Abstract
The epidermis and skin appendages are maintained by their resident epithelial stem cells, which undergo long-term self-renewal and multilineage differentiation. Upon injury, stem cells are activated to mediate re-epithelialization and restore tissue function. During this process, they often mount lineage plasticity and expand their fates in response to damage signals. Stem cell function is tightly controlled by transcription machineries and signalling transductions, many of which derail in degenerative, inflammatory and malignant dermatologic diseases. Here, by describing both well-characterized and newly emerged pathways, we discuss the transcriptional and signalling mechanisms governing skin epithelial homeostasis, wound repair and squamous cancer. Throughout, we highlight common themes underscoring epithelial stem cell plasticity and tissue-level crosstalk in the context of skin physiology and pathology.
Keywords: cutaneous squamous cell carcinomas, epigenetic regulators, lineage plasticity, signalling transduction, skin epithelial stem cells, transcription factors
1 ∣. INTRODUCTION
As the largest organ of the human body, our skin serves as a physical barrier between the individual and the environment. The skin preserves body fluid, guards against irradiation and pathogens and conducts sensations. The epidermis is composed of several epithelial layers. The basal cells attach to the basement membrane above the dermis, joined by hemidesmosomes and adherens junctions, and are home to the epidermal stem cells (EpdSCs), which undergo long-term self-renewal and continuously fuel the upward flux of differentiating cells, forming the skin barrier.1 EpdSC progenies transition through the multiple differentiated layers, starting with the suprabasal spinous layer, rich in desmosomes2; then the granular layer, containing keratohyalin and lamellar granules; and finally the stratum corneum, composed of flattened denucleated corneocytes with heavily cross-linked keratin cables and a cornified envelope, eventually sloughed off the skin surface.3 Connecting to the epidermis are many epidermal appendages, among which the most abundant are the sweat glands and the pilosebaceous units. Sweat glands are responsible for thermoregulation,4,5 while the pilosebaceous unit is composed of the sebaceous gland and hair follicle. The sebaceous glands secrete lipids (known as sebum) onto the skin surface for repulsion of water and protection against microbes,6,7 while the hair follicles produce hair to cover the body surface for thermoregulation and appearance.8,9 The stem cells of the hair follicles, particularly those in the bulge, undergo long-term self-renewal and cyclic bouts of multilineage differentiation to produce the hair, and, upon wounding, regenerate the entire pilosebaceous unit and the epidermis.10-17
Multiple cell types live with keratinocytes in the skin, establishing a closely intertwined community. Dermal fibroblasts comprise heterogeneous populations and secrete matrix proteins that provide skin with mechanical strength and coordinate wound healing, and some specialized fibroblasts associate with hair follicles to regulate the hair cycle.18 Adipocytes reside in the dermis and hypodermis, where they provide insulation and regulate wound repair.19 The blood and lymphatic vessels densely infiltrate the skin, exchanging nutrients and metabolites, and closely associate with the epidermis and its appendages.20,21 Immunocytes such as Langerhans cells, macrophages, neutrophils, mast cells, innate lymphoid cells, γδT cells and regulatory T cells together fulfil the responsibility of cutaneous surveillance and immunity.22-24 Skin is also heavily innervated.9 Epithelial-derived mechanosensory Merkel cells interact with sensory neurons and encode mechanical inputs.25,26 Melanocytes deliver skin pigmentations whose activities are critically dependent on their epithelial and neuronal niches.27,28 Sensory nerves,29 sympathetic nerves,30 and arrector pili muscles31 directly interact with HFSCs to coordinate tissue production. Therefore, the skin is essentially an organ hosting a “zoo” of cell types and tissue-level communications. Upon acute injury or infection, all units come together to dissipate danger, repair damage and restore tissue function, a captive choreography that goes awry in chronic or malignant diseases.
The murine skin, although notably different from that of humans, harbours almost every cell type of its human counterpart and employs highly conserved genes and pathways to execute similar biological functions.32 Additionally, the study of murine skin offers an arsenal of genetic tools and valuable experimental assays for examining molecular mechanisms of skin development and diseases.33 For example, we now appreciate the considerable plasticity in both the epithelial and mesenchymal compartments, especially under pathologic conditions, including cutaneous inflammation (eg dermatitis, psoriasis, acne, eczema), degeneration (eg blistering, alopecia) and malignancy (eg basal cell carcinoma, cutaneous squamous cell carcinoma [cSCC], melanoma).
