In this study, Su et al. investigate why ∼50% of Ewing sarcomas, driven by the EWS-FLI1 fusion oncogene, harbor chromosome 8 gains. Using an evolution approach, they show that trisomy 8 mitigates EWS-FLI1-induced replication stress through gain of a copy of RAD21, and deleting one copy of RAD21 in trisomy 8 cells largely neutralizes the fitness benefit of chromosome 8 gain and reduces tumorgenicity of a Ewing sarcoma cancer cell line in soft agar assays.
Keywords: DNA damage, Ewing sarcoma, RAD21, aneuploidy, cohesin, replication stress, trisomy 8
Abstract
Aneuploidy, defined as whole-chromosome gain or loss, causes cellular stress but, paradoxically, is a frequent occurrence in cancers. Here, we investigate why ∼50% of Ewing sarcomas, driven by the EWS-FLI1 fusion oncogene, harbor chromosome 8 gains. Expression of the EWS-FLI1 fusion in primary cells causes replication stress that can result in cellular senescence. Using an evolution approach, we show that trisomy 8 mitigates EWS-FLI1-induced replication stress through gain of a copy of RAD21. Low-level ectopic expression of RAD21 is sufficient to dampen replication stress and improve proliferation in EWS-FLI1-expressing cells. Conversely, deleting one copy in trisomy 8 cells largely neutralizes the fitness benefit of chromosome 8 gain and reduces tumorgenicity of a Ewing sarcoma cancer cell line in soft agar assays. We propose that RAD21 promotes tumorigenesis through single gene copy gain. Such genes may explain some recurrent aneuploidies in cancer.
Aneuploidy, defined as whole-chromosome gains and losses, causes many cellular stresses and cell proliferation defects in primary cells (Santaguida and Amon 2015). It is thus not surprising that aneuploidy is rare in normal somatic tissues (Santaguida and Amon 2015). Paradoxically, aneuploidy is a hallmark of cancer, a disease characterized by increased proliferation. Over 90% of solid tumors and 75% of hematopoietic malignancies are aneuploid (Weaver and Cleveland 2006; Beroukhim et al. 2010). Furthermore, some cancers harbor specific recurring chromosome gains and/or losses (Knouse and Amon 2013; Taylor et al. 2018). This observation suggests that although aneuploidy is generally detrimental, recurrent aneuploidies provide a selective advantage to the cancer. Here we investigate why chromosome 8 is so frequently gained in Ewing sarcoma.
Ewing sarcoma (ES) is the second most common pediatric bone and soft tissue cancer. While the cell of origin is still debated, it is generally thought to be a mesenchymal stem or progenitor cell (Grünewald et al. 2018). In 85% of patients, Ewing sarcomagenesis is initiated by the reciprocal t(11;22)(q24;q12) chromosomal translocation, which creates the EWS-FLI1 fusion (Brohl et al. 2014; Crompton et al. 2014; Tirode et al. 2014; Anderson et al. 2018). The EWS-FLI1 oncogene is a fusion of the transcriptional activator and RNA binding protein EWSR1 and the DNA binding domain of the ETS family transcription factor FLI1 to create a neomorphic transcription factor. The EWS-FLI1 protein recruits the BAF chromatin remodeling complex to tumor-specific enhancers (Boulay et al. 2017, 2018), thereby driving the transcriptional program that promotes Ewing sarcoma development. EWS-FLI1 also interferes with repair by homologous recombination and increases R-loop formation (Gorthi et al. 2018).
Besides the EWS-FLI1 translocation, Ewing sarcomas harbor relatively few other mutations (Lawrence et al. 2013; Tirode et al. 2014). However, ∼50% carry additional copies of chromosome 8, 21% show gains for chromosome 12, and 18% show gains for chromosome 1q (Crompton et al. 2014; Tirode et al. 2014). Given that the aneuploid state generally confers a fitness disadvantage due to aneuploidy-associated stresses, we hypothesized that specific genes encoded on the additional chromosomes are critical for Ewing sarcomagenesis. Here, we identify a cause of chromosome 8 gain in Ewing sarcoma. We show that expression of the EWS-FLI1 fusion in primary mesenchymal cell types accelerates S-phase entry and causes replication stress. Gain of additional copies of chromosome 8 mitigates this replication stress and improves proliferation of EWS-FLI1-expressing cells. Using a combination of computational and experimental evolution approaches, we show that cohesin subunit RAD21 contributes to these trisomy 8 benefits.
Results
EWS-FLI1 inhibits proliferation of primary mesenchymal cells
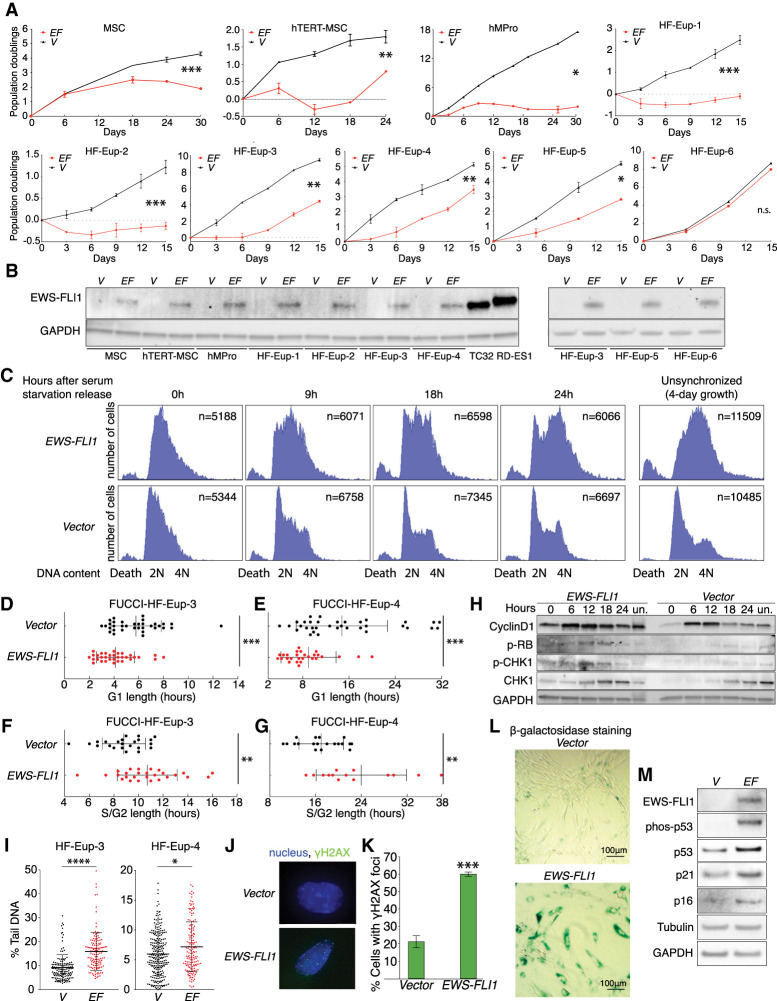
To determine whether and how chromosome 8 gain contributes to the initiation of Ewing sarcomagenesis, we transduced a constitutively expressed EWS-FLI1 fusion into early passage primary human mesenchymal stem cells (MSCs), hTERT-immortalized mesenchymal stem cells (hTERT-MSC), and mesenchymal progenitor cells (hMPro). Whole-genome sequencing confirmed that these cell lines started out as euploid and, with or without the presence of EWS-FLI1, maintained this karyotype at passages beyond the time frame of our experimental analyses (Supplemental Fig. S1A,B; Supplemental Table S1). The EWS-FLI1 fusion caused significant proliferation defects in all three mesenchymal cell types, even though the resulting EWS-FLI1 protein levels were lower than in Ewing sarcoma cell lines (Fig. 1A [top panel], B). Consistent with previous reports (Deneen and Denny 2001; Lessnick et al. 2002), EWS-FLI1 expression also interfered with proliferation of differentiated mesenchymal cells—primary human fibroblasts. Specifically, we expressed the EWS-FLI1 fusion from the doxycycline-inducible TET-ON promoter and found that induced, acute expression of EWS-FLI1 impaired the growth of five of the six tested fibroblast lines (Fig. 1A; with karyotype confirmation in Supplemental Fig. S1C,D; Supplemental Table S1). Thus, expression of the EWS-FLI1 fusion inhibits proliferation of euploid mesenchymal cells.
Figure 1.
EWS-FLI1 expression accelerates S-phase entry and causes replication stress. (A) Proliferation of indicated primary cells was measured after 2–3 d of selection for the presence of the lentivirus. (EF) EWS-FLI1 gene; (V) vector control. Error bars represent standard error of the mean (SEM) of biological duplicates. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (n.s.) not significant, linear regression. (B) EWS-FLI1 protein levels in various primary mesenchymal cell lines. GAPDH was used as a loading control. TC32 and RD-ES1 are Ewing sarcoma cell lines. N = 2. A representative picture is shown. (C) DNA content analysis following release of hMPro cells from a serum starvation-induced G1 arrest. (2N) G1 phase, (4N) G2 phase/mitosis, (unsynchronized) DNA content after 4 d of growth, (death) sub-G1 cell population, (n) number of cells analyzed. Experiments were repeated twice, and one set of representative graphs is shown. (D–G) Analysis of G1-phase (D,E) and S/G2-phase (F,G) length in human fibroblasts using the FUCCI system. Each dot represents a single cell. Average and standard deviation (SD) are shown. (***) P < 0.001, (**) P < 0.01, two-tailed Wilcoxon test. (H) Euploid human fibroblasts (HF-Eup-3) harboring the indicated lentivirus were released from a serum starvation-induced G1 arrest to determine the levels of cyclin D, phospho-Rb, and CHK1 phospho-CHK1 at the indicated times. Quantifications are shown in Supplemental Figure S2D. (Un) Unsynchronized. N = 3; a representative picture is shown. (I) Degree of DNA damage measured by comet assay in unsynchronized euploid fibroblasts expressing empty vector (V) or the EWS-FLI1 fusion (EF). Each dot represents a single comet. The middle line represents the mean; error bar: SD. (****) P < 0.0001, (*) P < 0.05, two-tailed Wilcoxon test. (J) Examples of EWS-FLI1-expressing hMPro cells harboring γH2AX foci and of control cells lacking foci. DNA in blue; γH2AX foci, green. (K–M) hMPro cells expressing the EWS-FLI1 fusion or a vector control were cultured for 8 d and the percentage of cells harboring >10 γH2AX foci (K) and senescence-associated β-galactosidase (L) was analyzed (quantification in Supplemental Fig. S2F). (***) P < 0.001, two-tailed t-test. (M) EWS-FLI1, phospho-p53, total p53, p21, and p16 levels were analyzed in control (V) and EWS-FLI1-expressing hMPro cells. GAPDH and tubulin were used as loading controls. N = 3. A representative picture is shown. Statistics and number of experiments are shown in Supplemental Table S2 (multiple tabs).
