.1146/annurev.arplant.57.032905.105329 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426: 147–153. 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- Friml J, Benfey P, Benková E, Bennett M, Berleth T, Geldner N, Grebe M, Heisler M, Hejátko J, Jürgens G, et al. 2006. Apical-basal polarity: why plant cells don't stand on their heads. Trends Plant Sci 11: 12–14. 10.1016/j.tplants.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M. 2004. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030. 10.1242/dev.01388 [DOI] [PubMed] [Google Scholar]
- Gao P, Xiang D, Quilichini TD, Venglat P, Pandey PK, Wang E, Gillmor CS, Datla R. 2019. Gene expression atlas of embryo development in Arabidopsis. Plant Reprod 32: 93–104. 10.1007/s00497-019-00364-x [DOI] [PubMed] [Google Scholar]
- Gooh K, Ueda M, Aruga K, Park J, Arata H, Higashiyama T, Kurihara D. 2015. Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev Cell 34: 242–251. 10.1016/j.devcel.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Haecker A, Groß-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668. 10.1242/dev.00963 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jurgens G. 1999. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387. [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615. 10.1101/gad.229402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Bartels A, Cheng X, Meyer A, An YC, Hsieh TF, Xiao W. 2019. Epigenetics regulates reproductive development in plants. Plants (Basel) 8: 564. 10.3390/plants8120564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411. 10.1093/emboj/17.5.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. 2004. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131: 1089–1100. 10.1242/dev.00925 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. 10.1016/S0092-8674(00)80865-X [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galván-Ampudia CS, Offringa R. 2010. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142. 10.1105/tpc.109.072678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Cooke TJ. 1997. Fundamental concepts in the embryogenesis of dicotyledons: a morphological interpretation of embryo mutants. Plant Cell 9: 1903. 10.2307/3870553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar A, Leydon AR, Lemmex AC, Klavins E, Nemhauser JL. 2018. Synthetic hormone-responsive transcription factors can monitor and re-program plant development. eLife 7: e34702. 10.7554/eLife.34702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. 2009. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169. 10.1105/tpc.109.068676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Würschum T, Breuninger H. 2004. Genetic regulation of embryonic pattern formation. Plant Cell 16: S190–S202. 10.1105/tpc.016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Estelle M. 2016. Mechanisms of auxin signaling. Development 143: 3226–3229. 10.1242/dev.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. 2006. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. 10.1371/journal.pbio.0040143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiol 176: 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Weijers D. 2018. A toolkit for studying cellular reorganization during early embryogenesis in Arabidopsis thaliana. Plant J 93: 963–976. 10.1111/tpj.13841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210. 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie C, Kelsom C, Wu X. 2012. WOX2 and STIMPY-LIKE/WOX8 promote cotyledon boundary formation in Arabidopsis. Plant J 72: 674–682. 10.1111/j.1365-313X.2012.05113.x [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Zhao J, Tang X, Tian S, Chen J, Shi C, Wang W, Zhang L, Feng X, et al. 2015. Direct evidence that suspensor cells have embryogenic potential that is suppressed by the embryo proper during normal embryogenesis. Proc Natl Acad Sci 112: 12432–12437. 10.1073/pnas.1508651112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K. 2013. Auxin metabolism and homeostasis during plant development. Development 140: 943–950. 10.1242/dev.086363 [DOI] [PubMed] [Google Scholar]
- Locascio A, Roig-Villanova I, Bernardi J, Varotto S. 2014. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: a focus on auxin. Front Plant Sci 5: 412. 10.3389/fpls.2014.00412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. 1991. Early embryogenesis in Arabidopsis thaliana. II: The developing embryo. Can J Bot 69: 461–476. 10.1139/b91-063 [DOI] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci 108: 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. 10.1016/S0092-8674(00)81703-1 [DOI] [PubMed] [Google Scholar]
