Abstract
Aims/Introduction
Given that mutations related to maturity‐onset diabetes of the young (MODY) are rarely found in Chinese populations, we aim to characterize the mutation spectrum of MODY pedigrees.
Materials and Methods
Maturity‐onset diabetes of the young candidate gene‐ or exome‐targeted capture sequencing was carried out in 76 probands from unrelated families fulfilling the clinical diagnostic criteria for MODY. MAF <0.01 in the GnomAD or ExAC database was used to filter significant variants. Sanger sequencing was then carried out to validate findings. Function prediction by SIFT, PolyPhen‐2 and PROVEAN or CADD was carried out in missense mutations.
Results
A total of 32 mutations in six genes were identified in 31 families, accounting for 40.79% of the potential MODY families. The MODY subtype detection rate was 18.42% for GCK, 15.79% for HNF1A, 2.63% for HNF4A, and 1.32% for KLF11, PAX4 and NEUROG3. Seven nonsense/frameshift mutations and four missense mutations with damaging prediction were newly identified novel mutations. The clinical features of MODY2, MODY3/1 and MODYX are similar to previous reports. Clinical phenotype of NEUROG3 p.Arg55Glufs*23 is characterized by hyperglycemia and mild intermittent abdominal pain.
Conclusions
This study adds to the emerging pattern of MODY epidemiology that the proportion of MODY explained by known pathogenic genes is higher than that previously reported, and found NEUROG3 as a new causative gene for MODY.
Keywords: Chinese, Maturity‐onset diabetes of the young, Pathogenic genes
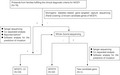
Flow chart of the procedures for screening of MODY mutations.
Introduction
Maturity‐onset diabetes of the young (MODY) is a monogenetic disease with the characteristics of autosomal dominant inheritance, early onset (<25 years) and non‐insulin dependence, accounting for 1–4% of patients with diabetes 1 , 2 . Mutations in at least 13 different genes (HNF4A, GCK, HNF1A, PDX1, HNF1B, NEUROD1, KLF11, CEL, PAX4, INS, BLK, ABCC8 and KCNJ11) have been shown to cause MODY subtypes 1–13 3 , 4 , 5 . Previous studies have shown that mutations in the GCK gene (MODY2), HNF1A (MODY3) and HNF4A gene (MODY1) represent the most common causes of MODY in the Western population, accounting for 80–90% of all MODY cases, but mutations in these genes account for just 10–20% of MODY cases in Asia (including China, Japan and Korea) 6 . Therefore, genetic causes of >80% of Asian patients with MODY remain as yet unidentified 7 , 8 . Accordingly, there is a clear unmet need to reassess the mutational spectrum of MODY in the Chinese population. In the current study, 76 families with possible MODY were first screened for mutations using a next‐generation sequencing strategy, and the results were then validated using Sanger sequencing.
Methods
Participants
A total of 76 unrelated families who participated in the “National Registration Project for Monogenetic Diabetes” initiated by the Chinese Diabetes Society from 2013 to 2016 were recruited from 35 hospitals (Table S1). All the families met the generally accepted clinical diagnostic criteria for MODY as follows: a positive familial history of diabetes, with at least two successive generations affected; early‐onset hyperglycemia (at least one family member diagnosed aged <25 years); and non‐insulin dependence (lack of a need for insulin treatment or a serum C‐peptide level of >0.60 ng/mL, even after 3 years of insulin treatment) 9 . The mean age at diagnosis of the probands was 22.30 ± 9.33 years, and the mean body mass index (BMI) was 22.31 ± 5.64 kg/m2. A total of 74 pedigrees were of Chinese Han ethnicity, and two were of Tibetan and Manchu ethnicity. The clinical data of all probands are shown in Table S2.
The study conforms to the provisions of the Declaration of Helsinki, and was approved by the Clinical Medical Research Ethics Committee of the Third Affiliated Hospital of Sun Yat‐sen University, Approval No. [2013]2‐106. All informed consent was obtained from all the participants(s) and/or guardian(s).
Target gene and whole‐exome capture sequencing
Genomic deoxyribonucleic acid (DNA) was extracted from peripheral blood leukocytes using the QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany)according to the manufacturer’s manual. The DNA concentration and purity were measured using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). A 260 : 280 ratio of ~1.8 is accepted to indicate purity of DNA.
The library was prepared, and the genes of interest or exome were captured using the BGI MGISEQ‐2000 (BGI, Shenzhen, Guangdong, China)platform. Briefly, genomic deoxyribonucleic acid was broken into 100–500 bp fragments by an enzyme kit (Segmentase; BGI), and the “A” base was then added at 3′ overhangs of 280–320 bp fragments, which were collected by magnetic beads. After adaptor ligation, the library was amplified by ligation‐mediated polymerase chain reaction. Capture enrichment was carried out by array hybridization of the library with specific capture probes designed to target genes of interest or exon regions (Roche NimbleGen, Madison, WI, USA), followed by elution and post‐capture amplification. Agilent 2100 bioanalyzer and BMG were used to determine the concentration of the products. Qualified single‐chain library products were circularized into DNA nanoballs and finally sequenced on the MGISEQ‐2000 platform of BGI.
Mutation screening and bioinformatics analysis
The flow chart of mutation screening is shown in Figure S1. Briefly, 76 probands from unrelated families with MODY were screened with the target gene panel of monogenic diabetes‐related disorders (DX0270; BGI‐Shenzhen Huada Gene Technology Co Ltd., Shenzhen, China), covering 13 genes previously associated with MODY (HNF4A, GCK, HNF1A, PDX1, HNF1B, NEUROD1, KLF11, CEL, INS, BLK, ABCC8, KCNJ11). All variants with a minor allele frequency <0.01 in the Genome Aggregation Database (GnomAD) v2.1.1 or ExAC (v1.0) database found by panel detection were further validated using Sanger sequencing for both probands and family members. For families without identifying genetic causes in the MODY‐target gene panel sequencing, whole‐exome sequencing was carried out to broaden the scope of genetic screening.
Functional prediction of new missense variants was carried out by SIFT, PolyPhen‐2, Protein Variation Effect Analyzer (PROVEAN) or combined annotation‐dependent depletion (CADD). A variant was predicted to be deleterious, with SIFT score <0.05, PolyPhen‐2 HumVar score >0.95 and PROVEAN score <−2.5 or CADD Phred score >20.