Our current essay summarizes the literature on the transcriptional and signalling aspects of epithelial plasticity in the skin. We first take a deep dive into several key squamous lineage transcription factors (TFs) that have been extensively characterized using genetically engineered mouse models and discuss their roles in skin development, wound repair and cSCC. We then survey a group of signalling transduction pathways whose effector TFs have been shown to genetically and functionally interact with squamous lineage TFs and impact dermatologic diseases. Finally, we review regulatory mechanisms impacting these TF levels and activities, including epigenetic regulators that shape skin epithelial stem cell function and direct fates upon niche stimulations. During our discussion, we touch upon a few skin diseases wherever applicable. We direct a main focus on cSCC, the second most common skin cancer,34 presenting a prototype for tumors as wounds that do not heal.35-37
2 ∣. SQUAMOUS LINEAGE TFS CONTROL SKIN EPITHELIA FORM, FUNCTION AND DISEASES
Several TFs known to govern skin epithelial lineage specification, stratification and barrier formation may be considered squamous lineage TFs. The deregulation of these TFs often leads to congenital, degenerative and malignant diseases in the skin. We discuss several TFs in this category, starting with a special focus on p63 (encoded by TP63), a highly studied, “poster child” squamous lineage TF in the context of skin development and squamous cancer. While p63 has multiple alternatively spliced forms38,39 with distinct functions, here we focus on ΔNp63 (referred to as p63 hereafter). Readers are referred to excellent reviews40-43 for related TFs.
p63 specifically labels the basal layers of stratified epithelia (skin, oral, oesophageal), transitional epithelia (cervix, anogenital, urothelial) and glandular epithelia (breast, prostate).44 Germline mutation of p63 in humans leads to a spectrum of congenital syndromes manifesting various defects of ectodermal derivatives.45,46 Recapitulating human diseases, p63−/− mice fail to stratify the squamous epithelia and lack epithelial appendages including hair follicles, teeth and mammary glands.47,48 Their limbs are truncated and often absent owing to failed differentiation of the apical ectodermal ridge.47 These mice die shortly after birth owing to dehydration and lack of a skin barrier.47,48 Fulfilling p63's role as the defining member of the squamous lineage TF family, the targets of p63 encompass almost all the known pathways regulating development and maintenance of the skin epithelia, as we elaborate below (Figure 1).
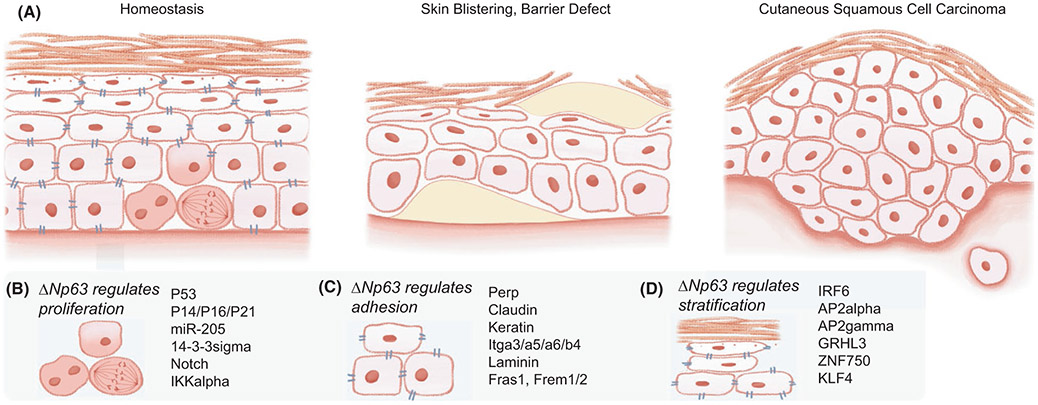
FIGURE 1.
