In this study, Tang et al. investigated how metastasis is regulated by a common hot spot mutation, p53R175H, in esophageal squamous cell carcinoma (ESCC). Using a carcinogen-induced approach in Trp53R172H/− mice to model ESCC, they demonstrate that expression of Survivin, an antiapoptotic protein encoded by BIRC5 increases in the presence of Trp53R172H and depletion of Survivin specifically decreases Trp53R172H-driven lung metastasis.
Keywords: mutant p53, metastasis, Survivin, YAP
Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most lethal cancers worldwide and evolves often to lung metastasis. P53R175H (homologous to Trp53R172H in mice) is a common hot spot mutation. How metastasis is regulated by p53R175H in ESCC remains to be investigated. To investigate p53R175H-mediated molecular mechanisms, we used a carcinogen-induced approach in Trp53R172H/− mice to model ESCC. In the primary Trp53R172H/− tumor cell lines, we depleted Trp53R172H (shTrp53) and observed a marked reduction in cell invasion in vitro and lung metastasis burden in a tail-vein injection model in comparing isogenic cells (shCtrl). Furthermore, we performed bulk RNA-seq to compare gene expression profiles of metastatic and primary shCtrl and shTrp53 cells. We identified the YAP-BIRC5 axis as a potential mediator of Trp53R172H-mediated metastasis. We demonstrate that expression of Survivin, an antiapoptotic protein encoded by BIRC5, increases in the presence of Trp53R172H. Furthermore, depletion of Survivin specifically decreases Trp53R172H-driven lung metastasis. Mechanistically, Trp53R172H but not wild-type Trp53, binds with YAP in ESCC cells, suggesting their cooperation to induce Survivin expression. Furthermore, Survivin high expression level is associated with increased metastasis in several GI cancers. Taken together, this study unravels new insights into how mutant p53 mediates metastasis.
Metastases contribute to the majority of cancer-associated deaths (Steeg 2016). The complex invasion-metastasis cascade involves multiple steps, including cancer cell invasion, intravasation into circulation, extravasation from circulation, and metastatic colonization into a distant organ (e.g., liver and lung). However, despite a large number of studies demonstrating key mechanisms and treatment strategies in primary tumors, the mechanisms underlying tumor metastasis are still in evolution (Lambert et al. 2017). Increasing evidence has shown that tumor intrinsic factors such as genomic, genetic, and epigenetic aberrations are important contributors to metastasis. Specifically, tumor intrinsic oncogenic events induce genomic instability, facilitate invasion to the stroma, and are crucial driving forces of metastases (Steeg 2016).
Across all human cancers, TP53 is the most frequently mutated gene, and alterations in p53 functions and its downstream signaling pathways have been validated in diverse cancers (Muller and Vousden 2014). Unlike other tumor suppressor proteins that harbor frameshift or nonsense mutations that lead to loss of expression, the majority of cancer-associated TP53 alterations encompass missense mutations (Olivier et al. 2010; Freed-Pastor and Prives 2012). Missense TP53 mutations occur frequently on six residues, namely R175, G245, R248, R249, R273, and R282, and are designated as “hot spot” mutations (Rivlin et al. 2011). Functionally, hot spot p53 mutations have been demonstrated to promote tumorigenesis (Freed-Pastor and Prives 2012). For example, mice with germline Trp53 (equivalent to TP53 in human) mutations have a different tumor spectrum compared with Trp53-null mice. Trp53R172H/− (equivalent to human p53R175H/−) mice have increased incidence of carcinomas, whereas Trp53−/− mice frequently develop lymphomas and sarcomas (Olive et al. 2004). Compared with Trp53+/− mice, Trp53R172H/+ mice harbor metastatic osteosarcoma and epithelial carcinomas, whereas tumors in Trp53+/− mice typically do not metastasize (Lang et al. 2004). Furthermore, in several types of cancers, mutant p53, especially p53R175H, promotes cell invasion, motility, and metastasis through the regulation of different pathways (Bossi et al. 2006; Dong et al. 2009; Muller et al. 2009, 2013; Tabach et al. 2010; Weissmueller et al. 2014). Furthermore, analysis of clinical data has validated these findings. For example, in head and neck squamous cell carcinoma (HNSCC), patients harboring p53R175H have poor clinical outcomes, including increased metastasis and decreased overall survival (Neskey et al. 2015). Taken together, mutant p53 contributes to an aggressive tumor phenotype characterized by enhanced cell motility, invasion, and metastatic capacity in several cancer types.
Esophageal cancer is a highly aggressive cancer. Globally, it was estimated that approximately 570,000 people were diagnosed with esophageal cancer in 2018 (Uhlenhopp et al. 2020). The major subtype of esophageal cancer worldwide is esophageal squamous cell carcinoma (ESCC), which typically has a poor prognosis, since many cases are not detected until late stages (Pennathur et al. 2013; Rustgi and El-Serag 2014). At the time of diagnosis, local invasion and distant metastasis, especially lung metastasis, are frequently detected (Rustgi and El-Serag 2014). Thus, understanding the mechanisms underlying metastasis in ESCC is crucial to improvements in treatment strategies and patient outcomes, with potential therapeutic applications to other genomically related squamous cell cancers (SCC) arising from the head and neck, lung, and anogenital tract (Dotto and Rustgi 2016; Campbell et al. 2018). The most frequent genetic alteration in ESCC is TP53 mutation, occurring in 83% of ESCC patients (Song et al. 2014), and as such, ESCC represents one of the top cancers that harbor TP53 mutations (Olivier et al. 2010). In ESCC, R175H mutation is one of the top three hot spot TP53 mutations, which leads to distortion of the global protein structure and is referred to as confirmation mutation (IARC database) (Olivier et al. 2010). How mutant p53, especially the R175H mutation, contributes to ESCC invasion and metastasis remains to be understood, and discoveries therein can be applied to other cancers, especially SCC.
In this study, we focused on the oncogenic role of mutant p53 in driving lung metastasis. Through in vitro and in vivo studies, we demonstrated that p53R175H promotes ESCC cell motility, invasion, and lung metastasis. To investigate the underlying mechanisms, we profiled gene expression signatures in metastatic and primary ESCC cells. We identified BIRC5 (encoding an antiapoptosis protein Survivin) to be highly enriched in metastatic mutant p53 ESCC cells. Furthermore, we determined that Survivin expression is dependent upon p53R175H and is necessary and specific to mediate p53R175H-driven lung metastasis. Mechanistically, we revealed that Trp53R172H, but not wild-type Trp53, interacts physically with YAP (Yes-associated protein) in ESCC cells. We also discovered that Trp53R172H enhances the binding of YAP on the BIRC5 promoter, suggesting their cooperation to induce Survivin expression. Additionally, we found that there is up-regulation of Survivin expression in ESCC based upon a human ESCC tissue microarray and TCGA analysis. There is correlation of high Survivin expression with p53 nuclear accumulation in ESCC. Furthermore, there is Survivin up-regulation with increased metastasis in other GI cancers (pancreatic and colon). Taken together, this study underscores the novel finding that BIRC5 is a novel mediator of p53R175H in promoting lung metastasis, thereby offering new insights into how mutant p53 promotes metastasis and opening up new avenues for translational therapeutics.
Results
Mutant p53 is required for invasion and metastasis in ESCC
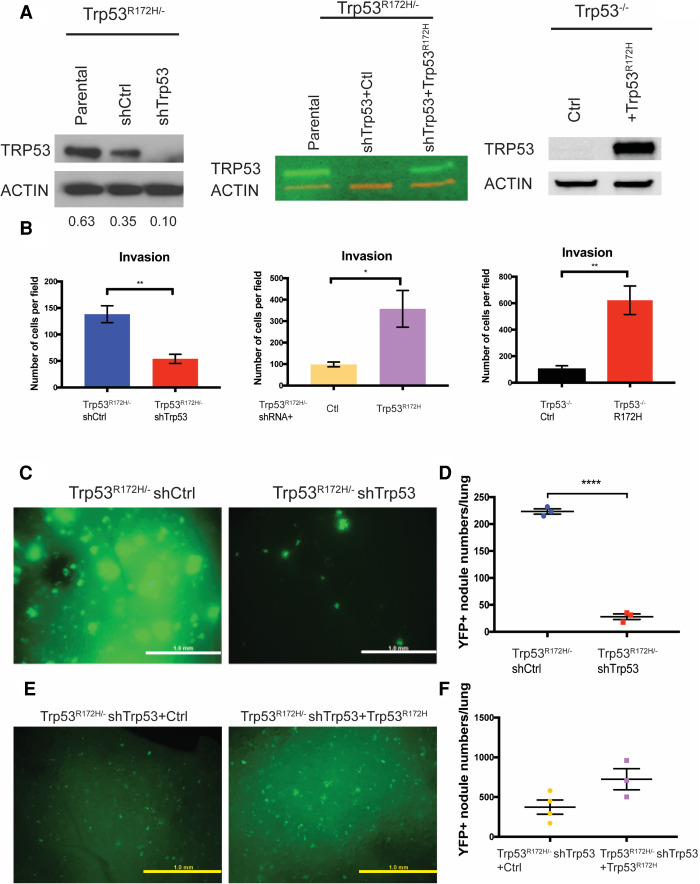
To understand the function of mutant p53 in ESCC, a mutant p53-driven ESCC mouse model was generated combining genetic and carcinogenic approaches. This model recapitulates human ESCC grossly and histologically (Tang et al. 2021). Additionally, we generated Trp53R172H/− and Trp53−/− cell lines from these primary tumors and only used early passage cells (Tang et al. 2021). To elucidate the role of mutant p53 in tumor cell invasion and metastasis, we generated an isogenic control system through stably expressing shRNA targeting Trp53 in Trp53R172H/− cells (shTrp53) and compared their biological behavior with control shRNA-expressing Trp53R172H/− cells (shCtrl) (Fig. 1A; Supplemental Fig. S1A). We also generated a “rescue” cell line by introducing shTrp53-resistant Trp53R172H (Fig. 1A). The CRISPR/Cas9 system was also used to deplete Trp53 in Trp53R172H/− cells (sgTrp53), comparing with an empty vector-expressing Trp53R172H/− cells (Vector) (Supplemental Fig. S1B). In addition, we ectopically expressed Trp53R172H in Trp53−/− cells (R172H) to compare with Trp53−/− cells expressing an empty vector (Ctrl) (Fig. 1A).
