Here, Seavey et al. developed a conditional epithelioid hemangioendothelioma (EHE) mouse model whereby Wwtr1-Camta1 is controlled by the endogenous transcriptional regulators upon Cre activation by using an approach to target the Wwtr1-Camta1 gene fusion to the Wwtr1 locus. Their results demonstrate that TAZ-CAMTA1 is sufficient to drive EHE formation and that this EHE mouse model is a valid preclinical model for understanding the role of TAZ dysregulation in cancer.
Keywords: endothelial cells, epithelioid hemangioendothelioma, FLEx system, fusion gene, Hippo pathway, mouse models of cancer, sarcoma, TAZ-CAMTA1, YAP/TAZ
Abstract
Epithelioid hemangioendothelioma (EHE) is a genetically homogenous vascular sarcoma that is a paradigm for TAZ dysregulation in cancer. EHE harbors a WWTR1(TAZ)-CAMTA1 gene fusion in >90% of cases, 45% of which have no other genetic alterations. In this study, we used a first of its kind approach to target the Wwtr1-Camta1 gene fusion to the Wwtr1 locus, to develop a conditional EHE mouse model whereby Wwtr1-Camta1 is controlled by the endogenous transcriptional regulators upon Cre activation. These mice develop EHE tumors that are indistinguishable from human EHE clinically, histologically, immunohistochemically, and genetically. Overall, these results demonstrate unequivocally that TAZ-CAMTA1 is sufficient to drive EHE formation with exquisite specificity, as no other tumor types were observed. Furthermore, we fully credential this unique EHE mouse model as a valid preclinical model for understanding the role of TAZ dysregulation in cancer formation and for testing therapies directed at TAZ-CAMTA1, TAZ, and YAP/TAZ signaling.
Epithelioid hemangioendothelioma (EHE) is a rare vascular sarcoma that harbors a reciprocal chromosomal t(1p36;3q25) translocation yielding a WWTR1(TAZ)-CAMTA1 gene fusion in 90% of cases (Mendlick et al. 2001; Errani et al. 2011; Tanas et al. 2011). Approximately 45% of EHE tumors carry only the WWTR1-CAMTA1 (WC) gene fusion with no other genetic alterations present (Seligson et al. 2019). The WC fusion transcript is generated by joining the 5′ end of WWTR1 at either exon 3 or 4 to the 3′ end of CAMTA1 at either exon 8 or 9 (Errani et al. 2011; Tanas et al. 2011; Patel et al. 2015). TAZ, the protein coded for by the WWTR1 gene, and its paralog YAP are the end effectors of the Hippo pathway (Piccolo et al. 2014). The main function of the Hippo pathway is to restrict YAP/TAZ localization to the cytoplasm where they are degraded (Zhao et al. 2007). As YAP/TAZ function primarily as coactivators of gene transcription, regulation of their localization is a key mechanism for modulating their biologic function (Kanai et al. 2000; Huang et al. 2005). As such, constituent serine/threonine kinases of the Hippo pathway act to inhibit YAP/TAZ nuclear translocation by phosphorylation, yielding both cytoplasmic retention by binding to 14-3-3 proteins and proteolytic degradation via polyubiquitinylation (Kanai et al. 2000; Liu et al. 2010). We have previously demonstrated the mechanism by which the TAZ-CAMTA1 fusion protein induces cell transformation in vitro is via its ability to function as a dysregulated TAZ-like protein (Tanas et al. 2016).
In a “Hippo off” state, whereby Hippo kinases are inactivated, YAP/TAZ remain unphosphorylated and are shuttled to the nucleus where they bind to transcription factors to alter gene transcription (Liu et al. 2010; Boopathy and Hong 2019). Importantly, the major transcriptional targets of YAP/TAZ are protumorigenic and enhance cell proliferation, survival, and motility (Cordenonsi et al. 2011; Zanconato et al. 2016). Furthermore, as YAP/TAZ are major drivers of chemotherapy resistance, metastasis, and cancer stem cell phenotypes in many cancers, these transcriptional coactivators are being investigated broadly as therapeutic targets (Zanconato et al. 2016; Warren et al. 2018).
Mechanistically, when compared with wild-type TAZ, we have previously demonstrated that TAZ-CAMTA1 gains a C-terminal nuclear localization signal from CAMTA1, which drives aberrant nuclear localization of the fusion protein (Tanas et al. 2016). Furthermore, expression of TAZ-CAMTA1 in NIH3T3 and HEK293 cells yields an up-regulation of a TAZ-like gene expression profile (Tanas et al. 2016). In addition, we found that TAZ-CAMTA1 relies on binding to the TEAD family of transcription factors (TEADs 1–4), DNA-binding proteins that tether YAP/TAZ to genic regulatory elements and allows them to function as transcriptional coactivators (Tanas et al. 2016). Overall, the current working model for the mechanism of this fusion protein is that the fusion of TAZ to CAMTA1 results in a constitutively activated TAZ-like protein (Tanas et al. 2016; Lamar et al. 2018).
While the WWTR1-CAMTA1 gene fusion is present in the majority of human EHE tumors and in 45% of cases is the only genetic abnormality, the ability of the resultant TAZ-CAMTA1 protein to sufficiently promote tumorigenesis in vivo has remained an open question. Addressing this and other important biological questions related to EHE has been hampered by a complete lack of EHE cell lines and mouse models. Therefore, we sought to develop a physiologically relevant EHE mouse model that will also serve as a platform to study other important biological questions in EHE and YAP/TAZ/Hippo pathway signaling. Here, we describe the successful development of such a model of EHE, define an EHE-specific gene signature, and also provide insights into the EHE cell of origin.
Results
Design and development of a conditional Wwtr1-Camta1 knock-in mouse allele
In order to model EHE in vivo, we sought to generate a conditional mouse in which the Wwtr1-Camta1 allele replaces the endogenous Wwtr1 allele—a genetic event that is consistently observed in human EHE (Errani et al. 2011; Tanas et al. 2011; Patel et al. 2015). When designing our model, we hypothesized that embryonic expression of Wwtr1-Camta1 would be lethal, thus necessitating temporal control of the transgene. Furthermore, we aimed to maintain the genic transcriptional regulation by having the fusion gene under the control of the endogenous Wwtr1 promoter as the human WWTR1-CAMTA1 gene fusion is similarly regulated by the WWTR1 promoter. We used a first of its kind adaptation of the FLEx (flip-excision) system, which converts the Wwtr1 gene locus into a Wwtr1-Camta1 fusion gene upon the action of Cre recombinase.
To produce the FLEx cassette, a construct containing the correctly oriented wild-type third exon of Wwtr1 and an inverted portion of the murine Wwtr1-Camta1 CDS (Wwtr1 exon 3 and Camta1 exon 9 through exon 24) was created (Supplemental Fig. S1A). These exons are flanked by both conventional LoxP and modified Lox2272 sites (Fig. 1A). This cassette was targeted to the endogenous Wwtr1 locus through homologous recombination (Supplemental Fig. S1A).
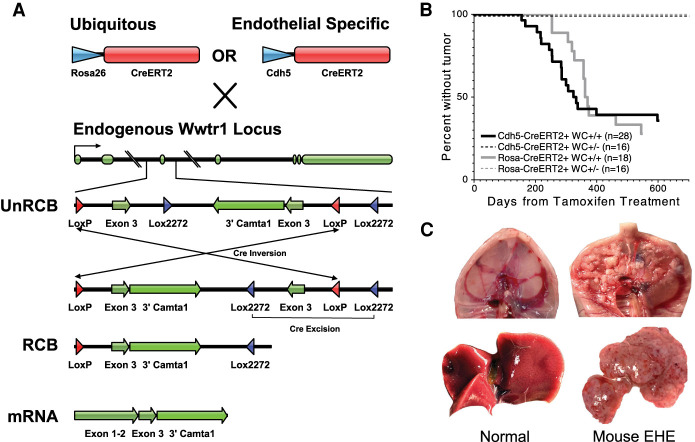
Figure 1.
The mouse Wwtr1-Camta1 model produces EHE tumors. (A) Schematic of the mouse model locus and CreERT2 crosses. (UnRCB) Unrecombined allele, (RCB) recombined allele, (exons 1–2) Wwtr1 exons 1–2, (exon 3) Wwtr1 exon 3, (3′Camta1) Camta1 exon 9-stop codon in exon 24 adjoined to a hGH terminator sequence). (B) Kaplan–Meier curve for the development of EHE subdivided based on the number of WC alleles and CreERT2 driver. No tumors developed after tamoxifen treatment in mice with only one WC fusion gene allele and one wild-type Wwtr1 allele. (n) Total number of mice of that genotype. (C) Representative gross pathology images of mouse EHE tumors (right) with corresponding normal tissue for comparison (left). (Top) Peritoneal surface/diaphragmatic lesions (CWC). (Bottom) Liver lesions (RWC).
Expression of Cre mediates an inversion via the inward facing loxP sites. This flips the previously inverted Wwtr1 (exon 3)-3′ Camta1 fusion cDNA into the Wwtr1 exon 3 position. As a result of the inversion event, the Lox2272 sites are oriented in the correct configuration to allow for the excision of the wild-type Wwtr1 exon 3. Finally, this excision event makes the whole process irreversible and prevents further inversions. Importantly, it is also possible that the initial inversion event occurs with the Lox2272 sites. However, as the Lox2272 inversion orients the LoxP sites available for Cre-Lox excision, the end result yields an equivalent recombined allele (Supplemental Fig. S1B). During mRNA transcription, the first two exons of Wwtr1 are spliced to the mutant Wwtr1 (exon 3)-3′ Camta1, thereby creating the full-length Wwtr1-Camta1 fusion transcript (Fig. 1A).
One concern when using this model at a genic locus is that the addition of the FLEx cassette would lead to significant derangement in the normal splicing of the endogenous portion of the Wwtr1 gene prior to Cre-mediated recombination. When compared with the wild-type Wwtr1 allele, mice that contain one or two copies of the Wwtr1-Camta1 allele in the unrecombined position do not show any significant differences in Wwtr1 splicing at the second and third exons, suggesting that the Wwtr1-Camta1 insert does not significantly alter the post-transcriptional modification of the endogenous transcript (Supplemental Fig. S1C).
