Abstract
Background
The role of miR-1 and miR-133 in regulating the expression of potassium and calcium ion channels, and mediating cardiomyocyte apoptosis in mice with viral myocarditis (VMC) is investigated herein.
Methods
Male Balb/c mice were randomly divided into groups: control group, VMC group, VMC + + miR-1/133 mimics group, or VMC + miR-1/133 negative control (NC) group. VMC was induced with coxsackievirus B3 (CVB3). MiR-1/133 mimics ameliorated cardiac dysfunction in VMC mice and was compared to the VMC+NC group.
Results
Hematoxylin and eosin staining showed a well-arranged myocardium without inflammatory cell infiltration in the myocardial matrix of the control group. However, in the VMC and VMC+NC groups, the myocardium was disorganized and swollen with necrosis, and the myocardial matrix was infiltrated with inflammatory cells. These changes were alleviated by miR-1/133 mimics. TUNEL staining revealed decreased cardiomyocyte apoptosis in the VMC + miR-1/133 mimics group compared with the VMC group. In addition, miR-1/133 mimics up-regulated the expression of miR-1 and miR-133, the potassium channel genes Kcnd2 and Kcnj2, as well as Bcl-2, and down-regulated the expression of the potassium channel suppressor gene Irx5, L-type calcium channel subunit gene α1c (Cacna1c), Bax, and caspase-9 in the myocardium of VMC mice. MiR-1/133 also up-regulated the protein levels of Kv4.2 and Kir2.1, and down-regulated the expression of CaV1.2 in the myocardium of VMC mice.
Conclusions
MiR-1 and miR-133 decreased cardiomyocyte apoptosis by mediating the expression of apoptosis-related genes in the hearts of VMC mice.
Keywords: miR-1, miR-133, viral myocarditis, ion channels, cell apoptosis
Introduction
MicroRNA (miR)-1 and miR-133 are members of a miRNA cluster expressed exclusively in skeletal and cardiac muscle cells [1], and are essential for heart development [2]. MiR-1 and miR-133 have been shown to regulate the expression of gap junctions [3], pacemaker channels [4] and potassium (K+) channels, including inward-rectifying Kir2.1, which stabilizes resting potential [5] and Kv4.2, which dominates the outward-transient current (Ito) [6]. In addition, miR-133 has been shown to target CaV1.2, the primary α-subunit for L-type calcium (Ca2+) channels underlying cardiac excitability [7]. Previous studies have shown that approximately 20% of infant deaths due to fatal arrhythmia and severe heart failure were caused by acute severe viral myocarditis (VMC), mainly caused by coxsackievirus B3 (CVB3) [8]. However, there is currently a lack of effective etiological treatment for severe VMC. Introducing miR-1/133 can reduce virus escape and mutation, and as such, one of the major treatments involves the use of RNA interference (RNAi) combined with miR mimics [9, 10]. A previous study showed that approximately 20% of miR-1/133 mimics, labeled with Dy547 and packaged in RNA-LANCEr II neutral ionic lipids, arrived in cardiomyocytes and exhibited intracellular Dy547 particle signals when administered by tail vein injection [10]. In addition, miR-133 has been shown to target caspase-9, a pro-apoptotic factor, and promotes cardiac cell apoptosis by interfering with the expression of mRNA [11].
With rapid advances in molecular biology, miRs have been widely studied in different fields. However, reports regarding the effects of miRs on VMC have been limited. Under investigation in the present study, was the impact of miR-1/133 mimics and the appropriate miR-1/133 negative control on a mouse model of acute VMC, and the hypothesis that miR-1 and miR-133 could etiologically help to treat severe VMC was examined.
Methods
This study was in accordance with the National Research Council Guide for the care and use of laboratory animals. The animal experiments in this study were performed with approval by the Animal Care Committee of the Medical School of Shandong University.
Mouse model of acute VMC and interference
CVB3 was passaged in HeLa cells by transfection as described previously [12]. Virus titer was determined using a tissue culture infective dose 50 (TCID50) assay at the beginning of the experiments. HeLa cells were cultured in Dulbecco’s Modified Eagle medium (DMEM, Life Technologies, USA) supplemented with fetal bovine serum (10%; Gibco, Australia) and penicillin-streptomycin (100 IU/mL; Invitrogen, USA).
