Abstract
The COVID-19 pandemic has sparked an urgent need to uncover the underlying biology of this devastating disease. Though RNA viruses mutate more rapidly than DNA viruses, there are a relatively small number of single nucleotide polymorphisms (SNPs) that differentiate the main SARS-CoV-2 lineages that have spread throughout the world. In this study, we investigated 129 RNA-seq data sets and 6928 consensus genomes to contrast the intra-host and inter-host diversity of SARS-CoV-2. Our analyses yielded three major observations. First, the mutational profile of SARS-CoV-2 highlights intra-host single nucleotide variant (iSNV) and SNP similarity, albeit with differences in C > U changes. Second, iSNV and SNP patterns in SARS-CoV-2 are more similar to MERS-CoV than SARS-CoV-1. Third, a significant fraction of insertions and deletions contribute to the genetic diversity of SARS-CoV-2. Altogether, our findings provide insight into SARS-CoV-2 genomic diversity, inform the design of detection tests, and highlight the potential of iSNVs for tracking the transmission of SARS-CoV-2.
Coronavirus (CoV) genomes are the largest among single-strand RNA (ssRNA) viruses, ranging from 26 to 32 kb. Although ssRNA viruses typically display very high mutation rates, coronaviruses encode an RNA polymerase with 3′-to-5′ proofreading activity that allows them to replicate their genome with high fidelity, lowering their mutation rate (Drake and Holland 1999; Gorbalenya et al. 2006; Denison et al. 2011; Peck and Lauring 2018). Additionally, SARS-CoV-2 contains a common 69-bp 5′ leader sequence fused to the body sequence from the 3′ end of the genome (Sola et al. 2015). Then, leader-to-body fusion occurs during negative-strand synthesis at short motifs called transcription-regulating sequences (TRSs), which are conserved 5- to 10-bp sequences that are adjacent to the ORFs (Wu et al. 2020).
On March 11, 2020, the WHO determined that an outbreak of a novel coronavirus SARS-CoV-2 that began in Wuhan, China in December 2019 had reached pandemic status. Initial consensus-level genomic data from the Global Initiative on Sharing All Influenza Data (GISAID) (Elbe and Buckland-Merrett 2017) indicated that the SARS-CoV-2 mutational rate (Shen et al. 2020) was similar to other CoVs (Eckerle et al. 2010). In order to properly assess the genomic diversity of any RNA virus, and specifically SARS-CoV-2, it is necessary to also consider the intra-host polymorphisms (Park et al. 2015; Poon et al. 2016; Barbezange et al. 2018; Borucki et al. 2019), including often overlooked structural variation. Recent studies have claimed that host-dependent RNA editing might be a key factor for understanding the mutational landscape of SARS-CoV-2 within hosts (Giorgio et al. 2020; Ramazzotti et al. 2021). However, these studies were based on a limited number of samples (<20). In order to explore both the intra-host and inter-host mutational landscape of SARS-CoV-2, we leveraged a data set consisting of 10 RNA-seq samples from the Baylor College of Medicine and 119 RNA-seq samples from the Weill Cornell College of Medicine, plus 6928 consensus genomes downloaded from GISAID.
Understanding the intra-host genomic diversity of SARS-CoV-2 is also important for other purposes. Most SARS-CoV-2 detection tests rely on oligonucleotide probes and primers that must be sensitive to SARS-CoV-2. In this setting, sensitivity determines how well the detection tests can capture the diversity of all SARS-CoV-2 variants. Lack of sensitivity leads to an increase in false-negative qRT-PCR results, because two or more mismatches can result in increases in CT values and degradation in accuracy of viral load estimates (Whiley and Sloots 2005; Farkas et al. 2020). Moreover, recent studies on Ebola and flu viruses (Park et al. 2015; Pauly et al. 2017) highlight the importance of intra-host variation for studying viral population dynamics and transmission scenarios. In this study, we investigate the intra-host diversity of SARS-CoV-2 by conducting a broad evaluation of (1) intra-host single nucleotide variants (iSNV), (2) consensus-level single nucleotide polymorphisms (SNPs), and (3) structural variants (SVs), across consensus genomes and RNA-seq data sets totaling over 7000 samples.
Results
We analyzed three SARS-CoV-2 genomic data sets: RNA-seq reads for 10 patient samples collected by the Baylor College of Medicine (Houston, TX, USA) (Doddapaneni et al. 2020), RNA-seq reads for 119 patient samples collected by Weill Cornell University (New York [NYC], NY, USA) (Butler et al. 2021), plus 6928 consensus genomes downloaded from GISAID. We evaluated structural variants across the 129 RNA-seq samples in both NYC and Houston; the inferred SVs are shown in Figure 1. We also evaluated single nucleotide polymorphisms in the GISAID genomes, whereas the variants analyzed in the Houston and NYC RNA-seq data sets include both SNPs and intra-host single nucleotide variants. The inferred phylogenetic tree for the GISAID genomes is shown in Supplemental Figure S1. We note that the major clades correspond to the geographic and time distribution of the samples, with clades 19A and 19B being common in Asia in the early months of the outbreak, clade 20A corresponding to the outbreak in Europe, and 20C to the North American outbreak (Hadfield et al. 2018). We also observe that some of the clade-defining SNPs occur intermittently outside of the main phylogenetic clades. We will now dive deep into three main results: (1) the intra-host structural variant landscape; (2) the intra-host single nucleotide variant landscape; and (3) exploratory analyses of shared SNPs and iSNVs within and across patients in NYC.
