Abstract
The COVID-19 outbreak has represented a challenge for the international scientific community and particularly for forensic sciences. The lack of Coronavirus post-mortem testing led the National Institute of Toxicology and Forensic Sciences (INTCF) from Spain to verify the performance and utility of a quantitative reverse transcription PCR (RT-qPCR) clinical diagnosis protocol for SARS-CoV-2 detection (TaqPath™ COVID-19 CE-IVD RT-PCR Kit), to shed light on the cause of death (COD) in potentially COVID-19 cases in judicial autopsies. Two different RNA extraction methods were also tested (EZ1® DSP Virus Kit on the EZ1® Advanced XL robot versus MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit) regarding extraction efficiency, precision and contamination. RT-qPCR was evaluated for precision, specificity, limit of detection and concordance. Both the automated and the manual RNA extraction procedures showed good efficiency, but the automated virus extraction by bio-robot produced more reproducible results than the manual extraction. The SARS-CoV-2 RT-qPCR assay showed high sensitivity with a detection limit up to 10 copies/reaction and high specificity, as no cross-reactivity was detected between any of the 12 different RNA viruses tested, including three types of coronaviruses (SARS-CoV, NL63 and 229E). Reproducibility and repeatability of the studied method as well as concordance with other SARS-CoV-2 molecular detection protocols were also demonstrated.
Keywords: Coronavirus Infectious Disease-19 (COVID-19), Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Quantitative reverse transcription PCR (RT-qPCR), Validation, Forensic microbiology, Forensic genetics
1. Introduction
The disease caused by the novel Coronavirus SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2), also called COVID-19 (Coronavirus Infectious Disease-19), represents a cluster of pneumonia that emerged at the end of 2019 in the Chinese city of Wuhan, Hubei province, from where it spread to China and other countries in the world [1], [2], [3], [4], [5]. The COVID-19 was declared by the World Health Organization (WHO) as a Public Health Emergency of International Concern (PHEIC) on January 30, 2020 [6], and was characterized as a pandemic on March 11, 2020 [7].
The availability of the complete genome of SARS-CoV-2 early in the epidemic [8], allowed the development of specific primers, targeting the viral genes (ORF1ab, E, S and N), to amplify the genetic material of SARS-CoV-2 by using the Reverse Transcription Quantitative Real Time Polymerase chain reaction (RT-qPCR).
Initially, several laboratories developed many in house molecular tests but shortly after different commercial assays started to appear. Currently there are more than 240 molecular assays to diagnose COVID-19, with CE-IVD certification [9], among them the TaqPath™ COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Inc., CA, USA) [10]. Other nucleic amplification/detection methods such as CRISPR or LAMP (loop-mediated isothermal amplification) are in the process of being commercialized [11], [12], [13], [14]. The WHO recommends molecular testing (RT-qPCR) as a confirmatory diagnostic test [15], [16], although laboratories undertaking COVID-19 virus testing should perform a previous validation and both external and internal controls PCR should be included to provide a reliable test result.
Since the beginning of the coronavirus pandemic, clinical laboratories have been validating and implementing molecular tests with the aim of establishing reliable COVID-19 diagnosis in patients and contacts. However, confirmation of COVID-19 as cause of death (COD) in previously undiagnosed patients, either at home or in hospitals, has not always been possible: the lack of adequate safety requirements has led to autopsy restrictions and therefore to underdiagnosis.
In the light of the importance of autopsy findings for understanding COVID-19 features and the establishment of the COD, the Instituto Nacional de Toxicología y Ciencias Forenses (INTCF, National Institute of Toxicology and Forensic Sciences), a technical body attached to the Ministry of Justice of Spain [17], decided to validate and implement a protocol for extracting and detecting the genetic material of the SARS-CoV-2 in post-mortem specimens from judicial autopsies. In this work, we present the results obtained from the performance characteristics studied.
2. Materials and methods
2.1. Samples/controls, RNA controls and experimental designs
Extraction efficiency, repeatability and reproducibility were conducted using the AMPLIRUN® TOTAL SARS-CoV-2 CONTROL (SWAB) (Vircell Microbiologists, Granada, Spain) [18], integral Control of the Process of Extraction and detection (hereinafter CPE), which simulates swabs. From this control, sub-samples were prepared to be extracted with 2 different known (and theoretical) viral copies (Genome Copy Equivalent, GCE) inputs at the end point (in the RT-qPCR reaction) of high and medium GCE (500 and 250 total copies). The GCE value is “theoretical” based on the value provided by the manufacturer. The CPE control was extracted/detected in triplicate within each extraction batch, for the two total GCE inputs per reaction, for each extraction system, and detected on two plates (i.e., 3 replicates × 2 inputs × 2nd extraction batch = 12 samples/controls for each extraction system × 2 extraction systems = 24 samples/controls × 2 detection plates = 48 samples/controls in total).
Repeatability test was also performed in triplicate with a quantified control of SARS-CoV-2 RNA (10,000 copies/µL) provided by the Respiratory Viruses and Flu Unit of the National Center for Microbiology of the Carlos III Health Institute (CNM-ISCIII, Majadahonda, Madrid, Spain). This control was introduced with a known final input of 5000 copies/reaction, and whose replicates were detected on two plates (i.e., 3 replicates × 2 detection plates = 6 samples in total).
