Abstract
The adult liver has an exceptional ability to regenerate, but how it maintains its specialized functions during regeneration is unclear. Here, we used partial hepatectomy (PHx) in tandem with single-cell transcriptomics to track cellular transitions and heterogeneities of ∼22,000 liver cells through the initiation, progression, and termination phases of mouse liver regeneration. Our results uncovered that, following PHx, a subset of hepatocytes transiently reactivates an early-postnatal-like gene expression program to proliferate, while a distinct population of metabolically hyperactive cells appears to compensate for any temporary deficits in liver function. Cumulative EdU labeling and immunostaining of metabolic, portal, and central vein–specific markers revealed that hepatocyte proliferation after PHx initiates in the midlobular region before proceeding toward the periportal and pericentral areas. We further demonstrate that portal and central vein proximal hepatocytes retain their metabolically active state to preserve essential liver functions while midlobular cells proliferate nearby. Through combined analysis of gene regulatory networks and cell–cell interaction maps, we found that regenerating hepatocytes redeploy key developmental regulons, which are guided by extensive ligand-receptor-mediated signaling events between hepatocytes and nonparenchymal cells. Altogether, our study offers a detailed blueprint of the intercellular crosstalk and cellular reprogramming that balances the metabolic and proliferative requirements of a regenerating liver.
The liver is a multifunctional organ critical for carrying out numerous biosynthetic, metabolic, and detoxification functions. Owing to its detoxification roles, the liver is frequently exposed to many hepatotoxins, resulting in tissue damage and cell death. Accordingly, it has evolved a unique ability to regenerate in response to a wide range of physical and toxic injuries (Diehl and Chute 2013), and mammalian livers can replenish up to 70% of the lost tissue mass and functionality within weeks of surgical resection (Michalopoulos and DeFrances 1997; Michalopoulos 2017; Bangru and Kalsotra 2020). However, hepatic regeneration in humans is often compromised following xenobiotic injuries, viral infections, chronic inflammation, or excessive alcohol consumption, which leads to irreparable damage, fibrosis, and fulminant liver failure (Forbes and Newsome 2016; Seitz et al. 2018). It is estimated that nearly two million people die from liver disease every year, making it a prominent cause of global morbidity and mortality (Asrani et al. 2019).
Because most liver injuries trigger hepatocyte death, the regenerative course is primarily devoted to replenishing the lost hepatocyte population. Several cell-fate and lineage-tracing studies have determined that—under normal circumstances—the majority of new hepatocytes are derived from pre-existing hepatocytes (Schaub et al. 2014; Yanger et al. 2014; Font-Burgada et al. 2015). Depending on the extent of the injury, surviving hepatocytes rely on hypertrophic growth, cellular proliferation, or both to restore normal liver function (Miyaoka et al. 2012). Consequently, in order to stimulate cell division and growth, the regenerating hepatocytes undergo global alterations in gene expression through dynamic changes in mRNA abundance, splicing, and translation (Bangru et al. 2018; Wang et al. 2018 2019; Hyun et al. 2020). Although many previous studies have focused on the proliferative capacity of hepatocytes, the exact mechanics of regeneration such as how quiescent hepatocytes transition into a proliferative state, how regenerating livers sustain normal metabolic activities while the tissue recovers from injury, or what cell–cell interactions initiate and terminate the regenerative response are unknown.
Here, we leveraged a single-cell RNA sequencing (scRNA-seq) strategy to capture all resident cell types from mouse livers and dissected their cellular heterogeneities and responses to 70% partial hepatectomy (PHx) through the initiation, progression, and termination phases of regeneration. We also combined cumulative EdU labeling with immunostaining experiments to validate the scRNA-seq results and localized the proliferating vis-à-vis metabolically active hepatocytes within regenerating livers. Furthermore, we identified a vast array of ligand–receptor interactions among hepatocytes, endothelial, Kupffer, stellate, and T cells that coordinate the overall time course of regeneration. Thus, our study offers a high-resolution view of liver regeneration while providing a rich resource for the identification of genes and signaling pathways that facilitate hepatic repair in response to injury.
Results
Cell type composition, heterogeneity, and metabolic dynamics of a regenerating liver
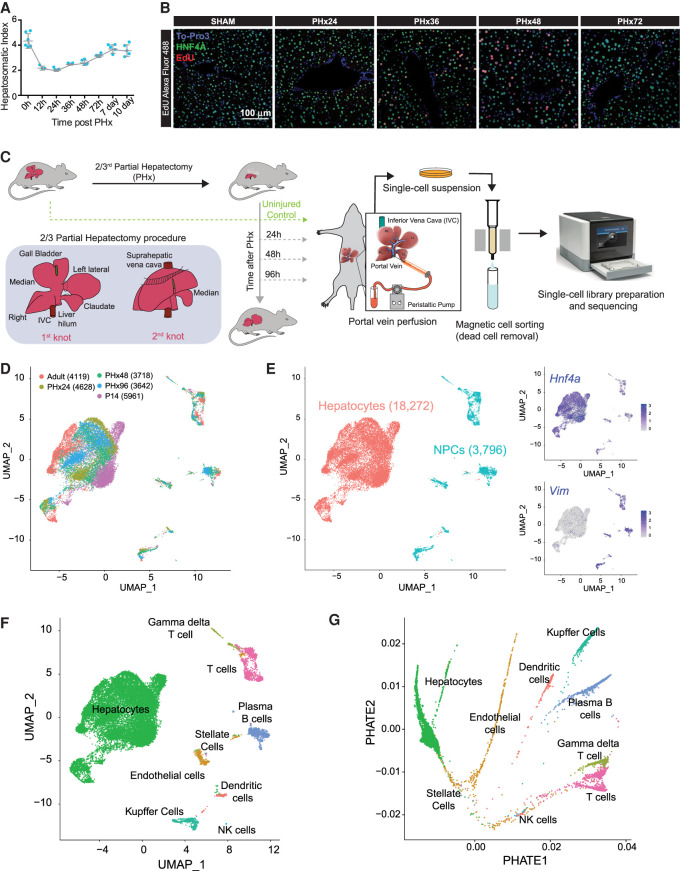
Surgical resection of the adult mouse liver by PHx induces rapid hyperplasia and hypertrophy in the remnant tissue, such that the liver recovers its original mass and function within 7 d (Fig. 1A; Supplemental Fig. S1A; Mitchell and Willenbring 2008). Hepatocytes, which constitute the bulk of liver parenchyma, are among the first cells to enter the cell cycle after PHx, followed by the proliferation of other stromal cells (Fausto et al. 2006). By labeling new DNA synthesis with 5-ethynyl-2′-deoxyuridine (EdU) and combining it with hepatocyte nuclear factor 4, alpha (HNF4A) immunostaining, we detected maximal proliferation activity between 24 and 72 h after PHx, which peaked at ∼36–48 h (Fig. 1B; Supplemental Fig. S1B). Therefore, to sample the cellular composition and diversity as well as profile their regenerative response at a single-cell resolution, we utilized a 10x Genomics-based scRNA-seq platform and studied the transcriptomes of all resident cell types isolated from mouse livers from uninjured adults as well as at 24, 48, and 96 h after PHx surgery (Fig. 1C; Supplemental Fig. S1C). In parallel, we also collected cells from postnatal day 14 (P14) livers—a midpoint between the neonatal period and weaning—and performed scRNA-seq to analyze the cellular transitions and gene programs associated with normal maturation of the liver. Single cells were isolated by two-step collagenase perfusion (Bhate et al. 2015), followed by magnetic-activated cell sorting that allows rapid and easy removal of dead cells (Supplemental Methods).
Figure 1.
Single-cell analysis of resident hepatic cell populations from immature, adult, and regenerating mouse livers. (A) Time course plot showing the restoration of liver-to-body weight ratio after partial hepatectomy (PHx). The liver recovers its original mass within 7 d after PHx (n = 5). (B) Fluorescent imaging of hepatocyte proliferation measured by in vivo EdU incorporation in post-PHx and Sham livers. White arrows indicate proliferating hepatocytes (colabeled for HNF4A in green, incorporated EdU in red, and nuclei in blue). Images taken under 20× resolution are shown. (C) Overview schematic demonstrating workflow for isolation of mouse liver cells for single-cell RNA sequencing (scRNA-seq). Portal vein perfusion of collagenase containing buffer was used to isolate single liver cells from uninjured P14 pups and adults as well as mice at 24, 48, and 96 h after 2/3rd PHx (n = 1/time point). Single-cell library preparation was performed with whole-cell suspensions individually for each mouse using a 10x Genomics Chromium Single Cell 3′ Reagent Kit (V3 chemistry) after magnetic-activated cell sorting to remove dead cells. The inset details our PHx procedure, showing the position of two knots before excision of the respective liver lobes. (D) Combined UMAP projection of all 22,068 cells identified after QC cutoffs and batch correction. Cells are colored by the batch of origin, and the total number of cells identified from each batch is given in parentheses. (E) Identification of hepatocyte and nonparenchymal cell (NPC) subpopulations. Graph-based clustering in Seurat v3.1 followed by marker gene analysis revealed broad epithelial and nonepithelial cellular identities. Feature plots shown as insets show higher expression of expression of Hnf4a (a hepatocyte marker) and Vim (a nonepithelial marker) especially in populations identified as hepatocytes and NPCs, respectively. (F) Combined UMAP projection of all cells, colored by the annotated cell type. (G) PHATE projection of the ∼22,000 cells from different stages of liver development and regeneration. Cells are colored by annotated cell types from F.
After appropriate quality control and normalization (Supplemental Fig. S2A–C), we captured a total of 22,068 cells that were evenly distributed among the five time points and had a mean UMI of 2148 and a median of 1097 genes expressed per cell (Supplemental Fig. S2B). A higher fraction of reads from the mitochondrial genome is often associated with low quality or dying cells. Because hepatocytes possess a very high mitochondrial content (MacParland et al. 2018) we used a relatively higher percentage of mitochondrial-read threshold (30%) in our analysis. A current limitation of the scRNA-seq technology is that it captures only ∼10% of the transcriptome per cell, which can influence the interpretation of underlying cell states and hinder the complete understanding of cellular processes. Therefore, we used stringent cutoffs and the gene imputation method MAGIC to minimize the potential influence of dropouts in our inferences (van Dijk et al. 2018). To further assess the reproducibility and faithful representation of the whole-liver transcriptome in our scRNA-seq experiments, we compared the summed expression of all genes in a given sample with corresponding bulk RNA-seq from published studies (Bhate et al. 2015; Wang et al. 2019). We found a high correlation with bulk RNA-seq measurements for all samples (Supplemental Fig. S3A), demonstrating good reliability of our scRNA-seq data sets. Next, to allow cross-sample comparisons, we integrated data sets from all samples and corrected their batch effects using the BEER algorithm (Zhang et al. 2019). Cell-type identity was assigned based on the top differentially expressed genes and from previously identified lists of canonical cell type–specific markers (MacParland et al. 2018; Xiong et al. 2019). Next, a graph-based clustering was performed to group cells according to their gene expression profiles. The integrated data set was projected into 2D-coordinates using Uniform Manifold Approximation and Projection (UMAP). Based on the relative expression of Hnf4a and the mesenchyme-derived cell marker vimentin (Vim), our time-resolved scRNA-seq data—after filtering, dead-cell removal, and batch-correction—yielded 18,272 hepatocytes and 3796 nonparenchymal cells (NPCs) (Fig. 1D,E).
