Abstract
Acute myeloid leukemia (AML) is characterized by attenuation of lineage differentiation trajectories that results in impaired hematopoiesis and enhanced self-renewal. To date, sequencing studies have provided a rich landscape of information on the somatic mutations that contribute to AML pathogenesis. These studies show that most AML genomes harbor relatively fewer mutations, which are acquired in a stepwise manner. Our understanding of the genetic basis of leukemogenesis informs a broader understanding of what initiates and maintains the AML clone and informs the development of prognostic models and mechanism-based therapeutic strategies. Here, we explore the current knowledge of genetic and epigenetic aberrations in AML pathogenesis and how recent studies are expanding our knowledge of leukemogenesis and using this to accelerate therapeutic development for AML patients.
Acute myeloid leukemia (AML) is a clonal disorder initiated in hematopoietic stem and progenitor cells (HSPCs) that is characterized by impaired myeloid differentiation and abnormal proliferation at the expense of the normal hematopoietic system. AML is the most common form of acute leukemia in adults; an estimated 19,940 people will be diagnosed in 2020, and 11,180 patients will die of the disease (Siegel et al. 2020). AML is the 10th leading cause of cancer deaths in the United States. AML is most common in older adults, with a median age at diagnosis of 68 yr. Treatment of AML remains a significant challenge owing to the diverse molecular mechanisms contributing to different AML subtypes, and limited therapeutic options. Even though two-thirds of fit patients with newly diagnosed AML achieve morphological remission with intensive chemotherapy consisting of cytarabine and an anthracycline, the majority of these patients eventually relapse (Döhner et al. 2017; Kishtagari et al. 2020).
The first whole-genome sequencing of a cancer patient was of a patient with cytogenetically normal AML, reported more than a decade ago (Ley et al. 2008). Since then, gene discovery studies have identified an extensive catalog of recurrent somatic mutations in different AML subtypes (Mardis et al. 2009; Patel et al. 2012; Cancer Genome Atlas Research et al. 2013; Papaemmanuil et al. 2016; Tyner et al. 2018). These discovery studies have led to a greater understanding of AML biology, including stepwise accumulation of genetic alterations and recurrent mutations in epigenetic modifiers in AML. The types of mutations found in AML include chromosomal rearrangements (such as translocations or inversion), gain or loss of chromosomes (aneuploidy), total or partial gene deletion, point mutation, insertion, or gene duplication/amplification. AML can develop after an antecedent myeloid malignancy (secondary AML [s-AML]), after leukemogenic therapy (therapy-related AML [t-AML]), or without an identifiable prodrome or known exposure (de novo AML). The therapeutic strategy in “fit” patients with AML (primarily influenced by patient-related factors such as advanced age, performance status, and pretreatment comorbidities) is divided into induction and postremission therapy. Induction refers to the use of cytotoxic therapy (combination of cytarabine and an anthracycline) to induce remission, preferably without measurable residual disease (MRD). Standard postremission strategies include lower-intensity cytotoxic therapy and hematopoietic stem cell transplantation (HSCT). The use of different therapies in the postremission treatment is largely determined by prognostic risk stratification (Table 1). The treatment of AML patients who are “unfit” for intensive therapy include low-dose cytarabine, hypomethylating agents, and combination treatment strategies with novel agents (e.g., venetoclax, glasdegib). Recent data suggests remarkable efficacy for venetoclax/azacytidine in older adults with AML who are not candidates for intensive therapy, such that this regimen is quickly emerging as a new alternative for first-line therapy for older adults with AML (Pollyea et al. 2018).
Table 1.
The European LeukemiaNET (ELN) 2017 risk stratification of acute myeloid leukemia (AML)
| Risk category | Genetic abnormality |
|---|---|
| Favorable |
|
| Intermediate |
|
| Adverse |
|
Data reprinted from Döhner et al. (2017), with permission from The American Society of Hematology, © 2017.
In this review, we provide an overview of AML pathophysiology through the prism of genomic and epigenomic alterations and discuss how these mechanisms are perturbed in leukemogenesis. Acute promyelocytic leukemia (APL), a distinct subtype of AML, will not be addressed in this review, as we would posit it is a unique malignancy with distinct mechanisms of transformation and therapeutic options. In essence, we will discuss the pathogenesis of AML by addressing the role of recurrent cytogenetic abnormalities in AML pathogenesis, the underlying molecular events that contribute to AML, how our evolving understanding of the molecular basis of AML is refining prognostic schema, and how these insights are leading to new therapeutic approaches.
CHROMSOMAL ABNORMALITIES
In AML, detection of chromosomal abnormalities by cytogenetic analysis is critically important for diagnosis, risk stratification, and therapeutic decision-making (Fig. 1). Chromosomal abnormalities are detected in ∼50% of patients with AML at the time of diagnosis. Chromosomal abnormalities most commonly contribute to leukemogenesis through the production of a fusion oncoprotein and/or by altering cis-regulatory elements. These large-scale copy-number alterations can serve as founding events and drive disease evolution in AML. Detection of t(8:21)(q22;q22.1), inv(16)(p13.1q22), t(16;16)(p13.1;q22), or translocations generating PML-RARA fusion transcripts (APL) allow the diagnosis of AML, regardless of the percentage of myeloblasts in the bone marrow or peripheral blood.
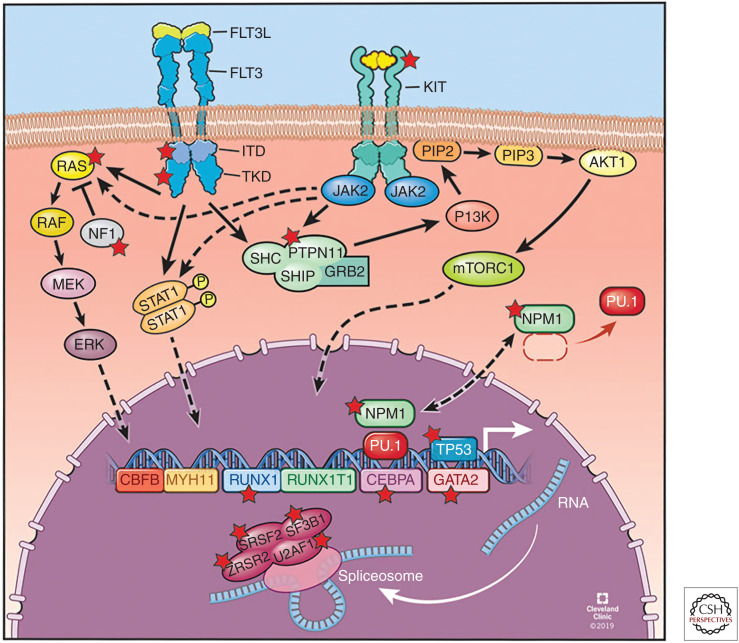
Figure 1.
Overview of frequently mutated cellular pathways in acute myeloid leukemia (AML). These include (1) CBFB-MYH11 and RUNX1-RUNX1T1 fusion oncogenes; (2) nucleophosmin 1 (NPM1); (3) transcription factors RUNX1, CEBPA, and GATA2; (4) tumor suppressors TP53; (5) signal transduction, including FLT3-ITD, FLT3-TKD, KIT, CBL, NRAS, KRAS, PTPN11, and NF1; and (6) RNA splicing: SF3B1, SRSF2, U2AF1, and ZRSR2. Mutated genes are highlighted with a red star. (Refer to the main text for detailed explanations of the depicted processes.) (Figure reprinted with permission, Cleveland Clinic Center for Medical Art & Photography © 2019–2020. All rights reserved.)
Core Binding Factor Rearrangements
The inv(16)(p13;q22) or t(16;16)(p13.1;q22) and t(8;21)(q22;q22.1) translocations lead to the expression of the CBFB-MYH11 and RUNX1-RUNX1T1 fusion oncogenes, respectively. In steady state hematopoiesis, core binding factor beta (CBFβ) and RUNX1 heterodimerize to bind DNA and recruit lineage-defining transcription factors to regulate hematopoietic differentiation (Tahirov et al. 2001). In CBF-AML, chromosomal rearrangements alter this transcriptional complex, thus dysregulating normal hematopoiesis. These rearrangements are present in ∼15% of AML cases. They are the most responsive AML subtypes to intensive chemotherapy, including postremission therapy with high-dose cytarabine (HiDAC) such that patients with these fusion oncoproteins are treated with induction chemotherapy followed by HiDAC consolidation as standard of care (Bloomfield et al. 1998; Byrd et al. 2004). Patients with CBF-AML usually have a relatively good outcome with overall survival rates of 60%–70% in adults. However, the expression of CD56 and the activating mutations in KIT have been correlated to greater risk of recurrence and shorter survival. The KIT mutations are found in 20%–25% of t(8;21)(q22;q22) and in ∼30% of inv(16) (p13;q22) patients (Paschka et al. 2006). However, the role of KIT mutations in prognostication and in molecularly targeted therapies for AML requires further prognostic and therapeutic studies. Both CBFB-MYH11 and RUNX1-RUNX1T1 fusion transcripts are well-established markers for MRD monitoring by real-time quantitative polymerase chain reaction (qRT-PCR) to guide postremission therapy (Schuurhuis et al. 2018).
KMT2A (MLL1) Rearrangements
The mixed-lineage leukemia gene (KMT2A, also known as MLL1) encodes a histone H3 lysine 4 (H3K4) methyltransferase that regulates the expression of target genes, including homoeobox (Hox) genes. In MLL1 fusions, MLL1 retains the DNA-binding amino-terminal domain, and the catalytic methyltransferase activity carboxy-terminal SET domain is replaced by a fusion partner of one of more than 70 partner genes, most commonly AF4 t(4;11), AF9 t(9;11), ENL t(11;19), AF10 t(10;11), and ELL t(11;19) (Krivtsov and Armstrong 2007). The most common fusion partners of MLL1 are members of the DOT1L complex and/or the super elongation complex (SEC), which are both involved in transcriptional control. MLL1-fusion leukemias are characterized by increased expression of HOXA9, MEIS1, and MEF2C, which encode for leukemogenic transcription factors (Armstrong et al. 2002). MLL1 fusions occur in as many as 10% of patients with AML and are common in t-AML following exposure to topoisomerase II inhibitors (Strissel et al. 1998). MLL1-fusion AML carries a poor prognosis because of refractoriness to chemotherapy and shorter period to relapse (Meyer et al. 2018). Aberrant recruitment of the DOT1L by MLL1-fusion leukemias provided a rationale for development of small-molecule inhibitors (e.g., pinometostat) targeting DOT1L with modest results (Stein et al. 2018). Moreover, recent data showing that MLL1 fusions, but not wild-type MLL1, requiring menin as a co-factor to activate downstream target gene expression has led to the development of small-molecule inhibitors of the MLL1/menin interaction, which are now entering clinical trials (Yokoyama et al. 2005; Grembecka et al. 2012; Borkin et al. 2015; Krivtsov et al. 2019; Klossowski et al. 2020). Another mechanism by which MLL1 plays a role in leukemogenesis is through an in-frame partial tandem duplication (MLL1-PTD) spanning exons 3 to 9, exons 3 to 10, or exons 3 to 11 (Steudel et al. 2003). MLL1-PTDs occur in 3.2%–11% of adult AML and more frequently present in AML with normal karyotype and trisomy 11 (Basecke et al. 2006). They confer a worse prognosis with shortened overall survival. MLL1-PTD AML has a distinct gene expression signature, and concomitant DNMT3A and NRAS mutations were associated with adverse clinical outcomes (Hinai et al. 2019).