EWS-FLI1 expression causes replication stress and senescence
To determine how EWS-FLI1 interfered with cell proliferation, we transduced the EWS-FLI1 fusion into mesenchymal progenitors (hMPros) at early passage and then arrested cells in G1 by serum starvation (Supplemental Fig. S2A). Upon serum addition, hMPro cells with EWS-FLI1 entered S phase faster than control cells but then delayed in S/G2 phase (Fig. 1C). The S/G2-phase accumulation was also observed in unsynchronized hMPro cultures (Fig. 1C; Supplemental Fig. S2B). EWS-FLI1 expression also caused a slight increase in cell death (Fig. 1C; Supplemental Fig. S2B).
Serum starvation was somewhat less effective in synchronizing the trisomy 8 fibroblasts than the euploid cells. Because of this, and to allow analyses of the effects of EWS-FLI1 in unsynchronized population, we also used the FUCCI (fluorescence ubiquitination cell cycle indicator) system (Sakaue-Sawano et al. 2008; Abe et al. 2013), in which G1 length is marked by the presence of a Cdt1-mCherry fusion and S + G2-phase length by a Geminin-Venus fusion (Supplemental Fig. S2C). EWS-FLI1-expressing fibroblasts, like hMPro cells expressing the fusion, spent significantly less time in G1 and were delayed in S/G2 phase compared with control cells (Fig. 1D–G).
To characterize the shortened G1 in EWS-FLI1-expressing fibroblasts, we analyzed cyclin D levels and Rb phosphorylation. Fibroblasts harboring EWS-FLI1 expressed cyclin D earlier upon release from the G1 block, and Rb phosphorylation was somewhat increased, an indication of its inactivation (Fig. 1H; Supplemental Fig. S2D). We conclude that EWS-FLI1 accelerates S-phase entry, like many other oncogenes. Given that EWS-FLI1 binds to the cyclin D promoter (Kennedy et al. 2015; Boulay et al. 2017), we propose that it does so in part by up-regulating cyclin D expression.
The S/G2 delay observed in EWS-FLI1-expressing cells was due to replication stress, as previously described (Lessnick et al. 2002; Nieto-Soler et al. 2016; Gorthi et al. 2018; Koppenhafer et al. 2020). Fibroblasts expressing the EWS-FLI1 fusion had increased DNA damage, as judged by the migration distance of DNA tails in comet assays (Fig. 1I). This increased DNA damage was likely due to replication stress, as CHK1 phosphorylation was elevated, which occurs in response to activation of the ATR-dependent replication stress checkpoint (Fig. 1H; Supplemental Fig. S2D). In contrast, Nbs1 was only slightly, if at all, phosphorylated in EWS-FLI1-expressing cells, indicating that the ATM-dependent double-strand break-responsive checkpoint was not highly active (Supplemental Fig. S2E).
hMPro cells expressing EWS-FLI1 also harbored more DNA damage. Furthermore, after 8 d of culture, hMPro cells underwent senescence as judged by persistent high levels of DNA damage, p53 phosphorylation, accumulation of p21, expression of senescence-associated β−galactosidase activity, and the presence of a senescence-associated secretory phenotype (SASP) cytokine signature (Fig. 1J–M; Supplemental Fig. S2F–H). We conclude that EWS-FLI1 induces rapid S-phase entry, which in turn leads to replication stress and oncogene-induced cellular senescence, like many classic oncogenes (Kotsantis et al. 2018). EWS-FLI1's ability to interfere with BRCA1-mediated DNA repair (Gorthi et al. 2018) likely further accelerates senescence. We note that previous studies reported that the EWS-FLI1 fusion can transform mesenchymal stem cells (Riggi et al. 2005, 2008). We believe that the difference between these reports and our findings is that we examined proliferation immediately after introduction of EWS-FLI1. At later passages, cells likely evolve mechanisms to suppress the EWS-FLI1-induced oncogenic stress.
Chromosome 8 copy number increase mitigates replication stress in EWS-FLI1-expressing cells
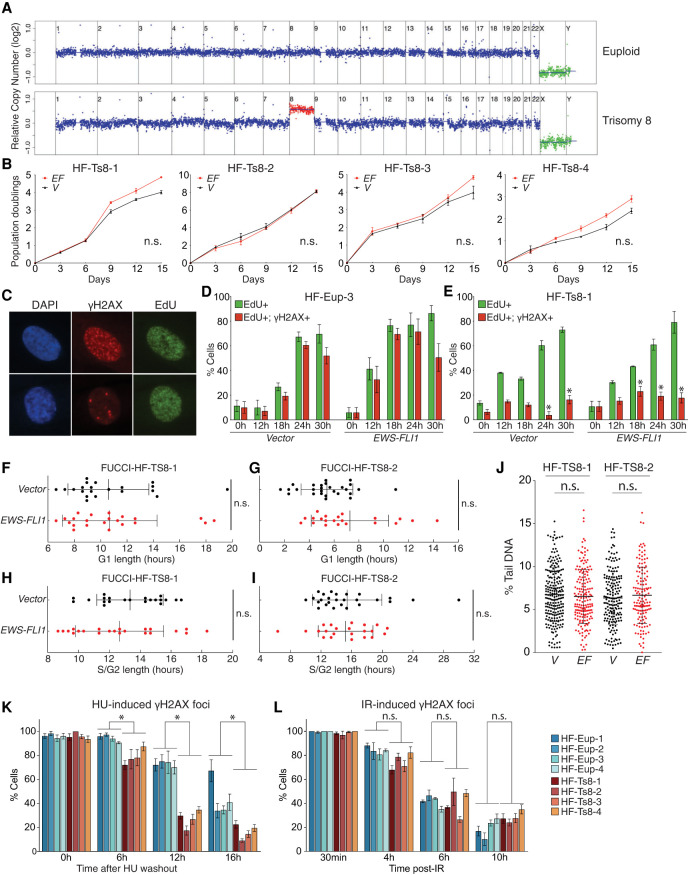
Fifty percent of Ewing sarcomas harbor additional copies of chromosome 8. Given that the aneuploid state generally confers a fitness disadvantage due to aneuploidy-associated stresses (Santaguida and Amon 2015), we hypothesized that additional copies of specific genes encoded on chromosome 8 act to mitigate this stress. To test this hypothesis, we expressed EWS-FLI1 in four different primary trisomy 8 fibroblast cell lines (for karyotype confirmation before and for the extent of our experiments, see Fig. 2A; Supplemental Fig. S1E,F, Supplemental Table S1; note that line HF-Ts8-3 was mosaic for trisomy 8). Although all four cell lines expressed EWS-FLI1 at levels similar to those observed in euploid fibroblasts (Supplemental Fig. S3A), their proliferation was not significantly affected (Fig. 2B). In contrast, EWS-FLI1 did severely impair the proliferation of trisomy 9 and trisomy 13 fibroblasts (Supplemental Fig. S3B–D; Supplemental Table S1). We conclude that gain of a copy of chromosome 8, and not aneuploidy in general, mitigates the adverse effects of EWS-FLI1 expression.
Figure 2.
Trisomy 8 mitigates EWS-FLI1- and HU-induced replication stress. (A) Representative fibroblast karyotypes (with additional examples shown in Supplemental Fig. S1C–F). (B) Proliferation of indicated primary cells measured 2–3 d after selection for the lentivirus. (EF) EWS-FLI1 gene, (V) vector control. Error bars represent SEM of biological duplicates. (n.s.) Not significant, linear regression. (C) Representative examples of EdU-positive, γH2AX-positive cells (>10 foci; top), and EdU-positive, γH2AX-negative cells (<10 foci; bottom). (Green) EdU, (red) γH2AX. (D,E) Analysis of EdU incorporation and γH2AX focus formation in EdU-positive cells in euploid (D) and trisomy 8 (E) fibroblasts following release from a serum starvation-induced G1 arrest. Error bar represents SEM of biological replicates. (*) P < 0.05, two-tailed t-test, comparison between euploid and trisomy 8 cells at the same timepoint; n > 2. (F–I) Analysis of G1- or S/G2-phase length in two trisomy 8 fibroblast cultures. Each dot represents a single cell. Average and SD are shown. (n.s.) Not significant, two-tailed Wilcoxon test. (J) Degree of DNA damage measured by comet assay in trisomy 8 fibroblasts expressing empty vector (V) or the EWS-FLI1 fusion (EF). Each dot represents a single comet. The middle line indicates the mean; error bar represents SD. (n.s.) Not significant by two-tailed Wilcoxon test. (K,L) Euploid and trisomy 8 cells were released from a starvation-induced G1 arrest into either medium containing hydroxyurea (HU; 2 mM) and EdU (10 µM), and HU was washed out after 24 h (K), or irradiated with 2 Gy (IR) (L). The percentage of cells harboring >10 γH2AX foci was determined at the indicated times. Error bars represent SEM of biological replicates, n > 2. (*) P < 0.05, (n.s.) not significant, two-tailed nonparametric two-group Mann–Whitney U-test. Statistics and number of experiments are shown in Supplemental Table S2 (multiple tabs).
