Abstract
Objectives
SARS-CoV-2 T-cell response characterization represents a crucial issue for defining the role of immune protection against COVID-19. The aim of the study was to assess the SARS-CoV-2 T-cell response in a cohort of COVID-19 convalescent patients and in a group of unexposed subjects.
Methods
SARS-CoV-2 T-cell response was quantified from peripheral blood mononuclear cells (PBMCs) of 87 COVID-19 convalescent subjects (range 7–239 days after symptom onset) and 33 unexposed donors by ex vivo ELISpot assay. Follow-up of SARS-CoV-2 T-cell response was performed in ten subjects up to 12 months after symptom onset. The role of SARS-CoV-2 specific CD4 and CD8 T cells was characterized in a group of COVID-19 convalescent subjects. Moreover, neutralizing antibodies were determined in serum samples.
Results
In 14/33 (42.4%) unexposed donors and 85/87 (97.7%) COVID-19 convalescent subjects a positive result for at least one SARS-CoV-2 antigen was observed. A positive response was observed up to 12 months after COVID-19 infection (median 246 days after symptom onset; range 118–362 days). Of note, SARS-CoV-2 T-cell response seems to be mainly mediated by CD4 T cells. A weak positive correlation was observed between Spike-specific T-cell response and neutralizing antibody titre (p 0.0028; r2 = 0.2891) and positive SARS-CoV-2 T-cell response was observed in 8/9 (88.9%) COVID-19 convalescent subjects with undetectable SARS-CoV-2 neutralizing antibodies.
Discussion
Cross-reactive SARS-CoV-2 T-cell response in uninfected patients may be due to previous infections with other common coronaviruses. Our data suggest that long-term SARS-CoV-2 T-cell response might accompany a waning humoral response.
Keywords: ELISpot, Humoral response, Neutralizing antibodies, SARS-CoV-2, T-cell response
Introduction
There is an urgent need to define the immunological mechanisms involved in SARS-CoV-2 infection. Data obtained in Rhesus macaques showed that immune response generated after first SARS-CoV-2 infection might be protective against reinfection [1], but no clear data are available in humans.
Studies on SARS-CoV-2 immune response revealed that neutralizing antibodies correlate with disease severity, being almost undetectable in patients with mild or asymptomatic infection [2,3], raising the question if humoral response might be sufficient to avoid severe disease [4]. Similarly, in the SARS-CoV-1 setting, the incidence of severe pulmonary disease was higher in those subjects showing higher levels of neutralizing antibody titres [5].
From common coronaviruses infection, including HCoV-HKU1, HCoV-NL63, HCoV-OC43 and HCoV-229E, we have learned that specific IgG and neutralizing antibodies disappear within 1 year [6] while SARS-CoV-1 and MERS-CoV antibody responses wane in a few years with a partial protection from reinfection [7], but less is known in terms of cell-mediated immune response. Studies reported highly activated T cells in patients who experienced COVID-19 [[8], [9], [10]] and the documented presence of T-cell reactivity against SARS-CoV-2 antigens in unexposed subjects raises interesting question about cross-reactivity and cross-protection [11].
Our goals were (a) to investigate and characterize SARS-CoV-2 specific T-cell response in both unexposed donors and a cohort of convalescent patients reporting mild and severe disease and (b) to define the correlation between humoral and cellular response in COVID-19 positive patients.
Materials and methods
Study setting
Mononuclear cells from heparinized whole blood samples (peripheral blood mononuclear cells (PBMCs)) derived from 87 convalescent subjects (median age 47 years; 43 males and 44 females) with RT-PCR proven SARS-CoV-2 infection [12] were retrospectively analysed. Sixty-four out of 87 (73.6%) were mostly asymptomatic or mild symptomatic, showing fever, cough, asthenia; 23/87 (26.4%) were hospitalized for moderate or severe SARS-CoV-2 infection and ten of them (43.5%) required assisted ventilation. The most frequently observed symptoms were fever, anosmia, asthenia, cough and dyspnoea (Table 1 ). Blood samples were obtained from day 7 to day 239 after symptom onset (median 30 days) according to Helsinki declaration and the study was approved by the Ethics Committee of IRCCS Policlinico San Matteo (P-20200041154 and P-20200029440). As controls, stored residual samples obtained from 33 healthy subjects (median age 40 years, range 26–62; 12 males/21 females) collected by August 2019 were used.