Wwtr1-Camta1 leads to postimplantation embryonic lethality
As YAP/TAZ plays a significant role in normal mammalian embryogenesis and organogenesis, we hypothesized that the Wwtr1-Camta1 fusion would lead to embryonic lethality (Varelas 2014; Zheng and Pan 2019). Mice heterozygous for the Wwtr1-Camta1 allele were paired with mice that were hemizygous for an embryonically active Cre recombinase. The progeny from this cross displayed no offspring with complete recombination of the Wwtr1-Camta1 (WC) allele (n = 38: 9 WC−/Cre−, 13 WC−/Cre+, 12 unrecombined WC+/Cre−, and 4 WC mosaic mice that contained both recombined and unrecombined alleles due to incomplete recombination by Cre). These mosaic mice were further crossed to segregate the WC recombined allele; however, no such progeny were identified. Overall, these results demonstrate that expression of Taz-Camta1 during development results in embryonic lethality.
To further identify the developmental stages that are altered by the Wwtr1-Camta1 allele, in vitro fertilization (IVF) was performed with donor oocytes from homozygous Cre transgene bearing mice and sperm from heterozygous unrecombined WC male mice. The generated blastocysts at E3.5 did not display any lethal effect of the recombined WC allele (n = 32: 13 WT, 9 unrecombined WC allele, 3 mosaic, and 7 recombined WC allele). The overall wild type to WC (both unrecombined and recombined) allelic frequencies were 41% versus 59% suggesting our observed WC-induced embryonic lethality occurs postimplantation. IVF was further repeated and the resultant blastocysts were implanted into wild-type pseudopregnant females and collected at E13.5. At E13.5, no embryos with the recombined WC allele remained (n = 31: 18 WT and 13 unrecombined WC). In sum, these findings suggest WC-induced lethality occurs between implantation and E13.5.
Wwtr1-Camta1 is sufficient to promote EHE tumorigenesis
In order to abrogate the aforementioned embryonic lethality, the WC-bearing mice were crossed with mice bearing temporally controllable CreERT2 recombinases that could be activated postdevelopment. WC mice were paired with either ubiquitously expressed Rosa26-CreERT2 [B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J, Jax] or endothelial-specific Cdh5-CreERT2 [C57BL/6-Tg(Cdh5-cre/ERT2)1Rha, Taconic] (Ventura et al. 2007; Sörensen et al. 2009). Tamoxifen was administered to Cdh5-CreERT2/WC (CWC) and Rosa26-CreERT2/WC (RWC) mice on three consecutive days at sexual maturity (7 wk) to initiate Cre-mediated recombination. After a minimum latency of 157 and 120 d, both CWC and RWC mice reliably produced tumors with a penetrance of 68% and 72.2%, respectively. For mice that develop tumors, the median time to tumor development is 285 d for CWC and 357 d for RWC mice; however the difference in time to tumor development is not statistically significant between these two models (Mantel-Cox P = 0.854) (Fig. 1B). Two copies of the WC transgene was necessary to generate tumors as no mice with one copy developed tumors (Fig. 1B).Tumor penetrance is not significantly different in males and females in both models (CWC: χ2 = 0.055, P = 0.81; RWC: χ2 = 1.23, P = 0.27).
Tumors develop along the peritoneal surface, and in the liver, lungs, and soft tissue (Supplemental Fig. S2A). Grossly, the tumors are tan and fibrotic, often with areas of punctate hemorrhage (Fig. 1C). Individual organ involvement ranges from microscopic tumors to subtotal replacement of the normal tissue parenchyma. Peritoneal surface tumors develop as small tan to erythematous confluent masses along the entire abdominal surface of the diaphragm and the retroperitoneum, often encasing abdominal organs including liver, kidney, pancreas, and spleen (Fig. 1C). Importantly, these anatomic locations correspond closely to those seen in human EHE (Sardaro et al. 2014).
WC mouse tumors are histologically identical to human EHE
As human EHE displays a highly distinctive histologic phenotype that differentiates it from the large constellation of vascular tumors including angiosarcomas, other hemangioendotheliomas, hemangiomas, etc., replication of this phenotype in a mouse model is essential for modeling EHE (Makhlouf et al. 1999). Human EHE is characterized by cords and nests of small epithelioid cells set in a distinctive myxohyaline stroma (Makhlouf et al. 1999). Tumor cells are often vacuolated as well. EHE can be histologically confused with poorly differentiated carcinomas, and therefore, the use of the endothelial cell markers CD31, CD34, and ERG are important in distinguishing EHE from other diagnostic mimics (Weiss and Enzinger 1982; Makhlouf et al. 1999).
Remarkably, WC mouse tumors display the characteristic cell morphology and stroma found in human EHE. Morphologically, mouse EHE displays the characteristic myxohyaline stroma and tumor cells with cytoplasmic vacuoles that are defining histological features of human EHE (Fig. 2A). These histologic hallmarks are further conserved across all anatomic locations (Supplemental Fig. S2B). The peritoneal based mouse EHE tumors display two distinct morphological phenotypes: (1) with microscopic papillary fronds emanating from the tumor base, or (2) as flat well-defined lesions (Fig. 2B). Finally, these tumors stain strongly and diffusely for CD31, CD34, and ERG by immunohistochemistry, which is identical to what is seen in human EHE (Fig. 2C). The use of CAMTA1 immunohistochemistry is often used diagnostically to confirm the diagnosis of EHE as polyclonal antibodies directed at CAMTA1 react with CAMTA1 component of TAZ-CAMTA1, showing a primarily nuclear staining pattern (Shibuya et al. 2015). This was also seen in mouse EHE (Fig. 2C).
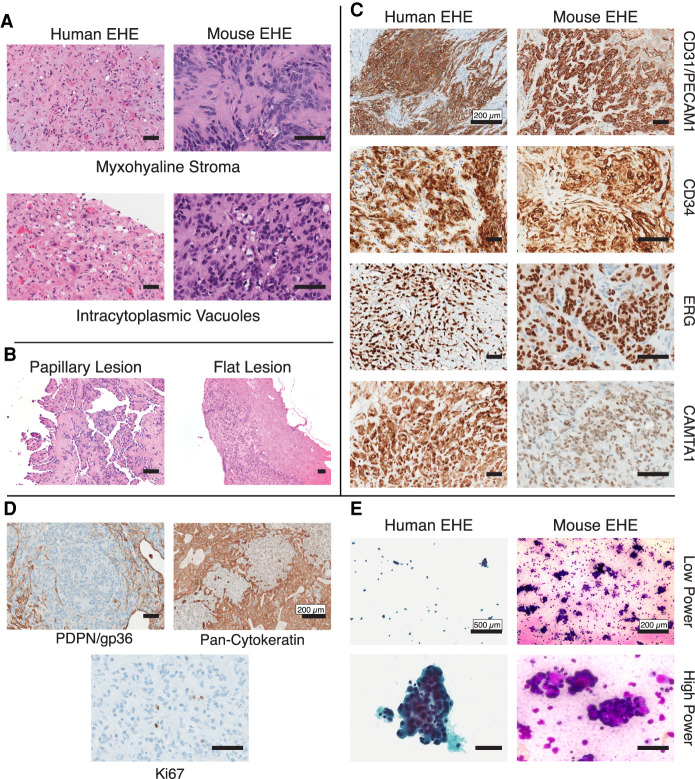
Figure 2.
Mouse EHE is histologically identical to human EHE. (A) Representative H&E photomicrographs of both human and mouse EHE displaying the hallmark histologic features of EHE myxohyaline stroma and intracytoplasmic vacuoles. Scale bars, 50 µm. (B) Representative H&E photomicrograph of murine EHE displaying the two morphological phenotypes of peritoneal surface tumors. Scale bars, 50 µm. (C) Immunohistochemical staining of mouse and human EHE for defining IHC stains. The primary antibodies used are listed at the right. Scale bars, 50 µm unless specifically listed. (D) Immunohistochemical staining of mouse EHE for the defining IHC stains. The primary antibodies used are listed below. Scale bars, 50 µm unless specifically listed. (E) Cytology of human and murine EHE at low and high magnification (Left) Human EHE from a pleural effusion (Papanicolaou stain). (Right) Cytopathology of malignant ascites from mouse EHE (Diff Quik staining). Scale bars, 50 µm unless specifically listed.
We have further observed these tumors have a predilection for growing along and within lymphatic vessels (Supplemental Fig. S2C). However, these tumors do not express the characteristic lymphatic endothelial marker, PDPN, suggesting they are not of mature lymphatic origin (Fig. 2D). Mouse EHE did not stain for cytokeratins, which is seen occasionally in human EHE (Fig. 2D). Finally, in comparison with more aggressive vascular tumors, EHE displays a low Ki67 index (Shiba et al. 2018). Concordantly, mouse EHE displays a similarly low mitotic index with low frequency of Ki67-positive cells (1%–3%) (Fig. 2D).
Similar to humans with peritoneal involvement by EHE, both CWC and RWC mice develop malignant ascites (Lau et al. 2011). These mice can present with severe abdominal distension from ascites, which ranges in color from milky white to turbid red–brown due to the presence of erythrocytes. This ascites fluid contains numerous clusters of malignant-appearing cells that display cytologic morphology similar to that of human EHE (Fig. 2E; Jebastin Thangaiah et al. 2020).
Murine EHE recapitulates the human EHE transcriptional landscape
To further validate our murine genetically engineered mouse model (GEMM) of EHE, we sought to determine whether the defining transcriptional features of human EHE are replicated in our mouse model. First, we identified a human EHE-specific gene signature by performing a cross endothelial tumor analysis in which we compared human EHE with other malignant vascular sarcomas. We performed whole transcriptome analysis of human EHE (n = 6) and used publicly available transcriptomes from angiosarcoma (AS; n = 24), hemangioblastoma (HAB; n = 5), and Kaposi sarcoma (KS; n = 4) for comparison (Tso et al. 2018; Lesluyes et al. 2019; Wang et al. 2021). Principal component analysis of these tumors demonstrates that each tumor forms their own independent cluster (Fig. 3A).