To determine how trisomy 8 improves proliferation of EWS-FLI1-expressing cells, we created a highly enriched G1 population of euploid and trisomy 8 fibroblasts by serum starvation and then examined the kinetics of S-phase entry by EdU incorporation following serum addition. EWS-FLI1 expression accelerated S-phase entry of euploid fibroblasts (Fig. 2C,D; Supplemental Fig. S3E), but this effect was ameliorated in trisomy 8 fibroblasts (Fig. 2E; Supplemental Fig. S3F). Similar results were obtained when we analyzed unsynchronized populations using the FUCCI system (Fig. 2F,G). We do not yet know whether trisomy 8 slows the accelerated G1-to-S-phase transition resulting from EWS-FLI1 expression or whether it accelerates the transition to such an extent that EWS-FLI1 cannot speed it up further. We favor the latter to be the case because trisomy 8 cells harbor higher levels of cyclin D (Supplemental Fig. S3G) and the trisomy 8 cells have a reduced predisposition to arrest in G0 in response to serum starvation.
Trisomy 8 also had a dramatic effect on S-phase events. The comet assay showed that EWS-FLI1 did not cause significant DNA damage in exponentially growing trisomy 8 fibroblasts (Fig. 2J). Trisomy 8 also suppressed DNA damage that occurs during normal DNA replication. Specifically, virtually all populations harbor γH2AX foci during S phase, but these were dramatically reduced in trisomy 8 cells compared with euploid cells, irrespective of whether they expressed EWS-FLI1 or not (Fig. 2D,E; Supplemental Fig. S3E,F). Furthermore, EWS-FLI1 expression did not cause the increase in S/G2 length in trisomy 8 cells, as it did in the euploid controls (cf. Figs. 2H,I and 1F,G), indicating that replication stress and the ensuing DNA damage are responsible for the dramatic S-phase delay caused by EWS-FLI1. We conclude that trisomy 8 either prevents EWS-FLI1-induced replication stress or enhances its resolution.
To distinguish between a role of trisomy 8 in protecting cells from replication stress or enhancing recovery therefrom, we examined the kinetics of recovery from hydroxyurea (HU) or aphidicolin (APH) treatment. These drugs cause replication stress by causing nucleotide depletion and inhibiting DNA replication, respectively. We released euploid and trisomy 8 fibroblasts from a serum starvation-induced G1 arrest into medium containing HU or APH and then determined the proportion of cells containing γH2AX foci following HU or APH washout (Supplemental Fig. S2A). γH2AX foci were resolved much faster in trisomy 8 cells compared with euploid controls (Fig. 2K; Supplemental Fig. S3H). Interestingly, this accelerated resolution of γH2AX foci in trisomy 8 cells was specific to replication stress, as we observed little or no difference in the kinetics of irradiation-induced or genotoxic agent-induced double strand break (DSB) repair between euploid and trisomy 8 fibroblasts (Fig. 2L; Supplemental Fig. S3I,J). We conclude that trisomy 8 enhances the resolution of HU-, aphidicolin-, and EWS-FLI1-induced replication stress.
Increased chromosome 8 copy number correlates with lower DNA damage in Ewing sarcoma
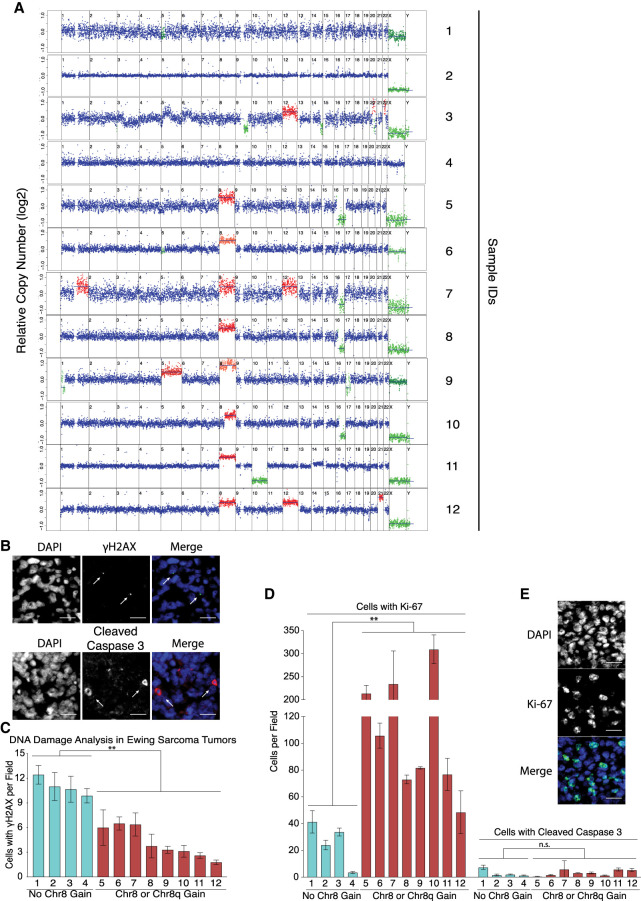
Does chromosome 8 gain also enhance resolution of replication stress in Ewing sarcomas? To address this question, we asked whether there was a relationship between the number of γH2AX foci and chromosome 8 gain in Ewing sarcoma specimens (Supplemental Table S1). We identified tumor regions in histological sections using the Ewing sarcoma cell surface marker CD99 and then examined degree of DNA damage and proliferation by γH2AX and Ki-67 staining, respectively. Only then did we assess chromosome 8 copy number in the tumors. We found that sarcomas disomic for chromosome 8 harbored significantly more γH2AX foci than tumors with three or more copies of chromosome 8 (Fig. 3A–C). Interestingly, gain of chromosome 8q appeared sufficient to reduce γH2AX foci (sample 10 in Fig. 3A,C). Consistent with our cell culture experiments, we found that decreased amounts of γH2AX foci correlated with increased proliferative capacity as judged by Ki-67 staining (Fig. 3D,E). We conclude that trisomy 8 is associated with decreased replication stress and/or DNA damage in Ewing sarcomas.
Figure 3.
Chromosome 8 gain correlates with reduced number of γH2AX foci and increased proliferation in Ewing sarcomas. (A) Karyotypes of 12 Ewing sarcomas. Median chromosome 8 copy number is shown in Supplemental Table S1. (B) Representative examples of the histological analysis of γH2AX foci (green) and apoptotic cells (cleaved caspase 3; red) in tumor regions, which were identified by the presence of the CD99 epitope on parallel slides by a certified pathologist. The white arrows highlight such cells. DNA is in blue. Scale bar, 100 µm. (C) Cells harboring γH2AX foci (one or more large focus) but not cleaved caspase 3 were quantified in Ewing sarcoma samples. Chromosome 8 copy number status (from A) is indicated below. Error bars represent SEM of at least three fields. At least 1500 nuclei were evaluated. (**) P < 0.01, two-tailed nonparametric two-group Mann–Whitney U-test. (D) Cells harboring Ki-67 or cleaved caspase 3 were determined in Ewing sarcoma samples. Error bars represent SEM of at least three fields. At least 1500 nuclei were evaluated. (**) P < 0.01, (n.s.) not significant, two-tailed nonparametric two-group Mann–Whitney U-test. (E) Example of cells expressing Ki-67 (green). Scale bar, 100 µm.
Gain of chromosome 8 is associated with poor prognosis in Ewing sarcoma
A previous study did not find chromosome 8 copy number to affect prognosis (Tirode et al. 2014). We re-examined these data, ranking patients according to the highest gain value of any chromosome 8 segment per patient (Supplemental Material; Supplemental Table S3) and then comparing the overall survival times of the top one-third of patients (all harboring at least one additional copy of chromosome 8) and the bottom third of patients (all euploid for chromosome 8) (Supplemental Table S3). This analysis showed that increased chromosome 8 copy number correlated with shorter survival (Supplemental Fig. S4A). The risk of succumbing to Ewing sarcoma was 2.4-fold higher for patients whose tumors harbor additional copies of chromosome 8 compared with patients whose tumors were disomic for the chromosome (Supplemental Fig. S4B). Moreover, increased chromosome 8 copy number was associated with disease relapse (Supplemental Fig. S4C). Given that chromosome 8 is gained in only 50% of Ewing sarcomas, the cancer can clearly develop in the absence of chromosome 8 gain, but our data suggest that trisomy 8 contributes to aggressive disease.
A method to identify genes that drive chromosome 8 gain in Ewing sarcoma
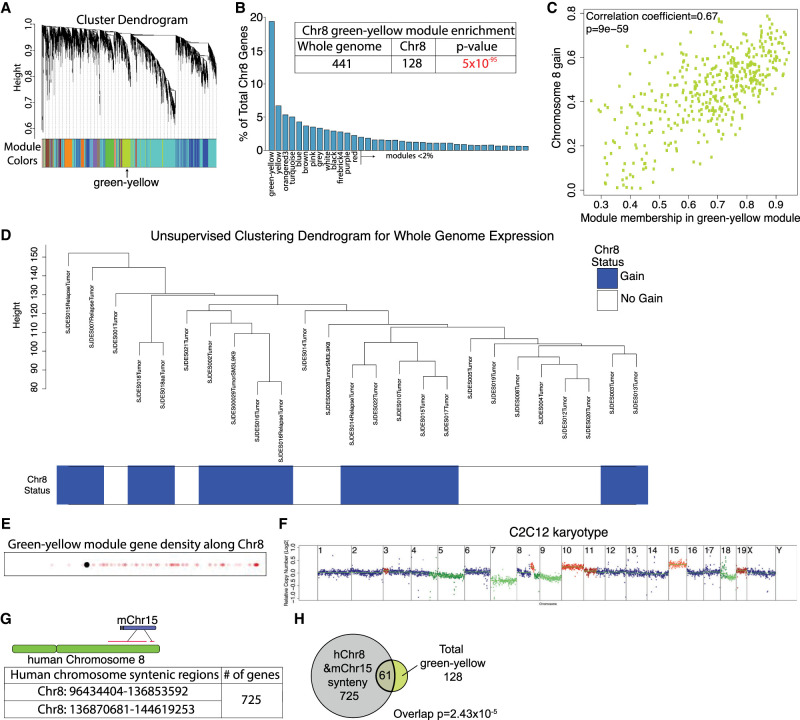
Our results show that chromosome 8 gain mitigates replication stress and improves proliferation in EWS-FLI1-expressing cells, raising the question of which of the 2372 genes encoded on chromosome 8 might be responsible. We hypothesized that the relevant gene(s) might be highly expressed even in tumors disomic for chromosome 8. To identify genes whose expression is high and correlated with each other across all Ewing sarcoma samples, we applied a weighted correlation network analysis (WGCNA) (Langfelder and Horvath 2008) to gene expression analyses obtained from 25 different Ewing sarcoma samples, of which 15 gained chromosome 8 and 10 were disomic (sample annotations in Supplemental Table S3; Crompton et al. 2014). This analysis revealed 76 clusters of coexpressed genes (referred to here as modules) (Fig. 4A). Interestingly one module, color-coded as green–yellow in Figure 4A, was highly enriched for chromosome 8-encoded genes (128 out of 441) (Fig. 4B; Supplemental Table S4). Within the green–yellow module, the consistency of increased gene expression across tumor samples (eigengene value) was much greater for chromosome 8-encoded genes than for genes encoded on other chromosomes, indicating that the module is defined by the chromosome 8-encoded genes. Furthermore, we found that being a member of the green–yellow module was significantly correlated with chromosome 8 gain (Fig. 4C), suggesting that this module could be a driver of chromosome 8 gain. We note, however, that while chromosome 8 gain drove relatedness in gene expression within the green–yellow module, it was not a driver of relatedness of gene expression patterns across the whole genome, because tumors with chromosome 8 gain did not group together in unsupervised clustering analyses of gene expression patterns (Fig. 4D). The location of chromosome 8-encoded members of the green–yellow module is also noteworthy. Of the 128 genes, 125 were located on chromosome 8q (Fig. 4E; Supplemental Table S4). The observation that 8q gain correlated with reduced DNA damage in Ewing Sarcoma (Fig. 3C) raised the possibility that one or several of the 125 green–yellow module genes located on 8q mediated the chromosome 8 benefits for cells expressing the EWS-FLI1 fusion.