Missense variants with a positive phyloP score and/or with a PhastCons value closer to 1 were predicted to be conserved 10 .
For pathogenic variants, the criteria for pathogenic or likely pathogenic in the American College of Medical Genetics/Association for Molecular Pathology Variant Interpretation Standards and Guidelines 11 were applied.
Clinical assessment and laboratory measurement
The following clinical data were obtained from patients and their family members: age, sex, nationality, native place, diagnostic approach, age at diagnosis, BMI, glycated hemoglobin (HbA1c), serum glucose, insulin and C‐peptide (CP), lipid profile (total cholesterol, low‐density lipoprotein, high‐density lipoprotein and triglycerides), past medical history (e.g., hypertension, diabetic microvascular and macrovascular complications), and treatment.
A 75‐g oral glucose tolerance test (OGTT) was carried out in probands and their family members without a prior history of diabetes to determine glucose tolerance and evaluate islet function. A steamed bread meal test (SBMT) was carried out for those with previously diagnosed diabetes. The steamed bread was made of 100 g flour containing carbohydrates equivalent to approximately 75 g glucose. Patients began fasting after dinner the day before the test. On the test day, after collecting fasting blood samples, patients ate the steamed bread completely within 15 min, and venous blood samples were collected at 30 and 120 min. SBMT‐ and OGTT‐ stimulated blood glucose, serum insulin and C‐peptide are equivalent 12 . In China, SBMT is often used as a simulation to replace OGTT to observe the postprandial changes in blood glucose, and islet function in patients with diabetes for safety and comfort reasons.
Glucose‐related indices were calculated, including incremental 2‐h glucose (∆2h‐G) during the OGTT or SBMT, the area under the curve (AUC) of glucose (AUCGlu), the AUC of CP (AUCCP), AUCCP/AUCGlu (AUCCP/G) and CP30’ increment/glucose30’ increment (∆CP30’/∆Glu30’). The AUC was calculated using the trapezoidal principle
Homeostatic model assessment of insulin resistance (HOMA‐IR) and homeostatic model assessment of β‐cell function (HOMA‐B) were calculated using the following formulas: fasting insulin (µIU/mL) × fasting plasma glucose (FPG; mmol/L) / 22.5 and 20 × fasting insulin (µIU/mL) / (fasting plasma glucose; mmol/L − 3.5) (%), respectively.
Statistical analysis
Statistical analysis was carried out using Statistical Package for Social Sciences (SPSS for Windows, version 25.0, SPSS Inc., Chicago, IL, USA). Clinical data and laboratory parameters are expressed as the mean ± standard deviation. Comparisons of numerical variables between or among groups were carried out by Student’s t‐test or the Kruskal–Wallis test; the χ2‐test was used for categorical data. The P‐value was corrected by the Bonferroni correction. All tests were two‐sided. In all statistical analyses, P < 0.05 was considered to show statistical significance.
Results
Mutation detection rate
Among 76 pedigrees, 32 mutations in six genes (Table 1) that were either previously described or novel likely pathogenic were identified in 31 pedigrees (Figure S2), accounting for 40.79% of MODY cases. The mutations in each proband, including 15 GCK mutations (MODY2), 12 HNF1A mutations (MODY3), two HNF4A mutations (MODY1), one KLF11 mutation (MODY7), one PAX4 mutation (MODY9) and one NEUROG3 mutation, are listed in Table 1. All mutations were inherited genetic alterations, except for two de novo mutations (HNF4A‐MODY1 p.Cys808Ser and HNF1A‐MODY3 p.Lys120Glu). Proband 16 carried compound GCK mutations inherited from her father. Thus, the frequency of MODY subtype was 18.42% for MODY2, 15.79% for HNF1A, 2.63% for HNF4A, and 1.32% for PAX4, KLF11 and NEUROG3. In addition, an ABCC8 variant is suspected to be causal of MODY12 (Table 1). All of these mutations are absent from or have a minor allele frequency <0.01 in the GnomAD and ExAC databases, and all missense mutations except HNF 1A p.Ile618Met affect a highly conserved residue with a positive PhyloP score and PhastCons score equal to or close to 1 (Table 1).
Table 1.
List of mutations selected to be pathogenic and suspected for maturity‐onset diabetes of the young
| Probands | Gene | Sex | Age at diagnosis (year) | BMI (kg/m2) | Mutation source | cDNA changes | AA changes | Reported |
GnomAD (v2.1.1) East Asian |
ExAC (v1.0) East Asian |
PhyloP | PhastCons |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic mutation | ||||||||||||
| 1 | HNF4A | Female | 16 | 20.00 | De novo | c.238T>A | p.Cys80Ser | No | / | / | 5.046 | 1 |
| 2 | HNF4A | Female | 27 | 21.56 | M | c.885_894delGAGCTTGGAG | p.Ser296Thrfs*31 | No | / | / | ||
| 3 | GCK | Female | 25 | 17.22 | P | c.480T>G | p.Asp160Glu | No | / | / | 3.277 | 1 |
| 4 | GCK | Female | 24 | 24.58 | M | c.505A>G | p.Lys169Glu | No | / | / | 5.034 | 1 |