Table 1.
Demography and clinics of 87 SARS-CoV-2 exposed donors
| Measured variable | n (%) |
|---|---|
| Sex | |
| Male | 43 (49.4) |
| Female |
44 (50.6) |
| Age (years; median and range) |
47 (19–88) |
| Symptoms | |
| Fever | 62 (71.3) |
| Anosmia | 19 (21.8) |
| Asthenia | 30 (34.5) |
| Cough | 24 (27.6) |
| Dyspnoea | 10 (11.5) |
| Diarrhoea | 5 (5.7) |
| Hospitalization |
23 (26.4) |
| Assisted ventilationa | 10 (11.5) |
High flow nasal cannulae, ventimask and/or continuous positive airway pressure therapy (CPAP).
Peptide pools and antigens
Peptide pools (15 mers 11 overlap) representative of Spike (S) (315 peptides), VME1 (53 peptides), NCAP (102 peptides), NS7B (8 peptides) and NS8 (28 peptides) were used (0.25 μg/mL per well). Additionally, whole lysate obtained from 105 PFU/mL SARS-CoV-2 viral strain isolated in our laboratory was inactivated at UV light and used as antigen.
Ex vivo enzyme-linked immunospot assay (ELISpot assay)
Membrane-bottomed 96-well plates (Multiscreen-IP) from Merck Millipore, Germany, were coated with anti-interferon (IFN)-γ monoclonal capture antibody against from Human IFN-γ ELISpot kits (Diaclone, France) and kept at 4°C overnight. Then, after 2-hr blocking with culture medium, 200 000 cells/100 μL per well were stimulated with antigens; phytohemagglutinin (PHA, 5 μg/mL, Sigma-Aldrich) and medium alone were used as positive and negative control, respectively. All the experiments were performed in duplicate. Plates were maintained overnight at 37°C in a 5% CO2 humidified atmosphere. After multiple wash, anti–IFN–γ biotinylated antibody was added and incubated overnight at 4°C. Finally, streptavidin–alkaline phosphatase conjugate was added, and after 60 min incubation at 37°C in a 5% CO2, substrate 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) was added for 20 min at room temperature. Plates were washed under running water kept overnight at room temperature before spot counting. AID ELISPOT reader system from Autoimmun Diagnostika GmbH (Strasburg, Germany) was used for count.
Results were given as IFN-γ spot forming units (SFU)/106 PBMC, after subtracting medium alone response. Valid results were considered when in presence of PHA higher than 100 IFN-γ SFU/200 000 cells and medium lower than 5 IFN-γ SFU/200 000 cells. Antigen responses higher than mean plus 2 standard deviation (SD) of unstimulated wells and higher than 10 IFN-γ SFU/106 PBMC were considered to be positive.
Depletion of CD4 and/or CD8 T cells
Human CD8 or CD4 MicroBeads from MiltenyiBiotec (Bergisch Gladbach, Germany) were used for depleting CD8 and CD4 cells, respectively in 11 COVID-19 positive subjects according to manufacturer's instruction. MS columns from Milteny Biotec were used for magnetic cell isolation and flow cytometry assay confirmed that the depleted fractions contained less than 5% of target cells. ELIspot assay was then performed in parallel using total PBMC, CD4-depleted PBMC and CD8-depleted PBMC.
Anti-SARS-CoV-2 neutralization assay
Neutralizing antibodies (NT-Abs) titres against SARS-CoV2 was defined according to reported protocol [2] (please see supplementary material for detailed protocol).
Statistical analysis
Quantitative data were reported as absolute number and frequency, while qualitative data were reported as prevalence of event. Descriptive statistics for quantitative data was reported as median and interquartile range (IQR). Comparison between groups was performed using the Mann–Whitney test while Spearman's test was used for the correlation analysis. The Wilcoxon test was used for paired samples analysis. All tests were two-tailed and p < 0.05 was considered statistically significant. GraphPad Prism 8.3.0 (GraphPad Software, La Jolla, CA, USA) was used for analyses.
