Abstract
Bcl-x, a member of the Bcl-2 family, plays a key role in apoptosis. Alternative splicing of Bcl-x pre-mRNA through alternative 5’ splice-site selection produces an anti-apoptotic mRNA isoform that includes exon 2b and a pro-apoptotic Bcl-x mRNA isoform that excludes exon 2b. Here we used Bcl-x minigene and identified SRSF2 and SRSF6 as two regulatory factors of 5’ splice-site selection of Bcl-x pre-mRNA. We selected binding clusters closer to 5’ splice-sites from multiple potential binding sites of SRSF2 and SRSF6 to perform loss of functions analysis through site-directed mutagenesis. Our results demonstrated that these mutations did not abolish regulatory functions of SRSF2 or SRSF6, indicating that a single binding motif or a cluster was not a functional target of these proteins in Bcl-x pre-mRNA splicing. Random deletion mutagenesis did not disrupt the role of SRSF2 and SRSF6. Importantly, mutagenesis of 5’ splice-site to a conserved or a weaker score demonstrated that the weaker strength of the target 5’ splice-site or higher strength of the other 5’ splice-site strength limited the role of SRSF2 and SRSF6 in 5’ splice-site activation.
Keywords: 5’ splice-site, Alternative splicing, Bcl-x, SRSF2, SRSF6
INTRODUCTION
Alternative splicing provides genome diversity. Defects of alternative splicing causes multiple human diseases such as cancer (1, 2). Bcl-x, a member of the Bcl-2 family, plays a key role in apoptosis (3). Nearby 5’ splice-site (5’L) selection of Bcl-x pre-mRNA produces a Bcl-x longer mRNA isoform that includes exon 2b, which subsequently directs translation of longer anti-apoptotic Bcl-x protein isoform (Bcl-xL) (Fig. 1A) (4). By contrast, distant 5’ splice-site (5’SS) activation produces a Bcl-x mRNA isoform that excludes exon 2b which subsequently directs translation of shorter pro-apoptotic Bcl-x protein isoform (Bcl-xS) (Fig. 1A). Anti-apoptotic Bcl-xL protein is predominantly expressed in many cancer cells to resist apoptotic stimuli such as chemotherapeutic agents (5). By contrast, pro-apoptotic Bcl-xS causes apoptosis and increase sensitivity to anti-cancer drugs (3). Thus, balanced alternative splicing of Bcl-x plays important roles in the decision of cell survival or cell death. Multiple splicing factors including Sam68, SRSF1, SRSF2, SRSF9 and hnRNP K are shown to regulate alternative splicing of Bcl-x (6).
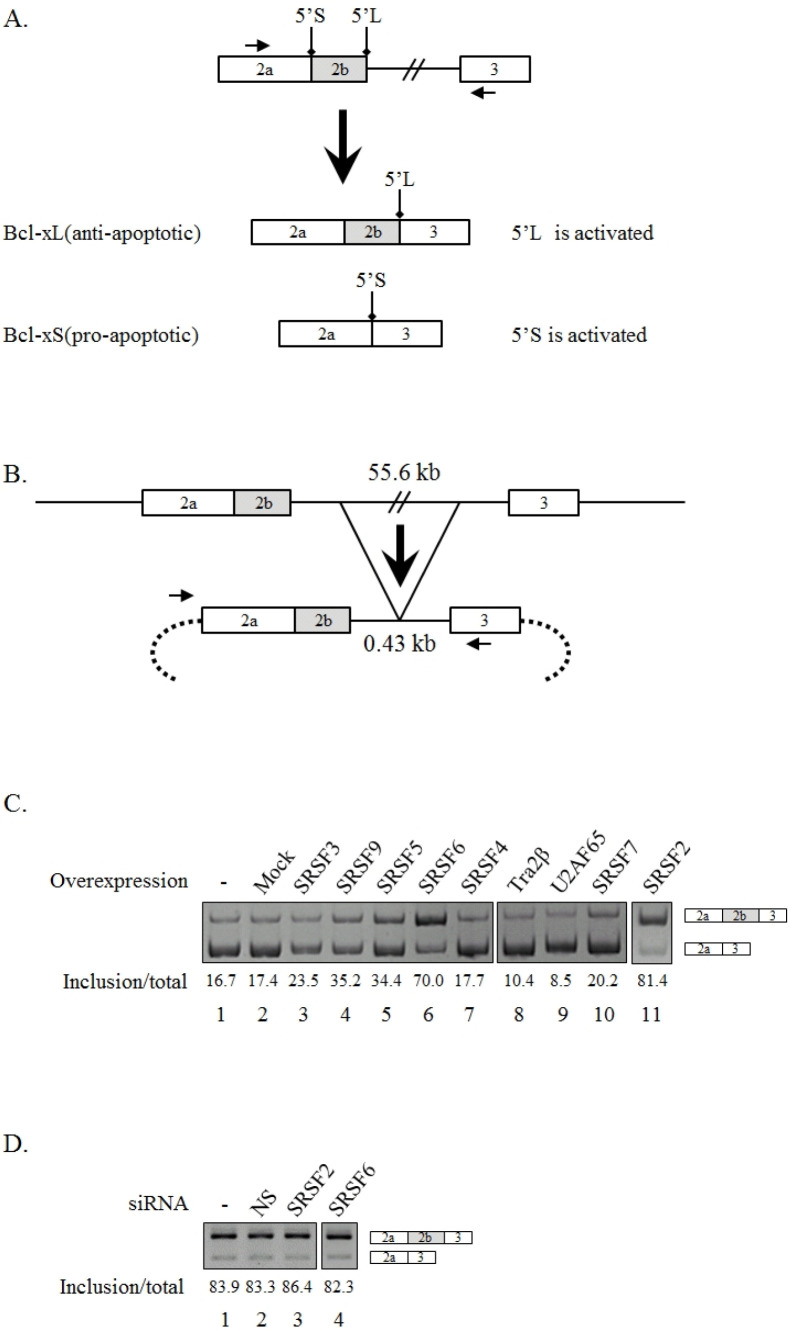
Fig. 1.
