Abstract
Objectives
Genotyping of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been instrumental in monitoring viral evolution and transmission during the pandemic. The quality of the sequence data obtained from these genotyping efforts depends on several factors, including the quantity/integrity of the input material, the technology, and laboratory-specific implementation. The current lack of guidelines for SARS-CoV-2 genotyping leads to inclusion of error-containing genome sequences in genomic epidemiology studies. We aimed to establish clear and broadly applicable recommendations for reliable virus genotyping.
Methods
We established and used a sequencing data analysis workflow that reliably identifies and removes technical artefacts; such artefacts can result in miscalls when using alternative pipelines to process clinical samples and synthetic viral genomes with an amplicon-based genotyping approach. We evaluated the impact of experimental factors, including viral load and sequencing depth, on correct sequence determination.
Results
We found that at least 1000 viral genomes are necessary to confidently detect variants in the SARS-CoV-2 genome at frequencies of ≥10%. The broad applicability of our recommendations was validated in over 200 clinical samples from six independent laboratories. The genotypes we determined for clinical isolates with sufficient quality cluster by sampling location and period. Our analysis also supports the rise in frequencies of 20A.EU1 and 20A.EU2, two recently reported European strains whose dissemination was facilitated by travel during the summer of 2020.
Conclusions
We present much-needed recommendations for the reliable determination of SARS-CoV-2 genome sequences and demonstrate their broad applicability in a large cohort of clinical samples.
Keywords: Amplicon, Coronavirus, Genome, Genotyping, Guidelines, Next-generation sequencing, NGS, Recommendations, SARS-CoV-2
Graphical abstract
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the family Coronaviridae, is at the origin of the pandemic that started at the end of 2019 [[1], [2], [3]]. Rapid worldwide spread of this pathogen, which has infected and killed millions, has led to an unprecedented global effort to characterize the viral genome and track its evolution. This genomic epidemiological work is indispensable for public health monitoring and for high-resolution contact tracing [4,5]. The unprecedented extent and speed of SARS-CoV-2 genome sequence submissions since the beginning of the pandemic—e.g. 386 000 SARS-CoV-2 genomes deposited as of January 2021 (https://www.gisaid.org)—is a testimony to how crucial genotyping has been in understanding and controlling viral transmission. These efforts have been reinforced with the emergence of viral strains with higher transmissibility, such as B.1.1.7 [6], and the need to ensure the efficacy of the vaccines currently deployed against the circulating strains [7].
The rapid increase in the amount of genomic information is inevitably associated with genome submissions of variable quality, depending on the viral load or integrity of the isolate, or data generation and analysis pipelines, for example. Genotyping errors introduced in this way can impact the conclusions of downstream analyses [[8], [9], [10]].
Broadly applicable guidelines which consider the sensitivity, specificity and limit of detection of different genotyping approaches are essential to reduce the number of miscalls in SARS-CoV-2 genotypes. Due to their relative low cost and simplicity, amplicon-based methods are the most widely used approach for SARS-CoV-2 genotyping [[11], [12], [13], [14]]. Despite their widespread use, these technologies are associated with artefacts and limitations that must be accounted for to ensure that the genotypes obtained are reliable [[8], [9], [10],15]. For example, the fraction at which a variant can be confidently separated from technical noise in Zika virus amplicon-based genotyping is no lower than 3% even when sufficient input material, sequencing depth and replicates are used [16]. However, some of the variants considered in the analysis of SARS-CoV-2 intra-host genome variability have been reported at lower fractions [[17], [18], [19]]. Furthermore, data generated by different laboratories might contain specific biases, compromising direct comparison and downstream analysis [8,9].
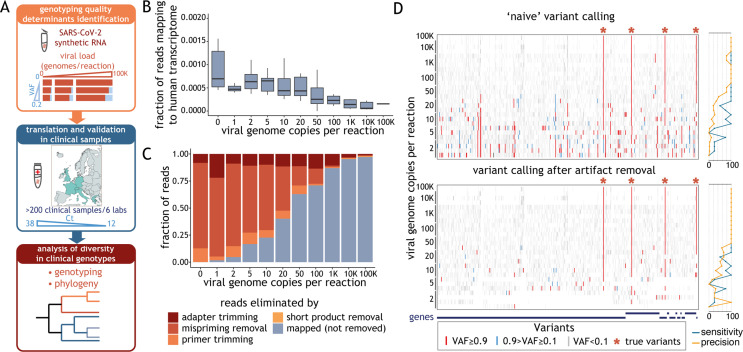
Herein we aim to establish much-needed guidelines for the implementation of amplicon-based SARS-CoV-2 genotyping. We have evaluated the impact of viral load, sequencing depth and coverage uniformity on assay performance using synthetic reference SARS-CoV-2 genomes. To ensure the wide applicability of our conclusions, we analysed over 200 clinical samples from six independent European laboratories (Fig. 1 A). Our study provides general recommendations for reliable determination of viral genome sequences using amplicon-based methods for SARS-CoV-2 genotyping.
