Abstract
Systemic lupus erythematosus (SLE) is an incurable autoimmune disease disproportionately affecting women. A major obstacle in finding targeted therapies for SLE is its remarkable heterogeneity in clinical manifestations as well as in the involvement of distinct cell types. To identify cell-specific targets as well as cross-correlation relationships among expression programs of different cell types, we here analyze six major circulating immune cell types from SLE patient blood. Our results show that presence of an interferon response signature stratifies patients into two distinct groups (IFNneg vs. IFNpos). Comparing these two groups using differential gene expression and differential gene coexpression analysis, we prioritize a relatively small list of genes from classical monocytes including two known immune modulators: TNFSF13B/BAFF (target of belimumab, an approved therapeutic for SLE) and IL1RN (the basis of anakinra, a therapeutic for rheumatoid arthritis). We then develop a multi–cell type extension of the weighted gene coexpression network analysis (WGCNA) framework, termed mWGCNA. Applying mWGCNA to RNA-seq data from six sorted immune cell populations (15 SLE, 10 healthy donors), we identify a coexpression module with interferon-stimulated genes (ISGs) among all cell types and a cross–cell type correlation linking expression of specific T helper cell markers to B cell response as well as to TNFSF13B expression from myeloid cells, all of which in turn correlates with disease severity of IFNpos patients. Our results demonstrate the power of a hypothesis-free and data-driven approach to discover drug targets and to reveal novel cross-correlation across cell types in SLE with implications for other autoimmune diseases.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that affects multiple organs including the skin, joints, the central nervous system, and the kidneys (Davidson 2016; Kaul et al. 2016). It is caused by an aberrant autoimmune response to produce antibodies against self-antigens, and these autoantibodies form immune complexes (ICs), which are then deposited into different organs and affect their normal function (Bayry 2016). SLE is a highly heterogeneous disease in terms of development, presentation, manifestations, and severity; also, the incidence and prevalence vary significantly (Carter et al. 2016). Thus, the time course of flares and remission is unpredictable (Obermoser and Pascual 2010). The diverse clinical manifestations of SLE present a challenge because of the involvement of several organs as well as diversified autoantibodies (Tsokos 2011). Therefore, accurate diagnosis and disease activity assessment is essential for managing SLE disease (Lam and Petri 2005). Even though the SLE Disease Activity Index (SLEDAI) score, which measures disease activity from 24 different clinical variables (Bombardier et al. 1992), is widely adopted, it still leaves out several manifestations because of the heterogeneous nature of SLE (Thanou et al. 2014). Such heterogeneity in manifestations and in assessing disease severity and activity has made it difficult to develop efficient therapeutics and, to date, only one drug, belimumab, which targets the TNF superfamily member 13b, TNFSF13B (also known as BAFF), has been approved for use in SLE (Kaul et al. 2016).
Clinical heterogeneity is the main obstacle in finding effective treatments for SLE (Touma and Gladman 2017); therefore, it is important to discover molecular subtypes and signatures that correlate to clinical phenotypes. One strong signature characterized to date is the heightened expression levels of type I interferon (IFN)-related genes in the blood transcriptome of SLE patients and in correlation with disease activity (Baechler et al. 2003; Feng et al. 2006; Crow 2014) and pathogenesis (Obermoser and Pascual 2010). Interferons induce the expression of canonical interferon-stimulated genes (ISGs) as well as a specific epigenetic signature (Barrat et al. 2019). Elevated levels of IFN in SLE patients was first reported over 40 years ago (Hooks et al. 1979), and the role of IFN signaling has been well studied ever since (Rönnblom and Leonard 2019), including in clinical studies (Chaichian et al. 2019; Oke et al. 2019). Additionally, a recent single-cell transcriptome study revealed the up-regulation of IFN-inducible or IFN-stimulated genes in lupus nephritis patients (Der et al. 2017). However, how this IFN response affects molecular programs of different immune cells and how it influences the cross-correlation among different types of immune cells is largely unknown, as most studies use mixed populations of immune cells (e.g., peripheral blood mononuclear cells, PBMCs) and some studies used only a few cell types (Kyogoku et al. 2013; Catalina et al. 2019). Therefore, transcriptome profiling with sorted populations of major immune cell types provides an opportunity to define cell-specific molecular programs using traditional differential expression analysis (Love et al. 2014) and coexpression analysis (Oldham et al. 2006; Fuller et al. 2007; Southworth et al. 2009; van Nas et al. 2009; de la Fuente 2010) and with multi–cell type analysis to identify cross-correlation among expression programs that, in turn, correlate with disease activity and severity.
In order to interrogate both cell-specific and cross–cell type correlates of SLE, we have generated gene expression profiles of six major human immune cell types from a cohort of SLE patients and healthy controls using RNA-seq (Stark et al. 2019). Our cohort consists of samples from 64 SLE patients and 24 controls as well as their demographic information, clinical features (e.g., SLEDAI), and measurements of the plasma levels of several relevant cytokines and chemokines. For classical monocytes, we generated RNA-seq data from all donors (64 SLE, 24 healthy). In addition, we profiled T cells (n = 24), neutrophils (n = 24), B cells (n = 20), and conventional (n = 20) and plasmacytoid (n = 22) dendritic cells from SLE patients and from healthy control subjects (n = 11 or 12 for each cell type).
Results
Transcriptional profile of classical monocytes reveals two molecular subtypes of SLE
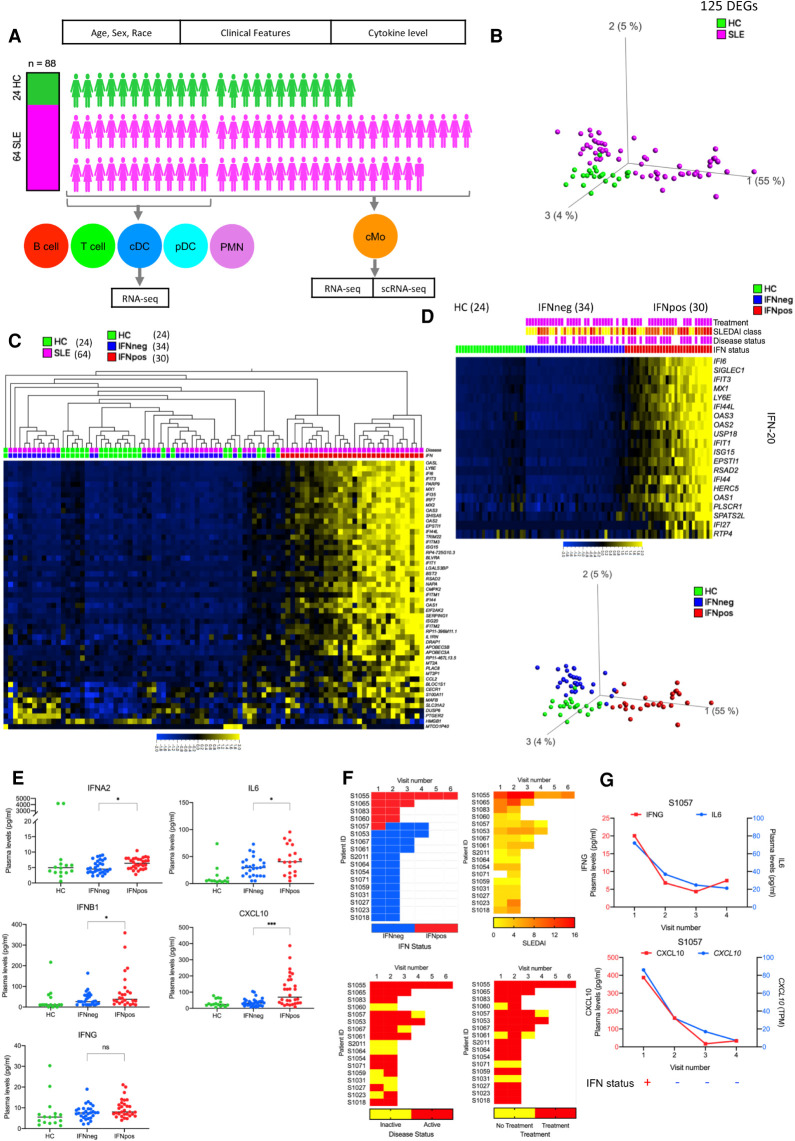
Because SLE is known for its heterogeneity, we first investigated the full set of patients in our cohort (Supplemental Table S1) for distinct molecular signatures. To this end, we analyzed the bulk RNA-seq-based transcriptome profiles of classical monocytes (cMos) from 64 SLE patients and compared them to that of 24 healthy controls (HCs) (Fig. 1A; Supplemental Fig. S1A,B). For this analysis, we only used the samples gathered from the first study visit of each patient, setting aside the longitudinal samples for validation. This analysis revealed a total of 125 differentially expressed genes (DEGs) (P.adj < 0.05) with 109 up- and 16 down-regulated genes (Supplemental Fig. S1C). The PCA plot of these 125 DEGs highlights the heterogeneity within SLE patients, with one group showing clear differences compared to HCs and the other showing no such separation (first principal component) (Fig. 1B). Further analysis of these 125 DEGs showed that most of these DEGs are enriched in interferon-related pathways, and a hierarchical clustering based on the top 50 most variable DEGs showed a clear distinction between patients with high (IFNpos) versus low (IFNneg) expression of IFN genes in SLE (Fig. 1C; Supplemental Fig. S1C; details in Supplemental Table S2). To further illustrate this point, we used 20 interferon signature genes (IFN-20) for Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005) and showed a significant enrichment of their expression in SLE compared to HC (normalized enrichment score [NES] = 0.962; FDR q-value = 0.023) (Supplemental Fig. S1C). Additionally, IFNpos versus IFNneg SLE patients showed a clear difference in their expression of ISGs (first visit for each patient) (Fig. 1D; Supplemental Fig. S1D), a pattern that was conserved when we included their longitudinal samples (Supplemental Fig. S1E). To better quantify these differences, we conducted pairwise comparisons of the resulting three groups (IFNpos, IFNneg, HC) identifying 1439 DEGs between IFNpos and IFNneg and 1288 DEGs between IFNpos and HC, whereas there were only five DEGs between IFNneg and HC (P.adj < 0.05) (Supplemental Table S2). These results suggest that it is important to investigate these two molecular subgroups of SLE (IFNpos and IFNneg) separately and that there is no clear cMo molecular signature that distinguishes IFNneg patients from HCs. This distinction is likely also critical for developing therapeutics for SLE, as their clinical testing may suffer from such high heterogeneity among the patient cohort.
Figure 1.
Transcriptional profiling of classical monocytes reveals two molecular subtypes of SLE. (A) Overview of the SLE cohort. Healthy control (HC) samples are highlighted in green color and SLE samples are highlighted in magenta color. Six different immune cell types, including classical monocytes (cMo), Polymorphonuclear Neutrophils (PMNs), conventional dendritic cells (cDC), plasmacytoid dendritic cells (pDC), T cells, and B cells are displayed in different colors. (B) The PCA plot based on 125 DEGs (P.adj < 0.05 from a Benjamini–Hochberg test in DESeq2) between SLE and HC in classical monocytes. Green and magenta colors represent HC and SLE patients, respectively. (C) Heat map of top 50 most variable DEGs (one per row) in a SLE versus HC comparison (P.adj < 0.05) clustered with hierarchical clustering and presented as row-wise z-scores (−2.0 [blue] to 2.0 [yellow]) of transcripts per million (TPM) in SLE (magenta) or HC (green); each column represents an individual patient. IFNpos (red) and IFNneg (blue) are shown in different colors. (D) SLE patients (first study visits) within each group (HC, IFNneg, IFNpos) are sorted with respect to the first principal component of IFN-20 gene expression. The SLEDAI CLASS panel shows the SLEDAI score and is divided into three different categories based on score: DA1 (0–2), DA2 (3–7), and DA3 (>7) (yellow, orange, and red, respectively). Patients with active disease status and those taking treatment are highlighted in purple color. This color scheme is used throughout all figures. (E) Scatter dot-plots show the expression level (pg/ml) in plasma of IFNG, IL6, and CXCL10. Only first visit samples (n = 76) were used in these scatter dot-plots. The difference in measurements from IFNpos and IFNneg patients has been calculated using unpaired t-test, two-tailed. (ns) Not significant, (*) <0.05, (***) <0.001. (F) Longitudinal IFN response status of 17 SLE patients (five patients were IFNpos and 12 patients were IFNneg at the first study visit) (top left). Only one patient (S1057) changed their IFN response status between study visits, changing from IFNpos to IFNneg between the first and second study visit and remained IFNneg in all follow-up visits. Similar heat maps of IFN status and different clinical parameters such as SLEDAI, disease status, and treatment for patients with longitudinal information. (G) Two plots with connecting lines (upper and lower) show expression changes of multiple cytokines and chemokines in the longitudinal data for patient S1057, where different analytes are plotted on different y-axes (IFNG on y1-axis in red color and IL6 on y2-axis in blue color) in different colors. The RNA-level expression of CXCL10 is also shown (y2-axis) with protein-level expression of CXCL10 (y1-axis). The lower panel of each plot shows their IFN response status over multiple longitudinal visits.