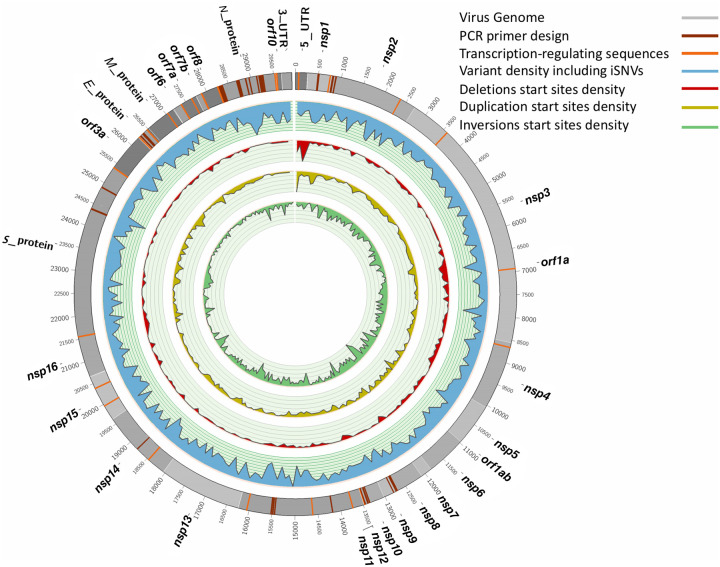
Figure 1.
Overview of general diversity of SARS-CoV-2. From outer to inner layers: Annotation of SARS-CoV-2 genome (gray), PCR primer designs (dark red), transcription-regulating sequences (TRS) (orange), intra-host variant density including iSNVs (blue), deletions start sites (red), duplication start sites (yellow), and inversion start sites (green) along the entire genome. For SNPs + iSNVs + SVs, we plotted the density scaled by their allele frequency across the population over 100-bp windows.
Structural variant landscape
We identified 3311 structural variants across the 129 RNA-seq samples, with the majority being inversions (1504) and tandem duplications (1157), followed by deletions (625) and a few insertions (25) (Fig. 1). Overall, because we are identifying SVs based on RNA-seq data, the majority of these SVs are likely to be highlighting variability in the SARS-CoV-2 transcriptome (Davidson et al. 2020), which is influenced by fusion, deletions, frame-shifts, and recombination. We observed a significant overlap (Kolmogorov–Smirnov [KS] test: P-value = 0.03, D = 0.32) for the 58 start and 18 end breakpoints with the annotated transcription-regulating sequences (dark red, Fig. 1). Subsequently, we focus on smaller SVs (<1 kb) that more likely indicate true underlying SVs rather than transcription signals. We identified 286 deletions and 25 insertions across all 129 SARS-CoV-2 genomes. The imbalance of insertions and deletions is likely due to the low ability to detect insertions using short reads (Mahmoud et al. 2019). Figure 1 shows the allele frequency (AF) of these SVs across all samples. We observed 16 deletions that are highly shared among 26 or more samples (AF: <20%) (see Supplemental File S1). These impact multiple genes of SARS-CoV-2, including M protein (two deletions), N protein (two deletions), S protein (four deletions), nsp15 (one deletion), nsp1 (two deletions), nsp3 (one deletion), nsp4 (one deletion), nsp 6 (one deletion), and orf1 (five deletions).
Next, we investigated where these SVs are mainly located with respect to the annotated regions. We identified an enrichment of SVs in nsp11 and nsp12 when taking the size of the annotated regions into account (Supplemental Fig. S2). In addition, it is interesting to see that a higher number of SVs are also clustering in E protein (five del), nsp7 (five del and one ins), nsp9 (seven del and one ins), orf6 (six del), and orf7b (three del).
We further compared our SV call set with single deletions reported by various groups. Davidson et al. (2020) reported a 24-bp deletion in the subgenomic mRNA encoding the spike (S) glycoprotein that played a role in removing a proposed furin cleavage site from the S glycoprotein. We were able to identify this deletion (position: 25,234 bp) in three of our samples. For the gene encoding spike (S) protein, we identified six deletions shared among samples. Only four out of the six had an allele frequency of 20% or higher: 21,740 bp (39 bp, AF: 49.61%), 21,984 bp (9 bp, AF: 25.58%), 23,558 bp (22 bp, AF: 41.86%), and at 24,014 bp (15 bp, AF: 27.91%). We further identified five deletions; one (at 28,245 bp) was present in 10 samples (AF: 7.52%) in orf8, a potentially important gene for viral adaptation to humans (Muth et al. 2018).
Intra-host single nucleotide variant landscape
We considered intra-host single nucleotide variants to be those with an AF between 2% and 50% in a sample. Above 50%, all single nucleotide variants were considered to be consensus-level single nucleotide polymorphisms, as it is a common threshold for consensus-calling in genome assembly (Wright et al. 2011; Quick et al. 2017). Figure 2A shows the iSNV AF distribution, with the peak occurring in the 2%–5% range. The predominant iSNVs observed are U > C and C > U (Fig. 2B). We also note that A > G, G > A, and G > U iSNVs are common. These findings, specific to the iSNV mutational profile and frequency, are highly concordant with the recent intra-host SARS-CoV-2 genomic analyses from COVID-19 positive patients in Austria (Popa et al. 2020).
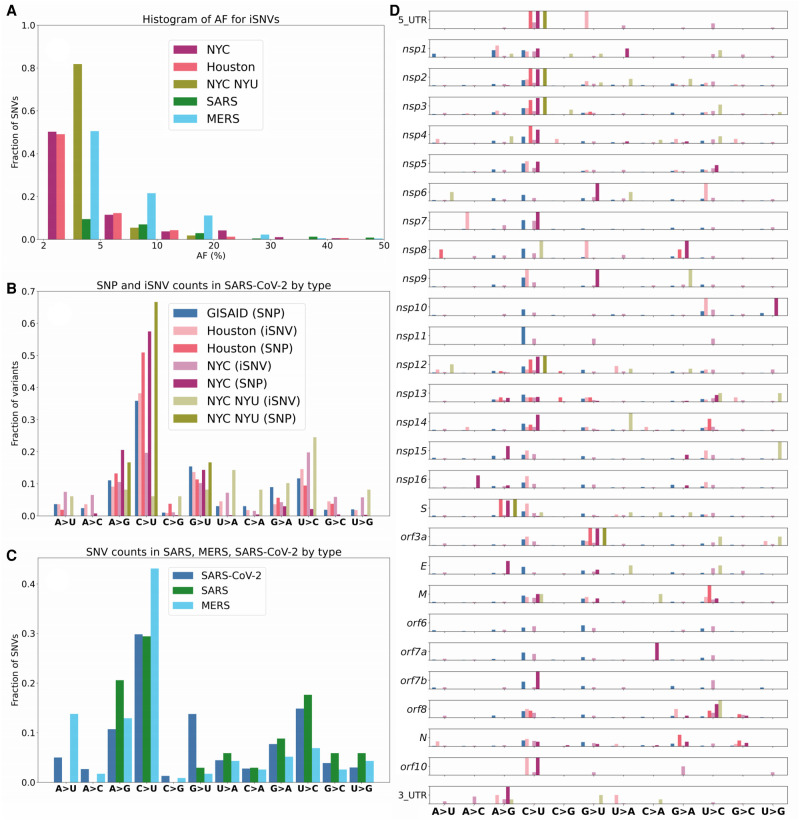
Figure 2.
