Abstract
RFamide-related peptides (RFRPs, mammalian orthologs of gonadotropin-inhibitory hormone) convey circadian, seasonal, and social cues to the reproductive system. They regulate gonadotropin secretion by modulating gonadotropin-releasing hormone (GnRH) neurons via the RFRP receptor. Mice lacking this receptor are fertile but exhibit abnormal gonadotropin responses during metabolic challenges, such as acute fasting, when the normal drop in gonadotropin levels is delayed. Although it is known that these food intake signals to the reproductive circuit originate in the nucleus tractus solitarius (NTS) in the brainstem, the phenotype of the neurons conveying the signal remains unknown. Given that neuropeptide FF (NPFF), another RFamide peptide, resides in the NTS and can bind to the RFRP receptor, we hypothesized that NPFF may regulate GnRH neurons. To address this question, we used a combination of techniques: cell-attached electrophysiology on GnRH-driven green fluorescent protein–tagged neurons in acute brain slices; calcium imaging on cultured GnRH neurons; and immunostaining on adult brain tissue. We found (1) NPFF inhibits GnRH neuron excitability via the RFRP receptor and its canonical signaling pathway (Gi/o protein and G protein–coupled inwardly rectifying potassium channels), (2) NPFF-like fibers in the vicinity of GnRH neurons coexpress neuropeptide Y, (3) the majority of NPFF-like cell bodies in the NTS also coexpress neuropeptide Y, and (4) acute fasting increased NPFF-like immunoreactivity in the NTS. Together these data indicate that NPFF neurons within the NTS inhibit GnRH neurons, and thus reproduction, during fasting but prior to the energy deficit.
Keywords: NPFFR1, NPFF, GnRH, fasting
In 2000, Tsutsui et al. isolated gonadotropin-inhibitory hormone (GnIH), a new member of the peptide family with a C-terminal Arg-Phe-NH2 (RFamide [RFa]) motif that inhibited pituitary gonadotropin release in birds (1). The gene coding for GnIH was discovered (2) and vertebrate orthologs were identified. The gene in mammals, Rfrp, encodes a precursor that is cleaved into active peptides: RFamide-related peptide (RFRP)-1 and RFRP-3 (3) (Fig. 1A). In rodents, whether RFRPs affect luteinizing hormone (LH) secretion by inhibiting pituitary gonadotrophs is controversial (4-7). But, RFRP neurons located in the dorsomedial hypothalamus are not hypophysiotropic (8). Instead, anatomical (4, 9, 10) and functional (11) evidence supports RFRPs inhibiting gonadotropin-releasing hormone (GnRH) neurons, which in turn control the gonadotrophs and gonadotropin release. However, the loss of RFRP receptor (neuropeptide FF [NPFF] receptor 1 [NPFFR1]) does not compromise puberty onset or fertility. Notably, 5 RFa peptide groups exist in mammals: RFRP/GnIH, NPFF, kisspeptin, prolactin-releasing peptide (PrRP), and pyroglutamylated RFa peptide (12). Each group has its cognate G protein–coupled receptor: GPR147/NPFFR1, GPR74/NPFFR2, GPR54/Kiss1R, GPR10/PrRP receptor and GPR103/pyroglutamylated RFa peptide receptor, respectively. Yet, most RFa peptides can trigger a signaling cascade through NPFFR1 and NPFFR2 (13) as well as other family member receptors. For example, intracerebroventricular RFRP-3 increases LH levels via Kiss1R (7) while intracerebroventricular RFRP-1 impairs the action of morphine (14) via GPR10/PrRP receptors (15, 16). Kisspeptin and RFRP-3 similarly excite the firing rate of unidentified neurons in Kiss1R knockout mice (17). The poor specificity of RFa receptors for its ligand makes the physiological role of each peptide difficult to assess.
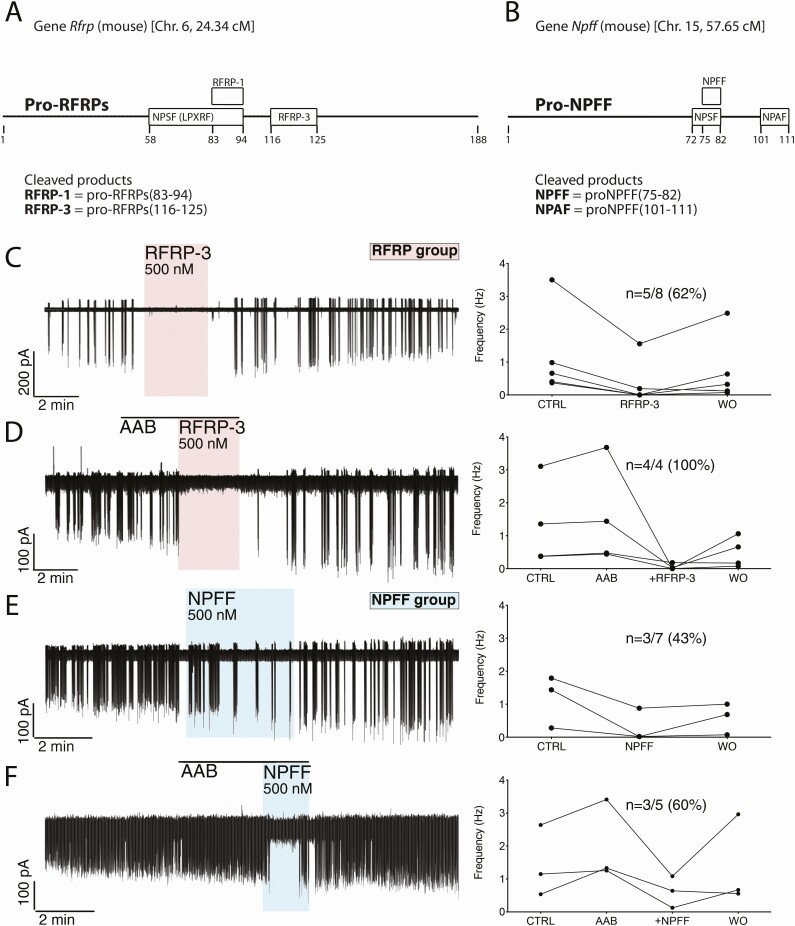
Figure 1.
RFRP-3 and NPFF inhibits GnRH neuronal activity. (A,B) Schematics of RFamide gene products and their position in the propeptides. The mouse Rfrf gene, on chromosome 6, translates into a propeptide, proRFRP, which is cleaved into 2 active peptides, RFRP-1 and RFRP-3 (A). The mouse Npff gene, on chromosome 15, translates into a propeptide, proNPFF, which is cleaved into 2 active peptides, NPFF and NPAF (B). Note that at the time of their discovery, both precursors of RFRP-1 and NPFF were named neuropeptide SF. However, 1 belongs to the LPXRF family (RFRP group), the other to the XPQRF family (NPFF group). (C-F) Left, electrophysiological recordings of adult GFP-tagged GnRH neurons in acute brain slices. Right, Summary data showing quantification of the firing rate (in Hz) in individual GnRH neurons tested during the control (CTRL), ± pretreatment AAB, treatment (RFRP-3 or NPFF), and washout (WO). The values represent the average of 1-second bins for the last 1 minute of each period. (C,E) RFRP-3 (500 nM) and NPFF (500 nM) evoked a potent decrease in GnRH neuron firing rate that was maintained in presence of amino acid blockers (AAB, D,F).
While the loss of NPFFR1 does not impair fertility, it delays the decrease in the LH level normally evoked by short-term fasting. This indicates the loss of an inhibitory pathway triggered by acute metabolic stress (18). Yet, short-term fasting does not impact Rfrp expression (19), suggesting that another member of RFa peptide family may modulate GnRH neuronal activity via NPFFR1.
The first RFa peptide, FMRFa, was isolated from clam (20). Antibodies against FMRFa expanded FMRFa-like peptides to the whole animal kingdom (21). Three mammalian FMRFa-like peptides derive from the Npff gene: NPFF, neuropeptide AF (NPAF) (15), and neuropeptide SF (22) (Fig. 1B). The best characterized role for NPFF is pain modulation (23, 24), but NPFF has many more functions, such as anti-inflammation, pain transmission, regulation of the neuroendocrine system, and energy homeostasis (25). Relevant for fertility, cell bodies immunoreactive for NPFF are present in the caudal part of the medial nucleus tractus solitarius (NTS) (26) and GnRH neurons receive inputs from the NTS, including the caudal region (27, 28). The NTS acts as a nutrient-sensing center (29), and fasting-induced inhibition of pulsatile LH secretion requires estrogen feedback in the caudal NTS (30). Yet, the identity of the NTS neurons that participate in this aspect of the reproductive circuit is unknown. Here, we investigated whether NPFF neurons may link energy homeostasis and fertility by regulating GnRH neurons directly.
Material and Methods
Animals
All procedures were approved by National Institute of Neurological Disorders and Stroke, Animal Care and Use Committee and performed in accordance with National Institutes of Health guidelines. For electrophysiology, immunostaining, and dot blot and hormone assay, male mice were selected to avoid hormonal fluctuations inherent to estrous cycle. For calcium imaging, both genders were recorded since the sex of embryos used to make explants was not determined.
Adult male mice (GnRH-green fluorescent protein [GFP], neuropeptide Y [NPY]-GFP, and C57BL/6), anesthetized with isoflurane then killed with an intraperitoneal overdose of ketamine (20 mg/20 g), were transcardially perfused with 0.1 M phosphate-buffered saline (PBS) then 4% formaldehyde in PBS. The brains were removed, postfixed in the same fixative (overnight at 4°C), then transferred to a 30% sucrose-PBS solution. The next day, the brains were frozen in dry ice and kept at –80°C until sectioning. Sectioning (40 µm) was done using a sliding freezing microtome. The hypothalamus was cut in 4 series (coronal) while the brainstem was cut in 3 series (either coronal or sagittal). Sections were kept at –20°C in cryoprotectant (31) until staining. Sections from 3 RFRP-GFP adult male rats were generously providing by Drs. Soga and Parhar (32). Each animal was divided in 3 series. For fasting experiments, sibling animals were housed together until the experiment, when they were separated into fed and fasting groups (3 animals each). To control for stress, all animals were moved to a fresh cage at 1 pm in the animal facility. Fed animals had regular chow and water while fasting animals had only water. At 5 pm, animals were brought from the animal facility to the laboratory, allowed to settle for 1 hour then switched to dark to mimic usual light extinction at 6 pm. The mice were perfused between 7 pm and 8 pm and processed as described above for staining. Another set of mice (6 animals each) underwent the same paradigm (fed vs fasting) and blood samples collected at 1 pm and 7 pm (33). The apex areas of the brainstems (~1 mm3) were collected after the last blood sample, and protein was extracted and used for dot blots (see below).
Explants were cultured as previously described (34). Briefly, embryonic day 11.5 embryos (undetermined sex) were obtained from timed pregnant NIH Swiss mice. Nasal pits were dissected under aseptic conditions in Gey’s balanced salt solution (Life Technologies, Inc., Grand Island, NY) supplemented with glucose (Sigma Chemical Co., St. Louis, MO). Explants were adhered onto coverslips by a plasma (Cocalico Biologicals, Reamstown, PA)/thrombin (Sigma) clot and maintained in a defined serum-free medium (SFM) in a humidified atmosphere at 37°C with 5% CO2. On culture day 3, SFM was replaced by fresh SFM containing fluorodeoxyuridine (2.3 µM; Sigma) for 3 days to inhibit proliferation of dividing olfactory neurons and non-neuronal explant tissue. On culture day 6, and every 2 days afterward, the medium was changed with fresh SFM. Explants were used between 6 and 11 days in vitro.
Electrophysiology
All electrophysiological experiments were undertaken using adult male GnRH-GFP mice (35). Mice were killed at approximately 10:30 am by cervical dislocation. The brain was removed from the skull and placed in ice-cold low [Ca]/high [Mg] (0.5/6 mM respectively) artificial cerebrospinal fluid (aCSF), equilibrated with 5% CO2 carbogen for at least 30 minutes. The brain was glued to the vibratome plate and conventional coronal sections (200 μm) were cut in low [Ca]/high [Mg] aCSF using a vibratome (Leica VT1000S). After sectioning, slices were incubated at 30°C in carbogenated normal aCSF containing (in mM): 118 NaCl, 3 KCl, 2.5 CaCl2, 1.2 MgCl2, 10 HEPES, 25 NaHCO3, and 11 D-glucose (pH 7.3). Individual slices were transferred into a recording chamber mounted on an upright microscope (Nikon Eclipse FN1, Tokyo, Japan) and continuously superfused with carbogenated normal aCSF maintained at 28-30°C, at a rate of approximately 2 mL/minute. Individual GnRH neurons were identified with fluorescence (20-nm narrow bandpass EGFP filter centered at 480 nm) using a 40× water immersion objective (Nikon 40×/0.80 W, WD 2.0). Visualized with a charge-coupled device camera (QImaging Retiga EXi Blue, Surrey, Canada) piloted by the open source software Micro-Manager version 1.4, the neurons were patched under fluorescence and differential interference contrast. The pipettes (3-5 MOhm) were backfilled with aCSF. Electrophysiological recordings were acquired at a rate of 10 kHz using a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA) and digitized by a Digidata (1550) analogic to digital converter (Molecular Devices).
Calcium Imaging
Experiments were performed as previously described (36). Briefly, Calcium Green-1 AM (Life Technologies) was dissolved at 2.7 mM in dimethyl sulfoxide containing 20% pluronic F-127 (Life Technologies), then diluted down to 13.5 µM in SFM, aliquoted, and kept frozen until use. Explants were incubated in warm loading solution for 20 minutes at 37°C in a 5% CO2 humidified incubator. After washes in fresh SFM, explants were mounted in a perfusion chamber (Warner Instruments, Hamden, CT) and continuously perfused at a rate of ~300 µL/minute. Calcium Green-1 was visualized using an inverted Nikon microscope, through a 20× fluorescence objective and a charge-coupled device camera (QImaging, Surrey, Canada) connected to a computer. Time-lapse recording was piloted by iVision imaging software (Scanalytics, Inc) and pictures acquired every 2 seconds. Light was generated by a SOLA light engine (Lumencor, Beaverton, OR). Excitation wavelength was defined by a medium-width excitation bandpass filter at 465 to 495 nm, and emission was monitored through a 40-nm bandpass centered on 535 nm. Calcium imaging recordings were divided into periods. When possible, the treatment period was preceded by a 5-minute control period in SFM to determine the basal level of GnRH neuronal activity. When a drug required a long pretreatment (ie, pertussis toxin, cesium), recordings started at the end of the incubation with a 5-minute period in the incubation drug. Drugs were sequentially added through multiple treatment periods. All recordings were terminated with a 40 mM KCl stimulation to ensure the viability of the cells. The phenotype of the analyzed cells was confirmed using a chromogenic immunocytochemistry against GnRH. The changes of fluorescence over time (peaks/minute) in single GnRH neurons were measured a posteriori with iVision and quantified with MATLAB (Mathworks, Natick, MA) as previously described (36). On average, explants (N = 70) contained 32.8 ± 1.9 identified GnRH neurons within the imaged field.