- Mironova VV, Omelyanchuk NA, Wiebe DS, Levitsky VG. 2014. Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genomics 15(Suppl 12): S4. 10.1186/1471-2164-15-S12-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova V, Teale W, Shahriari M, Dawson J, Palme K. 2017. The systems biology of auxin in developing embryos. Trends Plant Sci 22: 225–235. 10.1016/j.tplants.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Möller BK, ten Hove CA, Xiang D, Williams N, López LG, Yoshida S, Smit M, Datla R, Weijers D. 2017. Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc Natl Acad Sci 114: E2533–E2539. 10.1073/pnas.1616493114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Kubeš M, Bielach A, Gaykova V, Petrášek J, Skůpa P, Chand S, Benková E, Zažímalová E, Friml J. 2008. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135: 3345–3354. 10.1242/dev.021071 [DOI] [PubMed] [Google Scholar]
- Mroue S, Simeunovic A, Robert HS. 2018. Auxin production as an integrator of environmental cues for developmental growth regulation. J Exp Bot 69: 201–212. 10.1093/jxb/erx259 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. 10.1038/35095061 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. 2007. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134: 2959–2968. 10.1242/dev.006296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovaara J, Saiga S, Wendrich JR, van ‘t Wout Hofland N, van Schayck JP, Hater F, Mutte S, Sjollema J, Boekschoten M, Hooiveld GJ, et al. 2017. Transcriptome dynamics revealed by a gene expression atlas of the early Arabidopsis embryo. Nat Plants 3: 894–904. 10.1038/s41477-017-0035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci 106: 22540–22545. 10.1073/pnas.0911967106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Sun MX. 2018. The suspensor as a model system to study the mechanism of cell fate specification during early embryogenesis. Plant Reprod 31: 59–65. 10.1007/s00497-018-0326-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW. 2009. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517. 10.1242/dev.025932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Greenham K, Zhang Y, Santner A, Castillejo C, Mutka AM, O'Malley RC, Ecker JR, Kunkel BN, Estelle M. 2016. The Arabidopsis auxin receptor F-Box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 6: 1383–1390. 10.1534/g3.115.025585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: e54740. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. 2011. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68: 597–606. 10.1111/j.1365-313X.2011.04710.x [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Rios AF, Borst JW, Lukowitz W, Jürgens G, et al. 2012. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 22: 211–222. 10.1016/j.devcel.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan D, Shanmukhan AP, Kareem A, Aiyaz M, Varapparambathu V, Toms A, Kerstens M, Valsakumar D, Landge AN, Shaji A, et al. 2020. A coherent feed-forward loop drives vascular regeneration in damaged aerial organs of plants growing in a normal developmental context. Development 147: dev185710. 10.1242/dev.185710 [DOI] [PubMed] [Google Scholar]
- Radoeva T, Vaddepalli P, Zhang Z, Weijers D. 2019a. Evolution, initiation, and diversity in early plant embryogenesis. Dev Cell 50: 533–543. 10.1016/j.devcel.2019.07.011 [DOI] [PubMed] [Google Scholar]
- Radoeva T, Lokerse AS, Llavata-Peris CI, Wendrich JR, Xiang D, Liao CY, Vlaar L, Boekschoten M, Hooiveld G, Datla R, et al. 2019b. A robust auxin response network controls embryo and suspensor development through a basic helix loop helix transcriptional module. Plant Cell 31: 52–67. 10.1105/tpc.18.00518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoeva T, Albrecht C, Piepers M, De Vries SC, Weijers D. 2020. Suspensor-derived somatic embryogenesis in Arabidopsis. Development 147: dev188912. 10.1242/dev.188912 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J. 2013. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512. 10.1016/j.cub.2013.09.039 [DOI] [PubMed] [Google Scholar]
- Robert HS, Crhak Khaitova L, Mroue S, Benková E. 2015a. The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J Exp Bot 66: 5029–5042. 10.1093/jxb/erv256 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J. 2015b. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142: 702–711. 10.1242/dev.115832 [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, Wójcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, et al. 2018. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553. 10.1038/s41477-018-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. 10.1105/tpc.114.133744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916. 10.1038/nature08836 [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW. 1994. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245. [DOI] [PubMed] [Google Scholar]