Figure 1.
Mutant p53 promotes ESCC invasion and lung metastasis. (A, left) Trp53R172H is depleted by shRNA in Trp53R172H/− cells. shRNA-resistant Trp53R172H is reintroduced to Trp53R172H/− shTrp53 (middle), and Trp53R172H is ectopically expressed in Trp53−/− cells (right). Relative intensity densitometry of TRP53 results are shown at the bottom. (B) Depletion of Trp53R172H suppresses cell invasion, whereas reintroduction of shRNA-resistant Trp53R172H or ectopic expression of Trp53R172H promotes cell invasion in a Boyden chamber assay. Trp53R172H/− shCtrl versus shTrp53, n = 4 per group, P = 0.0035, unpaired t-test; Trp53R172H/− shRNA + Ctl versus + Trp53R172H, n = 3 per group, P = 0.0398, unpaired t-test; Trp53−/− Ctrl versus R172H, n = 3 per group, P = 0.0094, unpaired t-test. Error bars represent SEM. (C,D) YFP+ ESCC cells were injected into the tail vein to induce lung metastatic nodules. Mice were sacrificed 8 wk after injection. Lung metastasis colony number in Trp53R172H/− shCtrl injected mice was increased greatly compared with shTrp53 injected mice. Trp53R172H/− shCtrl versus shTrp53, n = 3 per group, P < 0.0001, unpaired t-test. Scale bar, 1 mm. (E,F) YFP+ Trp53R172H/− shTrp53 cells reintroduced with shRNA-resistant Trp53R172H were used to repeat the tail-vein injection assay. Mice were sacrificed 8 wk after injection. Trp53R172H/− shTrp53-rescued cell line-injected mice had a trend of increased lung metastatic colony number. Trp53R172H/− shTrp53+Ctrl (n = 4 per group) versus shTrp53+ Trp53R172H (n = 3 per group), P = 0.0709, unpaired t-test. Scale bar, 1 mm.
We showed that tumor cell invasion is dependent on Trp53R172H using a transwell invasion assay. Depletion of Trp53R172H abrogated cell invasion, whereas reintroduction of shTrp53-resistant Trp53R172H or ectopic expression of Trp53R172H enhanced invasion capacity (Fig. 1B; Supplemental Fig. S1C). To evaluate whether alterations in proliferation or apoptosis might influence invasion, we performed the carboxyfluorescein succinimidyl ester (CFSE) proliferation assay and observed no significant difference in proliferation upon Trp53R172H depletion (Supplemental Fig. S1D). Additionally, no significant change was observed in apoptosis (Supplemental Fig. S1E). The impact of Trp53R172H in cell invasion was determined further using 3D organotypic culture (OTC), an air–liquid interface model system (Kalabis et al. 2012). In OTC with epithelial Trp53R172H depletion, there is reduced cell invasion in the stroma (Supplemental Fig. S1F). Taken together, Trp53R172H promotes tumor cell invasion.
Next, we examined whether Trp53R172H is required for ESCC metastatic potential through the injection of shCtrl and shTrp53 cells into the lateral tail veins of athymic nude mice (NCr nude, Taconic). Lung metastases were detected at 2 wk, 5 wk, and 8 wk postinjection, and the number and size of metastases were scored. At each time point, the metastatic burden was decreased significantly upon Trp53R172H depletion (Fig. 1C,D; Supplemental Fig. S2A,B). In addition, we performed a “rescue” experiment and observed a partial rescue of the metastatic phenotype following introduction of shTrp53-resistant Trp53R172H (Fig. 1E,F; Supplemental Fig. S2C,D). As independent corroboration, we repeated the tail-vein injection assay with R172H and Ctrl cell lines and observed a trend of increased metastatic nodule number in the presence of ectopic Trp53R172H expression (Supplemental Fig. S2E,F). Taken together, these results underscore that Trp53R172H enhances ESCC metastatic capability to the lungs.
Mutant p53-mediated reprogramming of pathways in lung metastasis
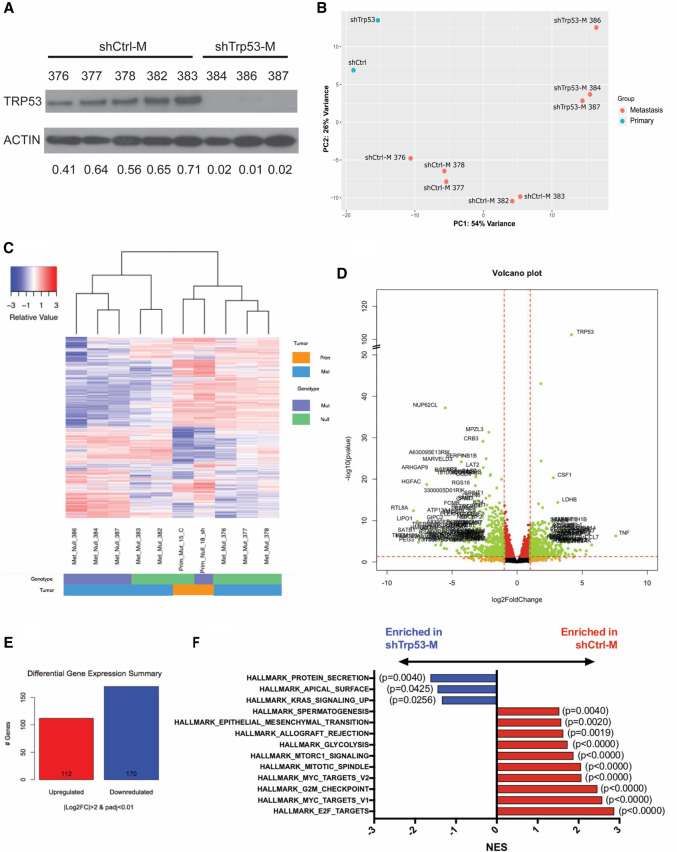
Following the generation and characterization of our lung metastatic model, we generated further a panel of metastatic cell lines (shTrp53-M and shCtrl-M; M indicates metastatic) from lung metastases at week 8 through YFP+ FACS sorting and confirmed that Trp53R172H depletion was maintained in shTrp53-M cells (Fig. 2A). To understand the molecular basis of Trp53R172H-driven ESCC lung metastasis, we performed RNA-seq analysis on shCtrl-M (n = 5), shTrp53-M (n = 3), shCtrl (n = 1), and shTrp53 (n = 1) cell lines. Principal component analysis (PCA) revealed that primary tumor cells (shCtrl and shTrp53) clustered away from metastatic cells (shCtrl-M and shTrp53-M) (Fig. 2B). Compared with two primary tumor cell lines shCtrl and shTrp53, metastatic cell lines shCtrl-M and shTrp53-M exhibited a distinct gene expression pattern, suggesting that gene expression signatures are changed during metastasis (Fig. 2C). In metastatic cells, we identified 112 genes significantly up-regulated and 170 genes down-regulated in shCtrl-M cells compared with shTrp53-M cells (Padj < 0.01 and |log2 fold change| > 2). Among these, Trp53 is the most significantly up-regulated gene, which served to validate our system (Fig. 2D,E). Gene set enrichment analysis (GSEA) Hallmark revealed 13 down-regulated pathways and 20 up-regulated pathways in shCtrl-M cells. Epithelial–mesenchymal transition (EMT), an up-regulated pathway, has been shown previously to play an important role in metastasis (Fig. 2F; Tsai and Yang 2013; Dongre and Weinberg 2019). In addition, other signaling pathways such as MYC, mTORC1, and glycolysis are also known to be involved in regulating metastasis (Fig. 2F; Wolfer and Ramaswamy 2011; Huang and Zhou 2012; Payen et al. 2016).