| 5 | GCK | Female | 9 | 10.30 | M | c.571 C>T | p.Arg191Trp | Yes 18 , 43 | 5.437e‐5 (1/18,392) | / | 0.54 | 0.974 |
| 6 | GCK | Male | 38 | 18.52 | P | c.571 C>T | p.Arg191Trp | Yes 18 , 43 | 5.437e‐5 (1/18,392) | / | 0.54 | 0.974 |
| 7 | GCK | Male | 36 | 23.89 | P | c.571 C>T | p.Arg191Trp | Yes 18 , 43 | 5.437e‐5 (1/18,392) | / | 0.54 | 0.974 |
| 8 | GCK | Male | 10 | 15.73 | P | c.571 C>T | p.Arg191Trp | Yes 18 , 43 | 5.437e‐5 (1/18,392) | / | 0.54 | 0.974 |
| 9 | GCK | Female | 17 | 15.06 | P | c.622 G>A | p.Ala208Thr | Yes 43 | 0 (0/1,560) | / | 6.234 | 1 |
| 10 | GCK | Female | 24 | 16.61 | M | c.683 C>T | p.Thr228Met | Yes 44 | 0 (0/18,372) | 0 (0/8,582) | 5.915 | 1 |
| 11 | GCK | Female | 6 | 13.77 | M | c.781 G>A | p.Gly261Arg | Yes 44 | 0 (0/18,388) | 0 (0/8,614) | 5.864 | 1 |
| 12 | GCK | Male | 12 | 17.19 | M | c.865delT | p.Tyr289Metfs*5 | No | / | / | ||
| 13 | GCK | Male | 5 | 14.86 | P | c.971T>C | p.Leu324Pro | Yes 45 | / | / | 3.107 | 1 |
| 14 | GCK | Female | 15 | 17.44 | M | c.1020‐7_c.1035del23 | p.Asp341‐Ser418 del | No | / | / | ||
| 15 | GCK | Male | 7 | 16.91 | M | c.1166T>A | p.Val389Asp | Yes 19 | / | / | 4.991 | 1 |
| 16 | GCK | Female | 7 | 17.36 | P | c.1239C>A | p.Tyr413Ter | No | / | / | 6.034 | 1 |
| GCK | Female | 7 | 17.36 | P | c.1241_1242insA | p.Leu415Alafs*44 | No | / | / | |||
| 17 | HNF1A | Male | 24 | ‐ | M | c.139G>A | p.Gly47Arg | No | 0 (0/19,720) | 0 (0/7,460) | 0.459 | 1 |
| 18 | HNF1A | Female | 13 | 20.54 | M | c.160C>T | p.Arg54Ter | Yes 29 , 46 | / | / | 0.383 | 0.297 |
| 19 | HNF1A | Female | 13 | 17.30 | P | c.160C>T | p.Arg54Ter | Yes 29 , 46 | / | / | 0.383 | 0.297 |
| 20 | HNF1A | Male | 13 | 18.90 | M? | c.358A>G | p.Lys120Glu | Yes 28 | / | / | 4.669 | 1 |
| 21 | HNF1A | Male | 17 | 19.88 | M | c.476 G>A | p.Arg159Gln | Yes 27 | / | / | 5.569 | 0.999 |
| 22 | HNF1A | Female | 30 | 21.00 | M | c.599G>A | p.Arg200Gln | Yes 27 | 0 (0/18,394) | / | 4.974 | 0.991 |
| 23 | HNF1A | Female | 12 | 20.20 | M | c.811 C>T | p.Arg271Trp | Yes 47 | 0 (0/18,342) | / | 1.277 | 1 |
| 24 | HNF1A | Female | 12 | 19.65 | M | c.811 C>T | p.Arg271Trp | Yes 47 | 0 (0/18,342) | / | 1.277 | 1 |
| 25 | HNF1A | Female | 24 | 18.07 | M | c.872dupC | p.Pro291fs*26 | Yes 26 , 48 | 4.850e‐4 (9/18,558) | 1.838e‐3 (11/5,984) | ||
| 26 | HNF1A | Female | 15 | 29.14 | M | c.872dupC | p.Pro291fs*26 | Yes 26 , 48 | 4.850e‐4 (9/18,558) | 1.838e‐3 (11/5,984) | ||
| 27 | HNF1A | Female | 22 | 19 | P | c.1136_1137insC | p.Val380Cysfs*39 | Yes 49 | 0 (0/18,356) | / | ||
| 28 | HNF1A | Male | 24 | 29.74 | M | c.1854C>G | p.Ile618Met | Yes 50 | 3.509e‐4 (7/19,950) | / | −2.829 | 0.012 |
| 29 | KLF11 | Male | 15 | 27.78 | M | c. 1357_1359 delAAG | p.Lys453del | No | 1.631e‐4 (3/18,394) | 1.156e‐4 (1/8,654) | ||
| 30 | PAX4 | Male | 12 | 18.83 | M | c.332 C> T | p.Pro111Leu | No | 6.524e‐4 (13/19,926) | 6.096e‐4 (5/8,202) | 5.625 | 1 |
| 31 | NEUROG3 | Female | 14 | 22.22 | P | c.163delC | p.Arg55Glufs*23 | No | / | / | ||
| Suspected mutation | ||||||||||||
| 32 | ABCC8 | Male | 5 | 19.44 |
M |
c.3344C>T | p.Thr1115Met | No | 0 (0/18,394) | 0 (0/8,644) | 6.424 | 1 |
The proband’s mother died, neither his father nor his mother’s sister carried the mutation.
AA change, amino acid change; EX, exon; Het, heterozygote; Hom, homozygote; IN, intron; M, maternal; OHA, oral hypoglycemic agents; P, paternal.
MODY2 mutations
A total of 12 different GCK mutations were identified (Table 1), including six previously described missense mutations. Four of the six new GCK mutations found in the present study are inactivating, as they caused truncation of proteins due to the introduction of a premature stop codon (p.Tyr289Metfs*5 and p.Tyr413Ter) or caused nonsense proteins due to the modification of the reading frame (p.Asp341‐Ser418del and p.Leu415Alafs*44). Two new missense mutations (p.Asp160Glu and p.Lys169Glu) were predicted to be deleterious by SIFT, PolyPhen‐2 and PROVEAN, with a CADD Phred score >20 (Table S3). The Arg191Trp, a previously reported and functionally verified mutation in white and Asian populations, was identified in four families (Table 1), making it the most common GCK mutation in the present study.
MODY3 and MODY1 mutations
Nine different HNF1A mutations were identified, all of which have been reported previously. Three known mutations (p.Arg54Ter, p.Arg271Trp, p.Pro291fs*26) were detected in two probands each. In addition, three mutations (p.Arg54Ter, p.Pro291fs*26, p.Val380Cysfs*39) are truncation mutations that represented 41.67% of the HNF1A mutations identified in our MODY3 population.
Two new HNF4A mutations were identified. p.Ser296Thrfs*31 is a truncation mutation, and p.Cys80Ser is a de novo mutation and suspected to be a harmful variant according to SIFT, PROVEAN and PolyPhen‐2, with a CADD Phred score of 26.4 (Table S3).