Fig. 1.
Artefact removal is a prerequisite for reliable variant calling. (A) Schematic representation of the study. In experiments using synthetic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA, we varied a number of experimental parameters—including viral load, variant allele fraction (VAF) and sequencing depth—and determined which of these factors critically impact(s) genotyping quality (top box). We validated these metrics using data obtained from clinical samples, whose viral load is reflected by the cycle threshold (Ct) value (middle box). We determined the phylogeny of all clinical samples that met our guidelines (bottom box). (B) Distribution of the fraction of raw reads aligning to human transcriptome (y-axis), obtained with STAR aligner, as a function of the number of synthetic viral genome in the sample (x-axis). The horizontal line in the boxplot indicates the median and the whiskers the 5% and 95% quantile. (C) Average fraction (from at least three replicates) of sequencing reads that mapped to the SARS-CoV-2 genome or were the result of different technical artefacts (y-axis) for samples with varying amounts of synthetic viral genomes (x-axis). (D) Ideogram depicting the location of variants detected in samples with a varying number of synthetic viral genomes (denoted on the left) before (top panel) and after (bottom panel) removal of reads labelled as technical artefacts. Variants with allele fraction <0.1, between 0.1 and 0.9, and >0.9 are shown in grey, blue and red, respectively. Expected SARS-CoV-2 variants present in the control are marked with asterisks. Plots on the right show sensitivity and precision of the variant calls.
Methods
For a detailed description of the methods refer to Supplementary Material: Methods.
Library preparation and sequencing
Synthetic SARS-CoV-2 strains were used at the indicated number of copies (Supplementary Material Table S1) and libraries were prepared with the CleanPlex SARS-CoV-2 Panel (Paragon Genomics #918011) according to the manufacturer's instructions [12]. The resulting libraries were quantified, mixed in equimolar amounts and sequenced with 150 bp-long paired-end reads using Illumina sequencers. Sequencing data from clinical isolates were generated independently with the same method and obtained from six European institutions (Supplementary Material Table S2). Only samples in which the presence of SARS-CoV-2 could be confirmed by qPCR were considered.
Sequencing data processing
Read alignment to the reference genome NC_045512.2, read filtering and variant calling were performed using the SOPHiA GENETICS proprietary analysis workflow detailed in Supplementary Material Fig. S1A. Briefly, after read mapping adaptors were trimmed, mispriming events were removed, read softclipped regions were realigned and primer sequences trimmed. Read fragments shorter than 21 bp were excluded, and the resulting alignment was used for variant calling using our pipeline (Supplementary Material Fig. S1A).
RT-qPCR calibration
RT-qPCR calibration was performed in two institutions. Source B performed the test using two types of reference material: synthetic SARS-CoV-2 RNA or plasmid bearing viral genes as described by Jacot et al. [20]. The SOPHiA GENETICS lab performed the test using synthetic SARS-CoV-2 RNA, CDC-USA assay targeting gene N (IDT # 10006713) and One Step PrimeScript™ III RT-PCR Kit (TaKaRa #RR600A) according to the manufacturer's instructions.
Results
Benchmarking of SARS-CoV-2 amplicon-based genotyping using synthetic viral genome
We established an analysis workflow that detects and removes technical artefacts from sequencing data, a step essential to reduce miscalls introduced by amplification errors and mispriming in amplicon-based approaches (Supplementary Material: Methods, Fig. S1A,B). Comparison of the results obtained by our analytical workflow or by the well-established and widely used pipeline iVar [16] demonstrated that the two pipelines have similar overall performance (Supplementary Material Fig. S1C) but our workflow removes more false-positive calls (Supplementary Material Fig. S1D) and detects additional low-frequency deletions (Supplementary Material Fig. S1E). Our analytical pipeline was used for the remainder of the analysis.
We utilized synthetic RNA controls to assess how the number of viral genome copies in a sample impacted the quality of the SARS-CoV-2 genotyping results obtained with a commonly used amplicon-based approach [12]. We spiked 50 ng of reference human RNA with varying amounts of synthetic SARS-CoV-2 genome and sequenced these samples to a median depth of 1.1M reads. The virtual absence of human reads (median 0.03%, Fig. 1B and Supplementary Material Fig. S1F) supports the specificity of the assay and alleviates the legal, ethical and technical concerns of other methods [3,21]. As expected, the total fraction of reads mapped to the SARS-CoV-2 genome after read filtering (hereafter ‘effective reads’) correlated with the number of viral copies in the sample and the yield of amplification of product of the expected size (Fig. 1C, Supplementary Material Fig. S1G,H).