Squamous lineage transcription factors (TFs) control skin epithelia form, function and diseases. (A) The stratified skin epidermis is composed of several epithelial layers, including the basal layer, harbouring epidermal stem cells (EpdSCs), suprabasal spinous and granular layers, and the stratum corneum (left). Skin blistering diseases occur when the epidermal adhesions and junctions are compromised, and barrier defects originate from disruptions of the stratification programme, including genes of cross-linking enzymes, cornified envelop and lipid metabolism. A common non-melanoma skin cancer, cutaneous squamous cell carcinoma (cSCC), arises from the skin epithelium and mimics wounds that never heal. (B) p63 maintains EpdSC proliferation and prevents precocious stratification by antagonizing p53, cell cycle inhibitors, 14-3-3σ, Notch, IKKα and activating miR-205. (C) p63 also regulates the expression of many cell adhesion and junction components, including Perp (desmosome), claudin (tight junction), keratins (intermediate filaments), integrins (adherens junctions), laminins (basement membrane), and Fras1 and Frem1/2 (extracellular matrix). (D) Squamous lineage TFs are p63 targets and join p63 to regulate epidermal stratification; many of these TFs are deregulated in congenital ectodermal conditions and skin inflammatory and malignant diseases
2.1 ∣. p63 coordinates epidermal proliferation, stratification and adhesion
A distinguishing feature of skin epithelial stem cells is their longevity and ability to maintain self-renewal over an individual's lifetime. A well-characterized function of p63 is its maintenance of epithelial stem cell proliferation49-51 (Figure 1B). As a p53 family TF, p63 is able to bind and activate the p53 response element in heterologous reporter assays.52,53 p63 antagonizes apoptosis induced by p5354,55 and another close family member, p73.56 Knockdown of p53 rescues the proliferative defect of p63-depleted cells in human organotypic epidermal culture,57 suggesting that a main consequence of p63 loss in this context is the aberrant activation of p53. p63 directly suppresses the cell cycle inhibitor p21(WAF1) in proliferating keratinocytes.58 Remarkably, the arrested epidermal development of p63-null mice can be rescued by inactivating either of the cell cycle inhibitors p16(INK4A) and p14(ARF),59 suggesting that cell cycle regulation is a major function of p63. Consistently, p63+/− mice exhibit premature ageing, correlated with increased p53 activity, elevated p16(INK4A) levels and senescence.60 p63 additionally induces the squamous miRNA miR-205, which suppresses epidermal differentiation genes and maintains proliferation of basal cells.61,62
Meanwhile, p63 suppresses epidermal differentiation through multiple mechanisms (Figure 1B). For example, p63 directly suppresses 14-3-3σ (also known as stratifin or Sfn), a member of the 14-3-3 family harbouring phosphoserine protein that binds the pleckstrin homology domain.58 Unlike other relatively ubiquitous 14-3-3 family members, Sfn is preferentially expressed in the epithelial lineage and promotes stem cell differentiation.50,63 Sfn is down-regulated in epithelial cancer, and its mutation leads to hyper-plasic epithelia.64 Besides regulating Sfn, p63 maintains EpdSCs by preventing the activation of Notch signalling,65-68 a master differentiation programme (discussed below).
p63 governs another key property of squamous epithelia, their cellular junctions and adhesions, to maintain epithelial integrity (Figure 1C). Initially identified as a p53 target gene regulating apoptosis, the tetraspan membrane protein Perp is a p63 target that influences cellular adhesion by localizing to and regulating desmosomes.69 In turn, Perp deletion leads to skin blistering disease69 and accelerates tumorigenesis.70 p63 targets also include components of tight junctions, adherens junctions, intermediate filaments and extracellular matrix, by which p63 helps maintain junction integrity and skin barrier function71-75 (Figure 1A). Remarkably, in a system orthogonal to the skin epidermis, p63−/− mammary epithelial cells undergoing anoikis can be rescued by Itgb4 expression,73 suggesting that certain p63-targeted adhesion components may play dominant roles in maintaining epithelial integrity.
2.2 ∣. Squamous TFs join efforts to drive epidermal stratification and hair follicle differentiation
As EpdSCs exit self-renewal and undergo lineage commitment, a stratification programme is activated in their progenies. In the coordination of this epidermal stratification and differentiation, p63 is joined by many squamous TFs (Figure 1D). For instance, 63 induces interferon regulatory factor 6 (IRF6),76 whose mutations underlie a group of human ectodermal dysplasia syndromes including cleft palate.77 While the IRF family is well known for their regulation of interferon responses upon viral infection, IRF6 appears to be an outlier specifically involved in epidermal stratification, and its deletion in mice leads to hyperproliferative epidermis.78-80 Interestingly, disruption of an IRF6 enhancer is also associated with cleft lip, and this enhancer is bound by the activating enhancer binding protein 2 (AP2).81 Notably, both AP2α and AP2γ are known to regulate squamous development.82-84 p63 and AP2 likely share many targets and coordinate their function during epidermal specification.62
p63 also targets the grainyhead–like transcription factor 3 (GRHL3),62 whose mutation again leads to human cleft lip and palate.85 GRHL3 is the mammalian homolog of Drosophila grainyhead (GRH), whose mutation leads to “grainy” and discontinuous head skeletons, hence its name.86 GRHL3 deficiency in mice leads to the curly tail phenotype and neural tube defects87; severe skin barrier defects due to the loss of a skin stratum corneum cross-linking enzyme, transglutaminase 188; exacerbated inflammatory response upon challenge89; and defects in epithelial wound repair.90,91 Grainyhead–like family member GRHL1 is similarly expressed in the suprabasal layer of the adult epidermis and regulates desmosomes.92
Another squamous lineage TFs integrated into the stratification programme is zinc finger protein 750 (ZNF750),93 which is down-regulated in human patients with cleft palate syndrome harbouring mutant p63.94 ZNF750 promotes epidermal differentiation by closely associating with Krüppel-like factor 4 (KLF4),93,95,96 which is critical for skin barrier formation.97,98 Of note, mutations of ZNF75099 and KLF4100 have been linked to psoriasis, an inflammatory skin disease strongly associated with defects in innate immunity and skin barrier function. These studies share a common theme in which germline mutations of squamous lineage TFs are frequently found in an overlapping spectrum of human ectodermal diseases, suggesting that these TFs are instrumental for early ectoderm specification and subsequently are repurposed to regulate squamous differentiation. Moreover, the squamous stratification programme, while essential for skin epidermal development and barrier formation, may, when compromised, predispose an individual to skin immunologic deregulations or malignant transformations (as we will discuss later).