The NPC population was further resolved into eight distinct clusters that represented liver sinusoidal endothelial cells (LSECs), stellate cells, Kupffer cells, dendritic cells, T cells, γ-δ T cells, Plasma B cells, and NK cells (Fig. 1F; Supplemental Figs. S3B,C, S4, S5A,B). We could not identify any cholangiocytes, and we attribute this absence to their relatively low numbers in quiescent and PHx-induced regenerating livers. Furthermore, we also projected BEER-corrected data using PHATE embedding (Moon et al. 2019) and found cells annotated as a cell type to cluster together in the projection (Fig. 1G; Supplemental Fig. S5C). To further assess the dependability of the BEER algorithm, we compared it with three other batch correction methods, including fastMNN, Conos, and Harmony (Supplemental Fig. S6A–D; Haghverdi et al. 2018; Barkas et al. 2019; Korsunsky et al. 2019; Tran et al. 2020). After batch correction, we labeled cells within the 2D projection according to their cell type (Supplemental Figs. S3B,C, S4). We found that all algorithms clustered the same cells across time points, indicating similar levels of correction success.
Metabolic adaptation due to hepatic insufficiency is a hallmark of liver tissue renewal and regeneration (Huang and Rudnick 2014; Caldez et al. 2018). Accordingly, we assessed changes in the strength of metabolic pathways among hepatocytes isolated from naive and regenerating (PHx) livers. Several pathways were coordinately down-regulated at 24 and 48 h after PHx, evident from a global shift in their pathway score distribution (Fig. 2A,B). Whereas the gene sets belonging to fatty acid, lipid, and amino acid metabolism showed the most drastic decrease through the initiation (24 h after PHx) and progression (48 h after PHx) phases of regeneration, other pathways such as glycolysis, gluconeogenesis, and pentose monophosphate shunt were only subtly muted (Fig. 2A). The dampening of liver metabolism was transient and largely reversed in the termination phase (96 h after PHx), revealing the dynamic nature of such changes evoked during regeneration. Moreover, while assessing the strength of different pathways/gene sets, we found trends wherein, at times when biosynthesis, detoxification, complement/coagulation, and other secretory functions associated with mature hepatocytes were down-regulated, the pathways related to cell cycle, proliferation, and growth such as ribosome biogenesis, RNA processing, splicing, and translation were up-regulated (Fig. 2B). Although the cell cycle and growth-related pathways had predominantly switched off at 96 h after PHx, some of the adult hepatocyte functions were not yet fully restored by this time. These results are in line with the bulk measurements that have explored how cell division and energy metabolism intersect to support liver regeneration (Bangru et al. 2018; Caldez et al. 2018; Wang et al. 2018, 2019), and they highlight the metabolic flexibility of regenerating hepatocytes.
Figure 2.
Specific hepatocyte population reversibly reprograms to an immature postnatal-like state during regeneration. (A) Ridge plots showing relative scoring on hepatocyte subpopulation using Seurat3.1 demonstrate extensive rewiring of metabolic genes during regeneration. Relative scores were computed based on the lists of genes for each pathway obtained from the Rat Genome Database (RGD). (B) Heat map showing relative scores of the top differentially regulated metabolic pathways. (C) Pseudotime plots demonstrating cellular trajectories during postnatal maturation (including P14 and adult hepatocytes), initiation, and progression (including adult, PHx24 and PHx48 hepatocytes), and termination and rematuration (including PHx48 and PHx96 hepatocytes). Single-cell trajectories were constructed and pseudotime values calculated using Monocle 2. Trajectories are colored by pseudotime (left) and sample identity (right). (D) Heat maps representing modules of genes that co-vary along the pseudotime during postnatal maturation, initiation–progression, and termination–rematuration phases. DAVID-based Gene Ontology (GO) analysis revealed that reversible reprogramming of developmentally regulated gene expression programs essentially reverts postnatal maturation, and this is followed by transitions that reinstate mature hepatic program. The top up-regulated and down-regulated GO terms are described below the respective heat maps. (E) Pseudotime plot indicating cellular trajectories of hepatocytes from all samples. Single-cell trajectories were constructed and pseudotime values were calculated using Monocle 2. Trajectories are colored by pseudotime. (F) Pseudotime plots showing distribution of each sample along combined cellular trajectories shown in C. The adult and P14 hepatocytes present distinct distributions along the trajectory; however, the distribution shifts toward P14 at PHx24–48 and back toward the adult at PHx96.
Regenerating hepatocytes reprogram to an early postnatal-like state
To map the cellular transitions as hepatocytes move from a quiescent to proliferative state and back, we performed pseudotime ordering of hepatocytes. Because we expected our data to follow a linear trajectory with some branching, we utilized the trajectory inference tools best suited for such analyses. We built individual trajectories using the Monocle2 DDRTree pipeline, which provides high accuracy and overall performance and is comparable to Slingshot or other existing methods (Saelens et al. 2019). The top differentially expressed genes among hepatocytes collected from different time points were used for ordering cells (Qiu et al. 2017). We constructed discrete cell-state trajectories for (1) the normal postnatal maturation (P14→Adult), (2) the initiation–progression phase of regeneration (Adult→PHx24→PHx48), and (3) the termination–rematuration phase of regeneration (PHx48→PHx96) (Fig. 2C). For both postnatal maturation and termination–rematuration trajectories, the cells from respective time points yielded distinct branches with a few transitioning cells (Fig. 2C, top and bottom). Conversely, for the initiation–progression phase trajectory, whereas a large number of regenerating hepatocytes diverged away from the adult state, a significant portion retained their mature identity, revealing a distributive model of regeneration (Fig. 2C, middle).
Next, to identify the functional pathways changing within individual trajectories, we determined the expression dynamics of the top 2000 genes that vary as a function of progress through the pseudotime. Along the postnatal maturation trajectory, the expression of genes encoding RNA processing, ribosome assembly, and translational regulation factors declined first, followed by a simultaneous increase in the expression of genes associated with various adult hepatocyte functions (Fig. 2D, left). Recently, bulk transcriptome analyses revealed that, in response to toxin-mediated injury, regenerating hepatocytes reprogram to an early postnatal-like state (Bangru et al. 2018). However, it is unclear whether regeneration after PHx involves a similar reprogramming event. We observed that along the initiation–progression trajectory of regeneration—opposite to postnatal maturation—many metabolic genes in hepatocytes were down-regulated prior to the up-regulation of genes encoding cell cycle, RNA processing, and translation regulation factors. Particularly, the genes involved in ribosome biogenesis and assembly were activated only briefly along the pseudotime, underscoring that a temporary boost in protein synthesis is needed to prime hepatocytes for cell cycle re-entry (Fig. 2D, middle). Meanwhile, along the termination–rematuration trajectory—similar to postnatal maturation—down-regulation of cell cycle-, RNA processing-, and translation-related genes preceded a synchronous increase in the expression of various metabolic and biosynthetic genes (Fig. 2D, right).
It has been postulated that most hepatocytes can re-enter the cell cycle and proliferate after 2/3rd PHx (Michalopoulos 2017). To determine if all hepatocytes after PHx reprogram to a postnatal-like state and proceed simultaneously toward the proliferative trajectory, we performed pseudotime ordering of cells from all time points (i.e., P14, Adult, PHx24, PHx48, and PHx96) (Fig. 2E). We found that while the P14 and adult hepatocytes resided on separate arms of the trajectory, regenerating hepatocytes were distributed among all three arms (Fig. 2F). The majority of hepatocytes at 24 h after PHx migrated away from their adult position at the beginning of pseudotime and occupied a position around the branch point (Fig. 2F). At 48 h after PHx, a large number of hepatocytes inhabited the far-right arm of the trajectory overlapping with the position of P14 hepatocytes. However, at 96 h after PHx, most hepatocytes had left the P14 state, returning back to their initial adult state (Fig. 2F). To assess the reliability of these findings, we reanalyzed our data using alternate pipelines. Briefly, we performed batch correction and projected data using Harmony and UMAP embedding (Seurat package), respectively (Supplemental Fig. S7A–D), following which we used the 2D-projection to identify trajectories with Slingshot and Monocle3 (Street et al. 2018). Pseudotime values derived from either methodology showed a high correlation in terms of the positioning of cells (Supplemental Fig. S7E,F). These findings were further corroborated with Wishbone (Supplemental Fig. S8; Setty et al. 2016). Furthermore, we also used PHATE projection on FastMNN corrected embedding of hepatocytes and combined it with Slingshot to validate bifurcation of cells toward the metabolic or proliferative lineages (Supplemental Fig. S9). Thus, our analysis captured the cellular plasticity of hepatocytes as they progress through different stages of regeneration. At all times, a considerable population (∼20%–30%) of cells remained adult-like and occupied their original position on the pseudotime axis, indicating that not all hepatocytes are reprogrammed after PHx (Fig. 2F; Supplemental Fig. S10A–C). Together, these findings illustrate that reversible postnatal-like reprogramming facilitates hepatocytes to transition from a quiescent to a proliferative state and back. The dampening of mature characteristics followed by a transient increase in protein synthesis is likely a prerequisite for cell cycle re-entry required for jump-starting the regenerative process.
