Abstract
Embryonic stem cells have the capacities of self-renewal and pluripotency. Pluripotency establishment (somatic cell reprogramming), maintenance, and execution (differentiation) require orchestrated regulatory mechanisms of a cell’s molecular machinery, including signaling pathways, epigenetics, transcription, translation, and protein degradation. RNA binding proteins (RBPs) take part in every process of RNA regulation and recent studies began to address their important functions in the regulation of pluripotency and reprogramming. Here, we discuss the roles of RBPs in key regulatory steps in the control of pluripotency and reprogramming. Among RNA binding proteins are a group of RNA helicases that are responsible for RNA structure remodeling with important functional implications. We highlight the largest family of RNA helicases, DDX (DEAD-box) helicase family and our current understanding of their functions specifically in the regulation of pluripotency and reprogramming.
1. Pluripotency and reprogramming
Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of the blastocyst and they can be maintained indefinitely in culture. ESCs have two main characteristics: self-renewal, the ability of a cell to propagate indefinitely in the same state; and pluripotency, the potential of a single ESC to develop into any cell types of an embryo or an adult animal (Young, 2011).
During the mouse embryo development, at around embryonic day 3.5 (E3.5), the blastomeres compact into a blastocyst and the blastocyst has two different cell populations: the outer layer cells or the trophectoderm which will develop into the extra embryonic tissues; and the ICM which will develop into the primitive endoderm (hypoblast) and primitive ectoderm (epiblast). The primitive endoderm will give rise to the secondary extra embryonic tissues while the primitive ectoderm will produce the three germ layers of the embryo: ectoderm, mesoderm and endoderm (Morris et al., 2010).
At around mouse embryonic day E4.5, the blastocyst implants into the uterus to undergo the further development. ESCs are derived from the ICM of the pre-implantation blastocyst. It is also found that some of the post-implantation epiblast cells are capable of giving rise to all three embryonic germ layers, like ESCs. Based on the definition of pluripotency, these cells would be also considered pluripotent (Young, 2011). However, there are many differences between the cells derived from the ICM of the pre-implantation blastocyst and the cells from the post-implantation epiblast, such as the capacity to contribute to the chimeras and germ line transmission, the signaling to support cell’s pluripotency. Besides, human ESCs, which are also derived from the ICM of human pre-implantation embryos (Thomson et al., 1998), display characteristics much closer to the mouse post-implantation epiblast stem cells (EpiSCs), than to the mouse ICM-derived mESCs. This observation suggests that hESCs correspond to a more differentiated developmental stage, or a primed pluripotency state (Brons et al., 2007; Tesar et al., 2007).
Research into molecules that control pluripotency has led to the landmark discovery in 2006 by Yamanaka’s group who found a way to convert the mouse fibroblasts into a pluripotent ESC-like state through over expression of four transcription factors Oct4, Sox2, Klf4, and c-Myc. These reprogrammed cells are called induced pluripotent stem cells (iPSCs) and are highly similar to ESCs (Takahashi & Yamanaka, 2006). In 2007, Yamanaka’s group also reported the reprogramming of human somatic cells to a pluripotent state with the same set of factors (Takahashi et al., 2007).
Human ESCs and iPSCs have tremendous therapeutic and regenerative potentials by providing a precious resource for drug testing, disease modeling, and cell replacement. A better understanding of the molecular regulatory mechanisms underlying pluripotency and reprogramming is a prerequisite for ESCs and iPSCs to be applied in disease therapeutics and regenerative medicine. An interplay of transcription factors, epigenetic factors, and signal transduction pathways are crucially important in the regulation of establishment, maintenance, and execution of pluripotency. While epigenetic and transcriptional regulation of pluripotency and reprogramming has been extensively studied and reviewed (reviewed by Chambers & Tomlinson, 2009; Gökbuget & Blelloch, 2019; Theunissen & Jaenisch, 2017; Yeo & Ng, 2013), post-transcriptional encompassing translational and posttranslational controls are relatively under-explored and are becoming the subjects of an ever increasing number of recent publications in the field of stem cell biology understanding pluripotency and reprogramming (Di Stefano et al., 2019; Freimer, Hu, & Blelloch, 2018; Li et al., 2017a; Yoffe et al., 2016; Zhang et al., 2020).
2. RNA binding proteins
RNA binding proteins (RBPs) are key factors in gene expression regulation by participating in every RNA-involved process, from transcription, RNA maturation, transport, stability, to translation and RNA degradation (reviewed by Guallar & Wang, 2014; Ye & Blelloch, 2014). RBPs are defined as proteins that contain one or multiple well-known RNA-binding domains (RBDs); or less commonly, proteins that reside within the ribonucleoproteins even if they don’t directly interact with RNA (Gerstberger, Hafner, & Tuschl, 2014).
RBPs can be classified based on their target RNAs: mRNA-binding, tRNA-binding, pre-rRNA-binding, small nucleolar RNA (snoRNA)-binding, small nuclear RNA (snRNA)-binding, and other non-coding RNA (ncRNA)-binding. Notably, some RBPs can interact with different RNA types, such as the RNA exosome that regulates general RNA turnover. In these cases, researchers usually group the RBPs into their predominant target groups. Also, some RBPs with well-known RBDs are without available RNA target information (Gerstberger et al., 2014). Many published studies are focused on the mRNA-binding proteins (mRBPs), and relatively less is known about other RBPs subclasses such as the ncRNA-binding proteins. In the Online Mendelian Inheritance in Man (OMIM) database that links the known diseases to the relevant genes in the human genome, there are around 150 RBPs listed. In this list, only one-third of the RBPs are mRNA-binding, with the rest mostly targeting diverse ncRNAs (Hamosh, 2004), supporting the significance of studying the latter group.
Owing to the important roles that RBPs play in the gene expression regulation, it is not surprising that RBP families are well conserved across eukaryotes. Previous studies show that there are at least 200 distinct RBPs which are also present in the lowest common animal ancestor (Anantharaman, Koonin, & Aravind, 2002). Gerstberger et al. reported that in human, 50% of the RBP families are conserved in S. cerevisiae and even more are conserved in higher eukaryotes. The relative percentage of each RBP subclass based on its RNA targets is also maintained across phylogenies, 38% for mRBPs and 12% for tRNA-binding proteins (Gerstberger et al., 2014). Gerstberger et al. also showed that in human, 98% of paralogous RBP families are ubiquitously expressed across tissues while only 2% of paralogous families have tissue-specific expression patterns (Gerstberger et al., 2014). As evolutionary origin and tissue-specificity of gene expression often correlate with the protein function, highly evolutionary conservation and ubiquitous expression of RBPs support their critical roles in basic cellular functions (Freilich et al., 2005; Ramsköld, Wang, Burge, & Sandberg, 2009; Winter, Goodstadt, & Ponting, 2004).
RBD determines the specificity of binding of a certain RBP to its targets (MacKay, Font, & Segal, 2011). The following RBDs are some of the best characterized domains described in the literature:
RNA-Recognition Motif (RRM): 90–100 amino acids in length, present in up to six copies per protein, the most abundant and the most extensively studied RBD in higher vertebrates (Maris, Dominguez, & Allain, 2005). The RRM-RNA interaction is specific to single-stranded RNA, with low sequence-specificity. RRM has been shown to be also capable to interact with DNA and proteins.