The bulk RNA-seq data provides average gene expression but lacks the resolution to determine the proportion of cMos that are positive for the expression of IFN signature genes. To this end, we used single-cell RNA-seq to test whether the IFNpos status is conferred by a subset of cells or is shared across each individual cMo cell. Our analysis of the transcriptome profiles of ∼156 single cells from three different IFNpos patients demonstrated that nearly all cMos are positive for the expression of at least several of the ISGs (Supplemental Fig. S1F). Particularly, IFI6 and LY6E, both well-known type I interferon-related genes (Zhang et al. 2015a), are highly expressed in most cMos across different patients (violin plot, Supplemental Fig. S1F). A recent high-throughput 10x Genomics–based single-cell study also showed an expansion of monocytes with enriched ISG expression in SLE patients (adult and pediatric), with the highest disease activity in both pediatric and adult SLE patients (Nehar-Belaid et al. 2020) confirming the results from our low-throughput single-cell data.
IFN response status of SLE patients correlates with the levels of pro-inflammatory cytokines and chemokines
For a subset of donors from our cohort, we measured the plasma level of different cytokines and chemokines such as IFNA2, IFNB1 IFNG, IL2, IL6, IL10, CXCL10, TNF, and TGFB1 and compared the levels from only the first study visit data across the three groups: HC (n = 16), IFNneg (n = 30), and IFNpos (n = 30). Out of these, IFNA2 (P = 0.0190), IFNB1 (P = 0.0460), IL6 (P = 0.0413), and CXCL10 (P = 0.0002) levels were higher in SLE subgroups in comparison to HCs. These three cytokines (IFNA2, IFNB1, IL6) as well as CXCL10 had a statistically significant difference between IFNpos and IFNneg SLE patients as well (Fig. 1E; Supplemental Fig. S2A). The plasma levels of different interferons, pro-inflammatory cytokines that are well studied in SLE (Banchereau et al. 2016; Rönnblom and Leonard 2019), all showed a positive but nonsignificant correlation with a transcriptome-derived IFN score derived using gene set variation analysis (IFN-20 GSVA; Spearman's correlation of 0.25, 0.22, and 0.16 with IFNA2, IFNB1, and IFNG, respectively). Another pro-inflammatory cytokine with a significant difference between IFNpos and IFNneg is IL6, which induces the maturation of B lymphocytes into plasma cells and increases Ig secretion. Increasing IL6 levels have been shown to correlate with increased disease activity and with anti-DNA autoantibody levels in human SLE (Lai and Yap 2010). It has also been shown that IL6-174G/C and IL6-572G/C polymorphisms are associated with the development of SLE (Cui et al. 2015). We also identified a significantly higher level of a pro-inflammatory chemokine, C-X-C motif chemokine ligand 10 (CXCL10) in IFNpos patients (Fig. 1E), the level of which has also been reported to correlate with SLE disease activity and with organ manifestations of this disease (Kong et al. 2009).
IFN response status molecular signature is mainly conserved in longitudinal samples
Next, we asked whether the IFN response status changes across different visits of the same patient and if this change is related to serum levels of pro-inflammatory molecules or other clinical features. We observe that, for the majority of the patients, the IFN response status is identical across all their follow-up visits, regardless of changes in disease activity, flares and/or treatment regimens (e.g., prednisone use and dose) across the multiple visits. Among the 17 SLE patients with longitudinal data from multiple visits that are at least 1 mo apart, 12 patients were IFNneg and five patients were IFNpos at their first study visit (Fig. 1F). Out of these, only one patient (S1057) showed a change in the IFN response status, with the first collection being IFNpos and the next three follow-up visits being IFNneg (Fig. 1F). To understand the variation across different IFNs, pro-inflammatory cytokines (IL6 and CXCL10), and clinical parameters such as SLEDAI, disease status, and treatment, we generated a series of heat maps (Fig. 1F; Supplemental Fig. S2B) highlighting that IFN levels in plasma do not always correlate with transcriptome-derived IFN status in classical monocytes. All three patients (S1057, S1055, and S1053) with ≥4 visits show uncoordinated changes in IFNs plasma level with respect to changes in IFN signature gene-based GSVA score (Supplemental Fig. S2C). Furthermore, a principal component-based clustering of the plasma levels of three different interferons is not able to distinguish either the IFNpos versus IFNneg patients or their SLEDAI score-based disease activity categories (DA1 [0–2], DA2 [3–7], and DA3 [>7]) (Supplemental Fig. S2D).
In the single patient with IFN response status change, the serum levels of CXCL10 significantly dropped (2.4-fold), in line with switching from IFNpos to IFNneg between the first and second sample collections (Fig. 1G). This sharp decrease is also reflected as a 2.7-fold decrease in the mRNA levels of CXCL10, which encodes CXCL10 protein, in classical monocytes between the same two visits (Fig. 1G). Although the differences in IFNG levels between IFNpos and IFNneg groups did not reach statistical significance (P = 0.0644) for the overall cohort (Fig. 1E), for this specific patient, IFNG levels also showed a threefold decrease in the second visit (Fig. 1G). Consistent with comparisons from the whole cohort, the level of IL6 also showed a twofold decrease for the second visit of this patient (Fig. 1G). Even though these results suggest a tight link between levels of pro-inflammatory molecules and IFN status for this patient, our analysis of two other patients (S1055 and S1053) with at least four visits shows that significant changes in plasma levels of pro-inflammatory molecules are not always accompanied by changes in the transcriptome-based IFN status (e.g., S1055 in Supplemental Fig. S2E). Overall, these results show that IFN-based stratification of SLE patients is relatively stable across their longitudinal samples and the relationship between changes in gene transcriptome-derived status and changes in the plasma levels of specific pro-inflammatory cytokines or chemokines is not one-to-one.
Weighted gene coexpression network analysis highlights important gene modules
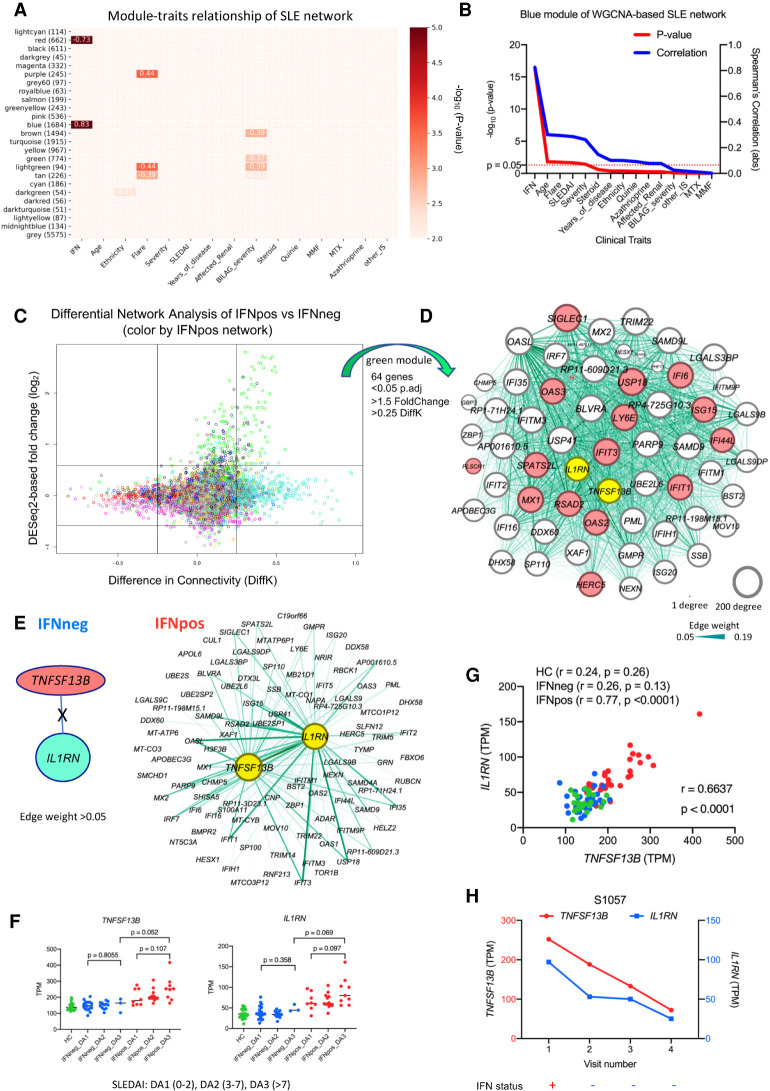
The major limitation with differential gene expression analysis is that it treats each gene individually while comparing expression profiles, whereas most biological functions are performed by a group of genes working together in coordination. The traditional approach is to study each cell type one-by-one and to find differentially expressed genes for those cells based on the gene-level expression estimates within and across different groups. This approach is complemented by analyzing the patterns of correlated gene expression (coexpression) which relies on the “guilt-by-association” principle (Chu et al. 1998), suggesting genes with coordinated changes in expression are more likely to be involved in similar biological functions. Therefore, we used weighted gene coexpression network analysis (WGCNA) to generate a network from classical monocyte gene expression profiles of 64 SLE donors (Langfelder and Horvath 2008). This coexpression network revealed a total of 25 different modules, represented by different colors, where each module is a cluster of coexpressed genes. Furthermore, the correlation of eigengene value of each module with external clinical features showed that IFN response status is the most-correlated feature in comparison to other available clinical features (module-trait relationships) such as age, ethnicity, flare, severity, SLEDAI, and treatment regimens (Fig. 2A). The blue module is positively correlated (r = 0.83; P-value = 5 × 10−17) with IFN response status (Fig. 2B) and it consists of 1684 genes. In addition to our IFN-20 gene set, we also manually curated a broader set of 363 IFN-related genes (IFN-363) from different sources and found that most of the hub genes (genes with highest connectivity in the coexpression network) in the blue module are part of this IFN-363 set (Supplemental Fig. S3A). The red module is negatively correlated (r = −0.73; P-value = 1 × 10−11) with IFN response status and it has 662 genes, most of which are related to protein translation (Supplemental Fig. S3B). This WGCNA suggests that the dominance of IFN response signature in the stratification of SLE patients using gene expression is conserved when we use gene coexpression relationships. From the same analysis, we also identified two modules that significantly correlate with flare in SLE patients: the purple module (r = 0.44; P-value = 0.0003) (Supplemental Fig. S3C) and the light green module (r = −0.44; P-value = 0.0003) (Supplemental Fig. S3D). Among the hub genes of the purple module was TNFRSF12A (Supplemental Fig. S3C), which is the receptor of TNFSF12 (also known as TWEAK), a pro-inflammatory cytokine targeted as a potential therapeutic for SLE (Leng et al. 2011). For the light green module, TLR5 (Supplemental Fig. S3D) was one of the hub genes, a stop codon polymorphism of which was shown to be involved in resistance to SLE (Hawn et al. 2005; Devarapu and Anders 2018).
Figure 2.
Combined analysis of differential network and gene expression of classical monocytes reveals two known immune modulators (TNFSF13B and IL1RN). (A) The module-traits correlation matrix heat map of WGCNA based on total 25 different modules and external clinical features such as assigned IFN response status, age, ethnicity, flare, severity, and other clinical parameters. The color shows significance (−log10 of P-value) of the Spearman's correlation between a trait and the module eigengene with the correlation values in each cell that corresponds to a significant correlation (P-value < 0.01). The number of genes in each module is also shown (in brackets) with the module name. IFN response status is the most correlated clinical feature, with the blue module being the top positively correlated (r = 0.83; P = 5 × 10−17) and the red module being the top negatively correlated (r = −0.73; P = 1 × 10−11). (B) The line plot shows significance (−log10 of P-value on the y1-axis in red color) and Spearman's correlation (y2-axis in blue color) of different clinical traits with blue module's eigengene. The clinical traits are sorted in decreasing order of statistical significance of the correlation for the blue module. (C) The plot shows differential expression and differential connectivity of genes between IFNpos and IFNneg patients (x-axis is DiffK = K1 − K2; K1 = connectivity in IFNpos network; K2 = connectivity in IFNneg network, and y-axis is the DESeq2-based log2 fold change). Colors of genes represent different modules based on the IFNpos (Network1) network. (D) Sixty-four genes were selected from the green module using significant DEGs (P.adj < 0.05; 1.5-fold change) as well as DCGs (|DiffK| > 0.25) and visualized by Gephi, where nodes are sized according to the number of edges (connections) and the edge thickness is proportional to the strength of coexpression. Available IFN signature genes (IFN-20) are highlighted in red colors, and two known immune modulators (IL1RN and TNFSF13B [BAFF]) are highlighted in yellow. (E) An example showing the importance of differential network analysis because IL1RN and TNFSF13B, which are in same green module, have a large number of connected genes in the IFNpos network but no connected genes (at a threshold of 0.05 edge weight) in the IFNneg network. The strength of coexpression is also varying and is presented by the width of the connection. TNFSF13B and IL1RN are in different modules (salmon and turquoise, respectively) in the IFNneg network. (F) Expression of TNFSF13B and IL1RN is plotted according to IFN response status and SLEDAI class (DA1, DA2, or DA3). Green, blue, and red colors represent HC, IFNneg, and IFNpos, respectively. The provided P-values are calculated using an unpaired t-test (two-tailed). (G) The Spearman's correlation between TNFSF13B and IL1RN expression (TPM) for HC, IFNneg, and IFNpos group as well as the whole cohort (r = 0.6637; P < 0.0001). (H) Gene expression changes for TNFSF13B and IL1RN in longitudinal data from patient S1057, where genes are plotted on different y-axes; TNFSF13B on y1-axis in red and IL1RN on y2-axis in blue. The lower panel of this plot shows corresponding IFN response status over multiple longitudinal visits.