SRSF2 and SRSF6 promote longer isoform of Bcl-x alternative splicing. (A) Schematic of alternative 5’ splice-site selection in Bcl-x pre-mRNA is shown. Exons are shown with boxes. Introns are shown with lines. Location of 5’ splice-sites of longer (5’L) and shorter (5’S) isoforms are shown. Primers used in endogenous RT-PCR are shown by arrows. (B) Schematic of Bcl-x minigene is shown. Deleted intron length is shown. Sequences from vector are shown as dotted arc. Primers used in RT-PCR are shown with arrows. (C) RT-PCR assays of 5’ splice-site selection in Bcl-x pre-mRNA within Bcl-x minigene in pcDNA (3.1), SRSF2, SRSF3, SRSF4, SRSF5, SRSF6, SRSF7, SRSF9, Tra2β and U2AF65 overexpressed cells are shown. Quantitation results are also shown. (D) RT-PCR analysis of Bcl-x alternative splicing in SRSF2 or SRSF6 knockdown, untreated or non-silencing shRNA treated 293T cells are shown.
SRSF2 and SRSF6 are members of arginine/serinerich (SR) protein family (7). SR proteins share a bipartite structure with two functional domains: an RNA binding domain with multiple RNA recognition motifs (RRMs) and an arginine/serinerich (RS) domain (7). SR proteins regulate constitutive splicing and alternative splicing. While RNA binding domains provide binding affinity to RNA, RS domain activates splicing (8). Binding motifs of SRSF2 is more degenerate than other SR proteins. Using in vitro functional SELEX, CLIP-seq and structure-based analysis, it was demonstrated that SRSF2 prefers GA- or purinerich and SSNG (S: C/G) (9, 10). SRSF6 prefers USCGKM (S: G/C, K: U/G, M: A/C) sequence in in vitro functional SELEX assay (11).
In the present study, using overexpression of various SR proteins with a Bcl-x minigene, we identified SRSF2 and SRSF6 as the regulatory factors of 5’ splice-site selection of Bcl-x pre-mRNA. Among multiple predicted potential binding sites of SRSF2 and SRSF6 in Bcl-x pre-mRNA, we selected binding clusters closer to 5’ splice-sites to perform loss of functions analysis through site-directed mutagenesis. Our results demonstrated that these mutations did not abolish regulatory functions of SRSF2 or SRSF6, indicating that single binding motif or cluster was not a functional target of these proteins in Bcl-x pre-mRNA splicing. Random deletion mutagenesis did not disrupt the function of SRSF2 and SRSF6. Importantly, mutagenesis of 5’ splice-site to a conserved or a weaker score demonstrated that weaker strength of target 5’ splice-site or higher strength of the other 5’ splice-site limited the role of SRSF2 and SRS6 in 5’ splice-site activation.