K-Homology Domain (KH): around 70 amino acids in length. KH can recognize four single-stranded nucleotides with rather weak affinity, the stronger affinity or longer than four nucleotides target can be achieved by synergy in multiple copies (Beuth, Pennell, Arnvig, Martin, & Taylor, 2005).
Double-Stranded RNA-Binding Domain (dsRBD): around 70–75 amino acids, present in up to five copies per protein. The dsRBD recognizes double-stranded RNA in a sequence-independent way. The recognition covers 15 nucleotides with two minor grooves separated by a major groove. The additional functional domains modulate the binding specificity for various RNA shapes (Stefl, Skrisovska, & Allain, 2005).
DEAD-Box Domain: the name of DEAD-box is coming from their characteristic Asp-Glu-Ala-Asp (DEAD) motifs. DEAD-box proteins form the largest helicase family and they utilize ATP to bind or remodel RNA and ribonucleoproteins (Linder & Jankowsky, 2011). A major focus of this review (see more in Section 3).
PUF RNA-Binding Repeats: the PUF (formed by Pumilio and FBF) domain is around 36 amino acids, present six to eight tandem repeats per protein, packed in a curved structure, bind to single-stranded RNAs (Guallar & Wang, 2014).
PAZ Domain: the PAZ (Piwi/Argonaute/Zwille) domain is around 110 amino acids, recognizes the two-base 3′ overhang of dsRNA and ssRNA. The PAZ-domain RBPs function in the post-transcriptional gene silencing (Tian, Simanshu, Ma, & Patel, 2011).
Zinc-Finger Domains (ZnF): Znf domain is a classical DNA-binding domain, but it is also able to interact with RNA (Teplova & Patel, 2008). ZnFs present alone or in multiple copies per protein, they can also work in combination with other RBDs.
Even though above RBDs are well characterized in their association with RNA, many proteins have now been shown to interact with RNA in the absence of known RBDs. It is still necessary to determine whether those candidates directly interact with their RNA targets, and to characterize potential new RBDs. The high number of proteins that have been shown to bind RNA underscores the importance of both RBPs and RNA regulation in cellular function (Guallar & Wang, 2014).
Because RBPs are involved in every process of RNA regulation, from transcriptional to post- transcriptional as well as translational regulation, it is not surprising that they also play important roles in pluripotency and reprogramming (reviewed by Guallar & Wang, 2014; Ye & Blelloch, 2014). Therefore, categorization of RBPs in pluripotent stem cells provides an inroad to understanding their biology in pluripotency and reprogramming. Kwon et al. identified 555 proteins, including 283 novel RBP candidates, to constitute the mESC mRNA interactome. In this interactome, 68 proteins are preferentially expressed in ESCs by comparison to differentiated cells (Kwon et al., 2013). Bao et al. developed an approach to capture the newly transcribed RNA interactome using click chemistry (RICK) and applied it in mESCs. They identified 518 high-confidence proteins, 160 of which are overlapped with Kwon et al.’s interactome and the rest 358 are defined as RICK-exclusive mESC RBPs with RNA binding and polyA-RNA binding capacities. Among these 358 proteins, expression levels of 95 proteins are higher in mESCs than in differentiated cells, suggesting their specific roles in ESC self-renewal and pluripotency (Bao et al., 2018). He et al. performed proteomic identification of RNA-binding regions in mESCs, and identified 803 nuclear RBPs, many of which are well-known transcriptional regulators and chromatin modifiers, such as NANOG and TET2 (He et al., 2016).
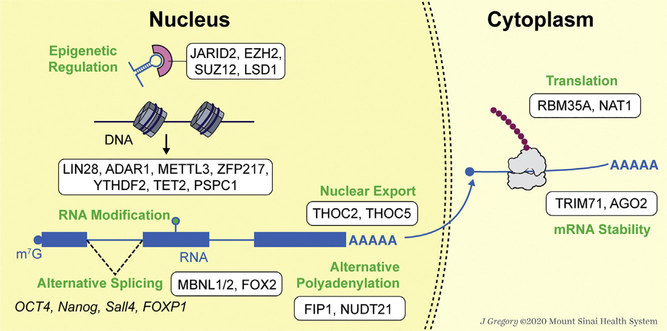
Mechanistically, RBPs participate in the regulation of pluripotency and reprogramming in many different regulatory layers (Fig. 1), which are discussed in detail below.
Fig. 1.
RBPs participate in multiple regulatory layers to control pluripotency and reprogramming. See Sections 2.1–2.7 for details.
2.1. Epigenetic regulation
RBPs can also interact with ncRNAs to control chromatin activation or repression, the epigenetic control that serves as another important regulatory layer in the embryonic development. Examples of such RBPs are JARID2, an Xist-interacting RBP that promotes PRC2 recruitment for X chromosome inactivation in early female development and also during female ESC differentiation in vitro (da Rocha et al., 2014; Kaneko et al., 2014); and EZH2 and SUZ12, two catalytic subunits of PRC2 (polycomb repressive complex 2) that interact with lncRNAs (such as HOTAIR lncRNA (Brockdorff, 2013)) to function during embryonic development. HOTAIR lncRNA is also bound by an epigenetic modifier LSD1, a histone demethylase. LSD1 plays important roles in ESC differentiation through its H3 demethylase activity (Adamo et al., 2011; Whyte et al., 2012). Studies by Tsai et al. showed that HOTAIR promoted the bridging between PRC2 and LSD1 to facilitate their cooperation in regulating gene repression (Kaya & Higuchi, 2010). In addition, HOTAIR is induced during differentiation and its expression is also required in epithelial-to-mesenchymal transition (EMT) and metastasis in cancer cell lines (Gupta et al., 2010; Pádua Alves et al., 2013).
2.2. RNA modification
Posttranscriptional RNA modification provides a new layer of gene regulation at the RNA level. RNA modifications can be separated into two types: the addition of untemplated nucleotides and the chemical modification of the template nucleotides. One example for the former is the uridylation of pre-let-7 miRNA: let-7 is an important miRNA in facilitating ESC differentiation and repressing reprogramming of somatic cells (Melton, Judson, & Blelloch, 2010). Its formation can be regulated through uridylation in two opposite ways: LIN28A can direct 3′ terminal uridylyl transferases (TUTases) ZCCHC11 and ZCCHC6 to add a string of around 11 uridines to the pre-let-7 miRNA (Hagan, Piskounova, & Gregory, 2009; Heo et al., 2008), then the oligouridylated pre-let-7 would be targeted and degraded by the DIS3L2 exoribonuclease (Ustianenko et al., 2013); on the contrary, the addition of only one uridine to pre-let-7 would facilitate the maturation of let-7 (Heo et al., 2012).
The latter type of RNA modification can take several forms. One major class of RNA modifications is editing by deamination (Bass, 2002). Classical examples are Adenosine-to-inosine, A-to-I, catalyzed by the adenosine deaminase acting on RNA (ADAR) family (Eggington, Greene, & Bass, 2011), and cytidine-to-uridine, C-to-U, catalyzed by the AID-APOBEC enzyme (Powell et al., 1987). A-to-I RNA editing catalyzed by ADAR1 is important in human embryogenesis and ADAR1 is required for hESC differentiation and neural induction (Chen et al., 2015; Shtrichman et al., 2012).