Guidelines for reliable detection of clonal variants
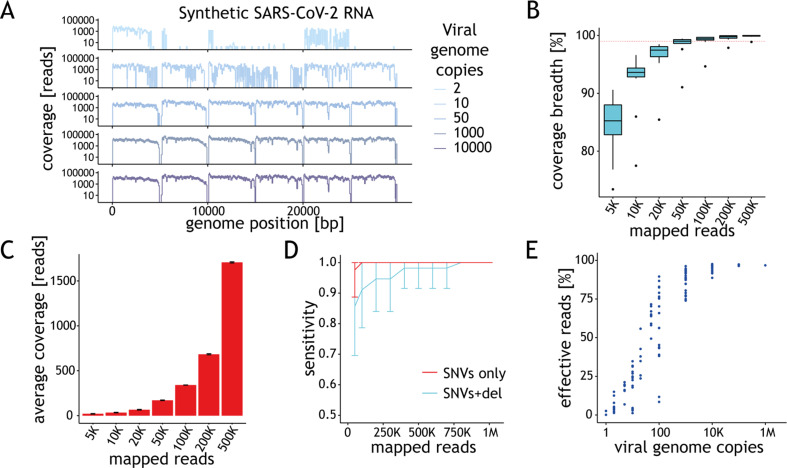
Breadth and depth of coverage are critical determinants of genotyping reliability. Both depend on the number of viral genome copies in the input (Fig. 2 A and Supplementary Material Fig. S2A). To establish the depth required to achieve >10x genome coverage across >98% of positions we randomly down-sampled reads from samples with 10 000 genome copies per reaction (g.c.p.r.). We found that at least 200K mapped, 150 bp paired-end reads are required to achieve these thresholds (Fig. 2B). At this read depth, base coverage was 683x on average (Fig. 2C). No significant improvement in breadth and uniformity of coverage (Fig. 2B and Supplementary Material Fig. S2B) was observed for read depths >200K.
Fig. 2.
Performance of the assay depends on the amount of starting material. (A) Ideograms depicting the genome coverage (y-axis) for representative samples with varying amount of synthetic viral genomes (x-axis). Signal drops every 5 kb are expected due to gaps in the reference material. (B) Distribution of the genome coverage breadth (y-axis) as a function of the number of mapped reads for samples with 10 000 genome copies per reaction (g.c.p.r.). Horizontal dashed line depicts 98% coverage breadth. The horizontal line in the boxplot indicates the median and the whiskers the 5% and 95% quantiles. (C) Average coverage depth across synthetic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome (y-axis) as a function of the number of mapped reads (x-axis) based on data from samples with 10 000 g.c.p.r. (D) Average sensitivity of variant calling for single nucleotide variants (SNVs) (red) or SNVs + 10 bp indel (cyan) in SARS-CoV-2-c1 (y-axis) as a function of the number of mapped reads based on the results obtained for samples with at least 98% genome coverage breadth. Error bars represent standard deviation. (E) Percentage of effective reads (y-axis) shown as a function of the viral load (g.c.p.r.) in the sample. Each point represents the data for one sample.
For single nucleotide variants (SNVs), 100% sensitivity in clonal variant calling was achieved for read depths >50K (Fig. 2D). The presence of a 10-nucleotide deletion, overlapping the annealing site of one of the amplicon primers and leading to a strong decrease in the PCR efficiency (Supplementary Material Fig. S2C), required as much as 800K read depth for detection with 100% sensitivity (Fig. 2D).
The fraction of effective reads is inversely proportional to the number of viral genome copies in the sample. To ensure confident variant calling, we recommend genotyping samples with 1000 or more g.c.p.r. Based on the minimal fraction of mapped reads (74%, Fig. 2E) observed for samples with this viral load, we advise sequencing to a depth of at least 280K reads to achieve the recommended minimal depth of >200K reads. The breadth (Supplementary Material Fig. S2A) and uniformity (Supplementary Material Fig. S2D) of coverage for samples with fewer than 1000 viral g.c.p.r. is often lower than recommended and highly variable, reflecting the technical challenges of handling samples with low input material. As further validation of these recommendations, we performed variant calling on data obtained with the synthetic control representing the B.1.1.7 strain. Except for one SNV adjacent to a gap in the synthetic genome, all the remaining 27 expected clonal variants were detected, with no false-positive calls, in samples with ≥1000 g.c.p.r. when 200K effective reads were used (Supplementary Material Fig. S2E).