Concordance was assessed using five eluates of nasopharyngeal swabs from SARS-CoV-2 positive patients. These samples were previously analyzed at the Clinical Microbiology Service of the San Carlos Clinical Hospital (HCSC, Madrid, Spain). These eluates were divided into 2 aliquots (1 for each extraction system). One aliquot of each of these 5 samples was extracted by both extraction systems and detected on 2 plates (i.e., 5 samples × 2 systems × 2 detection plates = 20 samples). Once analyzed by RT-qPCR, the Cts values obtained were compared between the extraction system and with the results obtained in the HCSC.
Specificity studies were drawn from 12 different RNA viruses extracts to assess possible cross-reactivity. The AMPLIRUN® CORONAVIRUS SARS (2003) RNA CONTROL (Vircell Microbiologists, Granada, Spain) (hereinafter COV1 control) [19] was used, RNA purified from the Coronavirus SARS-CoV (2003), of which two known inputs were analyzed: 5000 and 50 copies/reaction. Additionally, other respiratory viruses from the Proficiency Testing Nucleic acid Amplification, Respiratory (ID2) del College of American Pathologists (CAP) [20] were analyzed, including: two of the most frequent Coronaviruses, two extracts of Coronavirus NL63 (2017-ID2-06 and 2019-ID2-06) and one extract of Coronavirus 229E (2018-ID2-06); eight extracts of Influenza A virus H1N1/2009 (2018-ID2-01), Influenza A virus H3N2 (2019-ID2-01), Human metapneumovirus B (2019-ID2-05), Influenza B virus (2019-ID2-07), Parainfluenza virus 2 (2019-ID2-08), Respiratory syncytial virus (2019-ID2-09), and Rhinovirus (2019-ID2-12). All extracts were analyzed in duplicate on each of the two detection plates (i.e., 12 virus RNA × 2 replicates × 2 detection plates = 48 viruses RNA in total).
Limit of detection (sensitivity testing) was studied by using 5 serial dilutions (10,000; 5000; 500; 50 and 10 copies/reaction) of two SARS-CoV-2 RNA controls: the AMPLIRUN® CORONAVIRUS SARS-CoV-2 RNA CONTROL [21] (hereinafter C19 control), with an initial concentration of 14,500 copies/µL, and the TaqPath™ COVID-19 Control [10] (hereinafter CTP control), with an initial concentration of 10,000 copies/µL. The last dilution, 10 copies/reaction, is the limit established by the manufacturer [10]. Each dilution was analyzed in duplicate on two plates (i.e., 5 serial dilutions × 2 replicates × 2 controls × 2 detection plates = 40 controls in total).
To evaluate potential external or cross-contamination, eight negative extraction controls were analyzed. In each extraction batch 2 negative controls were incorporated (2 CN_EXT × 4 extraction batch = 8 CN_EXT), which were positioned so that they were close to those samples that contained the greatest number of copies.
The MS2 Phage Control, included in the TaqPath™ COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Inc., CA, USA) (hereinafter TPkit) [10], was added to the samples before extraction of the RNA, according to the manufacturer's recommendation [10]. The MS2 Phage Control allowed to verify the efficacy of the sample preparation and the absence of inhibitors in the PCR reaction [10]. By this way, the generic evaluation of this control was performed considering the Cts values obtained for the MS2 target of the RT-qPCR in each of the samples analyzed in this verification: different inputs of the CPE control [18]; clinical samples provided by the HCSC; negative extraction controls for each batch for each of the extraction systems evaluated.
2.2. Viral nucleic acids extraction
Viral nucleic acids from samples and controls were extracted with two different extraction systems according to the manufacturer's recommendation: (1) one automated, the EZ1® DSP Virus Kit [22] on the EZ1® Advanced XL robot [23] (QIAGEN, Hilden, Germany) (hereinafter QEZ1); (2) another manual, the MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Inc., CA, USA) [24], [25] (hereinafter TFS-MM). For both extraction systems, the following parameters were verified extraction efficiency, repeatability, reproducibility, contamination and concordance.
The extraction efficiency was evaluated through a “relative recovery index”, which provides a relative percentage value of the extraction efficiency between both systems. Based on the GCE value obtained for each theoretical input and the expected GCE value, this recovery index was calculated as follows: RI% = (GCE obtained) / (GCE expected).
2.3. SARS-CoV-2 nucleic acids detection
The TaqPath™ COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Inc., CA, USA) [10] was verified for the qualitative detection of nucleic acid from SARS-CoV-2. This kit with CE-marked In Vitro Diagnostics (CE-IVD) allows the virus detection in upper respiratory specimens (such as nasopharyngeal, oropharyngeal, nasal, and mid-turbinate swabs, as well as nasopharyngeal aspirate) and bronchoalveolar lavage (BAL) specimens from individuals suspected of COVID-19. In this kit, probes anneal to three target sequences that are specific to SARS-CoV-2, reducing the risk of detecting other coronaviruses. Each target is located between unique forward and reverse primers for the following genes: ORF1ab (open reading frame), N Protein (nucleocapsid protein) and S Protein (spike protein). All analyses were carried out on the QuantStudio 5 Real-Time PCR Systems (Thermo Fisher Inc., CA, USA) (hereinafter QS5) [26], although it was also set up for detection on the 7500 Real-Time PCR System [27]. The RT-qPCR amplification cycles were performed according to the manufacturer’s user guide [10]. For this detection system, the following parameters were verified with the specified samples / controls: selectivity / specificity, limit of detection (LoD), repeatability, reproducibility, concordance and false positives and negatives.