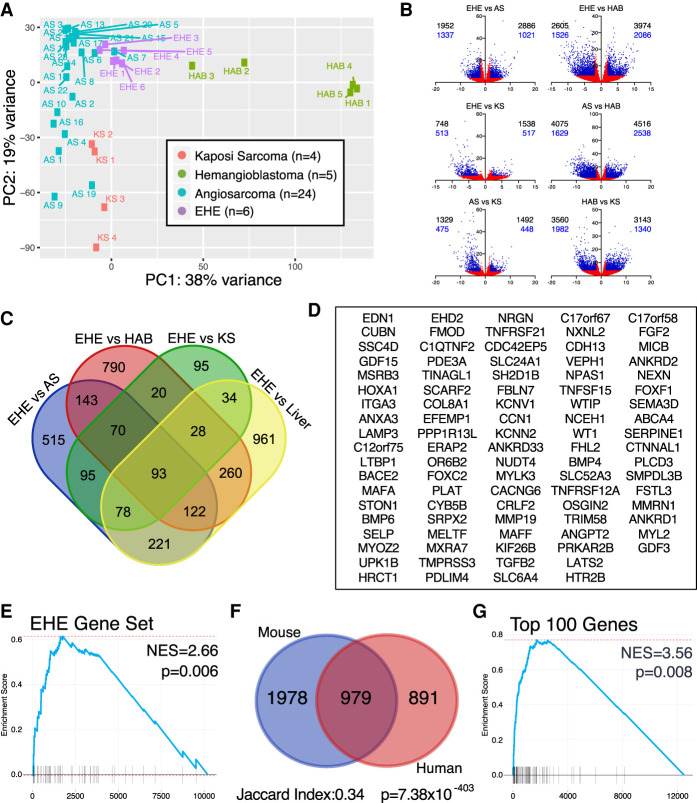
Figure 3.
Mouse EHE recapitulates the human transcriptional targets. (A) Principal component analysis of endothelial tumors via pairwise comparisons in DESeq2. (B) Volcano plots of pairwise comparisons between endothelial tumors. (Black numbers) All transcripts, (blue numbers) protein-coding genes, (blue points) log2FC ≥ 2 or log2FC ≤ −2 and FDR ≤ 0.05, (red points) −2 < log2FC < 2 or FDR > 0.05. (C) Venn diagram of genes overexpressed in EHE compared with the three other endothelial tumors and control liver; overexpressed is defined as log2FC ≥ 2 and FDR ≤ 0.05 from DESeq2. (D) Gene set of the 93 genes enriched in human EHE when compared with the three additional endothelial tumors and liver from Figure 4B. (E) GSEA shows that the EHE-specific gene set (90 mouse orthologs of the 93 EHE-specific human genes) is enriched in mouse EHE tumors. GSEA was performed on the gene set obtained after pairwise comparison of mouse EHE with control mouse livers. (NES) Normalized enrichment score. (F) Venn diagram showing the overlap of overexpressed genes in mouse and human EHE tumors compared with their control liver samples. Overexpression cutoff is defined as log2FC ≥ 2 and FDR ≤ 0.05. Jaccard index and hypergeometric P-values for enrichment are listed. (G) GSEA of mouse EHE transcripts in comparison with control mouse liver for the top 100 most overexpressed genes in human EHE in comparison with control human livers. (NES) Normalized enrichment score.
Pairwise comparisons between each tumor type were performed. These comparisons identified 1952, 2605, and 748 transcripts (of which 1337, 1526, and 513 were protein-coding genes) that were overexpressed in EHE (log2FC ≥ 2 and FDR ≤ 0.05) when compared with AS, HAB, and KS, respectively (Fig. 3B). Protein-coding genes that were overrepresented in EHE in all three of these comparisons were identified, thus yielding a set of 163 genes. However, this set is enriched in hepatocyte-specific transcripts, likely due to contamination by hepatic cells since the tissue of origin for the majority of the tumors is liver. To subtract liver tissue-specific transcripts, a four-way pairwise comparison was further performed that includes the three transcriptomes and control liver tissue resulting in a 93-EHE-specific gene set (Fig. 3C,D). Indeed, this gene set includes multiple well-validated YAP/TAZ target genes including CCN1 (CYR61), TGFB2, FGF2, and SERPINE1, among others (Cordenonsi et al. 2011).
Having identified a unique EHE-defining gene signature, we then aimed to test whether murine EHE mimics the transcriptional profile of human EHE. Murine EHE tumor samples were isolated and whole transcriptome RNA sequencing was performed. From the human EHE-specific gene set, we generated a mouse gene set of 90 mouse orthologs that correlated with the 93 human genes. We subsequently performed GSEA on mouse EHE in comparison with control liver. Our data indicate that our disease-defining human EHE transcripts are similarly enriched in murine tumors (NES = 2.66 P = 0.006) (Fig. 3E). To further demonstrate that murine EHE is not similar to the other endothelial tumors, we tested whether the signatures that are specific to other endothelial tumors are enriched in murine EHE. Consistently, mouse EHE demonstrated no enrichment for the AS- or KS-specific gene sets and demonstrated negative enrichment for the HAB-specific transcripts (Supplemental Fig. S3A). Altogether, our results demonstrate that our murine EHE tumors are not just endothelial tumors, but are transcriptionally defined by the same unique features that are characteristic of human EHE.
While the cross-tumor analysis demonstrates that murine EHE replicates the defining features of human EHE compared with other endothelial tumors, it is similarly necessary to demonstrate that the global transcriptional landscape of EHE is conserved in our model. To perform this validation, we used a bioinformatic technique that has reliably validated other mouse sarcoma models. This technique compares differential gene expression of a known human tumor against a human control tissue versus a candidate mouse tumor against the same control tissue in mice (Haldar et al. 2007; Goodwin et al. 2014). Testing whether transcriptional differences are conserved offers the ability to globally assay whether the transcriptional profiles of the tumors from different species are similar.
When compared with control livers, the genes overexpressed in human EHE showed significant overlap with the genes overexpressed in mouse EHE (Jaccard index 0.33, hypergeometric P = 1.78 × 10−359) (Fig. 3F). Furthermore, a gene set was created using the top 100 overexpressed genes in human EHE compared with human liver. Murine EHE demonstrated significant enrichment for these targets on GSEA (Fig. 3G). Finally, individual expression fold-changes of EHE over control liver correlated well between human and mouse samples (Pearson r = 0.538) (Supplemental Fig. S3B). Overall, the mouse EHE transcriptome displays significant similarity with human EHE at a global transcriptional level, conclusively demonstrating that the transcriptional identity present in human EHE is conserved in our EHE GEMM.
EHE shows enrichment in YAP/TAZ target genes and YAP/TAZ related pathways
Based on our previous work, we hypothesize that TAZ-CAMTA1 behaves like a dysregulated form of TAZ that drives expression of tumorigenic YAP/TAZ downstream targets (Tanas et al. 2016). However, the enrichment in YAP/TAZ target genes and pathways regulated by YAP/TAZ in human EHE has never been investigated.
When compared with the three other endothelial tumors described above, EHE is enriched in multiple canonical cancer-related YAP/TAZ targets including CCN1, SERPINE1, TGFB2, CRIM1, AMOTL2, and ANKRD1, among others (Fig. 4A; Cordenonsi et al. 2011). In addition, gene set enrichment analysis data indicate that the Cordenonsi signature of YAP/TAZ targets is more enriched in human EHE than in other endothelial tumors (Fig. 4B; Cordenonsi et al. 2011). Altogether, these findings demonstrate that human EHE is enriched in YAP/TAZ target genes.
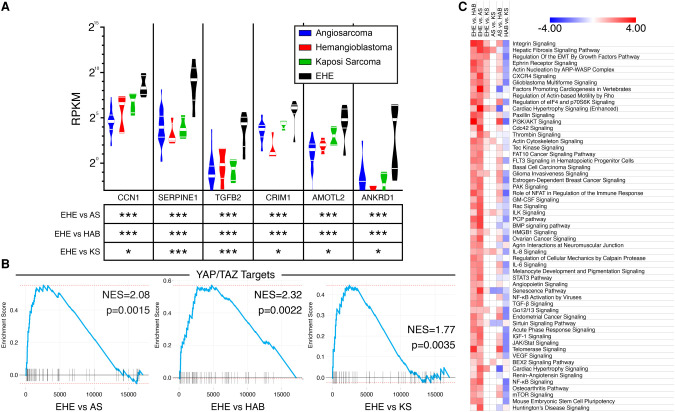
Figure 4.
Cross endothelial tumor analysis demonstrates enrichment in both YAP/TAZ targets and novel disease-defining pathways in EHE. (A) Violin plots of reads per kilobase per million mapped reads (RPKM) of canonical cancer-related YAP/TAZ target genes in different endothelial tumors. (Thin white bars) 25th percentile and 75th percentile, (thick white bar) median. The table shows false discovery rates from pairwise comparisons of EHE versus other endothelial tumors by DESeq2. (*) FDR < 0.05, (***) FDR < 0.001. (B) GSEA of YAP/TAZ target genes (Cordenonsi YAP targets) from pairwise comparisons of human EHE versus other endothelial tumors. (NES) Normalized enrichment score. (C) Heat map of Z-scores of enriched canonical pathways in EHE versus other endothelial tumors from IPA. Pathways are ranked by the sum of Z-scores across all three comparisons. Color key is listed above. Heat map is truncated. For the full list refer to Supplemental Figure S4.