Figure 2.
Depletion of Trp53R172H in ESCC cells alters the transcriptome during lung metastasis. (A) Depletion of Trp53R172H is maintained in isolated metastatic cell lines. Relative intensity densitometry of TRP53 results are shown at the bottom. (B) Principle component analysis (PCA) demonstrated distinct gene expression profiles depending upon p53 status and primary tumor versus metastatic tumor status. (C) Heat map of differentially regulated genes upon Trp53R172H depletion in primary and metastatic cells, profiled by RNA-seq. (D) Volcano plot of differentially regulated genes in metastatic cells. (E) Numbers of significantly (Padj < 0.01, |log2FC| > 2) changed genes in metastatic Trp53R172H/− cells (n = 5) compared with Trp53−/− cells (n = 3). (F) GSEA reveals hallmark pathways significantly enriched in shCtrl-M and shTrp53-M cells. (M) Metastatic.
Trp53R172H promotes BIRC5 function
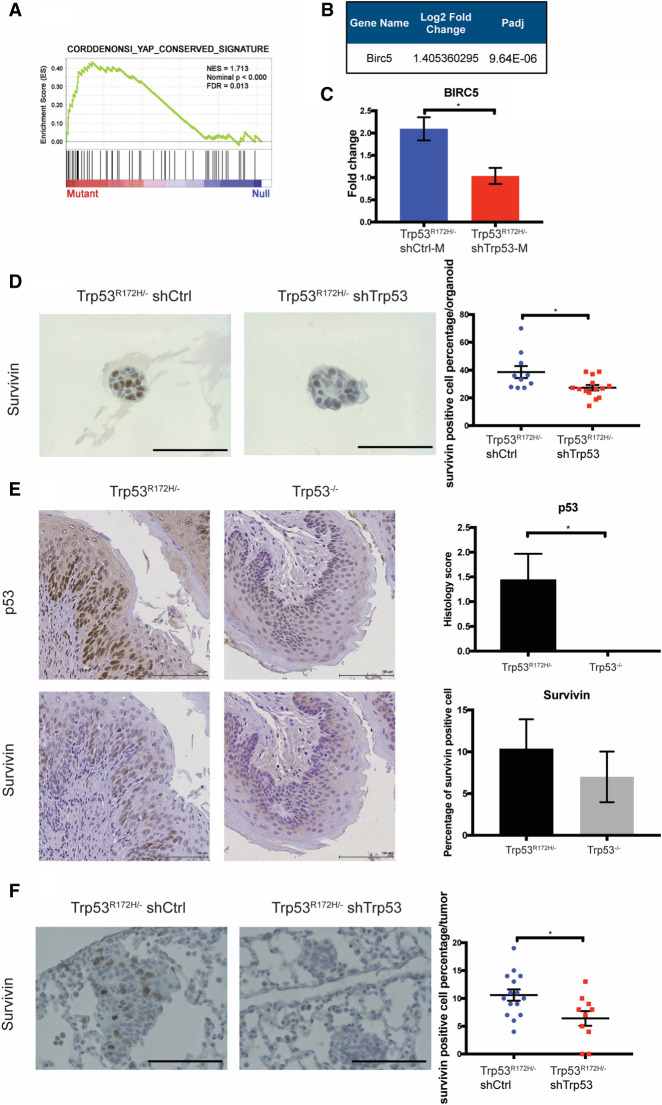
To identify and characterize candidate genes that mediate Trp53R172H/− functional effects in the promotion of lung metastasis, we focused on pathways and genes that are enriched in shCtrl-M cells. We found that YAP (Yes-associated protein) signaling was up-regulated in shCtrl-M cells compared with shTrp53-M cells (Fig. 3A). YAP signaling has been demonstrated as a crucial oncogenic pathway (Muramatsu et al. 2011; Cui and Li 2018). To investigate the role of YAP in ESCC, we depleted YAP with a pool of shRNAs in Trp53R172H/− ESCC cells (shYAP) and compared its oncogenic behaviors with nontarget control (shNT) (Supplemental Fig. S3A). Depletion of YAP in Trp53R172H/− ESCC cells significantly reduced invasion capacity (Supplemental Fig. S3B). In addition, YAP depletion also significantly decreased subcutaneous tumor growth (Supplemental Fig. S3C,D). Taken together, these data underscore YAP as an important oncogenic factor in ESCC. Oncogenic functions of YAP have also been investigated in several other cancers, including its effects in fostering metastasis (Wang et al. 2018; Lee et al. 2019). Furthermore, YAP has been suggested to interact with mutant p53 and other p53 family members in cell lines (Di Agostino et al. 2016; Mello et al. 2017; Furth et al. 2018). However, whether the interaction of mutant p53 and YAP promotes metastasis remains to be elucidated. Taken together, we hypothesize that YAP signaling may function as a mediator of mutant p53-driven metastasis.
Figure 3.
The expression of SURVIVIN depends upon Trp53R172H status. (A) GSEA reveals enrichment of the YAP signature in Trp53R172H/− shCtrl-M cells. (B) RNA-seq shows that BIRC5, a YAP target gene, is increased significantly in Trp53R172H/− shCtrl-M cells. (C) QRT-PCR of BIRC5 confirms its up-regulation in Trp53R172H/− shCtrl-M compared with shTrp53-M cells. Trp53R172H/− shCtrl-M (n = 5) versus shTrp53-M (n = 3), P = 0.0287, unpaired t-test. Error bars represent SEM. (D) Immunohistochemical staining of SURVIVIN in 3D organoids form by Trp53R172H/− shCtrl and shTrp53 cells. Trp53R172H/− shCtrl (n = 10 organoids) versus shTrp53 (n = 14 organoids), P = 0.016, unpaired t-test. Error bars represent SEM. Scale bar, 100 µm. (E) Immunohistochemical staining of p53 (top) and SURVIVIN (bottom) in the esophagus epithelium of Trp53R172H/− andTrp53−/− mice treated with 4NQO. P53 staining: Trp53R172H/− versus Trp53−/−, n = 8 each group. (*) P = 0.0168, unpaired t-test. SURVIVIN staining: Trp53R172H/− versus Trp53−/−, n = 8 each group. Error bars represent SEM. Scale bar, 100 µm. (F) Immunohistochemical staining of SURVIVIN in Trp53R172H/− shCtrl and shTrp53 in metastatic lung lesions. Trp53R172H/− shCtrl (n = 15 tumors) versus shTrp53 (n = 10 tumors), P = 0.0178, unpaired t-test. Error bars represent SEM. Scale bar, 100 µm.
To investigate further the activation of YAP signaling in shCtrl-M cells, we found that a YAP downstream target, BIRC5, was enriched significantly in shCtrl-M cells (Fig. 3B; Muramatsu et al. 2011). To validate the RNA-seq results, we confirmed by qRT-PCR that BIRC5 mRNA was up-regulated significantly in shCtrl-M cells compared with shTrp53-M cells (Fig. 3C). Next, we examined the expression of Survivin, the protein encoded by BIRC5, in 3D organoids generated from shCtrl and shTrp53 cells. Indeed, Survivin expression is up-regulated in shCtrl compared with shTrp53 3D organoids (Fig. 3D). In addition, we examined Survivin expression in the esophagi of Trp53R172H/− and Trp53−/− 4NQO mouse models. Survivin expression trended to be higher in the Trp53R172H/− esophagi (Fig. 3E). Finally, we assessed Survivin expression in metastatic lung lesions formed by shCtrl and shTrp53 cells. In metastatic lung lesions formed by shCtrl cells, Survivin expression is increased compared with metastatic lesions formed by shTrp53 cells (Fig. 3F). In summary, our complementary in vitro and in vivo results reveal for the first time that Trp53R172H enhances BIRC5 gene expression in lung metastasis.