Mutational frequencies of iSNVs and SNPs. (A) Distribution of iSNV AF. We note that the distribution of AF is strictly <50% as iSNVs are below consensus-level by definition. (B) Mutational spectrum of SARS-CoV-2. (C) Mutational spectra of SARS-CoV-1, SARS-CoV-2, and MERS. (D) Mutational spectrum of SARS-CoV-2 by orf/nsp.
When the distribution of iSNVs is mapped onto the SARS-CoV-2 genome, we observe that C > U is the dominant SNP in 10 out of 16 genes (Fig. 2D). The nsp6 and nsp10 genes stand out as having larger fractions of U > C iSNVs, and nsp7 has a large fraction of A > C iSNVs in the Houston data set (Fig. 2D). Additionally, nsp6 and orf3a have a high fraction of G > U SNPs, and orf8 and M genes have a high fraction of U > C SNPs. We also identified several interesting SNP and iSNV mutational patterns within the ORFs of SARS-CoV-2. Of note, SARS-CoV-2 encodes three tandem macrodomains within nonstructural protein 3 (nsp3). The nsp3 protein is essential for SARS-CoV-2 replication and represents a promising target for the development of antiviral drugs (Lin et al. 2018). The nsp3 protein is also one of the most diverged regions of SARS-CoV-2 compared to SARS-CoV-1 and MERS-CoV.
We note that the mutational spectra for SNPs matches the one observed for iSNVs, namely A > G, G > A, U > C, and G > U are most common (Fig. 2B). However, one difference is the relatively lower percentage of C > U changes in iSNVs from both NYC data sets (10%–20%) compared to 40% C > U iSNVs for the Houston samples and over 50% C > U in the Houston and NYC SNPs. The fraction of GISAID C > U SNPs is nearly identical to the fraction of Houston C > U iSNVs, clearly distinguishing GISAID SNPs and Houston iSNVs from Houston and NYC SNPs. We also note that the mutational spectra of SNPs across the genes of SARS-CoV-2 closely match the iSNV mutational spectra (Fig. 2D). The mutational spectrum of NYC SNPs is significantly different from both NYC iSNVs (Kolmogorov–Smirnov [KS] test: P-value ∼10−100) and GISAID SNPs (KS test: P-value ∼10−40) mutational spectra. When compared to SARS and MERS, SARS-CoV-2 has a larger proportion of G > U iSNVs (Fig. 2C). The other four major iSNV types (C > U, U > C, A > G, and G > A) are well represented in all three viruses. We also note that the SARS data sets do not contain any A > U nor A > C iSNVs.
To further investigate patterns of difference and similarity between SNPs and iSNVs, we analyzed the functional impact of the observed variants. First, in the GISAID SNPs, we observe 1191 (36.45%) synonymous, 2021 (61.86%) missense, and 40 (1.22%) stop gained variants. In the NYC iSNVs, we observed 586 (31.73%) synonymous, 1207 (65.35%) missense, and 54 (2.92%) stop gained variants. Finally, in the Houston iSNVs, we observed 43 (31.16%) synonymous, 86 (62.31%) missense, and five (3.62%) stop gained variants. Altogether, about two thirds of all observed variants are missense and about a third are synonymous, with good agreement for SNPs and iSNVs. Because SNPs and iSNVs can represent viral populations related by transmission (Fig. 3A) or arising independently, we have further investigated the patterns of the overlap between iSNV and SNPs (Fig. 3B). We note that there are 10 SNPs found in all three data sets (NYC, Houston, and GISAID). We also observed that 190 SNVs occur both as an iSNV in at least one sample and as SNPs in the GISAID data. Finally, there are three iSNVs shared between the Houston and NYC samples and that also occur as SNPs (Fig. 3B). The mutational spectrum of the variants that occur as both SNPs and iSNVs is similar to the general one outlined above, with ∼65% of the changes being C > U, followed by ∼15% of G > T, and ∼12% of U > C.
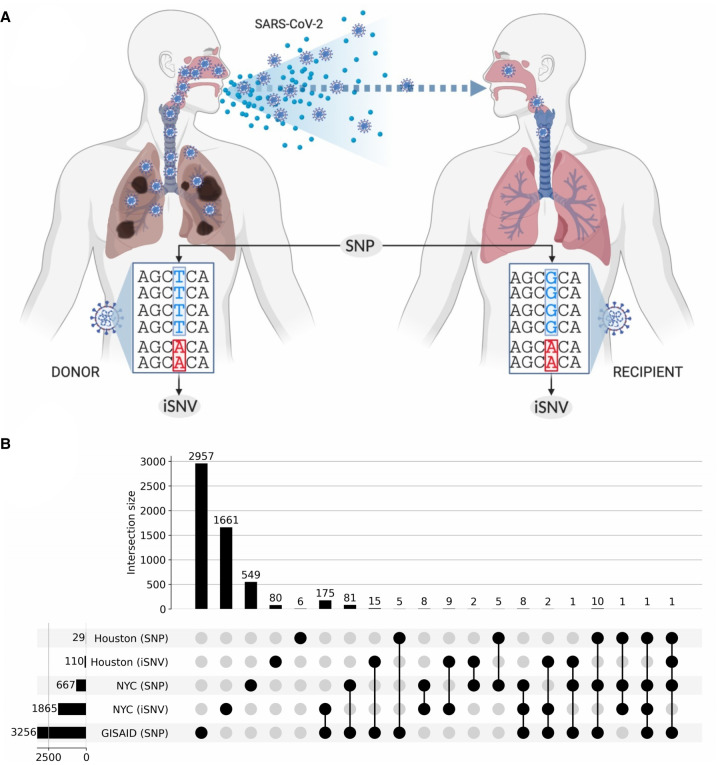
Figure 3.