A second class of RNA modifications is methylation of adenosine to form N6-methyladenosine (m6A), the most abundant modification of eukaryotic mRNA which is critical for pluripotency and reprogramming. METTL3, a m6A methyltransferase, is required for m6A in mRNAs of ESCs. While ESCs without Mettl3 can preserve their naïve pluripotent identity, Mettl3 knockout (KO) naïve ESCs cannot be transferred to primed state, and they lose differentiation competence, staying in a hyper-naïve pluripotency state. Such resistance to differentiation is because during the transition from naïve to primed pluripotent states, m6A is required to timely destabilize the transcripts of pluripotency factors, which is necessary for proper lineage differentiation (Batista et al., 2014; Geula et al., 2015). In iPSC reprogramming and naïve ESCs, ZFP217 interacts with and sequesters METTL3, inhibiting m6A deposition on the transcripts of the core stem cell network, such as Nanog, Sox2, and c-Myc (Aguilo et al., 2015). Apart from transcripts of these core pluripotency factors, a recent paper shows that in human pluripotent stem cells, m6A is also important in the regulation of R-loops, the tripartite nucleic acid structures that are formed during transcription with an RNA:DNA hybrid and a non-hybridized single-stranded DNA. During cell cycles, m6A-containing R-loops accumulate during G2/M phases and are drastically depleted during G0/G1 phases. An m6A reader, YTHDF2, interacts with RNA:DNA hybrids. The depletion of YTHDF2 or METTL3 leads to accumulation of RNA:DNA hybrids and increases γH2AX, a marker of DNA double-strand breaks, indicating genome instability (Abakir et al., 2019).
A third class of RNA modifications is oxidation of 5-methylcytidine (5mC) to 5-hydroxymethylcytidine (5hmC) on RNA that can be catalyzed by Tet enzymes (Delatte et al., 2016; Fu et al., 2014; Masiello & Biggiogera, 2017; Miao et al., 2016; Zhang, Xiong, Qi, Feng, & Yuan, 2016). In mESCs, TET2 can be recruited to actively transcribed MERVL RNAs through its physical association with another RBP PSPC1 and deposit 5hmC modification on MERVL RNAs, contributing to MERVL destabilization in mESCs (Guallar et al., 2018). Besides TET2, PSPC1 can also recruit HDAC1/2 (histone deacetylase complex) to silence MERVL transcriptionally (Guallar et al., 2018). Readers are encouraged to read this review (Frye & Blanco, 2016) to gain additional information on RNA modifications in development and stem cells.
2.3. Alternative splicing
In mouse and human, more than half of the genes can generate different transcripts through alternative splicing (Modrek, 2001). Many pluripotency-associated transcripts, including OCT4 and Nanog, two of the most important pluripotency factors, have different isoforms generated through alternative splicing: in human, OCT4A is the key pluripotency transcription factor in ESCs while OCT4B expresses in nonpluripotent cells without known functions (Wang & Dai, 2010); in mouse, the three isoforms of Nanog contributes with various efficacies to maintaining ESC pluripotency (Atlasi, Mowla, Ziaee, Gokhale, & Andrews, 2008; Das, Jena, & Levasseur, 2011). Another example is Sall4, a transcription factor essential for pluripotency. It has two isoforms, Sall4a and Sall4b, which can form either homodimers or a heterodimer with each other. The genomic binding loci of Sall4a and Sall4b are overlapped but not identical. Sall4b is relatively more important than Sall4a in the regulation of pluripotency as Sall4b, but not Sall4a, can partially rescue the loss-of-function phenotype of both isoforms (Rao et al., 2010). A fourth example is FOXP1. It has an ESC-specific isoform that promotes iPSC reprogramming and ESC maintenance by stimulating the expression of pluripotency factors (Gabut et al., 2011).
Apart from the RBPs involved in the core machinery of the spliceosome, specific RBPs are also needed to generate the ESC-specific splicing signature. For example, MBNL1 and MBNL2 are conserved negative regulators of cassette exon alternative splicing events that are differentially controlled among cell types. The alternative splicing event of aforementioned ESC-specific FOXP1 isoform is inhibited by MBNL1/2 and consistent with such inhibitory control of ESC-specific FOXP1 isoform, the depletion of MBNL1/2 enhances iPSC reprogramming (Han et al., 2013). Another splicing regulator example is FOX2, which is critical for pluripotency in hESCs as its depletion drives hESCs into differentiation and death. The CLIP-seq (crosslinking immunoprecipitation with high-throughput sequencing) from Yeo et al. showed that FOX2 binding to one intron induced the inclusion of the upstream flanking exon and the exclusion of the downstream flanking exon. In addition, FOX2 also acts as an upstream splicing master regulator because it targets and regulates the alternative splicing of several other splicing regulators, such as LIN28, FOX2 itself and serine/threonine kinases (Yeo et al., 2009). All these highlight the important functions of FOX2 in the splicing program to maintain the hESC pluripotency. For more examples, we direct readers to two related reviews of the subject (Chen, 2015; Cheong & Lufkin, 2011).
2.4. Alternative polyadenylation
Around 70% of the mammalian RNAs are subjected to alternative polyadenylation (APA), leading to different 3′UTR lengths of transcripts (Derti et al., 2012). The various 3′UTR lengthening can affect the stability, localization and translation of transcripts, leading to differential protein expression. Previous studies show that APA is closely related with cell states: somatic cell reprogramming is associated with 3′UTR shortening (Ji & Tian, 2009; Sandberg, Neilson, Sarma, Sharp, & Burge, 2008), whereas embryonic development and exit from pluripotency are accompanied by 3′UTR lengthening (Ji, Lee, Pan, Jiang, & Tian, 2009; Shepard et al., 2011). In transcript cleavage and polyadenylation, cleavage and polyadenylation specificity factor (CPSF) complex recognizes the polyadenylation signal flanking upstream of the cleavage site (Lackford et al., 2014). Lackford et al. demonstrated that Fip1, one subunit of CPSF, functioned as an mRNA 3′ processing factor in establishing ESC-specific APA profile. Fip1 knockdown resulted in partial differentiation in mESCs and inhibited MEF reprogramming. Deep sequencing showed that Fip1 depletion changed the APA profile of 374 genes with 3′UTR lengthening (Lackford et al., 2014). Further studies are needed to investigate how Fip1 regulates the 3′UTR length in contributing to pluripotency. Another protein complex, known as cleavage factor Im (CFIm) complex, acts as an activator of transcript cleavage and polyadenylation. Nudt21 (also called Cpsf5), a component of CFIm, regulates cell fates by manipulating alternative polyadenylation. Nudt21 is a barrier to reprogramming as its depletion dramatically increases iPSC reprogramming efficiency. Its depletion also enhances the transdifferentiation of MEFs to induced trophoblast stem cells but impairs ESC differentiation. Mechanistically, Nudt21 knockdown facilitates alternative polyadenylation of chromatin regulators, such as Rybp, Chd1, and Wdr5, that play important roles in reprogramming (Ang et al., 2011; Brumbaugh et al., 2018; Gaspar-Maia et al., 2009; Li et al., 2017b). For additional information on APA in stem cell biology, readers are referred to this review (Mueller, Cheung, & Rando, 2013).