Immunostaining
The antibodies used are listed in Table 1.
Table 1.
Antibodies
| Peptide/Protein target | Name of antibody (RRID) | Manufacturer, catalog number, and/or name of individual providing the antibody | Species raised in; monoclonal or polyclonal | Dilution used |
|---|---|---|---|---|
| rbGnRH | SW-1 (AB_2629221) | S. Wray | Rabbit; polyclonal | 1:3000 |
| mGnRH | F1D3C5 (not available) + SMI-41 (AB_10123893) | A. Karande + Abcam | Mouse; monoclonal | 1:4000 + 1:6000 |
| NPFFR1 | ab45350 (AB_944474) | Abcam | Rabbit; polyclonal | 1:350 |
| FMRFa | ab15348 (AB_805291) | EMD Millipore | Rabbit; polyclonal | 1:1000 |
| Green fluorescent protein (GFP) | ab13970 (AB_300798) | Abcam | Chicken; polyclonal | 1:2000 |
| White-crowned sparrow GnIH | PAC 123a (AB_2617160) | G. Bentley | Rabbit; polyclonal | 1:50 000 |
| Kisspeptin | Lot # 564 (AB_2314707) | A. Caraty | Rabbit; polyclonal | 1:5000 |
| Neuropeptide Y | ab10980 (AB_297635) | Abcam | Rabbit; polyclonal | 1:15 000 |
| Neurophysin-AVP | PS41, PS45, PS46 (not available) | H. Gainer | Mouse; monoclonal | 1:100 |
| Tyrosine hydroxylase (TH) | Anti-Tyrosine Hydroxylase (AB_10013440) | Aves | Chicken; polyclonal | 1:1000 |
| Cholecystokinin (CCK) | CCK-8 (AB_572224) | ImmunoStar | Rabbit; polyclonal | 1:4000 |
| Estrogen receptor alpha | C1355 (AB_310305) | Millipore | Rabbit; polyclonal | 1:5000 |
| c-fos | na | MJ Iadarola | Rabbit; polyclonal | 1:1000 |
| pERK (phospho-p44/42 MAPK) | 4370 (AB_2315112) | Cell Signaling Technology | Rabbit; polyclonal | 1:200 |
Abbreviations: DAB, 3,3′-diaminobenzidine; DAC, donkey antichicken;DAM, donkey antimouse; DAR, donkey antirabbit; TSA, tyramine signal amplification.
Explants
After calcium imaging, explants were immunostained for GnRH to verify the phenotype of the recorded cells (36). Briefly, explants were fixed (4% formaldehyde, 30 minutes at room temperature) and placed in cryoprotectant until staining. After removing cryoprotectant in PBS, the explants were incubated in blocking solution (10% normal horse serum + 0.3% Triton X-100 (TX-100), 1 hour), washed in PBS then in primary antibody (overnight at 4°C, rbGnRH SW1 (37); Table 1). The next day, explants were washed in PBS, incubated for 1 hour with biotinylated secondary donkey antirabbit antibody (1:500 in PBS/0.3% TX-100; Vector Laboratories, Inc.), washed in PBS, and processed for avidin–biotin horseradish peroxidase (HRP)/3,3′-diaminobenzidine (DAB). For labeling of NPFFR1 on GnRH cells, double fluorescence was performed. Explants were processed as described above with the following changes. The blocking solution contained 0.1% Tween-20, incubation in the first primary antibody (rabbit anti NPFFR1) was for 2 nights at 4°C, and Alexa Fluor 555-conjugated secondary donkey antirabbit antibody was used (1:1000 in PBS/0.1% Tween-20; Vector Laboratories, Inc.). Explants were refixed, washed, and incubated (2 nights at 4°C) in the second primary antibody (mGnRH: F1D3C5 + SMI 41; Table 1), which was visualized with Alexa Fluor 488-conjugated secondary donkey antimouse antibody (1:1000 in PBS/0.1% Tween-20; Vector Laboratories, Inc.) and coverslipped with an antifade mounting solution (Electron Microscopy Sciences, Hatfield, PA).
Brain sections
Tissues were removed from cryoprotectant, washed several times in PBS, incubated for 1 hour in a blocking solution (10% normal horse serum + 0.3% TX-100), washed several times in PBS, and incubated in primary antibodies. Series from at least 3 different mice were used for each staining. Single labeling of sections was done as described above using nickel-enhanced avidin–biotin HRP/Ni-DAB.
Double immunofluorescent staining (primary antibodies from different species)
Sections were incubated (2 nights, 4°C) in the first primary antibody (FMRFa, GnIH, or NPY). The next day, sections were washed in PBS and incubated (1.5 hours) with Alexa Fluor 555-conjugated secondary donkey antirabbit antibody (1:1000 in PBS/0.3% TX-100). After several PBS washes, sections were fixed with 0.1 M PBS + 4% formaldehyde, washed, and incubated (2 nights, 4°C) in the second primary antibody (GFP or tyrosine hydroxylase [TH]). The next day, sections were washed in PBS and incubated (1.5 hours) with Alexa Fluor 488-secondary donkey antichicken antibody (1:1000 in PBS/0.3% TX-100). After several washes in PBS and water, sections were coverslipped with an antifade mounting solution.
Double or triple immunofluorescent staining (2 primary antibodies from same species)
To avoid any cross-reactivity between the 2 primary antibodies from the same species (GnIH, kisspeptin, cholecystokinin [CCK], NPY, c-fos, pERK, and FMRFa), staining was completed using tyramide signal amplification (TSA) (31, 36, 38). All incubations in primary antibodies were for 2 nights at 4°C and all secondary antibodies were for 1.5 hours. Briefly, sections were incubated in the first primary antibody (GnIH, kisspeptin, CCK, NPY, c-fos, or pERK). The next day, sections were washed in PBS, incubated in biotinylated secondary donkey antirabbit antibody (1:500 in PBS/0.3% TX-100), washed in PBS, then incubated (45 minutes) in avidin–biotin complex solution (1:500 in PBS/0.3% TX-100; Vector Laboratories, Inc.). After washes, sections were incubated in 0.5% biotinylated tyramine + 0.005% H2O2 for 20 minutes (Perkin Elmer TSA kit, Waltham, MA), washed, and incubated (2-3 hours) in Texas Red-conjugated streptavidin (1:200; Perkin Elmer). Sections were washed, fixed (0.1 M PBS + 4% formaldehyde), washed, and incubated in the second primary antibody (FMRFa). The next day, sections were washed in PBS, incubated with Alexa Fluor 647-secondary goat or donkey antirabbit antibody (1:1000 in PBS/0.3% TX-100). If proceeding for triple labeling, after several washes, sections were incubated in a third primary antibody (GFP). The next day, sections were washed in PBS, incubated with Alexa Fluor 488–conjugated donkey antichicken antibody (1:1000 in PBS/0.3% TX-100). After several washes in PBS and water, sections were coverslipped with an antifade mounting solution.
Unless stated otherwise, all immunofluorescent pictures were taken using spinning disk confocal (Yokogawa, Sugar Land, Texas) microscopy (Nikon Eclipse TE-200) though a 60× water immersion objective (Nikon Plan Apo 60X, NA 1.2, WD 0.27), captured with a high sensitivity camera (EM-CCD, Hamamatsu Photonics, Japan) and presented as flatten confocal stack or single focal plan.
NPFF antibody specificity
Neurons in the NTS express NPFF (39) and not RFRP (12, 40). Thus, brainstem sections were used to determine the specificity of the FMRFa antibody for NPFF. One series was incubated in the first primary antibody (FMRFa, 2 nights, 4°C). The next day, sections were washed in PBS, incubated (1.5 hours) with Alexa Fluor 555-conjugated secondary donkey anti-rabbit antibody (1:1000 in PBS/0.3% TX-100). The other series was incubated with FMRFa preabsorbed with 10 µM NPFF and stained in parallel. After several washes in PBS and water, sections were coverslipped with an antifade mounting solution with DAPI counterstaining.
To control for immunostaining with primary antibodies from same species (rabbit) using the TSA method, brainstem sections stained with the first primary antibody, NPY, were incubated, or not, with the second primary antibody, FMRFa. Only the sections incubated in FMRFa displayed colocalization, ruling out cross-reactivity of the second secondary antibody with the first primary antibody.
Although cross-reactivity between first primary antibody and second secondary antibody was ruled out with the TSA method, some FMRFa antibodies can cross-react with NPY (41, 42). Coronal forebrain sections from NPY-GFP mice were then stained with FMRFa or NPY. While NPY-GFP cortical cells did not display NPFF-like immunoreactivity, they were immunoreactive for NPY, ruling out cross-reactivity of our FMRFa antibody with NPY. Thus, our anti-FMRFa antibody was NPFF specific.
Semi-quantification of NPFF-like immunoreactivity in the NTS
Fed and fasted animals, analyzed in pairs, were stained simultaneously. Three NTS sections from each fed animal were matched with 3 NTS sections from a fasted animal. Analysis was done in FIJI as follows (43). Paired images were aligned and converted into 8-bits (0 = black, 255 = white). The NTS was delimited manually using the NPFF-like immunoreactivity in a fed animal section. The NTS region was duplicated on the matched fasted animal section. The intensity threshold was defined in fed animals to eliminate the background and applied to the matched fasted animals. The mean gray value and the total area (below threshold) were measured. The mean gray value was converted into a mean staining intensity value (0 = white, 255 = black) by subtracting mean gray value to 255.
Dot Blot
Tissue samples were rinsed in chilled PBS, lysed in RIPA buffer (Millipore, Burlington, MA) containing a protease inhibitor cocktail (Roche Basel, Switzerland) via ice-cold sonication. Samples were then centrifuged at 13 000 rpm for 20 minutes at 4°C, and protein concentration was determined with a BCA protein assay (Pierce BCA protein assay kit, Thermo Fisher). Each lysate sample was diluted to 0.2 μg/μL and 0.1 μg/μL. Samples were aliquoted and stored at –20°C until blotting.
For dot blots, 1 μL of protein samples at both dilutions was pipetted onto a dry nitrocellulose membrane (Bio-rad). Each sample was dotted twice onto 2 separate membranes. As a control, 1 μL recombinant proNPFF peptide (50 μg/mL and 25 μg/mL, CloudClone [RPG003Mu01]) was dotted onto the nitrocellulose membrane. Membranes were then air-dried for 30 minutes and blocked with 5% milk-TBST (room temperature, 1 hour). After brief Tris-buffered saline with 0.1% Tween 20 washes, membranes were transferred into antibodies (rabbit anti-proNPFF, 1:500, CloudClone [PAG003Mu02]; mouse anti-β-actin, 1:1000, Sigma) diluted in 5% BSA-TBST buffer and incubated overnight at 4°C with gentle agitation. The next day, after TBST washes, the membranes were incubated (room temperature, 1 hour) in secondary antibodies conjugated with HRP with gentle agitation (goat antirabbit-HRP, 1:20 000 (Promega, Madison, WI); donkey antimouse-HRP, 1:5000 (Jackson Immunoresearch, West Grove, PA), respectively). Blots were reacted using standard ECL (Clarity Max, Bio-rad, Hercules, CA) procedure and visualized using a ChemiDoc imaging system (Bio-rad).
Quantification
The background was automatically subtracted in FIJI using a rolling ball radius of 25 pixels and the integrated density of each dot was measured (43). The linear range of detection for the target (proNPFF) and internal loading control (β-actin) was assessed by the ratio of integrated density (0.2 μg/μL/0.1 μg/μL) (2.0 ± 0.13 for β-actin and 1.5 ± 0.12 for proNPFF, n = 24 dots). The integrated density of proNPFF was then normalized to the integrated density of β-actin.
Hormone Assay
Blood samples were assayed for LH in the same enzyme-linked immunosorbent assay (ELISA) with a sensitivity of 0.016 ng/mL and intra-assay coefficient of variation <3.1%. The ultrasensitive ELISA used has been reported previously (33) and was performed by the University of Virginia Center for Research in Reproduction, Ligand Assay and Analysis Core, supported by the Eunice Kennedy Shriver NICHD/NIH (Grant R24HD102061).
Drugs
NPAF (4-11), NPFF, 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; AMPA/kainate receptor antagonist), D-2-amino-5-phosphono-pentanoate (AP5; NMDA receptor antagonist), (-)-bicuculline methiodide (BIC; GABAA receptor antagonist), tertiapin-Q (TPNQ; blocker of G protein–coupled inwardly-rectifying potassium channel), and pertussis toxin (PTX; uncoupling Gi/o protein coupled receptor) were purchased from Tocris Bioscience (Bristol, UK). RFRP-3 (3-10), RFRP-1 (4-15), and prolactin-releasing peptide-20 (PrRP-20) were purchased from Phoenix (Burlingame, CA). Cesium chloride (Cs; nonspecific blocker of G protein–coupled inwardly-rectifying potassium channel) and phorbol 12-myristate 13-acetate (PMA; protein kinase C activator) were purchased from Sigma-Aldrich. BIBP3226 (a mixed Y1R/NPFF receptor antagonist) were obtained from Bachem Bioscience Inc. (Torrance, CA). dNPA was a gift from Dr. J.-M. Zajac (Institut de Pharmacologie et de Biologie Structurale, CNRS, Université de Toulouse, France). All stock solutions (1000 or 500×) were stored at –20°C and diluted prior each experiment at the specified concentration in SFM. The peptide sequences are listed in Table 2.
Table 2.