- Smit ME, Llavata-Peris CI, Roosjen M, van Beijnum H, Novikova D, Levitsky V, Sevilem I, Roszak P, Slane D, Jürgens G, et al. 2020. Specification and regulation of vascular tissue identity in the Arabidopsis embryo. Development 147: dev186130. 10.1242/dev.186130 [DOI] [PubMed] [Google Scholar]
- Smith ZR, Long JA. 2010. Control of Arabidopsis apical–basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426. 10.1038/nature08843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Swarup R, Bhosale R. 2019. Developmental roles of AUX1/LAX auxin influx carriers in plants. Front Plant Sci 10: 1306. 10.3389/fpls.2019.01306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. 2001. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127. [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lanctot A, Nemhauser JL. 2016. As above, so below: auxin's role in lateral organ development. Dev Biol 419: 156–164. 10.1016/j.ydbio.2016.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugartechea-Chirino Y, Swarup R, Swarup K, Péret B, Whitworth M, Bennett M, Bougourd S. 2010. The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann Bot 105: 277–289. 10.1093/aob/mcp287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valuchova S, Mikulkova P, Pecinkova J, Klimova J, Krumnikl M, Bainar P, Heckmann S, Tomancak P, Riha K. 2020. Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy. eLife 9: e52546. 10.7554/eLife.52546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289. 10.1038/36856 [DOI] [PubMed] [Google Scholar]
- Verna C, Ravichandran SJ, Sawchuk MG, Linh NM, Scarpella E. 2019. Coordination of tissue cell polarity by auxin transport and signaling. eLife 8: e51061. 10.7554/eLife.51061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW. 1994. Embryogenic transformation of the suspensor in twin, a polyembryonic mutant of Arabidopsis. Dev Biol 165: 566–573. 10.1006/dbio.1994.1276 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC. 2003. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577. 10.1105/tpc.012203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO. 2002. Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci 115: 1345–1354. [DOI] [PubMed] [Google Scholar]
- Weijers D, Jürgens G. 2005. Auxin and embryo axis formation: the ends in sight? Curr Opin Plant Biol 8: 32–37. 10.1016/j.pbi.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R. 2003. Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133: 1882–1892. 10.1104/pp.103.030692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G. 2006. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270. 10.1016/j.devcel.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D. 2005. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15: 436–440. 10.1016/j.cub.2004.12.079 [DOI] [PubMed] [Google Scholar]
- Wu X, Chory J, Weigel D. 2007. Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev Biol 309: 306–316. 10.1016/j.ydbio.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Liu HH, Duan CY, Zhang BK, Wei G, Zhang Y, Li S. 2019. Arabidopsis JANUS regulates embryonic pattern formation through Pol II-mediated transcription of WOX2 and PIN7. iScience 19: 1179–1188. 10.1016/j.isci.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, de Reuille PB, Lane B, Bassel GW, Prusinkiewicz P, Smith RS, Weijers D. 2014. Genetic control of plant development by overriding a geometric division rule. Dev Cell 29: 75–87. 10.1016/j.devcel.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Van Der Schuren A, Van Dop M, Van Galen L, Saiga S, Adibi M, Möller B, Colette A, Marhavy P, Smith R, et al. 2019. A SOSEKI-based coordinate system interprets global polarity cues in Arabidopsis. Nat Plants 5: 160–166. 10.1038/s41477-019-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nodzyński T, Pěnčík A, Rolčík J, Friml J. 2010. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci 107: 918–922. 10.1073/pnas.0909460107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tucker E, Hermann M, Laux T. 2017. A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev Cell 40: 264–277.e4. 10.1016/j.devcel.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2014. Auxin biosynthesis. The Arabidopsis Book 12: e0173–e0173. 10.1199/tab.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Shi C, Zhao P, Sun M. 2019. Isolation of living apical and basal cell lineages of early proembryos for transcriptome analysis. Plant Reprod 32: 105–111. 10.1007/s00497-018-00353-6 [DOI] [PubMed] [Google Scholar]