Forty healthy male Balb/c mice (6–8 weeks old) were randomly divided into four groups: control group (n = 10), in which mice were injected intraperitoneally with Eagle’s medium (0.1 mL); VMC group (n = 10), in which VMC was induced by intraperitoneal injection of 104 TCID50 CVB3 (0.1 mL); VMC + miR-1/133 mimics group (n = 10), in which VMC mice were injected intraperitoneally with miR-1/133 mimics (1 μg/g, GenePharma, Shanghai, China) on the day following VMC induction; VMC + NC group (n = 10), in which VMC mice were injected intraperitoneally with miR-1/133 NC (1 μg/g, GenePharma, Shanghai, China) via the tail vein on the day following VMC induction.
MiR-1/133 mimics, which were composed of mmu-miR-1 mimics and mmu-miR-133 mimics, were packaged in Lipofectamine 2000 Reagent (Invitrogen, US), and then directly mixed in the ultrasonic field to improve targeting ability. Feeding conditions of mice in each group were the same. Following virus injection, the diet, activity level, and coat appearance of the mice were observed on a daily basis.
Cardiac functional analysis
On the 7th day following establishment of the VMC model, i.e. the 6th day after miR-1/133 intervention, mice were anesthetized with an intraperitoneal injection of chloral hydrate (10%, 4 mg/g). After the mice were fixed in a supine position, the chest was shaved, and an M-echocardiograph was performed to measure the left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) of the mice in each group.
Specimen collection and disposition
On the 7th day after modeling, after all mice in the groups were anesthetized, their hearts were removed under sterile conditions, washed with normal saline, and then weighed. Half of each heart of the mice were frozen with liquid nitrogen and stored at −80°C for use in molecular biological experiments (half of each heart was used for quantitative reverse transcription followed by polymerase chain reaction (qRT-PCR) and Western blot, which were carried out by two people), the other half of the heart were fixed in 10% neutral formaldehyde, embedded in paraffin, and sectioned at 4.5 μm intervals for histological study (H&E staining and TUNEL staining).
Hematoxylin and eosin staining
Tissue sections were stained by hematoxylin and eosin (H&E; Beyotime, Shanghai, China), and cardiomyocyte morphology changes were observed under a light microscope.
TUNEL staining
Cardiomyocyte apoptosis was detected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining according to the manufacturer protocol (Roche, USA). Ten randomly selected fields from each slide were scored at a high magnification. The ratio of the number of brown cells, i.e. apoptotic cells, to a total of 30 cardiomyocytes counted per field was defined as the myocardial cell apoptotic ratio.
Quantitative real time RT-PCR
Total RNA was extracted from a part of each of frozen mouse heart using a TRIzol reagent (Ambion, USA). According to the manufacturer protocol (Thermo Scientific, USA), qRT-PCR was performed to detect the expression of miR-1 and miR-133, inward rectifier potassium current (Ik1) channel gene Kcnd2, its transcription repressor Irx5, transient outward potassium current (Ito) channel gene Kcnj2, the L-type calcium channel subunit α1c (Cacna1c), and apoptosis-related genes Bax, Bcl-2 and caspase-9 (Casp9) using an ABI 7500 real-time PCR system. β-actin was used as a reference for gene expression and U6 was used as a reference for expression of miR-1 and miR-133. The relative expression of genes was calculated using the 2−ΔΔCt method. The primers were synthesized by Guangzhou Saibo Corp, China. The sequences of gene primers used in the present study are shown in Table 1.
Table 1.
Prime sequences for quantitative reverse transcription followed by polymerase chain reaction (qRT-PCR).