Combined analysis of differential network and gene expression of classical monocytes reveals known immune modulators
Our application of WGCNA on all cMo SLE samples highlights important gene modules and confirms our findings from the differential expression analysis. Differential network analysis uses these coexpression patterns across different conditions to identify differentially connected genes (DCGs), revealing irregularities in the transcriptome wiring in the disease state (Oldham et al. 2006; Fuller et al. 2007; Southworth et al. 2009; van Nas et al. 2009; de la Fuente 2010). In order to prioritize specific genes and upstream regulators driving differences in gene expression between IFNpos and IFNneg groups, we then applied a differential WGCNA, which uses differential connectivity to identify differentially regulated genes (Fuller et al. 2007). Conceptually, a DCG may have the same level of expression in two different groups but may be coexpressed with distinct sets of genes, suggesting a rewiring of the underlying biological pathways involving this gene. A gene that is both differentially expressed and differentially connected is also likely to have a broader impact in the molecular profile that distinguishes the compared groups from each other. With this motivation, we applied WGCNA to generate two separate coexpression networks, one for IFNpos and one for IFNneg, to identify DCGs between the two conditions. For each gene in each coexpression network, we computed network connectivity as the sum of connection strengths (based on coexpression) with the other genes in that network. We identify DCGs by computing the difference between the connectivity (DiffK) values for each gene between the IFNpos and IFNneg network. Using stringent criteria, we identified a total of 99 genes that are DEGs (1.5-fold change; P.adj < 0.05) as well as DCGs (>0.25 or <−0.25 DiffK) when IFNpos and IFNneg patients are compared. A majority of these genes (64/99) are from the green module which includes known IFN-related genes as well as several other genes previously implicated in the context of SLE (Fig. 2C; Supplemental Table S3). A joint visualization of differential expression and connectivity highlights that the green module genes are enriched in significant differences in both expression as well as connectivity (Supplemental Fig. S4A) with enrichment in IFN-related pathways (Supplemental Fig. S4B). Aside from 16 genes from the IFN-20 set, the green module also harbors several hub genes such as TNFSF13B, IL1RN, BLVRA, ZBP1, PARP9, APOBEC3G, LGALS9DP, GMPR, and USP41 (Fig. 2D). This short list of genes includes two known immune modulators, TNFSF13B (BAFF) and IL1RN (highlighted in Fig. 2D). TNFSF13B is the target of the only approved therapeutic (belimumab) for SLE to date (Kaul et al. 2016). A recombinant form (anakinra) of the other gene, IL1RN (interleukin 1 receptor antagonist), is a therapeutic agent marketed for the treatment of rheumatoid arthritis (Furst 2004), and it inhibits the binding of pro-inflammatory IL1A and IL1B to the IL1 receptor (Sims and Smith 2010).
TNFSF13B and IL1RN expression is dysregulated in SLE
TNFSF13B is significantly differentially expressed (P.adj = 8.14 × 10−16; 1.55-fold change) as well as differentially connected (DiffK = 0.401) between IFNpos and IFNneg patients (Supplemental Table S3). IL1RN is also significantly differentially expressed (P.adj = 3.20 × 10−11; 1.8-fold change) and differentially connected (DiffK = 0.330). Both genes are hubs in the green module of the IFNpos network (Fig. 2E) but are not connected/coexpressed with any other gene in the IFNneg network (edge weight threshold of 0.05). The expression levels of both TNFSF13B and IL1RN are elevated, with higher SLEDAI scores (DA3 class) for IFNpos in comparison to IFNneg patients (Fig. 2F). This correlation is valid when the continuous SLEDAI score is used or when this score is divided into three different disease activity categories: DA1 (0–2), DA2 (3–7), and DA3 (>7) (Fig. 2F) as previously described (Banchereau et al. 2016). The expression levels of TNFSF13B and IL1RN are also strongly correlated with each other (r = 0.6637; P < 0.0001) (Fig. 2G). In the longitudinal data, the expression levels of both genes go down when the IFN response status of patient S1057 changes from IFNpos to IFNneg (from the first to second collection) (Fig. 2H; Supplemental Fig. S4C). The DICE database (Schmiedel et al. 2018), which profiled gene expression of nearly 100 healthy donors for 13 distinct human primary immune cell populations (including cMos and T and B cells), also shows that the expression of TNFSF13B and IL1RN are specifically high in classical monocytes in the circulating blood (Supplemental Fig. S4D).
Additionally, this differential expression and connectivity analysis also revealed many interesting novel genes that have similar expression patterns to TNFSF13B and IL1RN, including the interferon-related genes IFI6, LY6E, and IFIT3 (Supplemental Fig. S4E). For example, BLVRA (biliverdin reductase A) has a canonical function to convert biliverdin to bilirubin, but it plays an anti-inflammatory function by activation of the PI3K–AKT-IL10 pathway as well as inhibition of TLR4 expression via direct binding to the TLR4 promoter (Wegiel and Otterbein 2012). ZBP1 (Z-DNA binding protein 1) encodes a cytosolic DNA sensor that can activate type I IFN response (Takaoka et al. 2007), and it also regulates programmed cell death and other inflammatory responses (Kuriakose and Kanneganti 2018). DDX60L (DExD/H-Box 60 like) is an IFN-stimulated gene and is involved in antiviral immunity (Grünvogel et al. 2015). APOBEC3G (apolipoprotein B mRNA editing enzyme catalytic subunit 3G) is related to RNA editing and also plays an important role in antiviral immunity (Wang et al. 2012). GMPR (guanosine monophosphate reductase) was recently reported as a potential therapeutic target for Alzheimer's disease (Liu et al. 2018). The revelation of these validated immune-related genes (Supplemental Fig. S4E) shows the robustness of our data-driven and unbiased bioinformatics analysis in identifying known and novel therapeutic targets for SLE and potentially for other autoimmune diseases, in general.
All of the above-mentioned genes were up-regulated and highly connected in the IFNpos network compared to IFNneg. We did not identify any genes showing the opposite effect (1.5-fold higher expression in IFNneg and DiffK < −0.25) (Fig. 2C). However, when we explored the IFNneg WGCNA network on its own (Supplemental Fig. S3E), we identified the dark green module that negatively correlated (r = −0.5; P-value = 0.002) with the SLEDAI score. However, we found several interesting hub genes in the dark green module, such as TLR2 that was shown to be up-regulated in PBMC from SLE patients, alongside two other Toll like receptors TLR7 and TLR9 (Komatsuda et al. 2008) and RBPJ, the variants of which were shown to be highly disease-specific for rheumatoid arthritis (RA) and SLE (Lim and Kim 2019). When we used all 56 genes from this module, we did not find any significant Gene Ontology (GO) term enrichment (Supplemental Fig. S3F). A future direction would be to reevaluate our findings in larger cohorts of IFNneg SLE patients to better characterize the molecular correlates of disease activity and severity for this group of SLE patients.
IFN response molecular signature is conserved across multiple different immune cell types
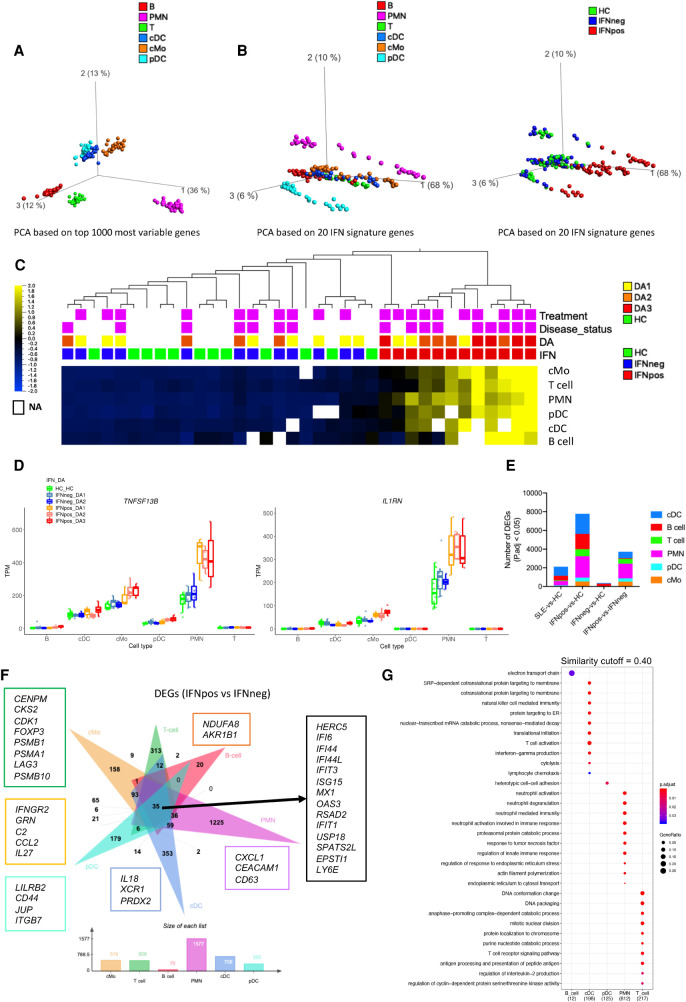
The recent studies highlight that multiple immune cell types are involved in the pathogenesis of SLE (Moulton et al. 2017). Therefore, it is important to systematically investigate distinct immune cell types from the same cohort of patients and to jointly study them in the context of disease heterogeneity and disease severity. Here, we used RNA from bead- or flow cytometry-based sorted populations of six different immune cell types including classical monocytes, polymorphonuclear neutrophils (PMNs), conventional dendritic cells (cDC), plasmacytoid dendritic cells (pDC), total T cells, and total B cells (Supplemental Fig. S1B) and performed bulk RNA-seq to understand the stability of IFN response signature as well as the importance of cell type–specific features in SLE pathogenesis. For this multi–cell type profiling, we used a matched set of HC, IFNneg, and IFNpos samples (n = 9–12 for each) from the first sample collections of different donors. The principal component analysis of these different cell types across each donor based on the top-1000 most variable genes reveals that each cell type clusters separately as expected (Fig. 3A). We then carried out PCA using only the expression of the 20 genes in the IFN-20 set and colored each sample either by its cell type (Fig. 3B, left panel) or by the IFN response status of the donor, which we determined from cMo data as previously described (Fig. 3B, right panel). This analysis highlights that, for each cell type, the IFNpos samples (red) are clearly separated from IFNneg and HC samples (green and blue, respectively) by the first principal component (Fig. 3B). The individual PCA plots for each cell type also show that the IFN-based annotations are cell type–independent and can be assigned at the patient level (Supplemental Fig. S5A–F). Heat map visualization using the IFN-20 gene set confirms this separation for each donor and for each cell type, both when the 20 genes are aggregated (Fig. 3C) and when visualized separately (Supplemental Fig. S5G). Out of 12 IFNpos patients, nine are classified into higher disease activity classes (DA2 or DA3), whereas for the IFNneg group, none of the patients were in the DA3 group, suggesting that IFN-based molecular stratification of SLE patients is also clinically relevant (Fig. 3C). Analysis of TNFSF13B and IL1RN gene expression, originally identified from our cMos analysis, shows that both are expressed at high levels in PMN cells and are specifically up-regulated in IFNpos samples (Fig. 3D), likely contributing to disease activity. Together with the results discussed before, we established that our classification with respect to IFN response status correlates with disease activity and is robust to the mode of gene expression profiling (bulk or single-cell) and to the immune cell type under investigation, supporting its clinical relevance in SLE.
Figure 3.
IFN response signature is present in all immune cell types. (A) PCA plot based on 1000 most variable genes across six different immune cell types represented by different colors. (B) PCA plots of different cell types based on IFN signature genes (IFN-20) colored either by cell types (left) or by IFN response status (right). (C) Average expression of IFN-20 genes clustered based on hierarchical clustering and presented as row-wise z-scores of TPM in IFNpos (red), IFNneg (blue), and HC (green) for each cell type separately. The DA panel shows disease activity categories determined by SLEDAI score. Patients with active disease at the time of visit and those taking treatments are highlighted in purple color. The detailed expression heat map with each one of the IFN-20 genes in all six cell types is provided in Supplemental Figure S5G. (D) Gene expression of TNFSF13B and IL1RN across six different cell types using IFN response status and SLEDAI-based categories. (E) A stacked column plot shows the number of RNA-seq-based differentially expressed genes with an adjusted P value threshold of 0.05 (Benjamini–Hochberg correction on DESeq2 P-values) in multiple cell type–specific data sets and different comparisons. (F) The flower plot (generated by jvenn [Bardou et al. 2014]) shows the overlap of DEGs from the IFNpos versus IFNneg comparison (Benjamini–Hochberg correction on DESeq2 P-values) in six different cell types. Some cell-specific genes of interest are highlighted in boxes with outline colors matching the color used for the relevant cell type. (G) Functional annotations (generated by clusterProfiler) of cell-specific DEGs from F. We used a similarity cutoff of 0.40 to remove similar Gene Ontology terms. The color shows the significance (in terms of P.adj) and the size is gene ratio of annotations.