2.4. Data analysis
All the data generated by the QS5 equipment were analyzed with the QuantStudio™ Design and Analysis Desktop Software v1.5.1 (Thermo Fisher Inc., CA, USA) (hereinafter DAv1.5.1) [28], which also allows controlling the equipment. With this software, the results were exported in spreadsheet format (XLSX) for subsequent analysis. Additionally, the Design & Analysis Software v2.3.3 (Thermo Fisher Inc., CA, USA) (hereinafter DAv2.3.3) [29] was used for data review and figure generation. Tables, assessment and statistical analyses were generated using Microsoft Excel. When appropriate, the Student's t-test (T-Student) was performed as a deductive statistic to determine if there was a significant difference between the means of two groups or series. Additionally, as internal parameters of this statistic, 2-tailed distributions were considered, appropriate for studies with hypotheses of difference in averages, and with two samples/series of different variance (heteroscedastic).
2.5. Workflow integration
Within the forensic genetics laboratories, Laboratory Information Management Systems (LIMS) are valuable tools, streamlining sample and analysis traceability, batching of multiple samples, integration and data transfer with genetic analyzers, monitoring of test results, or flexible reporting [30], [31], [32], [33].
Management of samples and test was performed using the templates of the LabWare LIMS v6 software (LabWare Inc., Wilmington DE, USA) [34]. LIMS Basic language (LabWare Inc., Wilmington DE, USA) was used to configure the automation scripts for communication with instruments.
3. Results and discussion
3.1. Viral nucleic acids extraction
In determining the most appropriate extraction method for obtaining the SARS-CoV-2 genetic material, the following results were obtained for the parameters evaluated:
3.1.1. Extraction efficiency
The evaluation of this parameter was performed with the CPE [18], at two known (and theoretical) inputs of copies per reaction (GCE). Based on these theoretical values, the relationship between the two extraction systems was assessed ( Table 1). Overall, the automatic method QEZ1 [22] was found to be more than 25% efficient for each probe compared to the manual method TFS-MM [24], [25]. These differences were greater the lower the GCE of the sample (for theoretical 250 GCEs, the differences between both systems would slightly exceed 29%, except in the case of the S gene probe). Considering the Cycles threshold (Cts) values, instead of the relative copy number value, the intra- and inter-system variability would be lower, taking into account the variability coefficient (Supplementary Table S1).
Table 1.
Relationship between the two extraction systems.
| 500 GCE |
250 GCE |
||||||
|---|---|---|---|---|---|---|---|
| ORF1ab | N gene | S gene | ORF1ab | N gene | S gene | ||
| QEZ1/TFS-MM Index | 0.997 | 1.199 | 1.873 | 2.052 | 1.299 | 0.872 | |
| Average QEZ1/TFS-MM index | 1.525 | 1.249 | 1.372 | ||||
Indices are collected between the automatic extraction system EZ1® DSP Virus Kit (QEZ1) and the manual system MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (TFS-MM) for each of the probes analyzed with the TaqPath™ COVID-19 CE-IVD RT-PCR Kit. These indices have been calculated for two theoretical inputs of the AMPLIRUN® TOTAL SARS-CoV-2 CONTROL. The averages of this index are collected for each input and for the set of both in each probe.
3.1.2. Repeatability
Repeatability was assessed with the CPE [18], for each extraction system and two different operators. In Supplementary Table S2, the results for each extraction system detected in two RT-qPCR plates are shown. For the two extraction batches performed for each system and detected in duplicate by RT-qPCR, in the three viral regions analyzed, a repeatability of 100% was obtained at a qualitative level.
In addition, it can be indicated that the average values of the coefficient of variation (CV%) for the quantitative data of Cts values did not exceed 3.5% (Supplementary Table S2). This fact shows that both extraction systems, at a quantitative level, have good repeatability, slightly better in the case of the automatic system QEZ1 [22].
3.1.3. Reproducibility
Reproducibility was assessed with the CPE [18], between extraction batches carried out by different operators for each extraction system. In the Supplementary Table S3, the results obtained in the two RT-qPCR detection plates are shown, in which the results were analyzed in duplicate. For the two extraction batches performed in each system and detected on both RT-qPCR detection plates, for the three regions analyzed, a reproducibility of 100% was obtained at a qualitative level.
For the quantitative data of Cts values, the average values of the CV% did not exceed 5%. At the quantitative level (based on Cts values), statistically significant differences were observed between both extraction batches for the manual system TFS-MM [24], [25] in both RT-qPCR detection plates for each viral region analyzed (Supplementary Table S3). This fact indicates that, although the reproducibility was 100% at the qualitative level (positive/negative results), the variability of this extraction system was statistically significant at the quantitative level. The clearest explanation is that it is a manual extraction system, with what this entails, and, although there may be quantitative differences between operators, these differences would not have any repercussions at a qualitative level. Consequently, the automatic extraction system QEZ1 [22] would be more robust, with no statistically significant differences at the quantitative level.