Rare Translocations and Chromosomal Alterations
inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2); GATA2, MECOM(EVI1)
These rearrangements result in overexpression of the proto-oncogene MECOM (EVI1) at 3q26.2, through juxtaposition of the EVI1 gene with a distal GATA2 enhancer, and simultaneously confers GATA2 haploinsufficiency (Gröschel et al. 2014). It is a distinct entity, which is present in de novo AML or AML arising from prior myelodysplastic syndromes (MDS), and is associated with aggressive disease with minimal response to standard therapies and short survival (Lugthart et al. 2010; Rogers et al. 2014). These rearrangements occur in 1%–2% of all AML cases. Mutations in genes activating RAS/receptor tyrosine kinase signaling pathways are reported in a significant proportion of the cases with these rearrangements; this may provide a target for a rational treatment strategy in this adverse-risk patient group (Gröschel et al. 2015).
t(6:9)(p23;q34.1); DEK-NUP214
This rearrangement results in a fusion of oncogene DEK on chromosome 6 with the nucleoporin gene NUP214 on chromosome 9. The resulting nucleoporin chimeric fusion protein interacts with exportin-1 (XPO1)/CRM1 and may affect the export of nuclear proteins (Saito et al. 2016). It is a distinct entity reported in ∼2% of AML cases and is associated with a very poor prognosis. The majority of the patients with these rearrangements have a concomitant mutation in FLT3-ITD. Outcomes are dismal with standard chemotherapy with the only chance of long-term remission achieved for patients who can undergo allogeneic hematopoietic stem cell transplantation in first complete remission (Díaz-Beyá et al. 2020; Kayser et al. 2020). Given the fusion chimeric protein interaction with XPO1/CRM1, it will be interesting to see whether this fusion imparts sensitivity to XPO1 inhibitors (e.g., selinexor).
Complex Karyotype, -5/5q, -7/7q, -17/17p, +8/8q
Somatically acquired chromosome copy or segment gains, chromosomal monosomies, as well as the accumulation of karyotypic abnormalities, classified as complex karyotype, defined as three or more chromosomal abnormalities, are reported in ∼5%–10% of patients with AML. These alterations carry an adverse prognosis and are associated with a poor response to cytotoxic chemotherapy. Common abnormalities include those involving chromosomes 5 and 7, particularly in AML arising out of the background of MDS, and in t-AML arising in patients treated with alkylating agents and/or radiation therapy for a previous cancer. About 50% of patients with complex karyotype abnormalities have mutations in TP53, and it is hypothesized that TP53 inactivation allows cells to tolerate and persist in a setting of ongoing chromosomal instability. Moreover, it remains unclear whether the poor prognosis of complex karyotype AML is due to chromosomal alterations, inactivation of the p53 pathway, or both molecular events acting in concert. Haploinsufficiency of the following genes have been implicated in myeloid malignancies with monosomy 5 or large deletions of 5q [-5/del(5q)], including RPS14, APC, CTNNA1, HSPA9, EGR1, and CSNK1A1 (Joslin et al. 2007; Liu et al. 2007; Ebert et al. 2008; Wang et al. 2010; Chen et al. 2011). Haploinsufficiency or loss of heterozygosity of candidate genes involved in -7/del(7q) include SAMD9/9L, DOCK4, EZH2, CUX1, MLL3, and LUC7L2, each of which has been suggested to contribute to myeloid transformation (Zhou et al. 2011; McNerney et al. 2013; Nagamachi et al. 2013; Chen et al. 2014; Hosono et al. 2014). Loss of chromosome 17p, including the TP53 locus at 17p13.1, is associated with complex karyotype as well as abnormalities in chromosomes 5 and 7. In contrast to AML with balanced translocations, leukemias that develop in the context of 17p alterations are characterized by greater genomic instability (Liu et al. 2016). It is important to note that NF1 is also on chromosome 17 and that some of the events involving 17q result in haploinsufficiency/loss of NF1 and augmented MAPK signaling (Parkin et al. 2010). Other rare rearrangements, such as t(3;17)(q26;q22), involving fusion of the EVI1 gene located on chromosome 3q26.2 and MSI2 (MUSASHI [MSI] family of RNA binding protein) located on 17q are found in AML (De Weer et al. 2008). Somatic acquisition of trisomy 8 is seen in ∼10% of patients with AML. They contribute to leukemogenesis, at least in part, via amplification of MYC, which is located at chromosome 8p24 and is implicated in a spectrum of malignancies including AML (Sloand et al. 2007).
RECURRENT MUTATIONS IN AML
NPM1
More than 50% of patients with normal karyotype AML harbor mutations in nucleophosmin 1 (NPM1) (Fig. 1). NPM1 is a multifunctional nucleus and nucleolar phosphoprotein involved in pleiotropic cellular functions (Falini et al. 2005). NPM1 gene mutations typically occur as insertions in exon 12, causing a frameshift mutation with an added nuclear export signal (NES) motif at the carboxy-terminus; NPM1c protein product thus accumulates aberrantly in the cytoplasm (Falini et al. 2005; Sportoletti et al. 2015). Multiple recent publications have demonstrated that mutant NPM1 is aberrantly localized in the cytoplasm (NPM1c) and this mislocalization is essential for maintaining active chromatin marks that may help maintain expression of key target genes including HOXA and HOXB genes and MEIS1, thereby inhibiting differentiation of leukemic cells (Brunetti et al. 2018; Gu et al. 2018). It has been suggested that NPM1c shuttles PU.1 (SPI1) into the cytoplasm, which abrogates PU.1 mediated repression of HOX/MEIS1 transcription (Gu et al. 2018). The World Health Organization (WHO) classification of hematopoietic tumors recognizes NPM1-mutant AML as a distinct entity (Table 1) (Arber et al. 2016; Döhner et al. 2017). Strikingly, the majority of NPM1-mutated AML have mutations in DNMT3A and FLT3. AML patients with NPM1 mutations without mutations in DNMT3A and FLT3 carry a favorable prognosis, but the co-mutations FLT3-ITD and DNMT3A confer an adverse prognosis (Döhner et al. 2017). In NPM1-mutant AML, prediction of relapse risk by MRD assessment by qRT-PCR demonstrated that persistent NPM1 positivity in blood was the sole predictor of relapse in multivariate analysis irrespective of the co-mutational status (Ivey et al. 2016; Dillon et al. 2020). This further supports its inclusion into daily clinical practice to better guide postremission therapy (Schuurhuis et al. 2018). NPM1 mutation is considered to be a leukemia-initiating event and thus represents an ideal therapeutic target. In a recent study, using Npm1c/Dnmt3a mutant knock-in mice and cell lines, a model of AML development, showed NPM1-mutant induced self-renewal properties in myeloid progenitor cells, which act as leukemia-initiating cells. In mice, oral administration of the small molecule inhibitor, which targets the interaction between the histone methyltransferase MLL1 and adaptor protein menin, eradicated preleukemic Npm1c-mutant myeloid progenitor cells, prevented AML development, and extended survival. This study demonstrates that the elimination of at-risk preleukemic cells using targeted inhibitors may be a promising strategy (Uckelmann et al. 2020).
Mutations Disrupting Transcription Factor Function in AML (Fig. 1)
Transcription factors tightly regulate normal hematopoiesis. Abrogation of specific transcription factor activity results in block in differentiation of hematopoietic progenitor cells and the development of hematologic malignancies, including AML.
RUNX1
RUNX1 encodes the sequence-specific master hematopoietic transcription factor Runt-related transcription factor 1 (RUNX1), which is an essential regulator of hematopoiesis by maintaining the balance between cell lineage specification, self-renewal, and proliferation (Ito et al. 2015). RUNX1 has a highly conserved “Runt” homology domain (RHD), the motif responsible for heterodimerization with CBFβ, which facilitates the specificity and affinity of RUNX1 binding to target genes (Crute et al. 1996). More than 50 mutations in RUNX1, both point mutations and chromosomal rearrangements, have been reported in various hematologic malignancies, including AML. Loss-of-function somatic mutations in RUNX1, which are mechanistically distinct from rearrangements, occur in ∼5%–15% of all patients with AML and confer a poor prognosis (Gaidzik et al. 2016). Mutations in RUNX1 are enriched in patients with intermediate-risk (including normal karyotype AML) disease (Patel et al. 2012; Papaemmanuil et al. 2016). RUNX1 mutations are usually secondary events that drive disease progression, and they are mutually exclusive with NPM1 and CEBPA mutations (Hirsch et al. 2016; Papaemmanuil et al. 2016). They are associated with older age and lower complete response (CR) rates and inferior overall survival (OS) (Mendler et al. 2012). Germline mutations in RUNX1 are associated with familial platelet disorder with propensity to myeloid malignancy (FPDMM), characterized by thrombocytopenia, platelet functional/ultrastructural defects, and a 20%–60% rate of transformation to overt myeloid malignancies (Song et al. 1999).