Depletion of BIRC5 expression reduces Trp53R172H- but not Trp53−/−-driven lung metastases
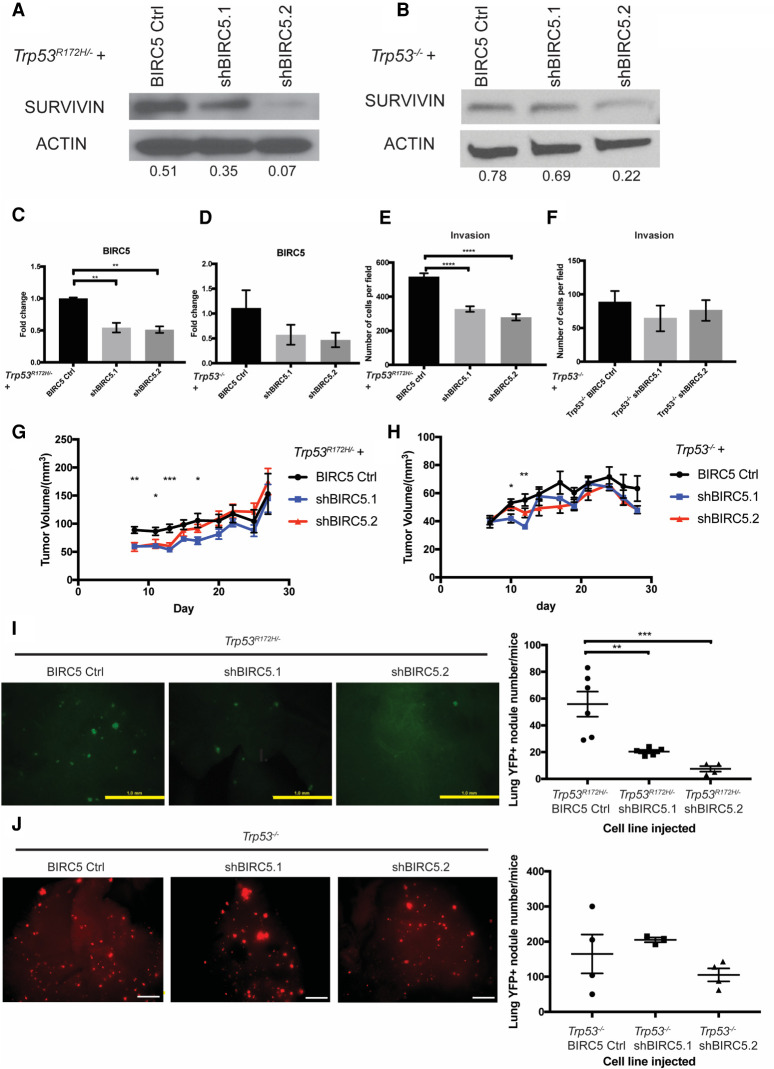
Given our finding that Trp53R172H induces BIRC5 gene expression, we next investigated the latter's direct role in mediating the phenotypic effects of Trp53R172H. As a result, we depleted BIRC5 with two different shRNAs in mouse primary Trp53R172H/− or Trp53−/− ESCC cells (Fig. 4A–D). Depletion of BIRC5 significantly reduced cell invasion in Trp53R172H/− but not Trp53−/− ESCC cells (Fig. 4E,F). Furthermore, depletion of BIRC5 significantly decreased early subcutaneous tumor growth (Fig. 4G,H). To assess the effect of BIRC5 in lung metastasis, we performed tail-vein injection assays with Trp53R172H/− BIRC5 Ctrl, Trp53R172H/− shBIRC5.1, or Trp53R172H/− shBIRC5.2 cell lines. At both 5 wk and 8 wk postinjection, the number and size of lung metastases were reduced drastically in Trp53R172H/− shBIRC5.1 or Trp53R172H/− shBIRC5.2 cell lines injected mice (Fig. 4I; Supplemental Fig. S4A). However, this effect is not observed in week 5 and week 8 lung metastases formed by Trp53−/− BIRC5 Ctrl, Trp53−/− shBIRC5.1, or Trp53−/− shBIRC5.2 cell lines (Fig. 4J; Supplemental Fig. S4B). Taken together, these results suggest that BIRC5 is a critical direct mediator of Trp53R172H-driven ESCC invasion and lung metastasis.
Figure 4.
Depletion of BIRC5 attenuates Trp53R172H-driven lung metastasis. (A,B) SURVIVIN expression is depleted or decreased significantly in Trp53R172H/− or Trp53−/− cells with two independent shRNAs (shBIRC5.1 and shBIRC5.2). Relative intensity densitometry of SURVIVIN results are shown at bottom. (C,D) BIRC5 depletion in Trp53R172H/− or Trp53−/− cells is confirmed by qPCR. Trp53R172H/− BIRC5 Ctrl versus shBIRC5.1 versus shBIRC5.2, n = 3 per group, (**) P = 0.0021 Trp53R172H/− BIRC5 Ctrl versus shBIRC5.1, (**) P = 0.0015 Trp53R172H/− BIRC5 Ctrl versus shBIRC5.2, one-way ANOVA. Trp53−/− BIRC5 Ctrl versus shBIRC5.1 versus shBIRC5.2, n = 3 per group. Error bars represent SEM. (E,F) Depletion of BIRC5 significantly decreased cell invasion in Trp53R172H/− but not Trp53−/− cells. Trp53R172H/− BIRC5 Ctrl versus shBIRC5.1 versus shBIRC5.2, n = 9 per group, (****) P < 0.0001 Trp53R172H/− BIRC5 Ctrl versus shBIRC5.1, (****) P < 0.0001 Trp53R172H/− BIRC5 Ctrl versus shBIRC5.2, one-way ANOVA. Error bars represent SEM. (G,H) BIRC5 depletion significantly reduces subcutaneous tumor growth at early stage. Trp53R172H/− BIRC5 Ctrl (n = 6 flanks) versus shBIRC5.1 (n = 8 flanks) versus shBIRC5.2 (n = 6 flanks). (**) P = 0.0042 day 8; (*) P = 0.00214 day 11; (***) P = 0.0004 day 13; (*) P = 0.0261 day 17, one-way ANOVA. Error bars represent SEM. Trp53−/− BIRC5 Ctrl (n = 6 flanks) versus shBIRC5.1 (n = 6 flanks) versus shBIRC5.2 (n = 6 flanks). (*) P = 0.0322 day 10; (**) P = 0.0016 day 12, one-way ANOVA. Error bars represent SEM. (I) Depletion of BIRC5 in Trp53R172H/− ESCC cells significantly reduces lung metastasis burden number detected at week 8 after tail-vein injection. BIRC5 Ctrl (n = 6) versus shBIRC5.1 (n = 5), P = 0.0057; BIRC5 Ctrl (n = 6) versus shBIRC5.2 (n = 4), P = 0.0009, one-way ANOVA. Error bars represent SEM. Scale bar, 1 mm. (J) Depletion of BIRC5 in Trp53−/− ESCC cells does not significantly reduce lung metastasis burden number detected at week 8 after tail-vein injection. BIRC5 Ctrl (n = 4) versus shBIRC5.1 (n = 3) versus shBIRC5.2 (n = 4). Error bars represent SEM. Scale bar, 1mm.
Trp53R172H, but not wild-type Trp53, binds with YAP and fosters Survivin expression in ESCC cells
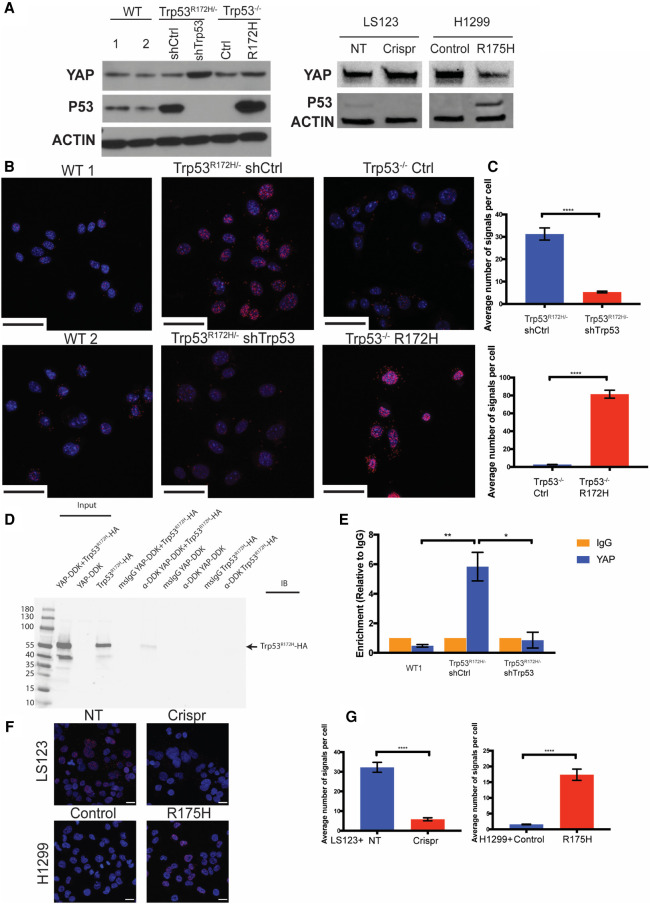
We next tested whether there might be direct interaction between Trp53R172H and YAP proteins in murine ESCC cells. First, we evaluated the endogenous expression of YAP and p53 in wild-type p53, Trp53R172H/− shCtrl /shTrp53, and Trp53−/− Ctrl/R172H cell lines (Fig. 5A). Then, we performed the proximity ligation assay (PLA) that is used to demonstrate direct protein–protein interactions in a quantitative fashion, representing a substantial enhancement for what has been reported (Di Agostino et al. 2016). This is based on the premise that YAP has been demonstrated to be an inducer of Survivin (Muramatsu et al. 2011). Both endogenously and ectopically expressed Trp53R172H associate with YAP in murine ESCC cells (Fig. 5B,C). However, in two murine ESCC cell lines expressing wild-type Trp53, binding of endogenous wild-type Trp53 with YAP is not detected (Fig. 5B). The binding of YAP and Trp53R172H is confirmed further by a coimmunoprecipitation (co-IP) assay (Fig. 5D). These results indicate the interaction with YAP is specific to Trp53R172H and suggests this specific YAP-Trp53R172H protein–protein interaction fosters Survivin expression. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) analysis in wild-type p53, Trp53R172H/− shCtrl, and shTrp53 cell lines. We verified binding of YAP to the BIRC5 promoter in Trp53R172H/− shCtrl, but not in wild-type p53 and shTrp53 cell lines (Fig. 5E). In addition, we used the human colorectal adenocarcinoma cell line LS123, which expresses p53R175H, and used the CRISPR/Cas9 system to deplete endogenous p53R175H. As an additional approach, we used a p53−/− non-small cell lung carcinoma cell line, H1299, with ectopic expression of p53R175H (Fig. 5A; Basu et al. 2018). We confirmed by PLA that YAP and p53R175H binding is also detected in these two human cancer cell lines (Fig. 5F,G). Taken together, these results suggest that mutant p53 might enable YAP to bind the BIRC5 promoter, thereby inducing Survivin expression.