Cell-specific transcriptional differences between two molecular subtypes of SLE
It is essential to study each sorted immune cell type separately, which unlike bulk analysis of PBMC, provides the resolution to identify cell type–specific transcriptional programs in relation to SLE pathogenesis. Therefore, we performed differential gene expression analysis comparing SLE-vs.-HC, IFNpos-vs.-HC, IFNneg-vs.-HC, and IFNpos-vs.-IFNneg for each cell type separately (Fig. 3E; Supplemental Table S4). We found only a few DEGs while comparing SLE-vs.-HC whereas the IFNpos-vs.-HC comparison provided hundreds of DEGs in most cell types (Fig. 3E). We also observed a large number of DEGs between IFNpos and IFNneg groups for all cell types except B cells. B cells also have the largest number of DEGs for IFNneg-vs.-HC analysis compared to all other cells. Most DEGs (P.adj < 0.05) are specific to one particular cell type, and only 35 DEGs, all of which are a part of the IFN-363 set, are present in all cell types when we performed the IFNpos-vs.-IFNneg comparison (Fig. 3F). The Gene Ontology–based functional annotation of DEGs revealed that all cell types were enriched for common IFN-related pathways (Supplemental Fig. S5H; Supplemental Table S5).
For the IFNpos-vs.-IFNneg comparison, a large fraction of DEGs from each cell type were differentially expressed only in that cell type (29%–78%) (Fig. 3F), suggesting that, aside from the common IFN response genes, each cell type has unique molecular differences between the two SLE subgroups. The functional enrichment analysis of these cell type–specific differences highlights several cell type–specific functions (Fig. 3G; Supplemental Table S5). The cDC-specific genes are involved in T cell activation (mainly XCL1, IL18, and PRDX2 genes) through antigen presentation. Expression of XCL1 (X-C motif chemokine ligand 1) by cross-presenting CD8+ dendritic cells has been reported to determine cooperation with CD8+ T cells in murine models (Dorner et al. 2009). IL18 is a pro-inflammatory cytokine that plays an important role in generating inflammation in lupus nephritis (Mohsen et al. 2013). PRDX2 (peroxiredoxin 2) is an antioxidant that plays a key role in inflammation (Knoops et al. 2016). The pDC-specific DEGs are enriched for heterotypic cell-cell adhesion (LILRB2, CD44, JUP, ITGB7), and B cells have an enrichment of electron transport chain (NDUFA8 and AKR1B1)-related genes. PMNs have genes enriched in neutrophil degranulation and neutrophil activation involved in immune response (CXCL1, CEACAM1, and CD63). T cell DEGs have an enrichment for annotations including cell-cycle (CENPM, CKS2, CDK1), T cell receptor signaling pathway (FOXP3, PSMB1, PSMA1), and antigen processing and presentation of peptide antigen (LAG3, PSMB10). Even though we only mention a few genes here (Fig. 3F), we also provide a complete list of genes and their GO-based functional enrichments for each cell type (Fig. 3G; Supplemental Fig. S5H, Supplemental Table S5). The number of DEGs is relatively low for each cell type in our SLE-vs.-HC comparisons (Fig. 3E), highlighting the necessity of stratifying patient groups prior to comparative transcriptomic analysis.
Integrated multi–cell type weighted gene coexpression network analysis (mWGCNA)
Multiple immune cell types are involved in immuno-pathogenesis of SLE, and they likely act in coordination (Tsokos et al. 2016; Moulton et al. 2017), which renders the approach to studying each cell type in isolation suboptimal. The availability of multi–cell type transcriptome profiles from a sufficient number of patients provides us the unprecedented opportunity to develop and apply data-driven and integrative bioinformatics approaches to study cross-correlation of SLE-related gene expression programs across these cells. The standard WGCNA (Langfelder and Horvath 2008) does not support multicell analysis; therefore, we developed a novel multi–cell type WGCNA (mWGCNA) approach, which combines transcriptomes of different cell types by representing each gene with its name plus a cell type identifier (e.g., cMo-IL1RN) to generate a single integrated weighted coexpression-based network. This integrated network resulted in 78 different modules from a total 101,282 “genes” (nearly 17,000 genes from each cell type), with most of the modules containing a substantial number of coexpressed genes from multiple cell types (Fig. 4A; Supplemental Table S6), with the exception of the largest module (∼19,000 genes), which consisted of over 70% B cell genes (Supplemental Fig. S6A). The existence of largely mixed modules suggests that genes from different cell types co-vary across patients and that a systematic quantification of such covariation could be useful for identifying disease-relevant signatures encompassing different cell types.
Figure 4.
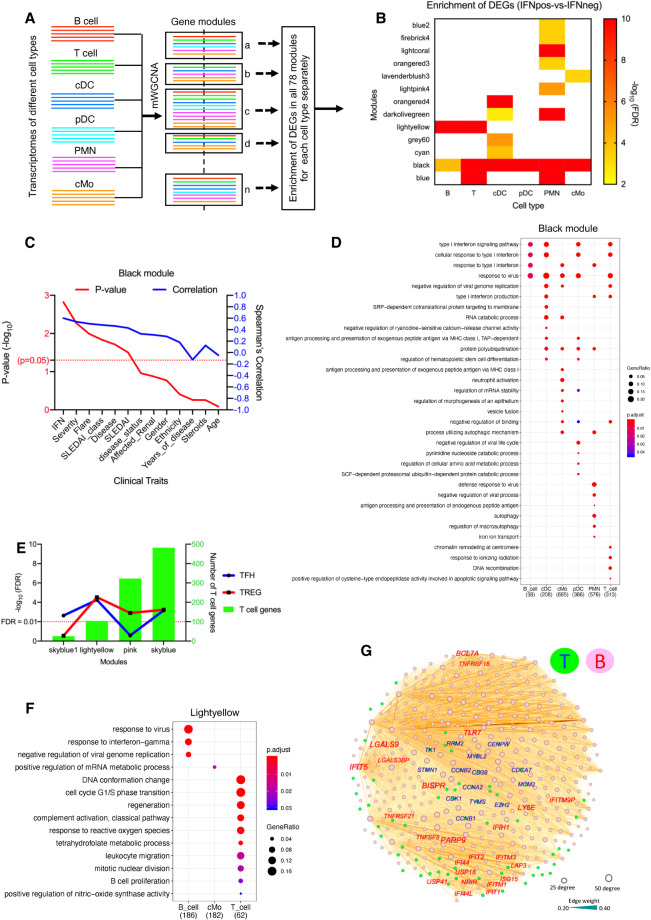
Multi–cell type WGCNA (mWGCNA) analysis reveals IFN-driven cross-talk between T cells and B cells. (A) A figure depicting the mWGCNA using combined transcriptomes from six different cell types. The mWGCNA has generated 78 different modules, and example modules show different proportion of genes from six different cell types. (B) We found 13 modules (out of total 78) that have significant enrichment of DEGs (IFNpos-vs.-IFNneg) in at least one cell type. The x-axis shows modules with their enrichment in each cell type and y1-axis shows −log10 of the adjusted P-value (based on a hypergeometric test). This includes four modules (black, blue, light yellow, and dark olive green) where the DEGs are significantly enriched in two or more cell types. (C) The line plot shows significance (−log10 of P-value) on the y1-axis in red and Spearman's correlation on the y2-axis in blue for the correlation of different traits with the black module eigengene. The clinical traits are sorted in decreasing order of statistical significance of the correlation for the black module. (D) Functional annotations (generated by clusterProfiler) of genes from each cell type in the black module. The color shows the significance (adjusted P-value), and the size of the dot corresponds to the proportion of genes with the corresponding annotation. (E) Enrichment of TFH and TREG gene sets in all 78 modules highlighting four modules with significant overlap for at least one of these gene sets. The x-axis is different modules, the y1-axis is −log10 of the adjusted P-value (based on a hypergeometric test), and the y2-axis shows the number of genes from T cells present in the corresponding module. (F) The functional annotations of significantly enriched genes from different cell types in the light yellow module. (G) Examples of T cell and B cell genes from the light yellow module as visualized by Gephi. The nodes are colored according to the cell of origin (T cells in green node with blue text and B cells in pink node with red text) and sized according to the number of edges (connections), and the edge thickness is proportional to the strength of coexpression.
To demonstrate the functional and biological relevance of our mWGCNA approach and the modules we report here, we first assessed whether absolute gene expression levels could be a confounding factor. To this end, we classified genes into five different categories based on TPM values to see their distributions in different modules. This analysis showed that all modules have genes from all expression classes with no clear clustering of genes according to their expression level categories (Supplemental Fig. S6B). We also looked at whether mWGCNA tends to put genes from different cell types with similar static gene annotations (e.g., GO terms) into the same module (e.g., all cell cycle genes from all cell types in a single module or a few modules), which would hinder its value in identifying disease-relevant cross-correlation patterns. Quantification of the fraction of genes that have multiple copies from different cell types (cMo-IL1RN, PMN-IL1RN, T-IL1RN) clustered in the same module showed that this is the case for only <1% of the genes for three or more cell types (Supplemental Fig. S6C). This co-clustering of the same genes across different cell types was most pronounced for the black module that mainly consists of highly shared IFN-related genes. However, even for the black module, 63% of genes came from only a single cell type and 18% came from two cell types (Supplemental Fig. S6D). These results suggest that coordinated variation of gene expression across patients (either within or across cell types), rather than gene expression category similarities or static gene annotations, drives the module identification by mWGCNA as aimed. Another implication of these findings is that gene coexpression relationships, hence modules, are highly cell type–specific.
To see whether our integration of multiple cell types in one coexpression framework caused this high cell type specificity, we also compared our results with the standard WGCNA for each cell type separately. To this end, we generated all possible coexpressed gene pairs in each module for each cell type (except the gray module that contains genes are not coexpressed with others), leading to a total of ∼148 million distinct pairs of genes that are in the same module for at least one cell type. A majority of these gene pairs (54%), however, were present in the same module only in one particular cell type and not the others (i.e., coexpressed only in one cell type) (Supplemental Fig. S6E). Only ∼3% of overall gene pairs were present in the same module for four or more cell types. These results indicate that the highly cell-specific nature of coexpression relationships and module assignments is not specific to mWGCNA and also is the case for the standard, per-cell-type WGCNA.
mWGCNA reveals IFN-driven cross-correlation among different cell types
Another important feature we implemented with mWGCNA to ease interpretation and to allow prioritization of modules is the calculation of the DEG enrichment for each module. The statistical significance of DEG-module overlaps for each cell type and for each module is computed using a hypergeometric test. Out of the 78 modules, we found 13 with a significant enrichment of DEGs (IFNpos-vs.-IFNneg) for at least one immune cell type and four (black, blue, light yellow, and dark olive green) with DEG enrichment from at least two cell types in the same module (Fig. 4B). Among them, the black module consisted of genes from each of the cell types and is relatively large, with 3940 genes (1216 from cMo, 1004 from PMN, 594 from pDC, 534 from T cells, 520 from cDC, and 72 from B cells) (Supplemental Table S6). Although there are several clinical features such as severity, flare, SLEDAI, and BILAG severity that are significantly correlated with the black module, the correlation with IFN response status is the dominant feature (Fig. 4C,D; Supplemental Fig. S7A; Supplemental Table S7). The TNFSF13B gene from three (cMo, PMN, and pDC) and the IL1RN gene from two different myeloid cell types (cMo and PMN) are also part of the black module showing coexpression with IFN-related genes across different immune cell types (Supplemental Fig. S7B). The black module also harbors four genes coming from all six cell types, including three that are IFN-related (IFI6, IFIT3, and RSAD2) and ODF3B, which is also likely regulated by IRF3 (Rouillard et al. 2016), a member of the interferon regulatory transcription factor (IRF) family. These results suggest that IFN response leads to coordination of gene expression changes across different cell types leading to a tightly connected mWGCNA module. Two other modules mentioned above (blue and dark olive green) (Fig. 4B) show enrichment in harboring DEGs from two different cell types (IFNpos-vs.-IFNneg comparison), but they lack clear functional enrichments for at least one of these two cell types (Supplemental Fig. S7C,D; Supplemental Table S7).
mWGCNA reveals IFN-driven cross-correlation between T cells and B cells
The fourth module (light yellow) mentioned above (Fig. 4B) shows a significant enrichment for both B cell and T cell DEGs. This is particularly striking given that most B cell genes are readily clustered into one large module (turquoise) as discussed (Fig. 4A; Supplemental Fig. S6A). Because the natural connection between T and B cells is the T cell help for B cell function, and B cells are critical targets for SLE, we further looked into specific T helper subtypes that may correlate with enriched B cell function. The follicular helper T cells (TFHs) are known to provide help and to play a central role in germinal center (GC) formation and development of high-affinity antibodies and memory B cells (Crotty 2014). A connection between follicular TFHs and IFN-γ response was established previously (Lee et al. 2012), and the expansion of circulating T cells resembling TFHs also has been shown in mouse models of SLE as well as a subgroup of patients with SLE (Simpson et al. 2010). TFH cells in the germinal centers as well as in circulation (cTFHs) (Locci et al. 2013) are known to promote B cell class-switching and help the development of high-affinity antibodies (Crotty 2014). More recently, a study described an expanded population of CD4+ helper T cells in SLE patient blood that are distinct from TFHs but still help activate B cells, further confirming heightened cross-correlation between circulating B cells and T cells in SLE (Caielli et al. 2019).