Figure 4.
A method to identify genes that mediate the chromosome 8 fitness benefit in EWS-FLI1-expressing cells. (A) Modules identified by hierarchical clustering and dynamitic tree cutting by WGCNA. Lines represent the hierarchical height of each gene in a cluster. Each color represents a module. The green–yellow module is identified. (B) Modules were rank-ordered based on the number of chromosome 8-encoded genes, shown as the percentage of total chromosome 8-encoded genes. The inset table shows the significant enrichment of chromosome 8-encoded genes in the green–yellow model (hypergeometric test). (C) Membership in the green–yellow module, as defined by relatedness of expression patterns in Ewing tumors, was correlated with gain of chromosome 8. (D) Unsupervised hierarchical clustering of expression patterns (WGCNA) in Ewing sarcoma samples using whole-genome expression data. Tumor sample names and annotations in the hierarchical tree are listed in Supplemental Table S3. (E) Location of genes of the green–yellow module on chromosome 8. Each semitransparent dot represents the location of a gene. The graph was made using the gene density visualizer tool (see the Supplemental Material). The black dot represents the centromere. (F) Karyotype of the C2C12 mouse myoblast cell line expressing the EWS-FLI1 fusion. (G) Synteny analysis between human chromosome 8 and mouse chromosome 15. Genes are listed in Supplemental Table S4. (H) Overlap between 128 chromosome 8-encoded genes of the green–yellow module and the 725 genes syntenic between human chromosome 8 and mouse chromosome 15 (hypergeometric test).
To further define which genes located on chromosome 8q promote proliferation in EWS-FLI1-expressing genes, we asked whether regions syntenic with chromosome 8 are amplified in the mouse C2C12 fibroblast cell line transformed with the EWS-FLI1 fusion (Li et al. 2010; Ben-David et al. 2016). Copy number analysis of this cell line revealed a gain of chromosome 15 (Fig. 4F). Seven-hundred-twenty-five genes encoded on mouse chromosome 15 are encoded on human chromosome 8 (Fig. 4G; Supplemental Table S4), and 61 of the 128 chromosome 8-encoded genes we identified as highly expressed in Ewing sarcomas are located on mouse chromosome 15 (Fig. 4H; Supplemental Table S4).
An evolution strategy identifies genes on chromosome 8 important for proliferation of EWS-FLI1-expressing cells
Among the 61 genes syntenic between human chromosome 8 and mouse chromosome 15 and highly expressed in Ewing sarcomas, we selected the 20 highest-expressed genes to create a lentivirus library (genes highlighted in Supplemental Table S4). We also added four genes implicated in connective tissue development as well as two highly expressed genes encoded elsewhere on chromosome 8 that served as negative controls. We then transduced this 26-ORF library together with the doxycycline-induced EWS-FLI1 construct into hMPro cells (Supplemental Fig. S5A). Introduction of the 26-ORF library significantly improved the growth of hMPro cells expressing EWS-FLI1 in two independent evolution experiments (Fig. 5A; Supplemental Fig. S5B). Notably, there were differences between the two experiments, which helped guide our subsequent analyses. In the first evolution, cells became immortalized as judged by their proliferation properties and morphological features (Fig. 5A; Supplemental Fig. S5D–G). Furthermore, DNA damage was decreased (Supplemental Fig. S5H). However, expression of the 26-ORF library did not cause transformation. Although growth on plates was enhanced, cells did not form clear foci (Supplemental Fig. S5I). Furthermore, EWS-FLI1-expressing cells transduced with the 26-ORF library neither produced colonies in soft agar nor formed tumors in immunocompromised mice (Supplemental Fig. S5J; data not shown). In the second evolution experiment, proliferation improved but growth eventually plateaued, indicating that immortalization did not occur (Supplemental Fig. S5B).
Figure 5.
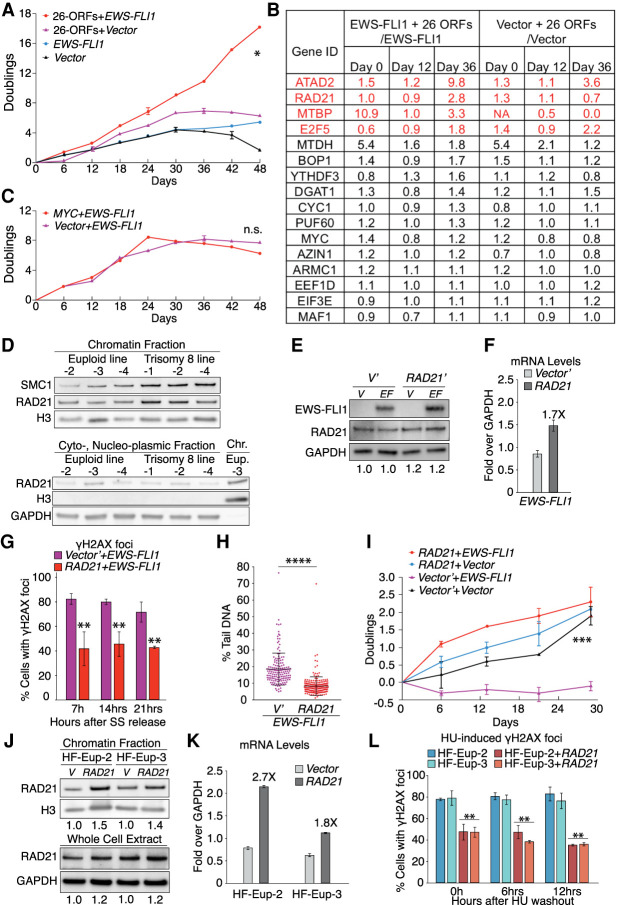
An evolutionary approach reveals RAD21 to be limiting for mitigating EWS-FLI1-induced replication stress. (A) Primary hMPro cells were infected at passage 7 (P7). Proliferation was measured 2–3 d after transduction of cells with the indicated lentiviral constructs. Error bars represent SEM of biological duplicates. Expression of genes in the 26-ORF library was measured at days 0, 12, and 36. (*) P < 0.05, linear regression. (B) Fold change expression of members of the 26-ORF library was determined in relation to cells that were not transduced with the library. Genes whose expression increased over time by greater than twofold are highlighted in red. (NA) Not analyzed (no detectable transcript in control cells). (C) Proliferation of hMPro cells harboring the indicated lentiviral constructs was measured 2–3 d after transduction. Error bar represents SEM of biological duplicates. (n.s.) Not significant, linear regression. (D) Chromatin association of the cohesin subunits RAD21 and SMC1 was determined by chromatin fractionation in the indicated fibroblast lines. The chromatin protein histone H3 (H3) and the cytoplasmic protein GAPDH in fractions served as controls. (Chr.) Chromatin fraction, (Eup.) euploid. N = 2; representative pictures are shown. (E,F) RAD21 protein (E) and mRNA (F) levels in control and EWS-FLI1-expressing euploid fibroblasts (HF-Eup-3) 8 d after transduction with a lentivirus expressing RAD21 from the CMV promoter. Numbers below the blot in E indicate degree of overexpression relative to vector transduced cells. Western blot: n = 2, with representative picture shown. RT-qPCR: error bar represents SEM of technical duplicates. We note that EWS-FLI1 protein levels were increased in cells overexpressing RAD21, presumably because increased RAD21 expression allows cells expressing higher levels of EWS-FLI1 to proliferate better. (G) Euploid fibroblasts (HF-Eup-3) were transduced with the indicated lentiviral constructs, and the percentage of EdU-positive cells harboring γH2AX foci (>10 foci/nucleus) was determined following release from a serum starvation-induced G1 arrest. Error bar represents SEM of biological replicates. (**) P < 0.01, two-tailed t-test; n > 2. (H) Assessment of DNA damage by comet assay in EWS-FLI1-expressing euploid fibroblasts (HF-Eup-3) expressing RAD21 or empty vector (V′). Each dot represents a single comet. The middle line represents the mean; error bars, SD. (****) P < 0.0001, two-tailed Wilcoxon test. (I) Proliferation of euploid fibroblasts (HF-Eup-3) transduced with the indicated lentiviral constructs was determined as described in Figure 1A. Error bars represent SEM of biological duplicates. (***) P < 0.001, linear regression. (J,K) Chromatin associated and total RAD21 protein (J) and mRNA (K) levels in control euploid cells and euploid fibroblasts expressing RAD21 from the CMV promoter. Numbers below the blots in J indicate degree of overexpression relative to vector transduced cells. Western blot: n = 2, with representative picture shown. RT-qPCR: error bar represents SEM of technical duplicates. (L) Cells characterized in J and K were released from a serum starvation-induced G1 arrest into medium containing 2 mM HU. The percentage of EdU-positive cells harboring γH2AX foci was determined at the indicated times following HU washout. Error bar represents SEM of biological replicates; n > 2. (**) P < 0.01, two-tailed t-test, comparison between the same types of fibroblasts. Statistics and number of experiments are shown in Supplemental Table S2 (multiple tabs).