Results
SARS-CoV-2 specific T-cell response in unexposed and exposed donors
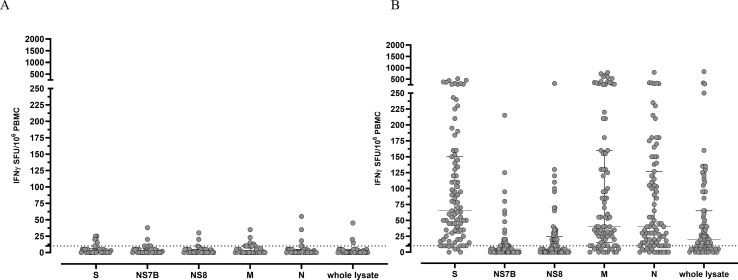
PBMCs from 33 SARS-CoV-2 unexposed donors against SARS-CoV-2 specific peptide pools and whole lysate were tested. Overall, median T-cell response was 2.0 (IQR 0–6.5) IFN-γ SFU/106 PBMCs and 1 (IQR 0–10) IFN-γ SFU/106 PBMC for Spike and Nucleoprotein peptide pools, respectively. Moreover, we detected a median of 1.0 (IQR 0.0–5.0) IFN-γ SFU/106 PBMC for membrane and of 1.0 (IQR 0.0–5.0) for NS8 peptide pools; median T-cell response was 1.0 (IQR 0.0–6.0) IFN-γ SFU/106 PBMC and 0.0 (IQR 0.0–6.0) IFN-γ SFU/106 PBMC for NS7B while the median T-cell response to the whole lysate was almost undetectable (median 0.0 IQR 0.0–4.0 IFN-γ SFU/106 PBMCs) (Fig. 1 A).
Fig. 1.
SARS-CoV-2 specific T-cell response against peptide pools and whole lysate in 33 unexposed donors (A) and 87 exposed donors. T-cell response was measured as IFN-γ SFU/106 PBMCs. The dotted horizontal line represent the chosen cut-off of positivity (10 IFN-γ SFU/106 PBMC).
Fourteen out of 33 (42.4%) unexposed donors showed a positive T-cell response for at least one SARS-CoV-2 peptide pool. Of them, 4/14 (28.6%) experienced a proven HCoV infection in the past while no information was available for the other ten subjects. Seven out of 33 (21.2%) were positive for spike-specific T-cell response, 9/33 (27.3%) showed a positive response T-cell response against nucleoprotein peptide pool, 4/33 (12.2%) were positive for membrane-specific T-cell response and 4/33 (12.2%) for NS8; in 7/33 (21.2%) we observed a positive T-cell response against NS7B peptide pool and 3/33 (9.1%) were positive for whole SARS-CoV-2 lysate.
In exposed donors, all the subjects except two showed a positive T-cell response against at least one SARS-CoV-2 antigen (97.7%). Eighty-two out of 87 (94.3%) subjects showed a positive response against Spike protein and, as expected, the median T-cell response was the highest observed (65 IFN-γ SFU/106 PBMCs, IQR 30–150). Seventy-four out of 87 (85.1%) patients developed a positive response for membrane-specific peptide pool (median 40 IQR 15–160 IFN-γ SFU/106 PBMCs); similarly 74/87 (85.1%) subjects showed a positive response against nucleoprotein and the median T-cell response was 40 IFN-γ SFU/106 PBMCs (IQR 12–127 IFN-γ SFU/106 PBMCs). In 65/87 (74.7%) subjects a positive T-cell response against the whole lysate was detected with a median T-cell response of 20 IFN-γ SFU/106 PBMC (IQR 5–65 IFN-γ SFU/106 PBMCs). Finally, 29/87 (33.3%) and 40/87 (46%) subjects showed a positive T-cell response against NS7B (median 5, IQR 0–25 IFN-γ SFU/106 PBMCs) and NS8 (median 5, IQR 0–25 IFN-γ SFU/106 PBMC) peptide pools was observed (Fig. 1B).