RESULTS
SRSF2 and SRSF6 promote longer isoform of Bcl-x alternative splicing
To identify the regulatory factors of alternative 5’ splice-site selection in Bcl-x, we applied a minigene-based analysis in 293T cells (Fig. 1A). We produced a Bcl-x minigene in which only 0.43 kilobases (kb) of intron 2 (55.6 kb) were included (Fig. 1B). As shown in Fig. 1C (lane 1), the minigene produced much more shorter Bcl-x isoform than longer isoform. Thus splicing factors that promote exon inclusion would be easier to be observed with this minigene. In addition, as endogenous Bcl-x had much more longer isoform than shorter isoform, we expected that splicing factors for promoting longer isoforms in minigene would reduce longer isoform in endogenous Bcl-x. Considering the fact that there are more longer Bcl-x isoforms in cells, detecting reduced longer isoform or increased shorted isoform would be much more apparent. To identify regulatory factors of Bcl-x 5’ splice-site selection, we carried out overexpression of various SR proteins along with Bcl-x minigenes (Fig. 1C). RT-PCR analysis showed that SRSF2 and SRSF6 promoted longer isoform splicing significantly (~81.4%, ~70%) (lanes 11 and 6), whereas SRSF3, SRSF4, SRSF5, SRSF7, SRSF9, Tra2β or U2AF65 did not (lanes 3, 7, 5, 10, 4, 8 and 9). To assess whether regulatory functions of SRSF2 and SRSF6 in Bcl-x minigene were also observable in endogenous Bcl-x, we performed splicing assays of endogenous Bcl-x following SRSF2- or SRSF6-targeting shRNA treatment. Fig. 1D shows that reduced expression of SRSF2 and SRSF6 could not alter endogenous Bcl-x splicing, suggesting an inconsistency of effects between minigene and endogenous alternative splicing. Thus we decided to focus our attention on the regulatory mechanisms of SRSF2 and SRSF6 in Bcl-x minigene.
Multiple potential binding motifs of SRSF2 and SRSF6 are predicted in the Bcl-x minigene
Given the fact that SR proteins would target specific RNA sequences to regulate RNA splicing, we first asked whether there might be potential binding sequences of SRSF2 and SRSF6. Functional SELEX (Systematic Evolution of Ligands by Exponential Enrichment) was applied to identify ESEs (Exon Splicing Enhancers) for SRSF2 and SRSF6 proteins. Sequences of ESEs were described in a web-based program called ESEfinder (10-12) (http://exon.cshl.edu/ESE/). In addition to SELEX based analysis, solution structure analysis and functional confirmation demonstrated that SRSF2 could also recognize SSNG (S = C/G) sequences. We then applied Bcl-x minigene RNA sequences into ESEfinder tool and SSNG to locate potential binding sequences of SRSF2 and SRSF6. We demonstrated that there are 72 potential SRSF2 (red) and 31 SRSF6 (blue) binding motifs/clusters in Bcl-x pre-mRNA (Supplementary Fig. 1). As the binding motifs of SR proteins especially SRSF2 are degenerate, some of the binding sequences are overlapped. We observed that there are 23 overlapped binding motifs of SRSF2 and SRSF6 (orange) in Bcl-x pre-mRNA. Thus, we conclude that multiple potential binding motifs/clusters exist in Bcl-x pre-mRNA.