Another T cell subset known as regulatory T cells (TREGs) also have been shown to directly suppress B cells in SLE (Iikuni et al. 2009) and in vitro in a TGFB1-dependent fashion (Xu et al. 2016). Therefore, we computed the enrichment of published gene sets for TFH (Locci et al. 2013) and TREG (Schmiedel et al. 2018) cells for each module identified from our mWGCNA. To account for overlap among TFH, TREG, and IFN response signatures, we removed genes from both the TFH and the TREG sets that overlap with any of the other two sets (Supplemental Fig. S7E). Using the remaining 265 TFH and 300 TREG signature genes, we identified a total of four modules with significant overlap with at least one of these gene sets (Fig. 4E), including the light yellow module we identified earlier (Fig. 4B). The functional annotation of this light yellow module showed a significant enrichment of B cell proliferation-related genes (CD38 and CD79A) expressed by T cells (Fig. 4F; Supplemental Table S7). Even though these genes are mainly expressed by B cells, they also have considerable transcription in other circulating immune cells including T cells (Schmiedel et al. 2018). The expression of these genes in T cells also showed correlation with TFH and cell cycle–related genes from T cells. Most of their coexpressed/connected genes from B cells are IFN response-related but include members of the tumor necrosis factor (TNF) and TNF receptor (TNFR) superfamilies’ (TNFSF8, TNFRSF18, and TNFRSF21) as well as galectins (LGALS9 and LGALS3BP), which are also well-connected in the light yellow module (Fig. 4G). We then revisited the DEGs from IFNpos-vs.-IFNneg comparisons and found that many genes that are characteristic of TFH cells (e.g., TIGIT, STMN1, TYMS, FABP5, LAG3, CCNA2, CDKN3, CDCA7, and KPNA2), and TREG cells (e.g., FOXP3 and HAVCR2/TIM3) are uniquely differentially expressed in T cells (Fig. 3F; Supplemental Table S4). Another module that emerged from the overlap analysis for TFH and TREG signatures was the sky blue module (Fig. 4E). Even though this module was not enriched in DEGs for any cell type, it harbored two genes (FADD and CRK) from cDCs that are involved in the regulation of T cell migration as well as T cell activation genes such as (CCL5, BATF, and RUNX3) from T cells, suggesting a potential connection to inflammatory response (Supplemental Fig. S7E).
Having discovered several multi–cell type modules with potential biological relevance through the mWGCNA approach and DEG enrichment, we then asked whether similar findings could have been possible by a post hoc integration of cell type–specific WGCNA modules. Our original mWGCNA approach has generated 78 modules, among which we have selected four modules based on the significant enrichment of DEGs (IFNpos-vs.-IFNneg) in more than one cell type. When we performed standard WGCNA for each cell type and used a similar DEG enrichment test, we found 15 modules total with a significant DEG enrichment across all six cell types, but none of them were from B cells (Supplemental Fig. S6F). This suggests that a post hoc integration of cell type–specific WGCNA modules would have missed the connection between T and B cells, which we discussed above. Furthermore, matching modules from one cell type to those from another is not a trivial task and will further complicate the effort to perform an integrative analysis. These highlight the value of mWGCNA in identifying biologically relevant cross-correlations (such as black and light yellow modules) through a unified framework.
IFNpos patients have higher expression levels of genes related to TFH and TREG subsets
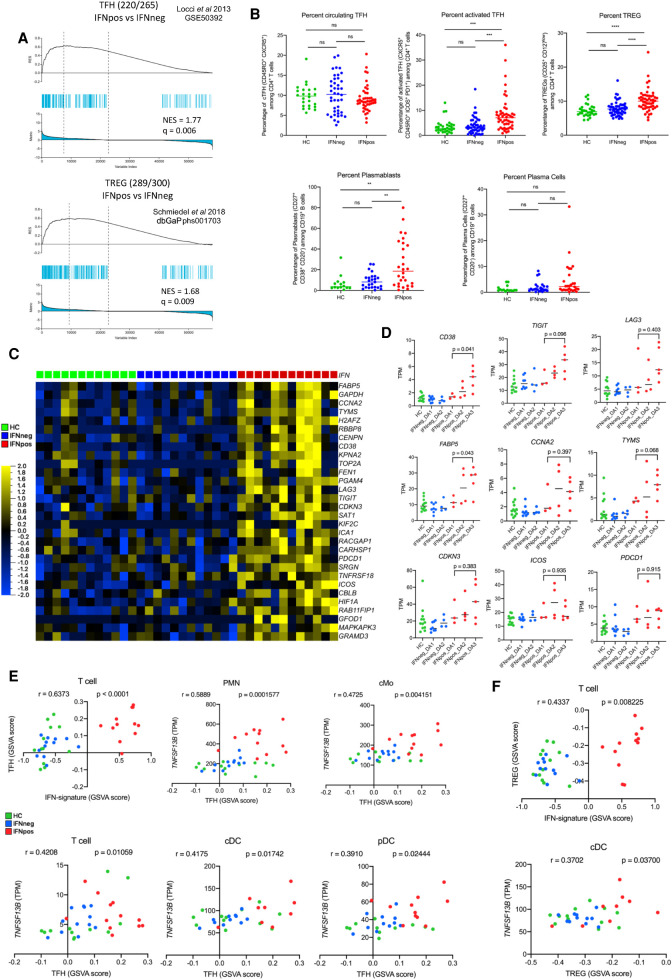
In order to better understand the influence of IFN response on the composition of different T cell subsets, we use signature gene sets of TH1 (Arlehamn et al. 2014), TH2 (Arlehamn et al. 2014), TH17 (Arlehamn et al. 2014; Hu et al. 2017), TFH (Locci et al. 2013), and TREG (Schmiedel et al. 2018) from published studies (Supplemental Table S8). As before, we remove the IFN response-related genes (IFN-363 set) (Supplemental Fig. S7E) from each T helper signature subset first. We then perform Gene Set Enrichment Analysis between IFNpos and IFNneg patients for each gene set. Whereas TFH (NES = 1.77, q = 0.006) and TREG (NES = 1.68, q = 0.009) gene sets show significant enrichment in IFNpos-vs.-IFNneg comparison (Fig. 5A), the other three subsets, TH1 (NES = 0.83, q = 0.694), TH2 (NES = 1.17, q = 0.326), and TH17 (NES = 1.47, q = 0.077), show no such enrichment (Supplemental Fig. S8A). In a confirmatory analysis, we used flow cytometry and found that there is a significant difference between IFNpos and IFNneg/HC patients in terms of their proportion of activated TFH cells and TREG but not for overall TFH cells in their blood (upper panel of Fig. 5B; Supplemental Fig. S9A).
Figure 5.
Enrichment of TFH and TREG signatures in IFNpos patients in correlation with TNFSF13B expression from myeloid cells. (A) Gene set enrichment analysis (GSEA) of TFH (top) and TREG (bottom) gene sets in the transcriptome of IFNpos versus IFNneg in T cells, presented as running enrichment score (RES) for the gene set, from most overrepresented genes on the left to the most underrepresented on the right. The values above the plot represent the normalized enrichment score (NES) and multiple testing-corrected significance value (Kolmogorov–Smirnov test). The source of the gene set is also provided. (B) Scatter dot-plots show the percentages of circulating TFH in CD3+ CD4+ T cells (top left), activated TFH in CD3+, CD4+, CXCR5+, CD45RO+ cTFH (top center), TREG in CD3+, CD4+ T cells (top right), plasmablasts (bottom left), and plasma cells in CD19+ B cells (bottom right) in healthy donor, IFNneg, and IFNpos patient blood. Data from all patient visits are included. Samples with <50 cells in the gate of interest were excluded. Differences between IFNpos and IFNneg were calculated using an unpaired t-test, and statistical significance (P-value) levels are shown in each plot. (ns) Not significant, (**) <0.005, (***) <0.0005, (****) <0.0001. The detailed gating strategies are provided in Supplemental Figure S9. (C) Expression of top 30 TFH-related genes within the total CD3+ T cell transcriptome, where each gene is presented as row-wise z-scores of TPM values in IFNpos (red), IFNneg (blue), and HC (green); each column represents an individual patient. (D) Individual expression plots for known TFH-related genes divided by IFN response status as well as SLEDAI categories. The provided P-values are based on an unpaired t-test (two-tailed). (E) The top left plot shows Spearman's correlation between GSVA scores derived either from IFN-20 genes (x-axis) or from the TFH gene set (y-axis) for T cell gene expression data. The rest of the plots show TFH gene set GSVA score versus TNFSF13B (BAFF) expression in five cell types with a significant correlation between the two axes. (F) Similar plots to those in E using GSVA scores from the TREG gene set.
We then investigated the top TFH-related genes and found that most have higher expression in the IFNpos group compared to no or very little expression in both the IFNneg and HC groups (Fig. 5C). Many important TFH-related genes (CD38, TIGIT, LAG3, FABP5, CCNA2, TYMS, and CDKN3) are significantly differentially expressed between T cells from IFNpos-vs.-IFNneg patients (Fig. 5D; Supplemental Table S4). In general, expression levels of these genes are elevated in higher disease activity groups (DA2 and DA3) within IFNpos patients. Similarly, TREG-related genes, including FOXP3 and HAVCR2/TIM3, also have higher expression in the IFNpos group (Supplemental Fig. S8B). These analyses show that both TFH-like and TREG-like molecular programs are enriched in IFNpos patients, though it is not clear whether the TREG-like signature is simply a response marker or plays a nontrivial role in SLE pathogenesis. Another important signature previously reported in SLE patient blood was the plasmablast activity (Banchereau et al. 2016). Even though we did not have DEG enrichment supporting this observation, which may be masked owing to a low proportion in plasmablasts in the total B cell compartment, we observed a significant difference between IFNpos and IFNneg/HC patients in the proportions of plasmablasts in their blood (lower panel of Fig. 5B; Supplemental Fig. S9B).
TNFSF13B expression from multiple cell types correlates with expression of TFH-related genes
An important factor in the context of B cell help is TNFSF13B, which promotes B cell survival and controls their maturation (Mackay and Browning 2002) while also regulating IFNG production by TFH cells in lupus-prone mice (Coquery et al. 2015). Increased expression of TNFSF13B induces expansion of activated B cells that produce autoantibodies and cause autoimmunity and is dependent, at least in part, on TFH (Chen et al. 2014). In our data, higher expression of TNFSF13B from all myeloid cells shows a strong correlation with the enrichment of the TFH gene signature in T cells, which itself is elevated in IFNpos SLE patients. Our identification of TNFSF13B from the combined DEGs and DCGs analysis of classical monocytes (Fig. 2D), together with the significant enrichment of TFH-related gene expression in T cells from IFNpos SLE patients, suggest that there could be a potential connection between TNFSF13B expression and TFH-like features in the SLE patient blood. Because multiple immune cells, mainly of myeloid lineage, produce TNFSF13B (Fig. 3D), it is also important to understand which cell types significantly contribute to the induction of TFH-like programs in T cells through the TNFSF13B axis.
To this end, we employed gene set variation analysis, which estimates variation of enrichment of a particular gene set over a sample population (Hänzelmann et al. 2013). GSVA provides enrichment scores for each sample for a given gene set, which can then be correlated with sample-specific features such as a gene expression measurement or another GSVA enrichment score. First, we found a strong positive correlation (r = 0.637; P < 0.0001) between GSVA scores from IFN-signature genes (IFN-20) and from TFH-related genes using expression profiles from T cells (Fig. 5E). Next, we computed the correlation between TNFSF13B expression from different immune cells and GSVA enrichment of TFH-related genes in T cells resulting in significant correlations for all cells, except B cells, with specifically strong correlations for PMNs (r = 0.589; P = 0.0001) and cMos (r = 0.4725; P = 0.004) (Fig. 5E; Supplemental Fig. S8C). Similar analysis for TREG-related genes also highlighted some statistically significant correlations, though to a lesser extent compared to TFH-related genes (Fig. 5F; Supplemental Fig. S8C). Although only correlative, these results suggest that TNFSF13B production from PMN and cMo cells, concordant with an increase in IFN response-related gene expression, may induce an extrafollicular T cell help program, akin to follicular B cell help by TFH cells, in IFNpos SLE patients. This in turn may lead to a robust autoantibody production by B cells leading to formation of immune complexes and SLE manifestations. Further mechanistic studies are needed to completely characterize this interplay among IFN response by all immune cells, TNFSF13B expression by myeloid cells, and the enrichment of B cell activation by turning on specific T cell help programs.