2.3 ∣. Squamous TF deregulation in cSCC
In parallel to their instrumental roles in skin epithelial development, squamous lineage TFs are critically involved in cSCC (Figure 1A). p63 is frequently amplified in SCCs of the head and neck, lung, oesophagus and cervix.101 Overexpression of p63 in the lung epithelia induces K5/K14 expression and squamous metaplasia in an otherwise simple epithelium.102,103 p63's oncogenic activity in squamous malignancies has been associated with various mechanisms, such as interaction with NF-κB104,105 and SOX2.106-108 Other squamous lineage TFs, such as GRHL2, ZNF750 and KLF4,109 have been associated with cSCC,101 further strengthening the notion that deregulation of squamous lineage TFs constitutes a signature for this type of skin malignancy.
In an unexpected twist to p63's tumor-promoting role, aged p63+/− mice undergo frequent loss of heterozygosity and exhibit increased tumorigenesis ranging from adenocarcinomas and sarcomas to, most intriguingly, SCCs,110 suggesting p63's tumor-suppressive function. Consistently, it has been observed that squamous cancer cells became more invasive when p63 was suppressed.111,112 It is intriguing to speculate that p63 loss may promote stem cell lineage infidelity (discussed below), where genes outside the squamous lineage become permissively induced,111 reversing the development trajectory.62,113,114 The tissue microenvironment is likely another major culprit, highlighted in human SCC patient samples where a similar loss of epithelial identity along with aberrant tumor stroma reaction and immune infiltration has been frequently documented.115-117
3 ∣. GROWTH AND STRESS SIGNALLING PATHWAYS DICTATE RESPONSIVENESS TO NICHE STIMULI DURING WOUNDING AND ARE HIJACKED IN SKIN MALIGNANCY
As important as lineage development and homeostatic turnover are, another key function of adult stem cells is coordinated wounding response and tissue repair.37 During tissue remodelling, many signalling pathways regulating growth are repurposed for damage control to restore organ function. In the context of wound repair, rather than homeostatic function, we generally refer to these regulators as stress signalling pathways and TFs. We discuss the roles in wound repair and cSCC of several extensively studied pathways in this category, including two pro-mitogenic and two pro-differentiation pathways in the skin (Figure 2).
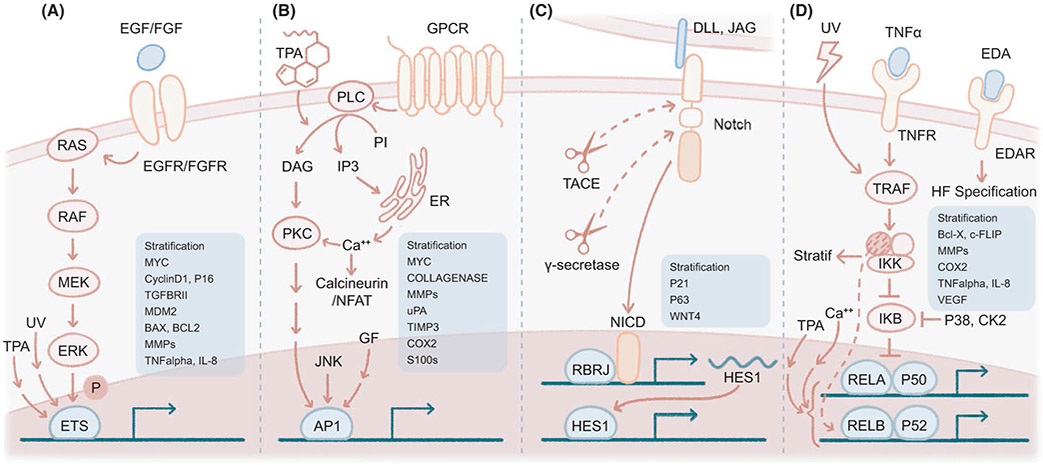
FIGURE 2.