2.5. Nuclear retention and export of RNAs
Most RNAs need to be exported from nucleus to cytoplasm to function, so gene expression can also be regulated through controlling the access of RNA to the cytoplasmic machineries (e.g., translation machinery). A study from Wang et al. showed this regulatory level played important roles in ESCs. The THO complex is a conserved complex regulating mRNA export from the nucleus to the cytoplasm. The depletion of two subunits of the THO complex, namely Thoc2 or Thoc5, didn’t change the overall transcripts level, however, resulted in the nuclear accumulation of a subset of pluripotency-related transcripts, including Nanog, Esrrb, Klf4, and Sox2. The interaction of THOC2 with these pluripotency-related mRNAs is THOC5-dependent. THOC5 is an adaptor protein, which is downregulated in normal development. The knockdown of Thoc5 promotes ESC differentiation and inhibits somatic cell reprogramming, while overexpression of Thoc5 delays the differentiation in ESCs (Wang et al., 2013). This example emphasizes the important role of RNA nuclear export control in pluripotency regulation (Saunders & Wang, 2014).
2.6. Translation
RBPs can also adjust the RNA/ribonucleoprotein structures to control the accessibility of the RNA to ribosomes or the movement of ribosomes along the mRNA to control protein synthesis. At this regulatory level, RBPs often bind to the 5′UTR of RNA and such 5′UTR-RBP interactions have been reported to regulate ESC proliferation and differentiation (Ye & Blelloch, 2014). For example, RBM35A was found to target the 5′UTR of Sox2 and Oct4 transcripts to prevent their loading into the polysomes, as demonstrated through RBM35A immunoprecipitation and polysome profiling. And Rbm35a depletion blocks ESC differentiation and facilitates somatic cell reprograming through promoting the expression of key pluripotency transcription factors, including Oct4 and Sox2 (Fagoonee et al., 2013). Another example is NAT1 (also known as EIF4G2, DAP5, and p97), which is homologous to the C-terminal of eukaryotic translation initiation factor 4G (EIF4G1). In both mESCs and hESCs, its depletion results in the resistance to differentiation induction due partly to the translational block of NAT1-mediated translation of a specific subgroup of proteins that are critical for ESC differentiation (Sugiyama et al., 2017; Yoffe et al., 2016). Translational control in pluripotency and reprogramming is being increasingly recognized (Tahmasebi, Amiri, & Sonenberg, 2019), although much more work needs to be done to further unravel this important regulatory layer.
2.7. mRNA stability and degradation
During quality surveillance of RNA, RBPs can bind to aberrant RNAs and export them in the cytoplasm for degradation (Reed & Hurt, 2002), as well as modify RNA in nucleus for degradation (Houseley, LaCava, & Tollervey, 2006; LaCava et al., 2005). Some RBPs that function in RNA quality control have been shown to be important for pluripotency and reprogramming. For example, TRIM71 can interact with miRNA-containing AGO2 and cooperate with ESC-specific miR-290 and miR-302 to target the 3′UTR of Cdkn1a, a repressor of the G1-S transition, inhibiting its activity to promote the cell cycle process for optimal ESC self-renewal (Chang et al., 2012). Loedige et al. also demonstrated the binding of TRIM71 to the 3′UTRs of a subset of prodifferentiation genes, leading to the downregulation of mRNA levels in an AGO2-independent way (Loedige, Gaidatzis, Sack, Meister, & Filipowicz, 2013). In addition, Worringer et al. showed that overexpression of TRIM71 promoted human somatic cell reprogramming, which was partly due to the post-transcriptional inhibition of the fibroblast-enriched EGR1 transcripts to which TRIM71 binds and negatively regulates (Worringer et al., 2014).
In sum, RBPs can function in multiple regulatory layers to control pluripotency and reprogramming. Some RBPs can even work multi-functionally by controlling various molecular layers of RNA regulation. One example is coming from the study by Dardenne et al., demonstrating the multiple functions of two RNA helicases DDX5 and DDX17 in various regulatory layers controlling myogenesis and EMT (epithelial-to-mesenchymal transition) (Dardenne et al., 2014). We will further discuss this particular family of RBPs below.
3. RNA helicases and DEAD-box helicase family
RNA is one of the most important biological macromolecules that function in many biological processes. To be functional, RNA must fold into specific secondary or tertiary structures in three dimensions, and many proteins are involved in the RNA folding/remodeling to regulate the physical characteristics of RNA or form ribonucleoprotein complexes for further function (Jarmoskaite & Russell, 2014). For example, in the spliceosome assembly, Sub2 and Prp5, two DEAD-box helicases, are required to promote the rearrangements allowing the recognition base-pairing between the branchpoint and U2 snRNA (Ruby, Chang, & Abelson, 1993).
Helicases are the enzymes responsible for nucleic acids remodeling by using the energy from nucleoside triphosphate binding and hydrolysis (Hardwick & Luisi, 2013). Helicases function in almost every cellular process in which nucleic acids are involved. Until now, at least two mechanisms have been reported: canonical translocation-based duplex unwinding and duplex unwinding by local strand separation, which are employed by some viral RNA helicases and DEAD-box helicases, respectively (Jankowsky, 2011). Helicases are classified into six superfamilies (SFs) and all eukaryotic RNA helicases are found in six families belonging to SFs 1 and 2; the remaining families in eukaryotes are composed of DNA helicases. Some families consist of both RNA and DNA helicases and some helicases work on both DNA and RNA (Fairman-Williams, Guenther, & Jankowsky, 2010; Putnam & Jankowsky, 2013). The DEAD-box (DDX) is the largest family of RNA helicases, belonging to SF2 (Hardwick & Luisi, 2013). The name of DEAD-box reflects their characteristic Asp-Glu-Ala-Asp (DEAD) motifs. This family is present in all eukaryotes and also in many Archaea and bacteria. These highly conserved helicases are involved in virtually every RNA metabolism step, from ribosome biogenesis, to transcription, RNA maturation, microRNA processing, translation, and RNA degradation. DDX proteins generally function as part of large multicomponent complex, like the spliceosome (Linder & Jankowsky, 2011). Because DDXs are widely involved in the RNA metabolism, it is not surprising that some DDX members also play important roles in pluripotency and reprogramming. To date, direct implication of DDX family members in stem cell pluripotency and somatic cell reprogramming came from the studies of following DDX factors.