Peptide sequences
| RFRP group (LPXRFamide) | ||
|---|---|---|
| Agonist [relative position in final peptide (Mouse)] | Absolute position in propeptide | Sequence |
| RFRP-3 (3–10) [SHFPSLPQRF-NH2] | Pro-RFRPs (116–125) | Phe-Pro-Ser-Leu-Pro-Gln-Arg-Phe-NH2 |
| RFRP-1 (4–15) [ANKVPHSAANLPLRF-NH2] | Pro-RFRPs (83–94) | Val-Pro-His-Ser-Ala-Ala-Asn-Leu-Pro-Leu-Arg-Phe-NH2 |
| NPFF group (XPQRFamide) | ||
| Agonist (relative position in final peptide [mouse]) | Absolute position in propeptide | Sequence |
| NPFF [FLFQPQRF-NH2] | Pro-NPFF (75–82) | Phe-Leu-Phe-Gln-Pro-Gln-Arg-Phe-NH2 |
| NPAF-like (4–11) [QFWSLAAPQRF-NH2] | Pro-NPFF (101–111) | Ser-Leu-Ala-Ala-Pro-Gln-Arg-Phe-NH2 |
| Other | ||
| Prolactin-releasing peptide-20 (PrRP-20) | Thr-Pro-Asp-Ile-Asn-Pro-Ala-Trp-Tyr-Thr-Gly-Arg-Gly-Ile-Arg-Pro-Val-Gly-Arg-Phe-NH2 | |
| dNPA (Synthetic NPFF2R agonist) | D.Asn-Pro-(N-Me)Ala-Phe-Leu-Phe-Gln-Pro-Gln-Arg-Phe-NH2 |
Bold represents either LPXRF or XPQRF sequence, whereas italics represents the X.
Statistical Analysis
For electrophysiology, loose-patch patch clamp configuration, the RFa-induced inhibition was determined as follows: action potentials (APs) were detected with Clampfit 10 on continuous recordings and the firing frequency (Hz) was determined by summing APs into 1-second bins for each cell (n, Fig. 1). The percentage of inhibition was calculated as [(average frequency before RFa) – (average frequency during RFa)]/(average frequency before RFa) × 100. Cells showing a change in average frequency greater than 25%, were considered as responders.
For calcium imaging, values from individual cells (n) originating from at least 3 independently recorded explants (N) were combined for each paradigm and used for statistical analysis. In Fig. 2 and Table 3, frequencies of calcium oscillations are expressed as mean ± standard error of the mean and Student’s paired t-test was used to assess the effectiveness of RFa between only 2 consecutives time periods. The inhibition was determined for each cell and expressed as % of the pre-RFa period as following: [(peaks/minute during RFa) – (peaks/minute before RFa)]/(peaks/minute before RFa) × 100 (Table 4). Comparisons of the strength of inhibition between 2 groups of cells were done with 1-way analysis of variance (ANOVA) and post hoc Dunnett’s multiple comparisons test, using each RFa peptide (RFRP or NPFF alone) as its reference for perturbations (RFa + PTX or + Cs or + PMA) (Figs. 3 and 4). The proportions of cells responding to RFa with inhibition >50% were compared using Fisher’s exact test.
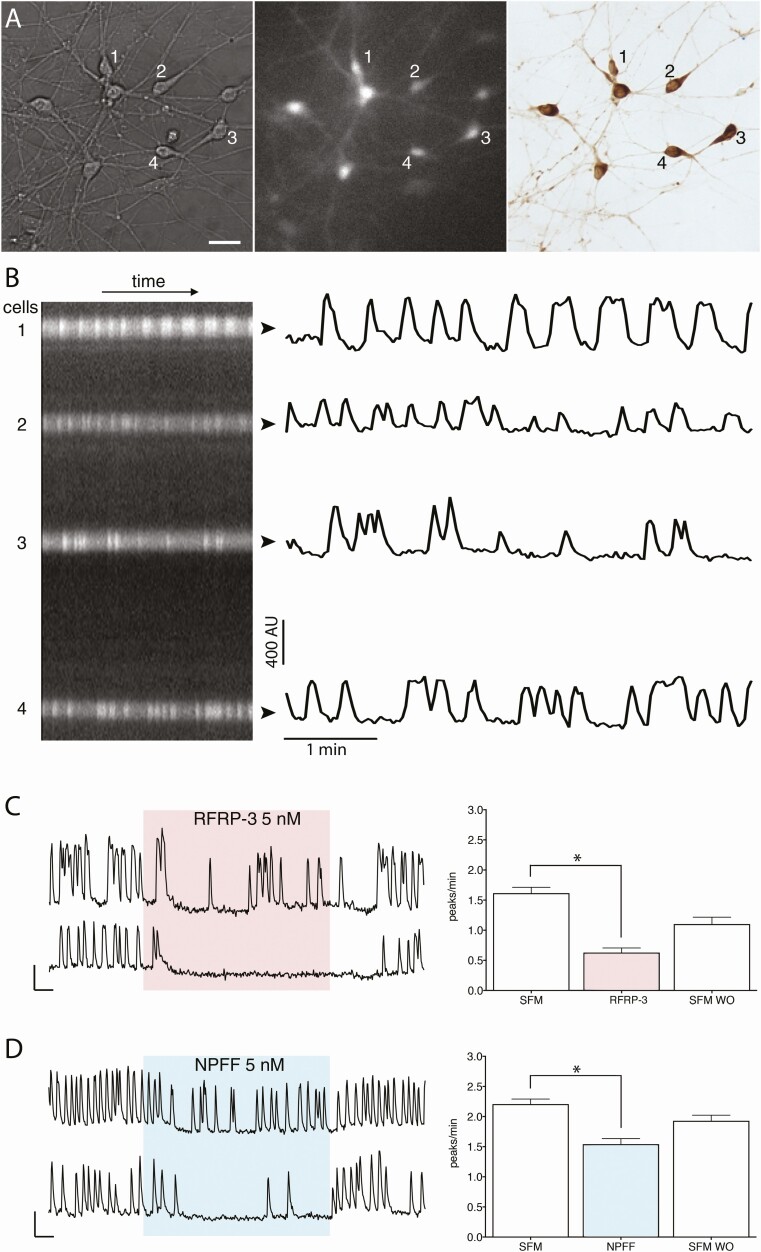
Figure 2.
Imaging of intracellular calcium oscillations in GnRH neurons in explants. (A) GnRH neurons maintained in explants were identified by their fusiform shape (left panel), loaded with calcium green 1 AM (middle panel) and phenotypically confirmed immunochemically post hoc (right panel)(bar 25 µm). (B) Left, kymograph from the cells numbered in A, and, right, corresponding fluctuating optical densities evoked by calcium oscillations. (C,D) Left, 2 representative traces showing calcium oscillations in GnRH neurons after application of RFRP-3 (5 nM) and NPFF (5 nM). Right, Quantification from independent experiments. Both RFRP-3 and NPFF evoked a decrease in the frequency of calcium oscillations. Asterisks indicate P < .05 (paired t-test).
Table 3.
Frequencies of calcium oscillations in GnRH neurons
| Paradigms with RFamides | Period 1 | Period 2 | Period 3 | Period 4 | P value (between consecutive bold cells) | Cells (n) | Explants (N) | |
|---|---|---|---|---|---|---|---|---|
| Frequencies in peaks/min | ||||||||
| RFRP group | ||||||||
| a | SFM – RFRP-3 – SFM | 1.62 ± 0.10 | 0.63 ± 0.08 | 1.10 ± 0.11 | — | <.0001 | 90 | 3 |
| b | SFM – RFRP-1– SFM | 1.53 ± 0.07 | 0.51 ± 0.05 | 0.81 ± 0.06 | — | <.0001 | 131 | 3 |
| c | SFM – AAB – AAB+ RFRP-3 – AAB | 1.81 ± 0.12 | 0.77 ± 0.08 | 0.43 ± 0.08 | 0.51 ± 0.09 | <.0001 | 72 | 3 |
| NPFF group | ||||||||
| d | SFM – NPFF – SFM | 2.21 ± 0.08 | 1.54 ± 0.09 | 1.93 ± 0.09 | — | <.0001 | 124 | 3 |
| e | SFM – NPAF – SFM | 1.99 ± 0.10 | 1.78 ± 0.10 | 2.01 ± 0.10 | — | .0002 | 96 | 3 |
| f | SFM – AAB – AAB+NPFF – AAB | 1.74 ± 0.10 | 1.07 ± 0.09 | 0.60 ± 0.07 | 0.87 ± 0.08 | <.0001 | 86 | 3 |
Bold indicates the 2 consecutive cells that were statistically analyzed.
Table 4.
Percentage of inhibition in GnRH neurons
| Paradigms (per ii –per i) | Inhibition (%) [(per ii –per i)/per i × 100] | P value | Cells (n) | Explants (N) | |
|---|---|---|---|---|---|
| GnIH group | |||||
| a | SFM – RFRP-3 | –58.70 ± 4.34 | Reference | 90 | 3 |
| b | BIBP – BIBP+ RFRP-3 | –36.36 ± 4.14* | .0021 | 127 | 3 |
| c | PTX – PTX+ RFRP-3 | 4.73 ± 6.17* | .0001 | 71 | 3 |
| d | Cs – Cs + RFRP-3 | 3.57 ± 10.83* | .0001 | 30 | 3 |
| e | TPNQ – TPNQ + RFRP-3 | –63.95 ± 2.73 | .8888 | 115 | 3 |
| f | PMA – PMA + RFRP-3 | –15 ± 3.78* | .0001 | 183 | 4 |
| NPFF group | |||||
| g | SFM – NPFF | –31.20 ± 4.41 | Reference | 122 | 3 |
| h | BIBP – BIBP + NPFF | –7.23 ± 4.29* | .0001 | 111 | 3 |
| i | PTX – PTX + NPFF | –8.36 ± 4.37* | .0002 | 99 | 3 |
| j | Cs – Cs+NPFF | 5.90 ± 3.50* | .0001 | 177 | 3 |
| k | TPNQ – TPNQ+NPFF | –28.24 ± 3.09 | .9759 | 108 | 3 |
| l | PMA – PMA+NPFF | –9.81 ± 5.26* | .0166 | 42 | 3 |
| m | PMA – PMA | –21.97 ± 4.16 | 128 | 3 |
*Statistical significance.
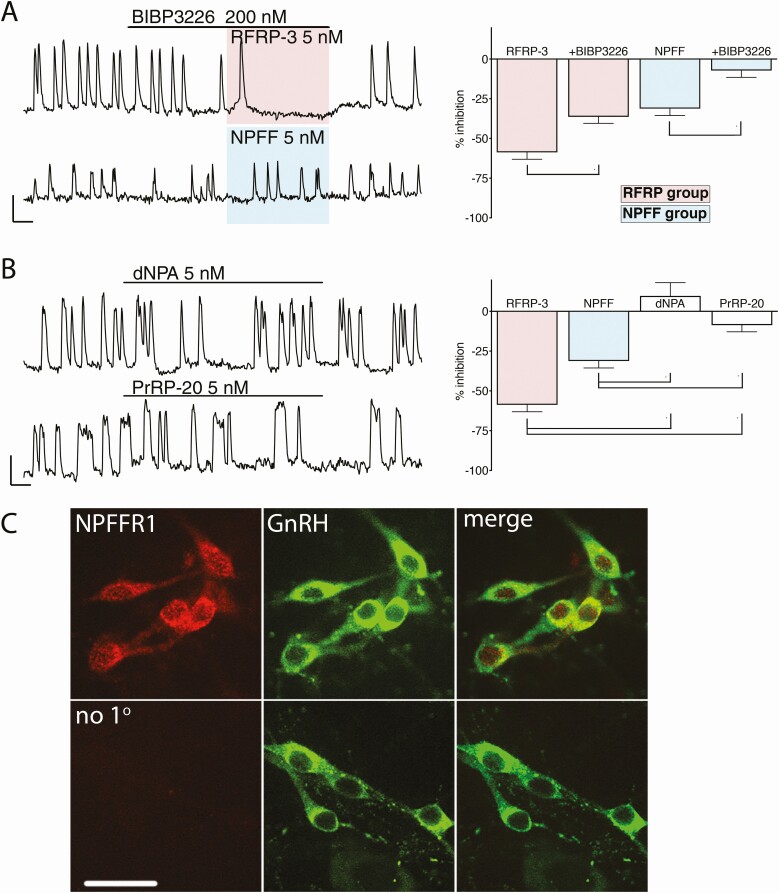
Figure 3.
Inhibition evoked by peptides from RFRP-3 and NPFF is mediated by NPFF1R. (A) Left, representative traces showing the inhibition of GnRH neuronal activity evoked by RFamide peptides from RFRP-3 and NPFF in presence of BIBP3226, a compound with preferential antagonistic activity for NPFF1R. Right, the magnitude of the inhibition with BIBP is compared with the magnitude of the inhibition without BIBP. Asterisks indicate P < .05 (1-way ANOVA, and post hoc Dunnett’s multiple comparisons test). However, the reduction in inhibition was more dramatic for NPFF than RFRP-3, indicating that the receptor involved has a greater affinity for RFPR-3 than NPFF, consistent with action via NPFFR1. (B) Left, representative trace showing the lack of effect of compounds with preferential agonistic activity for NPFF2R, dNPA, or PrRP-20, on GnRH neuronal activity. Right, The magnitude of the effect with dNPA and PrRP-20 is compared to the magnitude of the inhibition with RFRP-3 and NPFF. Asterisks indicate P < .05 (1-way ANOVA and post hoc Dunnett’s multiple comparisons test). Neither compound produced significant changes in GnRH neuronal activity, consistent with the lack of expression of NPFF2R on GnRH cells. (C) Upper panel, representative confocal images showing double immunofluorescent labeling for NPFF1R (left, red) and GnRH (middle, green). Images are merged on the right. Lower panel, omission of primary antibody against NPFF1R gave no staining in GnRH neurons (bar 20 µm).
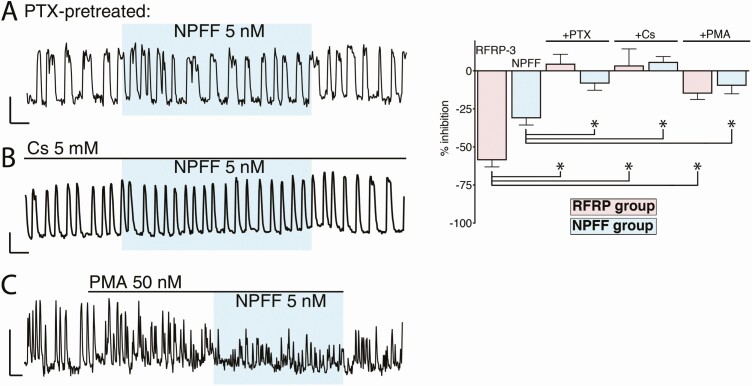
Figure 4.