CEBPA
The transcription factor CCAAT/enhancer-binding protein α (CEBPA) is a master regulator involved in cell fate decisions, including myeloid-lineage commitment (Avellino and Delwel 2017). CEBPA is expressed as two isoforms: the full-length (p42) and the amino-terminally truncated isoform (p30) that lacks the full trans-activation potential of p42. CEBPA mutations in AML harbor CEBPA amino-terminal nonsense or frameshift mutations, resulting in the increased expression of the dominant-negative p30 isoform, diminishing p42 availability to promote myeloid differentiation. Carboxy-terminal in-frame mutations interrupt DNA binding and homodimerization properties of the proteins and often co-occur on a separate allele from amino-terminal mutations. This pattern of mutations produces p30/p30 homodimers as the functional CEBPA transcription factor entity in CEBPA-mutant AML (Pabst et al. 2001; Kirstetter et al. 2008). These loss-of-function CEBPA mutations are reported in 5%–15% of AML patients and occur mostly in younger patients with normal karyotype AML (Fröhling et al. 2004; Cancer Genome Atlas Research et al. 2013). AML patients with biallelic CEBPA mutations, with one allele harboring an amino-terminus mutation and the other allele having a mutation in the carboxyl terminus, exhibit a distinct gene expression signature and a favorable prognosis with the majority of younger patients achieving a cure with standard chemotherapy alone (Table 1) (Wouters et al. 2009; Fasan et al. 2014). Biallelic CEBPA mutations have a JAK-STAT pathway activation signature, and a high percentage of these patients also harbor gain-of-function mutations in CSF3R and increased sensitivity to JAK kinase inhibitors (such as ruxolitinib) ex vivo (Lavallée et al. 2016; Maxson et al. 2016). Germline mutations in CEBPA are associated with autosomal dominant familial AML with 100% penetrance (Smith et al. 2004).
GATA2
GATA2, a member of the zinc finger transcription factor family, is a critical transcriptional regulator of hematopoietic stem and progenitor cell differentiation and self-renewal (Crispino and Horwitz 2017). Homozygous Gata2 knockout in mice are embryonically lethal because of the failure of definitive hematopoiesis (Tsai et al. 1994). Somatic mutations in GATA2 are infrequent in AML, occurring in <5% of AML overall, and are clustered in normal karyotype AML with concurrent biallelic CEBPA mutations (Greif et al. 2012). These mutations include frameshifts, amino acid substitutions, insertions, and deletions scattered throughout the gene but most often clustered in the region encoding the zinc finger domains, leading to impaired DNA binding and affecting transcriptional activity. In its capacity as a tumor suppressor, heterozygous germline mutations in GATA2 in coding or regulatory element regions cause a spectrum of hematopoietic disorders and a predisposition to MDS/AML, often with subsequent acquisition of somatic ASXL1 mutations (Hahn et al. 2011; Ostergaard et al. 2011; West et al. 2014).
Mutations Altering Signal Transduction (Fig. 1)
Deregulation of signaling pathway components is frequently reported in AML. This is associated with pro-proliferative states by constitutive activation.
FLT3-ITD and FLT3-TKD
Mutations in FMS-like tyrosine kinase 3 (FLT3) occur in approximately one-third of all patients with newly diagnosed AML. These mutations can occur either as an in-frame internal tandem duplication within the juxtamembrane domain of the receptor (FLT3-ITD), seen in ∼20%–25% of AML, or as point mutations most commonly in the activation loop of the catalytic domain (FLT3-TKD). Both mutations lead to autoactivation of FLT3-kinase activity and activation of downstream signaling pathways, which lead to proliferation and block in differentiation (Meshinchi and Appelbaum 2009). AML patients with mutations in FLT3-ITD have an adverse prognosis compared to patients with FLT3-TKD or wild-type FLT3, and the majority of these patients relapse after chemotherapy and allogeneic HSCT (Thiede et al. 2002; Sengsayadeth et al. 2012). The poor prognosis is particularly influenced by co-mutation status (DNMT3A mutations, absence of NPM1 mutations) and the allelic ratio of mutant FLT3-ITD to wild-type (Table 1) (Patel et al. 2012; Papaemmanuil et al. 2016). The advent of FLT3 inhibitors (midostaurin, gilteritinib, quizartinib, and crenolinib) in various stages of clinical development represents a paradigm of targeted therapy in AML and has changed practice. These agents act through the competitive inhibition of the ATP-binding site in the FLT3 receptor; however, they vary substantially in their inhibitory properties, pharmacokinetics, and toxicity profiles. There are currently two FDA-approved small-molecule inhibitors (midostaurin and gilteritinib) of FLT3-kinase activity. In randomized clinical trial, comparing midostaurin in combination with chemotherapy or placebo with chemotherapy has been shown to significantly improve survival of untreated FLT3-mutant AML patients (age 18–60 yr; 74.7 vs. 25.6 mo) (Stone et al. 2017). Gilteritinib, a next-generation tyrosine kinase inhibitor, is approved for the treatment of adults who have relapsed and/or have refractory AML with FLT3-ITD and FLT3-TKD mutations (Perl et al. 2019). Of note, therapy has substantive-agent activity, whereas midostaurin has not shown significant activity when given as monotherapy.
KIT
The KIT gene encodes a 145-kDa transmembrane glycoprotein that is a member of the type III receptor tyrosine kinase (RTK) family (Yarden et al. 1987). Upon binding of stem cell factor (KIT ligand), the monomeric KIT receptor dimerizes and becomes autophosphorylated at key tyrosine sites and activates diverse signal transduction cascades essential for cell proliferation/survival, self-renewal, and differentiation (Malaise et al. 2009). Somatic gain-of-function KIT mutations occur in 10% of AML patients, but they are clustered in patients with CBF-AML rearrangements (t(8;21)/RUNX1-RUNX1T1, inv(16)/CBFB-MYH11) and may confer a more adverse prognosis in this otherwise favorable AML subtype (Paschka et al. 2006).
CBL
Casitas B cell lymphoma (CBL) protein functions as E3 ligases that ubiquitinate and negatively regulate receptor tyrosine kinases, including FLT3 and KIT (Mohapatra et al. 2013). Mutations in the E3 ubiquitin ligase CBL have been reported in ∼1%–3% of AML patients. Mutations in CBL result in the loss of ubiquitin E3 ligase activity and resultant increased tyrosine kinase signaling by decreased proteasome-mediated degradation. In preclinical models, there is evidence that FLT3-ITD mutations cooperate with CBL to promote the rapid induction of AML (Taylor et al. 2015). Although, mutations in CBL cause biochemical loss of function, their functional consequences resemble canonical gain-of-function mutations (Sanada et al. 2009). The prognostic significance of these mutations in AML is unclear given the lack of robust data.
NRAS and KRAS
The Ras family of small GTPases, NRAS and KRAS, function as binary molecular switches by cycling between an active GTP-bound (RAS-GTP) and an inactive GDP-bound (RAS-GDP) confirmation that regulate downstream signaling effectors, such as Raf and PI3K (Schubbert et al. 2007). Somatic gain-of-function mutations in RAS are among the most common oncogenic drivers in cancers leading to constitutive activation of receptor tyrosine kinases (Li et al. 2018). NRAS (∼12%) and KRAS (∼5%) mutations are seen in a subset of AML patients at the time of diagnosis (Bacher et al. 2006). They are frequently acquired at the time of progression from MDS to AML and are associated with poor survival. In a recent study, the presence of NRAS and KRAS mutations in patients with AML receiving standard cytotoxic therapy was associated with decreased OS and event-free survival (EFS) (Ball et al. 2019). Mutations in epigenetic modifiers (TET2/IDH/WT1) often co-occur and cooperate with NRAS mutations to promote leukemogenesis (Papaemmanuil et al. 2016). Preclinical studies suggest this cooperativity and dependence on activated Ras signaling may be therapeutically exploited as they are preferentially sensitive to MAPK kinase (MEK) inhibition in mouse models and patient samples (Kunimoto et al. 2018). The emergence or co-occurrence of NRAS and KRAS mutant clones is a common and clinically relevant mechanism of resistance to FLT3 inhibitors, IDH inhibitors, and BCL-2 inhibitors (Amatangelo et al. 2017; McMahon et al. 2019; DiNardo et al. 2020).
PTPN11
The non–receptor protein tyrosine phosphatase PTPN11 (also known as SRC homology 2 domain–containing phosphatase 2 or SHP2) is implicated in several signaling pathways including RAS, JAK-STAT, PI3K, and others (Tartaglia et al. 2003). Mutations in PTPN11 are present in ∼5%–10% of AML cases and enriched in patients older than 60 yr with secondary AML (Makishima et al. 2017). These mutations co-occur and cooperate with AML1-ETO (Hatlen et al. 2016) and MLL-AF9 (Chen et al. 2015) rearrangements.
NF1
The neurofibromin 1 gene (NF1), encodes neurofibromin, a GTPase-activating protein and negative regulator of Ras. Inactivating mutations in NF1 leading to loss of neurofibromin and resulting in prolonged activation of the RAS/RAF/MAPK signaling pathway and ultimately an increased cellular proliferation (Cichowski and Jacks 2001). Recurrent somatic inactivating mutations in NF1 have been recently reported in ∼5% patients with AML (Eisfeld et al. 2018). In the same study, NF1-mutant AML patients had poor outcomes with lower complete remission rates and shorter overall survival when treated with standard chemotherapy.
Mutations in Epigenetic Modifiers: DNMT3A, TET2, IDH1/2, and Polycomb-Group Genes (Fig. 2)
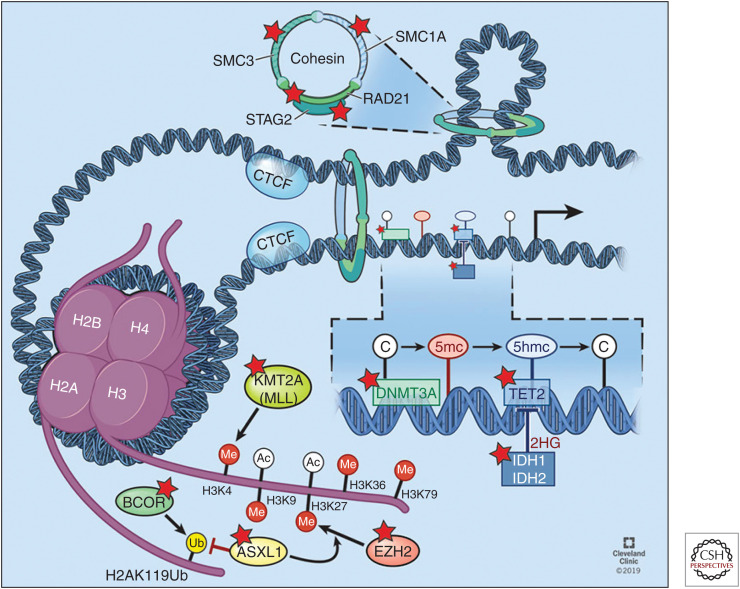
Figure 2.