Shared SNPs and SNVs across data sets. (A) Illustration differentiating what we define as an intra-host SNV (iSNV) and an inter-host consensus-level SNP. (B) UpSet plot captures the shared single nucleotide variants between iSNVs and consensus-level SNPs. The horizontal bars on the left show the total number of variants in the given category. Vertical bars indicate the size of the intersection between highlighted (with black circles) sets. Every variant contributes to exactly one intersection size to avoid double counting.
Prior studies have found iSNVs early in virus outbreaks that later establish as SNPs (Parameswaran et al. 2012; Rodriguez-Roche et al. 2016). Thus, we looked into whether clade-defining SNPs co-occur with iSNVs identified in the NYC and Houston data sets. We found that the C1059T and G25563T SNPs defining the 20C lineage co-occur with iSNVs in our RNA-seq samples. This indicates the emergence of an iSNV strongly correlated with the North American clade of the SARS-CoV-2.
Next, we estimated the genetic diversity (π) of SARS-CoV-2. We compared the genetic diversity computed using SNPs and iSNVs separately per SARS-CoV-2 NYC sample and observed that, when computing diversity using iSNVs, values are higher and more varied (KS test: P-value < 10−45). We observed a difference in the distribution of πN/πS ratios between iSNVs and SNPs in the NYC data set being lower for SNPs (median πN/πS: 0) than for iSNVs (median πN/πS: 0.4). The πN/πS ratios are consistent across the orfs/nsps of SARS-CoV-2 (Supplemental Fig. S3).
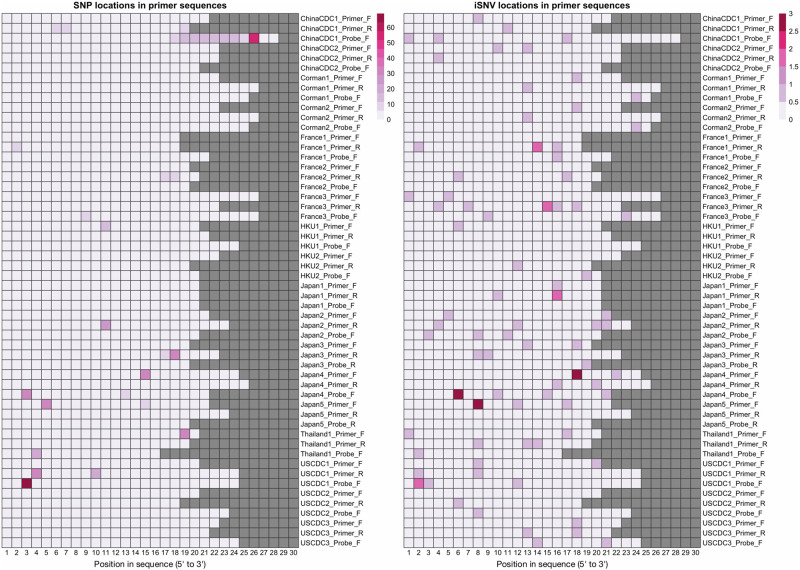
Finally, we analyzed the potential impact of iSNVs and SNPs on the probes and primers typically used for the detection of SARS-CoV-2 (Farkas et al. 2020; Khan and Cheung 2020) and also on the ARTIC primers used for SARS-CoV-2 amplicon sequencing. To evaluate this, we downloaded the set of probes and primers sequences available at the WHO website (54 sequences), as well as the ARTIC primers (218 sequences). Among these, 263 out of 272 contained at least one SNP or iSNV (Fig. 4 for WHO probes and primers and Supplemental File S2 for the ARTIC sequencing primers). On average, each probe/primer sequence contained 1.7 iSNVs and/or 3.1 SNPs. These results suggest the potential for a drop in the sensitivity of the affected probes and primers. We also note that, because the iSNV and SNP mutational profiles mimic each other for specific mutations, the potential impact of iSNVs on primer and probe binding should not be overlooked, given the possibility of iSNVs establishing as SNPs (Parameswaran et al. 2012).
Figure 4.
iSNV and SNP presence on widely used primers and probes. This figure shows the locations on WHO probes and primers that contain SNPs (left) and iSNVs (right). Columns correspond to base pair positions within the probe, and the sequences are 5′-aligned. Rows correspond to the oligonucleotide sequences, and squares are highlighted based on how many samples/genomes contain a variant in that position.
Exploratory transmission analysis of shared SNPs and iSNVs within and across patients
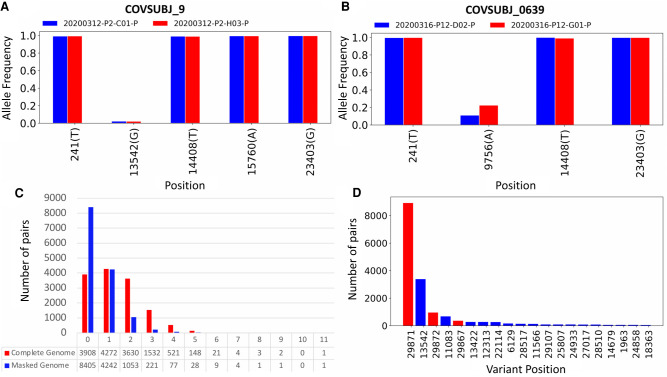
Shared viral genomic variants can be indicative of transmission events and routes (Worby et al. 2017), and iSNVs are a critically important tool for discerning direct transmission and for bottleneck calculations (Zwart and Elena 2015; Leonard et al. 2017). To assess our ability to identify shared iSNVs and SNPs across samples, we first compared all NYC longitudinal samples from the same patient taken within 24 hours (Fig. 5A,B). In Figure 5A, we show four shared SNPs and one shared iSNV that occur in longitudinal samples taken from patient 9. In Figure 5B, we show three shared SNPs and one shared iSNV.