| Gene | Primer Sequences (5′-3′) | |
|---|---|---|
| β-actin | ||
| sense | CCAGCCTTCCTTCTTGGGTAT | |
| antisense | TTGGCATAGAGGTCTTTACGG | |
| Kcnd2 | ||
| sense | TGACAACACTGGGGTATGGC | |
| antisense | CCGACTGAAGTTCGACACGA | |
| Kcnj2 | ||
| sense | TCTCACTTGCTTCGGCTCAT | |
| antisense | ACTTGTCCTGTTGCTGGTACA | |
| Irx5 | ||
| sense | GCCTTCTCTTACGTGGGCTC | |
| antisense | AGTGGCATTCTTCCGGTACG | |
| α1c (Cacna1c) | ||
| sense | TCCCGAGCACATCCCTACTC | |
| antisense | ACTGACGGTAGAGATGGTTGC | |
| Bax | ||
| sense | AAACTGGTGCTCAAGGCCC | |
| antisense | CTTGGATCCAGACAAGCAGC | |
| Bcl-2 | ||
| sense | GCTACCGTCGTGACTTCGC | |
| antisense | CCCACCGAACTCAAAGAAGG | |
| Casp9 | ||
| sense | TCAGGGGACATGCAGATATGG | |
| antisense | TTGGCAGTCAGGTCGTTCTTC | |
| U6 | ||
| sense | ATGACGTCTGCCTTGGAGAAC | |
| antisense | TCAGTGTGCTACGGAGTTCAG | |
| Mmu-miR-1a-3p | ||
| reverse | transcription | prime |
| CTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAGATACAT | ||
| sense | GCCGCTGGAATGTAAAGAAGT | |
| antisense | GTCCGAGGTATTCGCACTGGATA | |
| Mmu-miR-133a-3p | ||
| reverse | transcription | prime |
| GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGCTG | ||
| sense | GCTTTGGTCCCCTTCAAC | |
| antisense | GTCCGAGGTATTCGCACTGGATA | |
Western blot
To examine protein levels of Kv4.2, Kir2.1, and CaV1.2, total protein lysates were purified from the rest of frozen heart samples using RIPA buffer (Beyotime, Shanghai, China). A bicinchoninic acid (BCA) protein assay kit (Beyotime, Shanghai, China) was used to determine protein concentration. Protein (30–50 μg) was separated on an 8–15% gel with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was then blocked in 5% milk, followed by incubation at 4°C overnight with the primary antibodies of interest (rabbit polyclonal anti-Kv4.2 and anti-Kir2.1, Bioss, Beijing, China, diluted 1:300; mouse monoclonal anti-CACNA1C, Abcam, USA; and mouse anti-GAPDH, ZSGB-BIO Beijing, China, diluted 1:1000). Membranes were then washed (3 times) with tris-buffered saline with Tween (TBST), and incubated with the appropriate secondary antibodies (ZSGB-BIO Beijing, China, diluted 1:10000). Immunoreactive bands were detected by enhanced chemiluminescence (ECL; Millipore, USA) and quantified using C-digit (Model: 3600, Image Studio Digits Ver 4.0, Licor, Lincoln, NE, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a reference loading control. Data were collected from at least three independent experiments.
Data analysis
The experimental data are presented as mean ± standard error (SE). Statistical comparisons in multiple groups were performed using one-way analysis of variance (ANOVA), and the comparison between two groups was performed with the Fisher least significant difference t test. A probability value of p < 0.05 was considered statistically significant.
Results
MiR-1/133 mimics improved general status of VMC mice
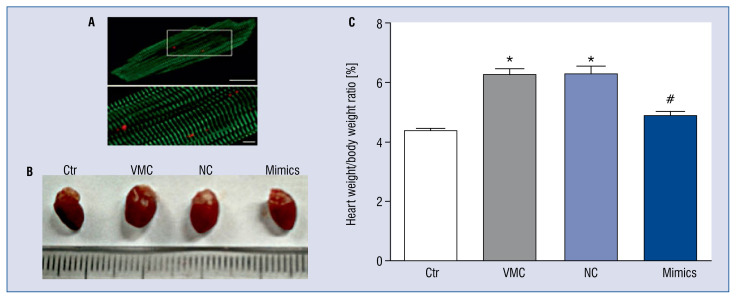
Compared with mice in the control group, on the third day following CVB3 injection, mice in the VMC group and in the VMC + NC group had disordered fur, were irritated easily or insensitive to irritation, ate little food and lost weight (data not shown). These conditions were improved in mice receiving the miR-1/133 mimics (data not shown). Figure 1A shows the specific myocardial staining, shown using antibodies against α-actinin (green) and miR-1/133 (red).
Figure 1.
Macroscopic view of hearts from different groups. A. Fluorescence showing specific staining of green (α-actinin) and red (Dy547) for miR-1/133; B. Global view of representative heart from each group; C. The statistical analysis of the heart weight/body weight ratio of mice in each group; Ctr — control group; VMC — viral myocarditis (VMC) group; NC — VMC + miR-1/133 negative control (NC) group; Mimics — VMC + miR-1/133 mimics (Mimics) group. Data are expressed as mean ± standard error (SE). N = 10, *p < 0.01 vs. control group, #p < 0.05 vs. VMC and VMC + NC group; for other abbreviations — see text.