Division of labor balances the proliferation and metabolic demands of a regenerating liver
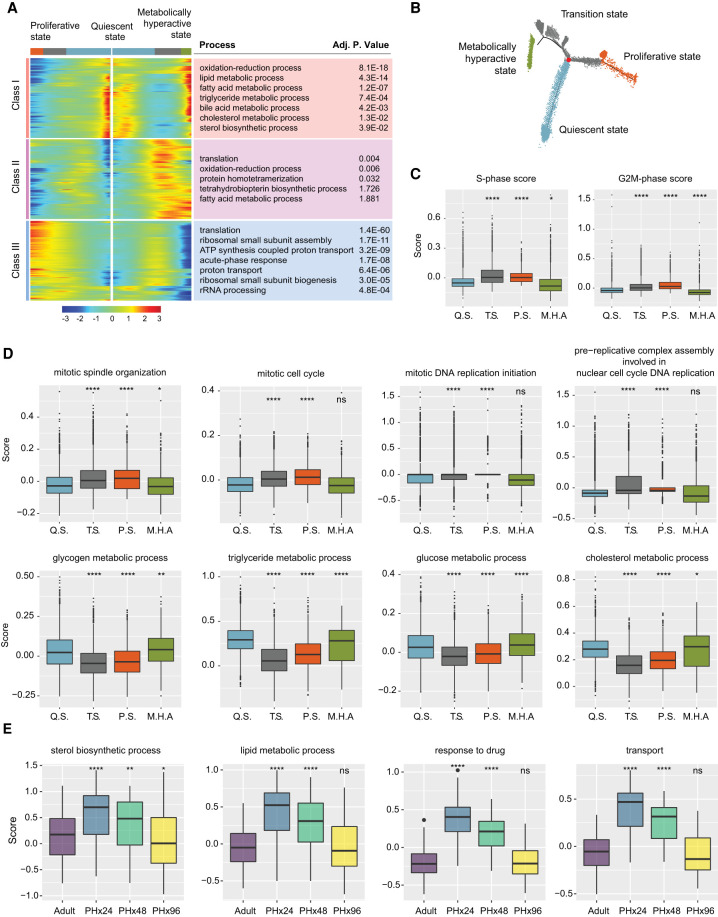
The DDRTree algorithm resolved our pseudotime trajectory into nine distinct hepatocyte populations (HEP1–HEP9) (Supplemental Fig. S10D). Consequently, to gain a deeper insight into the HEP1–HEP9 populations, we determined their gene expression profiles along the pseudotemporal trajectory. We found that HEP1 and HEP2 populations were enriched for genes expressed in immature hepatocytes and de-enriched for genes expressed in mature hepatocytes (Supplemental Fig. S10D,E). Contrary to HEP1 and HEP2, the HEP4 and HEP7 populations were enriched for genes expressed in mature and de-enriched for genes expressed in immature hepatocytes. HEP3, HEP5–6, and HEP8–9 populations showed intermediate enrichment for immature and de-enrichment for mature gene expression (Supplemental Fig. S10D,E). Based on these transcriptome signatures, we categorized HEP1–HEP9 populations into four broader clusters. The adult HEP4 cluster was designated as the “quiescent state.” The cluster near the branch point emerging after PHx and comprising HEP3, HEP5–6, and HEP8–9 populations was termed as the “transition state.” Further, the cluster formed by HEP1 and HEP2 populations was marked as the “proliferative state,” whereas the cluster formed by the HEP7 population was designated as the “metabolically hyperactive state” (Fig. 3A,B).
Figure 3.
Bifurcation of hepatocyte trajectory during regeneration produces hepatocytes enriched with complementary functions in proliferation and metabolism. (A) Heat map showing bifurcating of gene expression programs executed along the pseudotime after branching. The top GO terms enriched in each class of genes are listed with their corresponding adjusted P-values. (B) Trajectory demonstrating the three distinct states of hepatocytes. The branch point under evaluation is shown in red. (C) Box plots demonstrating cell cycle phase scores calculated from Seurat v3.1 for hepatocytes belonging to different cell states. Q.S., T.S., P.S., and M.H.A. denote quiescent, transition, proliferating, and metabolically hyperactive states, respectively. P-values were derived from a parametric t-test (unpaired). (*) P ≤ 0.05, (****) P ≤ 0.0001. (D) “Proliferating” and “metabolically hyperactive” states uniquely up-regulate proliferation- or metabolism-related functions, respectively. Box plots showing relative scoring of indicated pathways in hepatocytes belonging to different states. P-values were derived from a parametric t-test (unpaired). (*) P ≤ 0.05, (**) P ≤ 0.01, (****) P ≤ 0.0001, (ns) P > 0.05. (E) Metabolically hyperactive state transiently up-regulates metabolism-related functions during regeneration. Box plots showing time point-based scoring of hepatocytes from the metabolically hyperactive state for the indicated pathways. P-values were derived from a parametric t-test (unpaired). (*) P ≤ 0.05, (**) P ≤ 0.01, (****) P ≤ 0.0001, (ns) P > 0.05.
The quick emergence of the transition state following PHx suggested it is derived from the quiescent state, after which it bifurcates toward the proliferative or metabolically hyperactive states. To further examine how gene expression diverges after PHx and generates discrete clusters of proliferative and metabolically active hepatocytes, we used BEAM module analysis within the Monocle 2 pipeline. We identified genes changing along different arms of the DDRTree, and these were grouped into three main classes (Fig. 3A). Class I contained genes that were highly expressed in the quiescent state, down-regulated as the bifurcation point was approached and up-regulated again in the metabolically hyperactive state. In contrast, classes II and III contained genes that were poorly expressed in the quiescent state, up-regulated as cells progressed through the transition point but were then reciprocally up- or down-regulated in the proliferative and metabolically hyperactive states, respectively. Upon further gene set enrichment analysis, we confirmed that the metabolically hyperactive hepatocytes showed overrepresentation of functional categories related to biosynthesis and metabolism (Fig. 3A,D). Conversely, hepatocytes associated with the proliferative state showed an overrepresentation of cell cycle- and growth-related functional categories, including DNA replication, and mitosis. (Fig. 3A,C,D). Consistent with these results, recent histological analysis of regenerating mice livers after PHx detected intertwined sets of hepatocytes that could be distinguished according to elevated glycogen content with low mitotic activity or reduced glycogen content with high mitotic activity (Minocha et al. 2017).
Having demarcated the four cellular states along the trajectory, we reasoned that if metabolically hyperactive hepatocytes do actually compensate for any temporary deficits in liver function while other hepatocytes proliferate, they should exhibit a surge in the expression of metabolic genes after PHx. To test for this possibility, we analyzed the cells of metabolically hyperactive state in relation to their time point of origin. Relative to the healthy adults, metabolically active hepatocytes at 24 h after PHx showed significantly higher expression of biosynthetic, metabolic, detoxification, and transport related genes, which started to decline at 48 h and were essentially reversed by 96 h after PHx (Fig. 3E). Collectively, these data support a division-of-labor model wherein, after PHx, a subset of residual hepatocytes acquire the metabolically hyperactive state that up-regulates its adult gene programs to counteract regeneration-associated deficits in liver function.
Rewiring of gene regulatory networks activates cell state transitions during regeneration
To delineate the gene regulatory networks (GRNs) that might stimulate various cell state transitions in regeneration, we used the single-cell regulatory network inference and clustering (SCENIC) pipeline (Aibar et al. 2017) on our scRNA-seq data (see Methods). SCENIC computes the activity of transcription factors from individual cells by integrating co-expression data with transcription factor motif enrichment analysis, generating a “regulon,” which refers to an expressed transcription factor and all of its co-expressed target genes. We obtained the predicted regulon activities using AUCell, which ranks targets of each regulon among the expressed genes in each cell, yielding a regulon-by-cell activity matrix. The overarching function of SCENIC is to create regulon-driven clusters that are generated from the regulon-activity matrix through binarizing (by thresholding) the original AUCell score. We, however, discerned that instead of binarizing, maintaining the full AUCell score improves the inferences performed on the data.
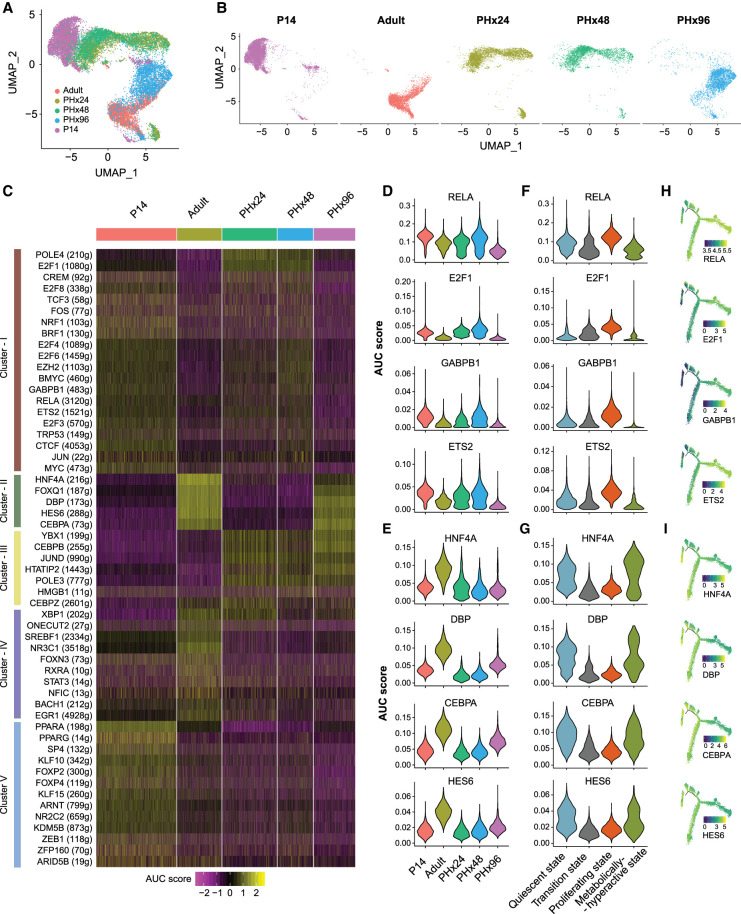
We hypothesized that the reimplementation of certain developmental GRNs might drive cellular transitions during regeneration. To test this hypothesis, we analyzed the AUCell score activity matrix of individual hepatocytes acquired from all time points (i.e., P14, Adult, PHx24, PHx48, and PHx96). Upon 2D UMAP visualization—built from the AUCell scores—we noted that hepatocytes from specific time points grouped together, underscoring a high degree of similarity in their regulon activity (Fig. 4A,B). The P14 and adult stage hepatocytes formed distinct nonoverlapping clusters at far ends of the UMAP plot, representing clear differences in their GRNs. The hepatocytes from 24 h and 48 h after PHx, however, clustered adjacent to P14 and away from the adult stage, demonstrating that similar regulons are active during postnatal development and initiation–progression phases of regeneration. Very few PHx96 hepatocytes overlapped with P14, PHx24, or PHx48 time points (Fig. 4A,B). Instead, they were clustered around adult cells, which indicates that, in the termination phase, hepatocytes restore their mature adult-like regulon activity. Based on these findings, we conclude that, in order to reprogram gene expression after PHx, the liver cells, in part, redeploy the same GRNs that are utilized for physiologic growth during the postnatal period of development.