Other MODY mutations
Other MODY mutations (MODY‐others) included those in KLF11 (MODY7), PAX4 (MODY9) and a novel mutation in NEUROG3 (Table 1). None of these mutations have been described previously. KLF11 p.Lys453del causes a single amino acid deletion, which might have a deleterious effect on the function of the protein, and has damaging predictions by PROVEAN (Table S3). PAX4 p.Pro111Leu is predicted to be harmful by SIFT, PolyPhen‐2 and PROVEAN, with a high CADD (Phred score = 28.7; Table S3). NEUROG3 p.Arg55Glufs*23 changes the reading frame, causes premature termination of translation and is considered a disease‐causing mutation.
Suspected causal variant
ABCC8 p.Thr1115Met was found in a pedigree. This variant is absent from the GnomAD and ExAC databases of East Asia, is evolutionally conserved (Table 1), and is predicted to be damaging by SIFT, PolyPhen‐2 and PROVEAN, and has a CADD phred score >30 (Table S3). Therefore, although the co‐segregation between mutation and phenotypes could not be clearly defined (Figure S2), we treat it as a suspected causal variant of MODY in our study.
Clinical characteristics of MODY subtypes and MODYX
The participants with MODY subtypes included probands and their mutation‐positive relatives, whereas participants with MODYX included only probands. As shown in Table 2, the carriers of MODY mutations had a mean age of 34.46 ± 17.91 years, a mean age at diagnosis of 28.79 ± 16.21 years, a mean BMI of 20.7 ± 3.58 kg/m2 and a mean HbA1c of 6.65 ± 1.20%.
Table 2.
Clinical characteristics of maturity‐onset diabetes of the young subtypes and maturity‐onset diabetes of the young X
| MODY total | MODY2 | MODY3/1 | MODY‐others | MODYX | P T vs X * | P 2 vs 3/1 vs O * | |
|---|---|---|---|---|---|---|---|
| Sex: M/F | 36/51 | 20/25 | 10/22 | 6/4 | 25/19 | 0.094 | 0.228 |
| Age (years) | 34.46 ± 17.91 | 33.43 ± 19.25 | 37.81 ± 15.29 | 28.60 ± 19.02 | 24.09 ± 7.89 | 0.003 | 0.268 |
| Age at diagnosis (years) | 28.79 ± 16.21 | 29.91 ± 18.28 | 28.29 ± 12.87 | 25.40 ± 16.86 | 19.70 ± 5.40 | 0.004 | 0.695 |
| Duration of diabetes (years) | 5.67 ± 6.48 | 3.52 ± 3.90 | 9.52 ± 8.25 | 3.20 ± 3.55 | 4.39 ± 4.75 | 0.483 | 0.003 |
| BMI (kg/m2) | 20.70 ± 3.58 | 19.82 ± 3.17 | 21.37 ± 3.13 | 22.60 ± 5.39 | 24.35 ± 5.59 | <0.001 | <0.001 |
| HbA1c (%) | 6.65 ± 1.20 | 6.35 ± 0.44 | 7.11 ± 1.64 | 6.58 ± 1.57 | 8.35 ± 2.12 | <0.001 | 0.051 |
| Glu0’ (mmol/L) | 7.34 ± 2.09 | 6.76 ± 0.90 | 8.23 ± 2.91 | 7.45 ± 2.41 | 8.44 ± 3.19 | 0.065 | 0.066 |
| CP0’ (ng/mL) | 1.08 ± 0.86 | 1.04 ± 0.94 | 1.02 ± 0.85 | 1.38 ± 0.36 | 2.45 ± 2.65 | 0.002 | 0.355 |
| AUCGlu (mmol/L min) | 1442 ± 417 | 1289 ± 232 | 1696 ± 533 | 1426 ± 398 | 1489 ± 460 | 0.001 | <0.001 |
| AUCCP (ng/mL min) | 388 ± 315 | 423 ± 329 | 324 ± 331 | 400 ± 182 | 601 ± 682 | 0.032 | 0.214 |
| AUCCP/G (µg/mmoL min) | 0.29 ± 0.25 | 0.33 ± 0.23 | 0.22 ± 0.31 | 0.31 ± 0.17 | 0.45 ± 0.56 | 0.095 | 0.552 |
| ∆2h‐G (mmol/L) | 6.63 ± 3.93 | 5.05 ± 2.99 | 8.87 ± 4.28 | 7.49 ± 3.66 | 6.22 ± 4.74 | 0.008 | <0.001 |
| ∆CP30’/∆G30’ (µg/mmol) | 0.40 ± 0.35 | 0.49 ± 0.32 | 0.22 ± 0.19 | 0.50 ± 0.57 | 0.68 ± 1.05 | 0.032 | 0.011 |
| HOMA‐IR | 1.75 ± 1.01 | 1.48 ± 0.85 | 1.78 ± 1.21 | 2.65 ± 0.72 | 2.51 ± 1.73 | 0.068 | 0.006 |
| HOMA‐B | 36.95 ± 28.18 | 30.86 ± 14.70 | 26.69 ± 20.23 | 74.83 ± 44.61 | 62.84 ± 73.61 | 0.027 | <0.001 |
| CHOL (mmol/L) | 3.84 ± 1.20, | 3.75 ± 0.96 | 4.45 ± 1.12 | 2.38 ± 1.14 | 3.99 ± 1.22 | 0.218 | <0.001 |
| TRIG (mmol/L) | 0.90 ± 0.71 | 0.81 ± 0.59 | 1.09 ± 0.91 | 0.76 ± 0.30 | 1.94 ± 2.65 | 0.015 | 0.169 |
| HDL (mmol/L) | 1.22 ± 0.37 | 1.24 ± 0.28 | 1.38 ± 0.38 | 0.68 ± 0.19 | 1.05 ± 0.35 | 0.032 | <0.001 |
| LDL (mmol/L) | 2.20 ± 0.85 | 2.15 ± 0.79 | 2.53 ± 0.82 | 1.46 ± 0.77 | 2.31 ± 0.81 | 0.426 | 0.011 |
| Overweight, n (%) | 13, 15.66% | 6, 13.64% | 3, 10.34% | 4, 40% | 18, 42.86% | 0.001 | 0.073 |
| Hypertension, n (%) | 11, 14.86% | 4, 9.76% | 5, 21.74% | 2, 20% | 7, 18.42% | 0.628 | 0.384 |
| Retinopathy, n (%) | 4, 4.60% | 0 | 4, 12.50% | 0 | 2, 4.55% | 1.000 | 0.027 |
| Nephropathy, n (%) | 2, 2.30% | 0 | 2, 6.25% | 0 | 3, 6.82% | 0.334 | 0.172 |
| Neuropathy, n (%) | 2, 2.30% | 0 | 2, 6.25% | 0 | 2, 4.55% | 0.602 | 0.172 |
| Macrovascular, n (%) | 2, 2.30% | 0 | 2, 6.25% | 0 | 0 | 0.550 | 0.172 |
| Treatment at time of visit (n) | 66 | 29 | 28 | 9 | 43 | ||
| Intensive insulin, n (%) | 2,3.03% | 0 | 2, 7.14% | 0 | 7, 16.28% | 0.027 | 0.247 |
| Insulin, n (%) | 19,28.79% | 3, 10.34% | 14, 50.00% | 2, 22.22% | 24, 55.81% | 0.005 | 0.004 |