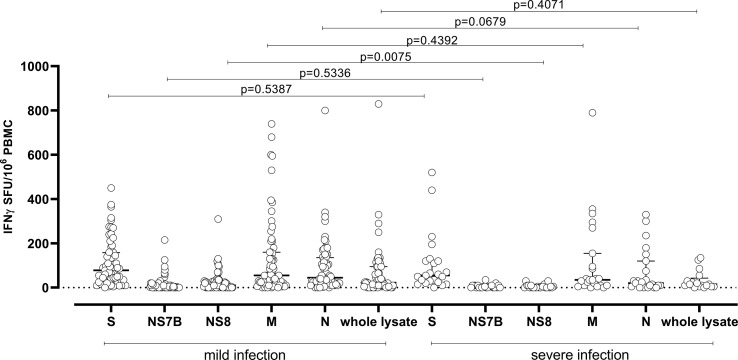
Furthermore, although lower median T-cell responses were observed in patients with severe COVID-19 infection, only a significant lower NS8-specific T-cell response was observed in those subjects than respect patients with mild infection (median T-cell response 10 IQR 0–30 IFN-γ SFU/106 PBMCs vs 0 IQR 0–10 IFN-γ SFU/106 PBMCs; p 0.0075) (Fig. 2 ).
Fig. 2.
Median T-cell responses against Spike (S), NS7B, NS8, Membrane (M), Nucleocapsid (N) and whole lysate were compared in subjects with mild (grey dots) and severe infection (white dots). p value are given for each comparison.
This difference in terms of antigen-specific T-cell response might be due to the number of peptides included in each peptide pool, since in Spike and NCAP peptide pools more than 100 peptides are included, while about 50 peptides are included in the VME1 pool. On the other side, only 28 and 8 peptides are included in NS8 and NS7B peptide pools.
Correlation between SARS-CoV-2 neutralizing antibodies and SARS-CoV-2 specific T-cell response
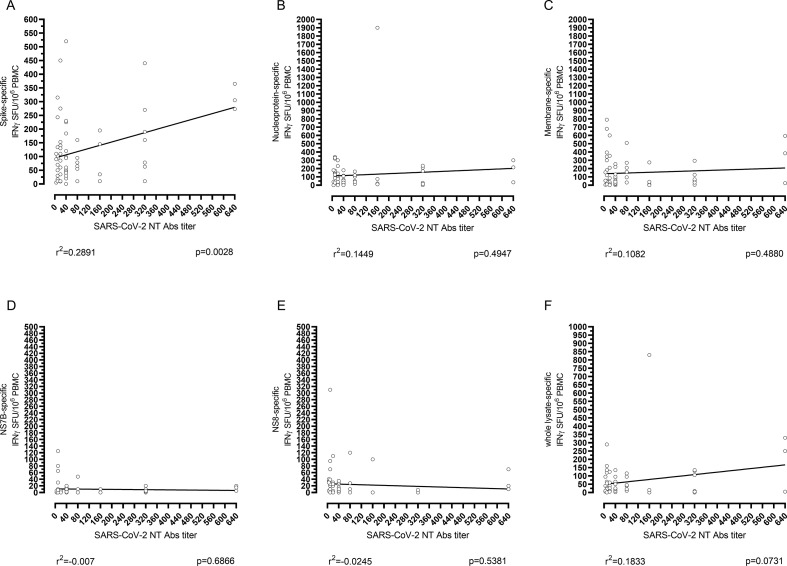
SARS-CoV-2 NT Abs were measured in 79/87 (90.8%) SARS-CoV-2 exposed subjects. Fifty-seven on 79 (72.2%) experienced mild infection while the remaining 22/79 (27.8%) developed severe COVID-19 infection. Nine out of 79 (11.4%) convalescent patients tested negative for SARS-CoV-2 NT Abs (NT Abs titre <1:10) while 70/79 (88.6%) tested positive (NT Abs ≥1:10). The large majority of NT Abs positive SARS-CoV-2 exposed patients showed a low level of SARS-CoV-2 NT Abs (37/70, 52.9%; NT Abs titre ≤1:40) while the remaining 33/70 (47.1%) reported medium-high level of SARS-CoV-2 NT Abs titres (ranging from 1:80 to 1:640).