The hearts from mice in the VMC group and NC group were bigger than those in the control and VMC + miR-1/133 mimics groups (Fig. 1B) and heart weight/body weight ratio was increased as well (Fig. 1C). There were also spot-like necrotic areas on the surface of the hearts of mice in the VMC group and the VMC + NC group (Fig. 1B).
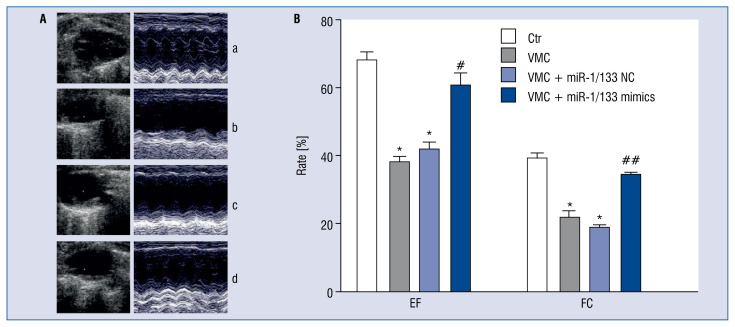
MiR-1/133 mimics improved cardiac function of VMC mice
Compared with those in the control group, EF and FS of mice in the VMC and VMC + NC groups were significantly decreased (p < 0.01), suggesting impaired cardiac function of VMC mice. However, treatment with miR-1/133 mimics improved EF and FS of VMC mice (p < 0.05, p < 0.01; Fig. 2).
Figure 2.
MiR-1/133 mimics improved cardiac function of viral myocarditis (VMC) mice. A. Echocardiography showed ejection fraction (EF) and fractional shortening (FS) of mice in each group; a. Control group (Ctr), b. VMC group, c. VMC + NC group, d. VMC + miR-1/133 mimics group. B. Statistical analysis of EF and FS of different groups. Data are expressed as mean ± standard error (SE). N = 10, *p < 0.01 vs. control group, #p < 0.05, ##p < 0.01 vs. VMC and VMC + NC group; for other abbreviations — see text.
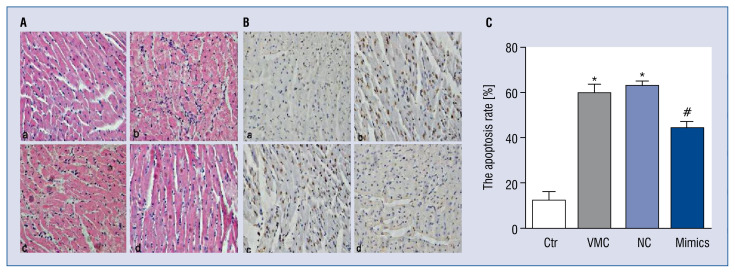
MiR-1/133 mimics attenuated pathological changes of VMC hearts
Compared with control group hearts, H&E staining showed that myocardia in VMC and VMC + NC groups were swollen and disordered, they were also infiltrated by inflammatory cells in the myocardial matrix. MiR-1/133 mimics reduced the edema of cardiomyocytes, improved arrangements, and diminished inflammatory cell infiltrate in the myocardial matrix (Fig. 3A).
Figure 3.
MiR-1/133 mimics improved pathological changes and reduced cardiomyocyte apoptosis in viral myocarditis (VMC) mouse hearts. A. H&E staining of myocardium in different group mice (magnification: 10 × 40); a. Control group (Ctr), b. VMC group, c. VMC + negative control (NC) group, d. VMC + miR-1/133 mimics group; B. TUNEL staining showed cardiomyocytes apoptosis of different group mice (10 × 40). Cells with brown nuclei were TUNEL positive, and cells with blue nuclei represented normal cells; a. Control group, b. VMC group, c. VMC+miR-1/133 NC group, d. VMC + miR-1/133 mimics group; C. Quantified cell apoptosis data. Data are expressed as mean ± standard error (SE). N = 10, *p < 0.01 vs. control, #p < 0.01 vs. VMC and VMC + NC group; for other abbreviations — see text.
MiR-1/133 mimics reduced cardiomyocyte apoptosis
TUNEL staining showed increased numbers of brown nuclei in the VMC and VMC + NC groups compared with those in the control group (p < 0.01), revealing increased cell death. MiR-1/133 mimics significantly reduced the number of brown nuclei (p < 0.01), indicating decreased cell death. The VMC + NC group had the highest rate of apoptosis while the control group had the lowest (Fig. 3B, C).