Overview of frequently mutated epigenetic modifiers. This includes (1) DNA cytosine modifications DNMT3A, TET2, and IDH1/2; (2) histone post-translational modifications ASXL1, BCOR/L1, and MLL1(KMT2A); and (3) Cohesin complex SMC1A, SMC3, and RAD21, bound to STAG1/2. Mutated genes are highlighted with a red star. (Refer to the main text for detailed explanations of the depicted processes.) (Figure reprinted with permission, Cleveland Clinic Center for Medical Art & Photography © 2019–2020. All rights reserved.)
One of the hallmarks of AML pathogenesis is disruption of epigenetic regulation. The mutations in genes encoding epigenetic modifiers are commonly acquired early and are present in the initiating clone. This has led to an increasing interest in the development of epigenetic therapies for AML.
DNMT3A
DNA methyltransferase 3A (DNMT3A) is a member of the DNA methyltransferase family, which includes DNMT1, DNMT3A, DNMT3B, and DNMT3L (Okano et al. 1998). It catalyzes de novo DNA methylation at the C-5 position of cytosine bases of unmethylated CpG dinucleotides. DNA methylation is a critical epigenetic modification and plays an essential role in maintaining hematopoietic homeostasis. Consistent with its function, Dnmt3a-null HSCs up-regulate genes implicated in HSC self-renewal and multipotency, while down-regulating expression of genes associated with HSC differentiation (Challen et al. 2011). Mutations in DNMT3A are observed in ∼30% of AML cases. DNMT3A-mutant AML patients are older and most commonly present with a normal karyotype (Ley et al. 2010; Cancer Genome Atlas Research et al. 2013). The mutations in DNMT3A include nonsense, frameshift, and missense alterations that are enriched within the methyltransferase domain. The recurrent mutation at codon R882 is the most frequent DNMT3A mutation in AML patients, the resultant protein exerts a dominant-negative effect on wild-type DNMT3A methyltransferase activity (Holz-Schietinger et al. 2012; Cancer Genome Atlas Research et al. 2013; Russler-Germain et al. 2014). Mutations in DNMT3A frequently co-occur with mutations in NPM1 and FLT3-ITD, which collectively confer adverse risk (Marcucci et al. 2012; Bezerra et al. 2020). DNMT3A mutations at codon R882 have been shown to promote anthracycline resistance through a proximal defect in nucleosome remodeling and resultant impaired DNA damaging sensing (Guryanova et al. 2016). Clonal hierarchy studies show that mutations in DNMT3A are one of the founder mutations shaping the course of leukemia evolution and progression (Welch et al. 2012), making this an attractive target for the development of novel therapeutic approaches. This is further underscored by studies showing that the DNMT3A is the most frequently mutated gene in clonal hematopoiesis (CH) (Genovese et al. 2014; Jaiswal et al. 2014; Shlush et al. 2014; Xie et al. 2014). Although essential insights into the role of DNMT3A mutations in leukemogenesis have emerged from human and preclinical studies, the fundamental mechanism(s) by which mutant DNMT3A leads to increased self-renewal in HSCs has not been delineated.
TET2
The mammalian ten-eleven-translocation (TET) family proteins TET1, TET2, and TET3, are α-ketoglutarate-dependent dioxygenases. The TET enzymes, in the presence of oxygen, reduced Fe2+, and ascorbic acid, catalyze the stepwise oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), with resultant loss of DNA methylation (Tahiliani et al. 2009; Ko et al. 2010; He et al. 2011; Ito et al. 2011). TET2, but not TET1 or TET3, loss-of-function mutations are common across a spectrum of hematologic malignancies (Delhommeau et al. 2009; Jankowska et al. 2009). Somatic mutations in TET2 are seen in ∼20% of patients with AML, especially in patients with a prior history of MDS or myeloproliferative neoplasm (MPN) (Abdel-Wahab et al. 2009). In AML, TET2 mutations co-occur with NPM1, FLT3-ITD, NRAS, and DNMT3A and are largely mutually exclusive with IDH1/2 neomorphic mutations (Papaemmanuil et al. 2016; Desai et al. 2018). TET2 mutations are among the most common mutations seen in clonal hematopoiesis (CH) (Busque et al. 2012).
Tet2−/− mice develop myeloid and lymphoid neoplasms, after long latencies, through the accumulation of numerous mutations, including Apc, Nf1, Flt3, Cbl, Notch1, and Mll2 (Pan et al. 2017). Increased production of inflammatory mediators (IL-6) and bacterial translocation across gut mucosa may enhance leukemogenesis in Tet2−/− mice (Zhang et al. 2015; Meisel et al. 2018). Recent studies also indicate aberrant expression of diverse metabolic pathways, such as BCAT1, can decrease TET function by inhibiting α-ketoglutarate (Raffel et al. 2017). Supplementation of vitamin C (ascorbate), a co-factor of α-ketoglutarate-dependent dioxygenases like TET enzymes, has recently been shown to enhance residual TET2 function in the heterozygous mutant state (Agathocleous et al. 2017; Cimmino et al. 2017). A clinical trial evaluating the safety and efficacy of high-dose vitamin C for TET2-mutated myeloid neoplasms is ongoing. Despite our mechanistic understanding of the function of TET proteins, how loss-of-function mutations in TET2 initiates and maintains leukemogenesis is largely unknown.
IDH1 and IDH2 Mutations
High-throughput sequencing of AML patient samples identified neomorphic mutations in cytosolic (IDH1 at R132) or mitochondrial (IDH2 at R140Q or R172) isoforms of isocitrate dehydrogenase, which are critical for the oxidative carboxylation of isocitrate to α-ketoglutarate. These mutations contribute to leukemogenesis through the production of the oncometabolite R enantiomer of 2-hydroxyglutratrate (R-2-HG), which, in turn, impedes epigenetic regulation via competitive inhibition of dioxygenases, including the TET family of enzymes and histone lysine demethylases (KDM), by competing with α-ketoglutarate (Mardis et al. 2009; Figueroa et al. 2010; Xu et al. 2011). This leads to a repressive chromatin landscape via DNA and histone hypermethylation, and consequent block in cellular differentiation (Lu et al. 2012; Losman et al. 2013). Mutations in IDH1 and IDH2 are present in ∼5%–10% and ∼15%–20% of patients with newly diagnosed AML, respectively. Mutations in IDH1 and IDH2 mutually exclusive and occur as early clonal events (Papaemmanuil et al. 2016). IDH1 and IDH2 mutations are more frequently observed in patients with trisomy 8 and normal karyotype AML (Medeiros et al. 2017). First-in-class, oral, selective inhibitors of mutant IDH1 (ivosidenib) and IDH2 (enasidenib) are U.S. Federal Drug Administration (FDA)-approved for the management of IDH1/2-mutant adults with refractory or relapsed AML (Stein et al. 2017; DiNardo et al. 2018). Several mechanisms of resistance to enasidenib (IDH2 inhibitor) leading to late relapse have already been described, including acquisition of IDH1-mutated subclones (Quek et al. 2018) or development of additional noncatalytic second-site mutations of IDH2 (Intlekofer et al. 2018).
ASXL1
Additional sex combs-like 1 (ASXL1), located at 20q11.21, is one of the three mammalian homologs of the Drosophila additional sex combs (Asx) gene. Asx functions as an epigenetic regulator that was originally identified as an enhancer of Trithorax (TrxG) and Polycomb group (PcG) genes (Micol and Abdel-Wahab 2016). ASXL1 interacts with BRCA1-associated protein 1 (BAP1) to form the Polycomb repressive deubiquitinase (PR-DUB) complex, which removes monoubiquitin from histone H2A at lysine 119 (H2AK119Ub) (a repressive mark) to activate genes targeted by PRC1 (Scheuermann et al. 2010). ASXL1 also interacts with the PRC2 complex to mediate histone H3 lysine 27 (H3K27) trimethylation (Abdel-Wahab et al. 2012; Inoue et al. 2013). It is yet to be determined if the observed changes in H3K27 methylation with ASXL1 loss are caused by direct interaction between ASXL1 and PRC2 or are secondary to depletion of H2AK119Ub and subsequent failure of PRC2 recruitment (Balasubramani et al. 2015). Somatic mutations in ASXL1 are frequently detected in clonal hematopoiesis and various myeloid malignancies, suggesting they are the initial events in the leukemic transformation. ASXL1 mutations occur in ∼10%–20% of patients with AML and are enriched in those with underlying myelodysplasia (Metzeler et al. 2011). ASXL1 mutations co-exist with those mutations in the splicing factor (SRSF2, U2AF1), signal transduction (NRAS, JAK2, NF1), and transcription factor (RUNX1) (Papaemmanuil et al. 2016). ASXL1 mutations are more common in older patients with AML and confer a poor prognosis (Schnittger et al. 2013). A mechanistic understanding of how ASXL1 mutations in myeloid malignancies drive leukemogenesis remains to be fully elucidated
BCOR and BCORL1
BCL6 corepressor (BCOR) and its closely related homolog, BCL6 corepressor like 1 (BCORL1), are located on the X-chromosome and are targeted by somatic mutations in AML, MDS, chronic myelomonocytic leukemia (CMML), and aplastic anemia (Damm et al. 2013; Yoshizato et al. 2015; Papaemmanuil et al. 2016). BCOR/BCORL1 function as components of PRC1.1, a noncanonical PRC1 complex. PRC1.1 is responsible for the deposition of monoubiquitination on lysine 119 of histone 2A (H2AK119ub1) and mediates transcriptional repression by recruitment of PRC2 and subsequent deposition of H3K27me3 mark (Blackledge et al. 2014; Di Carlo et al. 2019). In mice, loss of Bcor function results in enhanced self-renewal of myeloid progenitor cells and cooperates with KrasG12D to initiate leukemia in vivo (Kelly et al. 2019). Loss-of-function somatic mutations in BCOR and BCORL1 have been reported in ∼1%–4% of AML, and carry an unfavorable prognosis (Grossmann et al. 2011; Damm et al. 2013).