Figure 4.
Gene regulatory networks are rewired to a postnatal-like state during regeneration. (A) UMAP projection of all hepatocytes based on the AUC scores for each regulon calculated with SCENIC. Cells are colored according to the sample of origin. (B) AUC score-based UMAP projection, grouped according to the sample of origin. Cells are colored according to sample of origin. Adult and PHx96 hepatocytes cluster together, whereas PHx24 and PHx48 hepatocytes cluster together with P14 hepatocytes. (C) Heat map depicting the activities of different regulons that show time point-dependent variations. (D) Violin plot showing distribution of AUC scores for RELA, E2F1, GABP1, and ETS2 regulons across hepatocytes from each time point demonstrating their high activity in PHx24, PHx48, and P14 hepatocytes. (E) Violin plots showing distribution of AUC scores for HNF4A, DBP, CBPA, and HES6 regulons across hepatocytes from each time point demonstrating their high activity in adult and PHx96 hepatocytes. (F) Violin plots showing distribution of AUC scores of representative regulons across hepatocytes showing their up-regulation in the proliferative state. (G) Violin plots showing distribution of AUC scores of representative regulons across hepatocytes showing their up-regulation in quiescent and metabolically active states. (H) Pseudotime plots of hepatocyte cellular trajectories colored by the AUC scores of representative regulons showing high activity in the proliferative state. (I) Pseudotime plots of hepatocyte cellular trajectories colored by the AUC scores of representative regulons showing high activity in quiescent and metabolically active states.
Next, we explored regulon activities distinguishing the cellular features of normal and regenerating hepatocytes. Our goal was to identify GRNs that are selectively activated during the initiation–progression and the termination–rematuration stages of regeneration. We reasoned that regulon activities of the initiation–progression GRNs would typically be low in adult hepatocytes, stimulated at PHx24 and PHx48, and dissipated again at PHx96. Correspondingly, regulon activities of the termination–rematuration GRNs would normally be high in adult hepatocytes, muted at PHx24 and PHx48, and stimulated again at PHx96. Indeed, within the 56 differentially active regulons, we detected many that fit these criteria (Fig. 4C; Supplemental Fig. S11A–D). For instance, RELA, E2F1, GABPB1, and ETS2 regulons were active in hepatocytes from P14 and the initiation–progression stage of regeneration but were turned off in adult hepatocytes and at the termination–rematuration stages (Fig. 4C [cluster I],D), This indicates that these regulons likely play dual roles in regulating the hepatocyte hyperplastic response—that is, in normal liver development and following an injury in adult animals. In line with our results, previous studies have found that a rapid increase in the expression and/or DNA-binding activity of NF-kB (RELA and RELB), E2Fs (E2F1, 3, 4, 6, and 8), AP-1 (JUN and FOS), POLE4, TRP53, MYC, CREM, and ETS (ETS2, GABPB1) family of transcription factors is involved in the initiation of stress signaling, oxidative stress, DNA replication/repair, and cell-cycle entry at the early stages of liver regeneration (Bhat et al. 1987; Yang et al. 1991; Westwick et al. 1995; Iimuro et al. 1998; Servillo et al. 1998; Chaisson et al. 2002; Inoue et al. 2002; Baena et al. 2005; Stepniak et al. 2006; Beyer et al. 2008; Kurinna et al. 2013; Wu et al. 2013; Sladky et al. 2020).
In contrast, HNF4A, DBP, CEBPA, and HES6 regulons were highly active in adult hepatocytes, muted at P14 and the initiation–progression stages, but then reactivated at the termination–rematuration stage (Fig. 4C [cluster II], E), pointing toward their role in the termination of liver regeneration. The function of hepatocyte nuclear factor 4A (HNF4A), a nuclear receptor, in hepatocyte differentiation is well established (Parviz et al. 2003)—as it directs the expression of gene programs involved in xenobiotic, carbohydrate, and fatty acid metabolism as well as in bile acid synthesis, blood coagulation, and ureagenesis (Gonzalez 2008). Previous studies have described HNF4A's antiproliferative effects in hepatocytes (Hatziapostolou et al. 2011; Bonzo et al. 2012), and more recently, it was found that HNF4A is indispensable for terminating regeneration after PHx (Huck et al. 2019). Consistent with our regulon activity scores, HNF4A protein levels diminish rapidly after PHx, and this initial decrease followed by re-expression is needed for hepatocytes to timely enter and exit the cell cycle and to re-establish mature liver functions once regeneration is complete (Huck et al. 2019).
Like HNF4A, dynamic and temporally regulated activities of the CAAT/Enhancer-Binding Proteins (CEBPs) are critically important for coordinating gene expression changes through the shifting phases of regeneration (Greenbaum et al. 1995; Jin et al. 2015). CEBPA and CEBPB are basic region-leucine zipper-containing transcription factors that act as homo- or heterodimers and bind similar DNA sequences. We found that CEBPA and CEBPB regulon activities exhibit an opposing pattern through the initiation–progression and termination–rematuration stages of regeneration (Fig. 4C,E; Supplemental Fig. S11B). CEBPA regulon activity—comprising metabolic and liver homeostatic genes—is high in pre-PHx adult hepatocytes, suppressed at PHx24 and PHx48, then enhanced in PHx96 hepatocytes. Conversely, CEBPB regulon activity—comprising many acute phase- and cell cycle-related genes—is low in pre-PHx adult hepatocytes but rapidly up-regulated after PHx. Of note, CEBPA and CEBPB usually bind the same genomic locations in hepatocytes (Jakobsen et al. 2013), except with divergent temporal patterns during regeneration (Kuttippurathu et al. 2017). These dynamic shifts in genomic occupancies are tightly linked to the changes in the relative ratio of CEBP proteins such that a high CEBPA:CEBPB ratio promotes binding to cis-regulatory sequences boosting metabolic and suppressing acute phase response genes, whereas a low ratio directs binding to sequences that repress metabolic and activate cell cycle- and acute phase-related genes (Jakobsen et al. 2013). Although direct roles of D-box binding protein (DBP, a circadian PAR bZIP transcription factor) or the HES family of basic helix-loop-helix transcription factor 6 (HES6) in liver regeneration remain unexplored, their regulon activity patterns (Fig. 4E) hint that both are termination factors.
We also recognized several regeneration-specific regulons that became activated after PHx but were otherwise inactive in postnatal development (Fig. 4C [cluster III]; Supplemental Fig. S11A). Some developmental regulons that were redeployed following PHx maintained their activated/deactivated patterns up to 96 h, indicative of their extended roles in regeneration (Fig. 4C [cluster IV]; Supplemental Fig. S11B). Finally, we also detected some development-specific regulons that were not redeployed either in the initiation–progression or the termination–rematuration stages of regeneration (Fig. 4C [cluster V]; Supplemental Fig. S11C), suggesting that a portion of the genetic machinery critical for physiological liver growth and development is dispensable for regeneration. Hence, a variety of GRNs positively and negatively impact hepatocyte proliferation, and dynamic utilization of transcription factors instructs these regulons to synchronize the timely initiation, progression, and termination of liver regeneration.
We next studied the correlation of regulon activities with the pseudotemporal transition of hepatocytes and the four cellular states described earlier (Fig. 4F–I; Supplemental Fig. S12). By overlaying AUCell scores along the pseudotime trajectory, we noticed that RELA, E2F1, GABPB1, and ETS2 regulons—which were active in hepatocytes from P14 and initiation–progression stages of regeneration—displayed significantly higher activity in the proliferative state relative to quiescent, transition, or metabolically hyperactive states (Fig. 4F,H). In contrast, HNF4A, DBP, CEBPA, and HES6 regulons—all of which promote mature functions—were much more active in quiescent and metabolically hyperactive states relative to transition or proliferative states (Fig. 4G,I). Collectively, these data indicate that underlying changes in regulon activities are critical for determining hepatocyte identities and cell state transitions, which help preserve specialized liver functions while the regenerating tissue balances its metabolic and proliferation needs.
Discrete localization of proliferating and metabolically active hepatocytes during liver regeneration
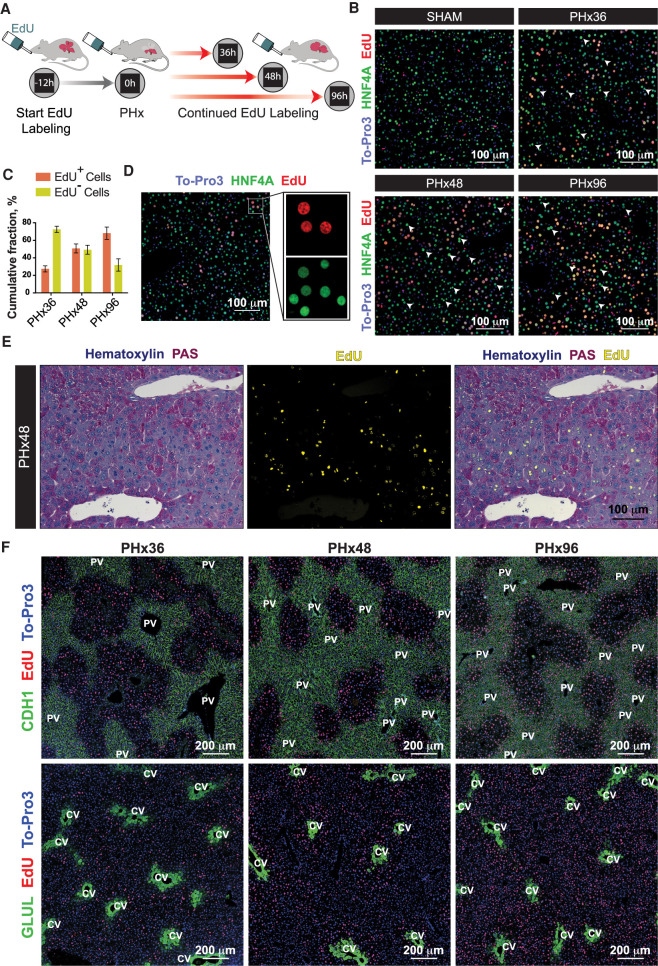
As described earlier, the trajectory analyses indicated that not all hepatocytes reprogram to a proliferative state after PHx and that a sizeable proportion maintains the adult state throughout the course of regeneration (Figs. 2, 3). To independently verify these observations, we fed EdU-supplemented water to mice ad libitum starting at 12 h before PHx or Sham surgery, continuing until the time of sacrifice (Fig. 5A). This provided cumulative labeling of all hepatocytes undergoing DNA synthesis and served as a proxy for proliferation through a given time frame. Although few EdU+ hepatocytes were detected after Sham surgery, a progressive increase in the number of EdU+ hepatocytes was evident during the course of regeneration (Fig. 5B,C). Nonetheless, a significant population of hepatocytes did not incorporate any EdU across all phases of regeneration, with nearly 30% remaining EdU− even at 96 h after PHx (Fig. 5B,C). These results corroborate our scRNA-seq data and demonstrate that a substantial number of hepatocytes do not proliferate after PHx (Supplemental Fig. S10B,C). Moreover, a side-by-side comparison of EdU+ and EdU− cells revealed that proliferating hepatocytes, in general, expressed lower amounts of HNF4A protein compared to the adjacent nonproliferating hepatocytes (Fig. 5D). These data are consistent with our GRN analysis of regenerating hepatocytes, wherein HNF4A regulon activity was significantly diminished in the proliferating state relative to the metabolically hyperactive state (Fig. 4G).