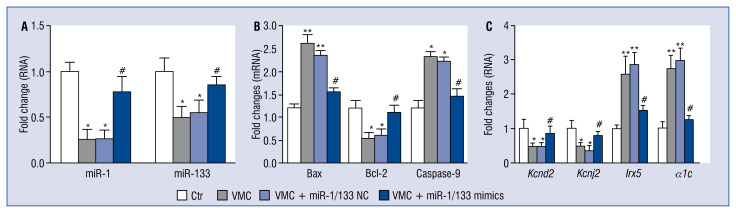
MiR-1/133 mimics improved expression of miR-1 and miR-133 in VMC hearts
qRT-PCR data showed that the relative expression of miR-1 and miR-133 in mouse hearts of the VMC and VMC + NC groups was significantly decreased compared to the control group (p < 0.05), but miR-1/133 mimics increased the expression of miR-1 and miR-133 (p < 0.05; Fig. 4A).
Figure 4.
MiR-1/133 mimics regulated the expressions of miR-1 and miR-133, apoptosis-related genes, potassium channel genes and a calcium channel gene. A. The fold changes of miR-1 and miR-133 expression. U6 was used as an internal reference and data was normalized to the control group (Ctr). N = 10, *p < 0.05 vs. control group, #p < < 0.05 vs. viral myocarditis (VMC) and VMC + NC groups; B, C. The fold changes of apoptosis-related genes Bax, Bcl-2, and Casp9, and potassium channel genes Kcnd2, Irx5, Kcnj2, and L-type calcium channel subunit α1c, Cacna1c. Values were normalized against β-actin as an endogenous control. Data are expressed as mean ± standard error (SE). The fold changes of qRT-PCR were determined using the 2−ΔΔCt method. N = 10, *p < 0.05, **p < 0.01 vs. control group, #p < 0.05 vs. VMC and VMC + NC groups; for other abbreviations — see text.
MiR-1/133 regulated the mRNA expression of apoptosis-related genes Bax, Bcl-2 and caspase-9 in VMC hearts
The relative expression of pro-apoptosis-related genes Bax (p < 0.01), and Casp9 (p < 0.05) mRNA in the VMC and VMC + NC groups were significantly up-regulated compared with those in the control group. However, miR-1/133 mimics greatly decreased their expression (p < 0.05). While the expression of the anti-apoptosis-related gene Bcl-2 mRNA was down-regulated in the VMC and VMC+NC groups, miR-1/133 mimics increased its expression (p < 0.05; Fig. 4B).
MiR-1/133 regulated the relative expressions of ion channel genes Kcnd2, Kcnj2, Irx5 and α1c in VMC hearts
Compared with those in the control group, the relative expression of Kcnd2 and Kcnj2 in the VMC and VMC + NC groups were significantly down-regulated as revealed by qRT-PCR data (p < 0.05), which was attenuated by miR-1/133 mimics interference (p < 0.05). Also, the relative expression of Irx5 and α1c (Cacna1c) was increased in VMC and VMC + NC groups compared with the control group (p < 0.01), and this increase was suppressed by miR-1/133 mimics interference (p < 0.05; Fig. 4C).
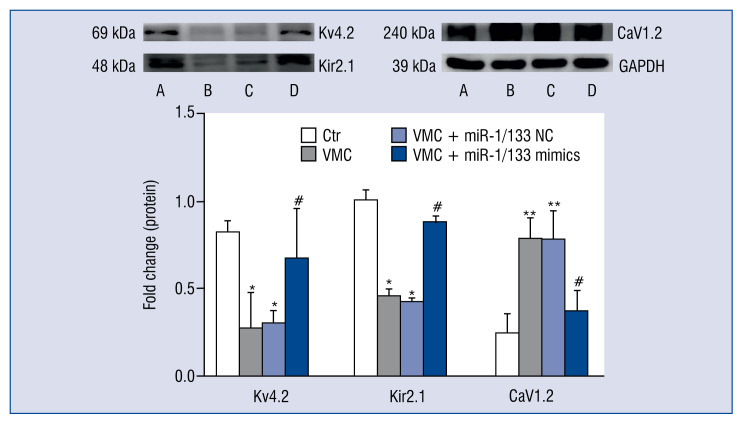
MiR-1/133 regulated the protein levels of Kv4.2, Kir2.1 and L-type calcium channel subunit CaV1.2 in VMC hearts
Compared with control mice, the protein levels of Ito target protein Kv4.2 and Ik1 target protein Kir2.1 in the VMC and VMC + NC groups were significantly decreased (p < 0.05), but this decrease was suppressed by miR-1/133 mimics. Also, the expression of the L-type calcium channel subunit protein CaV1.2 was significantly increased in the VMC and VMC + NC groups compared with the control group (p < 0.01), but again, this increase was attenuated by miR-1/133 mimics interference (p < 0.05; Fig. 5).