Single binding clusters of SRSF2/SRSF6 are not functional targets of SRSF2/SRSF6 proteins
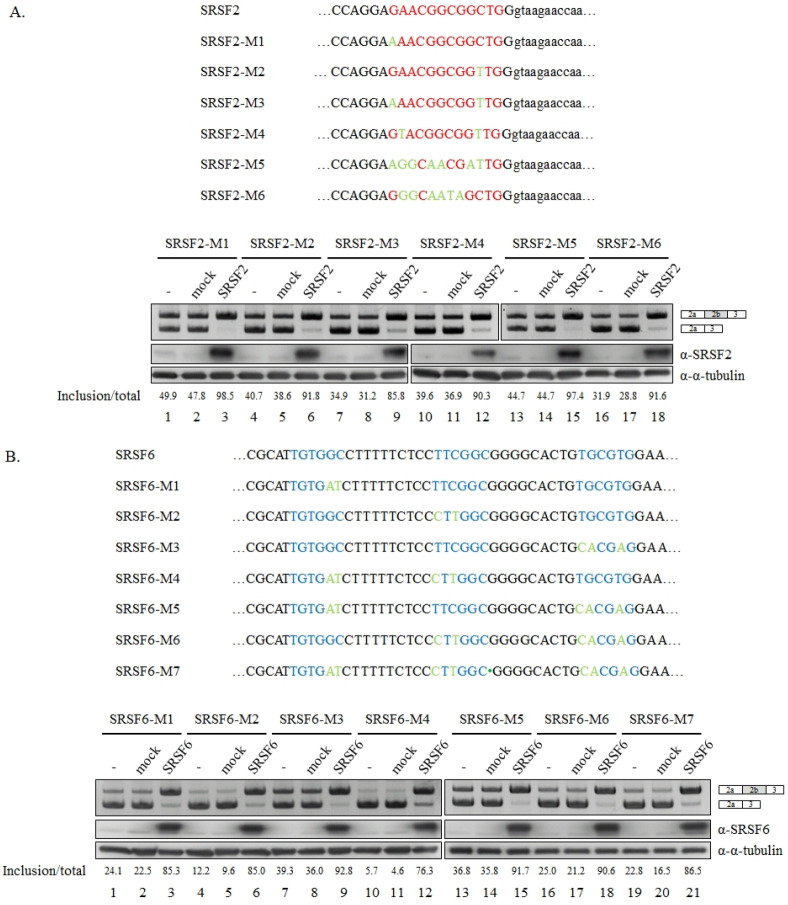
Our laboratory and other laboratories have demonstrated that single binding site or clusters are functional targets of SR proteins (13-15). Using previous approaches, we selected sequences located closer to 5’ splice-sites including several binding motifs of “SRSF2” and “SRSF6” in Fig. 2. We performed “loss-of-function” assay for these potential binding sites, in which mutation of potential binding sites of SRSF2 or SRSF6 in Bcl-x minigene should be able to disrupt the ability of SRSF2 or SRSF6 to promote the longer isoform splicing of Bcl-x. Potential SRSF2 binding sequences we analyzed were located immediately upstream of 5’ splice-site of longer isoform, containing several SRSF2 binding motifs to form a binding cluster (Supplementary Fig. 1 and Fig. 2A, red). We introduced single nucleotide (SRSF2-M1, SRSF2-M2), double nucleotides (SRSF2-M3, SRSF2-M4), and multiple nucleotides (SRSF2-M5, SRSF2-M6) into the Bcl-x minigene (green, Fig. 2A). As shown in Fig. 2A, all mutant minigenes still preserved the function of SRSF2 in promoting longer Bcl-x isoform, indicating that these mutations did not disrupt the function of SRSF2 (lanes 3, 6, 9, 12, 15 and 18). Thus the RNA sequences did not function as the target of SRSF2. We next carried out mutagenesis analysis of potential SRSF6 binding sites. The sequence we analyzed contained a cluster of three SRSF6 binding sequences sites (blue, Fig. 2B, upper). We produced one binding site (SRSF6-M1, SRSF6-M2, SRSF6-M3), two binding sites (SRSF6-M4, SRSF6-M5, SRSF6-M6) and three binding sites (SRSF6-M7) mutations in Bcl-x minigene (green, Fig. 3B, upper). As shown in Fig. 2B, none of these mutants disabled the function of SRSF6 in Bcl-x pre-mRNA splicing (lanes 3, 6, 9, 12, 15, 18 and 21). Taken together, these results indicate that binding clusters of SRSF2/SRSF6 did not play as functional targets of SRSF2/SRSF6.
Fig. 2.
Single binding clusters of SRSF2/SRSF6 are not functional targets of SRSF2/SRSF6 proteins. (A) (Upper) Mutated sequences in SRSF2–M1, SRSF2–M2, SRSF2–M3, SRSF2–M4, SRSF2–M5 and SRSF2– M6 minigenes are shown. Potential SRSF2 binding sequences are shown in red. Mutated sequences are shown in green. (Lower) RT-PCR analysis of 5’ splice-site selection in Bcl-x mutant minigenes, including SRSF2–M1, SRSF2–M2, SRSF2–M3, SRSF2–M4, SRSF2–M5, and SRSF2–M6 minigenes are shown. Quantitation results are shown. (B) (Upper) Mutated sequences in SRSF6–M1, SRSF6–M2, SRSF6–M3, SRSF6–M4, SRSF6–M5, SRSF6–M6, and SRSF6–M7 minigenes are shown. Potential SRSF6 binding sequences are shown in blue. Mutated sequences are shown in green. (Lower) RT-PCR analysis of 5’ splice-site selection in Bcl-x mutant minigenes are shown. Quantitation results are shown.