Figure 5.
Proliferating and metabolically active hepatocytes are discretely localized within regenerating livers. (A) Schematic showing the experimental strategy for cumulative labeling of proliferating cells by continued feeding of EdU during liver regeneration. (B) Representative immunofluorescence images of proliferating hepatocytes measured by cumulative EdU incorporation after PHx or Sham surgeries (n = 4 mice/time point). Proliferating hepatocytes were colabeled for HNF4A (green) and EdU (red). White arrowheads point to the hepatocytes without any EdU incorporation at each time point, representing cells that had not proliferated. Nuclei were stained with To-Pro-3 dye (blue). (C) Percentages of proliferated (EdU+) and nonproliferated (EdU−) hepatocytes in regenerating livers at PHx36, 48, and 96 h. Data are mean ± SD; n = 4 mice/time point. (D) Representative immunofluorescence image demonstrating down-regulation of HNF4A protein specifically in proliferating nuclei (EdU+) of a regenerating liver. Liver sections were colabeled for HNF4A in green, EdU in red, and nuclei in blue. (E) Representative images showing an overlay of hepatic glycogen content by PAS staining (purple) and fluorescently detected EdU incorporation (yellow) in PHx48 livers. Nonproliferating hepatocytes exhibited minimal glycogen depletion (n = 4 mice/time point). (F) Representative immunofluorescence images showing spatial segregation of metabolic and proliferating hepatocytes through different phases of liver regeneration. Periportal (CHD1+) or pericentral (GLUL+) hepatocytes were colabeled (green) along with EdU (red). Nuclei were stained with To-Pro-3 dye (blue) (n = 4 mice/time point).
To further confirm the mutually exclusive nature of proliferating and metabolically active hepatocytes in regenerating livers, we performed simultaneous labeling of intracellular glycogen (by PAS staining) and EdU after PHx. We found that (1) neighboring hepatocytes at 48 h after PHx could be either depleted or packed with glycogen, (2) EdU+ hepatocytes were essentially devoid of any glycogen, and (3) glycogen-depleted EdU+ hepatocytes were localized primarily in the midlobular region (Fig. 5E). This demonstrated clear segregation of proliferating and metabolically active hepatocytes—validating the division-of-labor model, wherein a subset of cells sustains their normal metabolic activities while others re-enter the cell cycle. Next, to ascertain the precise location of cycling and metabolic hepatocytes in regenerating livers, we probed for cumulative EdU incorporation over time along with staining for the portal (CDH1) and central (GLUL) vein-specific markers (Fig. 5F; Supplemental Fig. S13A–C). We noticed that, in both PHx36 and PHx48 livers, EdU+ hepatocytes were distributed around the midlobular zone, while glycogen-rich, periportal, and pericentral cells remained EdU− and maintained their zonated CDH1 and GLUL expression, respectively (Fig. 5E, left and middle). At 96 h, more EdU+ cells could be detected near the portal and central veins (Fig. 5E, right), indicating compensatory hepatocyte proliferation after PHx is initiated in the midlobular region before proceeding toward the periportal and pericentral areas. These results are particularly revealing and provide key evidence for the portal and central vein proximal regions as the likely depots of hepatocytes retaining a metabolically active state while midlobular cells proliferate to reconstitute the liver structure and function.
Transitions in hepatocyte states dictate the cell signaling dynamics of regeneration
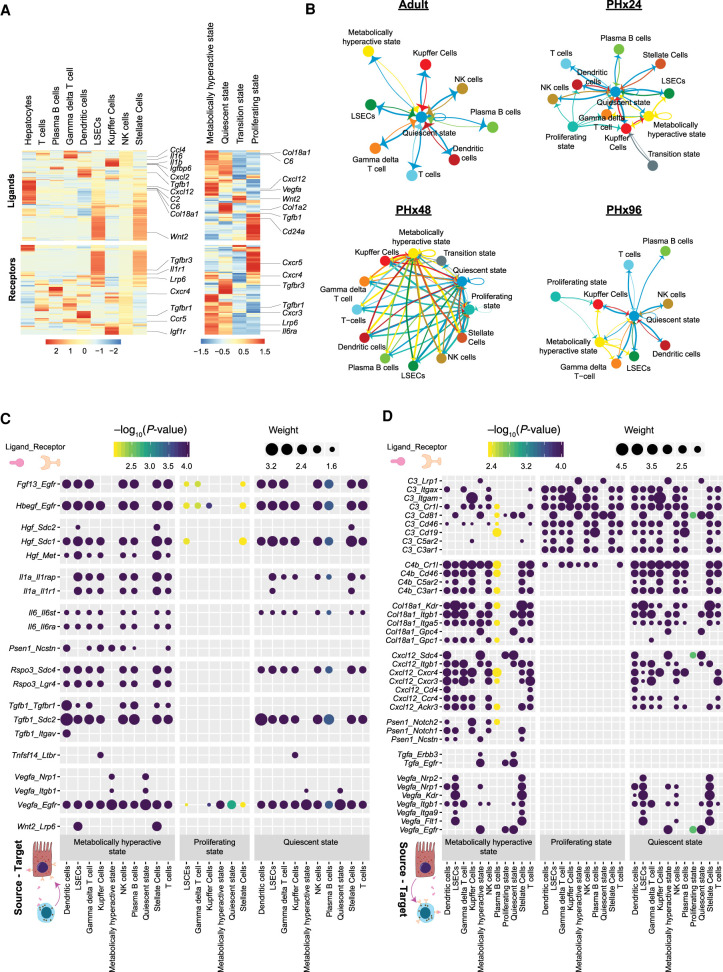
Next, we explored the dynamics of potential cell–cell communication networks at different stages of regeneration. We first inspected the cell type–specific RNA expression of various ligands in the liver secretome and their corresponding receptors, as previously described (Xiong et al. 2019). Our analysis mapped numerous unique clusters of ligand-receptor pairs with cell type–specific expression patterns, which highlights the distinct roles of hepatocytes and NPCs in shaping intrahepatic signaling topologies (Fig. 6A). Among NPCs, we noticed that LSECs and stellate cells comprised the largest clusters, underscoring their predominant roles in cell signaling (Fig. 6A, left). Significant differences in ligand-receptor expression profiles were also detected among hepatocytes belonging to different states, which indicates the remodeling of interaction landscapes with cell state transitions (Fig. 6A, right).
Figure 6.
Dynamics of cell–cell interactions during liver regeneration. (A) Heat map showing expression of various ligand molecules and cellular receptors from different liver cell types (left) and from hepatocytes belonging to different cell states (right). (B) Network diagrams showing cell–cell interactions indicated by arrows (edges) pointing in the source-to-target direction. Thickness indicates the sum of weighted paths between populations, and the color of arrows corresponds to the source. Network diagrams for Adult, PHx24, PHx48, and PHx96 are shown. (C) Dot plot of representative inbound signals to hepatocytes at PHx48. Size of each dot indicates the weight of the corresponding ligand–receptor interaction and the color indicates negative log10 P-value of the source-to-target interaction. Empirical P-values were calculated and Benjamini–Hochberg correction was performed. (D) Dot plot of representative outbound signals from hepatocytes to various liver cells at PHx48.
Particularly, we noted that many signaling molecules with established roles in liver regeneration—such as cytokines, chemotactic factors, secreted matrix proteins, growth factors, adhesion molecules, and mitogens—originate from specific cell types (Supplemental Fig. S14A–C). Expression of Wnt2, for instance, was predominantly seen in LSECs (Supplemental Fig. S14C), reaffirming results from previous studies of liver regeneration following PHx and acute CCl4 toxicity (Ding et al. 2010; Zhao et al. 2019). Pseudotime ordering revealed that up-regulation of Wnt2 and Hgf expression correlates with the transition of LSECs to an activated state (Supplemental Fig. S15A–C) and, as reported earlier, associates with KDR (VEGFR2)-ID1 activity (Ding et al. 2010). Furthermore, Wnt2 was expressed at much lower levels in Kupffer cells relative to LSECs (Supplemental Fig. S14C), consistent with their minor role in Wnt–beta-catenin signaling (Russell and Monga 2018).
To study the intracellular crosstalk among hepatic cell types and how it is modified during regeneration, we constructed cell–cell communication networks (Farbehi et al. 2019) for each time point from our data set (Fig. 6B; Supplemental Fig. S16). The edges of the network are directed from source to target cells, which express specific ligands and their corresponding receptors, respectively. The thickness of edges corresponds to weights representing fold-changes in the expression of ligand-receptor pairs (see Methods). Together, this generated a weighted and directed network of potential cell–cell interactions within normal and regenerating livers. Of note, the cell–cell communication networks exhibited significant rewiring, evoking a transient increase in overall cellular crosstalk during regeneration. We noticed an increase in interactions between hepatocytes and NPCs at PHx24 and particularly at PHx48, followed by a re-establishment of an adult-like communication network at PHx96. Hepatocytes displayed discrete profiles of interactions with other cell types in a state-dependent manner, as expected from the differences in ligand-receptor expression observed in Figure 6A. Throughout regeneration, quiescent hepatocytes consistently made strong inbound and outbound connections with most other cell types, whereas transition state hepatocytes were refractory to any crosstalk. The metabolically hyperactive hepatocytes exhibited an adaptable pattern, with prominent interactions at PHx48 but minimal interactions otherwise. The proliferating hepatocytes presented a unique interaction landscape with strong outbound connections and few to no inbound connections. It is noteworthy that cells transitioning to the proliferating state are more amenable to regenerative cues such that continued stimulation by pro-proliferative ligands would lead to excessive/uncontrolled proliferation. We postulate that the down-regulation of receptors related to pro-proliferative signals might be crucial for limiting the endless proliferation of hepatocytes and facilitating their timely exit from the cell cycle.