Figure 5.
MiR-1/133 mimics complex regulated the protein levels of Kv4.2, Kir2.1 and CaV1.2. Upper panel: Western blot. A. Control group (Ctr); B. Viral myocarditis (VMC) group; C. VMC + NC group; D. VMC + miR-1/133 mimics complex group. Lower panel: Statistical analysis of the upper panel. Data are expressed as mean ± standard error (SE). GAPDH was used as an internal reference, and data were normalized to the control group. N = 10, *p < 0.05, **p < 0.01 vs. control group, #p < 0.05 vs. VMC and VMC + NC groups; for other abbreviations — see text.
Discussion
MiRs have been shown to be implicated in a variety of human diseases, and may serve as potential bio-markers in certain diseases once released into the circulatory system [13]. Currently, more than 100 miRs have been identified in myocardial cells, and some, if not all, were dysregulated in heart diseases [14, 15]. MiR-1 and miR-133, which translate together in a bicistronic way [16], are specific to muscle and are abundant in the heart [17, 18]. MiR-1 and miR-133 play an important role in normal cardio genesis [19] and in the pathogenesis of several cardiovascular diseases including myocardial ischemia-reperfusion injury [11], cardiac hypertrophy [20, 21], arrhythmia [22], and myocardial infarction [23]. However, their involvement in VMC-linked cardiac phenotypes has not been explored. The major findings from the present investigation include: 1) miR-1/133 was downregulated in the hearts of VMC mice, 2) miR-1/133 mimics attenuated cardiac dysfunction and apoptosis in VMC mouse hearts, 3) miR-1/133 mimics mediated the expression of apoptosis-related genes in VMC mouse hearts, and 4) miR-1/133 mimics mediated the expression of potassium and calcium channels in VMC mouse hearts.
Viral invasion of myocardial cells causes persistent chronic inflammation, subsequently resulting in myocardial cell hypertrophy, myocardial cell apoptosis, and myocardial fibrosis [24–26]. Therefore, the progression of VMC will eventually lead to heart failure and fatal arrhythmias, which is linked to sudden death. In agreement with the above findings, in the present study, the VMC mice induced by CVB3 exhibited impaired cardiac function as evidenced by reduced EF and FS compared with control mice. Under further testing the effects of miR1/133 on cardiac function and pathology in VMC mice, and found that miR-1/133 mimics ameliorated cardiac function and pathological changes of VMC mice. This miR-1/133-induced improvement of cardiac function and pathology might be attributed to decreased cardiomyocyte apoptosis in VMC mouse hearts. To further understand the molecular basis underlying reduced apoptosis by miR-1/133 mimics, qRT-PCR was used to examine the relative expression of apoptosis-related genes Bax, Bcl-2 and Casp9 in VMC hearts of the different groups. The pro-apoptosis-related genes Bax and Casp9, as well as the anti-apoptosis-related gene Bcl-2 are the main genes involved in the progression of cell apoptosis. The present findings suggest that viral infection increased the expression of Bax and Casp9 but decreased the expression of Bcl-2, and that miR-1/133 mimics attenuated the changes in the expression of these three genes. Therefore, it is possible that these miR1/133-induced changes contributed to the improved cardiac function and pathology of VMC mice by miR1/133 mimics. In addition, the current findings demonstrate that miR-133 expression was inversely proportional with the expression of Casp9 and was in line with a previous study identifying caspase-9 as a target of miR-133 [27].