Limits of performance for intra-host variability measurement
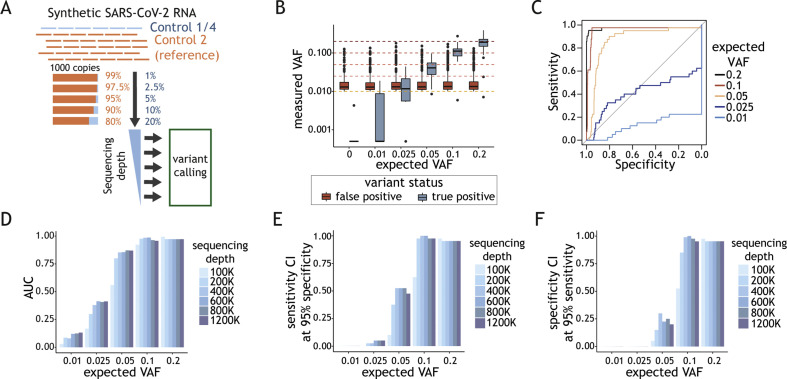
Next, we aimed to determine the lowest allele fraction which can be confidently measured. We spiked human RNA with predefined mixes of different SARS-CoV-2 synthetic genome strains (totalling 1000 g.c.p.r.) to obtain allele fractions between 0.01 and 0.2 (Fig. 3 A). At the recommended depth of 200K mapped reads, the variant allele frequency (VAF) of >95% of false-positive calls was lower than the VAF of true positives at the expected variant fractions of >0.1 (Fig. 3B). Variants at VAF >0.1 were detected with at least 95% sensitivity and 95% specificity (Fig. 3C). The impact of increasing read depth above 200K on the sensitivity and specificity of variant calling was modest (Figs. 3D–F). Experiments conducted with varying viral loads revealed that 100 g.c.p.r. is insufficient for reliable variant calling at frequencies in a range of 0.1–0.2. Increasing the viral to 10 000 g.c.p.r. did not significantly improve the performance (Supplementary Material Fig. S3).
Fig. 3.
Determination of assay parameters for reliable intra-host variability detection. (A) Schematic representation of the experimental design. Varying amounts of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) Control 1 or 4 (blue) were mixed with SARS-CoV-2 synthetic genome reference (Control 2) to obtain desired variant allele fractions (VAFs) (0.01–0.2). One thousand viral genome copy mixes (g.c.p.r.) were spiked into human RNA. Variant calling was performed at varying sequencing depths. (B) Distribution of variant fraction measured for known (true positives, blue) and background (false positives, red) variants (y-axis) as a function of the expected VAFs in the samples (x-axis). The black horizontal line in the boxplot indicates the median and the whiskers the 5% and 95% quantiles. (C) Sensitivity (y-axis) as a function of the specificity (x-axis) with VAF value used as a predictor for true variant calls. The ROC curves are colour-coded depending on the expected VAF of the known variants in each experiment. (D) Area under the ROC curve (AUC) (y-axis) as a function of the expected VAF of the variants (x-axis) at sequencing depth between 100K and 1200K reads. Colour code for analysis done with samples at different sequencing depth is depicted on the right. (E) Sensitivity CI (confidence interval) calculated at 95% specificity (y-axis) and (F) specificity CI at 95% sensitivity (y-axis) as a function the expected VAF for the variant (x-axis). Colour code for analysis done at different sequencing depths is depicted on the right.
In summary, variants at a VAF >0.1 can be confidently detected in samples with 1000 viral genomes or more sequenced to a depth of at least 200K mapped reads.
Multicentre study design for assessment of robust SARS-CoV-2 genotyping
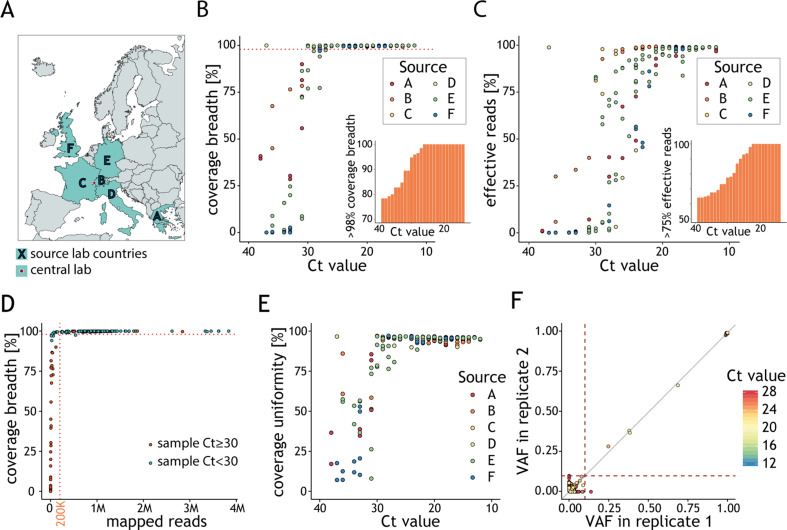
Since the exact number of viral genomes is often unknown for patient isolates, we next sought to make our guidelines broadly applicable to the analysis of clinical samples. We designed a multicentre study (Fig. 4 A and Supplementary Material Table S2) involving six independent European institutions. We left the implementation of the general amplicon-based genotyping protocol [12] to the discretion of each laboratory. The raw sequencing data for 227 clinical samples was processed using our analytical workflow (Supplementary Material Fig. S1A). The RT-qPCR cycle threshold (Ct) value for positive samples ranged between 12 and 38 (Fig. S4A). Independent calibration results suggest that samples with 1000 g.c.p.r. should yield Ct values between 29 and 31 (Supplementary Material Fig. S4B), consistent with those in previous reports [[22], [23], [24]]. Accordingly, 96% of clinical samples with Ct < 29 had at least 98% genome coverage breadth (Fig. 4B and Supplementary Material Fig. S4C), and 81% yielded at least 75% of effective reads (Fig. 4C). Similarly to what was observed in the experiments with synthetic RNA, no improvement in the genome coverage breadth was observed above the recommended value of 200K mapped reads for these samples (Fig. 4D). Despite this, and despite generally good coverage uniformity (Fig. 4E), samples with Ct values between 26 and 29 yield a highly variable fraction of effective reads (1.3–97.9%, median 60.8%, Fig. 4C), making it hard to estimate the depth required to ensure the recommended 200K mapped reads. For samples with Ct < 26, we typically obtained 70–90% of mapped reads. For these samples we recommend sequencing to a depth of ~280K reads to ensure sufficient genome coverage breadth and depth.