The inhibition evoked by RFamide peptides from RFRP-3 and NPFF relies on Gi protein and G protein–coupled inwardly-rectifying potassium channels. Left, representative traces showing the lack of inhibition with NPFF on GnRH neurons pretreated with pertussis toxin (PTX, in A), cesium (Cs, in B) or phorbol 12-myristate 13-acetate (PMA, in C). Right, the magnitude of the effect of RFRP-3 and NPFF on GnRH neurons pretreated with PTX, Cs and PMA compared with the magnitude of the inhibition without drugs. Asterisks indicate P < .05 (1-way ANOVA and post hoc Dunnett’s multiple comparisons test). The amplitude of calcium oscillations decreased with PMA leading to a failure in peak detection rather than a true decrease in neuronal activity.
For staining intensity, anatomically paired sections (fed/fasted) were used and values compared using a Wilcoxon matched-pairs signed rank test. For dot blots, values from brainstem proteins from fed/fasted mice, 2 concentrations, were compared using a 2-way ANOVA and Sidak’s multiple comparisons test. For ELISA, LH levels at 7 pm were compared using the nonparametric Mann–Whitney test.
In all experiments, statistically significant differences were defined as P < .05.
Results
RFRP-3 and NPFF Inhibit GnRH Neurons
To assess the effect of RFa peptides, electrophysiological recordings were performed on acute slices from adult mice treated with RFRP-3 (500 nM) and NPFF (1 μM and 500 nM). An arbitrary cut-off of a 25% decrease in firing was used to define inhibition. In agreement with Ducret et al. (11), RFRP-3 inhibited GnRH neurons by 87% (5/8; N = 4; Fig. 1C) and the inhibition persisted after the removal of GABAergic and glutamatergic inputs, the major inputs to GnRH neurons (44), with amino acid blockers (~90%; 4/4, N = 1; Fig. 1D). NPFF also inhibited GnRH neurons (3/3 at 1 μM (N = 2) and 3/7 at 500 nM (N = 3; Fig. 1E) by ~78% and ~81%, respectively). The NPFF-mediated inhibition (500 nM) also remained with AAB (~70%; 3/5, N = 1; Fig. 1F). To further characterize the NPFF-mediated inhibition of GnRH neurons, calcium imaging of primary GnRH neurons maintained in explants was performed (Fig. 2A, B).
As in acute slices, RFRP-3 (Fig. 2C) and NPFF (Fig. 2D) inhibited GnRH neurons in explants (Table 3, rows a and d, respectively; paired t-test, P < .05). The other members of the 2 families, RFRP-1 and NPAF, also inhibited GnRH neuronal activity (Table 3, rows b and e, respectively; paired t-test, P < .05). The inhibition evoked by RFRP-3 or NPFF was independent of GABAergic and glutamatergic inputs (Table 3, rows c and f; paired t-test, P < .05). The Rfrp-derived peptides appeared more effective in inhibiting GnRH neurons than Npff-derived peptides. To further examine this, every cell was normalized for the degree of inhibition evoked by a low dose (5 nM) of each RFa to its initial oscillation rate. The frequency distribution of these values was then used to determine the proportion of GnRH neurons showing an inhibition >50% for each treatment. Both Rfrp peptides produced a >50% inhibition of GnRH neuronal activity in the majority of cells (RFRP-3: 74% [64/87 cells]; N = 3; RFRP-1: 83% [106/127 cells]; N = 3: RFRP-3 vs RFRP-1; Fisher’s Exact Test, P > .05). NPFF inhibition was greater than NPAF inhibition (NPFF: 34% [40/119 cells]; N = 3; and NPAF: 16% [14/89 cells]; N = 3; NPFF vs. NPAF; Fisher’s Exact Test, P < .05), but the responses to either peptide were not as potent as the responses to the 2 Rfrp peptides (RFRP-3 vs NPFF; Fisher’s exact test, P < .05). Together, these data indicate a specific sensitivity of GnRH neurons to peptides from these 2 groups with RFRP-3 = RFRP-1 > NPFF > NPAF. To evaluate which receptor subtype peptides from the NPFF group inhibited GnRH neurons and the relevance of the inhibition, pharmacological and anatomical experiments were performed.
RFa Peptide from Both Groups Transduce Inhibition via NPFFR1
Pharmacological experiments were performed using primary GnRH cells in explants.
Unfortunately, NPFFR1/GPR147 and NPFF2/GPR74 lack specific agonists and antagonists. In fact, most RFa display binding affinity to NPFFR1 and NPFFR2 and can trigger a signaling cascade (13). Even RFRP-3 and NPFF, the endogenous ligands for NPFFR1 and for NPFFR2 respectively, only slightly prefer their receptors (45). The supposedly specific NPFFR2 antagonist RF9 (46) also acts as a ligand to Kiss1R (47). Since GnRH neurons express Kiss1Rs, RF9 potently excites the cells and is unusable (48). Thus, to identify the RFa receptor involved in the inhibition of GnRH neurons by NPFF peptides, 3 pharmacological agents were used for calcium imaging studies to examine the presence of NPFFR1.
For all pharmacological experiments, we used RFRP-3 and NPFF at 5 nM. BIBP3226 is a NPY receptor subtype 1 antagonist that also antagonizes NPFFR1 > NPFFR2 (45). While GnRH neurons in situ are regulated by NPY inputs, explants do not contain NPY therefore BIBP3226 has no effect on its own (49). First, BIBP3226 was applied alone (200 nM) and then a RFa ligand added. BIPB3226 reduced the inhibition of both RFas on GnRH neuronal activity (Fig. 3A and Table 4, rows a vs b and g vs. h, both P < .05; 1-way ANOVA, and post hoc Dunnett’s multiple comparisons test). However, BIBP3226 reduced the inhibition from NPFF by ~4 times and from RFRP-3 by ~1.6 times. Since the antagonism of BIBP3226 was weaker for RFRP-3 than NPFF, we can conclude that the receptor involved has a greater affinity for RFPR-3 than NPFF, consistent with action via NPFFR1. Next, 2 different agonists that favor NPFFR2: dNPA (5 nM (50)) and PrRP-20 (5 nM (51)) were used. Neither of them mimicked the inhibition of RFRP-3 or NPFF on GnRH neurons (Fig. 3B; 1-way ANOVA and post hoc Dunnett’s multiple comparisons test). These data are consistent with GnRH neurons lacking NPFFR2. To verify the pharmacological data, explants (N = 3) were stained using an antibody directed against NPFFR1. The majority of GnRH cells were immunoreactive (Fig. 3C). Together, these independent datasets indicate that NPFFR1 transduces signals from both RFRP and NPFF in GnRH neurons.
As predicted (36, 52-54), NPFF signaling, like RFRP signaling, required Gi/o protein and G protein–coupled inwardly-rectifying potassium channels. Both RFa inhibitions were attenuated by PTX (which uncouples Gi/o protein coupled receptors; Fig. 4A; Table 4, rows a vs c and g vs i; 1-way ANOVA, and post hoc Dunnett’s multiple comparisons test), Cs (a nonspecific blocker of inwardly rectifying potassium channels; Fig. 4B and Table 4, rows a vs d and g vs j; 1-way ANOVA and post hoc Dunnett’s multiple comparisons test) and PMA (protein kinase C activator; Fig. 4C and Table 4, rows a vs e and g vs l; 1-way ANOVA and post hoc Dunnett’s multiple comparisons test).
Anatomy of the NPFF–GnRH Circuit
Anatomical experiments were performed on brain slices from adult mice.
Due to the similarity of the peptides in this family, an antibody against the first RFa peptide isolated in mollusk, FMRFa, was used and the immunoreactivity referred to as NPFF-like (39, 55). NPFF-like immunoreactive fibers surrounded and apposed GnRH neurons that were caudal to the organum vasculosum laminae terminalis (Fig. 5A and 5B). While RFRP fibers surrounding GnRH neurons were detected with GnIH antiserum (consistent with (9)), the NPFF-like fibers were not immunostained with GnIH antiserum (Fig. 5C).
Figure 5.
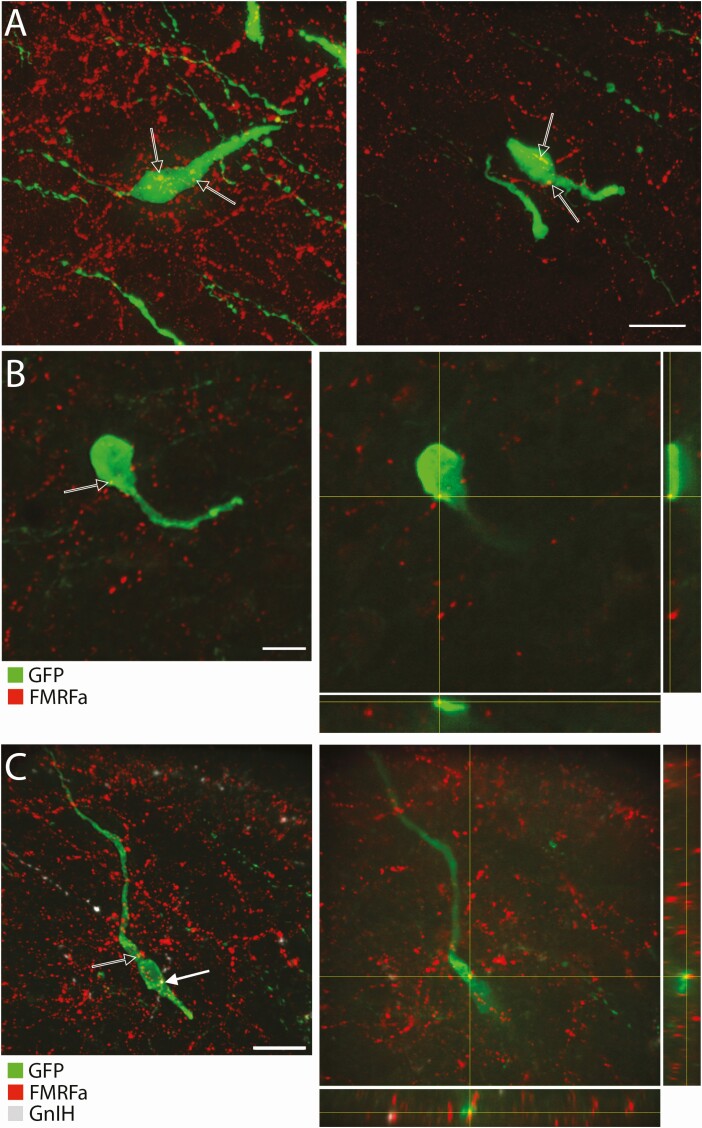
NPFF-like fibers appose to GnRH neuron cell bodies. (A) Flattened confocal z-stack images showing examples of NPFF-like (FMRFa, red) immunoreactive fibers around GnRH neurons (green)(bar 20 µm). Arrows point to varicosities close to GnRH cells. (B) Left, flattened confocal z-stack image corresponding to the focal plan on the right. Right, focal plan and orthogonal views where black arrow points to NPFF-like immunoreactive fiber (FMRFa, red) apposing a GnRH neuron (green) (bar 10 µm). (C) Left, flattened confocal z-stack image corresponding to the focal plan on the right. Right, focal plan and orthogonal views where black arrow and white arrow point, respectively, at NPFF-like (FMRFa, red) and RFRP (GnIH, white) immunoreactive fibers apposing a GnRH neuron (green) (bar 20 µm).
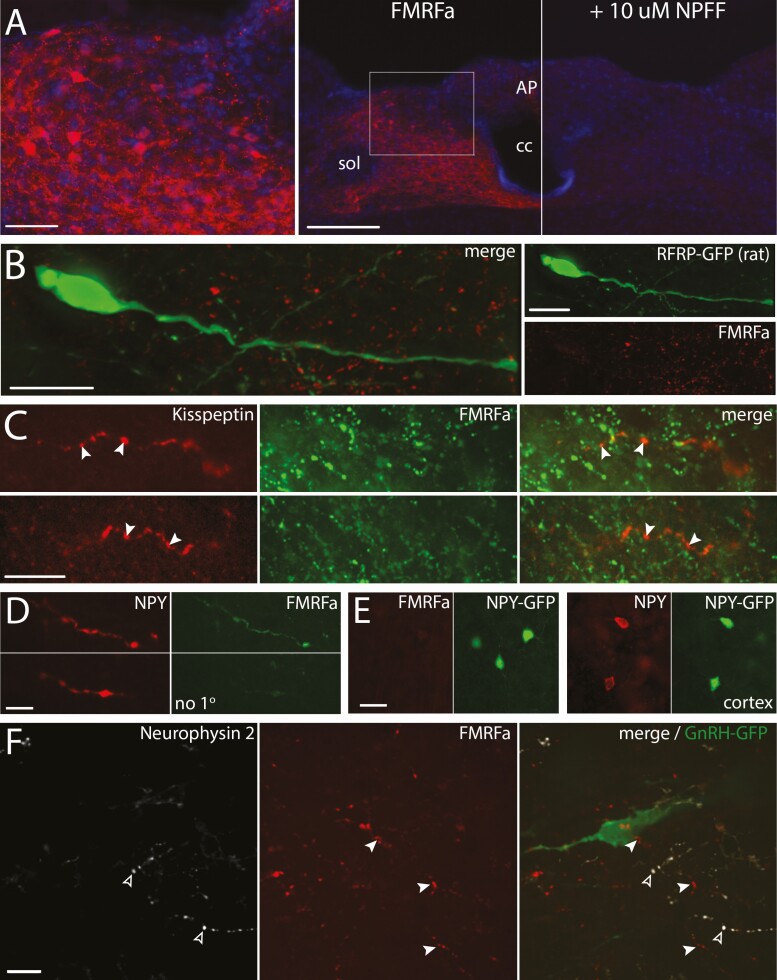
Antibodies directed against FMRFa are more specific than antibodies developed against the C-term of NPFF, which is shared among RFa peptides, but still require caution (56). As such, a series of controls was performed to establish the specificity of this antibody for NPFF. NPFF immunoreactive cell bodies were identified in the NTS and preabsorption of the antibody with NPFF prevented this immunoreactivity (Fig. 6A). In addition, the anti-FMRFa antibody did not highlight GFP-labeled RFRP neurons in the dorsomedial hypothalamus (Fig. 6B) nor kisspeptinergic fibers in the arcuate nucleus (Fig. 6C). Neither testis or kidney tissues were stained with anti-FMRFa antibody, both tissues showing the presence of mRNA for PrRP, but not NPFF (57). Technical antibody cross-reactivity with TSA (Fig. 6D) and anti-FMRFa antibody cross-reactivity with NPY (Fig. 6E) were also ruled out.
Figure 6.