3.1.4. Contamination
In Supplemental Table S4, the contamination study data are collected for each extraction batch and each system on each of the RT-qPCR detection plates run. Generally, the analysis of negative controls for both extraction systems detected in duplicate showed undetermined Ct values. Only in two of the probes a Ct result was detected (Supplementary Table S4, probe values in red). Nevertheless, these values can be considered as artifactual as they are not reproduced in the rest of the probes of the same negative extraction control (see images included in Supplementary Table S4). And the linear representation of the amplification plots for these logarithmic artifactual curves shows that there was no amplification process. Therefore, they should be considered as negative results at a qualitative level, according to the manufacturer's recommendations [10].
3.1.5. Concordance
In the Supplementary Table S5, the concordance results are collected for both extraction systems, for each of the RT-qPCR detection plates. As indicated (see Section 2.3.), the commercial TPkit [10] allows detecting three regions of the SARS-CoV-2. However, for the clinical samples previously analyzed in the HCSC, only the previous result of one of the genes (protein N gene) was available. For the clinical samples analyzed with each extraction system and detected in both RT-qPCR detection plates, the concordance for the protein N gene was 100% at a qualitative level, according to the HCSC’s results.
3.1.6. Internal extraction control (phage MS2)
The evaluation of the phage MS2 is essential due to its importance in the extraction process (as an efficiency control of this process). In Supplementary Table S6, the raw data of the Cts values obtained for each of the samples analyzed in this verification are collected: different inputs of the CPE control [18]; clinical samples provided by the HCSC; and negative extraction controls. In addition, at the Supplementary Table S6, the general statistical data (mean, standard deviation and coefficient of variation) of the Cts obtained for the probe MS2 are shown, for each extraction system in each detection plates of RT-qPCR, as well as for the joint data from both plates. Likewise, the expected Cts range for this MS2 probe is collected, calculated as the mean of the Cts values plus/minus 2 standard deviations (95% confidence interval) [35].
For the MS2 probe, the average variability between both extraction systems presented differences of almost double, this CV% being greater in the case of the manual system TFS-MM [24], [25]. After removing outlier values, the average ranges of expected Cts for the MS2 probe obtained for each type of extraction was 25.92–29.77 Cts, in the case of the QEZ1 extraction, and 25.77–31.22 Cts, in the case of the TFS-MM extraction. These ranges allow the quality parameters of the RT-qPCR detection system to be established, under laboratory conditions for each extraction system.
3.2. SARS-CoV-2 nucleic acids detection
3.2.1. Selectivity/specificity
During the developmental validation of the TPkit [10], in silico analysis of 43 microorganisms was performed. Although blast analysis showed ≥80% homology for one assay component (forward primer, reverse primer, or probe) for select isolates, there would be no anticipated amplification because hybridization of all three assay components are necessary to generate a signal [10]. The in silico analysis indicated that significant amplification of non-target sequences that result in cross-reactivity or potentially interfere with detection of SARS-CoV-2 was not likely to occur [10].
In this study, 12 real extracts from different RNA viruses were analyzed, including three types of coronaviruses (SARS, NL63 and 229E). All these extracts were amplified in duplicate on two different detection plates. In the Supplementary Table S7, the specificity results of the TPkit [10], for each RT-qPCR detection plate shows that there was no cross-reaction. Although, some samples presented Cts values, these values can be considered as artifactual as they were not reproduced in the rest of the probes of the same negative extraction control (see images included in Supplementary Table S7). And the linear representation of the amplification plots for these logarithmic artifactual curves showed that there was no amplification process. Therefore, they should be considered as negative results at a qualitative level, according to the manufacturer's recommendations [10].
3.2.2. Limit of detection (LoD)
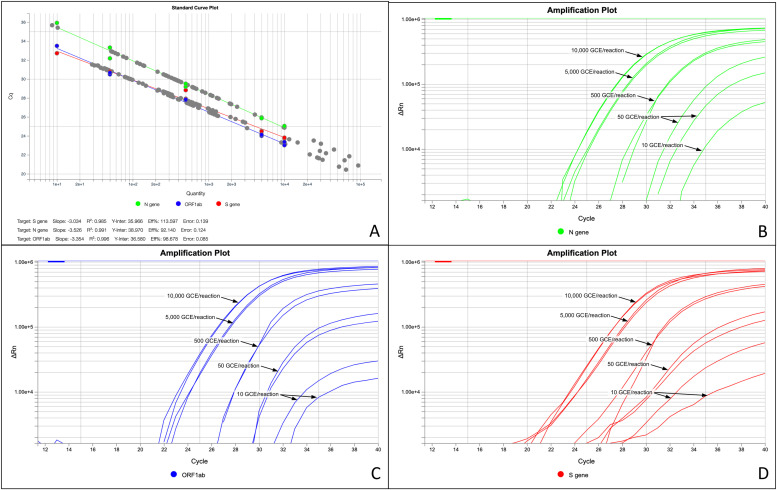
To assess this parameter, serial dilutions were used for controls C19 [21] and CTP [10], which were amplified in duplicate within each plate. Furthermore, the serial dilutions of the C19 control were used as standards to generate the standard curve in each of the RT-qPCR detection plates ( Fig. 1). As can be observed in the Supplementary Table S8, the LoD for the TaqPath™ COVID-19 CE-IVD RT-PCR Kit is 10 copies/reaction as established by the manufacturer.