Growth and stress signalling pathways dictate responsiveness to stimuli and are hijacked in skin malignancy. (A) ETS family TFs are phosphorylated by the RAS MAPK pathway, downstream of receptor tyrosine kinase (RTK) signalling, for example EGF/EGFR and FGF/FGFR. ETS is also stimulated by ultraviolet light and TPA exposure. Targets of ETS TFs include stratification genes (cross-linking enzymes, cornified envelop, lipid metabolism), cell cycle (MYC, Cyclin D1, P16, TGFBR2), apoptosis (MDM2, BAX, BCL2), matrix metalloproteases (MMPs) and cytokine/chemokine genes (IL-8, TNF-α). (B) AP-1 TFs are the principal effector TFs of TPA signalling. AP-1 is also activated by serum, growth factors and JNK signalling, and shares some common effectors with calcium signalling, such as protein kinase C (PKC). AP-1 induces stratification, matrix remodelling (collagenase, MMPs, uPA, TIMP3) and inflammation (COX2, S100), among others. uPA, urokinase-type plasminogen activator. (C) Notch receptor binds its ligands (DLL1, JAG1/2) in a juxtacrine or autocrine fashion and is activated by two consecutive protease activities (TACE, γ-secretase), resulting in activation of HES1, p21 and stratification genes and repression of p63 and WNT4. TACE, tumor necrosis factor-alpha–converting enzyme, also known as ADAM17 (ADAM metallopeptidase domain 17). NICD, notch intracellular domain. (D) NF-κB signalling has several skin-specific functions: EDA/EDAR regulates hair follicle (HF) specification, ultraviolet light activates TRAF2, IKKα regulates squamous stratification, and NF-κB functions as a tumor suppressor promoting epidermal differentiation. NF-κB is also activated by TPA and calcium, and its downstream targets include stratification, survival (Bcl-x, c-FLIP), inflammatory enzymes (COX2), matrix remodelling (MMPs), cytokines/chemokines (TNF-α, IL-1, IL-6, GM-CSF, IL-8, KC, MIP2) and angiogenesis (VEGF)
3.1 ∣. ETS transduces RAS MAPK signalling in the skin epithelia
The TF superfamily E-twenty-six (ETS) is activated by rat sarcoma (RAS) mitogen-activated protein kinase (MAPK) signalling118,119 (Figure 2A) and collectively recognizes the ETS motif.120 ETS proteins comprise several subfamilies based on structural similarities and divergences, and many of these proteins are rapidly phosphorylated and activated by MAPK signalling,121 constituting one of the earliest detectable events upon mitogen stimulation.122,123 Paralleling the function of their counterparts in mammals, ETS homologs in Drosophila124 and Caenorhabditis elegans125 are also key downstream effectors of RAS MAPK. ETS protein activation is further enabled through interaction with partner TFs, such as runt-related transcription factor 1 (RUNX1) in the context of haematopoiesis,126,127 and subsequent recruitment of cofactors such as the histone acetyltransferase EP300128,129 (Figure 2A).
Mammalian ETS family members are widely expressed in developing and adult tissues, including many epithelial lineages.130 Germline loss of ETS family members yields dramatic phenotypes in the developing immune and endothelial tissues.131 In the skin specifically, mice lacking ETS2 have wavy hair, curly whiskers and abnormal hair follicle shape and arrangement.132 These phenotypic changes highly resemble those of mutants lacking EGF signalling,133-135 consistent with the notion that ETS2 is the conserved “bona fide” downstream effector of MAPK signalling in the skin epithelia. ETS TFs also directly target epidermal stratification genes96,136-139 and hence are likely closely integrated into the squamous TF network.
In addition to stratification, ETS targets have been reported in pathways controlling the cell cycle, DNA damage, matrix remodelling, immune regulations and others (Figure 2A),140-143 closely linking its function to tumorigenesis. Notably, the ETS family is frequently bound by mutant p53, and they together promote malignant progression.144-147 While oncogenic fusion proteins involving ETS proteins are well documented in leukaemias,148,149 sarcomas150 and prostate cancers,151 ETS function is only starting to be revealed in cSCC. ETS1 is increased in malignant cSCC,152 and in transgenic mice, ETS1 overexpression in suprabasal layers induced dysplastic changes in the epidermis, accompanying elevated angiogenesis.153 ETS2, on the other hand, is functionally required for cSCC development in vivo.143,154 Interestingly, ETS2 is also essential for wound repair, and ectopic expression of constitutively active ETS2 in the otherwise homeostatic skin epithelia is sufficient to induce stem cell lineage infidelity (coexpression of and functional dependence on mixed lineage markers) in vivo, causing these epithelial cells to resemble activated stem cells during wound repair and tumorigenesis.37,154 This in vivo evidence is consistent with the view that RAS MAPK–mediated ETS2 phosphorylation and activation serve as an initiating event upon mitogen stimulation, kicking off a downstream signalling cascade in wounds and cancer.