Controls for FMRFa antibody specificity. (A) The FMRFa antisera highlights cell bodies in the dorsal part of the caudal NTS (AP, area postrema; cc, central canal; sol, solitary tract). Left, high magnification confocal image of the boxed area on the middle panel showing cell bodies immunoreacted with FMRFa antisera and counterstained with DAPI (bar 50 µm). Middle, low magnification image containing the left panel, obtained with conventional light microscope (bar 200 µm). Right, low magnification image corresponding to the section in the middle panel, in which the FMRFa antisera was preabsorbed with 10 µM NPFF. (B) Confocal imaging showing the NPFF-like immunoreactivity (FMRFa, red) does not stain RFRP-GFP neurons (green)(rat). Left, merged color channels. Right, individual color channels (bars 20 µm). (C) Confocal imaging showing that the NPFF-like immunoreactivity (FMRFa, green) does not stain kisspeptinergic fibers emerging from the side of the arcuate nucleus (green) (bars 20 µm). (D) Confocal images showing coimmunoreactivity between NPY (red) and NPFF-like (FMRFa, green) in the brainstem. The omission of FMRFa antibody gives no staining in NPY stained fibers with TSA method (bar 10 µm). (E) Confocal images showing no cross-reactivity of the FMRFa antibody with NPY. Left, absence of immunoreactivity with FMRFa in GFP positive neurons in the cortex. Right, presence of immunoreactivity with NPY in GFP positive neurons in the cortex. (bar 20 µm) (F) Flattened stack of confocal images showing no immunoreactivity for neurophysin 2, protein carrier for AVP (white, open arrowheads) in NPFF-like immunoreactive fibers (red, filled arrowheads) surrounding GnRH neuron (green). Left and Middle, individual color channels for neurophysin and FMRFa. Right, merge with GnRH staining.
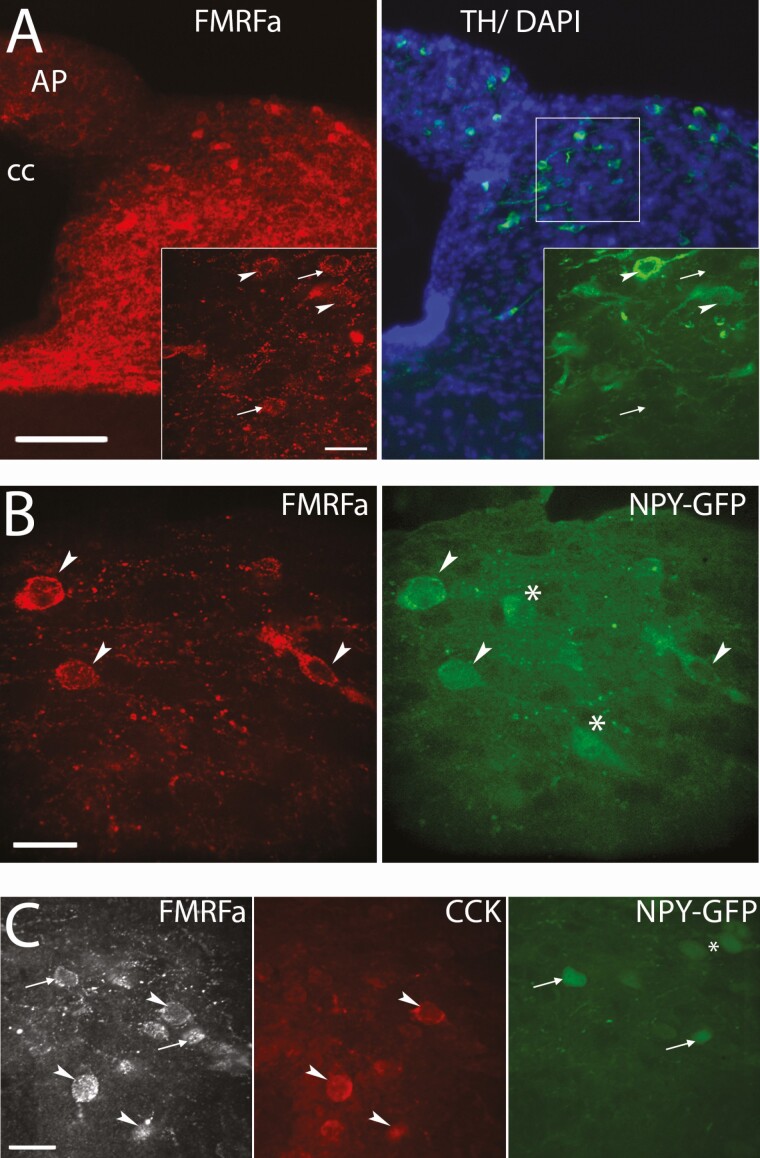
Since NPFF is coexpressed with vasopressin in magnocellular hypothalamic neurons (58), the origin of the NPFF-like fibers surrounding GnRH cells was tested using an antiserum against neurophysin 2, the vasopressin protein carrier (59). While the paraventricular nucleus (PVN) contained many NPFF-like immunoreactive fibers, double labeling for FMRFa and neurophysin failed to highlight colabeled fibers in the vicinity of GnRH neurons (Fig. 6F). To evaluate whether NPFF-like immunoreactive fibers to GnRH neurons arose from the NTS, containing primarily catecholaminergic (CAergic) neurons (60, 61), brainstem sections were stained for NPFF and TH. Immunostaining indicated the majority of NPFF-like immunoreactive neurons (~80%) coexpressed TH (Fig. 7A). Although the A2 CAergic cell group is centered within the caudal part of the NTS, they intermix with the trailing edge of the C2 CAergic cell group (62). Many C2 CAergic cells in this region coexpress NPY while A2 CAergic cells usually do not (63, 64). Staining for GFP in NPY-GFP mice showed that the majority of NPFF-like immunoreactive neurons (~80%) were GFP positive (Fig. 7B). To insure specificity, the potential cross-reactivity of our FMRFa antibody with NPY (41, 42) was tested and did not cross-react (Fig. 6D). CCK also labels a subpopulation of C2 CAergic cells (65). Triple immunofluorescence revealed that a minority of NPFF-like immunoreactive neurons (~20%) immunoreacted for CCK and these neurons were not tagged with GFP in NPY-GFP mice (Fig. 7C), indicating the NPFF-like immunoreactive neurons formed a heterogeneous cell population in the NTS, but that the majority coexpressed TH and NPY.
Figure 7.
NPFF-like cells in the NTS are largely colabeled with NPY. (A) Low magnification images showing NPFF-like (FMRFa, red) and tyrosine hydroxylase (TH; green) coimmunoreactivity in the NTS (AP, area postrema; cc, central canal; bar 100 µm). Insets, High magnification confocal images showing a close up of the boxed area on the left (bar 20 µm). Arrow heads point to NPFF-like cells colabeled with TH while arrows point to NPFF-like cells that are not colabeled with TH. (B) Confocal images showing NPFF-like (FMRFa, red) and GFP (green) coimmunoreactivity in the NTS from an NPY-GFP mouse (bar 20 µm). Arrow heads point to NPFF-like cells colabeled with TH while asterisks mark NPY cells that are not colabeled with NPFF-like. (C) Confocal images in the NTS from an NPY-GFP mouse showing cells that are NPFF-like (FMRFa, white) and CCK (red) coimmunoreactive (arrowheads) or NPFF-like (FMRFa, white) and NPY (green) coimmunoreactive (arrows, bar 20 µm). No cells were detected that expressed all 3 neuropeptides.
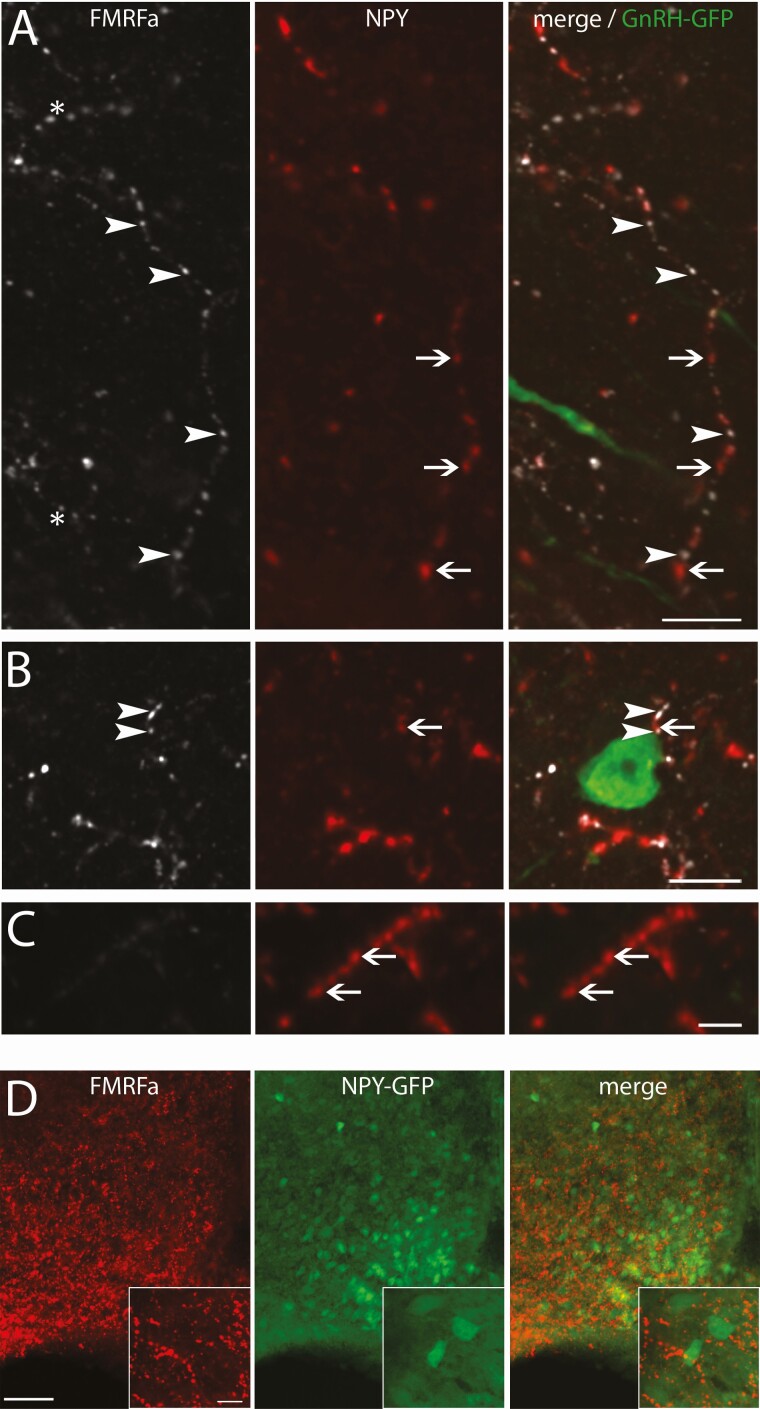
Since NPFF-like immunoreactive neurons in the NTS were mainly colabeled with NPY, a triple staining was performed on GnRH-GFP containing brain sections with FMRFa and NPY antisera. The majority of NPFF-like immunoreactive fibers in the vicinity of GnRH neurons were colabeled for NPY (Fig. 8A and 8B). As expected, some fibers were single labeled for NPY (Fig. 8C), most likely arising from NPY neurons in the arcuate nucleus that did not display NPFF-like immunoreactivity (Fig. 8D).
Figure 8.
NPFF-like fibers around GnRH neurons likely originate from the NTS. (A-C) Confocal images showing NPFF-like (FMRFa, white), NPY (red) and GnRH-GFP (green), cropped from the same confocal plan. (A) Confocal images showing example of a NPY and NPFF-like colabeled fiber showing alternating single-labeled NPFF-like (arrowheads) and NPY (arrows) varicosities. The asterisk marks a NPFF-like fiber that is not colabeled with NPY (bar 10 µm). (B) Confocal images showing example of such NPY / NPFF-like colabeled fiber passing in proximity of a GnRH neuron (bar 10 µm). (C) Confocal images showing example of a NPY fiber that is not immunoreactive for NPFF-like (bar 5 µm). (D) Low magnification images showing that the arcuate nucleus contains NPFF-like (red) fibers, NPY-GFP-tagged neurons (green) and that the NPY cells do not contain NPFF-like immunoreactivity, indicating the NPY fibers expressing NPFF-like immunoreactivity are not from the arcuate nucleus but the NTS (bar 50 µm). High magnification in insets: bar 10 µm.
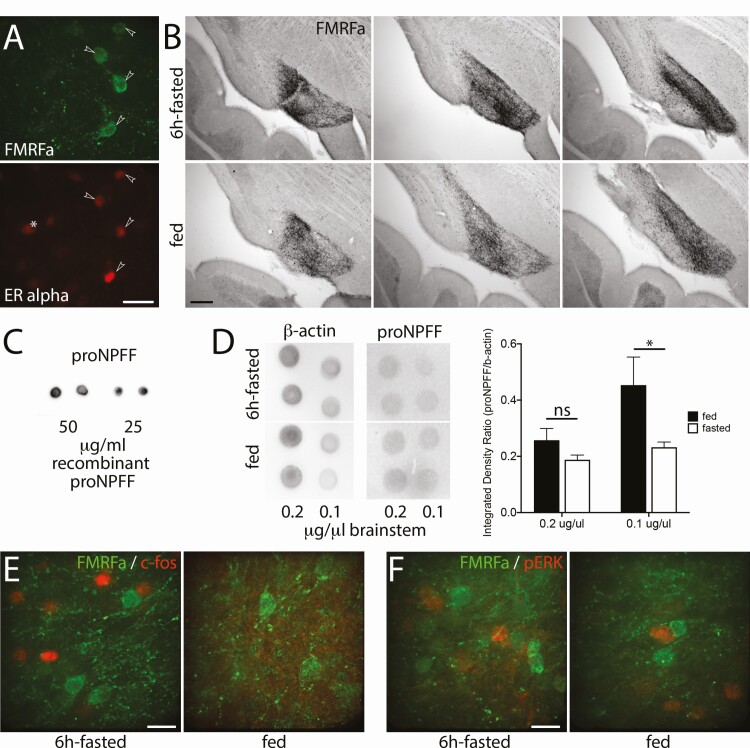
NTS NPFF-like immunoreactive neurons are ER alpha positive and respond to an acute fast, prior to the decrease in LH levels.