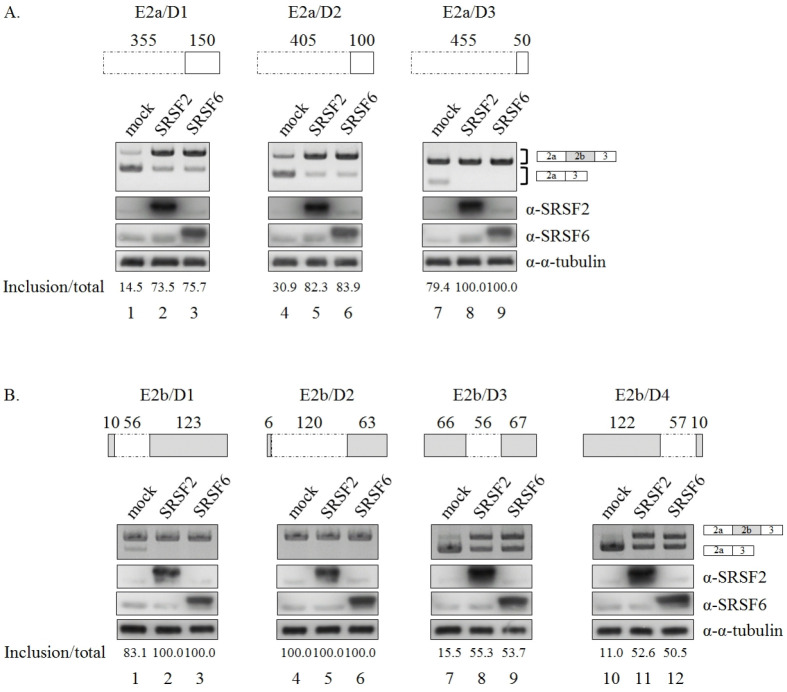
Fig. 3.
Random deletion of exons did not disrupt functions of SRSF2 or SRSF6 in Bcl-x pre-mRNA splicing. (A) (Upper) Deleted regions from exon 2a in E2a/D1, E2a/D2, and E2a/D3 minigenes are shown with dot lined boxes. Length of the deleted parts are also shown. (Lower) RT-PCR analysis of 5’ splice-site selection in E2a/D1, E2a/D2 and E2a/D3 minigenes are shown. Quantitation results are shown. (B) (Upper) Deleted regions from exon 2b in E2b/D1, E2b/D2, E2b/D3 and E2b/D4 minigenes are shown with dot lined boxes. Length of the deleted parts are also shown. (Lower) RT-PCR analysis of 5’ splice-site selection in E2b/D1, E2b/D2, E2b/D3 and E2b/D4 minigenes are shown. Quantitation results are shown.
Random deletion of exons did not disrupt functions of SRSF2/SRSF6 in Bcl-x pre-mRNA splicing
Considering the facts that there are multiple potential binding sequences in Bcl-x pre-mRNA and that site-directed mutagenesis of SRSF2/SRSF6 binding clusters could not identify functional targets of these proteins, we decided to test the possibility that SRSF2 and SRSF6 might function through more binding motifs or clusters. We first deleted 355 nt, 405 nt and 455 nt from exon 2a (E2a/D1, E2a/D2 and E2a/D3) (Fig. 3A, Upper). As shown in Fig. 3A, SRSF2 and SRSF6 were still able to promote longer form expression of E2a/D1 and E2a/D2 mutants (lanes 2, 3, 5 and 6, lower). E2a/D3 mutant produced E2b included isoform exclusively (lane 7), an increase of the inclusion isoform could not be observed (lanes 8 and 9). Thus, E2a deletion mutation was unable to identify the functional target of SRSF2 or SRSF6. We further performed deletion mutagenesis of exon 2b by deleting 56 nt, 120 nt at 5’ of exon 2b (E2b/D1 and E2b/D2), 56 nt from middle (E2b/D3), and 57 nt from 3’ of exon 2b (E2b/D4) (Fig. 3B, Upper). Among these mutants, E2b/D2 mutant produced E2b-included isoform exclusively (lane 4, Fig. 3B, lower), making it difficult to detect the abolishment of functions of SRSF2 and SRSF6 (lanes 5 and 6). Three other mutants showed strong activity of SRSF2 and SRSF6 on exon 2b inclusion (lanes 2, 3, 8, 9, 11 and 12). Thus, random deletions of exon 2b were unable to identify functional targets of SRSF2/SRSF6. These results imply that a combination of binding sites from different locations is needed to be functionally targeted by SRSF2 and SRSF6.