3.2 ∣. AP-1 mediates TPA and mitogen signals in cancer and inflammation
In its role as a physical barrier, skin directly responds to insults, such as carcinogens. The phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) is a well-known tumor promoter,155 acting by intercalating into the cell membrane, presumably altering phosphatidylinositol and diacylglycerol levels, leading to protein kinase C signalling activation156 (Figure 2B). Protein kinase C is typically activated downstream of the calcium-sensing G protein–coupled receptor and phospholipase C, or by intracellular calcium. Interestingly, both TPA and calcium treatment of cultured keratinocytes increased the expression of epidermal differentiation genes,157,158 even though their phenotypic outcomes differ significantly, with TPA promoting tumorigenesis155 and calcium inducing stratification,159 indicating diverging effectors downstream of TPA and calcium in the skin.
The transcriptional effector of TPA signalling is a group of TFs collectively known as the activator protein-1 (AP-1).160,161 Composed of core families Jun and Fos, plus the extended families Atf and Maf, AP-1 proteins form mono- or heterodimers to regulate gene expression via enhancers that contain an AP-1 motif.162-165 Many keratinocyte differentiation genes harbour AP-1–binding sites at their promoters,166 underlying their responsiveness to TPA. In addition to its activation by TPA, AP-1 is rapidly activated by serum and growth factors,167-172 placing AP-1 downstream of receptor tyrosine kinase MAPK signalling. TPA and growth factors, via AP-1, induce the expression of a plethora of targets,173-177 many of which are directly associated with tumorigenesis activity (Figure 2B).
cJun and JunB are among the most-studied family members of AP-1. Mice with either cJun or JunB single conditional knockout in the skin are born with a normal skin appearance. However, those with single cJun knockout have defective epithelial migration during development and open eye phenotype, likely due to reduced epidermal growth factor receptor (EGFR) signalling,178,179 and those with single JunB knockout become hyperproliferative and have elevated immune response upon further challenge.180-182 These observations suggest redundant yet often contrasting functions of AP-1 family members in the homeostatic skin. Strikingly, cJun and JunB double deletion in the embryonic skin epithelia leads to perinatal lethality owing to severe cachexia, caused by loss of the AP-1 target tissue inhibitor of metalloproteinase 3 (TIMP3), and consequently leads to uncontrolled activity of a disintegrin and metalloprotease domain 17 (ADAM17) and tumor necrosis factor-alpha (TNF-α) shedding. Double deletion of cJun and JunB in the adult skin epithelia results in a pleiotropic phenotype similar to psoriasis, including hyperkeratosis, massive immune infiltration and arthritic lesions.183 Downstream mediators of these psoriatic phenotypes include S100 calcium-binding proteins, which are direct AP-1 targets,183,184 as well as an indirect signalling axis involving miR-21,185 a potent oncogenic miRNA in cSCC.186
During skin tumorigenesis, both cJun179,187-189 and cFos190,191 have been shown to promote cSCC in vivo. Interestingly, cFos overexpression induces infiltration of CD4+ T cells,191 whereas loss of AP-1 induces infiltration of neutrophils and macrophages,183 highlighting the diverse mechanisms of epithelial-immune crosstalk mediated by epithelial AP-1 functions. Similarly, activated immune infiltration and barrier defects have been observed in epidermal-specific loss of fibroblast growth factor receptors,192 further associating MAPK signalling with AP-1 at the genetic level. Like EGF signalling,193,194 FGF signalling is heavily involved in epithelial growth, wound repair, inflammation and tumorigenesis,195,196 and both pathways are frequently altered in human SCCs.101
3.3 ∣. Notch signalling governs epithelial differentiation and mediates microenvironment crosstalk
Notch signalling regulates gene transcription through converting the recombination signal binding protein for immunoglobulin kappa J region (RBPJ) from a transcriptional repressor to an activator,197 leading to hairy and enhancer of split-1 (HES1)–mediated transcriptional activation (HES1 itself is an RBPJ target)198 (Figure 2C). The direct transcriptional output of Notch activation is to induce epidermal differentiation and suppress EpdSC self-renewal.65,197-199 At the cellular level, Notch signalling acts in either an autocrine or a juxtacrine fashion. In autocrine Notch signalling, the ligand and receptor are provided by a pair of juxtaposed cells. For instance, high expression of Delta-like 1 (DLL1, a Notch ligand) is restricted to the basal stem cells of human epidermis and directs