Fig. 4.
Viral genotype assignment in clinical samples reflects global genome diversity. (A) The multicentre study involved six laboratories, located in different European countries, which generated datasets analysed at a central location (SOPHiA GENETICS, Switzerland). (B) Fraction of viral genome covered by at least ten reads (y-axis) as a function of the cycle threshold (Ct) value (y-axis). Each point represents the results for a sample, colour-coded according to the source lab. The dashed line indicates 98% coverage breadth. The percentage of samples with at least 98% genome coverage breadth (y-axis) below a given Ct (x-axis) is represented in the inset. (C) Fraction of effective reads mapping to the genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (y-axis) as a function of the Ct value of the clinical samples (x-axis). Each point represents the results for a sample colour-coded according to the source lab. The percentage of samples with at least 75% effective reads (y-axis) below a given Ct (x-axis) is represented in the inset. (D) Fraction of viral genome covered by at least ten reads (y-axis) as a function of the number of reads mapping to the SARS-CoV-2 genome (x-axis). Each point represents a sample and is colour-coded according to its Ct value. The horizontal dotted line indicates 98% coverage breadth and vertical dotted line indicates 200K mapped reads. (E) Percentage of genome coverage uniformity (y-axis) as a function of the sample Ct value (x-axis). Each point represents the results for a sample colour-coded according to the source lab. (F) Relationship between variant fraction for variant calls in clinical samples processed in replicates and with genome coverage breadth >98%. Dotted lines demarcate variant allele fraction (VAF) = 0.1. Variants are coloured based on the Ct value of the replicate.
Taking advantage of replicates present in our dataset, we observed that >99% of variants with VAF ≥0.1 were reproducibly detected and their VAFs were strongly correlated (Pearson r = 0.996) (Fig. 4F). No correlation between replicates was detected for the remaining variants (Pearson r = –0.45). Clinical samples that did not fulfil our recommendations contained an excess of false-positive calls due to the increased background noise resulting from lower quantity/quality of the viral RNA (Supplementary Material Fig. S4D,E).
Viral genotype assignment in clinical samples reflects global genome diversity
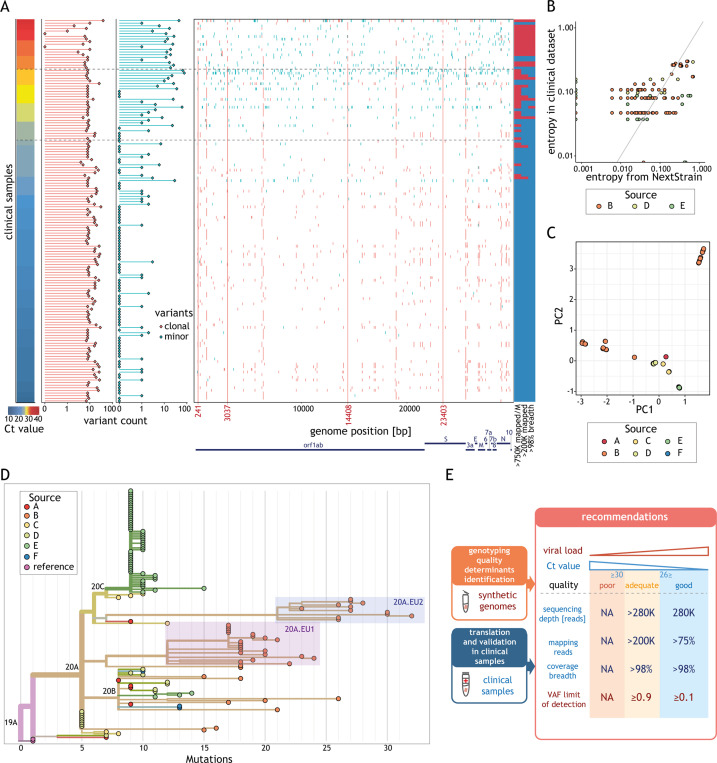
Most of the unique clinical isolates fulfilling our quality criteria (133/135, 98.5%) were characterized by the presence of four alternative alleles (Fig. 5 A) representative of the clade responsible for the European outbreak (C241T, C3037T, C14408T, A23403G) [8], as expected. The frequencies of the clonal variants identified matched well those of samples collected at the similar location/period (Pearson r = 0.701 and p < 10−15) (Fig. 5B). In addition, genetic diversity of the clonal variants reflects the location/time of collection (Fig. 5C). A large fraction of the variability is explained by the difference between samples collected before and after July (Supplementary Material Fig. S5B). Most clinical samples collected before July 2020 belong to clade 20A (G in GISAID nomenclature or lineage B.1 according to cov-lineages.org), characterized by the presence of variant D614G that seeded the SARS-CoV-2 outbreak in Europe, and its daughter clades 20B (GR in GISAID, B.1.1 lineage) and 20C (GH in GISAID, mostly B.1.329 lineage) (Fig. 5D). The increased diversity observed after July 2020 is explained by the presence of ~82% of clinical isolates belonging to subclades 20A.EU1 (lineage B.1.177) and 20A.EU2. These two strains are thought to have emerged in Europe during the early summer of 2020 and their spread across multiple European countries was facilitated by increased cross-border travelling during the summer holiday [25].