Figure 5.
Trp53R172H, not wild-type p53, binds to YAP and fosters Survivin expression in murine ESCC cells. (A, left) Endogenous expression level of YAP and p53 in wild type, Trp53R172H/− shCtrl, shTrp53, Trp53−/− Ctrl, and Trp53−/− R172H cells. (Right) Expression levels of YAP and p53 in LS123 nontarget control (NT), Crispr knockout (Crispr) cell lines, and H1299 empty vector control (control) and p53R175H (R175H) ectopic expressed cell lines. (B) Proximity ligation assay (PLA) for YAP and Trp53R172H. Scale bar, 50 µm. (C) Quantification of the average number of PLA signals shows significantly high levels of signals in Trp53R172H/− shCtrl cells (vs. shTrp53 cells, n > 100 cells per group, P < 0.0001, unpaired t-test, error bars represent SEM) and Trp53−/− R172H cells (vs. Ctrl cells, n > 100 cells per group, P < 0.0001, unpaired t-test, error bars represent SEM). (D) Immunoprecipitation-Western blot analysis of Trp53R172H-HA with YAP-DDK in 293 FT cells. (E) Chromatin immunoprecipitation (ChIP) with YAP antibody in WT1, Trp53R172H/− shCtrl, and shTrp53 cells demonstrates increased level of YAP on the BIRC5 promoter in the presence of Trp53R172H. shCtrl versus WT1 cells, n = 3 replicates per group, P = 0.0052, unpaired t-test; shCtrl versus shTrp53 cells, n = 3 replicates per group, P = 0.0107, unpaired t-test. Error bars represent SEM. (F) Proximity ligation assay for YAP and p53R175H. Scale bar, 20 µm. (G) Quantification of the average number of PLA signals shows significantly high levels of signals in LS123 NT cells (vs. Crispr cells, n > 100 cells per group, P < 0.0001, unpaired t-test, error bars represent SEM) and H1299 R175H cells (vs. control cells, n > 100 cells per group, P < 0.0001, unpaired t-test, error bars represent SEM).
High Survivin expression is detected in human ESCC and metastases in other GI cancers
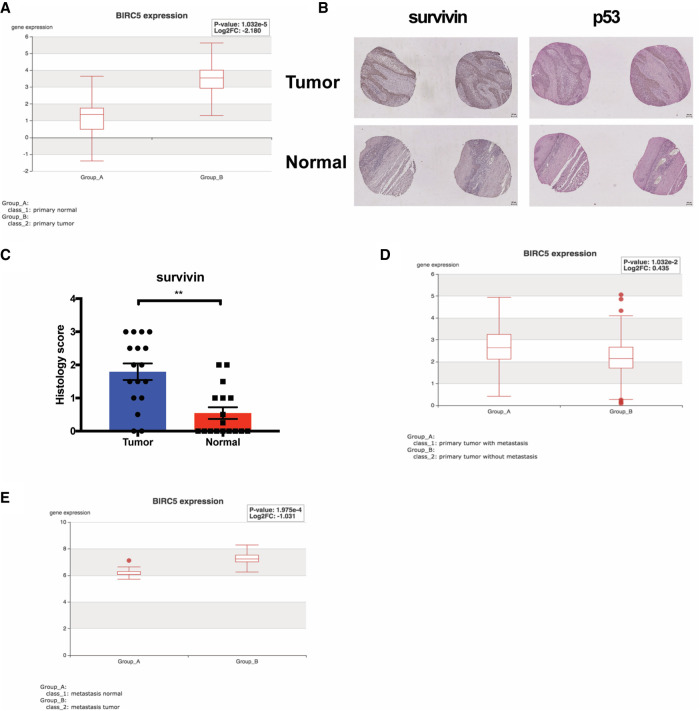
Analysis of TCGA revealed that Survivin expression is significantly elevated in human ESCC compared with adjacent normal tissues (P < 0.0001) (Fig. 6A). In addition, we examined Survivin expression in human ESCC tissue microarrays (TMAs). We observed that Survivin expression is elevated significantly in ESCC compared with adjacent normal tissues (P = 0.0014) (Fig. 6B,C). Next, we examined the correlation of Survivin expression level with p53 nuclear accumulation in ESCC. We found that high Survivin expression is associated significantly with p53 nuclear accumulation in ESCC (P = 0.027). Because TP53 mutations have been shown to cause aggregation and accumulation of mutant p53 protein in cells (Ano Bom et al. 2012; Silva et al. 2018), our observation indicates that up-regulation of Survivin may be correlated with TP53 mutations in ESCC patient tumor tissues. Finally, we investigated the association of Survivin with metastases in other gastrointestinal (GI) cancers. In pancreatic ductal adenocarcinoma (PDAC), Survivin expression is up-regulated significantly in primary PDAC tumors with metastases compared with primary PDAC tumors without metastases (P = 0.01) (Fig. 6D). Furthermore, in colorectal cancer (CRC) lung metastases, Survivin RNA expression is increased significantly compared with adjacent normal tissue (P = 0.0002) (Fig. 6E). Taken together, these results underscore the role of Survivin in ESCC tumorigenesis and indicate its potential contribution to metastasis of other GI cancers, such as PDAC and colorectal cancer.
Figure 6.
High survivin expression is detected in human ESCC and metastases and in other GI cancers. (A) Analysis of TCGA-ESCA database (n = 143) revealed significant increase of survivin expression in primary esophageal carcinoma tissue compared with normal tissue. P < 0.0001. (B) IHC staining of survivin and p53 in tissue microarrays (TMAs) of paired ESCC primary tumor and normal tissue. Scale bar, 100 µm. (C) Increased level of survivin is detected in ESCC primary tumor compared with paired normal tissue. n = 17 pairs, P = 0.0014, Wilcoxon matched-pairs signed rank test. Scoring standards: (0.5) <10% positive cells positive nuclei, (1) 10%–25% positive cells positive nuclei, (2) 25%–50% positive cells positive nuclei, (3) >50% positive cells positive nuclei. Error bars represent SEM. Increased expression of survivin is correlated with p53 nuclear accumulation in ESCC. n = 59, P = 0.027, Spearman's rank correlation test. (D) Analysis of TCGA pancreatic ductal adenocarcinoma (PDAC) data set (TCGA-PAAD, n = 185) showed that survivin expression is significantly elevated in PDAC tumors with metastasis. P = 0.01. (E) Analysis of RNA-seq (GSE68468, n = 27) in colorectal cancers reveals increased survivin levels in lung metastases compared with adjacent normal tissues. P = 0.0002.
Discussion
TP53 mutation is the predominant genetic alteration in ESCC (Song et al. 2014). In this study, we demonstrated the oncogenic role of a hot spot TP53 mutation, p53R175H, in invasion and lung metastasis. Through gene expression profiling of ESCC lung metastatic cells and primary tumor cells, we nominate BIRC5, an antiapoptotic gene, as a novel effector of p53R175H-driven lung metastasis. We demonstrated for the first time in both human and mouse cell lines that BIRC5 expression is dependent directly upon p53R175H, indicating that BIRC5 may be a novel mutant p53 target. We demonstrated that p53R175H, not wild-type TRP53, interacts directly with YAP, and this interaction induces BIRC5 gene expression. Survivin is also up-regulated in human ESCC, as well as metastatic PDAC and metastatic CRC when compared with their matched primary tumors, suggesting a possible basis for metastasis across common GI cancers.