In our cohort, 59% of subjects with severe infection developed high SARS-CoV-2 NT Abs titres (higher than 1:80) while only 35% of subjects with mild infection showed NT Abs titres higher than 1:80 (p 0.0748). Despite the absence of strong correlation between antigen-specific T-cell response and SARS-CoV-2 NT Abs titre, a weak positive correlation between SARS-CoV-2 NT Abs titre and Spike-specific T-cell response was observed, as well as between SARS-CoV-2 NT Abs and whole lysate-specific T-cell response (Fig. 3 ). Of note, in eight out of nine patients (88.9%) with negative SARS-CoV-2 NT Abs titre a sustained SARS-CoV-2 specific T-cell response was observed. No correlation was observed between SARS-CoV-2-specific T-cell response and days after symptom onset, since r calculated with Spearman's test ranged between 0.0464 and 0.2052; moreover, p values were always not significant (Fig. S1).
Fig. 3.
Correlation between SARS-CoV-2 NT Abs and SARS-CoV-2 specific T-cell response. SARS-CoV-2 specific T-cell response against Spike, NS7B, NS8, Membrane, Nucleoprotein and whole lysate (F) was correlated to the SARS-CoV-2 NT Abs using Spearman's test. The p value and r2 are given for each graph.
CD4 T cell response is mainly involved in SARS-CoV-2 T-cell response in COVID-19 positive patients
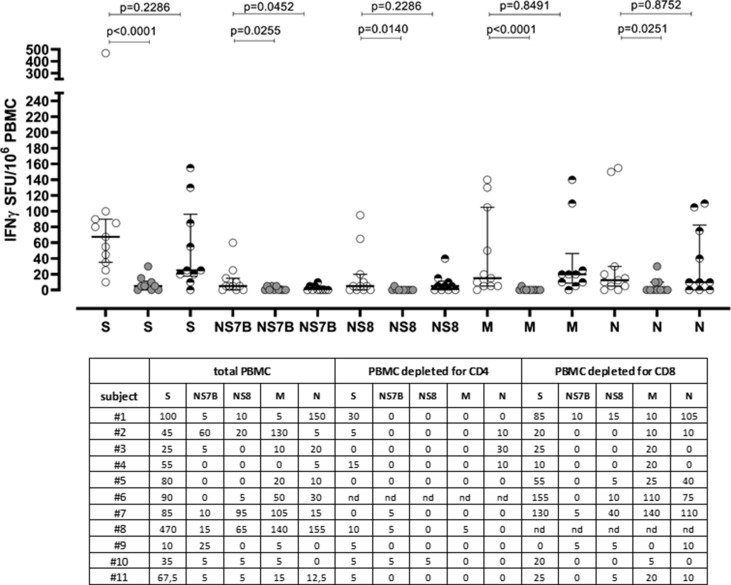
Due to the low amount of cells, we were able to perform pre-depletion assay in 11 COVID-19 positive subjects with sustained SARS-CoV-2-specific T-cell response. As reported in Fig. 4 , the overall SARS-CoV-2-specific T-cell response dramatically decreased by about 95% after CD4+ T-cell depletion. Additionally, Nucleoprotein Membrane and whole lysate-specific T-cell response were almost undetectable after CD4+ T-cell depletion, while the Spike-specific T-cell response was reduced to 7.4%. Otherwise, the overall SARS-CoV-2 T-cell response was reduced to about 80% after CD8+ T-cell depletion, suggesting that SARS-CoV-2 T-cell response was mainly mediated by the T helper response.
Fig. 4.
SARS-CoV-2-specific T-cell response of total PBMCs (white dots), PBMCs depleted for CD4 T cells (grey dots) and PBMCs depleted for CD8 T cells (black and white dots) was measured against Spike (S), NS7B, NS8, Membrane (M) and Nucleoprotein (N) peptide pools. Median T-cell responses were given (horizontal line) and compared using the Mann–Whitney test; p value for each comparison is given in the graph. Row data for total PBMC, CD4-depleted PBMC and CD8-depleted PBMC are provided.