Splicing Factor Mutations (SF3B1, SRSF2, U2AF1, ZRSR2, U2AF2, PRPF8, and LUC7L2)
Almost all human genes are subject to alternative pre-mRNA splicing, a primary source of diversity in messenger RNA species and protein isoforms, which is orchestrated by the macromolecular spliceosome complex (Fig. 1) (Lee and Rio 2015). Recent advances in high-throughput sequencing technologies have unexpectedly identified somatic mutations in the core spliceosomal proteins and associated RNA splicing factors in MDS, CMML, and AML (Anczuków and Krainer 2016). This includes mutations in SF3B1 (splicing factor 3b subunit 1), SRSF2 (serine/arginine rich splicing factor 2), U2AF1 (U2 small nuclear RNA auxiliary factor 1), and ZRSR2 (zinc finger RNA binding motif and serine/arginine rich 2). These mutations are mutually exclusive, heterozygous point mutations, suggesting synthetic lethal interactions if they were to co-occur (Yoshida et al. 2011; Lee et al. 2018). Splicing machinery mutations are most common in AML arising out of antecedent myeloid malignancy (secondary AML) (Lindsley et al. 2015). It has been shown that mutations in SRSF2 cooperate with IDH2, with resultant synergy between RNA splicing and epigenetic regulation in promoting leukemogenesis (Yoshimi et al. 2019). Several groups have developed therapeutic strategies targeting the core spliceosome (SF3B1 inhibitors), splicing regulatory factors (PRMT5 inhibitor, RBM39 degraders, and SRPKs/CLKs inhibitors), and modulating pathologic splicing events (oligonucleotide-based approaches) (Lee et al. 2016; Obeng et al. 2016; Seiler et al. 2018; Fong et al. 2019; Wang et al. 2019).
Cohesin Complex
The cohesin complex is a tripartite ring including three structural proteins, SMC1A, SMC3, and RAD21, bound to either STAG1 or STAG2 (Fig. 2). This complex has many functions, most notably an essential role in aligning and stabilizing sister chromatids during metaphase (Nasmyth and Haering 2009). Cohesin complex also plays a critical role in the regulation of gene expression by cooperating with the DNA-binding protein CTCF in maintaining topologically associated domains (TADs) and regulating three-dimensional (3D) genome organization (Wendt et al. 2008). Heterozygous loss-of-function mutations in the cohesin complex occur in 12%–20% of AML and MDS patients and are more prevalent in high-risk MDS and secondary AML (Kon et al. 2013; Thota et al. 2014). These mutations are not associated with aneuploidy. Several studies have reported that the haploinsufficiency induced by the cohesin mutations enhances HSPC self-renewal and block hematopoietic differentiation (Mazumdar et al. 2015; Mullenders et al. 2015; Viny et al. 2015). STAG2, which is X-linked, is the most commonly mutated cohesin gene across many hematopoietic and solid tumors. A recent study demonstrated a specific role for STAG2 (not by STAG1) in balancing self-renewal and differentiation in HSPCs (Viny et al. 2019).
Mutations in Tumor-Suppressor Genes: TP53, WT1, PHF6
TP53
The tumor-suppressor gene TP53 is the most frequently mutated gene in human cancer and has many cellular functions, which are discussed in detail elsewhere (Fig. 1) (Kastenhuber and Lowe 2017). Mutations in TP53 occur in 5%–20% of patients with newly diagnosed AML. There is an increased frequency of TP53 mutations in AML patients with therapy-related myeloid neoplasms, complex karyotype, monosomal karyotype, and relapsed disease. One possible postulation for TP53 pathogenesis in therapy-related myeloid neoplasms is that TP53 mutations are acquired in some HSPCs during normal aging (clonal hematopoiesis) and are then selected for upon exposure to cytotoxic chemotherapy, ultimately resulting in the expansion of HSPCs with TP53 mutations (Wong et al. 2015). The majority of TP53 mutations are missense mutations within the central DNA-binding domain, leading to loss of function. The resulting loss of function of p53 favors genomic instability and impaired apoptosis, which can contribute to resistance to chemotherapy. AML patients with TP53 mutations have been associated with an inadequate response to conventional cytotoxic therapies and inferior overall survival rates (median of 5–9 mo) (Stengel et al. 2017). Historically, the efforts to target p53 therapeutically have been challenging. The landscape has been changing with the development of novel therapeutic strategies targeting the TP53 pathway. One such promising approach is APR-246, a methylated derivative of PRIMA-1 that is converted to Michael acceptor methylene quinuclidinone (MQ), which can bind covalently to mutant p53 and restore wild-type transcriptional properties to the protein (Lambert et al. 2009). There is an ongoing multicenter phase III randomized study comparing the combination of APR-246 with azacytidine compared with azacytidine alone in TP53-mutant MDS and AML (NCT03745716).
WT1
Wilm's tumor 1 (WT1) encodes for a transcription factor that regulates cell proliferation, differentiation, apoptosis, and organ development (Call et al. 1990). Somatic loss-of-function mutations target WT1 in 6%–15% of AML patients. Also, overexpression of wild-type WT1 protein has been observed in AML, further challenging the mechanistic understanding of how WT1 contributes to leukemogenesis (Rampal and Figueroa 2016). Heterozygous loss of Wt1 enhances HSCs self-renewal and cooperates with Flt3-ITD to induce leukemic transformation (Pronier et al. 2018). Loss-of-function mutations in WT1 led to marked promoter hypermethylation pattern and reduction in 5hmC levels similar to AML patients with mutations in TET2 and IDH1/2 (Rampal et al. 2014). WT1 protein has also been shown to bind directly to TET2 and recruit TET2 to specific genomic sites to activate the gene expression of WT1-target genes (Wang et al. 2015). Mutations in WT1 are associated with younger age, often co-occur with FLT3-ITD, and confer adverse prognostic significance in AML with increased risk of primary refractory disease (Hou et al. 2010).
PHF6
Somatic loss-of-function mutations in the X-chromosome-linked tumor suppressor plant homeodomain factor 6 (PHF6) are present in ∼3% of AML patients (Van Vlierberghe et al. 2011). PHF6 is involved in chromatin-mediated transcriptional regulation by binding to the nucleosome remodeling and deacetylase (NuRD) chromatin remodeling complex (Todd and Picketts 2012; Liu et al. 2015). The presence of PHF6 mutations in preleukemic clonal hematopoiesis supports its role in leukemia initiation and HSC self-renewal (Yoshizato et al. 2015; Abelson et al. 2018). In line with this notion, in a mouse model of T-cell acute lymphoblastic anemia (T-ALL), loss of Phf6 enhances tumor initiation and transformation and leads to increased self-renewal in mouse HSCs (Wendorff et al. 2019). Mutations in PHF6 are associated with RUNX1 mutations and with reduced overall survival in AML patients (Patel et al. 2012).
CONCLUDING REMARKS
Mutational studies involving large cohorts of patients with AML have identified a complex mutational landscape, including patterns of significant co-occurrence and mutual exclusivity of mutations. The mechanistic interactions between different mutations have only begun to be elucidated, and we expect these insights can be used to better inform our understanding of molecular pathogenesis of the disease, improve risk stratification, and identify novel therapeutic vulnerabilities. In specific cases, such as co-occurring FLT3/TET2 (Shih et al. 2015) and SRSF2/IDH2 (Yoshimi et al. 2019) mutations, mechanisms of oncogenic synergy have been elucidated; however, the basis for cooperativity between AML disease alleles requires greater investigation. Moreover, bulk sequencing studies have given us a glimpse into the temporal and spatial sequence of somatic mutations that drive leukemogenesis, from early mutations, which induce clonal hematopoiesis (DNMT3A, TET2, ASXL1), to disease-defining mutations present in all cells at transformation (NPM1), to mutations that can occur in specific subclones and further enhance the fitness of AML cells (FLT3, RAS). However, the roles of all of the different mutations in this complex sequence of genetic events that contribute to AML are not known, and many AML patients are a complex amalgam of multiple branching clones such that the AML “ecosystem” has not been fully characterized. Moreover, the observations that relapse can come from rarer AML subclones (Ding et al. 2012; Shlush et al. 2017) suggest that a better understanding of the role of specific mutations in AML heterogeneity and drug response is urgently needed.
In addition to novel mechanistic insights, we need to ensure that this knowledge is translated into improved outcomes for our patients. Most AML patients do not present with mutations for which we have a molecularly targeted therapy, and targeted therapy has limited curative potential when given as monotherapy in AML. We need to identify additional mechanisms of leukemic transformation induced by genetic/epigenetic events in AML, such that we can uncover novel therapeutic targets that lead to genotype-based combination therapies for AML patients. It is our hope that the ever-evolving understanding of AML pathogenesis will continue to lead to novel therapies for our patients.
ACKNOWLEDGMENTS
This work was supported by National Cancer Institute R35 CA197594-01A1 (R.L.L.), National Cancer Institute R01 CA216421 (R.L.L.), National Cancer Institute PS-OC U54 CA143869-05 (R.L.L.), and Leukemia & Lymphoma Society Translational Research Foundation 6499-17 (R.L.L.). This work was supported in part by Memorial Sloan Kettering Cancer Center Core Grant P30 CA008748. A.K. is supported by the VeloSano Catalyst Award.