To evaluate whether pathways enriched in human EHE are regulated by YAP/TAZ, we performed ingenuity pathway analysis (IPA). We found that EHE is most highly enriched in integrin, hepatic fibrosis, regulation of EMT (epithelial–mesenchymal transition) by growth factor, and actin nucleation by ARP-WASP complex signaling pathways (Fig. 4C). Identification of enrichment of these signaling pathways in EHE supports the hypothesis that EHE is a YAP/TAZ-driven cancer because previous studies have shown that many of these pathways are associated with YAP/TAZ activation or are direct transcriptional targets of YAP/TAZ signaling (Totaro et al. 2018). Furthermore, the genes enriched in EHE were similarly transcriptionally regulated by the primary YAP/TAZ binding partners (TEADs 1–4) by IPA upstream analysis (Supplemental Fig. S4). Further validating this cross-tumor analysis, similar analyses were performed on the other endothelial tumors we evaluated, which accurately identified key pathways that mechanistically drive tumorigenesis in those cancers (Supplemental Fig. S4).
Interestingly, when analyzing regulators that influence the EHE signature, in addition to TEAD1-4, components of transforming growth factor β (TGFβ) and bone morphogenetic protein (BMP) signaling cascades displayed significant enrichment (Supplemental Fig. S4). Concordantly, TGFB2, BMP4, and BMP6 were identified in the EHE defining gene set and BMP and TGFβ signaling were similarly enriched in the canonical pathway analysis. As TGFβ and BMP molecules are both known upstream regulators of YAP/TAZ nuclear localization and downstream targets of YAP/TAZ transcriptional activity, this signaling pathway may play an unexplored role in EHE tumorigenicity (Cordenonsi et al. 2011; Lai and Yang 2013; Noguchi et al. 2018).
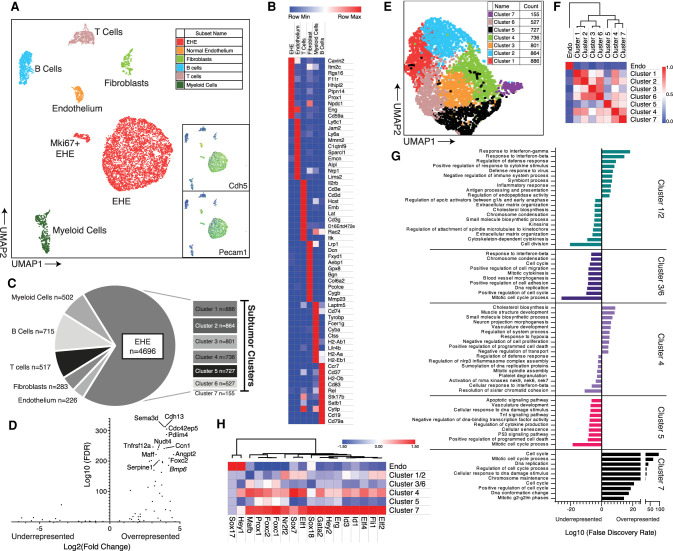
Single-cell analysis of EHE reveals enrichment in Taz-regulated transcripts and cellular pathways in EHE tumor cells
Single-cell RNA sequencing was performed on two diaphragmatic EHE tumors from the CWC model. Dimensionality reduction using principal component analysis (PCA)-guided uniform manifold and projection (UMAP) was performed, which identified seven cell populations (Stuart et al. 2019). Among these seven populations, two EHE cell populations were identified based on Cdh5, Pecam1, and Wwtr1 positivity, a large population with low Mki67 expression, and a much smaller mitotically active population that is Mki67-positive (Mki67+ EHE) (Fig. 5A). The other five cell populations identified were mature endothelial cells (Sparcl, Nrp1, and Emcn), T cells (Il2rb and Cd3 subunits), fibroblasts (Dcn, Col6a2, and Col1a1), myeloid cells (Tyrobp, H2-Ab1, and Cd74), and B cells (Cd19 and Cd79a), which were annotated based on the expression of the defining markers (Fig. 5B). Leukocytes, particularly B cells, accounted for the largest proportion of nontumor cells (Fig. 5C).
Figure 5.
Single-cell RNA sequencing of mouse EHE. (A) 2D UMAP plot of cells of filtered cells with cell population annotations. Insets demonstrate expression of Cdh5 and Pecam1. Color scale: high to low is shown by red to orange to yellow to green to blue. (B) Gene expression heat map of the top 10 most overexpressed genes from each group ranked by FDR values. Color key is listed above. (C) Pie chart of annotated cell populations with corresponding counts. The breakout chart shows counts of each of the individual tumor clusters as identified in E. (D) Volcano plot of EHE-defining genes from Figure 3D in EHE tumor cells. The top overenriched cell targets are labeled. (E) 2D UMAP plot of EHE cells in a tumor subgroup analysis, color-coded based on Seurat cluster. (F) Highly dispersed gene-guided similarity plot with unsupervised hierarchical clustering of endothelium and the seven tumor clusters from Seurat and displayed in E. (G) Enrichment plots of the top 10 most significantly enriched pathways (gene ontology, reactome, and KEGG) based on the top 500 significantly overexpressed and underexpressed genes in each cluster by differential gene expression (iCellR). Bars pointing to the right signify enrichment in the overexpressed gene set, and bars pointing to the left signify enrichment in the underexpressed gene set. (H) Gene expression heat map of normal endothelium and the tumor clusters for known transcription factors that regulate endothelial differentiation. Genes are aligned by unsupervised hierarchical clustering with the associated dendrogram. Color key listed is above.
To determine whether the EHE-defining gene set (93 genes) is enriched in the EHE tumor cell populations, both large and small, targeted expression profiling was performed between EHE and other cell populations, which demonstrated strong expression of the genes from the EHE gene set, most notably those genes that are canonical YAP/TAZ targets like Ccn1 and Serpine1 (Fig. 5D).
Having demonstrated above that the transcriptional identity of human EHE is maintained in our EHE GEMM, we aimed to determine whether EHE tumor cells in our model similarly display the YAP/TAZ signature observed in our cross-tumor analysis. In total, 4696 EHE cells were identified and used for subsequent analysis. In comparison with normal endothelium, 193 transcripts were overexpressed in EHE (FDR < 0.05, log2FC ≥ 1.7). Concurrent with the results from both our human bulk RNA-seq, EHE tumor cells are enriched in canonical YAP/TAZ targets by overrepresentation analysis (P = 3.09 × 10−5). To further identify whether YAP/TAZ-regulated pathways are similarly enriched, functional annotation of the overexpressed gene set was performed with the gene ontology and hallmark gene sets to identity functional pathways that are enriched in murine EHE tumor cells. EHE tumor cells demonstrated significant enrichment in blood vessel morphogenesis, positive regulation of cell migration, response to wounding, and regulation of cell adhesion by gene ontology and demonstrated enrichment in epithelial-to-mesenchymal transition, and hypoxia signaling by hallmark gene set analysis (Supplemental Fig. S5A). Importantly, these pathways have been previously demonstrated to be significantly regulated by YAP/TAZ activity (Mason et al. 2019). Overall, these results demonstrate that Taz-Camta1 induces a YAP/TAZ like transcriptional profile within tumor cells, further confirming our proposed mechanism of EHE tumorigenesis.
Intercluster analysis of EHE tumor cells and endothelial cells demonstrates multiple tumor cell subpopulations and an endothelial progenitor cell-like phenotype of EHE tumor cells
With the proliferation of single-cell RNA sequencing studies, there has been growing evidence that there are diverse tumor cell types, and the differentiation of the cell populations in the tumor into just tumor cells and stromal cells is oversimplistic (Patel et al. 2014). To further understand this diversity within our mouse model and to elucidate signaling cascades that are differentially enriched in tumor cells, we subclustered our EHE cell population with Seurat. This yielded seven total cell clusters (Fig. 5E). A similarity matrix of the generated EHE clusters and normal endothelium was created (Fig. 5F). While there was transcriptional diversity among tumor cell populations, each group was equally dissimilar to control endothelium, suggesting that the diversity of tumor clusters is not driven by a differentiation trajectory that incrementally transforms a normal endothelium to a malignant cell (Fig. 5F). Furthermore, subclusters 1 and 2, and 3 and 6 showed significant similarities and were thus grouped together for further analysis. Differential gene expression analysis was performed between these five groups (subclusters 1/2, 3/6, 4, 5, and 7) and the top 500 significantly overexpressed and underexpressed transcripts were functionally annotated by gene ontology (Fig. 5G). Of the five clusters analyzed, three had unique biological process signatures. Cluster 1/2 demonstrated a significant enrichment in targets of interferon signaling, specifically IFN-γ and IFN-β. Subcluster 4 demonstrated significant enrichment in morphogenic and developmental pathways in muscle and vasculature. Finally, subcluster 7 displayed profound enrichment in cell cycle/mitotic processes and this cluster corresponds to the Mki67+ EHE cells in the original analysis. In contrast, subclusters 3/6 and 5 functional annotations were primarily driven by their dissimilarity to the other subclusters and did not demonstrate their own unique enrichment profile. Overall, these results add to our understanding of the tumor cell heterogeneity within EHE and pathways that regulate its diversity.
As some of the top overexpressed genes between EHE and endothelial cells were transcription factors that regulated endothelial cell differentiation, most notably Prox1, which is a major transcription factor that drives the differentiation of lymphatic endothelial cells, we sought to identify other known transcription factors that similarly regulate endothelial cell differentiation and fate (Hong et al. 2002; Wigle et al. 2002). Of note, many of these developmental regulatory transcription factors were demonstrated to be enriched in endothelial precursor cells (Yu et al. 2016). Differential gene expression was performed with endothelial cells and the five subclusters of murine EHE cells. When compared with control endothelium, there was an overall increased expression of these transcriptional regulators in the EHE subclusters, subclusters 7 and 4 in particular (Fig. 5H). As transcription factor regulation is tightly controlled due to the fact that small changes in transcription factor abundance produces large changes in global gene expression, these multiple changes, in concert, suggest a profound alteration in cell identity. The EHE transcription factor expression signature similarity to endothelial progenitor cells demonstrates that EHE cells exhibit an endothelial precursor cell phenotype (Yu et al. 2016; Niwa 2018).
Discussion
We present here the first EHE mouse model that reliably reproduces the salient features that define human EHE. Tumors occur at similar anatomic locations and are histologically identical to human EHE. We observed significant overlap of tumor-specific genes between murine and human EHE, indicating that they have similar transcriptional profiles. Our model establishes WWTR1-CAMTA1 as the sole necessary oncogenic driver in EHE, indicating that EHE is “addicted” to TAZ-CAMTA1, and that TAZ-CAMTA1 is a strong therapeutic target for the treatment of EHE.