Discussion
The diagnostic complexity and heterogeneous nature of SLE disease require stratification of patients with the goal of correlating clinical features with molecular subgroups. Although we have used a variety of available clinical and demographic information (e.g., age, ethnicity, flare, severity, SLEDAI, and different treatments), IFN response status consistently provided the most striking relationships with gene expression differences and with gene coexpression modules (Fig. 2A,B). With this in mind, we analyzed SLE patients in two distinct groups, namely IFNpos and IFNneg, throughout this work. The joint analysis of DEGs and DCGs between IFNpos and IFNneg in classical monocytes (Fig. 2C) reveals two known immune modulators (TNFSF13B and IL1RN) within a small group of prioritized genes, as well as several other genes of interest that could be further explored as therapeutic targets or biomarkers for SLE (Fig. 2D). Although IFN response status was mainly stable across different visits of the same patient, our cohort had one patient whose IFN response status changed between visits, and this change was accompanied by changes in the serum levels and mRNA levels of pro-inflammatory cytokines (IL6, CXCL10, IFNG) (Fig. 1E). The expression and coexpression profiles of TNFSF13B and IL1RN also changed with IFN response status in this longitudinal data (Fig. 2H; Supplemental Fig. S4C). Recently, a phase III clinical trial (TULIP 2, NCT02446899) in SLE patients with a human monoclonal antibody (anifrolumab) that specifically blocks IFNAR1 (interferon alpha and beta receptor subunit 1) was completed (Morand et al. 2020). The trial results also suggested that knowing patients’ IFN response status before prescribing targeted therapy will be helpful in the clinical setting and in designing further targeted therapies for specific patient groups.
Here, we systematically analyzed the transcriptome profiles of multiple circulating immune cell types and found that IFN signature-based classification of SLE patients into two groups is very consistent across different immune cell types (Fig. 3C). The large and clear-cut transcriptional differences between gene expression profiles of IFNpos and IFNneg patient groups across different immune cell types (Fig. 3F), without such clear differences in IFN levels in the serum from the two groups, suggest that the cells might be pre-exposed to IFN signals prior to migrating into the blood. Beyond the shared IFN response signature, it has been challenging to infer biological functions of these cell-specific DEGs because GO-based functional annotations are generic and only provide information about canonical functions regardless of cell type. Recently, an integrated and multicohort analysis also has published a unified SLE MetaSignature of 93 genes in the blood, and most of these genes (85%) were IFN-related (Haynes et al. 2020). When these genes were compared to our work, we found that, out of the total 125 DEGs (SLE-vs.-HC) we report for classical monocytes, 44 are present in the SLE MetaSignature of 93 genes.
Another challenge was the involvement of multiple different immune cell types, each of which added another layer of complexity. Here, we proposed and demonstrated the use of a novel bioinformatics approach we named mWGCNA in exploring the cross-correlation among different immune cell types using coexpression relationships of gene pairs across different cell types. We then revisited the large number of DEGs identified from one-by-one DEG analysis of IFNpos versus IFNneg patients (Fig. 3F) using mWGCNA to explore potentially linked functional enrichments involving at least two different cell types. Out of four modules, which we narrowed down from a total of 78, one module corresponded mainly to the IFN response signature with an enrichment of IFNpos versus IFNneg DEGs from all six different cell types (Fig. 4B,C). Another module was significantly enriched for both T cell and B cell DEGs (Fig. 4B,G) and suggested that extrafollicular T cell help to B cells, that is, gene signatures in the blood that are reminiscent of TFH cells, could play an important role in heightened disease activity in the IFNpos patient group (Figs. 4E, 5A). Recently, multiple lines of evidence have accumulated that circulating follicular helper-like T cells are present in SLE blood and their expansion is associated with disease activity (Choi et al. 2015; Xu et al. 2015; Zhang et al. 2015b). Another recent work also identified a T helper subset that is distinct from TFHs but still promotes B cell activation and differentiation into plasmablasts extrafollicularly in SLE blood (Caielli et al. 2019). It has been also shown that T peripheral helper (Tph) cells (PD-1hiCXCR5−CD4+) stimulate B cell responses in lupus via MAF and IL21 (Bocharnikov et al. 2019). New evidence is also accumulating that T cell metabolism (Sharabi and Tsokos 2020) and epigenetics (Su et al. 2020) also regulate the TFH cell in SLE. Our results do not exclude either possibility but highlight that such B cell help-related T cell programs are likely more relevant targets for the treatment of IFNpos SLE patients and not for IFNneg SLE patients. In the present study, the systematic attempt to explore the connection between TFH cells and the interferon signature using SLE transcriptome profiles is a new discovery to the best of our knowledge. Furthermore, our finding that TNFSF13B from three different myeloid cell types come together in the black module with significant coexpression to IFN-related genes (Supplemental Fig. S7B) suggests that the effectiveness of a belimumab-based treatment could be impacted by the IFN response status of the patient.
In conclusion, we have found an IFN-based molecular heterogeneity in transcriptome profiles of SLE patients that is consistent across different immune cell types. We developed a novel computational approach (mWGCNA) to understand how this IFN-signal could affect the cross-correlation among different immune cells. We anticipate that the data source generated here will be very useful for further characterization of immune cell type–specific roles in SLE progression. We also believe that our data-driven bioinformatics approach to jointly analyze gene expression profiles from multiple immune cell types from the same tissue (i.e., blood) will be broadly applicable to studies of other heterogeneous autoimmune diseases where multiple cell types are implicated in disease pathogenesis.
Methods
Patient samples
Recruitment of subjects included in this study followed Institutional Review Boards’ La Jolla Institute for Immunology, University of California San Diego, The Scripps Research Institute, and Quorum Review approvals, and study participants gave written informed consent. SLE patients were previously diagnosed by a clinician according to the ACR SLE criteria (Petri et al. 2012) and classified as active if they had a SLEDAI score of at least four as well as a BILAG score of A or B at each study visit. A subset of patient samples (n = 10) were collected by Sanguine Bioscience and confirmed via their medical records to have a diagnosis of SLE by meeting the ACR SLE criteria. A modified SLEDAI score was determined based on patient self-reporting of active symptoms. Healthy volunteers were recruited by the Normal Donor Blood programs at La Jolla Institute for Immunology and The Scripps Research Institute. All demographics and clinical information of the SLE cohort are provided (Supplemental Table S1).
Sample collection and cell isolation
After the volunteers consented to donate their blood sample, 40 mL of blood were obtained via venipuncture. Thirty-six milliliters were collected for cellular analysis (6 × 6 mL BD vacutainer tubes with potassium EDTA; Becton Dickinson) and 4 mL for serum analysis. Whole blood was spun at 2000 rpm for 10 min. The plasma fraction was transferred to a fresh tube, aliquoted, and stored at −80°C until analyzed. The cell pellets were then resuspended in HBSS without calcium and magnesium and underlaid with Ficoll-Paque Plus (Sigma-Aldrich) for density gradient centrifugation purification of the PBMCs. The tubes were centrifuged at 2000 rpm at room temperature for 20 min in sealed carriers with the brake off. The top fraction (upper layer) was aspirated, leaving a minimum of 5-mL volume over the interface layer, which was collected using a sterile rubber bulb and Pasteur pipette into a new 50-mL conical tube. The collected PBMCs were washed in PBS by centrifuge at 1800 rpm at room temperature for 10 min. The PBMCs were passed through a 70-µm cell strainer, washed again, and then either frozen in 10% fetal bovine serum (HyClone US-origin defined FBS, GE Healthcare Life Sciences) in DMSO or processed for purification of immune cell subsets as described below. The cell pellets from the Ficoll gradients were used for isolation of PMNs. First, red blood cells were lysed by resuspending the cell pellet in Gey's solution. Following incubation on ice for 5 min, the solution was underlaid with 5 mL FBS and then centrifuged at 1800 rpm for 5 min. The cells were resuspended in PBS, passed through a 70-µm cell strainer, and washed twice with PBS by centrifugation at 1200 rpm for 5 min each. PMNs were resuspended in 30 mL HBSS without calcium or magnesium. Six milliliters of 6% dextran were added to the cells and mixed by inversion. Following incubation at room temperature for 30 min, the supernatant was transferred to a new tube which was spun for 5 min at 1200 rpm. The pellet was then resuspended with 5 mL H2O and mixed for 5 sec before addition of HBSS to a final volume of 50 mL. After a final spin at 1200 rpm for 5 min, the cells were resuspended in 5 mL HBSS, counted, and spun again at 1200 rpm for 5 min. One milliliter of TRIzol LS (Thermo Fisher Scientific) was added for each 5 × 106 cells and samples were stored at −80°C. Subsets of immune cells were further isolated by a combination of magnetic bead-based and flow cytometric-based cell sorting methods.
Cell sorting
Total T cells were isolated from PBMCs by magnetic bead-based separation, specifically using CD3 DynaBeads (Thermo Fisher Scientific) according to the manufacturer's instructions. The purified T cells were resuspended in TRIzol LS and stored at −80°C. The T-depleted PBMCs were resuspended in FACS buffer (PBS, 2% FBS, 2 mM EDTA, and 25 mM HEPES) containing mouse IgG (Jackson Immunochemicals) to block nonspecific binding. Following incubation for 10 min on ice, the cells were stained with a cocktail of antibodies against TCRαβ (BioLegend 306712), CD11c (BioLegend 337220), HLA-DR (BioLegend 307636), CD16 (BioLegend 302028), CD19 (BioLegend 302238), CD14 (BD Biosciences 555399), and CD303 (Miltenyi 130-090-511) (Supplemental Fig. S1B). B cells, classical, nonclassical, and intermediate monocytes, pDCs, and cDCs were sorted on either FACSAria or a BD Influx (Becton Dickinson). After sorting, the cells were washed, lysed in TRIzol LS, and frozen at −80°C immediately. Circulating classical CD14hi CD16− monocytes were isolated from PBMCs from 64 patients with systemic lupus erythematosus (SLE) and 24 healthy subjects (Fig. 1A). Longitudinal samples were collected from 17 SLE patients and two healthy individuals between September 2014 and August 2016 for one to five follow-up visits (Fig. 1F). Five additional immune cell types (B cells, T cells, cDCs, pDCs, and PMNs) were evaluated from a subset of the samples (24 SLE and 12 HC).
Flow cytometry
Cell staining of whole blood was performed for 25 min on ice in the dark in staining buffer composed of PBS, 0.5% BSA, and 0.05% sodium azide. Red blood cells were lysed by addition of 1× BD Pharm Lyse Buffer (BD Biosciences) and incubation in the dark at room temperature for 15 min. Following one wash, cells were fixed in 2% paraformaldehyde for 15 min at room temperature, washed again, and resuspended in staining buffer. A minimum of 300,000 total events were collected on a FACS Calibur using Cell Quest and analyzed with FlowJo software (TreeStar). Anti-human CD20 (clone 2H7, catalog #302304), CD19 (clone HIB19, catalog #302234), CD38 (clone HB7, catalog #356608), CD3 (clone UCHT1, catalog #300426), CD4 (clone RPA-T3, catalog #300506), CD25 (clone BC96, catalog #302632), CD127 (clone A019D5, catalog #351325), CCR4 (clone TG6, catalog #335405), CXCR5 (clone TG2, catalog #335001), CD45RO (clone UCHL1, catalog #304218), and PD-1 (clone EH12.2.H7, catalog #329907) antibodies were purchased from BioLegend, anti-human CD27 (clone O323, catalog #12-0279-42) from eBioscience, and anti-human ICOS antibody (clone DX29, catalog #557802) from BD Biosciences.
Cytokine profiling of plasma
Plasma was collected from patients as described above. Frozen plasma was shipped to Affymetrix and analyzed in a 34-protein vendor-defined multiplex Procarta Plex-2panel (Thermo Fisher Scientific) to profile differential plasma protein expression from healthy volunteers and patients with SLE. Analytes measured included soluble CD40 ligand, CXCL5, IFNA2, IFNB1, IFNG, IL10, IL23, IL12p70, IL15, IL17A, IL17F, IL18, IL1A, IL1B, IL1RA, IL2, IL21, IL4, IL6, CXCL10, CXCL11, CCL2, CCL5, TGFB1, TNF, VEGFA, IL13, leptin, PAI1, Resistin, Fas ligand, SDF1, IL22, and CSF2. GraphPad Prism8 was used to generate scatterplots and to perform statistical analyses of these data.
Bulk RNA sequencing
Total RNA was isolated from sorted cell populations using an miRNeasy Micro kit (Qiagen) and quantified. Three nanograms of total RNA were used to generate cDNA following the Smart-seq2 protocol. cDNA was purified using AMPure XP beads (0.8×, Beckman Coulter). Next, 1 ng of cDNA from each sample was used to generate a sequencing library (Nextera XT DNA sample preparation kit and index kit, Illumina). The libraries were pooled and sequenced on a HiSeq 2500 (Illumina) to obtain 50-bp single-end reads. Both full-transcriptome amplification and sequencing library preparations were performed in a 96-well format to reduce assay-to-assay variability. Quality control steps were included after each step to eliminate samples with low quality from the downstream process. A detailed protocol has been previously published (Rosales et al. 2018). Libraries were sequenced on a HiSeq 2500 Illumina to obtain a minimum of 10 million 50-bp single-end reads (HiSeq SR Cluster kit v4 cBot, HiSeq SBS kit v4).
Single-cell sequencing
Cells from three SLE (IFNpos) patients were sorted into plates to generate sequencing libraries following the Smart-seq2 protocol. cDNAs were purified twice with AmPure XT beads (0.8×). From each sample, 0.4 ng of cDNA was used to generate a sequencing library using the Nextera XT DNA sample preparation kit and index kit from Illumina. Quality control steps were included after each step to eliminate samples with low quality from the downstream process. A total of 156 single-cell libraries passed all quality control criteria. The libraries were pooled, then sequenced on a HiSeq 2500 to obtain a minimum of 50,000 50-bp single-end reads. A detailed protocol has been previously published (Rosales et al. 2018).