Footnotes
Editors: Michael G. Kharas, Ross L. Levine, and Ari M. Melnick
Additional Perspectives on Leukemia and Lymphoma: Molecular and Therapeutic Insights available at www.perspectivesinmedicine.org
REFERENCES
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. 2009. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114: 144–147. 10.1182/blood-2009-03-210039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R, et al. 2012. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 22: 180–193. 10.1016/j.ccr.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, Barda N, Zuzarte PC, Heisler L, Sundaravadanam Y, et al. 2018. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559: 400–404. 10.1038/s41586-018-0317-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, Cowin BL, Bruner E, Murphy MM, Chen W, et al. 2017. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549: 476–481. 10.1038/nature23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatangelo MD, Quek L, Shih A, Stein EM, Roshal M, David MD, Marteyn B, Farnoud NR, de Botton S, Bernard OA, et al. 2017. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 130: 732–741. 10.1182/blood-2017-04-779447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O, Krainer AR. 2016. Splicing-factor alterations in cancers. RNA 22: 1285–1301. 10.1261/rna.057919.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127: 2391–2405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 30: 41–47. 10.1038/ng765 [DOI] [PubMed] [Google Scholar]
- Avellino R, Delwel R. 2017. Expression and regulation of C/EBPα in normal myelopoiesis and in malignant transformation. Blood 129: 2083–2091. 10.1182/blood-2016-09-687822 [DOI] [PubMed] [Google Scholar]
- Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. 2006. Implications of NRAS mutations in AML: A study of 2502 patients. Blood 107: 3847–3853. 10.1182/blood-2005-08-3522 [DOI] [PubMed] [Google Scholar]
- Balasubramani A, Larjo A, Bassein JA, Chang X, Hastie RB, Togher SM, Lähdesmäki H, Rao A. 2015. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun 6: 7307. 10.1038/ncomms8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball BJ, Hsu M, Devlin SM, Famulare C, Cai SF, Dunbar A, Epstein-Peterson ZD, Menghrajani K, Glass JL, Taylor J, et al. 2019. RAS mutations are independently associated with decreased overall survival and event-free survival in patients with AML receiving induction chemotherapy. Blood 134: 18. 10.1182/blood-2019-125319 [DOI] [Google Scholar]
- Basecke J, Whelan JT, Griesinger F, Bertrand FE. 2006. The MLL partial tandem duplication in acute myeloid leukaemia. Br J Haematol 135: 438–449. 10.1111/j.1365-2141.2006.06301.x [DOI] [PubMed] [Google Scholar]
- Bezerra MF, Lima AS, Piqué-Borràs MR, Silveira DR, Coelho-Silva JL, Pereira-Martins DA, Weinhäuser I, Franca-Neto PL, Quek L, Corby A, et al. 2020. Co-occurrence of DNMT3A, NPM1, FLT3 mutations identifies a subset of acute myeloid leukemia with adverse prognosis. Blood 135: 870–875. 10.1182/blood.2019003339 [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157: 1445–1459. 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield CD, Lawrence D, Byrd JC, Carroll A, Pettenati MJ, Tantravahi R, Patil SR, Davey FR, Berg DT, Schiffer CA, et al. 1998. Frequency of prolonged remission duration after high-dose cytarabine intensification in acute myeloid leukemia varies by cytogenetic subtype. Cancer Res 58: 4173–4179. [PubMed] [Google Scholar]
- Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, et al. 2015. Pharmacologic inhibition of the menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27: 589–602. 10.1016/j.ccell.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, Gionfriddo I, Mezzasoma F, Milano F, Nabet B, et al. 2018. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 34: 499–512.e9. 10.1016/j.ccell.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. 2012. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 44: 1179–1181. 10.1038/ng.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Ruppert AS, Mrózek K, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Stamberg J, Koduru PR, Moore JO, et al. 2004. Repetitive cycles of high-dose cytarabine benefit patients with acute myeloid leukemia and inv(16)(p13q22) or t(16;16)(p13;q22): results from CALGB 8461. J Clin Oncol 22: 1087–1094. 10.1200/JCO.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH, et al. 1990. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell 60: 509–520. 10.1016/0092-8674(90)90601-A [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Laird PW, et al. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. New Engl J Med 368: 2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. 2011. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44: 23–31. 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TH, Kambal A, Krysiak K, Walshauser MA, Raju G, Tibbitts JF, Walter MJ. 2011. Knockdown of Hspa9, a del(5q31.2) gene, results in a decrease in hematopoietic progenitors in mice. Blood 117: 1530–1539. 10.1182/blood-2010-06-293167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z, Shroff AS, Dickins RA, Vakoc CR, Bradner JE, et al. 2014. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 25: 652–665. 10.1016/j.ccr.2014.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen W, Mysliwski M, Serio J, Ropa J, Abulwerdi FA, Chan RJ, Patel JP, Tallman MS, Paietta E, et al. 2015. Mutated Ptpn11 alters leukemic stem cell frequency and reduces the sensitivity of acute myeloid leukemia cells to Mcl1 inhibition. Leukemia 29: 1290–1300. 10.1038/leu.2015.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. 2001. NF1 tumor suppressor gene function: Narrowing the GAP. Cell 104: 593–604. 10.1016/S0092-8674(01)00245-8 [DOI] [PubMed] [Google Scholar]
- Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, et al. 2017. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170: 1079–1095.e20. 10.1016/j.cell.2017.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Horwitz MS. 2017. GATA factor mutations in hematologic disease. Blood 129: 2103–2110. 10.1182/blood-2016-09-687889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute BE, Lewis AF, Wu Z, Bushweller JH, Speck NA. 1996. Biochemical and biophysical properties of the core-binding factor α2 (AML1) DNA-binding domain. J Biol Chem 271: 26251–26260. 10.1074/jbc.271.42.26251 [DOI] [PubMed] [Google Scholar]
- Damm F, Chesnais V, Nagata Y, Yoshida K, Scourzic L, Okuno Y, Itzykson R, Sanada M, Shiraishi Y, Gelsi-Boyer V, et al. 2013. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood 122: 3169–3177. 10.1182/blood-2012-11-469619 [DOI] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. New Engl J Med 360: 2289–2301. 10.1056/NEJMoa0810069 [DOI] [PubMed] [Google Scholar]
- Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, Samuel M, Ritchie EK, Guzman ML, Ballman KV, et al. 2018. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 24: 1015–1023. 10.1038/s41591-018-0081-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weer A, Speleman F, Cauwelier B, Van Roy N, Yigit N, Verhasselt B, De Moerloose B, Benoit Y, Noens L, Selleslag D, et al. 2008. EVI1 overexpression in t(3;17) positive myeloid malignancies results from juxtaposition of EVI1 to the MSI2 locus at 17q22. Haematologica 93: 1903–1907. 10.3324/haematol.13192 [DOI] [PubMed] [Google Scholar]
- Díaz-Beyá M, Labopin M, Maertens J, Alijurf M, Passweg J, Dietrich B, Schouten H, Socié G, Schaap N, Schwerdtfeger R, et al. 2020. Allogeneic stem cell transplantation in AML with t(6;9)(p23;q34);DEK-NUP214 shows a favourable outcome when performed in first complete remission. Br J Haematol 10.1111/bjh.16433 [DOI] [PubMed] [Google Scholar]
- Di Carlo V, Mocavini I, Di Croce L. 2019. Polycomb complexes in normal and malignant hematopoiesis. J Cell Biol 218: 55–69. 10.1083/jcb.201808028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon R, Hills R, Freeman S, Potter N, Jovanovic J, Ivey A, Kanda AS, Runglall M, Foot N, Valganon M, et al. 2020. Molecular MRD status and outcome after transplantation in NPM1-mutated AML. Blood 135: 680–688. 10.1182/blood.2019002959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, Swords R, Collins RH, Mannis GN, Pollyea DA, et al. 2018. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. New Engl J Med 378: 2386–2398. 10.1056/NEJMoa1716984 [DOI] [PubMed] [Google Scholar]
- DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, Thijssen R, Pomilio G, Ivey A, Salmon JM, et al. 2020. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 135: 791–803. 10.1182/blood.2019003988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, et al. 2017. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129: 424–447. 10.1182/blood-2016-08-733196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. 2008. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451: 335–339. 10.1038/nature06494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld AK, Kohlschmidt J, Mrózek K, Mims A, Walker CJ, Blachly JS, Nicolet D, Orwick S, Maharry SE, Carroll AJ, et al. 2018. NF1 mutations are recurrent in adult acute myeloid leukemia and confer poor outcome. Leukemia 32: 2536–2545. 10.1038/s41375-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, et al. 2005. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. New Engl J Med 352: 254–266. 10.1056/NEJMoa041974 [DOI] [PubMed] [Google Scholar]
- Fasan A, Haferlach C, Alpermann T, Jeromin S, Grossmann V, Eder C, Weissmann S, Dicker F, Kohlmann A, Schindela S, et al. 2014. The role of different genetic subtypes of CEBPA mutated AML. Leukemia 28: 794–803. 10.1038/leu.2013.273 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JY, Pignata L, Goy PA, Kawabata KC, Lee SC, Koh CM, Musiani D, Massignani E, Kotini AG, Penson A, et al. 2019. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36: 194–209.e9. 10.1016/j.ccell.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, Tobis K, Döhner H, Döhner K. 2004. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol 22: 624–633. 10.1200/JCO.2004.06.060 [DOI] [PubMed] [Google Scholar]
- Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J, Wallrabenstein T, Kolbinger B, Köhne CH, Horst HA, et al. 2016. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia 30: 2160–2168. 10.1038/leu.2016.126 [DOI] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. New Engl J Med 371: 2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greif PA, Dufour A, Konstandin NP, Ksienzyk B, Zellmeier E, Tizazu B, Sturm J, Benthaus T, Herold T, Yaghmaie M, et al. 2012. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood 120: 395–403. 10.1182/blood-2012-01-403220 [DOI] [PubMed] [Google Scholar]
- Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM, Hartley T, et al. 2012. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 8: 277–284. 10.1038/nchembio.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, van der Velden VHJ, Havermans M, Avellino R, van Lom K, et al. 2014. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157: 369–381. 10.1016/j.cell.2014.02.019 [DOI] [PubMed] [Google Scholar]
- Gröschel S, Sanders MA, Hoogenboezem R, Zeilemaker A, Havermans M, Erpelinck C, Bindels EM, Beverloo HB, Döhner H, Löwenberg B, et al. 2015. Mutational spectrum of myeloid malignancies with inv(3)/t(3;3) reveals a predominant involvement of RAS/RTK signaling pathways. Blood 125: 133–139. 10.1182/blood-2014-07-591461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, Spanhol-Rosseto A, Klein HU, Dugas M, Schindela S, et al. 2011. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 118: 6153–6163. 10.1182/blood-2011-07-365320 [DOI] [PubMed] [Google Scholar]