One striking finding from modeling EHE in mice is that, in utero, universal activation of the Wwtr1-Camta1 locus yields postimplantation embryonic lethality at the time of organogenesis, while adult induction of the Wwtr1-Camta1 leads to EHE exclusively. Given that the Taz-Camta1 protein induces a protumorigenic YAP/TAZ transcriptional signature, one might expect that the ubiquitous presence of the Wwtr1-Camta1 allele might yield multiple different types of cancers in the RWC model (Cordenonsi et al. 2011; Tanas et al. 2016; Zanconato et al. 2016). However, no such phenotypes were observed. This leads us to conclude that the transformative effect of Wwtr1-Camta1 is either permissive only in endothelial cell lineage or its expression is very restricted to this cell lineage, which results in EHE exclusively. Indeed, WWTR1-CAMTA1 gene fusion is not found in any other human neoplasms (Tanas et al. 2011; Lamar et al. 2018).
Our hypothesis for the mechanism that dictates this specificity is via the epigenetic control of the endogenous Wwtr1 promoter. At physiological states, endothelial cells from different anatomic locations disproportionately possess the highest expression of Wwtr1 mRNA transcript based on single-cell RNA sequencing in mice (Supplemental Fig. S5B; The Tabula muris Consortium 2018). Coupled with the finding that two alleles of the Wwtr1-Camta1 locus are necessary in our mouse model to produce tumors, the tissue specificity in the RWC model is likely determined by a gene dosage effect, whereby transformation occurs only when there are sufficient levels of Wwtr1-Camta1 transcripts. As human EHE tumors are indolent and slow growing, a significant lead time is necessary to become macroscopically evident. As such, it is unsurprising and has been observed in our previous studies that in order to induce tumorigenesis in mice, larger gene dosage, via a second allele, appears to be necessary (Rubin et al. 2005). Another less probable hypothesis is that wild-type Taz in the heterozygous mice may inhibit tumorigenesis via an unexplored mechanism (i.e., competition for Tead binding, down-regulation of Wwtr1-Camta1 transcription, etc).
In this study, we needed to develop a mouse model that targeted the Wwtr1-Camta1 gene fusion to the endogenous Wwtr1 locus, such that the wild-type Wwtr1 locus could be converted to a Wwtr1-Camta1 gene fusion locus after embryogenesis, since we hypothesized that expression of Taz-Camta1 during embryogenesis was likely to be lethal, which did indeed turn out to be the case. We achieved our experimental aim by developing a first of its kind adaptation of the FLEx system. The FLEx system has been previously described as a genetic tool kit to expand the utility of the Cre-Lox system beyond the conventional Lox-Stop-Lox and exon deletion modalities (Schnütgen et al. 2003). Similar to conventional Cre-Lox models, the FLEx system maintains temporal control of Cre-mediated recombination when paired with an estrogen response element-Cre and tissue specificity when paired with a tissue-specific Cre. In addition, the FLEx system allows for exon replacement to either knock in a mutation or perform a complex knockout, whereby Cre activation yields loss of a genic exon that is replaced by a reporter gene (Schnütgen et al. 2003). While conventional Cre-Lox methods allow for inversion of transgenes using inward facing LoxP sites, inversion is reversible, especially when used with a non-tamoxifen-inducible Cre recombinase, as inversion events will continue indefinitely so long as Cre recombinase is active. As the FLEx system contains an irreversible step that “locks” the fusion in the recombined position, no further inversion events can occur. As a model that maintains a gene fusion under the control of the endogenous promoter serves as a better biologic replication of a human disease, this utilization of the FLEx system serves as an important proof of concept for a robust method of developing models of gene fusion-driven cancers. Presumably, this could be done with virtually any cancer that is thought to be driven by a single gene fusion.
Previously, using NIH3T3 cells and transient WWTR1-CAMTA1 overexpression, we demonstrated that the transcriptional profile induced by TAZ-CAMTA1 is statistically more similar to that of TAZ than CAMTA1 (Tanas et al. 2016). However, it was previously unknown whether human EHE had similar enrichment of TAZ signature genes. Here, through cross-vascular tumor analysis, we demonstrate a global enrichment in YAP/TAZ target genes and describe a gene signature that is specific to EHE that is not representative of other tumors of endothelial origin. This EHE-specific 93-gene set is further enriched in specific YAP/TAZ target genes that are known to be protumorigenic in other cancers. Overall, our results suggest that the therapies that inhibit YAP/TAZ activity could also be used to therapeutically target EHE.
In addition to identifying YAP/TAZ targets as highly enriched in EHE, BMP and TGFβ signaling were consistently found to be enriched in the EHE-defining gene set, in proteins that regulate the EHE transcriptional profile, and in genes up-regulated in EHE versus endothelial cells. These signaling molecules are well known secretory proteins from cancer cells that modulate cancer-associated fibroblast activity. Specifically, TGFβ is necessary for the differentiation of immature fibroblasts into highly active myofibroblasts (Evans et al. 2003). As other signaling molecules that either promote the activation and maturation of resident fibroblasts (FMOD, FGF2, and EDN1) or directly act to prevent ECM turnover (SERPINE1) were similarly enriched, the tumor cell/CAF/ECM axis presents an interesting pathway for future investigation (Kubala and DeClerck 2019; Pourhanifeh et al. 2019). Consistently, this enrichment of profibrotic targets correlates with the fact that human EHE tumors are densely fibrotic and microscopically contain a characteristic and abundant myxohyaline stroma (Fig. 2A).
Aided by the findings in Driskill et al. (2021) that demonstrate that the endogenous regulators of YAP/TAZ still have some functional role in modulating TAZ-CAMTA1 activity, we hypothesize that EHE's ECM may play a role in modulation of TAZ-CAMTA1 activity. One of the main mechanisms that regulate YAP/TAZ nuclear translocation is that of ECM stiffness via a well-described mechanotransduction pathway (Dupont et al. 2011). This pathway involves membrane integrins that activate Rho signaling, yielding F-actin polymerization. The end product of this cascade leads to the dephosphorylation of YAP/TAZ and YAP/TAZ transcriptional activity. As a stiff ECM has been shown to activate this cascade and a soft ECM has been demonstrated to inhibit this cascade, we hypothesize that the ECM may present a hospitable tumor niche that promotes TAZ-CAMTA1 transcriptional activity (Dupont et al. 2011). As many of the TAZ-CAMTA1 transcriptional targets that we identified in human EHE are direct stimulators of ECM production, we further hypothesize that this constitutes a feed-forward mechanism that promotes EHE tumorigenesis. This pathway involves TAZ-CAMTA1 up-regulation of key mediators that promote the activation of fibroblasts and increasing ECM stiffness with further augmentation of TAZ-CAMTA1 transcription activity. This feed-forward mechanism has been previously demonstrated with endogenous YAP/TAZ and has been implicated in many different cancers as well as fibrotic diseases of the lung, kidney, skin, and liver (Noguchi et al. 2018). However, this feed-forward mechanism has never been tested in TAZ-CAMTA1 or other YAP/TAZ fusion proteins such as are seen in cervical carcinoma (YAP1-SS18), pediatric CNS tumors (YAP1-MAMLD1 and YAP1-FAM118B), poroma and porocarcinoma (YAP1-MAML2 and YAP1-NUTM1), or pseudomyogenic hemangioendothelioma (TAZ-FOSB) (Hu et al. 2018; Panagopoulos et al. 2019; Sekine et al. 2019; Jünger et al. 2020; Schieffer et al. 2020)
Another fascinating finding in this study is that EHE tumor cells have an endothelial precursor cell phenotype as supported by the single-cell sequencing experiments where we identified multiple endothelial-specific developmental regulatory transcription factors in EHE cells. This has specific relevance to determining the identity of the cell of origin that gives rise to EHE. Broadly, two hypotheses have been posited that link the prospective cell of origin to the early steps of tumor initiation: a dedifferentiation hypothesis and a stem cell origin hypothesis (Reya et al. 2001; Friedmann-Morvinski and Verma 2014). In the first, mature differentiated endothelial cells undergo tumorigenesis as a result of the translocation/gene fusion and are able to re-enter the cell cycle (Friedmann-Morvinski and Verma 2014). Inherently, this would require activation of pioneer transcription factors and chromatin remodelers to make drastic changes in promoter accessibility of key mitotic genes (Carvalho 2020). Aided by the recent findings in Merritt et al. (2020) that demonstrated Taz-Camta1's potent ability to recruit chromatin remodelers to transcriptional start sites in comparison with the constitutively active form of the wild-type Taz (Taz4SA), such an epigenetic shift might be possible. This is coupled with the fact that mature endothelium is mitotically dormant yet has proliferative potential, especially under situations of hypoxia and wound healing (Potente et al. 2011).
In contrast, the stem cell hypothesis theorizes that an endothelial precursor cell subpopulation, which is mitotically active and has suitable chromatin conformation for cellular transformation, undergoes tumorigenesis as a result of expression of one or more oncogenes (Reya et al. 2001). As is seen in many other cancers, tumorigenesis proceeds as a result of cellular transformation in the correct cell type at the correct developmental stage. It is well established that both tissue-resident lineage-committed endothelial progenitor cell populations and bone marrow-resident trilineage (arterial, venous, and lymphatic) endothelial stem cells exist (Oswald et al. 2004; Yu et al. 2016; Wakabayashi et al. 2018). Specifically, these endothelial progenitor cell populations are important in both neovascularization and in hemostasis (Yu et al. 2016; Wakabayashi et al. 2018). Similar to what we observed in EHE tumor cells when compared with control endothelium, tissue resident endothelial progenitor cells are enriched in transcription factors that regulate endothelial differentiation and lineage (Yu et al. 2016). The central question that remains unknown is whether the progenitor cell phenotype seen in EHE represents dedifferentiation from mature cells along the normal developmental trajectory, as is suggested by the former hypothesis, or is as a result of a differentiation block of a progenitor cell, as is suggested by the latter hypothesis.