Viral myocarditis patients display different arrhythmias, and severe arrhythmia can cause sudden cardiac death. Electrical remodeling and myocardium structural remodeling are the main mechanisms leading to arrhythmia [28], which is associated with changes in the activity of ion channels present on the myocardial cell membrane [29], including potassium and calcium ion channels. Although many miRs are expressed abnormally in many heart diseases, there are few reports regarding the relationship between miR-1 and miR-133 and the activity of potassium ion channels and calcium ion channels in VMC. For example, miR-1 and miR-133 were shown to be involved in the regulation of Kv4.2 and Kir2.1 expression [30], the latter of which is the main component of Ik1 and is linked to arrhythmia [31]. Also, the aberrant expression of miR-1 influenced the activity of Ik1 and its expression [32, 33]. Kir2.1, which is the main potassium channel subunit, plays important roles in initiating and maintaining resting membrane potential of cardiomyocytes [34], is encoded by Kcnj2, and Kcnj2 was targeted by miR-1 [3]. Therefore, the aberrant expression of miR-1 could cause abnormality in the structure and function of potassium channels, influencing membrane potential stability, and result in arrhythmia. In addition, the potassium channel subunit Kv4.2, the main component of Ito, is rich in cardiomyocytes, and is encoded by the Kcnd2 gene. Irx5, a transcriptional repressor of Kcnd2, and regulated potassium ion current and repolarization of action potentials. MiR-1 was found to target Irx5 and thus influenced the expression of Kcnd2 [35]. In the present study, it was observed that the aberrant expression of Kcnd2 and Kcnj2 in VMC cardiomyocytes, which can result in a change of potassium ion concentration and influence membrane potential, and therefore may contribute to arrhythmia. In line with the above reports, it was also shown herein that there was a negative correlation between miR-1 and Irx5. Therefore, it was believed that miR-1 improved arrhythmia at least in part through mediating the expression of Kcnd2 and Kcnj2 and thus, regulating potassium concentrations. In addition, the L-type calcium channel subunit α1c is the main calcium ion channel in myocardium and plays an important role in stabilizing calcium concentration in the cell and maintaining action potential repolarization. α1c was identified as a potential target of miR-133 [7], which was consistent with the present findings that demonstrated a negative correlation between miR-133 and α1c. Taken together, it was speculated that miR-1 and miR-133 reduced arrhythmia by mediating the expression and activity of ion channels, and could serve as potential anti-arrhythmic targets.
Conclusions
In summary, this study reports that miR-1/133 mimics ameliorate cardiac function in VMC mice. Mechanistically, miR-1/133 mimics reduce cardiomyocyte apoptosis by regulating the expressions of apoptosis-related genes, improves arrhythmia through regulating the expressions of calcium and potassium ion channel genes of cardiomyocytes, and diminishes cardiac fibrosis in VMC mouse hearts. Therefore, miR-1/133 may serve as a potential therapeutic target for treatment of VMC in clinics.
Acknowledgements
This study was supported in part by grants from the National Natural Science Foundation (#21728701), The ministry of education postdoctoral fund (#2015M572044), Shandong Province Science and Technology Development Project (#GG201709260070).
Footnotes
Conflict of ineterest: None declared
References
- 1.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 3.Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13(4):486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 4.Li YD, Hong YF, Yusufuaji Y, et al. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol Med Rep. 2015;12(3):3243–3248. doi: 10.3892/mmr.2015.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girmatsion Z, Biliczki P, Bonauer A, et al. Changes in micro-RNA-1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm. 2009;6(12):1802–1809. doi: 10.1016/j.hrthm.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 6.da Costa Martins PA, Bourajjaj M, Gladka M, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118(15):1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Xiao J, Lin H, et al. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. 2009;37(3):e24. doi: 10.1093/nar/gkn1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman A, McNamara D. Myocarditis N Engl J Med. 2000;343(19):1388–1398. doi: 10.1056/nejm200011093431908. [DOI] [PubMed] [Google Scholar]
- 9.Xu F, Liu G, Liu Q, et al. RNA interference of influenza A virus replication by microRNA-adapted lentiviral loop short hairpin RNA. J Gen Virol. 2015;96(10):2971–2981. doi: 10.1099/jgv.0.000247. [DOI] [PubMed] [Google Scholar]