Follow-up of SARS-CoV-2 T cells in a subset of patients suggests a persistence of immunological memory over time
Persistence of SARS-CoV-2 specific T-cell response at median 56 days after symptom onset (range 20–84 days; T1) and at 246 days after symptom onset (range 118–362 days; T2) was evaluated in ten subjects (Fig. S2). Median T-cell response for Spike protein was stable and maintained at T1 and T2 since medians were 62.5 (IQR 34.8–110.8) and 65.0 (IQR 37.5–95.5) IFN-γ SFU/106 PBMCs (p 0.6074); the M (membrane) and N (nucleoprotein) median T-cell response at T1 were 65.0 (IQR 10–216.3) and 44.0 (IQR 12.5–103.5) IFN-γ SFU/106 PBMCs and declined to 25.5 (IQR 5–80) and to 26.5 (IQR 5.70.0) IFN-γ SFU/106 PBMCs, respectively; however, the differences were not statistically significant (p 0.6875 and p 0.1602, respectively). At T1, NS7B and NS8 median T-cell responses were 7.5 IQR (0–12.5) and 5.0 (IQR 0–10.7), respectively, while at T2 median T-cell responses were 5.0 (IQR 0–3.5) and 5 (IQR 0–10) and no statistical difference was reported (p 0.0625 and p 0.6875, respectively). Finally, a significant difference between T1 and T2 was observed only for whole lysate T-cell response, since the median level of response decreased from 29 (IQR 12–133.8) to 10 (10–27.5) IFN-γ SFU/106 PBMC (p 0.0313). However, for the latter antigen only eight paired samples were analysed, due to low amount of cells available.
Discussion
We observed that the SARS-CoV-2 T-cell response was detectable in more than 97% of convalescent COVID-19 positive subjects and in about 40% of unexposed donors sampled before the pandemic period, in agreement with previous observations [[13], [14], [15]]. The data obtained in healthy population could reflect the endemic circulation of common cold coronaviruses (HCoVs), since they account for about 20% of common cold cases and are ubiquitous [16,17]; thus, the possible cross-reactivity between HCoVs might be due to the recognition of conserved epitopes. According to this observation, it could be speculated that SARS-CoV-2 cross-reactive T-cell response derived from previous infection with common cold HCoVs might contribute to the understanding of protection mechanisms or COVID-19 disease severity. It is widely known that humoral immune response to coronaviruses is variable and commonly short-lived while coronavirus-specific T-cell response might be more sustained and long-term detectable [6,18].
Although 40–60% of unexposed individuals were positive for SARS-CoV-2 T-cell response, less is known about its relevance in the clinical outcome of a COVID-19-infected individual. In keeping with some authors, it could be speculated that the presence of cross-reactive T cells correlates with mild disease [19,20].
Furthermore, we demonstrated that SARS-CoV-2 reactive T cells were mainly CD4+, agreeing with other reports [9,11,13]. The induction of a SARS-CoV-2-specific CD4+ T cell response may be crucial in the generation of neutralizing antibodies, being T helper cell response and humoral response interdependent [21]. So far, we have evidenced a weak but significant linear correlation between Spike-specific T-cell response and SARS-CoV-2 NT Abs that suggested a strict relation between adaptive and humoral responses. A sustained number of COVID-19 positive subjects in convalescence showed a positive SARS-CoV-2 T-cell response in the absence of SARS-CoV-2 NT Abs, as confirmed by others [22]. From SARS, we learned that memory T cells could be detectable even 11 [23] and 17 years [14] after primary infection, suggesting a long-lasting memory response. This trend was also observed in other clinical settings, including HBV vaccination [24] and Flaviviruses infection [25]. According to our data, quantification of SARS-CoV-2-specific T-cell response might be useful in case of suspected COVID-19 infection in the absence of a detectable humoral response.
As reported in our paper and according to our previous observation [2], higher SARS-CoV-2 NT titres are detectable in those subjects who experienced severe COVID-19 infection. In terms of a cell-mediated immune response, mild cases have been reported to show a more prevalent T-cell response against Membrane and Nucleocapsid antigens than in patients with severe disease [26]. In our cohort, subjects with severe COVID-19 infection reported an overall lower SARS-CoV-2 T-cell response against all the peptide pools analysed. A significant difference was reported only for NS8 and the role of single proteins should be further explored, since evolutionary changes in functionality of NS7B and NS8 proteins seem to be involved in human infective characteristics of SARS-CoV-2 [27].