To determine which of the 26 genes were responsible for the accelerated growth of EWS-FLI1-expressing cells, we examined their expression levels after 12 and 36 d of culturing (Supplemental Fig. S5A). In the first evolution, the expression of four genes, ATAD2, RAD21, MTBP, and E2F5, increased by more than twofold during the 36 d of culturing (Fig. 5B; Supplemental Table S5). Whereas ATAD2 and E2F5 expression rose in both control and EWS-FLI1-expressing cells, RAD21 and MTBP RNA levels increased only in cells harboring the EWS-FLI1 fusion (Fig. 5B; Supplemental Table S5). In the second evolution experiment, expression of MTBP, ATAD2, and E2F5, but not RAD21, increased over time (Supplemental Fig. S5C). Notably, MYC, which was included in the 26-ORF library, was not among the most selected genes, indicating that higher levels of expression of this oncogene contribute little to the observed improvement in proliferation. Consistent with this conclusion, we found that introduction of a lentivirus harboring MYC did not improve proliferation of EWS-FLI1 transduced cells (Fig. 5C; Supplemental Fig. S5K). We conclude that expression of EWS-FLI1 selects for increased expression of ATAD2, RAD21, MTBP, and E2F5. RAD21 stands out in that it was selected for only in the evolution experiment where the growth properties of cells indicated that they had become immortalized.
RAD21 promotes cohesin binding to chromosomes and mitigates replication stress
RAD21 is a component of the cohesin complex, which holds the duplicated DNA strands together until the onset of chromosome segregation (Peters et al. 2008). The complex is conserved across eukaryotes and serves multiple functions, ranging from defining chromosome structure to mediating chromosome segregation, to regulating gene expression. Cohesin also plays a critical role in stabilizing stalled replication forks (Remeseiro et al. 2012; Tittel-Elmer et al. 2012; Mondal et al. 2019) and in facilitating postreplicative DNA repair by homologous recombination (Sjögren and Nasmyth 2001; Kim et al. 2002; Ström et al. 2004; Ünal et al. 2004). In fact, in mammals, RAD21 is rate-limiting for DNA repair (Xu et al. 2010). Consistent with the idea that RAD21 is rate-limiting for cohesion function, chromatin fractionation showed increased association of cohesin with chromosomes in trisomy 8 fibroblast lines (Fig. 5D).
Given Rad21's established role in stabilizing stalled replication forks and in postreplicative DNA damage repair, and our finding of increased binding of cohesin to chromosomes in trisomy 8 cells, we tested whether increased RAD21 levels mitigate replication stress. For this, we used lentiviral-mediated transduction to introduce a constitutively active CMV-RAD21 construct into control and EWS-FLI1 euploid fibroblasts. Surprisingly, RAD21 protein levels were only modestly increased in these cells (1.2-fold) even though RNA levels rose by 1.7-fold (note we were unable to assess the amount of RAD21 bound to chromatin because few cells expressing EWS-FLI1 survived) (Fig. 5E,F). We hypothesized that high levels of RAD21 might interfere with proliferation, leading to the selection of cells with only modest overexpression of the cohesin subunit, as previous studies in yeast showed that strong overexpression of MCD1/SCC1 inhibits cell proliferation (Sopko et al. 2006). To determine whether this was true in mammalian cells, we overexpressed RAD21 to varying degrees in euploid fibroblasts using the doxycycline-inducible TET-ON promoter. Increasing doxycycline levels led to a corresponding increase in RAD21 mRNA, up to 10-fold higher than wild-type cells, but RAD21 protein levels increased by less than twofold (Supplemental Fig. S6A–C). Presumably, RAD21 that is not incorporated into cohesin complexes is less stable. Notably, this small increase in RAD21 expression did slow cell proliferation (Supplemental Fig. S6D,E). Thus, RAD21 overexpression also impairs proliferation in mammalian cells.
Having established why ectopic RAD21 protein accumulates to such low levels upon overexpression, we used our CMV-RAD21 cells to determine whether constitutive overexpression of RAD21 enhanced DNA repair. Despite the modest 1.2-fold increase in levels, RAD21 caused a decrease in γH2AX foci in the EWS-FLI1-expressing cells (Fig. 5G,H). Importantly, ectopic RAD21 also improved proliferation and reduced senescence in EWS-FLI1-expressing hMPro cells (Fig. 5I; Supplemental Fig. S6F,G). We note that overexpression of RAD21 did not improve proliferation to the extent seen when cells were transduced with the 26-ORF library (Fig. 5, cf. A and I). We conclude that RAD21 is partly responsible for the fitness benefit that trisomy 8 exerts on EWS-FLI1-expressing cells, but it is not the only chromosome 8-encoded gene with such properties.
Ectopic RAD21 overexpression not only enhanced repair in EWS-FLI1-expressing cells, it did so also in fibroblasts experiencing hydroxyurea-induced replication stress. A mere 1.5-fold increase in chromatin-bound cohesin led to a significant reduction in γH2AX foci in HU-treated euploid fibroblasts (Fig. 5J–L). Given that cohesins associate with stalled replication forks and sites of DNA damage (Potts et al. 2006; Caron et al. 2012; Remeseiro et al. 2012; Tittel-Elmer et al. 2012), we presume that a global 1.5-fold increase in cohesin association with chromatin upon RAD21 overexpression is indicative of local high-level cohesin loading. We propose that this increased binding to stalled replication forks and sites of DNA damage stabilizes stalled forks and enhances repair, respectively.
MYC promotes the proliferation of EWS-FLI1-expressing cells when co-overexpressed with RAD21
The MYC oncogene plays a critical role in the etiology of many cancers (Dang 2012). Surprisingly, MYC was not selected for in our evolution experiments (Fig. 5B; Supplemental Fig. S5C) and did not affect the growth of EWS-FLI1-expressing cells when overexpressed on its own (Fig. 5C). However, it was possible that under conditions of mitigated replication stress, such as occurs when RAD21 is overexpressed, a tumorigenesis-promoting function of MYC overexpression might be revealed. This was indeed the case. When we overexpressed MYC and RAD21 together in EWS-FLI1-expressing hMPro cells, proliferation was enhanced compared with expression of each gene alone (Supplemental Fig. S6H,I). Notably, MYC expression did not significantly boost the γH2AX focus levels resulting from EWS-FLI1, and this DNA damage response was still effectively ameliorated by RAD21 (Supplemental Fig. S6J). We conclude that increased levels of MYC improve proliferation of EWS-FLI1-expressing cells but only when RAD21 is also overexpressed. We speculate that in our evolution experiments, cells harboring multiple different ORF library clones were rare, preventing us from identifying MYC as a promoter of Ewing sarcomagenesis.
RAD21 triplication is required for the trisomy 8 benefit to EWS-FLI1-expressing cells
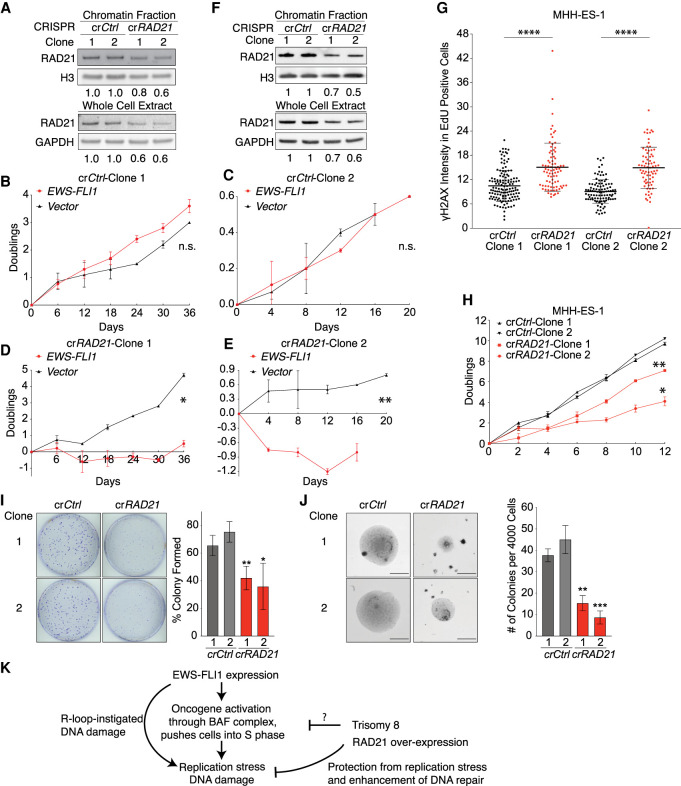
To determine whether RAD21 triplication contributed to the growth improvement of EWS-FLI1-expressing cells by trisomy 8, we deleted one of the three copies of RAD21 in trisomy 8 fibroblasts (Supplemental Fig. S7A,B). Whole-genome sequencing confirmed that cells remained trisomic for chromosome 8 and disomic for all other chromosomes, during the single-cell cloning procedure (Supplemental Fig. S7C). In clone 1, one copy of RAD21 was disrupted by generating a frameshift in exon 4 (Supplemental Fig. S7A), and this led to an expected decrease in RAD21 protein levels by ∼33% (Fig. 6A). In clone 2, the highly conserved phenylalanine 89 and arginine 90 within RAD21's SMC3 binding domain were replaced by leucine (Supplemental Fig. S7A), also resulting in a decline in RAD21 protein levels (Fig. 6A). We then introduced the EWS-FLI1 into these two clones, along with two trisomy 8 clones that retained three copies of RAD21. Remarkably, while trisomy 8 fibroblasts with three copies of RAD21 tolerated EWS-FLI1 expression well, the two trisomy 8 clones lacking one of the three RAD21 copies ceased to proliferate upon introduction of the oncogenic EWS-FLI1 fusion (Fig. 6B–E). We conclude that gain of an extra copy of RAD21 contributes to the growth-promoting properties of chromosome 8 gain in EWS-FLI1-expressing cells.