Fig. 5.
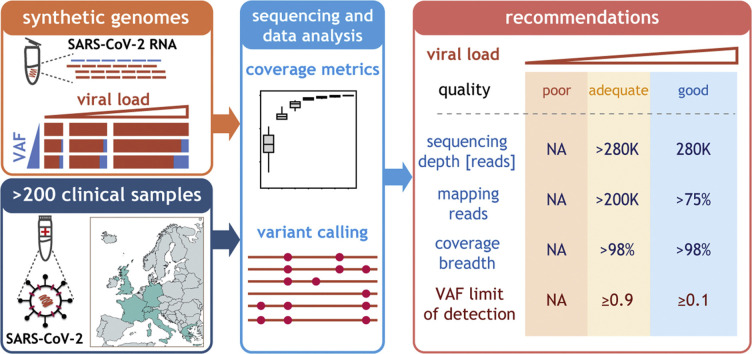
Variant frequencies found in the clinical dataset reflect global frequencies. (A) Summary of the variant calling analysis for all unique clinical samples (rows) sorted by the cycle threshold (Ct) value (left). The horizontal dashed lines indicate Ct values of 26 and 30. The numbers of clonal (variant allele fractions, VAF ≥ 0.9, red) and minor (0.1 < VAF < 0.9, cyan) variants for each sample are represented as horizontal bar-plots (middle left). The position of each clonal (red) and minor (cyan) variant is displayed along the genome (middle right). Coordinates marked in red indicate positions of the most prevalent variants. Classification of the samples relative to the different recommendations (listed below each column) (right): blue indicates the recommendation was fulfilled and red that it was not. (B) Relationship between the entropy estimated for all clonal variants in clinical samples (y-axis) and the entropy of the same variants in samples collected in the same country and during the same period according to Nextstrain [30] (x-axis). Only samples with >200 K effective reads and 98% coverage breadth from centres with data for more than 15 samples were considered in this analysis. (C) 2-D principal component analysis results of clonal variants in clinical isolates (points). Points are coloured based on the sample source. (D) Phylogenetic tree of all clinical isolates with >200 K effective reads and 98% coverage breadth criteria. Samples are coloured according to the source. Clades (according to Nextstrain) are indicated. Samples corresponding to subclade 20A.EU.1 and 20A.EU.2 are highlighted by red and blue boxes, respectively. Length of the branches reflects the number of mutations (x-axis). The tree visualization was generated using the Nextstrain platform [30]. (E) Schematic representation of the recommendations for reliable genotyping with amplicon-based approach. We used synthetic viral genomes to determine the minimal viral load and VAF. We validated these recommendations and made them broadly applicable using clinical samples by determining the minimal sequencing depth, fraction of mapped reads and coverage breadth. Samples were classified into three quality categories based on their viral load: good (≥1000 genome copies per reaction (g.c.p.r.)), adequate (uncertain g.c.p.r., Ct values in the range 26–30) and poor (<100 g.c.p.r., typically value Ct > 30).
Discussion
Sensitive, precise and high-throughput genotyping methods are central to monitoring SARS-CoV-2 transmission and evolution. Evaluation of the analytical performance of different approaches is required to safeguard the quality of the reported genomes and to prevent the inclusion of poor-quality sequences and miscalls in public repositories [[8], [9], [10],15]. We used synthetic RNA and clinical isolates from multiple centres to establish clear and widely applicable guidelines, ensuring reliable SARS-CoV-2 genotyping using an amplicon-based approach (Fig. 5E).
Our analysis shows that variants present at VAF >0.1 can be reliably detected in samples with at least 1000 g.c.p.r. (approximately 100 viral genomes per millilitre). Libraries generated from these samples are sufficiently complex to ensure at least 98% of genome coverage at a depth of ≥10 reads with >200K mapped reads. The large majority yield at least 75% effective reads.