the neighbouring cells expressing NOTCH1 to differentiate.200 Basal p63 activates Jagged-1 (JAG1, a Notch ligand) to induce neighbouring cells' Notch signalling activation and subsequent stratification201 (Figure 1B). JAG1/2 and NOTCH1/2 may also be coexpressed in differentiating keratinocytes at the suprabasal layer to reinforce terminal differentiation.197,202,203 Of note, EpdSCs expressing high levels of the ligand DLL1 are non-receptive to Notch signalling, as they lack receptors, although it is an interesting conundrum that the ligands, themselves being Notch signalling targets, manage to accumulate at high levels without detectable Notch activity in these EpdSCs. Several classic models are plausible to explain such cell fate segregations,204 all of which have supportive evidence in epidermal development, including cell sorting,200 lateral inhibition,205 intrinsic bias due to polarized localization of Notch signalling components206,207 and extrinsic bias owing to the distance to the cellular source emanating the stemness signal.208
Notch is among the top frequently altered pathways in cSCC.101,209 Although the oncogenic role of the Notch pathway is well established in many cancer types, its mutations are likely loss of function in SCCs.210,211 In the context of cSCC, the Notch pathway has been consistently revealed to be tumor suppressive in mouse models.212-216 Strikingly, treatment with an inhibitor of γ-secretase (Notch processing enzyme required for pathway activation) increased the frequency of SCCs in human patients,217 consistent with the well-established function of the Notch pathway in governing squamous stratification. Of interest, calcineurin inhibitors, a mainstream immunosuppressant used in organ transplant patients, increased cSCC in humans218 and in mice,219 phenocopying Notch suppression. Notch and calcineurin signalling may be closely associated given the role of Notch in epidermal stratification and the role of calcineurin in calcium responsiveness and the fact that a high calcium concentration induces epidermal differentiation in cultured keratinocytes.159 Indeed, Notch has been found to indirectly activate the calcineurin pathway, and loss of calcineurin in the skin epithelia leads to deregulation of Notch responsive genes and cyclic alopecia.220 Therefore, cSCC in these patients may originate indirectly, from suppression of immunocytes, as these immunosuppressant agents initially intended to do, or directly, from modification of tumor parenchyma by interfering with epidermal differentiation pathways.
Adding to the complexity of tissue-level crosstalk, epidermal-specific inactivation of Notch leads to transcriptional derepression of thymic stromal lymphopoietin (TSLP) and granulocyte colony-stimulating factor (G-CSF), followed by aberrant immune infiltrates.221-225 Additionally, Notch signalling in the dermal fibroblast compartment impacts epidermal development.226 Altogether, these findings highlight the tissue microenvironment as a likely major contributor in Notch-driven cSCC.
3.4 ∣. NF-κB integrates damage signals and immunity in squamous tissues
The nuclear factor of kappa light polypeptide gene enhancer in B-cell (NF-κB) pathway is essential for immune organ development and survival and is also a central player in mediating epithelial and immune crosstalk.227,228 In its epithelial role, NF-κB is essential for mounting innate immune responses upon infection229 or eliciting inflammation upon tissue damage230 while protecting tissues against apoptosis.230 In the meantime, within the haematopoietic lineage, NF-κB is required for immunocyte survival231 in addition to relaying inflammatory signals for further tissue remodelling.232-234
NF-κB functions in the skin not only follow similar paradigms, but also deviate with several interesting twists (Figure 2D). First, analogous to the conventional TNF-α/TNFR/NF-κB pathway, the strikingly parallel pathway ectodysplasin A (EDA)/EDAR/NF-κB is employed in ectoderm appendage development to initiate the specification of a group of hair follicles within a narrow window during embryogenesis.235-239 Second, ultraviolet irradiation, a well-known skin carcinogen, activates NF-κB via TNFR1 and its downstream partner TNF receptor-associated factor (TRAF2) independent of TNF-α,240 or by p38 MAPK and casein kinase II–mediated phosphorylation of inhibitor of NF-κB (IκB), independent of the IκB kinase (IKK).241 On the other hand, IKKα, a direct p63 target75,242,243 (Figure 1B), is critically required for epidermal differentiation but is uncoupled from many known components within the NF-κB pathway—it promotes squamous stratification independent of its kinase activity but is dependent on its nuclear localization.244-248 In sharp contrast, in other systems such as