Fig. 1.
Serial dilutions detection of the AMPLIRUN® CORONAVIRUS SARS-CoV-2 RNA CONTROL using the TaqPath™ COVID-19 CE-IVD RT-PCR Kit. Images from the Design & Analysis Software v2.3.3. A: Standard curves for each of the probes analyzed (ORF1ab, protein N and protein S genes) and their parameters (slope, R2, Y-Inter, Eff% and error). B: Serial dilutions amplification curves of the N protein gene probe. C: Serial dilutions amplification curves of the ORF1ab gene probe. D: Serial dilutions amplification curves of the protein S gene probe.
In the case of the CTP control, in the first detection plate, for one of the theoretical input replicates of 500 GCE/reaction, a discrepant Ct value was observed in the ORF1ab gene probe, which corresponded to an abnormal curve profile (see image included in Supplementary Table S8). Therefore, this Ct value was discarded from being considered as a probable outlier.
Table 2 shows the average Cts values for each probe tested with the TPkit [10]. For the lower input (10 GCE/reaction), Cts values were obtained that did not exceed 37 Cts on average on each probe tested. Based on the range of expected Cts (mean ± 2 sd.), it could be established that the detection limit of the technique with the TPkit [10] would be up to a Ct of 40 for this lower input. Above this Ct value, none of the probes detected with this kit could be considered positive.
Table 2.
General statistics of the average Cts values of the serial dilutions obtained for the AMPLIRUN® CORONAVIRUS SARS-CoV-2 RNA CONTROL and for the TaqPath™ COVID-19 Control of the TaqPath™ COVID-19 CE-IVD RT-PCR Kit.
| ORF1ab gene |
Range | Protein N gene |
Range | Protein S gene |
Range | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | s.d. | CV% | mean ± 2 sd. | mean | s.d. | CV% | mean ± 2 sd. | mean | s.d. | CV% | mean ± 2 sd. | |
| 10,000 GCE/reaction | 23.27 | 0.37 | 1.59 | 22.52 – 24.01 | 24.57 | 0.34 | 1.39 | 23.89 – 25.25 | 23.72 | 0.19 | 0.80 | 23.34 – 24.09 |
| 5000 GCE/reaction | 24.67 | 0.93 | 3.75 | 22.82 – 26.52 | 25.95 | 0.86 | 3.33 | 24.22 – 27.68 | 24.83 | 0.56 | 2.24 | 23.72 – 25.95 |
| 500 GCE/reaction | 29.73 | 2.12 | 7.12 | 25.50 – 33.96 | 30.22 | 1.25 | 4.13 | 27.72 – 32.71 | 29.54 | 1.29 | 4.37 | 26.95 – 32.12 |
| 50 GCE/reaction | 31.98 | 1.69 | 5.27 | 28.61 – 35.35 | 32.53 | 0.48 | 1.46 | 31.57 – 33.48 | 31.64 | 0.95 | 3.01 | 29.73 – 33.54 |
| 10 GCE/reaction | 35.08 | 1.32 | 3.76 | 32.44 – 37.72 | 35.60 | 0.28 | 0.78 | 35.05 – 36.16 | 36.72 | 1.78 | 4.86 | 33.15 – 40.29 |
The mean, standard deviation (s.d.) and coefficient of variation (CV%) are shown for each of the detection kit probes. The range of expected Cts values for each of the serial dilutions is also collected, calculated as the mean ± 2 sd.
In Fig. 1, the standard curves for each of the probes analyzed, obtained from the serial dilutions of the C19 control [30], are shown. Furthermore, in Fig. 1.A., the parameters for each standard curve of each probe are collected: slope, coefficient of determination (R2), y-axis intersection (Y-Inter), efficiency (Eff%) and error. All the probes presented optimal correlation coefficients (R2 > 0.980). At low GCE/reaction inputs (Fig. 1.C and 1.D), it was observed that the reproducibility of both replicates was noticeably worse (Supplementary Table S8). In any case, the TPkit [10] is a qualitative and non-quantitative detection system, although it could be considered as quantitative.
3.2.3. Repeatability
Those samples used for extraction repeatability were assessed in the detection repeatability (Supplementary Table S2). Additionally, three replicates of a quantified control of SARS-CoV-2 RNA provided by the CNM-ISCIII were evaluated (Supplementary Table S9). Overall, a repeatability of 100% was obtained at a qualitative level.
For the quantified control of RNA of the SARS-CoV-2 provided by the CNM-ISCIII, the average values of CV% for the quantitative data of Cts values did not exceed 1.5% (Supplementary Table S9) in each probe tested, consistent with the values obtained for the CPE control (Supplementary Table S2).
3.2.4. Reproducibility
The relative differences between the two RT-qPCR detection plates, performed on the same QS5 kit at two different times, were assessed. Supplementary Table S10 shows the results obtained for both RT-qPCR detection plates. The intra-group variability of samples did not exceed 20% (CV%) for each probe testes. Within each RT-qPCR detection plate, for each probe analyzed, the CV% did not exceed 15%.