Figure 5.
In-depth analysis of shared iSNVs. (A) Paired samples from patient COVSUBJ 9 in NYC. (B) Paired samples from patient COVSUBJ 0639 in NYC. (C) The distribution of the number of genomic pairs and their shared variants. (D) The number of pairs with variants at given nucleotide positions. Red color represents positions that were shown to be highly homoplasic and more likely to be affected by error (De Maio et al. 2020).
We next calculated the number of shared variants (SNPs + iSNVs) among all possible pairs of NYC samples (Fig. 5C). For each pair, we consider both possible assignments of donor and recipient, narrowing down the donor alleles to only include those with an AF between 0.02 and 0.5, and considering a site to be shared if the recipient also has that same variant present. We show these results on the raw data from the iSNV calls, as well as on the same data but after applying masking to sites near the ends of the genome. For the raw data before masking, most pairs have 0–3 shared variants, with about 500 pairs having four or more shared SNVs (Fig. 5C). After masking sites near the genome ends, these numbers drop substantially by reducing likely noise from the variant calls, and we see most pairs sharing 0–2 variants. When examining each possible pair, one immediately noticeable trend is that site 29,871 yields strong signals for shared SNVs between samples with large and similar AFs. We also observe that the number of samples with a variant at that site is unusually high (Fig. 5D).
Discussion
In this study, we have analyzed RNA-seq data sets from 129 COVID-19 positive patients plus 6928 SARS-CoV-2 genomes in depth to describe the intra-host variation in SARS-CoV-2. Our analyses yielded four major observations. First, the iSNV mutational spectra closely match the SNP mutational spectra inferred from the consensus genomes. In particular, the SARS-CoV-2 genome is enriched with C > U changes overall, both for iSNVs and SNPs. Genes nsp6 and nsp10 are particularly enriched for U > C mutations, whereas nsp7 has an enrichment of A > C SNVs. Second, the mutational profile of SARS-CoV-2 largely matches that of other Coronaviruses, but with some key differences. SARS-CoV-2 has a significantly larger proportion of G > U changes in both iSNVs and SNPs, when compared to SARS-CoV-1 and MERS. Additionally, we did not see A > U SNVs in SARS-CoV-1, as previously reported (Pavlović-Lažetić et al. 2004). Third, although the SV spectra are likely reflecting the transcriptome landscape of SARS-CoV-2, we detected a significant fraction of insertions and deletions that contributed to the genetic diversity of SARS-CoV-2. Fourth, the mutational spectra of the SNPs and iSNVs indicate that there is a complex interplay between endogenous SARS-CoV-2 mutational processes and host-dependent RNA editing. This observation is in line with several recent studies that propose APOBEC and ADAR deaminase activity as a likely driver of the C > U changes in the SARS-CoV-2 genomes (Giorgio et al. 2020). Of note, this recent study also reported that the number of observed transversions is compatible with mutation rates found in other Coronaviruses (Eckerle et al. 2010; Giorgio et al. 2020).We observed lower mutational complexity within the nsp3 region of the SARS-CoV-2, indicating that the mutations in this region tend to become SNPs. This agrees with previous reports that indicate that the nsp3 region has a stronger phylogenetic signal than the majority of the SARS-CoV-2 protein-coding regions (Pereson et al. 2020; Yuan et al. 2020).
We also investigated the potential impact of iSNVs and SNPs on probes and primers commonly used in RT-PCR-based detection and amplicon sequencing of SARS-CoV-2. Most probes we analyzed contain both SNPs and iSNVs. Although many platforms can tolerate a few single nucleotide mismatches without the loss of target hybridization, the overall diversity exhibited by SARS-CoV-2 presents potential challenges for probe and primer development. Because we observed an agreement in mutational profiles between the SNPs and iSNVs, for future probe and primer designs it could be useful to track the iSNVs to potentially predict and avoid variable regions of the genome. With the integration of these data into design processes at early stages, greater sensitivity could be achieved for hybridization primers and probes even as the virus evolves.
We analyzed longitudinal samples taken from the same COVID-19-positive patient within 24 hours of one another to analyze AFs of SNPs and iSNVs. We found that the SNP and iSNV profiles and AFs were concordant, indicating the potential of using shared SNPs and iSNVs and their respective AFs for tracking intra-host SARS-CoV-2 population dynamics. This agrees with the recent SARS-CoV-2 genomic epidemiology study in Austria, where iSNVs were found to be stable over time within the same patient (Popa et al. 2020). Although these analyses cannot confirm sample pairs as having been involved in direct transmissions without additional metadata, this exploratory analysis suggests the possible presence of such transmission pairs (Worby et al. 2017). We believe the analysis done here serves to highlight the potential of extracting possible events through sequence data alone.
Despite the potential for tremendous insight, the study of intra-host variation in viruses can be confounded by multiple factors. First, the estimated AFs are impacted by variable coverage and transcription patterns. Second, a low viral load (Ct values above 32) in samples can have an impact on downstream sequencing and analysis (Thorburn et al. 2015; Huang et al. 2019; Supplemental Fig. S4). Third, previous studies (De Maio et al. 2020) highlight SARS-CoV-2 sites marked as prone to high homoplasy that need to be taken into consideration for transmission analyses. Lastly, lack of additional metadata imposes a barrier to an in-depth study of transmission events. These factors should be addressed in future studies of intra-host variation in SARS-CoV-2.
In summary, our analysis of intra-host variation across 129 RNA-seq samples from COVID-19-positive patients revealed a complex landscape of within-host diversity that will likely shed additional light on the elusive mechanisms driving the rapid dissemination of SARS-CoV-2. Metatranscriptomic analysis is a powerful tool for interrogating the genomic and transcriptomic landscape of RNA viruses, as it provides a simultaneous peek into viral, bacterial, and host gene expression. Future studies able to integrate all three of these perspectives may hold the key to novel therapies and treatments of this devastating pandemic.