Mutant p53 has been demonstrated to promote tumor cell motility, invasion, and metastasis through gain-of-function mechanisms (Tang et al. 2020). For example, in vivo evidence has revealed that Trp53R172H/+ mice develop metastatic osteosarcomas and carcinomas. However, Trp53+/− mice do not harbor metastatic tumors (Lang et al. 2004). In different types of cancers, mutant p53 modulates certain pathways; for example, up-regulation of PDGFRβ and enhancement of mitochondrial function (Weissmueller et al. 2014; Basu et al. 2018). In addition, mutant p53 has been reported to promote lung metastasis through regulating glycoprotein folding (Vogiatzi et al. 2016). Furthermore, one recent study demonstrated that tumor cells harboring mutant p53 can establish proinvasive niches by secreting exosomes, indicating the oncogenic role of mutant p53 in altering the tumor microenvironment (Cooks et al. 2018). Here, we provided the first evidence that p53R175H regulates the YAP-BIRC5 axis to affect lung metastasis. Further investigation is required to determine whether the regulation of the YAP-BIRC5 axis by mutant p53 affects lung metastasis in other GI cancers.
In this study, we identified Survivin, encoded by BIRC5, as a novel downstream effector of mutant p53 to promote lung metastasis. We propose a model whereby p53R175H interacts with YAP and regulates survivin expression through binding of YAP to the BIRC5 promoter, which ultimately promotes lung metastasis. Survivin, encoded by BIRC5, is a key regulator of mitosis and programmed cell death. Survivin is expressed highly in transformed cells and cancers, such as ESCC, lung, breast, and pancreatic cancers (Mita et al. 2008). Several studies have revealed mechanisms underlying Survivin up-regulation, including gene amplification, exon demethylation, promoter activation, and phosphatidylinositol-3-kinase (PI3K) and MAPK signaling activation (Mita et al. 2008). Survivin has been suggested as a transcriptional target of YAP (Muramatsu et al. 2011). YAP overexpression is detected frequently in cells and primary tumors (Muramatsu et al. 2011). In breast cancer cell lines, YAP and mutant p53 cooperate to up-regulate proproliferative genes such as cyclin A, cyclin B, and CDK1 (Di Agostino et al. 2016). Survivin enhances tumor angiogenesis and chemoresistance (Mita et al. 2008). Previous studies demonstrated the role of Survivin in tumor cell invasion (Mehrotra et al. 2010; Rivadeneira et al. 2015). Given the important roles of Survivin in cancers, therapeutic strategies have been designed to target Survivin. These strategies include RNAi, antisense oligonucleotides, small molecule inhibitors, and developing a cancer vaccine (Mita et al. 2008; Santarelli et al. 2018). Notably, several Survivin peptide vaccine therapies are currently in clinical trials (NCT03349450, NCT03762291, and NCT01416038). Given the role of Survivin in ESCC lung metastasis, targeting Survivin in advanced ESCC may be a novel therapeutic approach.
In summary, we have demonstrated an oncogenic role of p53R175H in promoting invasion and lung metastasis through up-regulation of Survivin in ESCC. This study also provides a prism through which to view the molecular basis of p53R175H-driven lung metastasis and offers a platform to identify novel therapeutic targets for metastases harboring mutant p53.
Materials and methods
Cell culture
Murine ESCC cells were cultured in keratinocyte serum-free medium (KSFM; Gibco) without CaCl2, supplemented with 0.018 mM CaCl2, 50 µg/mL bovine pituitary extract (Gibco), 5 ng/mL human recombinant EGF (Gibco), and 1% penicillin/streptomycin (Gibco). Cells were cultured at 37°C in a 5% CO2 humidified incubator. To achieve Trp53 knockdown, cells were infected with mouse lentiviral shRNA against Trp53 (pMSCV puro, Clontech) or nontargeting control shRNA. Cells were selected in 2 µg/mL puromycin 48 h postinfection. Quantitative PCR, immunofluorescence staining, and Western blotting were performed to confirm knockdown efficiency. Alternatively, CRISPR/Cas9 against Trp53 (pLentiCRISPR v2, GenScript) or an empty vector control were used for infection, and puromycin selection and Trp53 expression validation were performed as described. For YAP knockdown, a pool of three to five YAP-specific shRNAs (Santa Cruz Biotechnology sc-38638-SH) or nontargeting control shRNA (Santa Cruz Biotechnology sc-108060) were used. For BIRC5 knockdown, mouse lentiviral shRNA against BIRC5 (pLKO.1-CMV-neo, Sigma) or nontargeting control shRNA was used to infect cells. Neomycin selection was performed at a concentration of 250 µg/mL 48 h postinfection. Knockdown validation was performed as described. To ectopically express Trp53R175H in Trp53−/− murine ESCC cells, we cloned Trp53R175H cDNA into the pUltra-Chili lentiviral construct (Addgene, plasmid 48687). pUltra-chili-Trp53R175H was then used to infect cells, followed by RFP flow cytometry sorting for cell selection and Western blotting to confirm expression level.
The human cell line LS123 was purchased from ATCC (ATCC CCL-255) and was cultured in Eagle's minimum essential medium (EMEM; ATCC 30-2003). LS123 cells were infected with the human TP53 CRISPR/Cas9 construct (pLentiCRISPR v2) or nontargeting control. Puromycin selection and p53 expression validation were performed as described. Human cell lines H1299 control and R175H overexpression were a gift from Dr. Maureen Murphy (Basu et al. 2018) and were cultured in RPMI-1640 medium (Thermo). The shRNA sequences used were mouse Trp53 (CCACTACAAGTACATGTGTAATAG), mouse Birc5 (TRCN0000054613; GAAGAACTAACCGTCAGTGAA), and mouse Birc5 (TRCN0000054616; CAAAGACTACCCGTCAGTCAA). The gRNA sequences used were mouse Trp53 (AGTGAAGCCCTCCGAGTGTC), mouse Trp53 (AACAGATCGTCCATGCAGTG), and human TP53 (CCATTGTTCAATATCGTCCG).
Western blot
Primary antibodies p53 (1:2000; Leica NCL-L-p53-CM5p), YAP (1:1000; Cell Signaling Technology 14074S), and β-actin (1:10,000; Sigma A5316), were incubated overnight at 4°C, and horseradish peroxidase (HRP)-conjugated secondary antibodies (anti-mouse: 1:10,000 [GE Healthcare LNA931V/AH]; and anti-rabbit: 1:10,000 [GE Healthcare LNA934V/AH]) were incubated for 1 h at room temperature. Western blots were imaged using ECL prime (Sigma GERPN 2232).
Invasion assay
Corning BioCoat Matrigel invasion chambers were used for invasion assays. Chambers were rehydrated in serum-free KSFM for 2 h at 37°C. Cells (105) resuspended in 500 µL of serum-free KSFM were seeded in the upper chamber, and 500 µL regular KSFM was added to the bottom of the well. Cells were incubated at 37°C for 24 h. After incubation, noninvading cells were removed from the upper surface with a cotton swab, and the remaining cells were fixed in 70% ethanol for 10 min at room temperature. Next, the invading cells were stained with 2% Crystal-Violet for 10 min at room temperature. Finally, the invading cell number was counted using a Keyence bright-field microscope at 10× with four fields per chamber. Invasion was calculated by the number of invaded cells per field. For 3D organotypic culture assays, invasion was determined by quantitating the area of invasive regions divided by the total epithelial length.
3D organotypic culture and 3D organoid culture assays
The protocol to culture esophageal cells in 3D organotypic culture was described previously (Kalabis et al. 2012). 3T3 cells were used to establish the fibroblast layer for mouse organotypic cultures. For 3D organoid culture, 3000 cells were resuspended in 50 µL of Matrigel in a 24-well dish followed by 1-h solidification to generate the organoids. After organoids solidified, the media (DMEM/F12, 1× Glutamax, 1× HEPES, 1× N2 supplement, 1× B27 supplement, 0.1 mM N-acetylcysteine, 50 ng/mL recombinant EGF, Noggin/R-Spondin conditioned media, 10 μM Y27632) was suppled at 500 µL per well and replaced every 2 d. Organoids were allowed to grow for 10–14 d. To harvest organoids, Matrigel was dissolved by pipetting several times. Recovered organoids were fixed in 4% PFA (Sigma 158127) overnight at 4°C and were embedded in 2% Bacto-Agar:2.5% gelatin.