A 6-hour fast (5 hour light + 1 hour dark) decreases LH levels (66), increases c-fos in the NTS (67), and requires an estrogen feedback from the NTS (30). To determine whether NTS NPFF neurons that project to GnRH neurons could be involved in this metabolic/reproductive circuit, a series of experiments was performed. Brainstem sections were stained with FMRFa and estrogen receptor alpha (ER alpha) antisera. The large majority, if not all, NPFF-like immunoreactive neurons in the NTS immunoreacted for ER alpha (Fig. 9A). To test whether fasting would affect NPFF-like immunoreactivity staining, brainstem sections from fed and 6-hour fasted animals were stained with FMRFa antiserum. The 6-hour fast increased the Ni-DAB staining intensity and area in the NTS by ~4.5 ± 1.0% and ~141 ± 32% (N = 3 in each group; P = .0078 for both, Wilcoxon matched-pairs signed rank test), respectively (Fig. 9B). Dot blots were used to examine the change in proNPFF protein levels in fed vs fasted animals (Fig. 9C). Using an antibody directed against proNPFF, a decrease in proNPFF/β-actin ratio subsequent to the 6-hour fasting was found (P = .0336, 2-way ANOVA; Sidak’s multiple comparisons test on Fig. 9D). Next, sections were double labeled for c-fos and NPFF-like immunoreactivity. Although a 6-hour fast increased the number of c-fos immunoreactive cells in the NTS, these cells were NPFF-like negative, though clearly within the same region (Fig. 9E). Since one might miss the temporal expression of c-fos, another marker for neuronal activation was used. Phosphorylated ERK (pERK) was chosen based on the facts that pERK increases in response to pain in spinal cord NPFF neurons (68) and to refeeding in NTS neurons (69). However, no changes were detected in pERK expression in the NTS between fed and fasted animals, and NPFF-like immunoreactivity was not colocalize with pERK (Fig. 9F). Finally, an ELISA for LH in fed and 6-hour fasted animals (n = 5-6) was performed but it did not detect a significant difference in LH levels between fed and fasted animals (in ng/mL, fed group: 1 pm 0.91 ± 0.23; 7 pm 1.20 ± 0.32; fasted group, 1 pm 2.00 ± 0.67; 7 pm 0.91 ± 0.21; nonparametric Mann–Whitney test at 7 pm: P = .7922).
Figure 9.
NPFF-like cells in the NTS express ERalpha and respond to acute fasting by changes in NPFF and proNPFF. (A) Confocal images showing NPFF-like cells (FMRFa, green) colocalizing with ERalpha (red, arrowheads). The asterisk indicates an ERalpha cell that is not stained with FMRFa (bar 20 µm). (B) Images of Ni-DAB staining (FMRFa) illustrate increased NPFF-like immunoreactivity in the NTS after 6-hour fasting (top row) compared with control (bottom row; bar 200 µm). (C) Images of a dot blot showing the detection of recombinant proNPFF with the antibody directed against proNPFF. (D) Left panel, images of a dot blot showing the detection of actin and proNPFF in the brainstem from 6-hour fasted and fed animals. Right panel, graph showing the quantification of the dot blot for both states normalized to actin signals. Asterisks indicate P < .05 (Sidak’s multiple comparisons test following the 2-way ANOVA). (E) Confocal images showing that 6-hour fasting (left) vs control (right) induces c-fos (red) immunoreactivity in the NTS but not in NPFF-like cells (FMRFa, green; bar 20 µm). (F) Confocal images showing that 6-hour fasting (left) vs control (right) does not induce pERK (red) immunoreactivity in the NTS and that pERK is not in NPFF-like cells (FMRFa, green; bar 20 µm).
Discussion
The present study identifies a new circuit to GnRH neurons from the NTS, NPFF cells. We found that Npff-like fibers are in the vicinity of GnRH neurons and that peptides derived from Npff directly inhibit GnRH neurons via NPFFR1 coupled to Gi/o protein and subsequent G protein–coupled inwardly-rectifying potassium channels. Notably, the NPFF-like fibers in close proximity to GnRH neurons coexpressed NPY, consistent with NPFF-like cells located in the NTS, an energy sensing nucleus. It is known that ERα-containing neurons in the caudal brainstem project to regions containing GnRH neurons (70) and estrogen feedback on cells in the caudal region of the NTS plays a role in the fast-induced decrease of LH secretion (30). NTS NPFF cells were positive for ERα and responded to short-term fasting by an increase in NPFF-like peptide as measured by immunoreactivity and a decrease in proNPFF as measured by dot blot analysis. Together, these data indicate that NTS NPFF neurons act via NPFFR1 receptors on a subpopulation of GnRH cells and that they are part of a circuit that conveys information during metabolic challenges to reproductive function, prior to the energy deficit. Although the cell signaling pathway was defined using GnRH neurons in explants obtained from unsexed embryos and the data obtained supported a generic inhibitory circuit in both sexes, circuitry and its functionality were studied in male mice to avoid variability driven by hormonal cycle.
Peptides derived from Rfrp and the NPFFR1 are linked to fertility (reviewed in 71-74), whereas peptides derived from Npff and the NPFF2 receptor are linked to pain modulation (reviewed in (23)). However, the line between each group of ligands and their cognate receptors is thin. All exogenously applied RFa peptides can modulate pain pathways (13, 45). In fact, the C-terminal Arg-Phe-NH2 motif seems sufficient to allow RFa peptides to bind with high affinity to both NPFF receptors (13, 45). While Rfrp- and Npff-derived peptides are more potent agonists for NPFFR1/R2 than other RFa peptides, neither group clearly discriminate its “own” receptor (75, 76). Thus, while GnRH neurons express NPFFR1, and not NPFFR2 (10), the promiscuous binding of NPFFR1, which is seen in the cortex (77), allows NPFF to modulate GnRH neurons.
It is clear that RFRP neurons modulate GnRH neurons and fertility but, to date, their role in regulating fertility appears minimal. RFRP fibers contact GnRH neurons (9) and RFRP-3 inhibits GnRH neuron firing rate (11). RFRP neurons are associated with external cues, either circadian with the timing of the ovulation (6, 7, 78), photoperiodic in seasonally breeding species (74, 79-82), or even social (83-85). However, under optimal breeding conditions, mice lacking NPFFR1 display normal fertility. Only acute metabolic stress (12-hour fast) that normally decreases circulating LH levels failed to do so in NPFFR1-null mice (18). Since this knockout mouse was designed to bypass pharmacological pitfalls and assess the importance of the “preferential” ligand to NPFFR1, namely RFRPs, León et al. concluded a role of RFRP neurons as mediators of metabolic cues (18). However, a 12-hour fast does not affect the level of RFRP mRNA (19). While RFRP neurons and RFRPs cannot be excluded in this metabolic stress (RFRP translation and release could increase), NPFF is a plausible alternative.
Both the NTS and PVN play a critical role in the fast-induced decrease in LH secretion (30, 86). The literature proposes a mechanism in which adrenergic neurons in the NTS sense energy deficiency, convey the signals to the PVN which in turns produces corticotropin releasing hormone (CRH) to inhibit GnRH neurons (87). However, fasting still decreases LH levels in mice lacking CRH receptors (88) and GnRH neurons do not respond to CRH (89), hence the need for other factor(s). Our attention turned to NPFF, since it is a RFa that displays agonistic activity with the NPFFR1 (75, 76) and is in anatomically relevant areas. In situ hybridization revealed a very limited expression of Npff. The supraoptic nucleus and PVN of the hypothalamus displayed a low signal whereas the medulla exhibited a strong signal in the NTS (22). The PVN population contributes to the neurohypophyseal NPFF (90). The NTS, containing CAergic, as well as diverse peptidergic cells, is known to project to the medial preoptic area, rich in GnRH neurons (62, 91-93). GnRH neurons receive noradrenergic inputs (94) and cell-specific retrograde-labeling identified afferents from the NTS to GnRH neurons (28).
To determine whether the NPFF-like fibers observed in the vicinity of GnRH neurons came from the NTS, we characterized this NPFF cell population and found that they co-expressed TH and NPY. Previous data support this observation: NPFF neurons are CAergic, in other words expressing TH (26); half of the NTS projections to GnRH neurons are labeled for TH (28); and some NPY afferents to GnRH neurons arise from the noradrenergic/adrenergic cell groups of the brainstem (95). While 1 study did not find NPY in TH-immunoreactive neurons (96), others showed some colocalization between NPY and dopamine-β-hydroxylase in the NTS (64, 97) and between NPY and PNMT or TH in the dorsal medulla (98). Here, we confirmed colocalization of NPFF-like and NPY immunoreactivity in fibers surrounding GnRH neurons, consistent with these NPFF-like fibers originating in the brainstem. Although contained in the same fibers, varicosities immunoreactive for NPFF-like and NPY were often single labeled. Examples of peptide-specific segregation in varicosities exist for both NPFF and NPY (99, 100).
The fasting-induced decrease in LH levels requires the gastric branch of the vagal nerve (101), the NTS and the PVN (30). In rats, a 6-hour fasting increases c-fos in the NTS (67) and decreases LH levels (66). In mice, we found a similar 6-hour fasting increased NPFF-like immunoreactivity in NTS cells and decreased proNPFF levels in brainstem (likely increased peptide processing via propeptide cleavage (102, 103)). Yet, the increased c-fos was in non-NPFF-like NTS cells. How these 2 NTS subpopulations relate to one another is unknown. The NTS forms a complex structure where somatic and visceral neurons commingle (104) and where emotional and physiological status integrates to regulate stress (105). Even within the PVN-projecting NTS subpopulation, largely second-order neurons from the vagal connection (106), emerge as an heterogenous population with distinct pathways (107).
Although the fasting protocol used in the present report resulted in a decrease in LH levels in female rats, as measured 1 hour after lights off, it did not change glucose levels (66). In contrast, a 3-hour fasting only at light off switch raises corticosterone levels (108). Since stress is known to inhibit episodic GnRH/LH secretion (109), one cannot rule out that the increased NPFF found in the NTS was stress-related, rather than due to a shift in energy homeostasis. This suggests that the decrease of LH in response to acute fasting may involve sequential events (stress then metabolic deficit) and that NPFF may convey stress, one of the first signals to tune down GnRH neurons during metabolic challenges. This might explain why the decrease in LH levels expected after overnight fasting is only delayed in NPFFR1-deficient mice (18). In fact, while proNPFF and NPFF levels were decreased and increased, respectively, we were unable to detect a decrease in LH levels. This is consistent with an increase in NPFF levels preceding a decrease in LH levels. However, it is certainly possible that the number of animals we used, combined with the pulsatile nature of LH levels, resulted in a failure to detect a decrease rather than the absence of decrease.
Kisspeptin/neurokinin B/dynorphin (KNDy) neurons drive GnRH secretion (110) and Wang et al. recently showed that GnRH/LH pulses are triggered by a local signal, kisspeptin, on the distal process of GnRH neurons, without the need of GnRH neuronal activity (111). Although acute inhibition on GnRH neuron firing does not impair LH pulses, the effect of a lasting one cannot be excluded. Ongoing GnRH neuronal activity might be permissive to the local signal on the distal process since the patterning of neuronal activity regulates gene expression and neuronal adaptation (112, 113). Yet, the relevance of regulating GnRH neuron firing and LH levels becomes questionable. After all, since KNDy neurons express NPFFR1 (114), receive inputs from the NTS (115) and fasting (≥12 hours) decreases hypothalamic Kiss1 mRNA (116), they might respond to NPFF. Unfortunately, the decrease of LH in response to acute fasting requires steroids (30) and steroids impede the immunohistological detection of KNDy neurons (117), hence the presence of NPFF-like fibers onto KNDy neurons was not evaluated in this study. The alternative, combined in situ hybridization and immunohistochemistry, is usually not used to assess contacts (118). Notably, the activation of KNDy neurons by another NTS neuropeptide, glucagon-like peptide 1, is not sufficient to reverse the LH decrease evoked by fasting (119). Similarly, while the PVN activation decreases LH levels, the inhibition is not dependent on the action of CRH on GnRH or KNDy neurons (120), emphasizing the need for another neuropeptide involved in the LH response to stress/metabolic changes.
In sum, this study identifies a direct input from the NTS to GnRH neurons, NPFF, that regulates GnRH neuronal activity, responds early to acute fasting and can integrate the steroid milieu, providing a new neuronal circuit to GnRH neurons that incorporates energy homeostasis to reproductive function.
Acknowledgments
We are grateful to Dr. Bentley for providing the GnIH antibody, and Dr. Zajac for the compound, dNPA. We also address special thanks to Drs. Soga and Parhar for providing brain sections from GnIH-GFP rats. We thank Dr. Roa-Rivas for his valuable input with the first draft of the manuscript and Dr. Sainani for her editorial advices. We thank David Shostak (Postbaccalaureate IRTA, CDNS) for his help collecting blood samples. We thank the University of Virginia Center for Research in Reproduction, Ligand Assay and Analysis Core (NICHD/NIH Grant R24HD102061) for their expertise with the ELISA assay.
Financial Support: This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke (ZIA NS002824).
Author Contributions : S.C. and S.W. designed research; S.C., K.P., K.M., Y.S., D.R. performed research; S.C. analyzed data; S.C. and S.W. wrote the paper.