We show that, in addition to the commonly used RT-qPCR Ct value, the fraction of effectively mapping reads and the breadth of coverage can be used to evaluate the quality and quantity of viral genome material in clinical samples. Based on the analysis of coverage breadth and depth we estimate that samples with Ct value of ≤29 contain the recommended viral load of >1000 g.c.p.r. The general applicability of this Ct threshold is supported by the qPCR analysis of serial dilutions of viral samples and the diverse array of chemistries and instrumentations used to estimate viral load by the different laboratories.
Even if several studies report variants found at frequencies <0.1 [17,19,[26], [27], [28]], only a few evaluated the confidence of such calls [18,29]. We found that the number of variants with VAF >0.1 detected in samples sequenced to the sufficient depth and with the recommended viral load is similar between samples collected during the same period and increases for samples collected later in the pandemic. In contrast, in clinical samples with low viral load, we observed an elevated number of low-frequency variants, consistent with increased background noise and false-positive calls. These observations illustrate the risk of not considering how technical factors impact the accuracy of the calls in SARS-CoV-2 genotyping and are a testament to the value of applying clear guidelines to select samples of sufficient quality to inform genomic epidemiology studies.
In summary, we demonstrate that at least 1000 viral copies per reaction are needed for reliable detection of variants with VAF >0.1 using amplicon-based approaches. Widespread implementation of technical guidelines for SARS-CoV-2 genotyping will improve the quality of reported genotypes and the reliability of downstream analysis.
Availability of data and materials
The datasets generated with synthetic SARS-CoV-2 genome and analysed during the current study are available in the Sequence Read Archive (SRA) repository under accession number PRJNA681574. The data generated with clinical samples and used in this study are available from each respective source institution (listed in Supplementary Material Table S2), but restrictions apply to the availability of these data, which were used under license for the current study and so are not publicly available. Requests must be directed to the owner of each dataset.
Author contributions
Study conception and design: SK, ACM, AW and ZX. Acquisition of data: SK, JS, CB, FDM, SP, TB, YD, CA, HS, AS, MM, EWS, ASM, JC, RS, PC, MS, GG and CT. Analysis and data interpretation: SK, ACM, XX, YW and LS. Study supervision: PM, AW and ZX. Drafting of manuscript: SK, ACM, LMS, VP and ZX. All authors read and approved the final manuscript.
Transparency declaration
The authors declare that they have no conflicts of interest. SK, ACM, XX, LS, YW, AS, MM, EWS, PM, MB, AW and ZX are employees of SOPHiA GENETICS. LMS is a co-founder of SOPHiA GENETICS. GG is co-director of JeuPRO, a game company distributing the Krobs game, and medical advisor of Resistell. Both have no impact on the present research project. VP reports grants from Sophia Genetics SA, during the conduct of the study. LMS reports other financial activities from Stanford University, European Molecular Biology Laboratory and Levitas Bio, USA, and personal fees from Recombia Biosciences, USA, and Danaher Corporation, USA, outside the submitted work. The work performed in this study was conducted outside of his duties with his primary employers Stanford University or the European Molecular Biology Laboratory.
Acknowledgements
We would like to thank Dr Frederic Michaud for discussion on how to visualize viral diversity; Dennis Deschka (LABCON-OWL GmbH, Bad Salzuflen) for his excellent advice and technical support during the project; Sébastien Aeby and Dr Katia Jaton (Institut de Microbiologie, CHUV) for their help in the acquisition of clinical sample data; Professor Christelle Thauvin and Professor Laurence Faivre (Département de Génétique, CHU Dijon); Hélène Giraudon, Catherine Manoha (Laboratoire de Virologie, CHU Dijon); Professor Lionel Piroth (Département des Maladies Infectieuses et Tropicales CHU Dijon); Professor Etienne Carbonnelle and Dr Ségolène Brichler (Service de Microbiologie, CHU Avicenne) for their help in acquisition of clinical sample data; Dr Stavroula Samara and Dr Katerina Oikonomaki for help in library preparation.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.03.029.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grubaugh N.D., Ladner J.T., Lemey P., Pybus O.G., Rambaut A., Holmes E.C. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Eurosurveillance; 2021. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020.https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.26.1.2002106 [Internet][cited 2021 Jan 29];26(1). Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E. Immunology; 2021. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants.http://biorxiv.org/lookup/doi/10.1101/2021.01.25.427948 [cited 2021 Feb 3]. Available from: [Google Scholar]
- 8.De Maio N., Walker C., Borges R., Weilguny L., Slodkowicz G., Goldman N. 2020. Issues with SARS-CoV-2 sequencing data.https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 [Internet] Available from: [Google Scholar]