Based on our current knowledge of this disease, we support a stem cell origin hypothesis. In a dedifferentiation model, it is unlikely that a single transformative event will lead to EHE formation. We know from this study that the Wwtr1-Camta1 fusion gene, on its own, can lead to EHE formation and it has been demonstrated that ∼45% of human EHE harbor only the WWTR1-CAMTA1 gene fusion (Seligson et al. 2019). Relative to mature endothelium, endothelial progenitor cells have enhanced replicative capacity and a more “primitive” stem cell-like phenotype. As no precursor lesions have been identified in humans and the single WWTR1-CAMTA1 mutation is sufficient to drive transformation, tumor initiation likely proceeds directly to tumorigenesis and exponential growth in the absence of a “priming/maturation stage” as is seen in other cancers (Fearon and Vogelstein 1990). The ability of mature endothelial cells to dedifferentiate in a single oncogenic step and proceed directly to exponential growth without a priming stage is a less likely possibility than transformation of a precursor cell.
Furthermore, EHE tumors demonstrate a uniform histologic phenotype and lack mature endothelial structures resembling blood vessels (vasoformative structures). If mature endothelial cells were the cell of origin, histologically, a variety of histological phenotypes would be expected due to the heterogeneity of cells along their dedifferentiation developmental trajectory from mature endothelial cells to primitive-stem cell like tumor cells. However, no such heterogeneity exists. This uniformity of EHE developmentally compared with control endothelium was further confirmed in our single-cell RNA seq experiments. While there was transcriptional diversity between tumor cell subclusters, each subcluster was equivalently dissimilar to control endothelium, suggesting that each cell subcluster was equivalently undifferentiated. If dedifferentiation were required to prime endothelial cells to become highly malignant, as is suggested by a dedifferentiation hypothesis, we would have expected to observe differential similarity with endothelial cells due to the nonuniformity of cells along the developmental trajectory.
We further hypothesize that the lack of apparent histological vascular differentiation is due to the fact that cells that give rise to EHE are intrinsically poorly differentiated as a reflection of their stem cell phenotype. Such stem cells would differentiate to become luminal forming mature endothelium during development. It would be reasonable to suggest that the loss of vascular differentiation is a necessary step in endothelial-specific tumorigenesis; however, angiosarcoma, a highly malignant and aggressive endothelial malignancy, retains vasogenic features, disproving this supposition. In the future, we plan to leverage the EHE mouse model described in this paper to investigate our hypothesis further.
Overall, this study presents a robust and faithful EHE mouse model, developed using an innovative genetic approach, which carefully replicates the expression of the WWTR1-CAMTA1 gene fusion in human EHE. This model demonstrates that Taz-Camta1 is sufficient to promote tumorigenesis and recapitulate all of the hallmark phenotypes of EHE. We further identify TAZ targets as highly enriched in both mouse and human EHE, proving that EHE is a cancer that is driven by dysregulation of the YAP/TAZ signaling mediated by TAZ-CAMTA1. These results also further confirm TAZ-CAMTA1 and downstream signaling as therapeutic targets in EHE. Indeed, as EHE is likely “addicted” to TAZ-CAMTA1, this tumor represents a fortunate opportunity to test therapies that target TAZ/YAP and downstream Hippo pathway mediators. Finally, we have identified that EHE cells possess an endothelial progenitor phenotype, providing strong evidence that EHE originate from a transformed endothelial precursor cell.
Materials and methods
Human samples
All human sample research was performed with the approval of the Cleveland Clinic Institutional Review Board (IRB 06-977). Samples were deidentified to the research team and were therefore exempt from participant consent for this specific study and in accordance with the HIPAA privacy rule.
Mouse strains
All mouse research was conducted with the approval of the Cleveland Clinic Institutional Animal Care and Use Committee. The Cdh5-CreERT2 [C57BL/6-Tg(Cdh5-cre/ERT2)1Rha, Taconic] and the Rosa26-CreERT2 [B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J, Jax] models have been previously described (Ventura et al. 2007; Sörensen et al. 2009). The methods for creation of the mouse bearing the WC allele are presented below. CreERT2-expressing mice were mated with WC allelic mice to heterozygosity and homozygosity for the WC allele. Control mice not treated with tamoxifen showed no abnormalities and normal fecundity. Mice were treated with tamoxifen (Sigma 74136) at 7–8 wk of age (4 mg/30 g body weight) via IP injection for three consecutive days. Genotyping primers for the WC, Cdh5-CreERT2, and Rosa26-CreERT2 alleles are listed in Supplemental Table S1.
Creation of the transgenic WC mouse allele
To generate a conditional Wwtr1-Camta1 conditional knock-in mouse allele, the flip excision (FLEx) system was used. Generation of this mouse was performed in collaboration with genOway. A targeting vector containing the correctly oriented third exon of Wwtr1 and the Wwtr1 exon 3/Camta1 exon 9-stop CDS in an inverted orientation was created. The Wwtr1 exon 3/Camta1 exon 9-stop CDS is fused to the hGH polyA terminator sequence. These coding sequences were double-flanked by two wild-type LoxP sites and two modified LoxP sites (Lox2272). This vector further contained a neomycin-negative selection cassette that was flanked by FRT excision sites. Finally, this insert was surrounded by homology domains to introns 2 and 3 to allow for homologous recombination. The generated targeting vector was then electroporated into mouse embryonic stem cells and treated with neomycin for negative selection. The generated colonies were screened for and demonstrated homologous recombination with the insertion of the FLEx cassette in the correct orientation by southern blot and PCR. These colonies were subsequently implanted into mouse E3.5 blastocysts, and the blastocysts were implanted into pseudopregnant females. The resultant chimeric mice were then bred with wild-type C57BL/6N mice to generate progeny containing the transgene allele. These progenies were then crossed with a C57BL/6N Flp deleter mouse to excise the neomycin cassette, thus yielding the mature unrecombined allele. The steps during the mouse allele generation are shown graphically in Supplemental Figure S1A.
Analysis of embryonic lethality of WC allele
To assess for overall embryonic lethality of the recombined WC allele, WC allele-bearing mice were mated with mice heterozygous for an embryonically active Cre recombinase (Cre deleter line). Resultant pups were genotyped for both the Cre recombinase, and both the unrecombined and recombined WC allele (primers are listed in Supplemental Table S1). Mosaicism for the unrecombined and recombined allele is signified by the presence of both the unrecombined and recombined alleles during genotyping due to the incomplete action of the Cre recombinase.
To assess for embryonic lethality at differing stages of development, sperm from WC heterozygous male mice were collected. In vitro fertilization (IVF) was performed with oocytes from Cre+ homozygous female mice. E3.5 blastocysts were either analyzed for the presence of the recombined and unrecombined WC allele for an early time point or implanted into pseudopregnant female mice. Pregnant female mice were then sacrificed at E13.5 to obtain a late, postimplantation, lethality time point. Fetuses from these mice were isolated and genotyped for the recombined and unrecombined WC alleles.
Histology, immunohistochemistry, and cytology
Murine histology preparations were performed with 10% formalin fixation, followed by paraffin-embedding and H&E staining. Immunohistochemistry was performed with the Roche Discovery Ultra automated stainer. Following antigen retrieval, staining was performed for Cd31 (1:50; Santa Cruz Biotechnology sc-1506-R), Cd34 (1:100; Abcam ab81289), Erg (1:250; Abcam ab92513), Pdpn (1:500; Abcam ab11936), Camta1 (1:75; Sigma SAB4301068), Ki67 (1:250; Abcam ab1580), and PanCK (1:75; Abcam ab9377). Visualization was performed by colorimetric detection with OmniMap anti-rabbit HRP (Roche 760-4311) and the ChromoMap DAB detection kit (Roche 760-1589). The Syrian hamster anti-Pdpn antibody required cross-linking with a secondary rabbit anti-Syrian hamster IgG antibody (Abcam ab6699) for use with the HRP-conjugated anti-rabbit antibody (Roche 760-4311). Slides were finally counterstained with hematoxylin and a bluing reagent.
For ascites cytology, 20 µL of ascites was smeared onto a slide and stained with the Diff-Quik stain (Jorgenson Laboratories J0322) according to the manufacturer's instructions. Slides were either scanned using a Leica slide scanner and visualized with the Leica web viewer version 12.4.3 or by bright-field microscopy with the DM6B-Z microscope using Leica application suite X, version 3.6.0.
Histology of human EHE samples were used for comparison, which had previously been both H&E-stained and IHC-stained for CD31, CD34, ERG, and CAMTA1 as a part of routine histopathologic diagnosis during clinical workup.
RNA extraction and bulk RNA sequencing
Whole RNA was isolated with the Qiagen RNeasy Plus minikit according to the manufacturer's instructions. RNA quality was verified with the Agilent 2100 bioanalyzer prior to library preparation. BGI DNB nanoball library preparation was performed. Briefly, polyA tail selection was performed with an oligo dT bead enrichment step. Selected RNA was reverse-transcribed and fragmented. The synthesized cDNA underwent end repair, and adapters were ligated to both ends of the sequence. This product was PCR-amplified and the resultant amplicon was circularized using DNA ligase and a split oligonucleotide directed at both tails of the adapters. The circularized ssDNA is replicated to generate a linear repetitive concatemer DNA nanoball. This DNA nanoball is then adsorbed onto a flow cell and sequenced on a DNBSEQ-G400 sequencer. FASTQ raw read files were obtained and transcriptomic analysis was performed as described below.
Bulk RNA sequencing transcriptome analysis
Paired-end reads were aligned to either the GRCh38.p13 (hg38) human genome or the GRCm38.p6 (mm10) mouse genome (Ensembl) with HISAT2 (Kim et al. 2019). Count files were generated with Featurecounts using the corresponding GTF references (Liao et al. 2014). DESeq2 was used for differential gene analysis while the Limma package with (<5 CPM filtering) was used for differential gene expression analysis when generating ranked gene sets for gene set enrichment analysis (Love et al. 2014). FGSEA was used for gene set enrichment using either gene sets stored in MSigDB or from gene sets developed in this paper (Korotkevich et al. 2019). T-scores from Limma were used as rank statistics for GSEA (Ritchie et al. 2015). Bioinformatic analysis was performed using the Galaxy project platform (Afgan et al. 2018).