Assessing the t-Student contrast statistic for the set of both RT-qPCR detection plates, no statistically significant differences between plates were observed, for each probe analyzed (Supplementary Table S10). In any case, making this assessment by group of samples, only statistically significant differences were observed for the quantified control of RNA of the SARS-CoV-2 provided by the CNM-ISCIII, but only for the N gene probe. These differences can be due to the fact that in this case only 3 controls were available, despite the t-Student statistic being indicated for small sample sizes. In any case, as indicated, the TPkit [10] is a qualitative not quantitative detection system, therefore the inter-plate reproducibility would be 100% at a qualitative level.
3.2.5. Concordance
The concordance for the TPkit [10] was assessed with the clinical samples analyzed in the extraction verification (see Section 3.1.5). Thereby, for the set of tests carried out and the working conditions of the laboratory, the concordance was estimated to be 100%, from a qualitative point of view (Supplementary Table S5).
3.2.6. False positives and negatives
The percentage of positive results was calculated for the genes analyzed by the TPkit [10] in viruses other than SARS-CoV-2 (used in the specificity parameter, see Section 3.2.1.) and in all those a priori negative samples, such as extraction and amplification controls. The Supplementary Table S11 shows the results for each RT-qPCR detection plate. Generally and qualitatively, the false positives were 0.00%. In any case, for the probes highlighted in red and blue in Supplementary Table S11, despite presenting a certain Ct value, this value corresponded to artifactual and/or non-reproducible curves in the replicas of those same samples (see images included in Supplementary Table S11). The linear representation of the amplification plots for these logarithmic artifactual curves shows that there was no amplification process. Therefore, they must be considered as negative results at a qualitative level.
On the other hand, the percentage of negative results was calculated for the genes analyzed by the TPkit [10] in samples that presumably containing the SARS-CoV-2 (controls C19, CTP and CPE; the quantified control of the SARS-CoV-2 virus RNA provided by CNM-ISCIII; and the clinical samples provided by HCSC). Supplemental Table S12 shows the results for each RT-qPCR detection plate. As can be seen, in a general and qualitative way, the false negatives were 0.00%.
3.3. General discussion
The RNA extraction method from post-mortem samples will be essential to obtain the greatest amount of genetic material from the SARS-CoV-2 (efficiency). Another key point for choosing the extraction method will be the speed and the least manipulation that it implies, but also, its accuracy and its reproducibility. Although the manual extraction method TFS-MM [24], [25] meets many of the requirements, the automatic extraction method QEZ1 [22] seems to be the most appropriate for the workflow of our laboratory.
Regarding the detection method, sensitivity and specificity, together with the low percentage or null of false positives and negatives, will be fundamental parameters to assess the utility of any diagnostic test and its predictive value [36], [37]. In the case of post-mortem samples, this predictive value will be relative since, except for anatomopathological studies, it will be difficult to confirm whether the RT-qPCR result is correct or is a false negative or positive result. As Watson et al. point out [38], no test gives a 100% accurate result and there is no “gold standard” test to compare with, so evaluating the accuracy of any test, even if it is RT-qPCR, will be challenging. Higher sensitivities are reported depending on which gene targets are used, and whether multiple gene tests are used in combination [39], [40].
Regarding the efficiency/specificity of the TPkit [10], it is important to highlight that its multi-target design (simultaneous analyses of three regions of the genome: ORF1ab, protein S and protein N genes), could allow to detect new variants of the SARS-CoV-2, such as the one discovered in the United Kingdom (B.1.1.7 lineage, 501Y.V1 variant) [41], already present in Spain. This variant has a specific mutation in the S-gene which results in a deletion of two amino acids at sites 69 (histidine) and 70 (valine), commonly referred to as 69–70del [41]. In this case, even if the mutation in the S-gene could produce a negative result in the protein S target (S-gene dropout), the other two probes results of the TPkit [10] reduce the number of false negatives (and/or positives), by requiring that at least two probes be positive to give a positive final result. Additionally, this kit includes an internal control of the extraction process, the phage MS2, which allows monitoring the entire process, from the efficiency of RNA extraction to the detection of possible amplification inhibitors.
Nevertheless, The European Centre for Disease Prevention and Control (ECDC) notes that, “Laboratories should review the PCR performance and drop-out of the S-gene. PCR could be used as an indicator for cases with the new variant for further sequencing and investigation” [42].
On the other hand, an important point in the accuracy of the detection method by RT-qPCR is that it probably also varies according to the stage of the disease [43] and the degree of viral multiplication or clearance of the virus [44]. In the case of post-mortem samples, this fact may have relevant consequences, since it is not possible to repeat the analyses at a later stage of the disease. Therefore, the sensitivity of the detection method will be essential. The TPkit [10] has very good sensitivity, up to 10 copies/reaction, which will mean up to 800 copies of the SARS-CoV-2 genome if it starts from a 400 µL sample, which is the initial volume allowed by the automatic extraction method QEZ1 [22].
4. Conclusions
The current health emergency caused by the COVID-19 pandemic has made it necessary for forensic genetics laboratories to contribute their experience in the molecular biology field. With this perspective, the INTCF made the decision to develop an integral workflow for the extraction and detection of genetic material from the SARS-CoV-2 with very specific purposes, fundamentally, to confirm the presence of the virus in suspicious post-mortem samples and to help establish the COD in judicial autopsies.