- 10.Myers R, Timofeyev V, Li N, et al. Feedback mechanisms for cardiac-specific microRNAs and cAMP signaling in electrical remodeling. Circ Arrhythm Electrophysiol. 2015;8(4):942–950. doi: 10.1161/CIRCEP.114.002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinali M, Arechavala-Gomeza V, Cirak S, et al. Muscle histology vs MRI in Duchenne muscular dystrophy. Neurology. 2011;76(4):346–353. doi: 10.1212/WNL.0b013e318208811f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Carthy CM, Cheung P, et al. Structural and functional analysis of the 5′ untranslated region of coxsackievirus B3 RNA: In vivo translational and infectivity studies of full-length mutants. Virology. 1999;265(2):206–217. doi: 10.1006/viro.1999.0048. [DOI] [PubMed] [Google Scholar]
- 13.Li LM, Cai WB, Ye Q, et al. Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World J Emerg Med. 2014;5(3):182–186. doi: 10.5847/wjem.j.1920-8642.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y, Ji R, Yue J, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170(6):1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busk PK, Cirera S. MicroRNA profiling in early hypertrophic growth of the left ventricle in rats. Biochem Biophys Res Commun. 2010;396(4):989–993. doi: 10.1016/j.bbrc.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 16.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 17.Liang Yu, Ridzon D, Wong L, et al. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo X, Zhang H, Xiao J, et al. Regulation of human cardiac ion channel genes by microRNAs: theoretical perspective and pathophysiological implications. Cell Physiol Biochem. 2010;25(6):571–586. doi: 10.1159/000315076. [DOI] [PubMed] [Google Scholar]
- 19.Lin CC, Chang YM, Pan CT, et al. Functional evolution of cardiac microRNAs in heart development and functions. Mol Biol Evol. 2014;31(10):2722–2734. doi: 10.1093/molbev/msu217. [DOI] [PubMed] [Google Scholar]
- 20.Sayed D, Hong C, Chen IY, et al. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100(3):416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 21.Diniz GP, Lino CA, Guedes EC, et al. Cardiac microRNA-133 is down-regulated in thyroid hormone-mediated cardiac hypertrophy partially via type 1 angiotensin II receptor. Basic Res Cardiol. 2015;110(5):49. doi: 10.1007/s00395-015-0504-7. [DOI] [PubMed] [Google Scholar]
- 22.Hedley PL, Carlsen AL, Christiansen KM, et al. MicroRNAs in cardiac arrhythmia: DNA sequence variation of MiR-1 and MiR-133A in long QT syndrome. Scand J Clin Lab Invest. 2014;74(6):485–491. doi: 10.3109/00365513.2014.905696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GK, Zhu JQ, Zhang JT, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31(6):659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 24.Woodruff JF. Viral myocarditis. A review. Am J Pathol. 1980;101(2):425–484. [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes MP, Lerner AM. Coxsackievirus myocarditis — with special reference to acute and chronic effects. Prog Cardiovasc Dis. 1985;27(6):373–394. doi: 10.1016/0033-0620(85)90001-5. [DOI] [PubMed] [Google Scholar]
- 26.Kishimoto C, Misaki T, Crumpacker CS, et al. Serial immunologic identification of lymphocyte subsets in murine coxsackievirus B3 myocarditis: different kinetics and significance of lymphocyte subsets in the heart and in peripheral blood. Circulation. 1988;77(3):645–653. doi: 10.1161/01.cir.77.3.645. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, Lu Y, Pan Z, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120(Pt 17):3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Dong Y, Zhu N, et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014;13:124. doi: 10.1186/1476-4598-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besser J, Malan D, Wystub K, et al. MiRNA-1/133a clusters regulate adrenergic control of cardiac repolarization. PLoS One. 2014;9(11):e113449. doi: 10.1371/journal.pone.0113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costantini DL, Arruda EP, Agarwal P, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123(2):347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gómez R, Caballero R, Barana A, et al. Structural basis of drugs that increase cardiac inward rectifier Kir2.1 currents. Cardiovasc Res. 2014;104(2):337–346. doi: 10.1093/cvr/cvu203. [DOI] [PubMed] [Google Scholar]
- 32.Wang Lu, Yuan Ye, Li J, et al. MicroRNA-1 aggravates cardiac oxidative stress by post-transcriptional modification of the anti-oxidant network. Cell Stress Chaperones. 2015;20(3):411–420. doi: 10.1007/s12192-014-0565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Zhang Y, Shan H, et al. MicroRNA-1 downregulation by propranolol in a rat model of myocardial infarction: a new mechanism for ischaemic cardioprotection. Cardiovasc Res. 2009;84(3):434–441. doi: 10.1093/cvr/cvp232. [DOI] [PubMed] [Google Scholar]
- 34.Diaz RJ, Zobel C, Cho HC, et al. Selective inhibition of inward rectifier K+ channels (Kir2.1 or Kir2.2) abolishes protection by ischemic preconditioning in rabbit ventricular cardiomyocytes. Circ Res. 2004;95(3):325–332. doi: 10.1161/01.RES.0000137727.34938.35. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]