3.1. DDX3
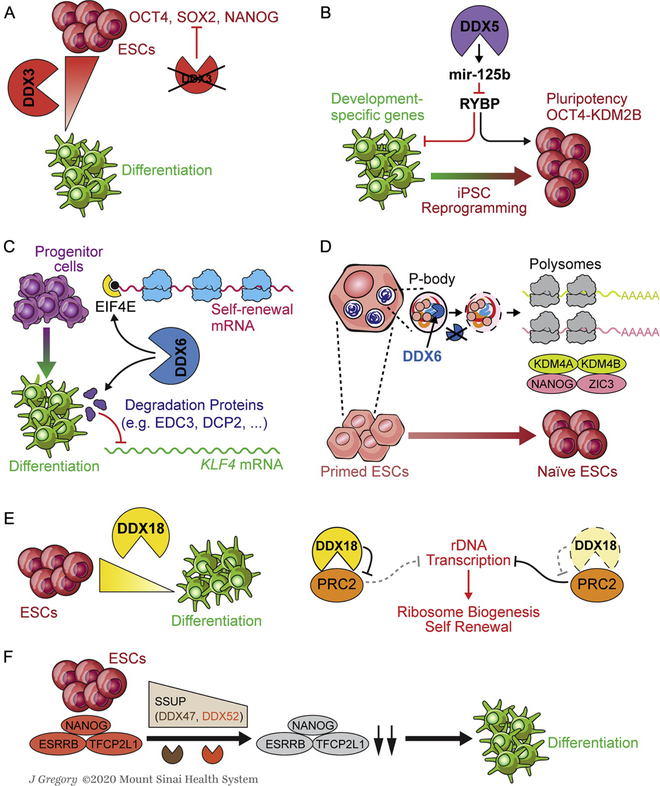
The expression level of DDX3 is highly enriched in human undifferentiated stem cells compared to differentiated cells. Inhibition of DDX3 reduces cellular proliferation in hESCs but doesn’t decrease proliferation of human embryonic fibroblast cells. In hESCs, inhibition of DDX3 also downregulates critical pluripotency markers (OCT4, SOX2, and NANOG) and facilitates differentiation (Kerr, Bol, Vesuna, & Raman, 2019) (Fig. 2A). Interestingly, Cruciat et al. reported that DDX3 could bind and activate the casein kinase 1 isoform epsilon (CK1ε), which leads to the Wnt-dependent phosphorylation of Disheveled, enabling β-catenin’s function in activating its target genes (Cruciat et al., 2013). As Wnt signaling plays important roles in both pluripotency and reprogramming, it remains to be determined whether DDX3’s regulatory role as the subunit of CK1ε defined by Cruciat et al. is part of the mechanism.
Fig. 2.
DDX proteins that are reported to be important in pluripotency and reprogramming. (A) DDX3 expression is highly enriched in human undifferentiated stem cells and decreases during differentiation. The inhibition of DDX3 in hESCs downregulates core pluripotency factors. (B) DDX5 inhibits iPSC reprogramming. Depletion of Ddx5 downregulates miRNA-125b, leading to the increase of RYBP. RYBP upregulation inhibits development-specific gene expression, and facilitates pluripotency through the activation of OCT4-KDM2B network. (C) DDX6 is necessary for the maintenance of adult progenitor cell functions, through facilitating the translation of proliferation and self-renewal transcripts, and the degradation of differentiation-inducing KLF4 mRNA. (D) Depletion of DDX6 promotes the reprogramming of primed hESCs to a naïve state. DDX6 interacts with P-body proteins and suppresses the target transcripts in P-bodies. Depletion of DDX6 leads to dissolvement of P-bodies, releasing target mRNAs that encode key cell fate transcription regulators and chromatin factors. The released mRNAs are translated to promote pluripotency and reprogramming. (E) Ddx18 expression is highly enriched in mESCs and decreases along differentiation. DDX18 interacts with PRC2, preventing it from accessing and marking rDNA with H3K27me3. DDX18 downregulation leads to inhibition of rDNA transcription and reduced ribosomal protein level as well as global translation level. (F) DDX47 and DDX52, as the components of SSUP, are highly expressed in ESCs and important for maintaining the protein levels of pluripotency factors. Downregulation of DDX47 or DDX52 leads degradation of pluripotency factors and consequent differentiation of mESCs. The references for each group are listed in the main text.
3.2. DDX5/DDX17
A notable example for DDX’s multi-functionality in the stem cell field comes from the study of DDX5 and DDX17 paralogs (Dardenne et al., 2014). Dardenne et al. show that DDX5 and DDX17 can cooperate with hnRNP (heterogeneous nuclear ribonucleoprotein) H/F splicing factors to express an epithelial- and myoblast-specific splicing subprogram. Also, DDX5 and DDX17 serve as transcriptional coregulators of key differentiation transcription factors to drive the transcription programs specific to the myogenesis and EMT (epithelial-to-mesenchymal transition), which in turn can produce differentiation-specific miRNAs resulting in the down-regulation of DDX5 and DDX17. Another example is the association of DDX3 and DDX5 with the Wnt/β-catenin signaling pathway (Cruciat et al., 2013; Yang, Lin, & Liu, 2006). This pathway is very important in the embryonic development and is directly linked to the pluripotency core transcription factors, playing essential roles in pluripotency and self-renewal regulation (Kim & Kimmel, 2006). In Yang et al. (2006) showed that the stimulated DDX5 could displace the inhibitor Axin from β-catenin, facilitating the transfer of β-catenin to the nucleus to activate the target gene expression instead of being phosphorylated and degraded.
DDX5 also inhibits iPSC reprogramming. The depletion of Ddx5 results in the dysregulation of dozens of miRNAs, including downregulation of microRNA-125b, which inhibits the expression of non-canonical polycomb complex 1 (PRC1) subunit Rybp. RYBP upregulation upon Ddx5 depletion not only facilitates the deposition of inhibitory H2AK119ub1 at lineage-specific genes through PRC1 but also activates the OCT4-KDM2B network to enhance pluripotency-associated gene expression independently of PRC1 (Fig. 2B) (Li et al., 2017b).
3.3. DDX6
DDX6 has been shown to be necessary for the maintenance of adult progenitor cell functions (Wang, Arribas-Layton, Chen, Lykke-Andersen, & Sen, 2015). On one hand, to maintain self-renewal, DDX6 facilitates the translation of proliferation and self-renewal transcripts by recruiting them to translation initiation factor EIF4E; on the other hand, to prevent differentiation of progenitor cells, through association with mRNA degradation proteins, DDX6 targets and destabilizes the mRNA of KLF4, a differentiation-inducing transcription factor that is required for the activation of the epidermal differentiation and the conversion of fibroblasts to keratinocyte-like cells (Fig. 2C) (Chen, Mistry, & Sen, 2014; Mistry, Chen, Wang, Zhang, & Sen, 2014; Segre, Bauer, & Fuchs, 1999; Wang et al., 2015). In mESCs, Ddx6 is required to maintain normal mESC cell morphology and proliferation. The loss of Ddx6 produces a similar downstream consequence as the depletion of Dgcr8, which is essential for miRNA biogenesis. Instead of miRNA-induced RNA degradation, Ddx6 is important in miRNA-induced translational repression in mESCs (Freimer et al., 2018). Recently, Di Stefano et al. showed that DDX6 is an important regulator of pluripotency in both human and mouse ESCs as its depletion leads ESCs to a differentiation-resistant state. Suppression of DDX6 also promotes the reprogramming of primed hESCs to a naïve state. DDX6 was also found to regulate the differentiation potential of adult somatic progenitors in a context-dependent manner. Mechanistically, DDX6 is associated with critical P-body proteins and mediates the translational suppression of the target transcripts in P-bodies. DDX6 loss results in dissolvement of P-bodies, which releases mRNAs encoding key cell fate transcription regulators and chromatin factors to the translational machinery in promoting pluripotency and reprogramming (Fig. 2D) (Di Stefano et al., 2019).