| OHA, n (%) | 30,45.45% | 11, 37.93% | 17, 60.71% | 2, 22.22% | 21, 48.84% | 0.729 | 0.072 |
| Sulfonylurea, n (%) | 12,18.18% | 5, 17.24% | 7, 25.00% | 0 | 5, 11.63% | 0.357 | 0.235 |
| Metformin, n (%) | 20,30.30% | 8, 27.59% | 11, 39.29% | 1, 11,11% | 18, 41.86% | 0.216 | 0.254 |
| Others, n (%) | 13,19.70% | 3, 10.34% | 9, 32.14% | 1, 11,11% | 11, 25.58% | 0.469 | 0.092 |
| Diet only, n (%) | 26,39.39% | 18, 62.07% | 3, 10.71% | 5, 55.56% | 10, 23.26% | 0.080 | <0.001 |
Continuous values are expressed as mean ± standard deviation.
The P‐value has been adjusted for age, duration and sex. The data of homeostatic model assessment of insulin resistance (HOMA‐IR) and homeostatic model assessment of β‐cell function (HOMA‐B) included only the patients who were not treated with insulin. ∆2h‐G, 2‐h glucose increment of oral glucose tolerance test and steamed bread meal test; ∆CP30’/∆G30’, CP30’ increment/glucose30’ increment; AUCCP, area under the curve of C‐peptide; AUCGlu, area under the curve of 2‐h glucose; BMI, body mass index; CHOL, cholesterol; HbA1c, glycated hemoglobin; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein; MODY, maturity‐onset diabetes of the young; OHA, oral hypoglycemic agents; T versus X, MODY total versus MODYX; 2 versus 3/1 versus O, MODY2 versus MODY3/1 versus MODY‐others; TRIG, triglycerides.
MODY2 showed mild fasting hyperglycemia (6.76 ± 0.90 mmol/L) and markedly elevated Glu120’ levels in OGTT or SBMT (11.76 ± 2.99 mmol/L). The mean 2‐h glucose increment of OGTT or SBMT (∆2h‐G) for MODY2 was 5.05 ± 2.99 mmol/L. A small increase in mean HbA1c was found in MODY2 (6.35 ± 0.44%). After dividing MODY2 participants into two groups on the basis of median ∆2h‐G (5.61 mmol/L), those with ∆2h‐G <5.61 mmol/L were significantly younger, younger at diagnosis, and had a shorter diabetes duration, lower BMI and higher ∆CP30’/∆G30’ than those with ∆2h‐G ≥5.61 mmol/L (Table S4). Among MODY2 participants, 62.07% were treated with diet alone, 37.93% with oral hypoglycemic agents (OHA) and 10.34% with insulin (Table 2). The mean duration of MODY2 was 3.52 ± 3.90 years, and no diabetic vascular complications were reported (Table 2). Further analysis showed that MODY2 patients treated with OHA were significantly older, older at diagnosis and had a longer duration with a relatively lower HOMA‐B value than patients treated with diet alone (Table S5).
MODY 3/1 participants had the longest duration (9.52 ± 8.25 years), the highest AUCGlu and the lowest ∆CP30’/∆G30’ after OGTT or SBMT among MODY subtypes. Microvascular and macrovascular complications of diabetes, including diabetic retinopathy, nephropathy, neuropathy and stroke, were only reported in the MODY3/1 group. A total of 25% of MODY3/1 patients were treated with sulfonylureas, and 50% were treated with insulin.
MODY‐others had the highest BMI, HOMA‐IR and HOMA‐B values among the MODY subtypes, and the frequency of overweight in MODY‐others tended to be higher than MODY2 and MODY3/1 (Table 2). MODY7 and MODY9 showed a high proportion of overweight or obesity with a mean HOMA‐IR of 2.86 ± 0.90 and a mean HOMA‐B of 74.24 ± 41.33 (Table S6). The clinical phenotype of NEUROG3 p.Arg55Glufs*23 is characterized by hyperglycemia with mild intermittent abdominal pain (the proband complained of abdominal pain, and diabetes was detected during her visit; her sister also showed intermittent mild abdominal pain and hyperglycemia). There was a lack of abdominal pressure pain, lack of rebound tenderness and no diarrhea in the NEUROG3 mutation carriers, and no abnormality was found on laboratory examination of the abdominal gastrointestinal tract. A total of 55.56% of patients with MODY‐others were treated with diet alone, and those treated with OHA and insulin accounted for 22.22% each.
MODYX patients had a significantly younger age and age at diagnosis; higher BMI, HbA1c and triglycerides; and lower high‐density lipoprotein than MODY patients, and also showed significantly higher AUCGlu, AUCCP, ∆CP30’/∆Glu30’ and HOMA‐B (Table 2). A significantly higher percentage of overweight was found in MODYX than in MODY patients. The proportions of MODY subtypes and MODYX patients treated with insulin were 28.79 and 55.81%, respectively (P = 0.005). A total of 16.28% of MODYX patients received initial intensive insulin treatment, whereas just 3.03% of MODY patients did. No statistically significant difference in other clinical features was found between MODY and MODYX patients.