The validation presented in this work shows that, the systems evaluated for the SARS-CoV-2 nucleic acids extraction from nasopharyngeal swabs are repetitive, reproducible and concordant with the automatic system showing slightly greater efficiency than the manual one. Regarding the virus detection system, the assessed kit is specific, sensitive, reproducible and concordant, presenting an acceptable predictive value for the required use.
The integral workflow for detecting the SARS-CoV-2 developed at INTCF is optimal for the required purpose. Furthermore, it allows a complete integration with the laboratory system, integrating the entire process with the internal LIMS system, from the generation of the samples to the generation of the final results report.
Conflict of interest statement
None declared.
Acknowledgments
This study was financially supported by the Ministry of Justice of the Government of Spain (NSU/2020/085). We would like to express our gratitude to Esther Culebras and Alberto Delgado-Iribarne, from the Clinical Microbiology Service of San Carlos Clinical Hospital (HCSC, Madrid, Spain) and Inmaculada Casas, from the Respiratory Viruses and the Flu Unit of the National Center for Microbiology of the Carlos III Health Institute (CNM-ISCIII, Majadahonda, Madrid, Spain) for providing some samples used in this validation. We would like to thank the members of the LIMS Administrators Team of the General Sub Directorate of New Technologies of Justice (SGNTJ) of the Ministry of Justice (Spain) for their helpful technical support. Lastly, we would like to thank the INTCF for their institutional support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.forsciint.2021.110775.
Appendix A. Supplementary material
Verification parameters of extraction systems.
.
. Verification parameters of detection system.
.
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K., Lau E., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.LM, Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol., 2020;5(4):536–544. doi: 10.1038/s41564–020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization, WHO director-general's statement on IHR Emergency Committee on Novel Coronavirus, (2019-nCoV). 〈https://www.who.int/dg/speeches/detail/who-director-general-s-statement-on-ihr-emergency-committee-on-novel-coronavirus-(2019-ncov)〉 (30 January 2020) (Accessed 2021.01.27).
- 7.World Health Organization, WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. 〈https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020〉, 11 March 2020 (Accessed 2021.01.27).
- 8.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. Erratum in: Nature. 580(7803) (2020) E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foundation for Innovative New Diagnostics (FIND), SARS-COV-2 DIAGNOSTIC PIPELINE. 〈https://www.finddx.org/covid-19/pipeline/〉, (Accessed 2021.01.27).
- 10.TaqPath™ COVID‑19 CE‑IVD RT‑PCR Kit, Instructions for use. 〈https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0019215_TaqPathCOVID-19_CE-IVD_RT-PCR%20Kit_IFU.pdf〉 (15 July 2020) (Accessed 2021.01.27).
- 11.Health Information and Quality Authority, Rapid health technology assessment of alternative diagnostic testing approaches for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), 〈https://www.hiqa.ie/sites/default/files/2020-05/Rapid_HTA_COVID-19_tests.pdf〉 (5 May 2020) (Accessed 2021.01.27).
- 12.Health Information and Quality Authority, Rapid health technology assessment (HTA) of alternatives to laboratorybased real-time RT-PCR to diagnose current infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), 〈https://www.hiqa.ie/sites/default/files/2020-10/Rapid-HTA-of-alternative-diagnostic-tests.pdf〉 (21 October 2020) (Accessed 2021.01.27).
- 13.Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26(7):771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H., Beeman M., Mahmood T., Nze U., Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021;413(1):49–71. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization, Advice on the use of point-of-care immunodiagnostic tests for COVID-19, Scientific Brief. 〈https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19〉 (8 April 2020) (Accessed 2021.01.27).
- 16.Pan American Health Organization (PAHO), Laboratory Guidelines for the Detection and Diagnosis of COVID-19 Virus Infection. 〈https://www.paho.org/en/documents/laboratory-guidelines-detection-and-diagnosis-covid-19-virus-infection〉 (30 March 2020) (Accessed 2021.01.27).
- 17.Instituto Nacional de Toxicología y Ciencias Forenses (Ministry of Justice of Spain). 〈https://www.mjusticia.gob.es/cs/Satellite/Portal/es/ministerio/organismos-ministerio-justicia/instituto-nacional〉, (Accessed 2021.01.27).
- 18.AMPLIRUN® TOTAL SARS-CoV-2 CONTROL (SWAB), L-MBTC030-R-EN-01. 〈https://en.vircell.com/products/amplirun-total-sars-cov-2-control/〉 (9 April 2020) (Accessed 2021.01.27).
- 19.AMPLIRUN® CORONAVIRUS SARS (2003) RNA CONTROL, L-MBC136-R-ES-01. 〈https://en.vircell.com/products/amplirun-coronavirus-rna-control/〉 (17 March 2020) (Accessed 2021.01.27).
- 20.College of American Pathologists (CAP), Proficiency Testing: Nucleic acid Amplification, Respiratory - ID2. 〈https://estore.cap.org/OA_HTML/xxCAPibeCCtpItmDspRte.jsp?section=10581&item=613558&sitex=10020:22372:US〉 (Accessed 2021.01.27).