3.4. DDX18
Zuo et al. (2009) constructed a protein interaction network encompassing hESC-enriched proteins in hESCs and found that DDX18 is among the top 5% highly connected nodes, suggesting that DDX18 may have important functions in hESCs. Very recently, Ddx18 was reported to be required for mESC maintenance and embryonic development. DDX18 directly interacts with PRC2 and modulates the formation of PRC2 complex. Such interaction prevents PRC2 from accessing and marking ribosomal DNA (rDNA) with repressive H3K27me3. rRNA (ribosomal RNA, the product of rDNA transcription) is highly expressed in ESCs and becomes downregulated upon differentiation. Ddx18 depletion increases PRC2 occupancy at rDNA loci to inhibit rDNA transcription, leading to reduced ribosomal protein level and global translation level (Fig. 2E) (Zhang et al., 2020). Owing to the alternative pluripotent states between mouse and human ESCs, it remains to be addressed whether the human ortholog DDX18 may play a similar or distinct role in regulating human pluripotency and reprogramming.
3.5. DDX21
RNA helicase DDX21 functions in multiple steps of ribosome biogenesis by coordinating transcription and rRNA processing. DDX21 was found associated with actively transcribed ribosomal genes as well as rRNAs and snoRNAs, facilitating rRNA modification (Calo et al., 2015). The multifaceted function of DDX21 in ribosome biogenesis suggests its potential role in ESCs as both ribosomal genes and rRNAs are highly expressed in ESCs and properly downregulated/repressed during early differentiation (Ingolia, Lareau, & Weissman, 2011; Savić et al., 2014; Woolnough, Atwood, Liu, Zhao, & Giles, 2016), although a definite functional contribution to pluripotency and reprogramming has yet to be tested.
3.6. DDX47 and DDX52
DDX47 and DDX52 are subunits of small subunit processome (SSUP), which mediates 18S rRNA biogenesis (Phipps, Charette, & Baserga, 2011; Tafforeau et al., 2013). The components of SSUP are highly expressed in stem cells and important to maintain the protein levels of pluripotency factors. As SSUP subunits, both DDX47 and DDX52 are validated to be necessary for ESC maintenance and efficient iPSC reprogramming: (1) depletion of either of them in mESCs induced differentiation; (2) they help to sustain the protein levels of labile pluripotency factors NANOG and OCT4 in mESCs; and (3) Both are required for efficient reprogramming of iPSCs (Fig. 2F) (You, Park, & Kim, 2015).
4. Conclusions
Understanding molecular mechanisms underlying pluripotency and reprograming is highly significant both scientifically and clinically. The posttranscriptional regulation by RBPs constitutes an important regulatory layer for controlling pluripotency and reprogramming. Although RBPs have been studied widely because of their involvement in a broad range of cellular processes, their regulatory functions in stem cell field are only just beginning to be appreciated. As post-transcriptional regulation enables cells to quickly respond by adjusting protein abundance, future studies are warranted to dissect mechanistic actions of RBPs during cell fate transitions and further our understanding of their roles in pluripotency and reprogramming. The potential multifaceted functions of RBPs on both RNA and DNA targets at transcriptome/epitranscriptome and genome/epigenome levels should be more carefully examined in light of their dual DNA/RNA binding capacities. Finally, as many RBPs contain intrinsically disordered regions, the roles of RBPs in the regulation of phase separations and gene expression, which are only recently recognized (A & Weber, 2019; Shorter, 2019; Xiao et al., 2019; Youn et al., 2019), await more future investigations at both physiological and pathological conditions.
Together, RBP studies would provide a platform and new framework for better understanding of molecular mechanisms underlying pluripotency and reprogramming, which would bring us closer to the practical applications of pluripotent ESCs/iPSCs for regenerative medicine, tissue engineering, and disease therapeutics.
Acknowledgments
We are grateful to Jill Gregory for the illustrations of Figs. 1 and 2 and the members of Wang lab for helpful discussions. We thank the following funding sources for support: National Institutes of Health (GM129157, HD095938, HD097268, and HL146664) and New York State Stem Cell Science (NYSTEM#C32583GG and C32569GG).
References
- A, & Weber. (2019). Evidence for and against liquid-liquid phase separation in the nucleus. Non-Coding RNA, 5(4), 50. 10.3390/ncrna5040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, et al. (2019). N 6-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nature Genetics, 52, 48–55. 10.1038/s41588-019-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Sesé B, Boue S, Castaño J, Paramonov I, Barrero MJ, et al. (2011). LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nature Cell Biology, 13(6), 652–661. 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, et al. (2015). Coordination of m 6 a mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell, 17(6), 689–704. 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, & Aravind L (2002). Comparative genomics and evolution of proteins involved in RNA metabolism. in survey and summary. Nucleic Acids Research, 30(7), 1427–1464. 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, et al. (2011). Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell, 145(2), 183–197. 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Mowla SJ, Ziaee SAM, Gokhale PJ, & Andrews PW (2008). OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells, 26(12), 3068–3074. 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- Bao X, Guo X, Yin M, Tariq M, Lai Y, Kanwal S, et al. (2018). Capturing the interactome of newly transcribed RNA. Nature Methods, 15(3), 213–220. 10.1038/nmeth.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL (2002). RNA editing by adenosine deaminases that act on RNA. Annual Review of Biochemistry, 71(1), 817–846. 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. (2014). m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell, 15(6), 707–719. 10.1016/J.STEM.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuth B, Pennell S, Arnvig KB, Martin SR, & Taylor IA (2005). Structure of a mycobacterium tuberculosis NusA–RNA complex. The EMBO Journal, 24(20), 3576–3587. 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N (2013). Noncoding RNA and Polycomb recruitment. RNA, 19, 429–442. 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva De Sousa Lopes SM, et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature, 448(7150), 191–195. 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Brumbaugh J, Di Stefano B, Wang X, Borkent M, Forouzmand E, Clowers KJ, et al. (2018). Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell, 172(1–2), 106–120.e21. 10.1016/j.cell.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, & Wysocka J (2015). RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature, 518(7538), 249–253. 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, & Tomlinson SR (2009, July 15). The transcriptional foundation of pluripotency. Development, 136, 2311–2322. 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, & Gregory RI (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nature Communications, 3, 923. 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K (2015). Alternative splicing: An important mechanism in stem cell biology. World Journal of Stem Cells, 7(1), 1. 10.4252/wjsc.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Mistry DS, & Sen GL (2014). Highly rapid and efficient conversion of human fibroblasts to keratinocyte-like cells. Journal of Investigative Dermatology, 134(2), 335–344. 10.1038/jid.2013.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Xiang J-F, Zhu S, Chen S, Yin Q-F, Zhang X-O, et al. (2015). ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Research, 25, 459–476. 10.1038/cr.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CY, & Lufkin T (2011). Alternative splicing in self-renewal of embryonic stem cells. Stem Cells International, 2011, 560261, 10.4061/2011/560261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat CM, Dolde C, De Groot REA, Ohkawara B, Reinhard C, Korswagen HC, et al. (2013). RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Science, 339(6126), 1436–1441. 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, et al. (2014). Jarid2 Is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Molecular Cell, 53(2), 301–316. 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Dardenne E, PolayEspinoza M, Fattet L, Germann S, Lambert MP, Neil H, et al. (2014). RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Reports, 7(6), 1900–1913. 10.1016/j.celrep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Das S, Jena S, & Levasseur DN (2011). Alternative splicing produces nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. Journal of Biological Chemistry, 286(49), 42690–42703. 10.1074/jbc.M111.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, et al. (2016). Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science, 351(6270), 282–285. 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, MacIsaac KD, Stevens RC, Sriram S, Chen R, et al. (2012). A quantitative atlas of polyadenylation in five mammals. Genome Research, 22(6), 1173–1183. 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Luo EC, Haggerty C, Aigner S, Charlton J, Brumbaugh J, et al. (2019). The RNA helicase DDX6 controls cellular plasticity by modulating P-body homeostasis. Cell Stem Cell, 25(5), 622–638.e13. 10.1016/j.stem.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington JM, Greene T, & Bass BL (2011). Predicting sites of ADAR editing in double-stranded RNA. Nature Communications, 2, 319. 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, et al. (2013). The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS One, 8(8), e72300. 10.1371/journal.pone.0072300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, & Jankowsky E (2010). SF1 and SF2 helicases: Family matters. Current Opinion in Structural Biology, 20(3), 313–324. 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Massingham T, Bhattacharyya S, Ponsting H, Lyons PA, Freeman TC, et al. (2005). Open access relationship between the tissue-specificity of mouse gene expression and the evolutionary origin and function of the proteins. Genome Biology, 6(7), 56. 10.1186/gb-2005-6-7-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer JW, Hu TJ, & Blelloch R (2018). Decoupling the impact of microRNAs on translational repression versus RNA degradation in embryonic stem cells. eLife 7, e38014. 10.7554/eLife.38014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, & Blanco S (2016). Post-transcriptional modifications in development and stem cells. Development (Cambridge, England), 143(21), 3871–3881. 10.1242/dev.136556. [DOI] [PubMed] [Google Scholar]
- Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, et al. (2014). Tetmediated formation of 5-hydroxymethylcytosine in RNA. Journal of the American Chemical Society, 136(33), 11582–11585. 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, et al. (2011). An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell, 147(1), 132–146. 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, et al. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature, 460(7257), 863–868. 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, & Tuschl T (2014). A census of human RNA-binding proteins. Nature Reviews Genetics, 15(12), 829–845. 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. (2015). m 6 A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science, 347(6225), 1002–1006. 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Gökbuget D, & Blelloch R (2019). Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development (Cambridge), 146, 1–16. 10.1242/dev.164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D, Bi X, Pardavila JA, Huang X, Saenz C, Shi X, et al. (2018). RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nature Genetics, 50(3), 443–451. 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D, & Wang J (2014). RNA-binding proteins in pluripotency, differentiation, and reprogramming. Frontiers in Biology, 9(5), 389–409. 10.1007/s11515-014-1326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464(7291), 1071–1076. 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, & Gregory RI (2009). Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature Structural and Molecular Biology, 16(10), 1021–1025. 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A (2004). Online Mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Research, 33(Database issue), D514–D517. 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, et al. (2013). MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature, 498(7453), 241–245. 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick SW, & Luisi BF (2013). Rarely at rest: RNA helicases and their busy contributions to RNA degradation, regulation and quality control. RNA Biology, 10, 56–70. 10.4161/rna.22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Sidoli S, Warneford-Thomson R, Tatomer DC, Wilusz JE, Garcia BA, et al. (2016). High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Molecular Cell, 64(2), 416–430. 10.1016/j.molcel.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, et al. (2012). Monouridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell, 151(3), 521–532. 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, & Kim VN (2008). Lin28 mediates the terminal Uridylation of let-7 precursor MicroRNA. Molecular Cell, 32(2), 276–284. 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, & Tollervey D (2006, July 17). RNA-quality control by the exosome. Nature Reviews Molecular Cell Biology, 7, 529–539. 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, & Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 147(4), 789–802. 10.1016/J.CELL.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E (2011). RNA helicases at work: Binding and rearranging. Trends in Biochemical Sciences, 36(1), 19–29. 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, & Russell R (2014). RNA helicase proteins as chaperones and remodelers. Annual Review of Biochemistry, 83(1), 697–725. 10.1146/annurevbiochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, & Tian B (2009). Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proceedings of the National Academy of Sciences of the United States of America, 106(17), 7028–7033. 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, & Tian B (2009). Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One, 4(12), e8419. 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, et al. (2014). Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Molecular Cell, 53(2), 290–300. 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya M, & Higuchi H (2010). Nonlinear elasticity and an 8-nm working stroke of single myosin molecules in myofilaments. Science, 329(5992), 686–689. 10.1126/science.1191484. [DOI] [PubMed] [Google Scholar]
- Kerr CL, Bol GM, Vesuna F, & Raman V (2019). Targeting RNA helicase DDX3 in stem cell maintenance and teratoma formation. www.Genes&Cancercom, Genes & Cancer, 10(1–2), 11–20. 10.18632/genesandcancer.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, & Kimmel A (2006). GSK3 at the edge: Regulation of developmental specification and cell polarization. Current Drug Targets, 7(11), 1411–1419. 10.2174/1389450110607011411. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, et al. (2013). The RNA-binding protein repertoire of embryonic stem cells. Nature Structural and Molecular Biology, 20(9), 1122–1130. 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. (2005). RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell, 121(5), 713–724. 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi E-A, et al. (2014). Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. The EMBO Journal, 33(8), 878–889. 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lai P, Jia J, Song Y, Xia Q, Huang K, et al. (2017a). Erratum: RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions (Cell Stem Cell (2017) 20(4) (462–477) (S1934590916304581) (10.1016/j.stem.2016.12.002)). Cell Stem Cell, 20(4), 571. 10.1016/j.stem.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Li H, Lai P, Jia J, Song Y, Xia Q, Huang K, et al. (2017b). RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell, 20(4). 10.1016/j.stem.2016.12.002, 462–477.e6. [DOI] [PubMed] [Google Scholar]
- Linder P, & Jankowsky E (2011). From unwinding to clamping—The DEAD box RNA helicase family. Nature Reviews Molecular Cell Biology, 12, 505–516. 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Loedige I, Gaidatzis D, Sack R, Meister G, & Filipowicz W (2013). The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Research, 41(1), 518–532. 10.1093/nar/gks1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay JP, Font J, & Segal DJ (2011, March). The prospects for designer single-stranded RNA-binding proteins. Nature Structural and Molecular Biology, 18, 256–261. 10.1038/nsmb.2005. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, & Allain FH-T (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS Journal, 272(9), 2118–2131. 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Masiello I, & Biggiogera M (2017). Ultrastructural localization of 5-methylcytosine on DNA and RNA. Cellular and Molecular Life Sciences, 74(16), 3057–3064. 10.1007/s00018-017-2521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, & Blelloch R (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature, 463(7281), 621–626. 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Xin N, Wei B, Hua X, Zhang G, Leng C, et al. (2016). 5-hydroxymethylcytosine is detected in RNA from mouse brain tissues. Brain Research, 1642, 546–552. 10.1016/j.brainres.2016.04.055. [DOI] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Zhang K, & Sen GL (2014). SNAI2 controls the undifferentiated state of human epidermal progenitor cells. Stem Cells, 32(12), 3209–3218. 