Discussion
In the current study, the molecular diagnosis of 31 out of 76 (40.79%) suspected MODY pedigrees was confirmed. MODY2 and MODY3 are common MODY subtypes, accounting for 18.42 and 15.79% of MODY cases, respectively, which was higher than that reported in Hong Kong (1% for MODY2 and 9% for MODY3) 14 and Shanghai (10.42% for MODY2 and 1.95% for MODY3) 15 , 16 .
Among MODY2 mutations, some have been reported to affect the enzyme affinity for adenosine triphosphate (ATP) (p.Val389Asp, p.Thr228Met) or glucose (p.Gly261Arg), or to decrease the thermal stability of the protein (p.Arg191Trp) 17 , 18 . The new missense mutations, p.Lys169Glu and p.Asp160Glu, are located in the glucose‐ and ATP‐binding sites of GCK 19 , 20 , respectively, and a different missense change at both sites had been reported to be pathogenic in in vitro kinetic assays 21 . With regard to compound mutations, two different loss‐of‐function mutations (p.Tyr413Ter and p. Leu415Alafs*44) on the same allele were identified, and the nonsense mutation is likely to be the causative variant, because it precedes the frameshift. The p.Arg191Trp, the most common mutation (28.57%, 4/14) in the present study, was also found to be common (15.79%, 9/57) in the study reported by Yorifuji et al. 22 in Japanese populations and another Chinese study reported by Wang et al. 18 (16.67%, 2/12), but was rare in Norwegian (0.82%, 1/122) and Italian (0%, 0/66) populations 23 , 24 .
Most MODY3 mutations found in the present study are located in the conserved domain of the gene 25 and lead to loss of function by inhibiting the activity of wild‐type HNF1A through dominant negative effects (p.Arg159Gln, p.Pro291fs*26) 26 , 27 or by reducing DNA binding (p.Lys120Glu, p.Arg271Trp) 27 , 28 . The p.Arg54Ter nonsense mutation results in a severely truncated HNF1A protein that lacks DNA binding and transcriptional activation domains based on analysis of the 3‐D structure 29 .
The clinical phenotypes of MODY2, MODY3/1 and MODYX reported herein are similar to those reported previously 30 , 31 , 32 . However, for MODY2, 57% had a high OGTT 2‐h glucose increment (>4.6 mmol/L), which was different from the previous reports in a large European study 13 that showed 71% of MODY2 having a small OGTT 2‐h increment (<3 mmol/L). Given that an increment of <4.6 mmol/L is often used as the criterion for priority screening of MODY2 9 , this finding showed that a missed diagnosis might occur using this criterion to identify when GCK testing is appropriate in the Chinese population.
Sulfonylureas are considered the best first‐line medications for patients with MODY3 and MODY1, and result in superior glycemic control than insulin 33 and metformin 34 . In the present study, approximately 25% of MODY3 and MODY1 patients were treated with sulfonylureas, indicating that most MODY3/1 patients did not receive the correct treatment. MODY2 is known to be a non‐progressive form of diabetes, which usually can be controlled by diet alone without any medication, except during pregnancy 35 . Nevertheless, approximately 40% of MODY2 patients were treated with OHA in the present study. Patients treated with OHA were significantly older and had worse β‐cell function than those treated with diet alone, suggesting that age‐related decline in islet function might be related to administration of OHA for MODY2.
MODY7 and MODY9 are very rare causes of MODY. Their clinical features have not been fully established 36 . In the present study, MODY7 and MODY9 have a high proportion of overweight or obesity, and with relatively higher HOMA‐IR than MODY2 and MODY3/1, suggesting a characteristic of metabolic syndrome, regardless, but no specific clinical characteristics other than common diabetes were found.
The newly reported gene, NEUROG3, is a key transcription factor in the differentiation of pancreatic and intestinal endocrine cells, and is responsible for transcriptional regulation of the NEUROD1 (MODY6 pathogenic gene) 37 . Previous studies have shown that homozygous mutations in the NEUROG3 gene lead to congenital malabsorptive diarrhea and neonatal diabetes 38 , suggesting that the NEUROG3 mutation has dual phenotypes regarding the gastrointestinal tract and blood glucose, which corresponds to the function of NEUROG3. This can also explain why the clinical phenotype of hyperglycemia and intermittent mild abdominal pain was observed in our patients with NEUROG3 mutation.
Some limitations of the study were as follows: (i) because some genes have highly repetitive complex regions or pseudogenes, the high‐throughput test cannot achieve 100% coverage, but the overall coverage can reach >95% (list link of low coverage regions http://db.bgidx.cn/); and (ii) MODY5 is a disease with low penetrant, and the most frequent mutations are monoallelic defects in all or some exons 39 . However, the NGS used in the present study cannot detect copy number variation in large genome fragments. Multiplex ligation‐dependent probe amplification is a gold standard for copy number variation detection 40 and considered to be more advantageous than comparative genomic hybridization array in the genetic diagnostics of MODY5 41 . Therefore, estimation of MODY5 prevalence without multiplex ligation‐dependent probe amplification analysis is a limitation of the present study. 17q12 deletion is the most common mutation of MODY5. As 70% of MODY5 caused by 17q12 deletion are sporadic 42 , the criteria for MODY screening used in the present study, such as the presence of family history, might also have affected the assessment of the prevalence of MODY5.
Finally, there was still 60% of MODY patients with no confirmed genetic causes in the present study. The major genes for MODY in the Chinese population remain to be identified. Hence, more extensive genetic studies are required on Chinese patients with MODY.
Disclosure
The authors declare no conflict of interest.
Supporting information
Table S1 | List of participating centers.
Table S2 | Clinical characteristic of probands.
Table S3 | Prediction scores from SIFT, PolyPhen‐2, Protein Variation Effect Analyzer and combined annotation‐dependent depletion for the novel mutations.
Table S4 | Clinical features of maturity‐onset diabetes of the young 2 stratified by the median of ∆2‐h glucose.
Table S5 | Clinical features of maturity‐onset diabetes of the young 2 patients treated with oral hypoglycemic agents and diet alone.
Table S6 | The clinical data of other maturity‐onset diabetes of the young mutations patients.
Figure S1 | Flow chart of the procedures for screening of maturity‐onset diabetes of the young mutations.