Glossary
Abbreviations
- aCSF
artificial cerebrospinal fluid
- ANOVA
analysis of variance
- AP
action potential
- CCK
cholecystokinin
- CRH
corticotropin releasing hormone
- DAB
3,3′-diaminobenzidine
- ELISA
enzyme-linked immunosorbent assay
- GFP
green fluorescent protein
- GnIH
gonadotropin-inhibitory hormone
- GnRH
gonadotropin-releasing hormone
- HRP
horseradish peroxidase
- LH
luteinizing hormone
- NPAF
neuropeptide AF
- NPFF
neuropeptide FF
- NPFFR1
neuropeptide FF receptor 1
- NPY
neuropeptide Y
- NTS
nucleus tractus solitarius
- PBS
phosphate-buffered saline
- PrRP
prolactin-releasing peptide
- PVN
paraventricular nucleus
- RFa
RFamide
- RFRP
RFamide-related peptide
- SFM
serum-free medium
- TH
tyrosine hydroxylase
- TSA
tyramide signal amplification
Additional Information
Disclosures : Authors report no conflict of interest.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References
- 1. Tsutsui K, Saigoh E, Ukena K, et al. A novel avian hypothalamic peptide inhibiting gonadotropin release. Biochem Biophys Res Commun. 2000;275(2):661-667. [DOI] [PubMed] [Google Scholar]
- 2. Satake H, Hisada M, Kawada T, Minakata H, Ukena K, Tsutsui K. Characterization of a cDNA encoding a novel avian hypothalamic neuropeptide exerting an inhibitory effect on gonadotropin release. Biochem J. 2001;354(Pt 2):379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hinuma S, Shintani Y, Fukusumi S, et al. New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nat Cell Biol. 2000;2(10):703-708. [DOI] [PubMed] [Google Scholar]
- 4. Johnson MA, Tsutsui K, Fraley GS. Rat RFamide-related peptide-3 stimulates GH secretion, inhibits LH secretion, and has variable effects on sex behavior in the adult male rat. Horm Behav. 2007;51(1):171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murakami M, Matsuzaki T, Iwasa T, et al. Hypophysiotropic role of RFamide-related peptide-3 in the inhibition of LH secretion in female rats. J Endocrinol. 2008;199(1):105-112. [DOI] [PubMed] [Google Scholar]
- 6. Anderson GM, Relf HL, Rizwan MZ, Evans JJ. Central and peripheral effects of RFamide-related peptide-3 on luteinizing hormone and prolactin secretion in rats. Endocrinology. 2009;150(4):1834-1840. [DOI] [PubMed] [Google Scholar]
- 7. Ancel C, Inglis MA, Anderson GM. Central RFRP-3 stimulates LH secretion in male mice and has cycle stage-dependent inhibitory effects in females. Endocrinology. 2017;158(9):2873-2883. [DOI] [PubMed] [Google Scholar]
- 8. Rizwan MZ, Porteous R, Herbison AE, Anderson GM. Cells expressing RFamide-related peptide-1/3, the mammalian gonadotropin-inhibitory hormone orthologs, are not hypophysiotropic neuroendocrine neurons in the rat. Endocrinology. 2009;150(3):1413-1420. [DOI] [PubMed] [Google Scholar]
- 9. Kriegsfeld LJ, Mei DF, Bentley GE, et al. Identification and characterization of a gonadotropin-inhibitory system in the brains of mammals. Proc Natl Acad Sci U S A. 2006;103(7):2410-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rizwan MZ, Poling MC, Corr M, et al. RFamide-related peptide-3 receptor gene expression in GnRH and kisspeptin neurons and GnRH-dependent mechanism of action. Endocrinology. 2012;153(8):3770-3779. [DOI] [PubMed] [Google Scholar]
- 11. Ducret E, Anderson GM, Herbison AE. RFamide-related peptide-3, a mammalian gonadotropin-inhibitory hormone ortholog, regulates gonadotropin-releasing hormone neuron firing in the mouse. Endocrinology. 2009;150(6):2799-2804. [DOI] [PubMed] [Google Scholar]
- 12. Sandvik GK, Hodne K, Haug TM, Okubo K, Weltzien FA. RFamide peptides in early vertebrate development. Front Endocrinol (Lausanne). 2014;5:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elhabazi K, Humbert JP, Bertin I, et al. Endogenous mammalian RF-amide peptides, including PrRP, kisspeptin and 26RFa, modulate nociception and morphine analgesia via NPFF receptors. Neuropharmacology. 2013;75:164-171. [DOI] [PubMed] [Google Scholar]
- 14. Liu Q, Guan XM, Martin WJ, et al. Identification and characterization of novel mammalian neuropeptide FF-like peptides that attenuate morphine-induced antinociception. J Biol Chem. 2001;276(40):36961-36969. [DOI] [PubMed] [Google Scholar]
- 15. Yang HY, Fratta W, Majane EA, Costa E. Isolation, sequencing, synthesis, and pharmacological characterization of two brain neuropeptides that modulate the action of morphine. Proc Natl Acad Sci U S A. 1985;82(22):7757-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laurent P, Becker JA, Valverde O, et al. The prolactin-releasing peptide antagonizes the opioid system through its receptor GPR10. Nat Neurosci. 2005;8(12):1735-1741. [DOI] [PubMed] [Google Scholar]
- 17. Liu X, Herbison A. Kisspeptin regulation of arcuate neuron excitability in kisspeptin receptor knockout mice. Endocrinology. 2015;156(5):1815-1827. [DOI] [PubMed] [Google Scholar]
- 18. León S, García-Galiano D, Ruiz-Pino F, et al. Physiological roles of gonadotropin-inhibitory hormone signaling in the control of mammalian reproductive axis: studies in the NPFF1 receptor null mouse. Endocrinology. 2014;155(8):2953-2965. [DOI] [PubMed] [Google Scholar]
- 19. Poling MC, Shieh MP, Munaganuru N, Luo E, Kauffman AS. Examination of the influence of leptin and acute metabolic challenge on RFRP-3 neurons of mice in development and adulthood. Neuroendocrinology. 2014;100(4):317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. 1977;197(4304):670-671. [DOI] [PubMed] [Google Scholar]
- 21. Boer HH, Schot LP, Veenstra JA, Reichelt D. Immunocytochemical identification of neural elements in the central nervous systems of a snail, some insects, a fish, and a mammal with an antiserum to the molluscan cardio-excitatory tetrapeptide FMRF-amide. Cell Tissue Res. 1980;213(1):21-27. [DOI] [PubMed] [Google Scholar]
- 22. Vilim FS, Aarnisalo AA, Nieminen ML, et al. Gene for pain modulatory neuropeptide NPFF: induction in spinal cord by noxious stimuli. Mol Pharmacol. 1999;55(5):804-811. [PubMed] [Google Scholar]
- 23. Yang HY, Tao T, Iadarola MJ. Modulatory role of neuropeptide FF system in nociception and opiate analgesia. Neuropeptides. 2008;42(1):1-18. [DOI] [PubMed] [Google Scholar]
- 24. Ayachi S, Simonin F. Involvement of mammalian RF-amide peptides and their receptors in the modulation of nociception in rodents. Front Endocrinol (Lausanne). 2014;5:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin YT, Chen JC. Neuropeptide FF modulates neuroendocrine and energy homeostasis through hypothalamic signaling. Chin J Physiol. 2019;62(2):47-52. [DOI] [PubMed] [Google Scholar]
- 26. Kivipelto L, Aarnisalo A, Panula P. Neuropeptide FF is colocalized with catecholamine-synthesizing enzymes in neurons of the nucleus of the solitary tract. Neurosci Lett. 1992;143(1-2):190-194. [DOI] [PubMed] [Google Scholar]
- 27. Wright DE, Jennes L. Origin of noradrenergic projections to GnRH perikarya-containing areas in the medial septum-diagonal band and preoptic area. Brain Res. 1993;621(2):272-278. [DOI] [PubMed] [Google Scholar]
- 28. Campbell RE, Herbison AE. Definition of brainstem afferents to gonadotropin-releasing hormone neurons in the mouse using conditional viral tract tracing. Endocrinology. 2007;148(12):5884-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blouet C, Schwartz GJ. Brainstem nutrient sensing in the nucleus of the solitary tract inhibits feeding. Cell Metab. 2012;16(5):579-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagatani S, Tsukamura H, Maeda K. Estrogen feedback needed at the paraventricular nucleus or A2 to suppress pulsatile luteinizing hormone release in fasting female rats. Endocrinology. 1994;135(3):870-875. [DOI] [PubMed] [Google Scholar]
- 31. Hoffman GE, Murphy KJ, Sita LV. The importance of titrating antibodies for immunocytochemical methods. Curr Protoc Neurosci. 2016;76:2.12.1-2.12.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soga T, Kitahashi T, Clarke IJ, Parhar IS. Gonadotropin-inhibitory hormone promoter-driven enhanced green fluorescent protein expression decreases during aging in female rats. Endocrinology. 2014;155(5):1944-1955. [DOI] [PubMed] [Google Scholar]
- 33. Steyn FJ, Wan Y, Clarkson J, Veldhuis JD, Herbison AE, Chen C. Development of a methodology for and assessment of pulsatile luteinizing hormone secretion in juvenile and adult male mice. Endocrinology. 2013;154(12):4939-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fueshko S, Wray S. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev Biol. 1994;166(1):331-348. [DOI] [PubMed] [Google Scholar]
- 35. Spergel DJ, Krüth U, Hanley DF, Sprengel R, Seeburg PH. GABA- and glutamate-activated channels in green fluorescent protein-tagged gonadotropin-releasing hormone neurons in transgenic mice. J Neurosci. 1999;19(6):2037-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Constantin S, Wray S. Nociceptin/Orphanin-FQ inhibits gonadotropin-releasing hormone neurons via G-protein-gated inwardly rectifying potassium channels. eNeuro. 2018;5(6):ENEURO.0161-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wray S, Nieburgs A, Elkabes S. Spatiotemporal cell expression of luteinizing hormone-releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Brain Res Dev Brain Res. 1989;46(2):309-318. [DOI] [PubMed] [Google Scholar]
- 38. Shindler KS, Roth KA. Double immunofluorescent staining using two unconjugated primary antisera raised in the same species. J Histochem Cytochem. 1996;44(11):1331-1335. [DOI] [PubMed] [Google Scholar]
- 39. Kivipelto L, Rubenstein J, Yang HY, Panula P. Ontogeny of the F8Famide-like (morphine-modulating) peptides in the central nervous system of rats. J Comp Neurol. 1991;304(1):14-33. [DOI] [PubMed] [Google Scholar]
- 40. Lein ES, Hawrylycz MJ, Ao Net al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168-176. [DOI] [PubMed] [Google Scholar]
- 41. Kivipelto L, Majane EA, Yang HY, Panula P. Immunohistochemical distribution and partial characterization of FLFQPQRFamidelike peptides in the central nervous system of rats. J Comp Neurol. 1989;286(2):269-287. [DOI] [PubMed] [Google Scholar]
- 42. Hung MY, Shen CL. Localization of fmrfamide-like immunoreactivity in the peripheral organs and its cross reaction with neuropeptide Y in the rat. Gaoxiong Yi Xue Ke Xue Za Zhi. 1994;10(10):550-557. [PubMed] [Google Scholar]
- 43. Crowe AR, Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio-Protoc. 2019;9(24):e3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Constantin S, Klenke U, Wray S. The calcium oscillator of GnRH-1 neurons is developmentally regulated. Endocrinology. 2010;151(8):3863-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mollereau C, Mazarguil H, Marcus D, et al. Pharmacological characterization of human NPFF(1) and NPFF(2) receptors expressed in CHO cells by using NPY Y(1) receptor antagonists. Eur J Pharmacol. 2002;451(3):245-256. [DOI] [PubMed] [Google Scholar]
- 46. Simonin F, Schmitt M, Laulin JP, et al. RF9, a potent and selective neuropeptide FF receptor antagonist, prevents opioid-induced tolerance associated with hyperalgesia. Proc Natl Acad Sci U S A. 2006;103(2):466-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Min L, Leon S, Li H, et al. RF9 acts as a KISS1R agonist in vivo and in vitro. Endocrinology. 2015;156(12):4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu X, Herbison AE. RF9 excitation of GnRH neurons is dependent upon Kiss1r in the adult male and female mouse. Endocrinology. 2014;en20141517. [DOI] [PubMed] [Google Scholar]
- 49. Klenke U, Constantin S, Wray S. Neuropeptide Y directly inhibits neuronal activity in a subpopulation of gonadotropin-releasing hormone-1 neurons via Y1 receptors. Endocrinology. 2010;151(6):2736-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roussin A, Serre F, Gouardères C, et al. Anti-analgesia of a selective NPFF2 agonist depends on opioid activity. Biochem Biophys Res Commun. 2005;336(1):197-203. [DOI] [PubMed] [Google Scholar]
- 51. Engström M, Brandt A, Wurster S, Savola JM, Panula P. Prolactin releasing peptide has high affinity and efficacy at neuropeptide FF2 receptors. J Pharmacol Exp Ther. 2003;305(3):825-832. [DOI] [PubMed] [Google Scholar]
- 52. Stevens EB, Shah BS, Pinnock RD, Lee K. Bombesin receptors inhibit G protein-coupled inwardly rectifying K+ channels expressed in Xenopus oocytes through a protein kinase C-dependent pathway. Mol Pharmacol. 1999;55(6):1020-1027. [PubMed] [Google Scholar]
- 53. Kelly MJ, Qiu J, Wagner EJ, Rønnekleiv OK. Rapid effects of estrogen on G protein-coupled receptor activation of potassium channels in the central nervous system (CNS). J Steroid Biochem Mol Biol. 2002;83(1-5):187-193. [DOI] [PubMed] [Google Scholar]
- 54. Constantin S, Wray S. Galanin activates G protein gated inwardly rectifying potassium channels and suppresses Kisspeptin-10 activation of GnRH neurons. Endocrinology. 2016;157(8):3197-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Majane EA, Panula P, Yang HY. Rat brain regional distribution and spinal cord neuronal pathway of FLFQPQRF-NH2, a mammalian FMRF-NH2-like peptide. Brain Res. 1989;494(1):1-12. [DOI] [PubMed] [Google Scholar]
- 56. Oehlmann VD, Korte H, Sterner C, Korsching SI. A neuropeptide FF-related gene is expressed selectively in neurons of the terminal nerve in Danio rerio. Mech Dev. 2002;117(1-2):357-361. [DOI] [PubMed] [Google Scholar]
- 57. Nieminen ML, Nystedt J, Panula P. Expression of neuropeptide FF, prolactin-releasing peptide, and the receptor UHR1/GPR10 genes during embryogenesis in the rat. Dev Dyn. 2003;226(3):561-569. [DOI] [PubMed] [Google Scholar]
- 58. Boersma CJ, Sonnemans MA, Van Leeuwen FW. Immunocytochemical localization of neuropeptide FF (FMRF amide-like peptide) in the hypothalamo-neurohypophyseal system of Wistar and Brattleboro rats by light and electron microscopy. J Comp Neurol. 1993;336(4):555-570. [DOI] [PubMed] [Google Scholar]
- 59. Ben-Barak Y, Russell JT, Whitnall MH, Ozato K, Gainer H. Neurophysin in the hypothalamo-neurohypophysial system. I. Production and characterization of monoclonal antibodies. J Neurosci. 1985;5(1):81-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Armstrong DM, Pickel VM, Joh TH, Reis DJ, Miller RJ. Immunocytochemical localization of catecholamine synthesizing enzymes and neuropeptides in area postrema and medial nucleus tractus solitarius of rat brain. J Comp Neurol. 1981;196(3):505-517. [DOI] [PubMed] [Google Scholar]