- 21.AMPLIRUN® CORONAVIRUS SARS-CoV-2 RNA CONTROL, L-MBC137-R-EN-01. 〈https://www.vircell.com/media/filer_public/ec/5f/ec5f0e3d-534f-4d58-a9d5–50024042615e/amplirun_sars_cov-2_rna_control_mbc137-r_ruo_en.pdf〉 (31 March 2020) (Accessed 2021.01.27).
- 22.EZ1® DSP Virus Kit Handbook, version 4. 〈https://www.qiagen.com/us/resources/download.aspx?id=6185be15-b095-4c9c-b1ce-9915300b89bf&lang=en〉 (March 2015) (Accessed 2021.01.27).
- 23.EZ1® Advanced XL, User Manual. 〈https://www.qiagen.com/us/resources/download.aspx?id=c9ecd500-147b-4a8e-ae71-3dc86cd3d17a&lang=en〉 (November 2017) (Accessed 2021.01.27).
- 24.MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit, User Guide. 〈https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0018072_MagMAXViralPathoNuclAcidIsolatKit_Manually_UG.pdf〉 (6 December 2019) (Accessed 2021.01.27).
- 25.Procedures for viral nucleic acid isolation, User bulletin. 〈https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0019332_Procedures_viral_nucleic_acid%20isolation_UB.pdf〉 (13 May 2020) (Accessed 2021.01.27).
- 26.QuantStudio™ 3 and 5 Real-Time PCR Systems, Installation, use, and maintenance. 〈https://tools.thermofisher.com/content/sfs/manuals/MAN0010407_QuantStudio3_5_InstallUseMaint_UG.pdf〉 (December 2015) (Accessed 2021.01.27).
- 27.Applied Biosystems 7500/7500 Fast Real-Time PCR Systems, System Maintenance. 〈http://tools.thermofisher.com/content/sfs/manuals/4387777d.pdf〉 (June 2020) (Accessed 2021.01.27).
- 28.QuantStudio™ Design and Analysis Desktop Software. User Guide. Getting started with design and analysis of experiments in the desktop software v1.5.x. 〈https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0010408_QuantStudioDesign_Analysis_Desktop_Software_UG.pdf〉 (19 October 2018) (Accessed 2021.01.27).
- 29.Design & Analysis Software v2.3.3. Release Notes. 〈https://www.thermofisher.com/content/dam/LifeTech/Documents/PDFs/100091492_RN_Design_And_Analysis_Software_v2.3.0.pdf〉, 2020 (Accessed 2021.01.27).
- 30.Steinlechner M., Parson W. Automation and high through-put for a DNA database laboratory: development of a laboratory information management system. Croat. Med. J. 2001;42:252–255. http://www.cmj.hr/2001/42/3/11387633.htm [PubMed] [Google Scholar]
- 31.Leclair B., Scholl T. Application of automation and information systems to forensic genetic specimen processing. Expert Rev. Mol. Diagn. 2005;5:241–250. doi: 10.1586/14737159.5.2.241. [DOI] [PubMed] [Google Scholar]
- 32.Hedman J., Albinsson L., Ansell C., Tapper H., Hansson O., Holgersson S., Ansell R. A fast analysis system for forensic DNA reference samples. Forensic Sci. Int. Genet. 2008;2:184–189. doi: 10.1016/j.fsigen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Barrio P.A., Martin P., Alonso A. The DNASEQEX consortium. LIMS configuration to fit new massively parallel sequencing workflows in forensic genetics. Forensic Sci. Int. Genet. Suppl. Ser. 2017;6:e104–e106. doi: 10.1016/j.fsigss.2017.09.040. [DOI] [Google Scholar]
- 34.Laboratory Information Management System (LIMS) provided by LabWare. 〈http://www.labware.com/en/p/Industries/Forensics〉, (Accessed 2021.01.27).
- 35.Altman D.G., Bland J.M. Standard deviations and standard errors. BMJ. 2005;331:903. doi: 10.1136/bmj.331.7521.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonelli P., Chiumello D., Cesana B.M. Statistical methods for evidence-based medicine: the diagnostic test. Part II. Minerva Anestesiol. 2008;74(9):481–488. https://www.minervamedica.it/en/journals/minerva-anestesiologica/article.php?cod=R02Y2008N09A0481 [PubMed] [Google Scholar]
- 37.Safari S., Baratloo A., Elfil M., Negida A. Evidence based emergency medicine part 2: positive and negative predictive values of diagnostic tests. Emergency. 2015;3(3):87–88. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4608333/ [PMC free article] [PubMed] [Google Scholar]
- 38.Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020;369:m1808. doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- 39.Vogels C.B.F., Brito A.F., Wyllie A.L., et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv. 2020 doi: 10.1101/2020.03.30.20048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan J.F.W., Yip C.C.Y., To K.K.W., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W., Choi G.K., Tam A.R., Cheng V.C., Chan K.H., Tsang O.T., Yuen K.Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies N.G., Barnard R.C., Jarvis C.I., et al. Estimated transmissibility and severity of novel SARS-CoV-2 variant of concern 202012/01 in England. medRxiv. 2020 doi: 10.1101/2020.12.24.20248822. [DOI] [Google Scholar]
- 42.European Centre for Disease Prevention and Control (ECDC), Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. 〈https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom〉 (20 December 2020) (Accessed 2021.01.27).
- 43.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 44.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Verification parameters of extraction systems.
. Verification parameters of detection system.