Methods
Data sets
We analyzed available RNA-seq data from 10 patient samples collected by Baylor College of Medicine (Doddapaneni et al. 2020), and from 140 patient samples collected by Weill Cornell College of Medicine (Butler et al. 2021). Both data sets consist of Illumina NovaSeq 6000 paired-end reads. Samples were first tested for the presence of the SARS-CoV-2 genetic material with CDC RT-PCR-based tests, and then metagenomic RNA-seq was performed (library preparation and sequencing details in Doddapaneni et al. 2020 and Butler et al. 2021). Host and bacterial genetic material have been removed from the data sets, and we performed all analyses on the viral read data. Additionally, we removed 21 samples from the Weill Cornell data sets due to either high Ct values (>32, 5 samples) in the RT-PCR tests or low read counts (<20,000 reads, 16 samples) for the SARS-CoV-2 reads.
We also used additional 147 RNA-seq samples obtained by the NYU Langone Sequencing center (PRJNA650245) (Maurano et al. 2020) in order to compare mutational profiles observed in two different NYC data sets (Fig. 2B,D) and investigate overlaps between NYC iSNVs and SNPs and Houston and GISAID data (Fig. 3B).
In addition, we downloaded 6928 SARS-CoV-2 consensus genomes from the GISAID database, available on April 18th, 2020. We only selected high-quality, complete (<29 kb) genomes.
Furthermore, we analyzed 42 samples of SARS-CoV-1 and 53 samples of MERS viral read data (PRJNA233943) (M Frieman, C Coleman, and SC Daugherty, pers. comm.) sequenced by the University of Maryland School of Medicine (Baltimore, MD, USA).
In total, we analyzed 6928 SARS-CoV-2 consensus sequences and 129 SARS-CoV-2 (119 NYC and 10 Houston RNA-seq samples), 42 SARS-CoV-1, and 53 MERS samples. The summary of all data used in the paper can be found in Supplemental File S3.
Read QC and mapping
We processed the Illumina paired-end reads using Trimmomatic ver. 0.39 (Bolger et al. 2014) to remove adapter sequences and trim low-quality base pairs. We used a universal set of Illumina adapters as a reference for the adapter removal. We set the maximum mismatch count to 2, palindrome clip threshold to 30, and simple clip threshold to 10. We also trimmed leading and trailing low-quality (quality value <3) and ambiguous (N) base pairs. Finally, we applied sliding window trimming cutting the read if the quality score of four contiguous bases made the average score drop below 15. After trimming in the final read set, we included the reads above the length of 36 with both reads from a pair passing quality control.
We aligned the trimmed reads to the reference genome using Burrows-Wheeler Alignment tool (BWA) ver. 0.7.17 (Li and Durbin 2009; Li 2013). We used paired-end mode for mapping reads to the SARS-CoV-2 reference genome (NC_045512).
We used SAMtools ver. 1.9 to convert the output of BWA from SAM to BAM format and to sort and generate indices for the BAM files (Li et al. 2009).
SNV calling and annotation
We used LoFreq ver. 2.1.4 to perform variant calling on the trimmed and mapped reads (Wilm et al. 2012). We filtered the variants with the default LoFreq parameters: minimum coverage was set to 10, Phred quality-score set to Q20 (99%), and strand-bias FDR correction P-value is greater than 0.001. We also filtered out the variants occurring below 0.02 AF threshold for the subsequent analyses (in accordance with the values used for SARS-CoV-2 iSNV analyses in other studies) (Popa et al. 2020) and required all iSNVs to be supported by 10× minimum coverage. We annotated the SNVs found in each of the data sets with SnpEff ver. 4.3 (Cingolani et al. 2012). We used SNPGenie (Nelson et al. 2015) with the default set of parameters to estimate the genetic diversity and nonsynonymous to synonymous diversity ratios in SARS-CoV-2.
SV calling
Structural variations were identified using Manta (ver. 1.6.0) (Chen et al. 2016). Subsequently, the SV calls were merged using SURVIVOR (ver. 1.0.7) (Jeffares et al. 2017) using a 100-bp maximum distance between the breakpoints and requiring that the SV types are in agreement in order to merge two SVs across the samples. We annotated the SV using a simple 1-bp overlap method using BEDTools (ver. 2.27.1) (Quinlan and Hall 2010) intersect using the annotations. The same method was used to establish if the start or stop breakpoints of an SV are overlapping with the TRS sites. To test the significance of the overlap, we used a permutation test where we randomized the TRS sites (using BEDTools random) to generate random TRSs with length of 5 bp, 1000 times and calculated per TRS the number of start/stop breakpoints of the SV catalog. Subsequently we used this together with the observed overlap using a Kolmogorov–Smirnov with an alternative set to “two.sided” in R (ver. 3.2.2).
To generate SV and SNV densities, we computed the number of variations per type within a 100-bp window. For each variant, we counted 1/AF where AF is the frequency of that variant across the samples. This was done based on a custom script (Supplemental Code). The plot was generated using Circos (ver. 0.69-8) (Krzywinski et al. 2009).
Phylogenetic tree construction
We used Parsnp (ver. 1.2) (Treangen et al. 2014) to align the GISAID genomes. We set the maximal cluster D value to 30,000, and the rest of the parameters were set to the default values. We used RAxML (Stamatakis 2014) to infer a phylogenetic tree from the GISAID alignment. We ran RAxML with default parameters using GTRCAT approximation model for tree scoring.
Probe and primer mapping
Primer and probe sequences were derived from the WHO website and hCoV-2019/nCoV-2019 version 3 amplicon set (Artic network). We mapped probes and primers against the SARS-CoV-2 reference genome (NC_045512) with Bowtie 2 (Langmead and Salzberg 2012). Analysis of the primer and probe mapping regions was performed with a custom R script (Supplemental Code), and visualizations were done with R-3.6.1 (R Core Team 2020).