Figure 6.
Three copies of RAD21 are required for mitigating EWS-FLI1-induced replication stress in primary cells and in a Ewing sarcoma cell line. (A,F) Chromatin associated and total RAD21 protein in trisomy 8 fibroblasts (HF-Ts8-1) (A) and the MHH-ES1 cancer cell line (F) targeted with control (crCtrl) or RAD21 (crRAD21) CRISPR constructs. Numbers underneath the blot indicate down-regulation relative to vector transduced cells. (crCtrl) Transfected with control CRISPR construct, (crRAD21) transfected with RAD21 CRISPR targeting construct, resulting in one of three copies of RAD21 deletion. Same nomenclature is used for all figures below. (B–E) Trisomy 8 fibroblasts (HF-Ts8-1) were transfected with control or RAD21 CRISPR targeting constructs, resulting in the generation of trisomy 8 fibroblasts with three (B,D) or two (C,E) copies of RAD21. Cells were then transduced with control or EWS-FLI1-expressing lentiviruses, and proliferation was measured after 3–4 d of selection for lentiviral constructs. Error bars represent SEM of biological duplicates. (*) P < 0.05, (**) P < 0.01, (n.s.) not significant, linear regression. (G) Exponential growing MHH-ES1 cells were pulse-labeled with EdU for 1 h. The intensity of γH2AX signal in EdU-positive cells was determined relative to the background signal. Note that 10-foci criteria were not applied here because MHH-ES1 cells harbored an extremely high number of γH2AX foci. Each dot represents a single cell. The middle line represents the mean; error bar represents SD. (****) P < 0.00001, by two-tailed Wilcoxon test. (H,I) Two independent MHH-ES1 clones ([1] clone 1, [2] clone 2) carrying two copies of RAD21 (crRAD21) show reduced proliferation (H) and form fewer colonies (I) than MHH-ES1 cells carrying three copies. (crCtrl) Control. (H) (*) P < 0.05, (**) P < 0.01, linear regression. (I) Error bars represent SEM of biological replicates. (**) P < 0.01, (*) P < 0.05, two-tailed t-test, comparison between crCtrl and crRAD21 cells within the same clone number; n = 4. (J) Two independent MHH-ES1 clones ([1] clone 1, [2 clone 2) carrying two copies of RAD21 (crRAD21) show reduced anchorage-independent proliferation, forming smaller (left pictures; scale bars, 300 µm) and fewer colonies (right graph). Error bars represent SEM of biological replicates. (**) P < 0.01, (***) P < 0.001, by two-tailed t-test, comparison between crCtrl and crRAD21 within the same clone number; n = 3. (K) A model for how trisomy 8 contributes to Ewing sarcomagenesis. R-loop-instigated DNA damage is from Gorthi et al. (2018). See the text for details. Statistics and number of experiments are shown in Supplemental Table S2 (multiple tabs).
RAD21 triplication enables the anchorage-independent growth of a Ewing sarcoma cell line
Does an extra copy of RAD21 continue to be important even when EWS-FLI1-expressing cells have developed into Ewing sarcomas? To determine the cancer-relevance of RAD21 triplication, we deleted one of the three copies of RAD21 in the Ewing sarcoma cell line MHH-ES1, which expresses EWS-FLI1 and harbors three copies of chromosome 8q (for karyotype confirmation, see Supplemental Fig. S1G; Supplemental Table S1). We isolated two single clones (clones 1 and 2), each containing an adenine insertion in exon 4 of one copy of RAD21, which reduced RAD21 protein levels by ∼30%–40% (Fig. 6F). Inactivation of the third copy of RAD21 did not affect the kinetics of S-phase entry in these tumor cells, but replication stress was significantly increased (Fig. 6G; Supplemental Fig. S7D). Most remarkably, restoring the copy number of RAD21 to the normal 2 significantly reduced the proliferative capacity of the MHH-ES1 Ewing sarcoma cell line and significantly impaired their ability to form foci and anchorage-independent colonies in soft agar assays (Fig. 6H–J; Supplemental Fig. S7E,F). We conclude that a third copy of RAD21 is critical for the tumorigenic growth of the MHH-ES1 Ewing sarcoma cell line.
Discussion
A model to explain chromosome 8 gain in Ewing sarcoma
We believe that our study explains at least in part why chromosome 8 is so frequently gained in Ewing sarcoma. The tumor-initiating EWS-FLI1 fusion accelerates entry into S phase, resulting in replication stress. Inhibition of BRCA1-mediated DNA repair and R-loop-instigated DNA damage caused by the EWS-FLI1 fusion (Gorthi et al. 2018) are likely to generate further DNA damage, leading to cellular senescence (Fig. 6K). Trisomy 8, while detrimental in wild-type cells, becomes advantageous in the context of EWS-FLI1-induced replication stress because it acts to mitigate this (Fig. 6K). These findings serve as a paradigm for why clonal aneuploidies exist in cancers despite the fitness penalties associated with whole-chromosome gains and losses. This concept also fits with prior observations that the aneuploid state enables drug resistance (Selmecki et al. 2006; Pavelka et al. 2010; Rutledge et al. 2016; Replogle et al. 2020).
Precedence for the idea that gain of a chromosome, and thus a specific gene, can create a fitness advantage exists in budding yeast. Eight percent of strains in the budding yeast knockout collection have gained a specific chromosome, which typically harbors a paralog of the deleted gene (Hughes et al. 2000). Under conditions that cause significant growth inhibition, specific aneuploidies are selected for because the fitness gain of duplicating or deleting a specific gene outweighs the fitness penalty associated with gain or loss of the entire chromosome, respectively. In the yeast example, the benefit of increasing the copy number of the paralog in the deletion strain outweighs the fitness penalty of the chromosome gain. In cells expressing the EWS-FLI1 fusion, which causes high levels of replication stress, replication fork stabilization and enhanced DNA repair provided by an extra copy of RAD21 (and likely other genes) (see below) outweighs the adverse effects of increasing the dosage of the other 2371 chromosome 8-encoded genes.
Multiple genes encoded by Chr8q24 provide a fitness advantage to EWS-FLI1-expressing cells
Chr8q24 is amplified in a large number of human tumor types. It harbors the famous oncogene MYC, which is widely presumed to be the sole driver of this amplification. Our data argue that suppression of oncogene-induced replication stress is an additional cause for why this genomic region is so frequently amplified in cancer. Excess RAD21 mitigates oncogene-induced replication stress. MTBP, another gene our evolution experiment selected for, is also located in Chr8q24. MTBP is required for DNA replication initiation (Boos et al. 2013), raising the possibility that it could help resolve stalled forks by initiating replication at dormant origins. Other genes located within Chr8q24 could also promote cell cycle entry. Our evolution strategy identified ATAD2, a coactivator of MYC and interactor of E2F (Ciró et al. 2009; Revenko et al. 2010). Perhaps excess ATAD2 accelerates the G1-to-S-phase transition.
We propose that the genes we identified in our evolution experiment to enhance the proliferation of EWS-FLI1-expressing cells promote Ewing sarcomagenesis when in excess. Consistent with this conclusion is the observation that high expression of RAD21, MTBP, and ATAD2 is correlated with poor prognosis in Ewing sarcoma (Supplemental Fig. S4D). The correlation between high-level expression of E2F5 and prognosis was less striking (Supplemental Fig. S4D). Notably, MYC overexpression, which was not selected for in our evolution experiment, is in fact not correlated with poor prognosis (Supplemental Fig. S4D). However, it clearly promoted proliferation of EWS-FLI1-expressing cells when it co-occurred with RAD21 gain. MYC by itself is also a potent inducer of replication stress (Kotsantis et al. 2018), and thus, we speculate that up-regulation of RAD21 may be required to mitigate the replication stress resulting from MYC overexpression. Interestingly, when RAD21 was overexpressed, we observed an increased amount of endogenous MYC (Supplemental Fig. S6H) and also up-regulation of EWS-FLI1 (Fig. 5E), suggesting that RAD21 enables higher levels of these stress-inducing proteins. We propose that co-overexpression of MYC and RAD21 could provide a potent combination in many cancer types by inappropriately driving cells into S phase, and enabling survival of the adverse consequences, by mitigating the consequent replication stress and enhancing repair. Consistent with this idea, we found that 70% of the 38 cancer types found in the TCGA database harbor recurrent chromosome 8q gains (defined as >25% of the tumor specimens) (Supplemental Fig. S4E; Supplemental Table S6).
RAD21 drives Ewing sarcomagenesis through single copy gain
Perhaps our most striking finding was that increasing global cohesin association with chromosomes by 50%, which likely reflects local high-level cohesin loading at stalled forks and sites of DNA damage, mitigating replication stress. RAD21 likely does so by multiple mechanisms. In budding yeast, cohesin is recruited to stalled replication forks, where it is thought to exert fork-stabilizing functions (Tittel-Elmer et al. 2012; Delamarre et al. 2020) and to promote equal sister chromatid exchange (Cortés-Ledesma and Aguilera 2006). Curiously, in mammalian cells, the cohesin remover WAPL is also recently reported to be required for the repair and restart of stalled replication forks, which was interpreted to mean that removal of cohesin from sites of DNA damage was required for fork repair and restart (Benedict et al. 2020). In light of our observation that overexpression of the rate-limiting cohesin subunit RAD21 mitigates oncogene-induced replication stress, we favor the idea that WAPL is required to mobilize cohesin elsewhere in the genome to enable cohesins to accumulate at stalled forks to facilitate their repair and restart. Increased accumulation of cohesins at sites of DNA damage could keep sister chromatids more tightly paired, favoring equal sister chromatid exchange or recruit repair factors to sites of DNA damage (Wu et al. 2012). In light of the fact that cohesins also regulate gene expression (Peters et al. 2008), it is also possible that increased RAD21 levels could affect the expression of fork-stabilizing and/or repair factors.