10.1002/stem.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B (2001). Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Research, 29(13), 2850–2859. 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Teo RTY, Li H, Robson P, Glover DM, & Zernicka-Goetz M (2010). Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America, 107(14), 6364–6369. 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AA, Cheung TH, & Rando TA (2013). All’s well that ends well: Alternative polyadenylation and its implications for stem cell biology. Current Opinion in Cell Biology, 25, 222–232. 10.1016/j.ceb.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pádua Alves C, Fonseca AS, Muys BR, de Barros e Lima Bueno R, Bürger MC, de Souza JES, et al. (2013). Brief report: The lincRNA Hotair is required for epithelialto-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells, 31(12), 2827–2832. 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- Phipps KR, Charette JM, & Baserga SJ (2011). The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdisciplinary Reviews: RNA, 2(1), 1–21. 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, & Scott J (1987). A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell, 50(6), 831–840. 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Putnam AA, & Jankowsky E (2013). DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochimica et Biophysica Acta—Gene Regulatory Mechanisms, 1829(8), 884–893. 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, & Sandberg R (2009). An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Computational Biology, 5(12), e1000598. 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan G-C, & Orkin SH (2010). Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Molecular and Cellular Biology, 30(22), 5364–5380. 10.1128/mcb.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, & Hurt E (2002, February 22). A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell, 108, 523–531. 10.1016/S0092-8674(02)00627-X. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, & Abelson J (1993). Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes and Development, 7(10), 1909–1925. 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, & Burge CB (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science, 320(5883), 1643–1647. 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, & Wang J (2014, May 1). Export and expression: MRNAs deliver new messages for controlling pluripotency. Cell Stem Cell, 14, 549–550. 10.1016/j.stem.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Savić N, Bär D, Leone S, Frommel SC, Weber FA, Vollenweider E, et al. (2014). LncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell, 15(6), 720–734. 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, & Fuchs E (1999). Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature Genetics, 22(4), 356–360. 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, & Shi Y (2011). Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA, 17(4), 761–772. 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J (2019). Phase separation of RNA-binding proteins in physiology and disease: An introduction to the JBC reviews thematic series. Journal of Biological Chemistry, 294, 7113–7114. 10.1074/jbc.REV119.007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtrichman R, Germanguz I, Mandel R, Ziskind A, Nahor I, Safran M, et al. (2012). Altered A-to-I RNA editing in human embryogenesis. PLoS One, 7(7), e41576. 10.1371/journal.pone.0041576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Skrisovska L, & Allain FH (2005). RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Reports, 6(1), 33–38. 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H, Takahashi K, Yamamoto T, Iwasaki M, Narita M, Nakamura M, et al. (2017). Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America, 114(2), 340–345. 10.1073/pnas.1617234114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, et al. (2013). The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Molecular Cell, 51(4), 539–551. 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Amiri M, & Sonenberg N (2019). Translational control in stem cells. Frontiers in Genetics, 9, 709. 10.3389/fgene.2018.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131(5), 861–872. 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, & Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teplova M, & Patel DJ (2008). Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nature Structural and Molecular Biology, 15(12), 1343–1351. 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, et al. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature, 448(7150), 196–199. 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, & Jaenisch R (2017). Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development (Cambridge, England), 144(24), 4496–4509. 10.1242/dev.157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science (New York, N.Y.), 282(5391), 1145–1147. 10.1126/SCIENCE.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, & Patel DJ (2011). Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proceedings of the National Academy of Sciences of the United States of America, 108(3), 903–910. 10.1073/pnas.1017762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, et al. (2013). Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA, 19(12), 1632–1638. 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Arribas-Layton M, Chen Y, Lykke-Andersen J, & Sen GL (2015). DDX6 orchestrates mammalian progenitor function through the mRNA degradation and translation pathways. Molecular Cell, 60(1), 118–130. 10.1016/J.MOLCEL.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, & Dai J (2010). Isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells, 28(5), 885–893. 10.1002/stem.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, et al. (2013). The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell, 13(6), 676–690. 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. (2012). Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature, 482(7384), 221–225. 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, & Ponting CP (2004). Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Research, 14(1), 54–61. 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolnough JL, Atwood BL, Liu Z, Zhao R, & Giles KE (2016). The regulation of rRNA gene transcription during directed differentiation of human embryonic stem cells. PLoS One, 11(6), e0157276. 10.1371/journal.pone.0157276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, et al. (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell, 14(1), 40–52. 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Chen J, Wan Y, Gao Q, Jing N, Zheng Y, et al. (2019). Regulation of zebrafish dorsoventral patterning by phase separation of RNA-binding protein Rbm14. Cell Discovery, 5, 37. 10.1038/s41421-019-0106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, & Liu ZR (2006). P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing axin from β-catenin. Cell, 127(1), 139–155. 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Ye J, & Blelloch R (2014). Regulation of pluripotency by RNA binding proteins. Cell Stem Cell, 15, 271–280. 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, & Gage FH (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature Structural and Molecular Biology, 16(2), 130–137. 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo JC, & Ng HH (2013, January). The transcriptional regulation of pluripotency. Cell Research, 23, 20–32. 10.1038/cr.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe Y, David M, Kalaora R, Povodovski L, Friedlander G, Feldmesser E, et al. (2016). Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes & Development, 30(17), 1991–2004. 10.1101/gad.285239.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KT, Park J, & Kim VN (2015). Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes & Development, 29(19), 2004–2009. 10.1101/gad.267112.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, et al. (2019, October 17). Properties of stress granule and P-body proteomes. Molecular Cell, 76, 286–294. 10.1016/j.molcel.2019.09.014. [DOI] [PubMed] [Google Scholar]
- Young RA (2011, March 18). Control of the embryonic stem cell state. Cell, 144, 940–954. 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu Z, Lu JY, Huang B, Zhou H, Xie W, et al. (2020). DEAD-box helicase 18 counteracts PRC2 to safeguard ribosomal DNA in pluripotency regulation. Cell Reports, 30(1), 81–97.e7. 10.1016/j.celrep.2019.12.021. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Xiong J, Qi BL, Feng YQ, & Yuan BF (2016). The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chemical Communications, 52(4), 737–740. 10.1039/c5cc07354e. [DOI] [PubMed] [Google Scholar]
- Zuo C, Liang S, Wang Z, Li H, Zheng W, & Ma W (2009). Enriching protein-protein and functional interaction networks in human embryonic stem cells. International Journal of Molecular Medicine, 23(6), 811–819. 10.3892/ijmm_00000197. [DOI] [PubMed] [Google Scholar]