As major limitations we reported the low number of samples used for pre-depletion assays and the lack of a phenotypical characterization of the SARS-CoV-2 T-cell response. We designed an ex vivo ELISpot assay for characterization of the SARS-CoV-2 T-cell response in convalescent subjects, demonstrating the presence of SARS-CoV-2-reactive T cells in more than 97% of convalescent COVID-19 patients and about 40% of healthy unexposed donors. The presence of detectable SARS-CoV-2 specific T cell response despite the absence SARS-CoV-2 NT Abs suggests that cellular immunity might be more sustained and long-lived than humoral response. Larger prospective studies are mandatory for understanding the mechanisms of immune protection against SARS-CoV-2.
Author contributions
Responsible for the study's conception and design: I.C., E.P. and F.Ba. Acquisition of data: I.C., F.Be, M.V., G.C., M.S., M.C., V.Z., A.C., E.M., A.F., G.C., R.B and M.B. Analysis and interpretation of data: A.P., I.C., E.P. and F.Ba. Drafting the article: I.C., E.P. Critical revision of the manuscript: all authors. Final approval of the version to be submitted: all authors.
Transparency declaration
The authors have no conflicts of interest. This study was supported by the European Commission Horizon 2020 Program (EU Project no 101003650-ATAC).
Acknowledgments
We thank Daniela Sartori for careful preparation of the manuscript.
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.03.010.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Percivalle E., Cambiè G., Cassaniti I., Nepita E.V., Maserati R., Ferrari A. Prevalence of SARS-CoV-2 specific neutralising antibodies in blood donors from the Lodi red zone in Lombardy, Italy, as at 06 april 2020. Euro Surveill. 2020;25:2001031. doi: 10.2807/1560-7917.ES.2020.25.24.2001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Q., He Y. Challenges of convalescent plasma therapy on COVID-19. J Clin Virol. 2020;127:104358. doi: 10.1016/j.jcv.2020.104358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L., Peng H., Zhu Z., Li G., Huang Z., Zhao Z. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J Gen Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 10.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 15.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H. SSRN; 2020. Single-cell transcriptomic analysis of SARS-CoV-2 reactive CD4 + T cells. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 18.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson T.M., Li C.K., Chui C.S., Huang A.K., Perkins M., Liebner J.C. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 20.Hancock K., Veguilla V., Lu X., Zhong W., Butler E.N., Sun H. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 21.Mitchison N.A. T-cell-B-cell cooperation. Nat Rev Immunol. 2004;4:308–312. doi: 10.1038/nri1334. [DOI] [PubMed] [Google Scholar]
- 22.Schwarzkopf S., Krawczyk A., Knop D., Klump H., Heinold A., Heinemann F.M. Cellular immunity in COVID-19 convalescents with PCR-confirmed infection but with undetectable SARS-CoV-2-specific IgG. Emerg Infect Dis. 2021:122–129. doi: 10.3201/2701.203772. [DOI] [PubMed] [Google Scholar]
- 23.Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassaniti I., Calarota S.A., Adzasehoun K.M., Chiesa A., Comolli G., Parea M. Memory T cells specific for HBV enumerated by a peptide-based cultured enzyme-linked immunospot assay in healthy HBV-vaccinated subjects. Hum Vaccin Immunother. 2016;12:2927–2933. doi: 10.1080/21645515.2016.1204500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percivalle E., Cassaniti I., Sarasini A., Rovida F., Adzasehoun K.M.G., Colombini I. WestNile or Usutu Virus? A three-year follow-up of humoral and cellular response in a group of asymptomatic blood donors. Viruses. 2020;12:157. doi: 10.3390/v12020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D. Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. Nat Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahmi M., Kubota Y., Ito M. Nonstructural proteins NS7b and NS8 are likely to be phylogenetically associated with evolution of 2019-nCoV. Infect Genet Evol. 2020;1:104272. doi: 10.1016/j.meegid.2020.104272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.