Relative strength of 5’ splice-site strength determines splices-site selection functions of SRSF2 and SRSF6
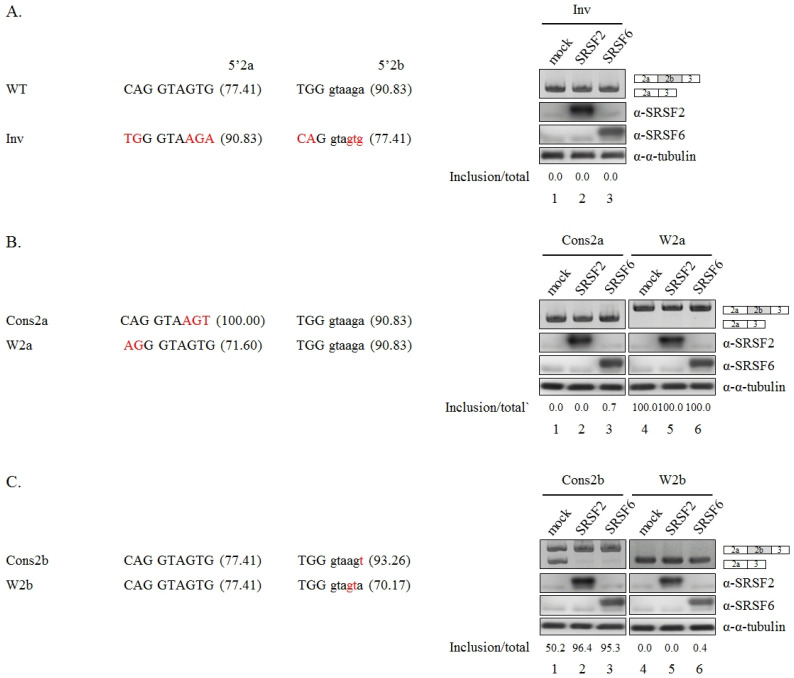
We observed that 5’ splice-site of E2b (5’2b) (90.83) had a much higher strength than E2a (5’2a) (77.41) (Human Splicing Finder, http://umd.be/Redirect.html) (16). We thus wondered whether 5’ splice-site strength might play important roles in the 5’ splice-site selection function of SRSF2 and SRSF6. We first produced a mutant minigene in which 5’2a and 5’2b were inverted (Inv) (Fig. 4A, left). Strikingly, activities of SRSF2 and SRSF6 in promoting exon 2b inclusion were almost abolished (lanes 2 and 3, right). Thus, 5’ splice-site strength plays a role in functions of SRSF2 and SRSF6. We next performed mutagenesis for the 5’ splice-site to assess effects of the strength of 5’ splice-site on splice-site selection. We first mutated 5’2a sequences without changing 5’2b sequences. We produced a conservative or a weaker score (71.60) mutations in 5’2a (Cons2a and W2a) (Fig. 4B, left). As shown in Fig. 4B, Cons2a almost completely abolished activities of SRSF2 and SRSF6 (lanes 2 and 3), indicating that 5’ splice-site with higher score could overcome activation of the other 5’ splice-site by SRSF2 and SRSF6. W2a mutant was spliced to form the longer Bcl-x isoform exclusively. Thus increase of longer isoform by SRSF2 and SRSF6 could not be observed. We further mutated 5’2b sequences without mutating 5’2a sequences. SRSF2 and SRSF6 promoted E2b inclusion of Cons2b mutant (Fig. 4C, right, lanes 2 and 3), in which 5’2b was mutated to a conserved sequence (Fig. 4C, left), suggesting that better splice-site did not interrupt its activation by SRSF2 or SRSF6. By contrast, W2b mutant abolished SRSF2 and SRSF6 (lanes 5 and 6), indicating that weaker 5’ splice-site was not able to support its activation by SRSF2 and SRSF6. Taken together, we conclude that, for 5’ splice-site selection activity of SRSF2 and SRSF6, mutagenesis of 5’ splice-site to a conserved or a weaker score demonstrated that an activated 5’ splice-site with weaker score or the other 5’ splice-site with a higher score could limit the role of 5’ splice-site.
Fig. 4.
Relative 5’ splice-site strength determines splices-site selection functions of SRSF2 and SRSF6. (A) (Upper) Two 5’ sequences of 5’ splice-site switched mutant minigene (Inv) are shown. Mutated sequences are shown in red. (Lower) RT-PCR analysis results of 5’ splice-site selection in Inv minigenes are shown. (B) (Upper) Two 5’ sequences of Cons2a and W2a minigenes are shown. Mutated sequences are shown in red. (Lower) RT-PCR analysis results of 5’ splice-site selection in Cons2a and W2a minigenes are shown. (C) (Upper) Two 5’ sequences of Cons2b and W2b minigenes are shown. Mutated sequences are shown in red. (Lower) RT-PCR analysis of 5’ splice-site selection in Cons2b and W2b minigenes are shown.