For cross-tumor analysis, FASTQ files from GEO data sets of RNA sequencing of samples of angiosarcoma (GSE102055), hemangioblastoma (GSE148216), and Kaposi sarcoma (GSE100684) were used (Tso et al. 2018; Lesluyes et al. 2019; Wang et al. 2021). RNA sequencing of human liver transcriptomes from ENCODE were similarly used for comparisons (libraries ENCLB173ZZZ, ENCLB648FPF, and ENCLB490UAX) (Supplemental Table S2; The ENCODE Project Consortium 2012; Lin et al. 2014; Zhang et al. 2019).
Ingenuity pathway analysis on bulk RNA sequencing was performed by inputting the log2FC and FDR values for each gene across all comparisons from the cross-tumor analysis. Log2FC ≥ 2 and FDR ≤ 0.01 were used to signify differentially expressed gene filtering. Canonical pathway and upstream analyses were performed. Z-scores were then outputted for generation of pathway and regulator heat maps (Supplemental Table S2).
Both the raw and processed files from bulk RNA sequencing of human and mouse EHE have been deposited in the GEO repository (GSE168493, GSE168494). The raw and processed files from single-cell RNA sequencing of mouse EHE files have similarly been deposited in the GEO repository (GSE168354).
Cross-species analysis of human and mouse EHE
When performing the cross-species analysis, differential gene expression was performed between human EHE and human control livers and mouse EHE and mouse control livers (DESeq2). Lists of genes overexpressed in mouse and human EHE (log2FC ≥ 2 and FDR ≤ 0.05) were created (Supplemental Table S2). The murine EHE gene set was filtered for genes that have known human orthologs and the human EHE gene set was filtered for genes that have known mouse orthologs (Ensembl). A Venn diagram was then created to identify the overlap between the murine EHE overexpressed genes and the murine ortholog of the human EHE overexpressed genes. A Jaccard index and a hypergeometic test were further calculated from the overlap in this analysis. Log2 fold changes between human EHE versus human liver and murine EHE versus murine liver were further plotted on a scatter plot for each orthologous gene. A Pearson correlation coefficient was created from the human and murine log fold changes in this analysis.
Mouse orthologs from the top 100 genes overexpressed in human EHE versus human liver were further used to create a gene set. This gene set was then used for gene set enrichment analysis to probe the mouse EHE versus mouse liver comparison.
Single-cell transcriptome library creation
Two samples from the Cdh5-CreERT2+ WC+/+ mouse were used for single-cell RNA sequencing. Mice were euthanized, the abdomen was opened, and the tumor-involved area of the diaphragm was isolated. A portion of the tumor was saved for H&E staining to confirm the histologic diagnosis of EHE. The remaining tumor was quickly transferred to chilled 1× PBS and mechanically dissociated. The dissociated tissue was then centrifuged and suspended in 10% DMSO in FBS and cooled 1°C per minute to −80°C. After rewarming, single-cell suspensions were created using the MACS tumor dissociation kit. The dissociated cells were then run through the 10X Genomics library preparation platform. Barcoded library preparations were then sequenced using the DNBSEQ-G400 sequencer.
Single-cell RNA sequencing analysis
Raw barcoded reads were processed through the Cell Ranger pipeline to yield 5850 and 4108 cells from each sample. Processed h5 count files were loaded into SeqGeq for analysis. The two samples were combined into a single file for analysis. Cell filtering was performed by removing low gene-count cells and doublet cells. Dimensionality reduction and cell clustering was performed using the Seurat pipeline to produce clustered uniform manifold and projection (UMAP). Cell annotations were performed by both defining genes from Seurat and similarity matrices based on highly dispersed genes. Multigroup differential gene expression (three or more simultaneous comparisons) was performed with iCellR and pairwise differential gene expression analysis was performed with the integrated differential gene expression algorithm in SeqGeq (Mann–Whitney U-test) (Khodadadi-Jamayran et al. 2020). Overrepresentation analysis was performed both with the integrated enrichment feature in SeqGeq with gene sets from mSigDB and with Metascape (Supplemental Table S3). Enrichment plots and volcano plots were created in Graphpad Prism v8.
Graphics and statistical analysis
Gene diagrams were produced in IBS v1.0.3. All heat maps were generated using the Morpheus heat map applet (Broad Institute). Statistical analyses were performed in Galaxy, SeqGeq v1.6, or in GraphPad Prism v8. Kaplan–Meier survival analysis was performed by the log-rank test (Mantel-Cox). χ2 tests were performed where noted. P-values and FDR values of <0.05 were considered statistically significant across all analyses unless otherwise stated.
Supplementary Material
Acknowledgments
We thank Dr. Cassandra Talerico (Cleveland Clinic) for her comments on the manuscript. C.N.S. is supported in part by a Crile Research Fellowship, Cleveland Clinic, and the Department of General Surgery, Cleveland Clinic. C.N.S. is a predoctoral student in the Molecular Medicine Ph.D. Program of Cleveland Clinic and Case Western Reserve University. This work received generous financial support from the Center for Research and Analysis of Vascular Tumors Foundation (http://www.cravatfoundation.org), the Epithelioid Hemangioendothelioma Foundation (http://www.fightehe.org), the Margie and Robert E. Petersen Foundation (http://www.pgpf.org), and a VeloSano grant provided by the Cleveland Clinic. We also acknowledge the Cleveland Clinic Robert J. Tomsich Pathology and Laboratory Institute Pathology Research Core, the Lerner Research Institute Histology Core and Genomics Core, and the Biological Resources Unit. J.M.L. is independently supported by the EHE fund (provided by the Epithelioid Hemangioendothelioma Foundation [http://www.fightehe.org], EHE Rare Cancer Charity [UK] [https://www.ehercc.org.uk], and EHE Rare Cancer Charity [Australia] [http://www.ehefoundation.com.au]).
Author contributions: C.N.S. and B.P.R. conceived of the presented project. C.N.S. and A.H. carried out the animal experiments. C.N.S., A.H., S.M., and J.P.R. performed in vitro experiments. C.N.S. performed the bioinformatic analysis with the guidance of A.V.P., R.K, J.M.L., and B.P.R. Writing of the final manuscript was performed by C.N.S., A.V.P., and B.P.R. with suggestions from other authors. B.P.R. supervised the project.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348220.120.
Competing interest statement
The authors declare no competing interests.
References
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46: W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathy GTK, Hong W. 2019. Role of hippo pathway-YAP/TAZ signaling in angiogenesis. Front Cell Dev Biol 7: 49. 10.3389/fcell.2019.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J. 2020. Cell reversal from a differentiated to a stem-like state at cancer initiation. Front Oncol 10: 541. 10.3389/fonc.2020.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. 2011. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147: 759–772. 10.1016/j.cell.2011.09.048 [DOI] [PubMed] [Google Scholar]
- Driskill JH, Zheng Y, Wu B-K, Wang L, Cai J, Rakheja D, Dellinger M, Pan D. 2021. WWTR1(TAZ)-CAMTA1 Rreprograms endothelial cells to drive epithelioid hemangioendothelioma. Genes Dev (this issue). 10.1101/gad.348221.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium . 2012. an integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, Healey JH, Antonescu CR. 2011. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer 50: 644–653. 10.1002/gcc.20886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RA, Tian YC, Steadman R, Phillips AO. 2003. TGF-β1-mediated fibroblast-myofibroblast terminal differentiation—the role of Smad proteins. Exp Cell Res 282: 90–100. 10.1016/s0014-4827(02)00015-0 [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767. 10.1016/0092-8674(90)90186-i [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Verma IM. 2014. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep 15: 244–253. 10.1002/embr.201338254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin ML, Jin H, Straessler K, Smith-Fry K, Zhu J-F, Monument MJ, Grossmann A, Randall RL, Capecchi MR, Jones KB. 2014. Modeling alveolar soft part sarcomagenesis in the mouse: a role for lactate in the tumor microenvironment. Cancer Cell 26: 851–862. 10.1016/j.ccell.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. 2007. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell 11: 375–388. 10.1016/j.ccr.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Hong Y-K, Harvey N, Noh Y-H, Schacht V, Hirakawa S, Detmar M, Oliver G. 2002. Prox1 is a master control gene in the program specifying lymphatic endothelial cell fate. Dev Dyn 225: 351–357. 10.1002/dvdy.10163 [DOI] [PubMed] [Google Scholar]
- Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, Lang FM, Martinez-Ledesma E, Lee SH, Zheng S, et al. 2018. Tumorfusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 46: D1144–D1149. 10.1093/nar/gkx1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122: 421–434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Jebastin Thangaiah J, Hanley K, Nomani L, Policarpio-Nicolas ML. 2021. Cytologic features and immunohistochemical findings of epithelioid hemangioendothelioma (EHE) in effusion: a case series. Diagn Cytopathol 49: E24.–. 10.1002/dc.24565 [DOI] [PubMed] [Google Scholar]
- Jünger ST, Andreiuolo F, Mynarek M, Dörner E, Zur Mühlen A, Rutkowski S, von Bueren AO, Pietsch T. 2020. Ependymomas in infancy: underlying genetic alterations, histological features, and clinical outcome. Childs Nerv Syst 36: 2693–2700. 10.1007/s00381-020-04655-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. 2000. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 19: 6778–6791. 10.1093/emboj/19.24.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadadi-Jamayran A, Pucella J, Zhou H, Doudican N, Carucci J, Heguy A, Reizis B, Tsirigos A. 2020. Icellr: combined coverage correction and principal component alignment for batch alignment in single-cell sequencing analysis. bioRxiv 2020.03.31.019109. 10.1101/2020.03.31.019109 [DOI] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Sergushichev A. 2019. Fast gene set enrichment analysis. BioRxiv 10.1101/060012 [DOI] [Google Scholar]