Figure S2 | Pedigrees of families with identified or suspected genetic causes of maturity‐onset diabetes of the young.
Acknowledgments
We thank the participants and their families for kindly agreeing to participate in this study, and the participating centers for providing patient information. This study was funded by the National Natural Science Foundation of China (81941022) and the Strategic Priority Research Program of Chinese Academy of Sciences (XDB 38010100).
J Diabetes Investig. 2021
References
- 1. Gardner DS, Tai ES. Clinical features and treatment of maturity onset diabetes of the young (MODY). Diabetes Metab Syndr Obes 2012; 5: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Timsit J, Saint‐Martin C, Dubois‐Laforgue D, et al. Searching for maturity‐onset diabetes of the young (MODY): when and what for? Can J Diabetes 2016; 40: 455–461. [DOI] [PubMed] [Google Scholar]
- 3. Bonnefond A, Philippe J, Durand E, et al. Whole‐exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS One 2012; 7: e37423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao R, Liu Y, Gjesing AP, et al. Evaluation of a target region capture sequencing platform using monogenic diabetes as a study‐model. BMC Genet 2014; 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prudente S, Jungtrakoon P, Marucci A, et al. Loss‐of‐function mutations in APPL1 in familial diabetes mellitus. Am J Hum Genet 2015; 97: 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kleinberger JW, Pollin TI. Undiagnosed MODY: time for action. Curr Diab Rep 2015; 15: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shields BM, Hicks S, Shepherd MH, et al. Maturity‐onset diabetes of the young (MODY): how many cases are we missing. Diabetologia 2010; 53: 2504–2508. [DOI] [PubMed] [Google Scholar]
- 8. Shim YJ, Kim JE, Hwang SK, et al. Identification of candidate gene variants in Korean MODY families by whole‐exome sequencing. Horm Res Paediatr 2015; 83: 242–251. [DOI] [PubMed] [Google Scholar]
- 9. Ellard S, Bellanné‐Chantelot C, Hattersley AT. Best practice guidelines for the molecular genetic diagnosis of maturity‐onset diabetes of the young. Diabetologia 2008; 51: 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pollard KS, Hubisz MJ, Siepel A, et al. Detection of non‐neutral substitution rates on mammalian phylogenies. Genome Res 2010; 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collaborative Study Groups for islet B‐cell function . The observation of glucose, insulin and C peptide of steam bread test among healthy population. Chin Med J 1982; 62: 643–647. [Google Scholar]
- 13. Stride A, Vaxillaire M, Tuomi T, et al. The genetic abnormality in the beta cell determines the response to an oral glucose load. Diabetologia 2002; 45: 427–435. [DOI] [PubMed] [Google Scholar]
- 14. Xu JY, Dan QH, Chan V, et al. Genetic and clinical characteristics of maturity‐onset diabetes of the young in Chinese patients. Eur J Hum Genet 2005; 13: 422–427. [DOI] [PubMed] [Google Scholar]
- 15. Liu L, Liu Y, Ge X, et al. Insights into pathogenesis of five novel GCK mutations identified in Chinese MODY patients. Metabolism 2018; 89: 8–17. [DOI] [PubMed] [Google Scholar]
- 16. Fang QC, Zhang R, Wang CR, et al. Scanning HNF‐1 alpha gene mutation in Chinese early‐onset and/or multiplex diabetes pedigrees. Chin J Med Gen 2004; 21: 329–334. [PubMed] [Google Scholar]
- 17. Beer NL, van de Bunt M, Colclough K, et al. Discovery of a novel site regulating glucokinase activity following characterization of a new mutation causing hyperinsulinemic hypoglycemia in humans. J Biol Chem 2011; 286: 19118–19126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z, Diao C, Liu Y, et al. Identification and functional analysis of GCK gene mutations in 12 Chinese families with hyperglycemia. J Diabetes Investig 2019; 10: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Osbak KK, Colclough K, Saint‐Martin C, et al. Update on mutations in glucokinase (GCK), which cause maturity‐onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat 2009; 30: 1512–1526. [DOI] [PubMed] [Google Scholar]
- 20. Kamata K, Mitsuya M, Nishimura T, et al. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure 2004; 12: 429–438. [DOI] [PubMed] [Google Scholar]
- 21. Raimondo A, Chakera AJ, Thomsen SK, et al. Phenotypic severity of homozygous GCK mutations causing neonatal or childhood‐onset diabetes is primarily mediated through effects on protein stability. Hum Mol Genet 2014; 23: 6432–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yorifuji T, Higuchi S, Kawakita R, et al. Genetic basis of early‐onset, maturity‐onset diabetes of the young‐like diabetes in Japan and features of patients without mutations in the major MODY genes: dominance of maternal inheritance. Pediatr Diabetes 2018; 19: 1164–1172. [DOI] [PubMed] [Google Scholar]
- 23. Sagen JV, Bjørkhaug L, Molnes J, et al. Diagnostic screening of MODY2/GCK mutations in the Norwegian MODY Registry. Pediatr Diabetes 2008; 9: 442–449. [DOI] [PubMed] [Google Scholar]
- 24. Capuano M, Garcia‐Herrero CM, Tinto N, et al. Glucokinase (GCK) mutations and their characterization in MODY2 children of southern Italy. PLoS One 2012; 7: e38906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamagata K. Regulation of pancreatic beta‐cell function by the HNF transcription network: lessons from maturity‐onset diabetes of the young (MODY). Endocr J 2003; 50: 491–499. [DOI] [PubMed] [Google Scholar]
- 26. Yamagata K, Yang Q, Yamamoto K, et al. Mutation P291fsinsC in the transcription factor hepatocyte nuclear factor‐1alpha is dominant negative. Diabetes 1998; 47: 1231–1235. [DOI] [PubMed] [Google Scholar]
- 27. Gu N, Suzuki N, Takeda J, et al. Effect of mutations in HNF‐1alpha and HNF‐1beta on the transcriptional regulation of human sucrase‐isomaltase in Caco‐2 cells. BiochemBiophys Res Commun 2004; 325: 308–313. [DOI] [PubMed] [Google Scholar]