- Gu X, Ebrahem Q, Mahfouz RZ, Hasipek M, Enane F, Radivoyevitch T, Rapin N, Przychodzen B, Hu Z, Balusu R, et al. 2018. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub that regulates granulomonocytic fates. J Clin Invest 128: 4260–4279. 10.1172/JCI97117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, Ganzel C, Durham BH, Mohanty A, Hoermann G, et al. 2016. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med 22: 1488–1495. 10.1038/nm.4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC, Babic M, Lin M, Carmagnac A, Lee YK, et al. 2011. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 43: 1012–1017. 10.1038/ng.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen MA, Arora K, Vacic V, Grabowska EA, Liao W, Riley-Gillis B, Oschwald DM, Wang L, Joergens JE, Shih AH, et al. 2016. Integrative genetic analysis of mouse and human AML identifies cooperating disease alleles. J Exp Med 213: 25–34. 10.1084/jem.20150524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinai A, Pratcorona M, Grob T, Kavelaars FG, Bussaglia E, Sanders MA, Nomdedeu J, Valk PJM. 2019. The landscape of KMT2A-PTD AML: Concurrent mutations, gene expression signatures, and clinical outcome. Hemasphere 3: e181. 10.1097/HS9.0000000000000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch P, Zhang Y, Tang R, Joulin V, Boutroux H, Pronier E, Moatti H, Flandrin P, Marzac C, Bories D, et al. 2016. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat Commun 7: 12475. 10.1038/ncomms12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz-Schietinger C, Matje DM, Reich NO. 2012. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J Biol Chem 287: 30941–30951. 10.1074/jbc.M112.366625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono N, Makishima H, Jerez A, Yoshida K, Przychodzen B, McMahon S, Shiraishi Y, Chiba K, Tanaka H, Miyano S, et al. 2014. Recurrent genetic defects on chromosome 7q in myeloid neoplasms. Leukemia 28: 1348–1351. 10.1038/leu.2014.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou HA, Huang TC, Lin LI, Liu CY, Chen CY, Chou WC, Tang JL, Tseng MH, Huang CF, Chiang YC, et al. 2010. WT1 mutation in 470 adult patients with acute myeloid leukemia: Stability during disease evolution and implication of its incorporation into a survival scoring system. Blood 115: 5222–5231. 10.1182/blood-2009-12-259390 [DOI] [PubMed] [Google Scholar]
- Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, Kagiyama Y, Kawabata KC, Nakahara F, Izawa K, et al. 2013. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest 123: 4627–4640. 10.1172/JCI70739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Shih AH, Wang B, Nazir A, Rustenburg AS, Albanese SK, Patel M, Famulare C, Correa FM, Takemoto N, et al. 2018. Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature 559: 125–129. 10.1038/s41586-018-0251-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Bae SC, Chuang LS. 2015. The RUNX family: Developmental regulators in cancer. Nat Rev Cancer 15: 81–95. 10.1038/nrc3877 [DOI] [PubMed] [Google Scholar]
- Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, Patel Y, Bhudia N, Farah H, Mason J, et al. 2016. Assessment of minimal residual disease in standard-risk AML. New Engl J Med 374: 422–433. 10.1056/NEJMoa1507471 [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. New Engl J Med 371: 2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O'Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP. 2009. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 113: 6403–6410. 10.1182/blood-2009-02-205690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. 2007. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood 110: 719–726. 10.1182/blood-2007-01-068809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber ER, Lowe SW. 2017. Putting p53 in context. Cell 170: 1062–1078. 10.1016/j.cell.2017.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser S, Hills RK, Luskin MR, Brunner AM, Terré C, Westermann J, Menghrajani K, Shaw C, Baer MR, Elliott MA, et al. 2020. Allogeneic hematopoietic cell transplantation improves outcome of adults with t(6;9) acute myeloid leukemia: Results from an international collaborative study. Haematologica 105: 161–169. 10.3324/haematol.2018.208678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MJ, So J, Rogers AJ, Gregory G, Li J, Zethoven M, Gearhart MD, Bardwell VJ, Johnstone RW, Vervoort SJ, et al. 2019. Bcor loss perturbs myeloid differentiation and promotes leukaemogenesis. Nat Commun 10: 1347. 10.1038/s41467-019-09250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, Theilgaard-Mönch K, Månsson R, Pedersen TA, Pabst T, et al. 2008. Modeling of C/EBPα mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell 13: 299–310. 10.1016/j.ccr.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Kishtagari A, Levine RL, Viny AD. 2020. Driver mutations in acute myeloid leukemia. Curr Opin Hematol 27: 49–57. 10.1097/MOH.0000000000000567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klossowski S, Miao H, Kempinska K, Wu T, Purohit T, Kim E, Linhares BM, Chen D, Jih G, Perkey E, et al. 2020. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J Clin Invest 130: 981–997. 10.1172/JCI129126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843. 10.1038/nature09586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. 2013. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45: 1232–1237. 10.1038/ng.2731 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7: 823–833. 10.1038/nrc2253 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Evans K, Gadrey JY, Eschle BK, Hatton C, Uckelmann HJ, Ross KN, Perner F, Olsen SN, Pritchard T, et al. 2019. A Menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell 36: 660–673.e11. 10.1016/j.ccell.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto H, Meydan C, Nazir A, Whitfield J, Shank K, Rapaport F, Maher R, Pronier E, Meyer SC, Garrett-Bakelman FE, et al. 2018. Cooperative epigenetic remodeling by TET2 loss and NRAS mutation drives myeloid transformation and MEK inhibitor sensitivity. Cancer Cell 33: 44–59.e8. 10.1016/j.ccell.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JM, Gorzov P, Veprintsev DB, Söderqvist M, Segerback D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ. 2009. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15: 376–388. 10.1016/j.ccr.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Lavallée VP, Krosl J, Lemieux S, Boucher G, Gendron P, Pabst C, Boivin I, Marinier A, Guidos CJ, Meloche S, et al. 2016. Chemo-genomic interrogation of CEBPA mutated AML reveals recurrent CSF3R mutations and subgroup sensitivity to JAK inhibitors. Blood 127: 3054–3061. 10.1182/blood-2016-03-705053 [DOI] [PubMed] [Google Scholar]
- Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84: 291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, Durham BH, Yoshimi A, Kim YJ, Thomas M, et al. 2016. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med 22: 672–678. 10.1038/nm.4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, North K, Kim E, Jang E, Obeng E, Lu SX, Liu B, Inoue D, Yoshimi A, Ki M, et al. 2018. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34: 225–241.e8. 10.1016/j.ccell.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72. 10.1038/nature07485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. New Engl J Med 363: 2424–2433. 10.1056/NEJMoa1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Balmain A, Counter CM. 2018. A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat Rev Cancer 18: 767–777. 10.1038/s41568-018-0076-6 [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, Pigneux A, Wetzler M, Stuart RK, Erba HP, et al. 2015. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 125: 1367–1376. 10.1182/blood-2014-11-610543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TX, Becker MW, Jelinek J, Wu WS, Deng M, Mikhalkevich N, Hsu K, Bloomfield CD, Stone RM, DeAngelo DJ, et al. 2007. Chromosome 5q deletion and epigenetic suppression of the gene encoding α-catenin (CTNNA1) in myeloid cell transformation. Nat Med 13: 78–83. 10.1038/nm1512 [DOI] [PubMed] [Google Scholar]
- Liu Z, Li F, Zhang B, Li S, Wu J, Shi Y. 2015. Structural basis of plant homeodomain finger 6 (PHF6) recognition by the retinoblastoma binding protein 4 (RBBP4) component of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem 290: 6630–6638. 10.1074/jbc.M114.610196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J, Spitzer B, Bosbach B, Kastenhuber ER, Baslan T, et al. 2016. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531: 471–475. 10.1038/nature17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG Jr. 2013. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339: 1621–1625. 10.1126/science.1231677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugthart S, Gröschel S, Beverloo HB, Kayser S, Valk PJ, van Zelderen-Bhola SL, Jan Ossenkoppele G, Vellenga E, van den Berg-de Ruiter E, Schanz U, et al. 2010. Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol 28: 3890–3898. 10.1200/JCO.2010.29.2771 [DOI] [PubMed] [Google Scholar]
- Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, Przychodzen B, Nagata Y, Meggendorfer M, Sanada M, et al. 2017. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 49: 204–212. 10.1038/ng.3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaise M, Steinbach D, Corbacioglu S. 2009. Clinical implications of c-Kit mutations in acute myelogenous leukemia. Curr Hematol Malig Rep 4: 77–82. 10.1007/s11899-009-0011-8 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrózek K, Radmacher MD, Kohlschmidt J, Nicolet D, Whitman SP, et al. 2012. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol 30: 742–750. 10.1200/JCO.2011.39.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. New Engl J Med 361: 1058–1066. 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson JE, Ries RE, Wang YC, Gerbing RB, Kolb EA, Thompson SL, Guidry Auvil JM, Marra MA, Ma Y, Zong Z, et al. 2016. CSF3R mutations have a high degree of overlap with CEBPA mutations in pediatric AML. Blood 127: 3094–3098. 10.1182/blood-2016-04-709899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM, et al. 2015. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 17: 675–688. 10.1016/j.stem.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, Pellegrino M, Durruthy-Durruthy R, Watt CD, Asthana S, et al. 2019. Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov 9: 1050–1063. 10.1158/2159-8290.CD-18-1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNerney ME, Brown CD, Wang X, Bartom ET, Karmakar S, Bandlamudi C, Yu S, Ko J, Sandall BP, Stricker T, et al. 2013. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood 121: 975–983. 10.1182/blood-2012-04-426965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. 2017. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia 31: 272–281. 10.1038/leu.2016.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel M, Hinterleitner R, Pacis A, Chen L, Earley ZM, Mayassi T, Pierre JF, Ernest JD, Galipeau HJ, Thuille N, et al. 2018. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557: 580–584. 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendler JH, Maharry K, Radmacher MD, Mrózek K, Becker H, Metzeler KH, Schwind S, Whitman SP, Khalife J, Kohlschmidt J, et al. 2012. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol 30: 3109–3118. 10.1200/JCO.2011.40.6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshinchi S, Appelbaum FR. 2009. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin Cancer Res 15: 4263–4269. 10.1158/1078-0432.CCR-08-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrózek K, Nicolet D, Whitman SP, Wu YZ, Schwind S, et al. 2011. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood 118: 6920–6929. 10.1182/blood-2011-08-368225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MS, et al. 2018. The MLL recombinome of acute leukemias in 2017. Leukemia 32: 273–284. 10.1038/leu.2017.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol JB, Abdel-Wahab O. 2016. The role of additional sex combs-like proteins in cancer. Cold Spring Harb Perspect Med 6: a026526. 10.1101/cshperspect.a026526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, et al. 2013. Protein tyrosine kinase regulation by ubiquitination: Critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta 1833: 122–139. 10.1016/j.bbamcr.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, et al. 2015. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med 212: 1833–1850. 10.1084/jem.20151323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamachi A, Matsui H, Asou H, Ozaki Y, Aki D, Kanai A, Takubo K, Suda T, Nakamura T, Wolff L, et al. 2013. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell 24: 305–317. 10.1016/j.ccr.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, Schneider RK, Lord AM, Wang L, Gambe RG, et al. 2016. Physiologic expression of Sf3b1K700E causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30: 404–417. 10.1016/j.ccell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220. 10.1038/890 [DOI] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P, et al. 2011. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 43: 929–931. 10.1038/ng.923 [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. 2001. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat Genet 27: 263–270. 10.1038/85820 [DOI] [PubMed] [Google Scholar]