Proliferation assay
Carboxyfluorescein diacetate succinimidyl ester (CFSE) is a fluorescent dye to measure cell division. Cells were labeled with 5 µM CFSE (Life Technologies C34554) for 30 min at 37°C. Then, cells were centrifuged at 1000 rpm for 5 min, the supernatant was removed, and cells were seeded in six-well plates in KSFM overnight at 37°C or 4°C. On the following day, cells were trypsinized and fluorescence was measured with BD Accuri C6. FlowJo was used to analyze results.
Apoptosis assay
To examine apoptosis, cells were plated in 12-well plates. Apoptosis was detected with the APC-AnnexinV and 7-AAD kit (BioLegend 640930), following the manufacturer's instructions. BD Accuri C6 was used to acquire data and results were analyzed with FlowJo.
Animal models
All animal studies were approved by our Institutional Animal Care and Use Committee. To establish the lung metastasis model, 5-wk-old female athymic nude mice were purchased (Taconic). At the age of 7 wk, 106 cells were resuspended in 100 µL of cold Dulbecco's phosphate-buffered saline (DPBS) and injected into the distal end of the lateral tail vein with a 30-gauge 0.5-in needle. After injection, mice were monitored daily for signs of pain or discomfort and weight loss. To detect the formation of lung metastatic lesions, mice were euthanized at 2, 5, and 8 wk by CO2 and cervical dislocation. To identify lung metastases, the mouse lungs were flushed with 20 mL of Heparin-PBS to deplete red blood cells and were imaged with an Olympus IX71 fluorescent microscope at 4×.
Lung metastatic cell isolation
Prior to cell isolation, all tools and solutions were autoclaved or filter-sterilized. Mouse lungs were harvested and placed in cold DPBS and were physically dissociated with scissors. To digest, dissociated tissues were mixed with 50 mL of 1 mg/mL solution of collagenase type V (Sigma Aldrich 9263), dissolved in prewarmed KSFM, and the mixture was stirred with a magnet for 20 min at 37°C. After digestion, the mixture was centrifuged twice at 1000 rpm for 5 min., resuspended in KSFM, and plated on a 10-cm cell culture dish. Cultured cells were washed with DPBS and replaced with new media every other day. After expansion, cells were trypsinized and flow cytometry-sorted for YFP, followed by Western blotting to confirm p53 expression.
RNA isolation and qRT-PCR
Total RNA was extracted with the GeneJET RNA purification kit (Thermo Fisher K0731) or RNAqueous kit (Invitrogen AM1912). cDNA was synthesized with a High-Capacity cDNA reverse Transcription kit (Thermo Fisher 4374966). qRT-PCR was performed with the StepOnePlus real-time PCR system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems 4367659). Primer sequences used were as follows: Trp53 (forward: CCCCGCAAAAGAAAAAACCAC; reverse: AGCTGGAGTGAGCCCTGC), Birc5 (forward: GAACCCGATGACAACCCGAT; reverse: TGGTCTCCTTTGCAATTTTGTTCT), and Actin (forward: ACCCAGATCATGTTTGAGACC; reverse: AGAGCATAGCCCTCGTAGAT).
RNA-seq
RNA was extracted from early passage cells as described above. Libraries were prepared with Illumina TruSeq stranded mRNA sample preparation kits from 500 ng of purified total RNA according to the manufacturer's protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and qRT-PCR using the Kapa Biosystems library quantification kit, according to the manufacturer's instructions. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on an Illumina NextSeq500 with single-end 75-bp reads at the Dana-Farber Cancer Institute Molecular Biology Core Facility. Sequenced reads were aligned to the UCSC mm10 reference genome assembly, and gene counts were quantified using STAR (v2.5.1b). Differential gene expression testing was performed by DESeq2 (v1.10.1) and normalized read counts (FPKM) were calculated using cufflinks (v2.2.1).
RNA-seq data were deposited at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo). All data sets are available on request.
Immunohistochemistry and immunofluorescence
Formalin-fixed, paraffin-embedded tissue was sectioned, and antigen retrieval was performed through pressure cooking in citric acid buffer (pH 6). Endogenous peroxidases were quenched by 3% peroxide, followed by avidin (Sigma Aldrich A9275), biotin (Sigma Aldrich B4501) blocking and protein blocking buffer (Thermo Fisher) at room temperature. Primary antibodies were incubated overnight at 4°C, and biotinylated secondary antibodies (1:600; Vector Laboratories) for 30 min at 37°C. Sections were then incubated with the ABC reagent (Vector Laboratories PK-6100) for 30 min at 37°C, and slides were treated with DAB substrate (Vector Laboratories SK4100) and counterstained with hematoxylin. The primary antibody was against Survivin (1:400; Cell Signaling Technology 2808S), p53 (1:200; Abcam ab1101).
For immunofluorescence, cells were cultured on cover slips, rinsed with PBS, and fixed with 4% PFA for 15 min at room temperature. Permeabilization was performed with 100% methanol for 10 min at −20°C. Cells were blocked with blocking solution (PBS, 1% BSA, 0.3% Triton X, 5% donkey serum) for 1 h at room temperature. Cells were then incubated with primary antibodies overnight at 4°C, washed, incubated with secondary antibody (1:600; JacksonImmunoresearch) for 1 h at room temperature in the dark, stained with DAPI (Invitrogen D1306) for 3 min, and mounted (KPL 71-00-16). Fluorescent imaging was performed with a Nikon E600 fluorescent microscope. The primary antibody was against p53 (1:200; Leica NCL-L-p53-CM5p).
Proximity ligation assay (PLA)
A Duolink In Situ Red Starter kit (Sigma DUO92101) was used to perform PLA assays, according to the manufacturer's instructions. ImageJ was used to quantify PLA signals. Primary antibodies included the following: p53(1C12) (1:2000; Cell Signaling Technology 2524), YAP (1:100; Cell Signaling Technology 14074).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (Chiarella et al. 2018) with ESCC cells using 10 million cells per replicate and experimental condition. YAP (Novus Biologicals NB110-58358) and IgG (Cell Signaling 5415S) antibodies were used. Following ChIP, qPCR was performed with SYBR Green (Thermo Scientific 4367659). Enrichment levels were normalized to the background levels detected with IgG. For each experimental type, three biological replicates were analyzed. The following Survivin primers (5′–3′) were used: (+)TGGACTGGTGAGGTTTAGGA and (−)ACATAGGCAGCTGGAACAAG.
Human tissues and databases
Well-annotated tissue microarrays (TMAs) representing paired primary ESCC tumors and adjacent normal mucosa from IRB-approved and de-identified therapy-naïve patients (n = 39) have been previously described (Liu et al. 2013; Natsuizaka et al. 2017). TCGA-ESCA, TCGA-PAAD, and GEO68468 were accessed and analyzed through the Human Cancer Metastasis Database (https://hcmdb.i-sanger.com).
Statistical analysis
Sample size and replicate number have been described in the text and figure legends. All data were presented as mean ± standard error of the mean (SEM). Statistical analysis including paired Student's t-test, unpaired Student's t-test, and ANOVA was performed by GraphPad Prism 7 (GraphPad Software). P < 0.05 was reported as statistically significant.
Data availability
RNA-seq data from this article have been deposited at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143696).
Supplementary Material
Acknowledgments
We thank the Flow Cytometry and Cell Sorting Core at the University of Pennsylvania Perelman School of Medicine; the Imaging Facility at the Wistar Institute; the Molecular Pathology, Genetically Engineered Mouse Model, Flow Cytometry, and Biostatistics Shared Resources at the Herbert Irving Comprehensive Cancer Center (HICCC) of Columbia University Irving Medical Center; and the Molecular Biology Core Facility (MBCF) at the Dana-Farber Cancer institute (DFCI). This work was funded by National Institutes of Health grant P01-CA098101 (A.K.R.) and the American Cancer Society Research Professorship (A.K.R.).