Bulk and single-cell RNA-seq analysis
Bulk RNA-seq data (FASTQ files) were mapped against the hg38 genome (GRCh38.p7) reference using TopHat (Trapnell et al. 2009) v2.0.9 (‐‐max-multihits 1 ‐‐microexon-search ‐‐bowtie1) with FastQC (v0.11.2), and SAMtools v0.1.19.0 (Li et al. 2009). Trimmomatic (v0.36) was used to remove adapters (Bolger et al. 2014). We employed htseq-count -m union -s no -t exon -i gene_name (part of the HTSeq framework, version v0.7.1 [Anders et al. 2015]) for calculating read counts. To identify differentially expressed genes between two groups, we used raw read counts and performed negative binomial tests for unpaired comparisons using DESeq2 (v1.14.1) with the package from Bioconductor (Love et al. 2014). We disabled the default options of DESeq2 for independent filtering and Cooks cutoff. Nonexpressed (no reads in all samples) genes were filtered out before running DESeq2. All genes with Benjamini–Hochberg-adjusted P value of <0.05 (based on DESeq2 results) were considered as differentially expressed genes in any comparison. The MAplot was generated by using the ggmaplot function of the ggpubr R package. Gene expression values were normalized as transcripts per million (TPM) and applied in the Qlucore Omics Explorer 3.3 software package for visualization and representation (heat maps, principal component analysis, and GSEA) of RNA-seq data. The top 30 TFH/TREG genes were selected based on filtering by variance from the statistics function of Qlucore. Different box plots and Spearman's correlation plots were generated by GraphPad Prism8 (v8.3.0). For the analysis of our plate-based single-cell RNA-seq data, we used the same pipeline as mentioned above for bulk RNA-seq to process, map, and quantify read counts. We used normalized TPM counts for analysis and plotting violin plots.
Weighted gene coexpression network analysis
The R package WGCNA (v1.61) was used to generate coexpression network from the TPM data matrix (Langfelder and Horvath 2008). To develop a standard WGCNA network for SLE (Fig. 2A), we used 16,444 well-expressed genes with TPM > 1 in at least 25% of the samples, and modules were generated using the blockwiseModules function (parameters: checkMissingData = TRUE, power = 5, TOMType = unsigned, minModuleSize = 30, maxBlockSize = 16444, mergeCutHeight = 0.25). The pickSoftThreshold function was used to optimize soft-thresholding power (β) by choosing the lowest power for which the scale-free topology fit index reaches 0.90. The default ‘gray’ module was generated by WGCNA for non-coexpressed genes. As each module by definition is comprised of highly correlated genes, their combined expression may be usefully summarized by eigengene profiles, effectively the first principal component of a given module. A small number of eigengene profiles may therefore effectively ‘summarize’ the principle patterns within the cellular transcriptome with minimal loss of information. This dimensionality-reduction approach also facilitates correlation of ME with traits. Different clinical features (IFN-status, age, ethnicity, flare, severity, SLEDAI score, years of disease, affected renal, BILAG severity, and different treatments) were used as a trait and correlated with MEs. Significance of correlation between this trait and MEs was assessed using Spearman's correlation and P-values.
In the differential network analysis (Fig. 2C), we used a standard WGCNA-based approach to generate coexpression networks for IFNpos and IFNneg separately. For each network, genes were clustered into a dendrogram and modules were assigned by blockwiseModules function (parameters: checkMissingData = TRUE, power = 6, TOMType = unsigned, minModuleSize = 30, maxBlockSize = 16444, mergeCutHeight = 0.25). In order to make both IFNpos and IFNneg networks comparable, we used the same soft-thresholding power (β = 6). Furthermore, network connectivity values using the softConnectivity function (power = 6) have been calculated for each gene, where connectivity (also known as degree) is defined as the sum of connection strengths (based on coexpression) with the other genes in the network. The difference between the connectivity (DiffK = K1 – K2) for each gene between IFNpos (K1) and IFNneg (K2) was calculated (as described in Fuller et al. 2007). Genes with at least ±0.25 difference (K1 − K2) in connectivity (DiffK) were considered as differentially connected genes.
Multi–cell type weighted gene coexpression network analysis
In the multi–cell type WGCNA (mWGCNA) approach (Fig. 4A), we have generated a single WGCNA network by merging transcriptome profiles of six different cell types (patient-matched) together. We used a total of 25 samples (10 HC, eight IFNneg, and seven IFNpos) from six different cell types. Highly correlated genes from combined transcriptomes across six immune cell types were identified, and a total of 78 modules were generated using blockwiseModules function (parameters: checkMissingData = TRUE, power = 3, TOMType = unsigned, minModuleSize = 50, maxBlockSize = 101282, mergeCutHeight = 0.25). In order to find gene set–specific important modules, we have measured the significance of a particular gene set (e.g., DEGs between IFNpos and IFNneg) by a hypergeometric test using phyper R function and, further, P-values were adjusted for multiple test correction using the p.adjust R function (method = fdr).
To visualize coexpression networks, we used the function exportNetworkToCytoscape at weighted = true, threshold = 0.05. A soft-thresholding power was chosen based on the criterion of approximate scale-free topology. Networks were generated in Gephi (v0.9.2) (Bastian et al. 2009) using Fruchterman Reingold and Noverlap functions (Clarke et al. 2019). The size and color were scaled according to the average degree as calculated in Gephi, while the edge width was scaled according to the WGCNA edge weight value.
Gene set enrichment analysis and gene set variation analysis
GSEA determines whether an a priori defined ‘set’ of genes (such as a signature) show significant cumulative changes in gene expression between phenotypic subgroups (Subramanian et al. 2005). We applied GSEA using the Qlucore Omics Explorer 3.3 software package for assessing significant enrichment of specific gene sets (e.g., IFN signatures or T cell subtypes) in one group relative to that in another group (e.g., IFNpos vs. IFNneg). In summary, first, all genes are ranked on the basis of their differential expression (TPM-based) in one group versus their expression in another group. Thereafter, a running enrichment score (RES) is calculated for a provided gene set on the basis of how often its genes appear at the top or bottom of the already ranked differential list. A default of 1000 random permutations of the phenotypic subgroups is used to establish a null distribution of RES against which a normalized running enrichment score and false-discovery-rate-corrected q values are calculated using the Kolmogorov–Smirnov statistic. We ran GSEA with different gene sets of TH1 (Arlehamn et al. 2014), TH2 (Arlehamn et al. 2014), TH17 (Arlehamn et al. 2014; Hu et al. 2017), TFH (Locci et al. 2013), and TREG (Schmiedel et al. 2018) from published studies (Supplemental Table S8) after removing IFN-20- and IFN-363-related genes (Supplemental Fig. S7F) to uncover only T cell subtypes–specific enrichments. These gene signatures were selected to test the null hypothesis that IFN-based subgroups (IFNpos and IFNneg) did not show significant enrichment for different T cell subtypes.
In order to establish correlation between two different gene sets or groups, we need to calculate enrichment score for each sample. The GSVA (Hänzelmann et al. 2013) estimates variation of enrichment of particular gene set over a sample population and provides an enrichment score for each sample. GSVA was implemented using the gsva function of the R package GSVA (v1.20.0) with rnaseq = TRUE parameter, and it provided GSVA scores that we used to correlate different gene sets with TNFSF13B expression (Fig. 5E,F; Supplemental Fig. S5C).
Gene Ontology–based functional annotations
The biological relevance of important genes from different analyses was further investigated using the clusterProfiler (Yu et al. 2012). To functionally annotate genes (e.g., DEGs or module genes) from one cell type, we used the enrichGO function (parameters: OrgDb = org.Hs.eg.db, ont = BP). We also removed redundant GO-terms (parameters: cutoff=0.40, by = p.adjust, select_fun = min, measure = Wang) with a more than 40% similarity cut-off. In the plots (Supplemental Figs. S1D, S4B), color shows the significance (in terms of P.adj), size is gene counts in annotation, and the x-axis shows gene ratio. To compare multiple cell types, the compareCluster function (parameters: fun = enrichGO, OrgDb = org.Hs.eg.db, ont = BP) was used to generate Gene Ontology–based comparative functional annotations of different cell types. We used all available genes from different cell types in a module to run clusterProfiler and only displayed cell types that have significant enrichment of any GO-term. In multicell plots (Figs. 3G, 4D,F), color displays the significance (in terms of P.adj) of particular GO-terms and size shows the gene ratio of annotations.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism8 (v8.3.0). The Spearman's correlation coefficient (r value) was used to access the significance of correlations between the levels of any two components of interest. R packages were applied with R version 3.3.3 (R Core Team 2020) using the x86_64-pc-linux-gnu (64-bit) platform under CentOS Linux 7 (Core).
Data access
All raw and processed RNA sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE149050. All data and gene-specific expression patterns across different cell types are also available online at https://ay-lab-tools.lji.org/sle. The scripts used to perform all the analyses can be found at GitHub (https://github.com/ay-lab/SLE-mWGCNA) and as Supplemental Codes 1 and 2.
Competing interest statement
R.S., A.J.M., and E.R. are employed by Kyowa Kirin Pharmaceutical Research, Inc. This does not alter the authors’ adherence to this journal's policies on sharing data and materials. There are no patents, products in development, or marketed products associated with this research to declare.
Supplementary Material
Acknowledgments
We acknowledge an Interactive Fund grant from Kyowa Kirin Pharmaceutical Research, Inc. to P.V. The HiSeq 2500 (Illumina) instrument was purchased with the National Institutes of Health S10OD016262 grant. We thank Anne-Laure Perraud for providing useful insights in this study.