- 9.Turakhia Y., Thornlow B., Gozashti L., Hinrichs A.S., Fernandes J.D., Haussler D. Genomics; 2020. Stability of SARS-CoV-2 phylogenies.http://biorxiv.org/lookup/doi/10.1101/2020.06.08.141127 [Internet] [cited 2020 Nov 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayko M., Komissarov A. Genomics; 2020. Quality control of low-frequency variants in SARS-CoV-2 genomes.http://biorxiv.org/lookup/doi/10.1101/2020.04.26.062422 [Internet] [cited 2020 Nov 3]. Available from: [Google Scholar]
- 11.St Hilaire B.G., Durand N.C., Mitra N., Pulido S.G., Mahajan R., Blackburn A. Genomics; 2020. A rapid, low cost, and highly sensitive SARS-CoV-2 diagnostic based on whole genome sequencing.http://biorxiv.org/lookup/doi/10.1101/2020.04.25.061499 [Internet] [cited 2020 Nov 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C., Debruyne D.N., Spencer J., Kapoor V., Liu L.Y., Zhou B. Genomics; 2020. Highly sensitive and full-genome interrogation of SARS-CoV-2 using multiplexed PCR enrichment followed by next-generation sequencing.http://biorxiv.org/lookup/doi/10.1101/2020.03.12.988246 [Internet] [cited 2020 Nov 3]. Available from: [Google Scholar]
- 13.Resende P.C., Motta F.C., Roy S., Appolinario L., Fabri A., Xavier J. SARS-CoV-2 genomes recovered by long amplicon tiling multiplex approach using nanopore sequencing and applicable to other sequencing platforms. Mol Biol. 2020 http://biorxiv.org/lookup/doi/10.1101/2020.04.30.069039 [Internet] [cited 2020 Nov 3]. Available from: [Google Scholar]
- 14.McNamara R.P., Caro-Vegas C., Landis J.T., Moorad R., Pluta L.J., Eason A.B. High-density amplicon sequencing identifies community spread and ongoing evolution of SARS-CoV-2 in the Southern United States. Cell Rep. 2020;33:108352. doi: 10.1016/j.celrep.2020.108352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karamitros T., Papadopoulou G., Bousali M., Mexias A., Tsiodras S., Mentis A. Genomics; 2020. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies.http://biorxiv.org/lookup/doi/10.1101/2020.03.27.009480 [Internet] [cited 2020 Nov 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Wang D., Zhang L., Sun W., Zhang Z., Chen W. Genomics; 2020. Intra-host variation and evolutionary dynamics of SARS-CoV-2 population in COVID-19 patients.http://biorxiv.org/lookup/doi/10.1101/2020.05.20.103549 [Internet] [cited 2020 Nov 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno G.K., Braun K.M., Halfmann P.J., Prall T.M., Riemersma K.K., Haj A.K. Microbiol; 2020. Limited SARS-CoV-2 diversity within hosts and following passage in cell culture.http://biorxiv.org/lookup/doi/10.1101/2020.04.20.051011 [Internet] [cited 2020 Nov 3]. Available from: [Google Scholar]
- 20.Jacot D., Greub G., Jaton K., Opota O. Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbe. Infect. 2020 doi: 10.1016/j.micinf.2020.08.004. S1286457920301519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houldcroft C.J., Beale M.A., Breuer J. Clinical and biological insights from viral genome sequencing. Nat Rev Microbiol. 2017;15:183–192. doi: 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opota O., Brouillet R., Greub G., Jaton K. Comparison of SARS-CoV-2 RT-PCR on a high-throughput molecular diagnostic platform and the cobas SARS-CoV-2 test for the diagnostic of COVID-19 on various clinical samples. Pathog Dis. 2020;78 doi: 10.1093/femspd/ftaa061. ftaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodcroft E.B., Zuber M., Nadeau S., Comas I., González Candelas F., SeqCOVID-SPAIN consortium . Epidemiol; 2020. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020.http://medrxiv.org/lookup/doi/10.1101/2020.10.25.20219063 [Internet] [cited 2020 Nov 19]. Available from: [Google Scholar]
- 26.Andrés C., Garcia-Cehic D., Gregori J., Piñana M., Rodriguez-Frias F., Guerrero-Murillo M. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg Microbe. Infect. 2020;9:1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sashittal P., Luo Y., Peng J., El-Kebir M. Bioinformatics; 2020. Characterization of SARS-CoV-2 viral diversity within and across hosts.http://biorxiv.org/lookup/doi/10.1101/2020.05.07.083410 [Internet] [cited 2020 Nov 3]. Available from: [Google Scholar]
- 28.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L. Genomic diversity of severe acute respiratory syndrome–coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lythgoe K.A., Hall M., Ferretti L., de Cesare M., MacIntyre-Cockett G., Trebes A. Shared SARS-CoV-2 diversity suggests localised transmission of minority variants. Genomics. 2020 http://biorxiv.org/lookup/doi/10.1101/2020.05.28.118992 [Internet] [cited 2020 Nov 3]. Available from: [Google Scholar]
- 30.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Nextstrain: real-time tracking of pathogen evolution. Kelso J, editor. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated with synthetic SARS-CoV-2 genome and analysed during the current study are available in the Sequence Read Archive (SRA) repository under accession number PRJNA681574. The data generated with clinical samples and used in this study are available from each respective source institution (listed in Supplementary Material Table S2), but restrictions apply to the availability of these data, which were used under license for the current study and so are not publicly available. Requests must be directed to the owner of each dataset.