Next, we studied the individual ligand–receptor interactions among various cell types. We constructed dot plots for each time point demonstrating all ligand–receptor interactions with a minimum path weight of 1.5, for all significant cell–cell relationships (Padj < 0.01) (Supplemental Figs. S17–S21). This provided comprehensive visualization of potential cellular interactions, divulging time point-specific differences in outbound and inbound signaling from/to hepatocytes. Even at PHx48, when maximal intercellular crosstalk was observed, proliferative state hepatocytes appeared to receive a distinctly low number of incoming signals (Fig. 6C; Supplemental Figs. S17–S21), which matched with the lower inbound edges to these cells seen in Figure 6B. Contrary to this, we detected significant inbound signaling toward quiescent and metabolically hyperactive state hepatocytes, which was mediated by several growth factors, interleukins, and the Wnt signaling pathway (Fig. 6C; Supplemental Figs. S17–S21). For instance, consistent with earlier reports (Schirmacher et al. 1992; Maher 1993; LeCouter et al. 2003), our analysis predicted hepatocytes to receive prominent HGF/MET signaling from LSECs and stellate cells. We did not capture an EGF-signaling network among different cell types, which is in agreement with low EGF expression in hepatic cells (Supplemental Fig. S14C) and its predominantly exogenous origin (St Hilaire et al. 1983; Olsen et al. 1985). However, we detected prominent heparin binding (HB)-EGF signaling from Kupffer cells and LSECs and other NPC populations (Kiso et al. 2003). Although TGFB protein levels in hepatocytes are debatable (Braun et al. 1988; Bissell et al. 1995), we found that Tgfb1 RNA is abundant in regenerating hepatocytes and most NPCs but without any significant autocrine TGFB activity within hepatocytes (Fig. 6C; Supplemental Fig. S14C). As expected, we detected RNA for many known mitogenic signals inbound to hepatocytes such as Fgf, Tnf, and Il6, which were high between 24 and 48 h after PHx but had declined by 96 h (Supplemental Figs. S17–S21).
Outbound signals from hepatocytes at PHx48 involved pathways such as Tgfa, Vegfa, collagen, complement, and chemokine signaling (Fig. 6D; Supplemental Figs. S17–S21). We noticed contrasting ligand-receptor nodes corresponding to outbound signals in metabolically hyperactive and proliferating cells, indicating opposing expression of ligands between these cell types. Certain ligands like TGFA produced by hepatocytes seemed to target hepatocytes themselves. This observation is supported by the previously proposed autocrine mode of mitogenic TGFA action (Mead and Fausto 1989; Webber et al. 1993). On the other hand, VEGFA ligands were directed more toward LSECs and stellate cells, in line with their known roles in the activation of these cell populations (LeCouter et al. 2003; Ding et al. 2010), whereas complement system ligands appeared to target diverse intrahepatic cell populations (Strey et al. 2003; Thorgersen et al. 2019). The cellular interactome analysis also predicted signaling events with uncharacterized roles in liver regeneration. For instance, Col18a1_Kdr/Itgb1/Itga5/Gpc1/4, Tgfa_Egfr/Errb3, and/or Psen1_Notch1/2 signaling events are excellent candidates to evaluate in the context of their function in emergence/stabilization of the metabolically hyperactive state of hepatocytes. Altogether, we identified a vast array of ligand–receptor interactions among hepatocytes and NPCs, provided a network-level portrait of intercellular crosstalk in normal and regenerating livers, and offered the first glimpse into how cell state transitions shape intrahepatic signaling at different stages of liver regeneration.
Discussion
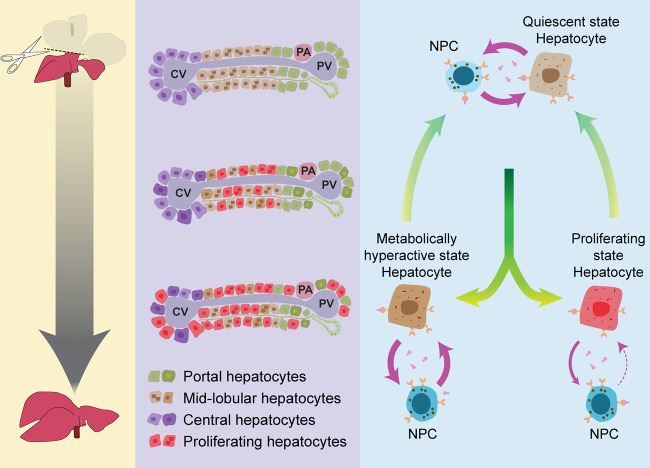
Regeneration requires simultaneous proliferation and maintenance of highly specialized cellular functions, and a regenerating liver continues to perform its crucial metabolic, biosynthetic, and detoxification roles (Michalopoulos and DeFrances 1997; Michalopoulos 2017; Bangru and Kalsotra 2020). However, how a regenerating tissue sustains its normal functions when large numbers of cells are proliferating is still a mystery. Based on our findings, we propose a division-of-labor model wherein, after PHx, residual hepatocytes undergo defined cellular transitions to support essential metabolic activities as the regenerating liver restores its original mass (Fig. 7). Five principal observations support this model. First, our single-cell trajectory analyses captured the plasticity of hepatocytes, identifying four distinct subpopulations representing the quiescent, transition, proliferative, and metabolically hyperactive states. Second, we discovered that, after PHx, quiescent hepatocytes promptly adopt an intermediate transition state from which they branch into either proliferative or metabolically hyperactive states. Third, we noticed visibly divergent regulon activities of proliferative and metabolically hyperactive hepatocytes. Cells transitioning into a proliferative state silenced regulons directing mature hepatocyte functions while stimulating regulons that support cell growth and proliferation. Conversely, cells transitioning into a metabolically hyperactive state activated multiple regulons supporting biosynthetic, metabolic, detoxification, and transport-related functions. Fourth, cumulative EdU labeling experiments combined with immunostaining of metabolic markers demonstrated that the portal and central vein proximal regions harbor the metabolically active hepatocytes, while glycogen-deficient midlobular hepatocytes proliferate nearby (Fig. 7). Fifth, we observed that the metabolically hyperactive hepatocytes develop transient but strong inbound and outbound connections with NPCs, whereas proliferating hepatocytes selectively down-regulate the receptors for inbound signals. Elimination of receptors for inbound pro-proliferative signals likely constrains the endless proliferation of hepatocytes and enables their timely cell-cycle exit. Altogether, these observations illustrate that dynamic shifts in regulon activities and cell–cell interactions broaden the hepatocellular plasticity to balance the metabolic and proliferation needs of a regenerating liver. It remains to be seen whether the up-regulation of metabolic genes in a subset of hepatocytes after PHx is a mouse-specific phenomenon or a conserved feature of regeneration. Future single-cell analyses of different liver regeneration models that incorporate multiple biological replicates in mice, as well as additional studies in rats and zebrafish, will shed light on how well the division-of-labor model applies to other species.
Figure 7.
Division-of-labor model for liver regeneration. Following surgical resection (PHx), the remnant liver tissue regenerates quickly and restores its original size and function. Hepatocyte proliferation initiates in the midlobular region before proceeding toward the periportal and pericentral areas. We propose that a subset of residual hepatocytes in the midlobular area reversibly activate an early-postnatal-like gene program to enter a proliferative state. Simultaneously, a distinct population near the portal and central vein proximal regions up-regulates their metabolic gene program to offset any regeneration-induced deficits in liver function. These reversible cell state transitions are guided by distinct ligand-receptor-mediated signaling events between hepatocytes and nonparenchymal cells. Thus, the division of labor maximizes the benefit-cost ratio of regeneration for an organism, ensuring quick and robust replenishment of the hepatic parenchyma while sustaining adequate metabolic and detoxification activities. (NPCs) PA: portal artery; PV: portal vein; CV: central vein.
Previous studies indicated that distinctly located pools of mature hepatocytes with progenitor-like features might serve specialized roles in liver regeneration (Miyajima et al. 2014). Although hepatocytes expressing stem/progenitor-like markers such as LGR5+, SOX9+, AXIN2+, TERT+, or MFSD2A+ are detectable, their overall requirement for normal maintenance and renewal after acute or chronic liver damage is debatable (Huch et al. 2013; Font-Burgada et al. 2015; Lu et al. 2015; Wang et al. 2015; Pu et al. 2016; Lin et al. 2018). For instance, a series of recent reports questioned the notion of a dedicated regenerative cell population in the liver and demonstrated that randomly distributed hepatocytes throughout the lobule repopulate the liver under both homeostatic and/or injury-related conditions (Chen et al. 2020; Matsumoto et al. 2020; Sun et al. 2020). Our single-cell transcriptomic data are consistent with these findings as we did not detect enrichment for such markers within the proliferating pool of hepatocytes. Alternatively, we found that, after PHx, a subset of remaining hepatocytes dedifferentiates to an early postnatal-like state before proceeding toward the proliferative trajectory. We also uncovered that a sizeable proportion of hepatocytes does not proliferate and instead maintains their mature metabolic state through the entire course of regeneration. Our cumulative EdU incorporation experiments, along with immunostaining of portal- and central vein-specific markers, confirmed the mutually exclusive nature of proliferating and metabolically active cells in regenerating livers. We demonstrated that hepatocyte proliferation after PHx initiates in the midlobular region before proceeding toward the periportal and pericentral areas and that the portal and central vein proximal regions are the likely depots of hepatocytes retaining the metabolically active state (Fig. 7).
Our understanding of molecular events that induce mature, quiescent hepatocytes to dedifferentiate and transition toward a proliferative state is incomplete. In this study, we combined systematic analyses of gene regulatory networks and intercellular interactions via ligand-receptor signaling on a compensatory model of regeneration in an otherwise healthy liver. We discovered that, following PHx, residual hepatocytes reversibly activate an early-postnatal-like gene expression program to transition from a quiescent to proliferative state and back. Our analysis revealed that transient dampening of adult gene programs followed by a brief surge in ribosome biogenesis precedes cell-cycle activation. We further determined that rewiring of developmental GRNs orchestrates cell-cycle entry during initiation of regeneration while facilitating rematuration of the newly generated hepatocytes so that they can resume their functions once regeneration is complete. In the future, it will be important to determine whether the hepatocyte subpopulations identified here reprogram similarly or differently in response to other types of periportal and/or pericentral liver injuries. Also, the changes in regulon activities within individual cells reflect the integration of many transcriptional and post-transcriptional events (Bangru et al. 2018; Bangru and Kalsotra 2020). Therefore, measuring the impact of various post-transcriptional events—such as mRNA stability, splicing, polyadenylation, and translation—within single cells would offer a more thorough understanding of the contribution of multiple modes of gene regulation in liver regeneration. Lastly, recent single-cell studies have surveyed the changes in cell–cell communication between healthy, NASH, and fibrotic livers (Dobie et al. 2019; Xiong et al. 2019). Comparing these disease data sets to the compendium of intercellular interactions documented here would expand our understanding of normal versus aberrant cell–cell communication networks, revealing defects in intrahepatic cell signaling that compromise regeneration in diseased livers. Such lines of investigations will not only map the signaling events that regulate hepatocellular plasticity but also help identify targets that may be leveraged to optimize hepatic repair and function after acute liver failure or in end-stage liver disease.