Author contributions: B.P., P.V., and F.A. conceived the work and designed and analyzed the experiments; B.P. performed RNA-seq, mWGCNA, and data analysis under the supervision of P.V. and F.A. The bulk and single-cell RNA-seq experiments were done by S.L., B.W., and G.S. The SLE cohort was conceived by R.S. and was generated by K.K., A.J.M., R.S., and E.R., including patient recruitment, obtaining consent, disease activity assessment, and sample collection. E.R. developed and performed the cell sorting strategy and flow cytometry analyses. B.P. wrote the first draft of the manuscript that was revised and edited by P.V., F.A., and R.S. All authors read and approved the final text of the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.265249.120.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B. 2014. Transcriptional profile of tuberculosis antigen–specific T cells reveals novel multifunctional features. J Immunol 193: 2931–2940. 10.4049/jimmunol.1401151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, et al. 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci 100: 2610–2615. 10.1073/pnas.0337679100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, Cepika A-M, Acs P, Turner J, Anguiano E, et al. 2016. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 165: 551–565. 10.1016/j.cell.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. 2014. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15: 293. 10.1186/1471-2105-15-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat FJ, Crow MK, Ivashkiv LB. 2019. Interferon target-gene expression and epigenomic signatures in health and disease. Nat Immunol 20: 1574–1583. 10.1038/s41590-019-0466-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media. San Jose, CA. [Google Scholar]
- Bayry J. 2016. Lupus pathogenesis: role of IgE autoantibodies. Cell Res 26: 271–272. 10.1038/cr.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharnikov AV, Keegan J, Wacleche VS, Cao Y, Fonseka CY, Wang G, Muise ES, Zhang KX, Arazi A, Keras G, et al. 2019. PD-1hiCXCR5– T peripheral helper cells promote B cell responses in lupus via MAF and IL-21. JCI Insight 4: e130062. 10.1172/jci.insight.130062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH, Austin A., Bell A., Bloch DA, Corey PN, Decker JL, et al. 1992. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum 35: 630–640. 10.1002/art.1780350606 [DOI] [PubMed] [Google Scholar]
- Caielli S, Veiga DT, Balasubramanian P, Athale S, Domic B, Murat E, Banchereau R, Xu Z, Chandra M, Chung CH, et al. 2019. A CD4+ T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat Med 25: 75–81. 10.1038/s41591-018-0254-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EE, Barr SG, Clarke AE. 2016. The global burden of SLE: prevalence, health disparities and socioeconomic impact. Nat Rev Rheumatol 12: 605–620. 10.1038/nrrheum.2016.137 [DOI] [PubMed] [Google Scholar]
- Catalina MD, Bachali P, Geraci NS, Grammer AC, Lipsky PE. 2019. Gene expression analysis delineates the potential roles of multiple interferons in systemic lupus erythematosus. Commun Biol 2: 140. 10.1038/s42003-019-0382-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichian Y, Wallace DJ, Weisman MH. 2019. A promising approach to targeting type 1 IFN in systemic lupus erythematosus. J Clin Invest 129: 958–961. 10.1172/JCI127101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lin X, Liu Y, Li Q, Deng Y, Liu Z, Brand D, Guo Z, He X, Ryffel B, et al. 2014. The function of BAFF on T helper cells in autoimmunity. Cytokine Growth Factor Rev 25: 301–305. 10.1016/j.cytogfr.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Ho JHE, Pasoto SG, Bunin V, Kim ST, Carrasco S, Borba EF, Gonçalves CR, Costa PR, Kallas EG, et al. 2015. Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity. Arthritis Rheumatol 67: 988–999. 10.1002/art.39020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. 10.1126/science.282.5389.699 [DOI] [PubMed] [Google Scholar]
- Clarke J, Panwar B, Madrigal A, Singh D, Gujar R, Wood O, Chee SJ, Eschweiler S, King EV, Awad AS, et al. 2019. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J Exp Med 216: 2128–2149. 10.1084/jem.20190249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquery CM, Loo WM, Wade NS, Bederman AG, Tung KS, Lewis JE, Hess H, Erickson LD. 2015. BAFF regulates follicular helper T cells and affects their accumulation and interferon-γ production in autoimmunity. Arthritis Rheumatol 67: 773–784. 10.1002/art.38950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41: 529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK. 2014. Type I interferon in the pathogenesis of lupus. J Immunol 192: 5459–5468. 10.4049/jimmunol.1002795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YX, Fu CW, Jiang F, Ye LX, Meng W. 2015. Association of the interleukin-6 polymorphisms with systemic lupus erythematosus: a meta-analysis. Lupus 24: 1308–1317. 10.1177/0961203315588971 [DOI] [PubMed] [Google Scholar]
- Davidson A. 2016. What is damaging the kidney in lupus nephritis? Nat Rev Rheumatol 12: 143–153. 10.1038/nrrheum.2015.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente A. 2010. From ‘differential expression’ to ‘differential networking’ – identification of dysfunctional regulatory networks in diseases. Trends Genet 26: 326–333. 10.1016/j.tig.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, et al. 2017. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2: e93009. 10.1172/jci.insight.93009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarapu SK, Anders HJ. 2018. Toll-like receptors in lupus nephritis. J Biomed Sci 25: 35. 10.1186/s12929-018-0436-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Güttler S, Hutloff A, Mages HW, Ranke K, Schaefer M, et al. 2009. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31: 823–833. 10.1016/j.immuni.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS, Dong X, Chen W, Kim MH, Weng HH, et al. 2006. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum 54: 2951–2962. 10.1002/art.22044 [DOI] [PubMed] [Google Scholar]
- Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. 2007. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome 18: 463–472. 10.1007/s00335-007-9043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst DE. 2004. Anakinra: review of recombinant human interleukin-I receptor antagonist in the treatment of rheumatoid arthritis. Clin Ther 26: 1960–1975. 10.1016/j.clinthera.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Grünvogel O, Esser-Nobis K, Reustle A, Schult P, Müller B, Metz P, Trippler M, Windisch MP, Frese M, Binder M, et al. 2015. DDX60L is an interferon-stimulated gene product restricting hepatitis C virus replication in cell culture. J Virol 89: 10548–10568. 10.1128/JVI.01297-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J. 2013. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14: 7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawn TR, Wu H, Grossman JM, Hahn BH, Tsao BP, Aderem A. 2005. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematosus. Proc Natl Acad Sci 102: 10593–10597. 10.1073/pnas.0501165102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes WA, Haddon DJ, Diep VK, Khatri A, Bongen E, Yiu G, Balboni I, Bolen CR, Mao R, Utz PJ, et al. 2020. Integrated, multicohort analysis reveals unified signature of systemic lupus erythematosus. JCI Insight 5: e122312. 10.1172/jci.insight.122312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. 1979. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med 301: 5–8. 10.1056/NEJM197907053010102 [DOI] [PubMed] [Google Scholar]
- Hu D, Notarbartolo S, Croonenborghs T, Patel B, Cialic R, Yang TH, Aschenbrenner D, Andersson KM, Gattorno M, Pham M, et al. 2017. Transcriptional signature of human pro-inflammatory TH17 cells identifies reduced IL10 gene expression in multiple sclerosis. Nat Commun 8: 1600. 10.1038/s41467-017-01571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iikuni N, Lourenço EV, Hahn BH, La Cava A. 2009. Cutting edge: regulatory T cells directly suppress B cells in systemic lupus erythematosus. J Immunol 183: 1518–1522. 10.4049/jimmunol.0901163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul A, Gordon C, Crow MK, Touma Z, Urowitz MB, van Vollenhoven R, Ruiz-Irastorza G, Hughes G. 2016. Systemic lupus erythematosus. Nat Rev Dis Prim 2: 16039. 10.1038/nrdp.2016.39 [DOI] [PubMed] [Google Scholar]
- Knoops B, Argyropoulou V, Becker S, Ferté L, Kuznetsova O. 2016. Multiple roles of peroxiredoxins in inflammation. Mol Cells 39: 60–64. 10.14348/molcells.2016.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuda A, Wakui H, Iwamoto K, Ozawa M, Togashi M, Masai R, Maki N, Hatakeyama T, Sawada K. 2008. Up-regulated expression of Toll-like receptors mRNAs in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin Exp Immunol 152: 482–487. 10.1111/j.1365-2249.2008.03646.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KO, Tan AW, Thong BYH, Lian TY, Cheng YK, Teh CL, Koh ET, Chng HH, Law WG, Lau TC, et al. 2009. Enhanced expression of interferon-inducible protein-10 correlates with disease activity and clinical manifestations in systemic lupus erythematosus. Clin Exp Immunol 156: 134–140. 10.1111/j.1365-2249.2009.03880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T, Kanneganti TD. 2018. ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol 39: 123–134. 10.1016/j.it.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku C, Smiljanovic B, Grün JR, Biesen R, Schulte-Wrede U, Häupl T, Hiepe F, Alexander T, Radbruch A, Grützkau A. 2013. Cell-specific type I IFN signatures in autoimmunity and viral infection: what makes the difference? PLoS One 8: e83776. 10.1371/journal.pone.0083776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KN, Yap DYH. 2010. Cytokines and their roles in the pathogenesis of systemic lupus erythematosus: from basics to recent advances. J Biomed Biotechnol 2010: 365083. 10.1155/2010/365083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam GKW, Petri M. 2005. Assessment of systemic lupus erythematosus. Clin Exp Rheumatol 23: S120–S132. [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. 2012. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity 37: 880–892. 10.1016/j.immuni.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Leng RX, Pan HF, Qin WZ, Wang C, Chen LL, Tao JH, Ye DQ. 2011. TWEAK as a target for therapy in systemic lupus erythematosus. Mol Biol Rep 38: 587–592. 10.1007/s11033-010-0144-9 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Kim K. 2019. Genetic variants differentially associated with rheumatoid arthritis and systemic lupus erythematosus reveal the disease-specific biology. Sci Rep 9: 2739. 10.1038/s41598-019-39132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Luo K, Luo D. 2018. Guanosine monophosphate reductase 1 is a potential therapeutic target for Alzheimer's disease. Sci Rep 8: 2759. 10.1038/s41598-018-21256-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. 2013. Human circulating PD-1+CXCR3−CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39: 758–769. 10.1016/j.immuni.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay F, Browning JL. 2002. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol 2: 465–475. 10.1038/nri844 [DOI] [PubMed] [Google Scholar]
- Mohsen MA, Abdel Karim SA, Abbas TM, Amin M. 2013. Serum interleukin-18 levels in patients with systemic lupus erythematosus: relation with disease activity and lupus nephritis. Egypt Rheumatol 35: 45–51. 10.1016/j.ejr.2012.09.005 [DOI] [Google Scholar]
- Morand EF, Furie R, Tanaka Y, Bruce IN, Askanase AD, Richez C, Bae SC, Brohawn PZ, Pineda L, Berglind A, et al. 2020. Trial of anifrolumab in active systemic lupus erythematosus. N Engl J Med 382: 211–221. 10.1056/NEJMoa1912196 [DOI] [PubMed] [Google Scholar]
- Moulton VR, Suarez-Fueyo A, Meidan E, Li H, Mizui M, Tsokos GC. 2017. Pathogenesis of human systemic lupus erythematosus: a cellular perspective. Trends Mol Med 23: 615–635. 10.1016/j.molmed.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehar-Belaid D, Hong S, Marches R, Chen G, Bolisetty M, Baisch J, Walters L, Punaro M, Rossi RJ, Chung CH, et al. 2020. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat Immunol 21: 1094–1106. 10.1038/s41590-020-0743-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G, Pascual V. 2010. The interferon-α signature of systemic lupus erythematosus. Lupus 19: 1012–1019. 10.1177/0961203310371161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke V, Gunnarsson I, Dorschner J, Eketjäll S, Zickert A, Niewold TB, Svenungsson E. 2019. High levels of circulating interferons type I, type II and type III associate with distinct clinical features of active systemic lupus erythematosus. Arthritis Res Ther 21: 107. 10.1186/s13075-019-1878-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. 2006. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci 103: 17973–17978. 10.1073/pnas.0605938103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri M, Orbai AM, Alarcõn GS, Gordon C, Merrill JT, Fortin PR, Bruce IN, Isenberg D, Wallace DJ, Nived O, et al. 2012. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 64: 2677–2686. 10.1002/art.34473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rönnblom L, Leonard D. 2019. Interferon pathway in SLE: one key to unlocking the mystery of the disease. Lupus Sci Med 6: e000270. 10.1136/lupus-2018-000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales SL, Liang S, Engel I, Schmiedel BJ, Kronenberg M, Vijayanand P, Seumois G. 2018. A sensitive and integrated approach to profile messenger RNA from samples with low cell numbers. Methods Mol Biol 1799: 275–302. 10.1007/978-1-4939-7896-0_21 [DOI] [PubMed] [Google Scholar]
- Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma'ayan A. 2016. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016: baw100. 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, et al. 2018. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175: 1701–1715.e16. 10.1016/j.cell.2018.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi A, Tsokos GC. 2020. T cell metabolism: new insights in systemic lupus erythematosus pathogenesis and therapy. Nat Rev Rheumatol 16: 100–112. 10.1038/s41584-019-0356-x [DOI] [PubMed] [Google Scholar]
- Simpson N, Gatenby PA, Wilson A, Malik S, Fulcher DA, Tangye SG, Manku H, Vyse TJ, Roncador G, Huttley GA, et al. 2010. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum 62: 234–244. 10.1002/art.25032 [DOI] [PubMed] [Google Scholar]
- Sims JE, Smith DE. 2010. The IL-1 family: regulators of immunity. Nat Rev Immunol 10: 89–102. 10.1038/nri2691 [DOI] [PubMed] [Google Scholar]
- Southworth LK, Owen AB, Kim SK. 2009. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet 5: e1000776. 10.1371/journal.pgen.1000776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R, Grzelak M, Hadfield J. 2019. RNA sequencing: the teenage years. Nat Rev Genet 20: 631–656. 10.1038/s41576-019-0150-2 [DOI] [PubMed] [Google Scholar]
- Su C, Johnson ME, Torres A, Thomas RM, Manduchi E, Sharma P, Mehra P, Le Coz C, Leonard ME, Lu S, et al. 2020. Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells. Nat Commun 11: 3294. 10.1038/s41467-020-17089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505. 10.1038/nature06013 [DOI] [PubMed] [Google Scholar]
- Thanou A, Chakravarty E, James JA, Merrill JT. 2014. Which outcome measures in SLE clinical trials best reflect medical judgment? Lupus Sci Med 1: e000005. 10.1136/lupus-2013-000005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touma Z, Gladman DD. 2017. Current and future therapies for SLE: obstacles and recommendations for the development of novel treatments. Lupus Sci Med 4: e000239. 10.1136/lupus-2017-000239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos GC. 2011. Systemic lupus erythematosus. N Engl J Med 365: 2110–2121. 10.1056/NEJMra1100359 [DOI] [PubMed] [Google Scholar]
- Tsokos GC, Lo MS, Reis PC, Sullivan KE. 2016. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol 12: 716–730. 10.1038/nrrheum.2016.186 [DOI] [PubMed] [Google Scholar]
- van Nas A, GuhaThakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, et al. 2009. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 150: 1235–1249. 10.1210/en.2008-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ao Z, Chen L, Kobinger G, Peng J, Yao X. 2012. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol 86: 3777–3786. 10.1128/JVI.06594-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel B, Otterbein LE. 2012. Go green: the anti-inflammatory effects of biliverdin reductase. Front Pharmacol 3: 47. 10.3389/fphar.2012.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu J, Cui X, Zuo Y, Zhang Z, Li Y, Tao R, Li Y, Pang J. 2015. Increased frequency of circulating follicular helper T cells in lupus patients is associated with autoantibody production in a CD40L-dependent manner. Cell Immunol 295: 46–51. 10.1016/j.cellimm.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Xu A, Liu Y, Chen W, Wang J, Xue Y, Huang F, Rong L, Lin J, Liu D, Yan M, et al. 2016. TGF-β-induced regulatory T cells directly suppress B cell responses through a noncytotoxic mechanism. J Immunol 196: 3631–3641. 10.4049/jimmunol.1501740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao J, Wu X, Mao Q, Wang Y, Gao F, Kong W, Liang Z. 2015a. Type I interferon related genes are common genes on the early stage after vaccination by meta-analysis of microarray data. Hum Vaccines Immunother 11: 739–745. 10.1080/21645515.2015.1008884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lindwall E, Gauthier C, Lyman J, Spencer N, Alarakhia A, Fraser A, Ing S, Chen M, Webb-Detiege T, et al. 2015b. Circulating CXCR5+CD4+helper T cells in systemic lupus erythematosus patients share phenotypic properties with germinal center follicular helper T cells and promote antibody production. Lupus 24: 909–917. 10.1177/0961203314567750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.