DISCUSSION
In this study, we identified SRSF2 and SRSF6, as regulatory proteins for 5’ splice-site selection of Bcl-x pre-mRNA using a minigene system. We applied site-directed mutagenesis to potential binding motifs or clusters that located closer to the 5’ splice-site to identify target RNA sequences of SRSF2 and SRSF6. We further performed random deletion mutagenesis of exons 2a and 2b to locate targets of these proteins. Our results indicate that both site-directed and deletion mutations could not abolish activities of Bcl-x. Remarkably, the strength of 5’ splice-site of the one activated by SRSF2 and SRSF6 and the other one play roles in functions of SRSF2 and SRSF6. Less conserved sequences of activated 5’ splice-site or more conserved sequences of the other one abolished activities of SRSF2 and SRSF6 in Bcl-x splicing, suggesting that the strength of 5’ splice-site could regulates the function of SRSF2 and SRSF6.
SR proteins have a single RNA binding motif or cluster in splicing regulation (15, 17). We observed that mutations of single binding motif or cluster in Bcl-x pre-mRNA were unable to disrupt functions of SRSF2 or SRSF6. Deletion mutations of exon 2a and 2b were unable to abolish their functions either, suggesting that multiple binding motifs or clusters from different locations might function as functional targets of SRSF2 and SRSF6. It has been shown that SR proteins collaborate in the regulation of alternative splicing (18, 19). Splicing machineries in multiple binding motifs could collaborate to regulate Bcl-x splicing. In Bcl-x pre-mRNA, many binding motifs or clusters of SRSF2 and SRSF6 were predicted. From these motifs or clusters, we selected sequences closer to the 5’ splice-site to perform functional analysis. These mutations were not functional targets of SRSF2 or SRSF6. Our previous results have demonstrated that single binding targets of SR proteins are located close to splice-sites (13, 17). Although we could not test all predicted binding motifs for functional analysis, distant sequences from splice-sites might also be functional targets of SRSF2 and SRSF6.
Although SRSF2 and SRSF6 regulate Bcl-x alternative splicing in a minigene assay with an overexpression approach, these proteins could not modulate endogeneous Bcl-x pre-mRNA using shRNA-mediated knockdown approach. Differences in the overexpression of exogenous SR proteins and endogenous knockdown can explain the disparity in results using different approaches. Such disparity might also come from differences of Bcl-x RNA. The Bcl-x minigene we tested did not include the whole intron (55.6 kb), but only partial intron (0.43 kb). It has been shown that overexpressed SRSF2 can regulate endogenous Bcl-x pre-mRNA splicing in A549 cells (20). However, we could not observe effects of SRSF2 effects on endogenous Bcl-x in the present study. Such differences between the previous study and the present study could be due to experimental differences and different cell lines tested.
Our results demonstrated the importance of 5’ splice-site strength in the function of SRSF2 and SRSF6 in regulating Bcl-x pre-mRNA splicing. This is consistent with previous reports showing that alternative 5’ splice-site is affected by the strength of 5’ splice-site and the distance between two 5’ splice-sites (21-23). 5’ splice-site selection has various selection mechanisms. One study has shown that distant 5’ splice-site affects proximal 5’ splice-site, while another report has shown that spliceosome prefers most proximal 5’ splice-site to distal 5’ splice-site (24, 25). Although strength of both distant and proximal 5’ splice-sites play important roles in the function of SRSF2 and SRSF6, they have opposite directions. How the strength of 5’ splice-site affects functions of SRSF2 and SRSF6 need to be clarified through further studies.
MATERIALS AND METHODS
Plasmid construction
Bcl-x minigene was produced by deleting part of exon 2a and intron from the minigene that we constructed previously (26). Nhe I and EcoR I restriction enzymes were used to clone Bcl-x minigene into pCI-neo vector. All mutant constructs were generated by site-directed mutagenesis using Bcl-x minigene as a template. All primer sequences are listed in Supplementary Table 1.
Cell culture, plasmid transfection and immunoblotting
HEK293T cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) media (HyClone) supplemented with 10% Fetal Bovine Serum (FBS) in a 5% CO2 incubator at 37°C. 2 mM Glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin were also added. To transfect DNA into cells, cells are treated with 100 µl DMEM containing 0.5 µg DNA mixed with 2 µg polyethyleneimide (PEI). Culture media were changed after 4 h of incubation. RNAs were extracted from cells at 48 h after transfection. Immunoblotting was performed as previously described (27) using anti-SC35 (Millipore), anti-SRp55 (Millipore) and anti-Tubulin (Abcam) antibodies.
RT-PCR
RT-PCR was performed as previously described (28). MLV reverse transcriptase (ELPISBIO) and oligo-dT18 primer were used to reverse transcribe RNA. In the PCR reaction, primer set bm1/bm2 was used analyze Bcl-x minigene splicing while primer set be1/be2 was used to detect endogenous Bcl-x splicing.