Methods
Animal care and surgeries
For all experimental procedures, 8- to 10-wk-old C57BL/6J male mice obtained from the Jackson Laboratories were used. National Institutes of Health (NIH) and UIUC institutional guidelines for the use and care of laboratory animals were followed, and all experimental protocols were performed as approved by the Institutional Animal Care and Use Committee at the University of Illinois, Urbana-Champaign (UIUC). We performed a 2/3rd partial hepatectomy procedure adapted from a previously reported protocol (Boyce and Harrison 2008; Mitchell and Willenbring 2008). Single-cell libraries were prepared from one mouse for each time point, and 4–5 animals per time point were used for histology, immunofluorescence staining, and liver function tests.
Immunofluorescence staining
EdU labeling and immunofluorescence staining were carried out as described before (Bangru et al. 2018). 5-Ethynyl-2′-deoxyuridine was administered intraperitoneally (100 µg/g body weight) 4 h before tissue collection to label nascent DNA synthesis. Alternatively, for cumulative EdU labeling experiments, mice were provided ad libitum drinking water supplemented with 1 mg/mL EdU starting 12 h before PHx or Sham surgery and continuing until sacrifice. This enabled EdU incorporation in replicating nuclei across the time period under consideration.
Tissue dissociation and isolation of liver cells
We adapted protocols from previously published reports by others, as well as in house protocols to dissociate and collect whole-cell suspensions from the liver (Bhate et al. 2015; Bangru et al. 2018).
Dead cell removal, single-cell library preparation, and sequencing
A MACS Dead Cell Removal Kit (Miltenyl Biotec) was used according to the manufacturer's protocol to remove dead cells and obtain unbiased single-cell suspensions of liver cells with high viability. Following this, single-cell sequencing libraries were prepared individually from each time point using the 10x Genomics Chromium Single Cell 3′ Kit v3 and sequenced with Illumina NovaSeq 6000 on a SP/S4 flow cell to obtain 150-bp paired reads.
Raw sequencing data processing and cell type identification
Single-cell libraries produced over four billion reads. We used Cell Ranger v3.1 pipelines from 10x Genomics to align reads to the mm10 genome assembly and produce feature matrices. Seurat v3.1 (Butler et al. 2018) was used for QC and analysis of individual feature matrices. Batch-effects across samples were removed using BEER v0.1.7 (Zhang et al. 2019). Data were log-normalized, scaled, and clustered using “FindNeighbors” and “FindClusters” functions within the Seurat package after PCA analysis. Hepatocyte and NPC clusters were identified based on the expression of various known marker genes. To identify cell types within the subset of NPCs, they were further subjected to unsupervised clustering as described above. Monocle v2.0 was used to perform pseudotime analysis, according to the online documentation (Qiu et al. 2017).
Gene regulatory network analysis
The SCENIC pipeline (Aibar et al. 2017) was used to estimate the AUCell Score activity matrix from the log-normalized Seurat object containing the subset of hepatocytes. Unlike the standard SCENIC workflow where this AUCell score activity matrix is binarized by thresholding to generate a binary regulon-activity matrix, we retained the full AUCell score for all further analysis. UMAP plots, heat maps, and violin plots demonstrating regulon activities based on AUCell scores were made in Seurat v3.1. AUCell scores were plotted over pseudotime cell-trajectories using Monocle 2.0.
Imputation and cell–cell communication analysis
Imputation using the MAGIC algorithm (van Dijk et al. 2018) was performed to correct for any missing values in the NPC data set before interpreting cell–cell interactions. We constructed cell–cell communication networks and performed statistics of interactions using methods previously described in detail by Farbehi et al. (2019). Additional information about mouse surgeries, liver cell isolation, and other experimental or bioinformatics analysis can be found in the Supplemental Methods.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE151309.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
Work in the Kalsotra laboratory is supported by National Institutes of Health (NIH) (R01HL126845, R01AA010154), Muscular Dystrophy Association (MDA514335), Planning Grant Award from the Cancer Center @ Illinois, and the Beckman Fellowship from the Center for Advanced Study at the University of Illinois Urbana-Champaign. U.V.C. is supported by the Herbert E. Carter fellowship in Biochemistry, UIUC. S.B. is supported by the NIH Tissue microenvironment training program (T32-EB019944) and Scott Dissertation Completion Fellowship, UIUC.
Author contributions: U.V.C., S.B., and A.K. conceived the project and designed the experiments. U.V.C. and S.B. performed experiments and analyzed the data. M.H. provided guidance with bioinformatics analyses. U.V.C., S.B., and A.K. interpreted results and wrote the manuscript. All authors discussed the results and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.267013.120.
References
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. 2017. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14: 1083–1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrani SK, Devarbhavi H, Eaton J, Kamath PS. 2019. Burden of liver diseases in the world. J Hepatol 70: 151–171. 10.1016/j.jhep.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Baena E, Gandarillas A, Vallespinós M, Zanet J, Bachs O, Redondo C, Fabregat I, Martinez-A C, de Alborán IM. 2005. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc Natl Acad Sci 102: 7286–7291. 10.1073/pnas.0409260102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangru S, Kalsotra A. 2020. Cellular and molecular basis of liver regeneration. Semin Cell Dev Biol 100: 74–87. 10.1016/j.semcdb.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangru S, Arif W, Seimetz J, Bhate A, Chen J, Rashan EH, Carstens RP, Anakk S, Kalsotra A. 2018. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat Struct Mol Biol 25: 928–939. 10.1038/s41594-018-0129-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkas N, Petukhov V, Nikolaeva D, Lozinsky Y, Demharter S, Khodosevich K, Kharchenko PV. 2019. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat Methods 16: 695–698. 10.1038/s41592-019-0466-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. 2008. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J 27: 212–223. 10.1038/sj.emboj.7601950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat NK, Fisher RJ, Fujiwara S, Ascione R, Papas TS. 1987. Temporal and tissue-specific expression of mouse ets genes. Proc Natl Acad Sci 84: 3161–3165. 10.1073/pnas.84.10.3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhate A, Parker DJ, Bebee TW, Ahn J, Arif W, Rashan EH, Chorghade S, Chau A, Lee J-H, Anakk S, et al. 2015. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat Commun 6: 8768. 10.1038/ncomms9768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell DM, Wang SS, Jarnagin WR, Roll FJ. 1995. Cell-specific expression of transforming growth factor-β in rat liver. evidence for autocrine regulation of hepatocyte proliferation. J Clin Invest 96: 447–455. 10.1172/JCI118055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzo JA, Ferry CH, Matsubara T, Kim J-H, Gonzalez FJ. 2012. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem 287: 7345–7356. 10.1074/jbc.M111.334599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce S, Harrison D. 2008. A detailed methodology of partial hepatectomy in the mouse. Lab Anim (NY) 37: 529–532. 10.1038/laban1108-529 [DOI] [PubMed] [Google Scholar]
- Braun L, Mead JE, Panzica M, Mikumo R, Bell GI, Fausto N. 1988. Transforming growth factor β mRNA increases during liver regeneration: a possible paracrine mechanism of growth regulation. Proc Natl Acad Sci 85: 1539–1543. 10.1073/pnas.85.5.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldez MJ, Van Hul N, Koh HWL, Teo XQ, Fan JJ, Tan PY, Dewhurst MR, Too PG, Talib SZA, Chiang BE, et al. 2018. Metabolic remodeling during liver regeneration. Dev Cell 47: 425–438.e5. 10.1016/j.devcel.2018.09.020 [DOI] [PubMed] [Google Scholar]
- Chaisson ML, Brooling JT, Ladiges W, Tsai S, Fausto N. 2002. Hepatocyte-specific inhibition of NF-κB leads to apoptosis after TNF treatment, but not after partial hepatectomy. J Clin Invest 110: 193–202. 10.1172/JCI0215295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Jimenez RJ, Sharma K, Luu HY, Hsu BY, Ravindranathan A, Stohr BA, Willenbring H. 2020. Broad distribution of hepatocyte proliferation in liver homeostasis and regeneration. Cell Stem Cell 26: 27–33.e4. 10.1016/j.stem.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AM, Chute J. 2013. Underlying potential: cellular and molecular determinants of adult liver repair. J Clin Invest 123: 1858–1860. 10.1172/JCI69966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B-S, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, et al. 2010. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 468: 310–315. 10.1038/nature09493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, Wallenborg K, Picelli S, Zagorska A, Pendem SV, et al. 2019. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep 29: 1832–1847.e8. 10.1016/j.celrep.2019.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE, Harvey RP. 2019. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife 8: 1241. 10.7554/eLife.43882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausto N, Campbell JS, Riehle KJ. 2006. Liver regeneration. Hepatology 43: S45–S53. 10.1002/hep.20969 [DOI] [PubMed] [Google Scholar]
- Font-Burgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A, Taniguchi K, Nakagawa H, Valasek MA, Ye L, et al. 2015. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162: 766–779. 10.1016/j.cell.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SJ, Newsome PN. 2016. Liver regeneration – mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 13: 473–485. 10.1038/nrgastro.2016.97 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ. 2008. Regulation of hepatocyte nuclear factor 4 α-mediated transcription. Drug Metab Pharmacokinet 23: 2–7. 10.2133/dmpk.23.2 [DOI] [PubMed] [Google Scholar]
- Greenbaum LE, Cressman DE, Haber BA, Taub R. 1995. Coexistence of C/EBP α, β, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration. Implications for maintenance of the differentiated state during liver growth. J Clin Invest 96: 1351–1365. 10.1172/JCI118170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, Marioni JC. 2018. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36: 421–427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, et al. 2011. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147: 1233–1247. 10.1016/j.cell.2011.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rudnick DA. 2014. Elucidating the metabolic regulation of liver regeneration. Am J Pathol 184: 309–321. 10.1016/j.ajpath.2013.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. 2013. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494: 247–250. 10.1038/nature11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck I, Gunewardena S, Español-Suñer R, Willenbring H, Apte U. 2019. Hepatocyte nuclear factor 4 α activation is essential for termination of liver regeneration in mice. Hepatology 70: 666–681. 10.1002/hep.30405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J, Sun Z, Ahmadi AR, Bangru S, Chembazhi UV, Du K, Chen T, Tsukamoto H, Rusyn I, Kalsotra A, et al. 2020. Epithelial splicing regulatory protein 2–mediated alternative splicing reprograms hepatocytes in severe alcoholic hepatitis. J Clin Invest 130: 2129–2145. 10.1172/JCI132691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimuro Y, Nishiura T, Hellerbrand C, Behrns KE, Schoonhoven R, Grisham JW, Brenner DA. 1998. NFkappab prevents apoptosis and liver dysfunction during liver regeneration. J Clin Invest 101: 802–811. 10.1172/JCI483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Tomiya T, Yanase M, Arai M, Ikeda H, Tejima K, Ogata I, Kimura S, Omata M, Fujiwara K. 2002. p53 may positively regulate hepatocyte proliferation in rats. Hepatology 36: 336–344. 10.1053/jhep.2002.34942 [DOI] [PubMed] [Google Scholar]
- Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, Porse BT. 2013. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res 23: 592–603. 10.1101/gr.146399.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Hong I-H, Lewis K, Iakova P, Breaux M, Jiang Y, Sullivan E, Jawanmardi N, Timchenko L, Timchenko NA. 2015. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology 61: 315–325. 10.1002/hep.27295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso S, Kawata S, Tamura S, Inui Y, Yoshida Y, Sawai Y, Umeki S, Ito N, Yamada A, Miyagawa J-I, et al. 2003. Liver regeneration in heparin-binding EGF-like growth factor transgenic mice after partial hepatectomy. Gastroenterology 124: 701–707. 10.1053/gast.2003.50097 [DOI] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16: 1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW, Barton MC. 2013. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology 57: 2004–2013. 10.1002/hep.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttippurathu L, Patra B, Cook D, Hoek JB, Vadigepalli R. 2017. Pattern analysis uncovers a chronic ethanol-induced disruption of the switch-like dynamics of C/EBP-β and C/EBP-α genome-wide binding during liver regeneration. Physiol Genomics 49: 11–26. 10.1152/physiolgenomics.00097.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCouter J, Moritz DR, Li B, Phillips GL, Liang XH, Gerber H-P, Hillan KJ, Ferrara N. 2003. Angiogenesis-independent endothelial protection of liver: role of VEGFR-1. Science 299: 890–893. 10.1126/science.1079562 [DOI] [PubMed] [Google Scholar]
- Lin S, Nascimento EM, Gajera CR, Chen L, Neuhöfer P, Garbuzov A, Wang S, Artandi SE. 2018. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556: 244–248. 10.1038/s41586-018-0004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W-Y, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, et al. 2015. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 17: 971–983. 10.1038/ncb3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, et al. 2018. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9: 4383–4321. 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JJ. 1993. Cell-specific expression of hepatocyte growth factor in liver. Upregulation in sinusoidal endothelial cells after carbon tetrachloride. J Clin Invest 91: 2244–2252. 10.1172/JCI116451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Wakefield L, Tarlow BD, Grompe M. 2020. In vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell 26: 34–47.e3. 10.1016/j.stem.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead JE, Fausto N. 1989. Transforming growth factor α may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc Natl Acad Sci 86: 1558–1562. 10.1073/pnas.86.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. 2017. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology 65: 1384–1392. 10.1002/hep.28988 [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. 1997. Liver regeneration. Science 276: 60–66. 10.1126/science.276.5309.60 [DOI] [PubMed] [Google Scholar]
- Minocha S, Villeneuve D, Rib L, Moret C, Guex N, Herr W. 2017. Segregated hepatocyte proliferation and metabolic states within the regenerating mouse liver. Hepatol Commun 1: 871–885. 10.1002/hep4.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Willenbring H. 2008. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 3: 1167–1170. 10.1038/nprot.2008.80 [DOI] [PubMed] [Google Scholar]
- Miyajima A, Tanaka M, Itoh T. 2014. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14: 561–574. 10.1016/j.stem.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. 2012. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 22: 1166–1175. 10.1016/j.cub.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, Yim K, Elzen AVD, Hirn MJ, Coifman RR, et al. 2019. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol 37: 1482–1492. 10.1038/s41587-019-0336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PS, Poulsen SS, Kirkegaard P. 1985. Adrenergic effects on secretion of epidermal growth factor from Brunner's glands. Gut 26: 920–927. 10.1136/gut.26.9.920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. 2003. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34: 292–296. 10.1038/ng1175 [DOI] [PubMed] [Google Scholar]
- Pu W, Zhang H, Huang X, Tian X, He L, Wang Y, Zhang L, Liu Q, Li Y, Li Y, et al. 2016. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat Commun 7: 13369. 10.1038/ncomms13369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14: 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JO, Monga SP. 2018. Wnt/β-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol 13: 351–378. 10.1146/annurev-pathol-020117-044010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W, Cannoodt R, Todorov H, Saeys Y. 2019. A comparison of single-cell trajectory inference methods. Nat Biotechnol 37: 547–554. 10.1038/s41587-019-0071-9 [DOI] [PubMed] [Google Scholar]
- Schaub JR, Malato Y, Gormond C, Willenbring H. 2014. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep 8: 933–939. 10.1016/j.celrep.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmacher P, Geerts A, Pietrangelo A, Dienes HP, Rogler CE. 1992. Hepatocyte growth factor/hepatopoietin A is expressed in fat-storing cells from rat liver but not myofibroblast-like cells derived from fat-storing cells. Hepatology 15: 5–11. 10.1002/hep.1840150103 [DOI] [PubMed] [Google Scholar]
- Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C, Mathurin P, Mueller S, Szabo G, Tsukamoto H. 2018. Alcoholic liver disease. Nat Rev Dis Primers 4: 16–22. 10.1038/s41572-018-0014-7 [DOI] [PubMed] [Google Scholar]
- Servillo G, Della Fazia MA, Sassone-Corsi P. 1998. Transcription factor CREM coordinates the timing of hepatocyte proliferation in the regenerating liver. Genes Dev 12: 3639–3643. 10.1101/gad.12.23.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe'er D. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 34: 637–645. 10.1038/nbt.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladky VC, Knapp K, Soratroi C, Heppke J, Eichin F, Rocamora-Reverte L, Szabo TG, Bongiovanni L, Westendorp B, Moreno E, et al. 2020. E2F-family members engage the PIDDosome to limit hepatocyte ploidy in liver development and regeneration. Dev Cell 52: 335–349.e7. 10.1016/j.devcel.2019.12.016 [DOI] [PubMed] [Google Scholar]
- Stepniak E, Ricci R, Eferl R, Sumara G, Sumara I, Rath M, Hui L, Wagner EF. 2006. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev 20: 2306–2314. 10.1101/gad.390506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Hilaire RJ, Hradek GT, Jones AL. 1983. Hepatic sequestration and biliary secretion of epidermal growth factor: evidence for a high-capacity uptake system. Proc Natl Acad Sci 80: 3797–3801. 10.1073/pnas.80.12.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19: 477–416. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strey CW, Markiewski M, Mastellos D, Tudoran R, Spruce LA, Greenbaum LE, Lambris JD. 2003. The proinflammatory mediators C3a and C5a are essential for liver regeneration. J Exp Med 198: 913–923. 10.1084/jem.20030374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Pikiolek M, Orsini V, Bergling S, Holwerda S, Morelli L, Hoppe PS, Planas-Paz L, Yang Y, Ruffner H, et al. 2020. AXIN2+ pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell 26: 97–107.e6. 10.1016/j.stem.2019.10.011 [DOI] [PubMed] [Google Scholar]
- Thorgersen EB, Barratt-Due A, Haugaa H, Harboe M, Pischke SE, Nilsson PH, Mollnes TE. 2019. The role of complement in liver injury, regeneration, and transplantation. Hepatology 70: 725–736. 10.1002/hep.30508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HTN, Ang KS, Chevrier M, Zhang X, Lee NYS, Goh M, Chen J. 2020. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol 21: 12–32. 10.1186/s13059-019-1850-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. 2018. Recovering gene interactions from single-cell data using data diffusion. Cell 174: 716–729.e27. 10.1016/j.cell.2018.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, Nusse R. 2015. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 524: 180–185. 10.1038/nature14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AW, Wangensteen KJ, Wang YJ, Zahm AM, Moss NG, Erez N, Kaestner KH. 2018. TRAP-seq identifies cystine/glutamate antiporter as a driver of recovery from liver injury. J Clin Invest 128: 2297–2309. 10.1172/JCI95120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang C, Hasson D, Desai A, SenBanerjee S, Magnani E, Ukomadu C, Lujambio A, Bernstein E, Sadler KC. 2019. Epigenetic compensation promotes liver regeneration. Dev Cell 50: 43–56.e6. 10.1016/j.devcel.2019.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber EM, FitzGerald MJ, Brown PI, Bartlett MH, Fausto N. 1993. Transforming growth factor-α expression during liver regeneration after partial hepatectomy and toxic injury, and potential interactions between transforming growth factor-α and hepatocyte growth factor. Hepatology 18: 1422–1431. 10.1002/hep.1840180622 [DOI] [PubMed] [Google Scholar]
- Westwick JK, Weitzel C, Leffert HL, Brenner DA. 1995. Activation of Jun kinase is an early event in hepatic regeneration. J Clin Invest 95: 803–810. 10.1172/JCI117730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xiao Y, Zhang S, Ji S, Wei L, Fan F, Geng J, Tian J, Sun X, Qin F, et al. 2013. The Ets transcription factor GABP is a component of the hippo pathway essential for growth and antioxidant defense. Cell Rep 3: 1663–1677. 10.1016/j.celrep.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao X-Y, Ji Y, Li C, Guo L, et al. 2019. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol Cell 75: 644–660.e5. 10.1016/j.molcel.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CL, Zhang SJ, Toomey NL, Palmer TN, Lee MY. 1991. Induction of DNA polymerase activities in the regenerating rat liver. Biochemistry 30: 7534–7541. 10.1021/bi00244a024 [DOI] [PubMed] [Google Scholar]
- Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H, Pikarsky E, Stanger BZ. 2014. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15: 340–349. 10.1016/j.stem.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu Y, Tian W. 2019. A novel approach to remove the batch effect of single-cell data. Cell Discov 5: 46. 10.1038/s41421-019-0114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Jin Y, Donahue K, Tsui M, Fish M, Logan CY, Wang B, Nusse R. 2019. Tissue repair in the mouse liver following acute carbon tetrachloride depends on injury-induced Wnt/β-catenin signaling. Hepatology 69: 2623–2635. 10.1002/hep.30563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.