- 28. Malikova J, Kaci A, Dusatkova P, et al. Functional analyses of HNF1A‐MODY variants refine the interpretation of identified sequence variants. J Clin Endocrinol Metab 2020; 105: e1377–e1386. [DOI] [PubMed] [Google Scholar]
- 29. Fang C, Huang J, Huang Y, et al. A novel nonsense mutation of the HNF1α in maturity‐onset diabetes of the young type 3 in Asian population. Diabetes Res Clin Pract 2015; 109: e5–e7. [DOI] [PubMed] [Google Scholar]
- 30. Costa A, Bescós M, Velho G, et al. Genetic and clinical characterisation of maturity‐onset diabetes of the young in Spanish families. Eur J Endocrinol 2000; 142: 380–386. [DOI] [PubMed] [Google Scholar]
- 31. Siddiqui K, Musambil M, Nazir N. Maturity onset diabetes of the young (MODY) – history, first case reports and recent advances. Gene 2015; 555: 66–71. [DOI] [PubMed] [Google Scholar]
- 32. Li X, Ting TH, Sheng H, et al. Genetic and clinical characteristics of Chinese children with Glucokinase‐maturity‐onset diabetes of the young (GCK‐MODY). BMC Pediatr 2018; 18: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raile K, Schober E, Konrad K, et al. Treatment of young patients with HNF1A mutations (HNF1A‐MODY). Diabet Med 2015; 32: 526–530. [DOI] [PubMed] [Google Scholar]
- 34. Pearson ER, Starkey BJ, Powell RJ, et al. Genetic cause of hyperglycemia and response to treatment in diabetes. Lancet 2003; 362: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 35. Hattersley AT, Pearson ER. Minireview: pharmacogenetics and beyond: the interaction of therapeutic response, beta‐cell physiology, and genetics in diabetes. Endocrinology 2006; 147: 2657–2663. [DOI] [PubMed] [Google Scholar]
- 36. Agata Juszczak KO. Identifying subtypes of monogenic diabetes. Diabetes Manage 2014; 4: 46–61. [Google Scholar]
- 37. Wang J, Cortina G, Wu SV, et al. Mutant neurogenin‐3 in congenital malabsorptive diarrhea. N Engl J Med 2006; 355: 270–280. [DOI] [PubMed] [Google Scholar]
- 38. Rubio‐Cabezas O, Jensen JN, Hodgson MI, et al. Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3 . Diabetes 2011; 60: 1349–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Horikawa Y. Maturity‐onset diabetes of the young as a model for elucidating the multifactorial origin of type 2 diabetes mellitus. J Diabetes Investig 2018; 9: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stuppia L, Antonucci I, Palka G, et al. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int J Mol Sci 2012; 13: 3245–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Omura Y, Yagi K, Honoki H, et al. Clinical manifestations of a sporadic maturity‐onset diabetes of the young (MODY) 5 with a whole deletion of HNF1B based on 17q12 microdeletion. Endocr J 2019; 66: 1113–1116. [DOI] [PubMed] [Google Scholar]
- 42. Murray PJ, Thomas K, Mulgrew CJ, et al. Whole gene deletion of the hepatocyte nuclear factor‐1beta gene in a patient with the prune‐belly syndrome. Nephrol Dial Transplant 2008; 23: 2412–2415. [DOI] [PubMed] [Google Scholar]
- 43. Sagen JV, Odili S, Bjørkhaug L, et al. From clinicogenetic studies of maturity‐onset diabetes of the young to unraveling complex mechanisms of glucokinase regulation. Diabetes 2006; 55: 1713–1722. [DOI] [PubMed] [Google Scholar]
- 44. Stoffel M, Froguel P, Takeda J, et al. Human glucokinase gene: isolation, characterization, and identification of two missense mutations linked to early‐onset non‐insulin‐dependent (type 2) diabetes mellitus. Proc Natl Acad Sci USA 1992; 89: 7698–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McKinney JL, Cao H, Robinson JF, et al. Spectrum of HNF1A and GCK mutations in Canadian families with maturity‐onset diabetes of the young (MODY). Clin Invest Med 2004; 27: 135–141. [PubMed] [Google Scholar]
- 46. Lambert AP, Ellard S, Allen LI, et al. Identifying hepatic nuclear factor 1alpha mutations in children and young adults with a clinical diagnosis of type 1 diabetes. Diabetes Care 2003; 26: 333–337. [DOI] [PubMed] [Google Scholar]
- 47. Chèvre JC, Hani EH, Boutin P, et al. Mutation screening in 18 Caucasian families suggest the existence of other MODY genes. Diabetologia 1998; 41: 1017–1023. [DOI] [PubMed] [Google Scholar]
- 48. Alkorta‐Aranburu G, Carmody D, Cheng YW, et al. Phenotypic heterogeneity in monogenic diabetes: the clinical and diagnostic utility of a gene panel‐based next‐generation sequencing approach. Mol Genet Metab 2014; 113: 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vaxillaire M, Rouard M, Yamagata K, et al. Identification of nine novel mutations in the hepatocyte nuclear factor 1 alpha gene associated with maturity‐onset diabetes of the young (MODY3). Hum Mol Genet 1997; 6: 583–586. [DOI] [PubMed] [Google Scholar]
- 50. Ng MC, Cockburn BN, Lindner TH, et al. Molecular genetics of diabetes mellitus in Chinese subjects: identification of mutations in glucokinase and hepatocyte nuclear factor‐1alpha genes in patients with early‐onset type 2 diabetes mellitus/MODY. Diabet Med 1999; 16: 956–963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 | List of participating centers.
Table S2 | Clinical characteristic of probands.
Table S3 | Prediction scores from SIFT, PolyPhen‐2, Protein Variation Effect Analyzer and combined annotation‐dependent depletion for the novel mutations.
Table S4 | Clinical features of maturity‐onset diabetes of the young 2 stratified by the median of ∆2‐h glucose.
Table S5 | Clinical features of maturity‐onset diabetes of the young 2 patients treated with oral hypoglycemic agents and diet alone.
Table S6 | The clinical data of other maturity‐onset diabetes of the young mutations patients.
Figure S1 | Flow chart of the procedures for screening of maturity‐onset diabetes of the young mutations.
Figure S2 | Pedigrees of families with identified or suspected genetic causes of maturity‐onset diabetes of the young.