- Kubala MH, DeClerck YA. 2019. The plasminogen activator inhibitor-1 paradox in cancer: a mechanistic understanding. Cancer Metastasis Rev 38: 483–492. 10.1007/s10555-019-09806-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Yang X. 2013. BMP4 is a novel transcriptional target and mediator of mammary cell migration downstream of the hippo pathway component TAZ. Cell Signal 25: 1720–1728. 10.1016/j.cellsig.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Lamar JM, Motilal Nehru V, Weinberg G. 2018. Epithelioid hemangioendothelioma as a model of YAP/TAZ-driven cancer: insights from a rare fusion sarcoma. Cancers 10: 229. 10.3390/cancers10070229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K, Massad M, Pollak C, Rubin C, Yeh J, Wang J, Edelman G, Yeh J, Prasad S, Weinberg G. 2011. Clinical patterns and outcome in epithelioid hemangioendothelioma with or without pulmonary involvement: insights from an internet registry in the study of a rare cancer. Chest 140: 1312–1318. 10.1378/chest.11-0039 [DOI] [PubMed] [Google Scholar]
- Lesluyes T, Baud J, Pérot G, Charon-Barra C, You A, Valo I, Bazille C, Mishellany F, Leroux A, Renard-Oldrini S, et al. 2019. Genomic and transcriptomic comparison of post-radiation versus sporadic sarcomas. Mod Pathol 32: 1786–1794. 10.1038/s41379-019-0300-2 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, et al. 2014. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci 111: 17224–17229. 10.1073/pnas.1413624111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yu F-X, Kim YC, Meng Z, Naipauer J, Looney DJ, Liu X, Gutkind JS, Mesri EA, Guan K-L. 2015. Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene 34: 3536–3546. 10.1038/onc.2014.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhlouf HR, Ishak KG, Goodman ZD. 1999. Epithelioid hemangioendothelioma of the liver: a clinicopathologic study of 137 cases. Cancer 85: 562–582. [DOI] [PubMed] [Google Scholar]
- Mason DE, Collins JM, Dawahare JH, Nguyen TD, Lin Y, Voytik-Harbin SL, Zorlutuna P, Yoder MC, Boerckel JD. 2019. YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J Cell Biol 218: 1369–1389. 10.1083/jcb.201806065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendlick MR, Nelson M, Pickering D, Johansson SL, Seemayer TA, Neff JR, Vergara G, Rosenthal H, Bridge JA. 2001. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am J Surg Pathol 25: 684–687. 10.1097/00000478-200105000-00019 [DOI] [PubMed] [Google Scholar]
- Merritt N, Garcia K, Rajendran D, Lin Z-Y, Zhang X, Mitchell KM, Borcherding N, Fullenkamp C, Chimenti M, Gingras A-C, et al. 2020. TAZ-CAMTA1 and YAP-TFE3 modulate the basal TAZ/YAP transcriptional program by recruiting the ATAC histone acetyltransferase complex. bioRxiv 2020.09.07.286633. 10.1101/2020.09.07.286633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. 2018. The principles that govern transcription factor network functions in stem cells. Development 145: dev157420. 10.1242/dev.157420 [DOI] [PubMed] [Google Scholar]
- Noguchi S, Saito A, Nagase T. 2018. YAP/TAZ signaling as a molecular link between fibrosis and cancer. Int J Mol Sci 19: 3674. 10.3390/ijms19113674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Boxberger S, Jørgensen B, Feldmann S, Ehninger G, Bornhäuser M, Werner C. 2004. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells 22: 377–384. 10.1634/stemcells.22-3-377 [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Lobmaier I, Gorunova L, Heim S. 2019. Fusion of the genes WWTR1 and FOSB in pseudomyogenic hemangioendothelioma. Cancer Genomics Proteomics 16: 293–298. 10.21873/cgp.20134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NR, Salim AA, Sayeed H, Sarabia SF, Hollingsworth F, Warren M, Jakacky J, Tanas M, Oliveira AM, Rubin BP, et al. 2015. Molecular characterization of epithelioid haemangioendotheliomas identifies novel WWTR1-CAMTA1 fusion variants. Histopathology 67: 699–708. 10.1111/his.12697 [DOI] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94: 1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Potente M, Gerhardt H, Carmeliet P. 2011. Basic and therapeutic aspects of angiogenesis. Cell 146: 873–887. 10.1016/j.cell.2011.08.039 [DOI] [PubMed] [Google Scholar]
- Pourhanifeh MH, Mohammadi R, Noruzi S, Hosseini SA, Fanoudi S, Mohamadi Y, Hashemzehi M, Asemi Z, Mirzaei HR, Salarinia R, et al. 2019. The role of fibromodulin in cancer pathogenesis: implications for diagnosis and therapy. Cancer Cell Int 19: 157. 10.1186/s12935-019-0870-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105–111. 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Antonescu CR, Scott-Browne JP, Comstock ML, Gu Y, Tanas MR, Ware CB, Woodell J. 2005. A knock-in mouse model of gastrointestinal stromal tumor harboring kit K641E. Cancer Res 65: 6631–6639. 10.1158/0008-5472.CAN-05-0891 [DOI] [PubMed] [Google Scholar]
- Sardaro A, Bardoscia L, Petruzzelli MF, Portaluri M. 2014. Epithelioid hemangioendothelioma: an overview and update on a rare vascular tumor. Oncol Rev 8: 259. 10.4081/oncol.2014.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieffer KM, Agarwal V, LaHaye S, Miller KE, Koboldt DC, Lichtenberg T, Leraas K, Brennan P, Kelly BJ, Crist E, et al. 2020. YAP1-FAM118B fusion defines a rare subset of childhood and young adulthood meningiomas. Am J Surg Pathol 45: 329–340. 10.1097/PAS.0000000000001597 [DOI] [PubMed] [Google Scholar]
- Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, Ghyselinck NB. 2003. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. united states. Nat Biotechnol 21: 562–565. 10.1038/nbt811 [DOI] [PubMed] [Google Scholar]
- Sekine S, Kiyono T, Ryo E, Ogawa R, Wakai S, Ichikawa H, Suzuki K, Arai S, Tsuta K, Ishida M, et al. 2019. Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J Clin Invest 129: 3827–3832. 10.1172/JCI126185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson ND, Awasthi A, Millis SZ, Turpin BK, Meyer CF, Grand'Maison A, Liebner DA, Hays JL, Chen JL. 2019. Common secondary genomic variants associated with advanced epithelioid hemangioendothelioma. JAMA Netw Open 2: e1912416. 10.1001/jamanetworkopen.2019.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba S, Imaoka H, Shioji K, Suzuki E, Horiguchi S, Terashima T, Kojima Y, Okuno T, Sukawa Y, Tsuji K, et al. 2018. Clinical characteristics of japanese patients with epithelioid hemangioendothelioma: a multicenter retrospective study. BMC Cancer 18: 993. 10.1186/s12885-018-4934-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya R, Matsuyama A, Shiba E, Harada H, Yabuki K, Hisaoka M. 2015. CAMTA1 is a useful immunohistochemical marker for diagnosing epithelioid haemangioendothelioma. Histopathology 67: 827–835. 10.1111/his.12713 [DOI] [PubMed] [Google Scholar]
- Sörensen I, Adams RH, Gossler A. 2009. DLL1-mediated notch activation regulates endothelial identity in mouse fetal arteries. Blood 113: 5680–5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tabula muris Consortium . 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula muris. Nature 562: 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, et al. 2011. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med 3: 98ra82. 10.1126/scitranslmed.3002409 [DOI] [PubMed] [Google Scholar]
- Tanas MR, Ma S, Jadaan FO, Ng CKY, Weigelt B, Reis-Filho JS, Rubin BP. 2016. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 35: 929–938. 10.1038/onc.2015.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Panciera T, Piccolo S. 2018. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol 20: 888–899. 10.1038/s41556-018-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso FY, Kossenkov AV, Lidenge SJ, Ngalamika O, Ngowi JR, Mwaiselage J, Wickramasinghe J, Kwon EH, West JT, Lieberman PM, et al. 2018. RNA-seq of Kaposi's sarcoma reveals alterations in glucose and lipid metabolism. PLoS Pathog 14: e1006844. 10.1371/journal.ppat.1006844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X. 2014. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141: 1614–1626. 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665. 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- Wakabayashi T, Naito H, Suehiro J-I, Lin Y, Kawaji H, Iba T, Kouno T, Ishikawa-Kato S, Furuno M, Takara K, et al. 2018. CD157 marks tissue-resident endothelial stem cells with homeostatic and regenerative properties. Cell Stem Cell 22: 384–397.e6. 10.1016/j.stem.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu W, Zhang S, Liang Z, Jiang L, Xue A, Cen X, Bu Q. 2021. Combined transcriptomic and lipidomic analysis reveals aberrant lipid metabolism in central nervous system hemangioblastomas. Sci Rep 11: 1314. 10.1038/s41598-020-80263-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JSA, Xiao Y, Lamar JM. 2018. YAP/TAZ activation as a target for treating metastatic cancer. Cancers 10: 115. 10.3390/cancers10040115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SW, Enzinger FM. 1982. Epithelioid hemangioendothelioma: a vascular tumor often mistaken for a carcinoma. Cancer 50: 970–981. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Harvey N, Detmar M, Lagutina I, Grosveld G, Gunn MD, Jackson DG, Oliver G. 2002. An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J 21: 1505–1513. 10.1093/emboj/21.7.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QC, Song W, Wang D, Zeng YA. 2016. Identification of blood vascular endothelial stem cells by the expression of protein C receptor. Cell Res 26: 1079–1098. 10.1038/cr.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Cordenonsi M, Piccolo S. 2016. YAP/TAZ at the roots of cancer. Cancer Cell 29: 783–803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-O, Gingeras TR, Weng Z. 2019. Genome-wide analysis of polymerase III-transcribed Alu elements suggests cell-type-specific enhancer function. Genome Res 29: 1402–1414. 10.1101/gr.249789.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Pan D. 2019. The hippo signaling pathway in development and disease. Dev Cell 50: 264–282. 10.1016/j.devcel.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.