- Pan F, Wingo TS, Zhao Z, Gao R, Makishima H, Qu G, Lin L, Yu M, Ortega JR, Wang J, et al. 2017. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat Commun 8: 15102. 10.1038/ncomms15102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. 2016. Genomic classification and prognosis in acute myeloid leukemia. New Engl J Med 374: 2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin B, Ouillette P, Wang Y, Liu Y, Wright W, Roulston D, Purkayastha A, Dressel A, Karp J, Bockenstedt P, et al. 2010. NF1 inactivation in adult acute myelogenous leukemia. Clinical Cancer Res 16: 4135–4147. 10.1158/1078-0432.CCR-09-2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW, Carroll AJ, et al. 2006. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A cancer and Leukemia Group B Study. J Clin Oncol 24: 3904–3911. 10.1200/JCO.2006.06.9500 [DOI] [PubMed] [Google Scholar]
- Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. 2012. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. New Engl J Med 366: 1079–1089. 10.1056/NEJMoa1112304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, Montesinos P, Baer MR, Larson RA, Ustun C, et al. 2019. Abstract CT184: Gilteritinib significantly prolongs overall survival in patients with FLT3-mutated (FLT3mut+) relapsed/refractory (R/R) acute myeloid leukemia (AML): Results from the Phase III ADMIRAL trial. Cancer Res 79: CT184. [Google Scholar]
- Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, D'Alessandro A, Culp-Hill R, Riemondy KA, Gillen AE, et al. 2018. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 24: 1859–1866. 10.1038/s41591-018-0233-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronier E, Bowman RL, Ahn J, Glass J, Kandoth C, Merlinsky TR, Whitfield JT, Durham BH, Gruet A, Hanasoge Somasundara AV, et al. 2018. Genetic and epigenetic evolution as a contributor to WT1-mutant leukemogenesis. Blood 132: 1265–1278. 10.1182/blood-2018-03-837468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek L, David MD, Kennedy A, Metzner M, Amatangelo M, Shih A, Stoilova B, Quivoron C, Heiblig M, Willekens C, et al. 2018. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat Med 24: 1167–1177. 10.1038/s41591-018-0115-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, Bullinger L, Poschet G, Nonnenmacher Y, Barnert A, et al. 2017. BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551: 384–388. 10.1038/nature24294 [DOI] [PubMed] [Google Scholar]
- Rampal R, Figueroa ME. 2016. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica 101: 672–679. 10.3324/haematol.2015.141796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. 2014. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9: 1841–1855. 10.1016/j.celrep.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ, Vardiman JW, Anastasi J, Raca G, Savage NM, Cherry AM, Arber D, Moore E, Morrissette JJ, Bagg A, et al. 2014. Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv(3)(q21q26.2)/t(3;3)(q21;q26.2): A Bone Marrow Pathology Group study. Haematologica 99: 821–829. 10.3324/haematol.2013.096420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, et al. 2014. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25: 442–454. 10.1016/j.ccr.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Cigdem S, Okuwaki M, Nagata K. 2016. Leukemia-associated Nup214 fusion proteins disturb the XPO1-mediated nuclear-cytoplasmic transport pathway and thereby the NF-κB signaling pathway. Mol Cell Biol 36: 1820–1835. 10.1128/MCB.00158-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, et al. 2009. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460: 904–908. 10.1038/nature08240 [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247. 10.1038/nature08966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger S, Eder C, Jeromin S, Alpermann T, Fasan A, Grossmann V, Kohlmann A, Illig T, Klopp N, Wichmann HE, et al. 2013. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia 27: 82–91. 10.1038/leu.2012.262 [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. 2007. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer 7: 295–308. 10.1038/nrc2109 [DOI] [PubMed] [Google Scholar]
- Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, Grimwade D, Haferlach T, Hills RK, Hourigan CS, et al. 2018. Minimal/measurable residual disease in AML: A consensus document from the European LeukemiaNet MRD Working Party. Blood 131: 1275–1291. 10.1182/blood-2017-09-801498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, Thomas M, Agrawal AA, Caleb B, Csibi A, Sean E, et al. 2018. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat Med 24: 497–504. 10.1038/nm.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengsayadeth SM, Jagasia M, Engelhardt BG, Kassim A, Strickland SA, Goodman S, Lucid C, Vnencak-Jones CL, Greer JP, Savani BN. 2012. Allo-SCT for high-risk AML-CR1 in the molecular era: Impact of FLT3/ITD outweighs the conventional markers. Bone Marrow Transplant 47: 1535–1537. 10.1038/bmt.2012.88 [DOI] [PubMed] [Google Scholar]
- Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. 2015. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 27: 502–515. 10.1016/j.ccell.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506: 328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, Medeiros JJF, Rao-Bhatia A, Jaciw-Zurakowsky I, Marke R, et al. 2017. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547: 104–108. 10.1038/nature22993 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2020. Cancer statistics, 2020. CA Cancer J Clin 70: 7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- Sloand EM, Pfannes L, Chen G, Shah S, Solomou EE, Barrett J, Young NS. 2007. CD34 cells from patients with trisomy 8 myelodysplastic syndrome (MDS) express early apoptotic markers but avoid programmed cell death by up-regulation of antiapoptotic proteins. Blood 109: 2399–2405. 10.1182/blood-2006-01-030643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. 2004. Mutation of CEBPA in familial acute myeloid leukemia. New Engl J Med 351: 2403–2407. 10.1056/NEJMoa041331 [DOI] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, et al. 1999. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 23: 166–175. 10.1038/13793 [DOI] [PubMed] [Google Scholar]
- Sportoletti P, Varasano E, Rossi R, Mupo A, Tiacci E, Vassiliou G, Martelli MP, Falini B. 2015. Mouse models of NPM1-mutated acute myeloid leukemia: Biological and clinical implications. Leukemia 29: 269–278. 10.1038/leu.2014.257 [DOI] [PubMed] [Google Scholar]
- Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW, et al. 2017. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130: 722–731. 10.1182/blood-2017-04-779405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, Jongen-Lavrenic M, Altman JK, Thomson B, Blakemore SJ, et al. 2018. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131: 2661–2669. 10.1182/blood-2017-12-818948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. 2017. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: An analysis of 3307 cases. Leukemia 31: 705–711. 10.1038/leu.2016.263 [DOI] [PubMed] [Google Scholar]
- Steudel C, Wermke M, Schaich M, Schäkel U, Illmer T, Ehninger G, Thiede C. 2003. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer 37: 237–251. 10.1002/gcc.10219 [DOI] [PubMed] [Google Scholar]
- Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Döhner K, Marcucci G, et al. 2017. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. New Engl J Med 377: 454–464. 10.1056/NEJMoa1614359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ. 1998. An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood 92: 3793–3803. 10.1182/blood.V92.10.3793 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, et al. 2001. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104: 755–767. 10.1016/S0092-8674(01)00271-9 [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hählen K, Hasle H, Licht JD, Gelb BD. 2003. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet 34: 148–150. 10.1038/ng1156 [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Thien CB, Dagger SA, Duyvestyn JM, Grove CS, Lee BH, Gilliland DG, Langdon WY. 2015. Loss of c-Cbl E3 ubiquitin ligase activity enhances the development of myeloid leukemia in FLT3-ITD mutant mice. Exp Hematol 43: 191–206.e1. 10.1016/j.exphem.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, Wermke M, Bornhäuser M, Ritter M, Neubauer A, et al. 2002. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99: 4326–4335. 10.1182/blood.V99.12.4326 [DOI] [PubMed] [Google Scholar]
- Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, Sekeres MA, Levine RL, Maciejewski JP. 2014. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 124: 1790–1798. 10.1182/blood-2014-04-567057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd MA, Picketts DJ. 2012. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res 11: 4326–4337. 10.1021/pr3004369 [DOI] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371: 221–226. 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, Long N, Schultz AR, Traer E, Abel M, et al. 2018. Functional genomic landscape of acute myeloid leukaemia. Nature 562: 526–531. 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckelmann HJ, Kim SM, Wong EM, Hatton C, Giovinazzo H, Gadrey JY, Krivtsov AV, Rücker FG, Döhner K, McGeehan GM, et al. 2020. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science 367: 586–590. 10.1126/science.aax5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF, Tallman MS, et al. 2011. PHF6 mutations in adult acute myeloid leukemia. Leukemia 25: 130–134. 10.1038/leu.2010.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A, et al. 2015. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med 212: 1819–1832. 10.1084/jem.20151317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, Bowman RL, Liu Y, Lavallée VP, Eisman SE, Xiao W, Durham BH, Navitski A, Park J, Braunstein S, et al. 2019. Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell 25: 682–696.e8. 10.1016/j.stem.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fernald AA, Anastasi J, Le Beau MM, Qian Z. 2010. Haploinsufficiency of Apc leads to ineffective hematopoiesis. Blood 115: 3481–3488. 10.1182/blood-2009-11-251835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. 2015. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 57: 662–673. 10.1016/j.molcel.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, Hockemeyer K, Ghebrechristos YE, Yoshimi A, Inoue D, et al. 2019. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 35: 369–384.e7. 10.1016/j.ccell.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. 2012. The origin and evolution of mutations in acute myeloid leukemia. Cell 150: 264–278. 10.1016/j.cell.2012.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendorff AA, Quinn SA, Rashkovan M, Madubata CJ, Ambesi-Impiombato A, Litzow MR, Tallman MS, Paietta E, Paganin M, Basso G, et al. 2019. Phf6 loss enhances HSC self-renewal driving tumor initiation and leukemia stem cell activity in T-ALL. Cancer Discov 9: 436–451. 10.1158/2159-8290.CD-18-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801. 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- West RR, Hsu AP, Holland SM, Cuellar-Rodriguez J, Hickstein DD. 2014. Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. Haematologica 99: 276–281. 10.3324/haematol.2013.090217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, et al. 2015. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 518: 552–555. 10.1038/nature13968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. 2009. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 113: 3088–3091. 10.1182/blood-2008-09-179895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. 2014. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20: 1472–1478. 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A. 1987. Human proto-oncogene c-kit: A new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J 6: 3341–3351. 10.1002/j.1460-2075.1987.tb02655.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. 2005. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123: 207–218. 10.1016/j.cell.2005.09.025 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- Yoshimi A, Lin KT, Wiseman DH, Rahman MA, Pastore A, Wang B, Lee SC, Micol JB, Zhang XJ, de Botton S, et al. 2019. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574: 273–277. 10.1038/s41586-019-1618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, et al. 2015. Somatic mutations and clonal hematopoiesis in aplastic anemia. New Engl J Med 373: 35–47. 10.1056/NEJMoa1414799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, Zhao D, Liu Y, Wang C, Zhang X, et al. 2015. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525: 389–393. 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Opalinska J, Sohal D, Yu Y, Mo Y, Bhagat T, Abdel-Wahab O, Fazzari M, Figueroa M, Alencar C, et al. 2011. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem 286: 25211–25223. 10.1074/jbc.M111.235028 [DOI] [PMC free article] [PubMed] [Google Scholar]