Irrespective of the mechanism whereby higher levels of RAD21 mitigate replication stress, we propose that it is this function of RAD21 that promotes Ewing sarcomagenesis. Whether expression of other cohesin subunits has a similar effect on replication stress remains to be determined. We note that ∼17%–22% of Ewing sarcomas harbor loss-of-function mutations in the gene encoding the cohesin subunit STAG2 (Brohl et al. 2014; Crompton et al. 2014; Tirode et al. 2014), which can cause transformation of EWS-FLI1-expressing murine mesenchymal stem cells lacking p53 (El Beaino et al. 2020). Reduction in STAG2 function is known to impair fork progression (Mondal et al. 2019), and it reduces binding of RAD21 to chromatin in primary mesenchymal cells (our unpublished observations). We wondered whether RAD21 copy number gain might be a hallmark of Ewing sarcomas with STAG2 mutation, but our examination of TGCA data showed STAG2 mutant tumors have a similar percentage of RAD21 gains (and also EWS-FLI1 or EWS-ERG fusions) compared with their STAG2 wild-type counterparts (Tirode et al. 2014; TCGA: cBioportal). We do not believe that these observations are at odds with our findings, as we think that RAD21 is only one of several genes that can mitigate oncogene-induced replication stress, including that resulting from STAG2 mutation.
Many oncogenes accelerate S-phase entry and hence cause replication stress. Our data indicate that this replication stress must be dampened without simultaneously antagonizing the proproliferative effects of the oncogenic mutation. Without this mitigation, replication stress is too severe, and cells with oncogenic mutations undergo senescence. Higher levels of RAD21 does just that. It alleviates replication stress without affecting EWS-FLI1's ability to drive cell proliferation. Given that many oncogenes cause replication stress, it is conceivable that increased RAD21 copy number promotes tumorigenic growth in many cancers. We further propose that RAD21 only confers a fitness advantage to oncogene-expressing cells and when overexpressed at low level. We predict that aneuploidies prevalent in specific cancers are driven by genes such as RAD21. The methodology described here could help in their identification.
Materials and methods
Human cells, tumor samples, and whole-genome sequence analyses
All primary human cells, their sources, and growth conditions are listed in Supplemental Table S1. Ewing sarcoma tumor samples were acquired from ProteoGenex and Asuragen (Supplemental Table S1). Karyotypes of cells and tumors were confirmed by low-coverage whole-genome sequencing. For MSC, hTER-MSC, hMPro, HF-Eup-3, HF-Eup-4, HF-Ts8-1, and HF-Ts8-2 cells, this was conducted within the same or two postpassages of lentiviral transduction, and subsequently at later passages, to confirm that the karyotypes did not change over the timeline of our study. DNA from ∼100,000 cells for most lines, and at least 40,000 cells for lines with limited cell numbers, such as some EWS-FLI1-expressing cells, was extracted by using a QIAmp DNA mini kit (Qiagen). Standard Nextera XT Illumina library preparation was applied and the libraries sequenced with 40-bp read length on an Illumina HiSeq2000. Sequence reads were trimmed to 40 nt and aligned to the mouse (mm9) or human (hg19) reference genomes using the BWA (0.7.12) backtrack algorithm (Li and Durbin 2009). HMMcopy (0.1.1) was used to detect copy number alterations by estimating DNA copy number in 500-kb bins, controlling for mappability and GC content (calculated by HMMcopy gcCounter) (Ha et al. 2012). Simulation using HMMcopy suggests that in an euploidy population, if >30% harbor a common chromosome gain, karyotypic difference will be shown in this analysis.
Lentiviral transduction and transfection
Lentivirus was produced using 293FT packaging cells (Thermo Fisher Scientific) and transduction conducted as described in the Supplemental Material.
Cell proliferation and tumorigenicity analyses
The proliferation analysis procedure is described in the Supplemental Material. For focus formation assays, 2000 cells (for hMPro) or 500 cells (for MHH-ES1) were plated in replicates on 10-cm plates and cultured for 21 d (for hMPro) or 14 d (for MHH-ES1) before being fixed with ice-cold methanol (10 min) and stained with a solution of 0.5% crystal violet in 25% methanol for 10 min. Plates were scanned to assess colony formation. For soft agar growth assays, 4000 cells (for MHH-ES1) were seeded in 0.7% Difco noble agar (BD Biosciences) in medium at 37°C. The 0.7% soft agar with cell solution was solidified on top of 1% Difco noble agar medium at room temperature. The cells were supplemented with medium (changed every 3 d) and cultured for 14 d. Colonies were imaged and counted using Nikon light microscope with 4× objective lens. Xenograft assays are described in the Supplemental Material.
DNA damage analysis by comet assay
Cells expressing the EWS-FLI1 fusion for 3–5 d (grown to 60%–80% confluency) were subjected to single-cell gel electrophoresis (Trevigen Comet assay kit, R&D Systems) under alkaline comet assay conditions. DNA was then stained with SYBR Gold nucleic acid gel stain (Invitrogen) for 30 min at room temperature in the dark and washed twice with H2O. Images were taken in the FITC channel with a Plan Apo 40×/0.2 objective, ORCA-ER camera, and NIS-Elements software. Exposure times (same for the same cell type) were determined using the autoexposure function of the NIS-Element software. The percentage of DNA in comet tails was analyzed using the automated OpenComet tool (Gyori et al. 2014) in FIJI (ImageJ). At least 150 cells per sample were analyzed.
DNA damage analysis by γH2AX focus analysis
Cells were grown on a coverslip and fixed, and (if relevant) EdU was visualized (as described in the Supplemental Material). Fixed cells were washed twice with PBS + BSA (3%), incubated with a γH2AX antibody (1:400 dilution) in PBS + BSA (3%) overnight at 4°C, and subjected to immunofluorescent staining (described below). Cells containing >10 clear γH2AX foci were defined as γH2AX-positive cells. At least 50 cells (for some slow-growing cell types), and in most cases, >100 cells, were analyzed.
Cell cycle analysis
Supplemental Figure S2A depicts the general cell cycle analysis scheme. For serum starvation synchronization, cells were cultured in their optimal medium (Supplemental Table S1), but FBS was omitted. hMPro cells constitutively expressing the EWS-FLI1 fusion or control vector were plated at the same density into six-well plates for 12 h and then switched to serum-free medium for 2.5 d, causing most cells to arrest in G1 (validated by flow cytometry). Human fibroblasts with doxycycline-inducible EWS-FLI1 fusion or the control vector were seeded onto glass coverslips. One day later, cells were serum-starved for 3.5–4 d in the presence of 1 µg/mL doxycycline. Notably, human fibroblasts, especially trisomy 8 fibroblasts, were not completely arrested in serum-free medium. For other cell types, >90% of cells were arrested in G1 after 3.5–4 d of serum starvation. Cells were then released into the cell cycle by switching to medium containing serum and 10 µM EdU. Cells on coverslips were fixed and analyzed at the indicated times.
Flow cytometry
Cells were harvested using 0.25% trypsin in a versene solution and resuspended in normal growth medium. DNA was stained with Vybrant DyeCycle Violet (5 µM; Thermo Fisher Scientific) for 30 min at 37°C. Alternatively, cells were fixed with 70% ethanol overnight at −20°C and then stained with an Alexa fluor 647 conjugated anti-phospho-histone H3 antibody in PBS containing 2% FBS and 0.05% Triton X-100 for 2 h at 4°C. Samples were then washed twice with PBS and 0.05% Triton X-100, and DNA was stained using 1 g/mL DAPI in PBS for 10 min at room temperature. Flow cytometry analysis was performed immediately using either a BD FACSCelesta or a BD FACS LSR cytometer with data acquisition through BD FACSDIVA software through BV 421/DAPI and APC (Alexa Fluor 647) channels. Five-thousand to 10,000 or all gated cells were analyzed.
Fluorescence microscopy and live cell imaging
Procedures for EdU analyses and fluorescence microscopy for cells and FFPE-fixed tumor samples are described in the Supplemental Material. All primary and secondary antibodies, along with their dilutions (in PBS + 0.1%Triton X-100, 1% BSA), are shown in Supplemental Table S7.
For live-cell imaging of the FUCCI system, cells were seeded on glass-bottom 12-well plates (MatTek) for 2 d in the presence of 1 µg/mL doxycycline, and the media was changed 1 h before imaging. Cells were monitored in a humid tissue culture chamber at 37°C with CO2, and multipoint images were taken using a Nikon Elipse Ti fluorescent microscope with a Plan Fluor 10×/0.3 objective, ORCA-R2 camera, and MetaMorph software. Color composite time-lapse images were exported as movies using MetaMorph and viewed and evaluated using Fiji (ImageJ) software.
Western blot analysis
Cells were lysed in RIPA buffer (Thermo Fisher Scientific) containing protease inhibitor and phosphatase inhibitor (PhosStop) cocktails (Roche). Protein extracts were quantified by Bradford (Bio-Rad) and equal amounts subjected to SDS-PAGE (4%–12%; Bio-Rad or Invitrogen). Proteins were transferred to a PVDF or a nitrocellulose membrane using a semidry transfer method. Blots were blocked using TBS-T (0.1% Tween 20) with 3% fat-free milk and 1% BSA and incubated with primary antibodies (listed in Supplemental Table S7 with dilutions in TBST with 1%BSA) overnight at 4°C. Horseradish peroxidase (HRP)-conjugated or fluorophore-conjugated antibodies were used as secondary antibodies (Supplemental Table S7) in TBST with 1% BSA for 1 h at room temperature and detected by a ImageQuant LAS400 system or ChemiDoc MP imaging system (Bio-Rad), respectively. Signals were quantified using the Fiji-ImageJ gel analysis software.
RNA sequencing and analysis
For the 26-ORF library evolution experiment 5, experimental replicates per time point were analyzed. Total RNA was extracted using the RNeasy kit (Qiagen). Procedures for RNA sequencing and analysis are listed in the Supplemental Material.