- 61. Okada T, Tashiro Y, Kato F, Yanagawa Y, Obata K, Kawai Y. Quantitative and immunohistochemical analysis of neuronal types in the mouse caudal nucleus tractus solitarius: focus on GABAergic neurons. J Chem Neuroanat. 2008;35(3):275-284. [DOI] [PubMed] [Google Scholar]
- 62. Rinaman L. Hindbrain noradrenergic A2 neurons: diverse roles in autonomic, endocrine, cognitive, and behavioral functions. Am J Physiol Regul Integr Comp Physiol. 2011;300(2):R222-R235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sawchenko PE, Swanson LW, Grzanna R, Howe PR, Bloom SR, Polak JM. Colocalization of neuropeptide Y immunoreactivity in brainstem catecholaminergic neurons that project to the paraventricular nucleus of the hypothalamus. J Comp Neurol. 1985;241(2):138-153. [DOI] [PubMed] [Google Scholar]
- 64. Härfstrand A, Fuxe K, Terenius L, Kalia M. Neuropeptide Y-immunoreactive perikarya and nerve terminals in the rat medulla oblongata: relationship to cytoarchitecture and catecholaminergic cell groups. J Comp Neurol. 1987;260(1):20-35. [DOI] [PubMed] [Google Scholar]
- 65. Kawai Y, Takagi H, Tohyama M. Co-localization of neurotensin- and cholecystokinin-like immunoreactivities in catecholamine neurons in the rat dorsomedial medulla. Neuroscience. 1988;24(1):227-236. [DOI] [PubMed] [Google Scholar]
- 66. Komatsu H, Miyamura A, Shibano T, Ôta K. Chronological study on changes in luteinizing hormone release during fasting and refeeding in ovariectomized estrogen-primed rats. Nihon Chikusan Gakkaiho. 2000;71(3):239-249. [Google Scholar]
- 67. Reyes BAS, Yamada S, Estacio MAC, Maeda K-I, Tsukamura H. Effect of fasting on c-Fos expression in the hypothalamic nuclei and nucleus of the solitary tract in male rats: time course study and the role of testosterone. J Reprod Dev. 2001;47(1):53-62. [Google Scholar]
- 68. Gutierrez-Mecinas M, Bell A, Polgár E, Watanabe M, Todd AJ. Expression of neuropeptide FF defines a population of excitatory interneurons in the superficial dorsal horn of the mouse spinal cord that respond to noxious and pruritic stimuli. Neuroscience. 2019;416:281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ritter RC. A tale of two endings: modulation of satiation by NMDA receptors on or near central and peripheral vagal afferent terminals. Physiol Behav. 2011;105(1):94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simonian SX, Spratt DP, Herbison AE. Identification and characterization of estrogen receptor alpha-containing neurons projecting to the vicinity of the gonadotropin-releasing hormone perikarya in the rostral preoptic area of the rat. J Comp Neurol. 1999;411(2):346-358. [DOI] [PubMed] [Google Scholar]
- 71. Ubuka T, Parhar I. Dual actions of mammalian and piscine gonadotropin-inhibitory hormones, RFamide-related peptides and LPXRFamide peptides, in the hypothalamic-pituitary-gonadal axis. Front Endocrinol (Lausanne). 2018;8:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hu KL, Chang HM, Li R, Yu Y, Qiao J. Regulation of LH secretion by RFRP-3 - from the hypothalamus to the pituitary. Front Neuroendocrinol. 2019;52:12-21. [DOI] [PubMed] [Google Scholar]
- 73. Tsutsui K, Ubuka T. How to contribute to the progress of neuroendocrinology: discovery of GnIH and progress of GnIH research. Front Endocrinol (Lausanne). 2018;9:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Angelopoulou E, Quignon C, Kriegsfeld LJ, Simonneaux V. Functional implications of RFRP-3 in the central control of daily and seasonal rhythms in reproduction. Front Endocrinol (Lausanne). 2019;10:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yoshida H, Habata Y, Hosoya M, Kawamata Y, Kitada C, Hinuma S. Molecular properties of endogenous RFamide-related peptide-3 and its interaction with receptors. Biochim Biophys Acta. 2003;1593(2-3):151-157. [DOI] [PubMed] [Google Scholar]
- 76. Gouardères C, Mazarguil H, Mollereau C, et al. Functional differences between NPFF1 and NPFF2 receptor coupling: high intrinsic activities of RFamide-related peptides on stimulation of [35S]GTPgammaS binding. Neuropharmacology. 2007;52(2):376-386. [DOI] [PubMed] [Google Scholar]
- 77. Buffel I, Meurs A, Portelli J, et al. Neuropeptide FF and prolactin-releasing peptide decrease cortical excitability through activation of NPFF receptors. Epilepsia. 2015;56(3):489-498. [DOI] [PubMed] [Google Scholar]
- 78. Beymer M, Henningsen J, Bahougne T, Simonneaux V. The role of kisspeptin and RFRP in the circadian control of female reproduction. Mol Cell Endocrinol. 2016;438:89-99. [DOI] [PubMed] [Google Scholar]
- 79. Gibson EM, Humber SA, Jain S, et al. Alterations in RFamide-related peptide expression are coordinated with the preovulatory luteinizing hormone surge. Endocrinology. 2008;149(10):4958-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Revel FG, Saboureau M, Pévet P, Simonneaux V, Mikkelsen JD. RFamide-related peptide gene is a melatonin-driven photoperiodic gene. Endocrinology. 2008;149(3):902-912. [DOI] [PubMed] [Google Scholar]
- 81. Smith JT, Coolen LM, Kriegsfeld LJ, et al. Variation in kisspeptin and RFamide-related peptide (RFRP) expression and terminal connections to gonadotropin-releasing hormone neurons in the brain: a novel medium for seasonal breeding in the sheep. Endocrinology. 2008;149(11):5770-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Talbi R, Klosen P, Laran-Chich MP, El Ouezzani S, Simonneaux V. Coordinated seasonal regulation of metabolic and reproductive hypothalamic peptides in the desert jerboa. J Comp Neurol. 2016;524(18):3717-3728. [DOI] [PubMed] [Google Scholar]
- 83. Soga T, Teo CH, Cham KL, Idris MM, Parhar IS. Early-life social isolation impairs the gonadotropin-inhibitory hormone neuronal activity and serotonergic system in male rats. Front Endocrinol (Lausanne). 2015;6:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jennings KJ, Chang J, Cho H, Piekarski DJ, Russo KA, Kriegsfeld LJ. Aggressive interactions are associated with reductions in RFamide-related peptide, but not kisspeptin, neuronal activation in mice. Horm Behav. 2016;78:127-134. [DOI] [PubMed] [Google Scholar]
- 85. Peragine DE, Pokarowski M, Mendoza-Viveros L, et al. RFamide-related peptide-3 (RFRP-3) suppresses sexual maturation in a eusocial mammal. Proc Natl Acad Sci U S A. 2017;114(5):1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Estacio MA, Yamada S, Tsukamura H, Hirunagi K, Maeda K. Effect of fasting and immobilization stress on estrogen receptor immunoreactivity in the brain in ovariectomized female rats. Brain Res. 1996;717(1-2):55-61. [DOI] [PubMed] [Google Scholar]
- 87. Matsuyama S, Kimura K. Regulation of gonadotropin secretion by monitoring energy availability. Reprod Med Biol. 2015;14(2):39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jeong KH, Jacobson L, Widmaier EP, Majzoub JA. Normal suppression of the reproductive axis following stress in corticotropin-releasing hormone-deficient mice. Endocrinology. 1999;140(4):1702-1708. [DOI] [PubMed] [Google Scholar]
- 89. Raftogianni A, Roth LC, García-González D, et al. Deciphering the contributions of CRH receptors in the brain and pituitary to stress-induced inhibition of the reproductive axis. Front Mol Neurosci. 2018;11:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Majane EA, Zhu J, Aarnisalo AA, Panula P, Yang HY. Origin of neurohypophyseal neuropeptide-FF (FLFQPQRF-NH2). Endocrinology. 1993;133(4):1578-1584. [DOI] [PubMed] [Google Scholar]
- 91. Ricardo JA, Koh ET. Anatomical evidence of direct projections from the nucleus of the solitary tract to the hypothalamus, amygdala, and other forebrain structures in the rat. Brain Res. 1978;153(1):1-26. [DOI] [PubMed] [Google Scholar]
- 92. Ito H, Seki M. Ascending projections from the area postrema and the nucleus of the solitary tract of Suncus murinus: anterograde tracing study using Phaseolus vulgaris leucoagglutinin. Okajimas Folia Anat Jpn. 1998;75(1):9-31. [DOI] [PubMed] [Google Scholar]
- 93. Rinaman L. Ascending projections from the caudal visceral nucleus of the solitary tract to brain regions involved in food intake and energy expenditure. Brain Res. 2010;1350:18-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hoffman GE, Wray S, Goldstein M. Relationship of catecholamines and LHRH: light microscopic study. Brain Res Bull. 1982;9(1-6):417-430. [DOI] [PubMed] [Google Scholar]
- 95. Turi GF, Liposits Z, Moenter SM, Fekete C, Hrabovszky E. Origin of neuropeptide Y-containing afferents to gonadotropin-releasing hormone neurons in male mice. Endocrinology. 2003;144(11):4967-4974. [DOI] [PubMed] [Google Scholar]
- 96. Simonian SX, Herbison AE. Differential expression of estrogen receptor and neuropeptide Y by brainstem A1 and A2 noradrenaline neurons. Neuroscience. 1997;76(2):517-529. [DOI] [PubMed] [Google Scholar]
- 97. Kourtesis I, Kasparov S, Verkade P, Teschemacher AG. Ultrastructural correlates of enhanced norepinephrine and neuropeptide Y cotransmission in the spontaneously hypertensive rat brain. ASN Neuro. 2015;7(5):1759091415610115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98..Everitt BJ, Hökfelt T, Terenius L, Tatemoto K, Mutt V, Goldstein M. Differential co-existence of neuropeptide Y (NPY)-like immunoreactivity with catecholamines in the central nervous system of the rat. Neuroscience. 1984;11(2):443-462. [DOI] [PubMed] [Google Scholar]
- 99. Vega A, Luther JA, Birren SJ, Morales MA. Segregation of the classical transmitters norepinephrine and acetylcholine and the neuropeptide Y in sympathetic neurons: modulation by ciliary neurotrophic factor or prolonged growth in culture. Dev Neurobiol. 2010;70(14):913-928. [DOI] [PubMed] [Google Scholar]
- 100. Kueh D, Jellies JA. Targeting a neuropeptide to discrete regions of the motor arborizations of a single neuron. J Exp Biol. 2012;215(Pt 12):2108-2116. [DOI] [PubMed] [Google Scholar]
- 101. Cagampang FR, Maeda K, Ota K. Involvement of the gastric vagal nerve in the suppression of pulsatile luteinizing hormone release during acute fasting in rats. Endocrinology. 1992;130(5):3003-3006. [DOI] [PubMed] [Google Scholar]
- 102. Muller L, Lindberg I. The cell biology of the prohormone convertases PC1 and PC2. Prog Nucleic Acid Res Mol Biol. 1999;63:69-108. [DOI] [PubMed] [Google Scholar]
- 103. Harno E, Gali Ramamoorthy T, Coll AP, White A. POMC: the physiological power of hormone processing. Physiol Rev. 2018;98(4):2381-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nomaksteinsky M, Kassabov S, Chettouh Z, et al. Ancient origin of somatic and visceral neurons. BMC Biol. 2013;11:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Myers B, Scheimann JR, Franco-Villanueva A, Herman JP. Ascending mechanisms of stress integration: Implications for brainstem regulation of neuroendocrine and behavioral stress responses. Neurosci Biobehav Rev. 2017;74(Pt B):366-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bailey TW, Hermes SM, Andresen MC, Aicher SA. Cranial visceral afferent pathways through the nucleus of the solitary tract to caudal ventrolateral medulla or paraventricular hypothalamus: target-specific synaptic reliability and convergence patterns. J Neurosci. 2006;26(46):11893-11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Affleck VS, Coote JH, Pyner S. The projection and synaptic organisation of NTS afferent connections with presympathetic neurons, GABA and nNOS neurons in the paraventricular nucleus of the hypothalamus. Neuroscience. 2012;219(1-2):48-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jensen TL, Kiersgaard MK, Mikkelsen LF, Sorensen DB. Fasting of male mice - effects of time point of initiation and duration on clinical chemistry parameters and animal welfare. Lab Anim. 2019;53(6):587-597. [DOI] [PubMed] [Google Scholar]
- 109. McCosh RB, Breen KM, Kauffman AS. Neural and endocrine mechanisms underlying stress-induced suppression of pulsatile LH secretion. Mol Cell Endocrinol. 2019;498:110579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Han SY, McLennan T, Czieselsky K, Herbison AE. Selective optogenetic activation of arcuate kisspeptin neurons generates pulsatile luteinizing hormone secretion. Proc Natl Acad Sci U S A. 2015;112(42):13109-13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang L, Guo W, Shen X, et al. Different dendritic domains of the GnRH neuron underlie the pulse and surge modes of GnRH secretion in female mice. eLife. 2020;9:e53945.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Brosenitsch TA, Katz DM. Physiological patterns of electrical stimulation can induce neuronal gene expression by activating N-type calcium channels. J Neurosci. 2001;21(8):2571-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yap EL, Greenberg ME. Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron. 2018;100(2):330-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Poling MC, Quennell JH, Anderson GM, Kauffman AS. Kisspeptin neurones do not directly signal to RFRP-3 neurones but RFRP-3 may directly modulate a subset of hypothalamic kisspeptin cells in mice. J Neuroendocrinol. 2013;25(10):876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Deura C, Minabe S, Ikegami K, et al. Morphological analysis for neuronal pathway from the hindbrain ependymocytes to the hypothalamic kisspeptin neurons. J Reprod Dev. 2019;65(2):129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Luque RM, Kineman RD, Tena-Sempere M. Regulation of hypothalamic expression of KiSS-1 and GPR54 genes by metabolic factors: analyses using mouse models and a cell line. Endocrinology. 2007;148(10):4601-4611. [DOI] [PubMed] [Google Scholar]
- 117. Moore AM, Coolen LM, Porter DT, Goodman RL, Lehman MN. KNDy cells revisited. Endocrinology. 2018;159(9):3219-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Grabinski TM, Kneynsberg A, Manfredsson FP, Kanaan NM. A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS One. 2015;10(3):e0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Heppner KM, Baquero AF, Bennett CM, et al. GLP-1R signaling directly activates arcuate nucleus kisspeptin action in brain slices but does not rescue luteinizing hormone inhibition in ovariectomized mice during negative energy balance. eNeuro. 2017;4(1):ENEURO.0198-16.2016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yip SH, Liu X, Hessler S, Cheong I, Porteous R, Herbison AE. Indirect suppression of pulsatile LH secretion by CRH neurons in the female mouse. Endocrinology. 2021;162(3):bqaa237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.