Author contributions: Q.T., G.E., A.M.C., M.C., and J.L. performed experiments and data analysis. T.Y., Z.S., T.K., and J.L. assisted with experiments. A.J.K.-S. performed pathology scoring. J.R.P., M.I., S.P., and J.H. performed RNA-seq analysis. S.N. collected TMA tissues. W.G., B.Z.S., K.-K.W., J.A.D., A.J.B., M.E.M., and H.N. designed the experiments and reviewed the manuscript. A.K.R. designed the experiments, performed data analysis, and wrote the manuscript. Q.T. also wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.340505.120.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Ano Bom APD, Rangel LP, Costa DCF, De Oliveira GAP, Sanches D, Braga CA, Gava LM, Ramos CHI, Cepeda AOT, Stumbo AC, et al. 2012. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J Biol Chem 287: 28152–28162. 10.1074/jbc.M112.340638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Gnanapradeepan K, Barnoud T, Kung C, Tavecchio M, Scott J, Watters A, Chen Q, Kossenkov AV, Murphy ME. 2018. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1α. Genes Dev 32: 230–243. 10.1101/gad.309062.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. 2006. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene 25: 304–309. 10.1038/sj.onc.1209026 [DOI] [PubMed] [Google Scholar]
- Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, Taylor AM, Wang C, Walter V, Akbani R, et al. 2018. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep 23: 194–212.e6. 10.1016/j.celrep.2018.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarella AM, Quimby AL, Mehrab-Mohseni M, Velasco B, Kasoji SK, Davis IJ, Dayton PA, Hathaway NA, Pattenden SG. 2018. Cavitation enhancement increases the efficiency and consistency of chromatin fragmentation from fixed cells for downstream quantitative applications. Biochemistry 57: 2756–2761. 10.1021/acs.biochem.8b00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Forshew T, Appella E, Gorgoulis VG, Harris CC, Morris J. 2018. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun 9: 771. 10.1038/s41467-018-03224-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Li Z. 2018. Downregulation of YAP inhibits proliferation and induces apoptosis in Eca‐109 cells. Exp Ther Med 15: 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Sorrentino G, Ingallina E, Valenti F, Ferraiuolo M, Bicciato S, Piazza S, Strano S, Del Sal G, Blandino G. 2016. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep 17: 188–201. 10.15252/embr.201540488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Xu Z, Jia N, Li D, Feng Y. 2009. Elevated expression of p53 gain-of-function mutation R175H in endometrial cancer cells can increase the invasive phenotypes by activation of the EGFR/PI3K/AKT pathway. Mol Cancer 8: 103. 10.1186/1476-4598-8-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre A, Weinberg RA. 2019. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20: 69–84. 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- Dotto GP, Rustgi AK. 2016. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell 29: 622–637. 10.1016/j.ccell.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. 2012. Mutant p53: one name, many proteins. Genes Dev 26: 1268–1286. 10.1101/gad.190678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Aylon Y, Oren M. 2018. P53 shades of Hippo. Cell Death Differ 25: 81–92. 10.1038/cdd.2017.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H. 2012. Role of mTOR signaling in tumor cell motility, invasion and metastasis. Curr Protein Pept Sci 12: 30–42. 10.2174/138920311795659407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M, Nakagawa H, Rustgi AK. 2012. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 7: 235–246. 10.1038/nprot.2011.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, Weinberg RA. 2017. Emerging biological principles of metastasis. Cell 168: 670–691. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119: 861–872. 10.1016/j.cell.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Lee Ck, Jeong Sh, Jang C, Bae H, Kim YH, Park I, Kim SK, Koh GY. 2019. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 363: 644–649. 10.1126/science.aav0173 [DOI] [PubMed] [Google Scholar]
- Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, et al. 2013. Sox2 cooperates with inflammation-mediated stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell 12: 304–315. 10.1016/j.stem.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. 2010. IAP regulation of metastasis. Cancer Cell 17: 53–64. 10.1016/j.ccr.2009.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello SS, Valente LJ, Raj N, Seoane JA, Flowers BM, McClendon J, Bieging-Rolett KT, Lee J, Ivanochko D, Kozak MM, et al. 2017. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14-Yap axis in pancreatic cancer. Cancer Cell 32: 460–473.e6. 10.1016/j.ccell.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita AC, Mita MM, Nawrocki ST, Giles FJ. 2008. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res 14: 5000–5005. 10.1158/1078-0432.CCR-08-0746 [DOI] [PubMed] [Google Scholar]
- Muller PAJ, Vousden KH. 2014. Perspective mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25: 304–317. 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PAJ, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell 139: 1327–1341. 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Muller PAJ, Trinidad AG, Timpson P, Morton JP, Zanivan S, Berghe PVD, Nixon C, Karim SA, Caswell PT, Noll JE, et al. 2013. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 32: 1252–1265. 10.1038/onc.2012.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T, Imoto I, Matsui T, Kozaki KI, Haruki S, Sudol M, Shimada Y, Tsuda H, Kawano T, Inazawa J. 2011. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis 32: 389–398. 10.1093/carcin/bgq254 [DOI] [PubMed] [Google Scholar]
- Natsuizaka M, Whelan KA, Kagawa S, Tanaka K, Giroux V, Chandramouleeswaran PM, Long A, Sahu V, Darling DS, Que J, et al. 2017. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat Commun 8: 1758. 10.1038/s41467-017-01500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neskey DM, Osman AA, Ow TJ, Katsonis P, McDonald T, Hicks SC, Hsu TK, Pickering CR, Ward A, Patel A, et al. 2015. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck cancer. Cancer Res 75: 1527–1536. 10.1158/0008-5472.CAN-14-2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119: 847–860. 10.1016/j.cell.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. 2010. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a001008. 10.1101/cshperspect.a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen VL, Porporato PE, Baselet B, Sonveaux P. 2016. Metabolic changes associated with tumor metastasis, part 1: tumor pH, glycolysis and the pentose phosphate pathway. Cell Mol Life Sci 73: 1333–1348. 10.1007/s00018-015-2098-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennathur A, Gibson MK, Jobe BA, Luketich JD. 2013. Oesophageal carcinoma. Lancet 381: 400–412. 10.1016/S0140-6736(12)60643-6 [DOI] [PubMed] [Google Scholar]
- Rivadeneira DB, Caino MC, Seo JH, Angelin A, Wallace DC, Languino LR, Altieri DC. 2015. Survivin promotes oxidative phosphorylation, subcellular mitochondrial repositioning, and tumor cell invasion. Sci Signal 8: ra80. 10.1126/scisignal.aab1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin N, Brosh R, Oren M, Rotter V. 2011. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2: 466–474. 10.1177/1947601911408889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustgi AK, and El-Serag H. 2014. Esophageal carcinoma. N Engl J Med 371: 2499–2509. 10.1056/NEJMra1314530 [DOI] [PubMed] [Google Scholar]
- Santarelli A, Mascitti M, Lo Russo L, Sartini D, Troiano G, Emanuelli M, Lo Muzio L. 2018. Survivin-based treatment strategies for squamous cell carcinoma. Int J Mol Sci 19: 971. 10.3390/ijms19040971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JL, Cino EA, Soares IN, Ferreira VF, De Oliveira G. 2018. Targeting the prion-like aggregation of mutant p53 to combat cancer. Acc Chem Res 51: 181–190. 10.1021/acs.accounts.7b00473 [DOI] [PubMed] [Google Scholar]
- Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al. 2014. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509: 91–95. 10.1038/nature13176 [DOI] [PubMed] [Google Scholar]
- Steeg PS. 2016. Targeting metastasis. Nat Rev Cancer 16: 201–218. 10.1038/nrc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Buganim Y, Molchadsky A, Solomon H, Madar S, Kamer I, Stambolsky P, Shelly A, Goldfinger N, Puisieux A, et al. 2010. Mutant p53 R175H upregulates Twist1 expression and promotes epithelial – mesenchymal transition in immortalized prostate cells. Cell Death Differ 18: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Su Z, Gu W, Rustgi AK. 2020. Mutant p53 on the path to metastasis. Trends Cancer 6: 62–73. 10.1016/j.trecan.2019.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Lento A, Suzuki K, Efe G, Karakasheva T, Long A, Giroux V, Islam M, Wileyto EP, Klein-Szanto AJ, et al. 2021. Rab11-FIP1 mediates epithelial–mesenchymal transition and invasion in esophageal cancer. EMBO Rep 22: e48351. 10.15252/embr.201948351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Yang J. 2013. Epithelial–mesenchymal plasticity in carcinoma metastasis. Genes Dev 27: 2192–2206. 10.1101/gad.225334.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. 2020. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol 13: 1010–1021. 10.1007/s12328-020-01237-x [DOI] [PubMed] [Google Scholar]
- Vogiatzi F, Brandt DT, Schneikert J, Fuchs J, Grikscheit K, Wanzel M, Pavlakis E, Charles J, Timofeev O, Nist A, et al. 2016. Mutant p53 promotes tumor progression and metastasis by the endoplasmic reticulum UDPase ENTPD5. PNAS 113: E8433–E8442. 10.1073/pnas.1612711114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Mao B, Cheng C, Zou Z, Gao J, Yang Y, Lei T, Qi X, Yuan Z, Xu W, et al. 2018. YAP promotes breast cancer metastasis by repressing growth differentiation factor-15. Biochim Biophys Acta Mol Basis Dis 1864: 1744–1753. 10.1016/j.bbadis.2018.02.020 [DOI] [PubMed] [Google Scholar]
- Weissmueller S, Manchado E, Saborowski M, Morris JP, Wagenblast E, Davis CA, Moon S, Pfister NT, Tschaharganeh DF, Kitzing T, et al. 2014. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 157: 382–394. 10.1016/j.cell.2014.01.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A, Ramaswamy S. 2011. MYC and metastasis. Cancer Res 71: 2034–2037. 10.1158/0008-5472.CAN-10-3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data from this article have been deposited at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143696).