Computational analysis and tools
Survival analysis, χ2 test for studying disease-risks, weighted correlation network analysis (WGCNA) for gene module identification, mouse synteny analysis, and development of the gene density visualizer (GDV) tool are described in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank the Swanson Biotechnology Center, especially the Integrated Genomics and Bioinformatics Core, for technical assistance; Alejandro Sweet-Cordero, Christy Chao, David Gordon, Simran Kaushal, Konrad Stopsack, James Haber, David Pellman, and Angelique Whitehurst for reagents and advice; Pei-hsin Hsu for help with photography; Amon laboratory members for suggestions and critical reading of the manuscript; and Jacqueline Lees for advice and guidance for revision of the manuscript. This work was supported by the Koch Institute Support (core) grant from the National Cancer Institute (P30-CA14051) and awards to A.A, an investigator of the Howard Hughes Medical Institute, from the National Institutes of Health (NIH) grant (CA206157), the Paul F. Glenn Center for Biology of Aging Research at Massachusetts Institute of Technology (MIT), and the Ludwig Center at MIT; X.A.S., from a MIT-Ludwig Oncology Fellowship and a Jane Coffin Childs Memorial Fellowship; J.M.R., from an MIT School of Science Fellowship in Cancer Research; K.S., from the NIH (R35 CA210030 and R01 CA204915) and Curing Kids Cancer; and J.F.A., from the Cancer Prevention and Research Institute of Texas (RP120685), the 1 Million 4 Anna Foundation, and Curing Kids Cancer.
Author contributions: X.A.S. and A.A. conceived and designed the research and wrote the manuscript. X.A.S. conducted the study and performed the majority of the experiments and part of the computational analysis. D.M. performed most of the computational analysis. J.V.P analyzed EWS-FLI1's effection proliferation in trisomy 9 and 13 cells and facilitated several experiment setups. J.M.R. performed the mouse injection experiment and monitored tumor growth. C.A.W. provided technical support for computational analysis and sequencing data submission. J.F.A. provided technical support. K.S. provided patient RNA, DNA sequencing data, and technical support. All authors read and approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.345454.120.
Competing interest statement
The authors declare no competing interests.
References
- Abe T, Sakaue-Sawano A, Kiyonari H, Shioi G, Inoue K, Horiuchi T, Nakao K, Miyawaki A, Aiszawa S, Fujimori T. 2013. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development 140: 237–246. 10.1242/dev.084111 [DOI] [PubMed] [Google Scholar]
- Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND, Hajjar S, Layeghifard M, Novokmet A, Kowalski PE, et al. 2018. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 361: eaam8419. 10.1126/science.aam8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Ha G, Khadka P, Jin X, Wong B, Franke L, Golub TR. 2016. The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat Commun 7: 12160. 10.1038/ncomms12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict B, van Schie JJM, Oostra AB, Balk JA, Wolthuis RMF, Riele HT, de Lange J. 2020. WAPL-dependent repair of damaged DNA replication forks underlies oncogene-induced loss of sister chromatid cohesion. Dev Cell 52: 683–698.e7. 10.1016/j.devcel.2020.01.024 [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Yekezare M, Diffley JF. 2013. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 340: 981–984. 10.1126/science.1237448 [DOI] [PubMed] [Google Scholar]
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. 2017. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171: 163–178.e19. 10.1016/j.cell.2017.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Volorio A, Iyer S, Broye LC, Stamenkovic I, Riggi N, Rivera MN. 2018. Epigenome editing of microsatellite repeats defines tumor-specific enhancer functions and dependencies. Genes Dev 32: 1008–1019. 10.1101/gad.315192.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, Patidar R, Hurd L, Chen L, Shern JF, et al. 2014. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 10: e1004475. 10.1371/journal.pgen.1004475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, Massip L, Losada A, Legube G. 2012. Cohesin protects genes against γH2AX induced by DNA double-strand breaks. PLoS Genet 8: e1002460. 10.1371/journal.pgen.1002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciró M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, et al. 2009. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res 69: 8491–8498. 10.1158/0008-5472.CAN-09-2131 [DOI] [PubMed] [Google Scholar]
- Cortés-Ledesma F, Aguilera A. 2006. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep 7: 919–926. 10.1038/sj.embor.7400774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, Kiezun A, Carter SL, Shukla SA, Mehta SS, et al. 2014. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 4: 1326–1341. 10.1158/2159-8290.CD-13-1037 [DOI] [PubMed] [Google Scholar]
- Dang CV. 2012. MYC on the path to cancer. Cell 149: 22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delamarre A, Barthe A, de la Roche Saint-André C, Luciano P, Forey R, Padioleau I, Skrzypczak M, Ginalski K, Géli V, Pasero P, et al. 2020. MRX increases chromatin accessibility at stalled replication forks to promote nascent DNA resection and cohesin loading. Mol Cell 77: 395–410.e3. 10.1016/j.molcel.2019.10.029 [DOI] [PubMed] [Google Scholar]
- Deneen B, Denny CT. 2001. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 20: 6731–6741. 10.1038/sj.onc.1204875 [DOI] [PubMed] [Google Scholar]
- El Beaino M, Liu J, Wasylishen AR, Pourebrahim R, Migut A, Bessellieu BJ, Huang K, Lin PP. 2020. Loss of Stag2 cooperates with EWS-FLI1 to transform murine mesenchymal stem cells. BMC Cancer 20: 3. 10.1186/s12885-019-6465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, Iniguez AB, Bernard X, Masamsetti VP, Roston S, et al. 2018. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 555: 387–391. 10.1038/nature25748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, Sorensen PH, Delattre O, Dirksen U. 2018. Ewing sarcoma. Nat Rev Dis Primers 4: 5. 10.1038/s41572-018-0003-x [DOI] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV. 2014. Opencomet: an automated tool for comet assay image analysis. Redox Biol 2: 457–465. 10.1016/j.redox.2013.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha G, Roth A, Lai D, Bashashati A, Ding J, Goya R, Giuliany R, Rosner J, Oloumi A, Shumansky K, et al. 2012. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res 22: 1995–2007. 10.1101/gr.137570.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25: 333–337. 10.1038/77116 [DOI] [PubMed] [Google Scholar]
- Kennedy AL, Vallurupalli M, Chen L, Crompton B, Cowley G, Vazquez F, Weir BA, Tsherniak A, Parasuraman S, Kim S, et al. 2015. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget 6: 30178–30193. 10.18632/oncotarget.4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K. 2002. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem 277: 45149–45153. 10.1074/jbc.M209123200 [DOI] [PubMed] [Google Scholar]
- Knouse KA, Amon A. 2013. The many sides of CIN. Nat Rev Mol Cell Biol 14: 611. 10.1038/nrm3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenhafer SL, Goss KL, Terry WW, Gordon DJ. 2020. Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells causes DNA damage and apoptosis via the CDK2-mediated degradation of RRM2. Mol Cancer Res 18: 91–104. 10.1158/1541-7786.MCR-19-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsantis P, Petermann E, Boulton SJ. 2018. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov 8: 537–555. 10.1158/2159-8290.CD-17-1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessnick SL, Dacwag CS, Golub TR. 2002. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 1: 393–401. 10.1016/S1535-6108(02)00056-9 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, McGee-Lawrence ME, Decker M, Westendorf JJ. 2010. The Ewing's sarcoma fusion protein, EWS-FLI, binds Runx2 and blocks osteoblast differentiation. J Cell Biochem 111: 933–943. 10.1002/jcb.22782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal G, Stevers M, Goode B, Ashworth A, Solomon DA. 2019. A requirement for STAG2 in replication fork progression creates a targetable synthetic lethality in cohesin-mutant cancers. Nat Commun 10: 1686. 10.1038/s41467-019-09659-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Soler M, Morgado-Palacin I, Lafarga V, Lecona E, Murga M, Callen E, Azorin D, Alonso J, Lopez-Contreras AJ, Nussenzweig A, et al. 2016. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget 7: 58759–58767. 10.18632/oncotarget.11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114. 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H. 2006. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J 25: 3377–3388. 10.1038/sj.emboj.7601218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A. 2012. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2076–2089. 10.1038/emboj.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle JM, Zhou W, Amaro AE, McFarland JM, Villalobos-Ortiz M, Ryan J, Letai A, Yilmaz O, Sheltzer J, Lippard SJ, et al. 2020. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc Natl Acad Sci 117: 30566–30576. 10.1073/pnas.2009506117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW. 2010. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol Cell Biol 30: 5260–5272. 10.1128/MCB.00484-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Provero P, Suvà ML, Kaloulis K, Garcia-Echeverria C, Hoffmann F, Trumpp A, Stamenkovic I. 2005. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 65: 11459–11468. 10.1158/0008-5472.CAN-05-1696 [DOI] [PubMed] [Google Scholar]
- Riggi N, Suvà ML, Suvà D, Cironi L, Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V, et al. 2008. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 68: 2176–2185. 10.1158/0008-5472.CAN-07-1761 [DOI] [PubMed] [Google Scholar]
- Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesús M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D. 2016. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep 6: 22828. 10.1038/srep22828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498. 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16: 473–485. 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370. 10.1126/science.1128242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren C, Nasmyth K. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol 11: 991–995. 10.1016/S0960-9822(01)00271-8 [DOI] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330. 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Ström L, Lindroos HB, Shirahige K, Sjögren C. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16: 1003–1015. 10.1016/j.molcel.2004.11.026 [DOI] [PubMed] [Google Scholar]
- Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, et al. 2018. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33: 676–689.e3. 10.1016/j.ccell.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, Zhang Z, Lapouble E, Grossetête-Lalami S, Rusch M, et al. 2014. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 4: 1342–1353. 10.1158/2159-8290.CD-14-0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, François P, Hohl M, Petrini JHJ, Pasero P, Cobb JA. 2012. Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol Cell 48: 98–108. 10.1016/j.molcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16: 991–1002. 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. 2006. Does aneuploidy cause cancer? Curr Opin Cell Biol 18: 658–667. 10.1016/j.ceb.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Wu N, Kong X, Ji Z, Zeng W, Potts PR, Yokomori K, Yu H. 2012. Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev 26: 1473–1485. 10.1101/gad.193615.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Balakrishnan K, Malaterre J, Beasley M, Yan Y, Essers J, Appeldoorn E, Tomaszewski JM, Vazquez M, Verschoor S, et al. 2010. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS One 5: e12112. 10.1371/journal.pone.0012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.