Transmission analyses
We counted the number of shared variants (SNPs + iSNVs) within individual pairs. For each pair, we consider both combinations of one sample as a putative donor and one sample as a putative recipient. Shared variants were then defined as variants that share the same variant nucleotide between the two samples and where the variant frequencies in the assigned donor sequences are from 0.02 to 0.5. We examined variants with frequencies ≥ 0.02 as the cutoff to avoid including variants caused by sequencing errors. For the 119 samples from New York, given that we consider each pair twice, there are 14,042 pairs. Note that, because we are looking for putative transmission events, we can only consider samples within the same geographic region, so we limited our analyses to the 119 samples that came from New York. We masked the variants that occur between positions 1 and 55 and 29,804 and 29,903 in the genome. Additionally, we masked 25 nucleotide positions between 56 and 29,804 that are highly homoplasic (De Maio et al. 2020). These positions are more prone to sequencing and mapping errors and therefore were not used in the transmission analyses.
We used variants supported by at least 10 reads. We also consider the case where the variant base is the same as the reference sequence base. In this case, for instance, when a variant is called at a site with 0.7 AF and no other variants are present, we take the reference base as a variant with 0.3 AF if there are no other reads present with an alternate allele and there are at least 10 reads mapping to the reference base.
Data access
All variant calling files generated in this study are available at https://rice.box.com/v/SARS-COV-2-SNV-Study and in Supplemental Files S4–S7. Scripts used for data analysis are available at https://gitlab.com/treangenlab/covirt_scripts. Scripts used for probe and primer analysis and visualization are available at GitHub (https://github.com/COV-IRT/microbial/tree/master/manuscript_references). All custom scripts used in this study are also available as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
The authors thank Jamie Purcell for feedback and discussion contributions on the effects of variants on the qRT-PCR detection methods. The authors also thank Dr. Luay Nakhleh for suggestions specific to comparative genomic analyses of SARS-CoV-1 and MERS-COV. The authors thank the GISAID contributors who provided the SARS-CoV-2 assemblies. Finally, the authors also thank all members of the COVID-19 International Research Team (www.cov-irt.org) for their helpful feedback during weekly meetings. N.S. and Y.L. are supported by the Department of Computer Science, Rice University. D.A., Q.W., N.S., R.A.L.E., T.J.T., and Y.L. are supported by startup funds from Rice University. N.S. and T.J.T. are partially supported by a C3.ai Digital Transformation Institute COVID-19 award (C3.ai DTI). M.D.J. is supported under National Institutes of Health (NIH) award No. R01HD091731 from the National Institute of Child Health and Human Development. F.J.S. acknowledges funding and part of the data was produced by Baylor College of Medicine under the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (U19AI144297-01). A.B. is supported by supplemental funds for COVID-19 research from Translational Research Institute for Space Health through National Aeronautics and Space Administration Cooperative Agreement NNX16AO69A (T-0404) and further funding was provided by KBR, Inc. D.P. is supported by the European Research Council (ERC-617457-PHYLOCANCER), Spanish Ministry of Economy and Competitiveness (PID2019-106247GB-I00), Fondo Supera COVID19 (EPICOVIGAL), and Xunta de Galicia (CT850A-2). This material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The views and conclusions expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Army, Department of the Navy, Department of Defense, Office of the Director of National Intelligence, Army Research Office, or U.S. Government.
Author contributions: N.S. led the iSNV and SNP analyses, interpreted the results, generated the figures, and wrote the manuscript. M.D.J. analyzed the impact of polymorphisms on probes and primers and generated figures. Y.L. analyzed single nucleotide variant data and generated the figures. D.A. analyzed phylogenetic data and generated the figures. M.D.L. analyzed genomic data and generated figures. Q.W. analyzed and interpreted viral transmission data, generated figures, and wrote the manuscript. R.A.L.E. interpreted the viral transmission and phylogenetic data and wrote and edited the manuscript. S.V. edited the manuscript, provided exchange of ideas, and generated figures. C.E.M. provided the RNA-seq data and contributed to the manuscript. T.J.T. supervised the analyses, interpreted the data, and edited and wrote the manuscript. M.M. and F.J.S. lead the SV analysis, interpretation of the data, and edited and wrote the manuscript. A.B. edited the manuscript and provided exchange of ideas. K.T. reviewed the SNV commands and called variants in public COVID-19 metatranscriptomes for comparison. D.P. proposed some of the analyses, helped with their interpretation, and contributed to manuscript writing. All co-authors read and edited the manuscript and provided constructive feedback.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.268961.120.
Freely available online through the Genome Research Open Access option.
References
- Barbezange C, Jones L, Blanc H, Isakov O, Celniker G, Enouf V, Shomron N, Vignuzzi M, van der Werf S. 2018. Seasonal genetic drift of human influenza A virus quasispecies revealed by deep sequencing. Front Microbiol 9: 2596. 10.3389/fmicb.2018.02596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki MK, Collette NM, Coffey LL, Rompay KKAV, Hwang MH, Thissen JB, Allen JE, Zemla AT. 2019. Multiscale analysis for patterns of Zika virus genotype emergence, spread, and consequence. PLoS One 14: e0225699. 10.1371/journal.pone.0225699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D, Mozsary C, Meydan C, Foox J, Rosiene J, Shaiber A, Danko D, Afshinnekoo E, MacKay M, Sedlazeck FJ, et al. 2021. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat Commun 12: 1660. 10.1038/s41467-021-21361-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AD, Williamson MK, Lewis S, Shoemark D, Carroll MW, Heesom KJ, Zambon M, Ellis J, Lewis PA, Hiscox JA, et al. 2020. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med 12: 68. 10.1186/s13073-020-00763-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N, Walker C, Borges R, Weilguny L, Slodkowicz G, Goldman N. 2020. Issues with SARS-CoV-2 sequencing data. https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 [accessed December 10, 2020].
- Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. 2011. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol 8: 270–279. 10.4161/rna.8.2.15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddapaneni H, Cregeen SJ, Sucgang R, Meng Q, Qin X, Avadhanula V, Chao H, Menon V, Nicholson E, Henke D, et al. 2020. Oligonucleotide capture sequencing of the SARS-CoV-2 genome and subgenomic fragments from COVID-19 individuals. bioRxiv 10.1101/2020.07.27.223495 [DOI] [PMC free article] [PubMed]
- Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc Natl Acad Sci 96: 13910–13913. 10.1073/pnas.96.24.13910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, et al. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6: e1000896. 10.1371/journal.ppat.1000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall 1: 33–46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas C, Fuentes-Villalobos F, Garrido JL, Haigh J, Barría MI. 2020. Insights on early mutational events in SARS-CoV-2 virus reveal founder effects across geographical regions. PeerJ 8: e9255. 10.7717/peerj.9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio SD, Martignano F, Torcia MG, Mattiuz G, Conticello SG. 2020. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv 6: eabb5813. 10.1126/sciadv.abb5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117: 17–37. 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34: 4121–4123. 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jennison A, Whiley D, McMahon J, Hewitson G, Graham R, De Jong A, Warrilow D. 2019. Illumina sequencing of clinical samples for virus detection in a public health laboratory. Sci Rep 9: 5409. 10.1038/s41598-019-41830-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8: 14061. 10.1038/ncomms14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KA, Cheung P. 2020. Presence of mismatches between diagnostic PCR assays and coronavirus SARS-CoV-2 genome. R Soc Open Sci 7: 200636. 10.1098/rsos.200636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AS, Weissman DB, Greenbaum B, Ghedin E, Koelle K. 2017. Transmission bottleneck size estimation from pathogen deep-sequencing data, with an application to human influenza A virus. J Virol 91: e00171-17. 10.1128/JVI.00171-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997 [q-bio.GN].
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-H, Moses DC, Hsieh C-H, Cheng S-C, Chen Y-H, Sun C-Y, Chou C-Y. 2018. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res 150: 155–163. 10.1016/j.antiviral.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. 2019. Structural variant calling: the long and the short of it. Genome Biol 20: 246. 10.1186/s13059-019-1828-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Ramaswami S, Zappile P, Dimartino D, Boytard L, Ribeiro-dos-Santos AM, Vulpescu NA, Westby G, Shen G, Feng X, et al. 2020. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City region. Genome Res 30: 1781–1788. 10.1101/gr.266676.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, Gloza-Rausch F, Balboni A, Battilani M, Rihtarič D, et al. 2018. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep 8: 15177. 10.1038/s41598-018-33487-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CW, Moncla LH, Hughes AL. 2015. SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31: 3709–3711. 10.1093/bioinformatics/btv449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, et al. 2012. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J Virol 86: 8546–8558. 10.1128/JVI.00736-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DJ, Dudas G, Wohl S, Goba A, Whitmer SLM, Andersen KG, Sealfon RS, Ladner JT, Kugelman JR, Matranga CB, et al. 2015. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell 161: 1516–1526. 10.1016/j.cell.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly MD, Procario MC, Lauring AS. 2017. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. eLife 6: e26437. 10.7554/eLife.26437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlović-Lažetić GM, Mitić NS, Beljanski MV. 2004. Bioinformatics analysis of SARS coronavirus genome polymorphism. BMC Bioinformatics 5: 65. 10.1186/1471-2105-5-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck KM, Lauring AS. 2018. Complexities of viral mutation rates. J Virol 92: e01031-17. 10.1128/JVI.01031-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereson MJ, Mojsiejczuk L, Martínez AP, Flichman DM, Garcia GH, Lello FAD. 2020. Phylogenetic analysis of SARS-CoV-2 in the first few months since its emergence. J Med Virol https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.26545 [accessed December 13, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LLM, Song T, Rosenfeld R, Lin X, Rogers MB, Zhou B, Sebra R, Halpin RA, Guan Y, Twaddle A, et al. 2016. Quantifying influenza virus diversity and transmission in humans. Nat Genet 48: 195–200. 10.1038/ng.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa A, Genger J-W, Nicholson MD, Penz T, Schmid D, Aberle SW, Agerer B, Lercher A, Endler L, Colaço H, et al. 2020. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci Transl Med 12: eabe2555. 10.1126/scitranslmed.abe2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, et al. 2017. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12: 1261–1276. 10.1038/nprot.2017.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazzotti D, Angaroni F, Maspero D, Gambacorti-Passerini C, Antoniotti M, Graudenzi A, Piazza R. 2021. VERSO: a comprehensive framework for the inference of robust phylogenies and the quantification of intra-host genomic diversity of viral samples. Patterns 10.1016/j.patter.2021.100212 [DOI] [PMC free article] [PubMed]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rodriguez-Roche R, Blanc H, Bordería AV, Díaz G, Henningsson R, Gonzalez D, Santana E, Alvarez M, Castro O, Fontes M, et al. 2016. Increasing clinical severity during a dengue virus type 3 Cuban epidemic: deep sequencing of evolving viral populations. J Virol 90: 4320–4333. 10.1128/JVI.02647-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, Zhou Z, Yang J, Zhong J, Yang D, et al. 2020. Genomic diversity of severe acute respiratory syndrome–coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis 71: 713–720. 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Almazán F, Zúñiga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2: 265–288. 10.1146/annurev-virology-100114-055218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn F, Bennett S, Modha S, Murdoch D, Gunson R, Murcia PR. 2015. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J Clin Virol 69: 96–100. 10.1016/j.jcv.2015.06.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15: 524. 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley DM, Sloots TP. 2005. Sequence variation in primer targets affects the accuracy of viral quantitative PCR. J Clin Virol 34: 104–107. 10.1016/j.jcv.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40: 11189–11201. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CJ, Lipsitch M, Hanage WP. 2017. Shared genomic variants: identification of transmission routes using pathogen deep-sequence data. Am J Epidemiol 186: 1209–1216. 10.1093/aje/kwx182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Morelli MJ, Thébaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP. 2011. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J Virol 85: 2266–2275. 10.1128/JVI.01396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579: 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Wang L, Fang Y, Wang L. 2020. Global SNP analysis of 11,183 SARS-CoV-2 strains reveals high genetic diversity. Transbound Emerg Dis https://onlinelibrary.wiley.com/doi/abs/10.1111/tbed.13931 [accessed December 13, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Elena SF. 2015. Matters of size: genetic bottlenecks in virus infection and their potential impact on evolution. Annu Rev Virol 2: 161–179. 10.1146/annurev-virology-100114-055135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.