Supplementary Materials
ACKNOWLEDGEMENTS
This work was supported by the following grants: NRF-2020R1A2C2004682 grants to Haihong Shen, NRF-2019R1I 1A1A01057372 grant to Xuexiu Zheng, and Cell Logistics Research Center (grant No. 2016R1A5A1007318) funded by the Ministry of Education and National Research Foundation of Korea. This work was also supported by “GIST Research Institute (GRI) ARI” grant funded by the GIST in 2020.
Footnotes
AUTHOR CONTRIBUTIONS
H.S., X.Z. and C.G. designed the concept of the present study and supervised it. N.C., Y.L., J.O., J.H. acquired, analyzed and interpreted the data. X.Z. and H.S. wrote the manuscript. All authors read and approved the manuscript.
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Wang L, Brooks AN, Fan J, et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell. 2016;30:750–763. doi: 10.1016/j.ccell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin S, Gambe RG, Sun J, et al. A Murine Model of Chronic Lymphocytic Leukemia Based on B Cell-Restricted Expression of Sf3b1 Mutation and Atm Deletion. Cancer Cell. 2019;35:283–296. doi: 10.1016/j.ccell.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sumantran VN, Ealovega MW, Nunez G, Clarke MF, Wicha MS. Overexpression of Bcl-XS sensitizes MCF-7 cells to chemotherapy-induced apoptosis. Cancer Res. 1995;55:2507–2510. [PubMed] [Google Scholar]
- 4.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 5.Espana L, Fernandez Y, Rubio N, Torregrosa A, Blanco J, Sierra A. Overexpression of Bcl-xL in human breast cancer cells enhances organ-selective lymph node metastasis. Breast Cancer Res Treat. 2004;87:33–44. doi: 10.1023/B:BREA.0000041579.51902.89. [DOI] [PubMed] [Google Scholar]
- 6.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 7.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/S1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20:1063–1071. doi: 10.1128/MCB.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon H, Cho S, Loh TJ, et al. SRSF2 promotes splicing and transcription of exon 11 included isoform in Ron proto-oncogene. Biochim Biophys Acta. 2014;1839:1132–1140. doi: 10.1016/j.bbagrm.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang HN, Lee M, Loh TJ, et al. Exon 9 skipping of apoptotic caspase-2 pre-mRNA is promoted by SRSF3 through interaction with exon 8. Biochim Biophys Acta. 2014;1839:25–32. doi: 10.1016/j.bbagrm.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 16.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang HN, Liu Y, Choi N, et al. Binding of SRSF4 to a novel enhancer modulates splicing of exon 6 of Fas pre-mRNA. Biochem Biophys Res Commun. 2018;506:703–708. doi: 10.1016/j.bbrc.2018.10.123. [DOI] [PubMed] [Google Scholar]
- 18.Crawford JB, Patton JG. Activation of alpha-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F. Mol Cell Biol. 2006;26:8791–8802. doi: 10.1128/MCB.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch KW, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 20.Merdzhanova G, Edmond V, De Seranno S, et al. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ. 2008;15:1815–1823. doi: 10.1038/cdd.2008.135. [DOI] [PubMed] [Google Scholar]
- 21.Yu Y, Maroney PA, Denker JA, et al. Dynamic regulation of alternative splicing by silencers that modulate 5' splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisette JF, Toutant J, Dugre-Brisson S, Desgroseillers L, Chabot B. hnRNP A1 and hnRNP H can collaborate to modulate 5' splice site selection. RNA. 2010;16:228–238. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bielli P, Bordi M, Di Biasio V, Sette C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection. Nucleic Acids Res. 2014;42:12070–12081. doi: 10.1093/nar/gku922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 25.Hicks MJ, Mueller WF, Shepard PJ, Hertel KJ. Competing upstream 5' splice sites enhance the rate of proximal splicing. Mol Cell Biol. 2010;30:1878–1886. doi: 10.1128/MCB.01071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Zhou J, Zheng X, et al. Identification of a novel cis-element that regulates alternative splicing of Bcl-x pre-mRNA. Biochem Biophys Res Commun. 2012;420:467–472. doi: 10.1016/j.bbrc.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Cho S, Moon H, Loh TJ, et al. Splicing inhibition of U2AF65 leads to alternative exon skipping. Proc Natl Acad Sci U S A. 2015;112:9926–9931. doi: 10.1073/pnas.1500639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon H, Jang HN, Liu Y, et al. RPM but not the Asp/Glu domain of hnRNP C1/C2 is required for splicing regulation of Ron exon 11 pre-mRNA. BMB Rep. 2019;52:641–646. doi: 10.5483/BMBRep.2019.52.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.