B-cell maturation249 and mammary gland development,250,251 downstream NF-κB effector TFs are essential to IKKα function. Consistent with its pro-differentiation function in the skin epidermis, IKKα is decreased in human SCCs,252-254 and its forced expression suppressed tumorigenesis in vivo.252 Mechanistically, IKKα has been suggested to cooperate with transforming growth factor β (TGF-β) signalling to induce the expression of MYC antagonists and suppress SCCs.254,255 Lastly, opposite to its oncogenic transformation activity in haematopoietic systems,256,257 NF-κB restrains hyperproliferation in the skin.258-260 In contrast to the loss of TNF-α, which renders skin resistant to tumorigenesis,261 loss of NF-κB promotes skin tumorigenesis,262-264 consistent with NF-κB function in suppressing p63 and promoting epidermal stratification.65
Of significance, the growth and mitogenic pathways discussed above interact extensively to regulate oncogenic responses and skin tumorigenesis (Figure 2). For example, ultraviolet B–induced genes highly overlap with those induced by TPA in the skin.265 The AP-1 inducer TPA has been reported to induce NF-κB266 and ETS,127 and conversely, ultraviolet radiation has been shown to induce ETS proteins.267,268 NF-κB can be induced by calcium or the Notch pathway.203 As aforementioned, calcineurin inhibition leads to enhanced SCCs in vivo, and this effect could be mediated by the activity of the extended AP-1 family member ATF3.219 Downstream of mitogens, both ETS and AP-1 are essential effectors and are known to cooperate on the chromatin level to regulate target transcription.154,269-273 Further illustrating the downstream convergence of these TFs is the regulation of angiogenesis and inflammation—for example, the induction of interleukin-8 by ETS2143 and NF-κB274 and the induction of matrix metalloproteinases by AP-1, ETS and NF-κB.177,191,275-277 These findings altogether depict the intertangled relationships between these TFs (Figure 2), positioning epithelial cells at the centre stage for secreting signalling molecules and directing the epithelial-immune interactions.278
4 ∣. COOPERATION OF SQUAMOUS, STRESS AND EPIGENETIC REGULATORS IN DRIVING SKIN EPITHELIAL STEM CELL PLASTICITY
Both the levels and the activity of TFs are tightly regulated to achieve precise cell signalling and stem cell fate decisions in skin development and diseases (Figure 3A). For example, p63 protein levels are subjected to degradation closely coupled to DNA damage,76,279-281 and its transcripts are modulated by miRNAs282-284 and by alternative splicing,38,39 all of which impact p63's activity in shaping the cellular transcriptional landscape. Upon serum and growth factor stimulation, the immediate early response is independent of transcription and is instead transduced at the level of post-translational modifications of signalling proteins followed by the activation of TFs, for instance, the phosphorylation of ETS285,286 and AP-1.168 Subsequently, signals are amplified and stabilized by feedback mechanisms. For example, both p63287 and AP-1288-290 are able to bind to their own promoters and further induce their own transcriptional activation, forming a positive feedback loop.
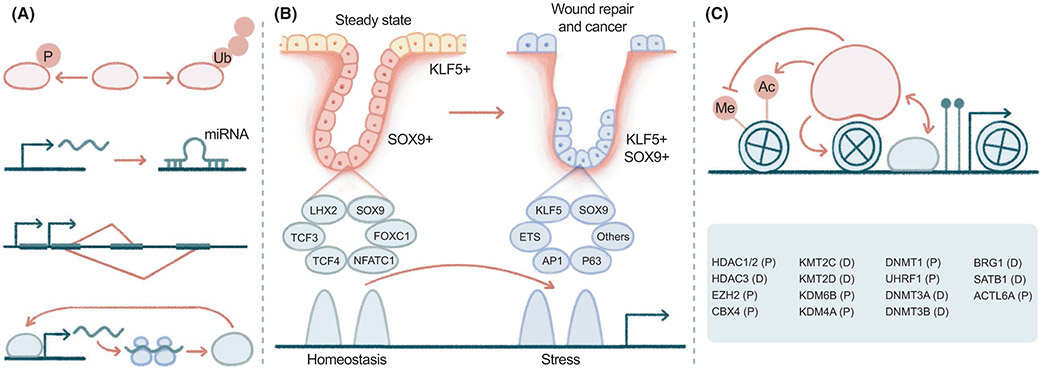
FIGURE 3.
Cooperation of squamous, stress and epigenetic regulators in driving skin epithelial stem cell plasticity. (A) TFs such as p63 are regulated at multiple levels, including post-translational modifications by phosphorylation and ubiquitination, post-transcriptional regulation by miRNAs, alternative splicing and positive feedback regulations. (B) A group of TFs govern hair follicle stem cell (HFSC) quiescence, including SOX9, LHX2, TCF3, TCF4, NFATC1 and FOXC1, while KLF5 is specifically enriched in EpdSCs. Upon wounding, HFSCs induce the expression of KLF5, which is coexpressed with SOX9, ETS2, AP-1 and p63, among others, resulting in stem cell lineage infidelity, an epigenetically rewired state that is functionally required for wound repair. (C) Several groups of epigenetic factors are integrated in the regulation of EpdSC proliferation (P) and differentiation (D), including histone deacetylases (HDAC1/2, HDAC3), histone methyltransferases, demethylases, and their regulators (EZH2, CBX4, KMT2C/D, KMT4A, KMT6B), chromatin remodelers (BRG1, SATB1, ACTL6A), and DNA methyltransferases and regulators (DNMT1, DNMT3A/B, UHRF1). KMT2C is also known as MLL3, KMT2D as MLL2, KDM4A as JMJD2, and KDM6B as JMJD3. The hypothetical epigenetic factor is depicted arbitrarily with multiple activities
The spatial and temporal distributions of TFs are dynamically regulated during development and wounding to direct changes in stem cell fate. For example, GRHL3 and the LIM domain TF LMO4 localize to basal and suprabasal epithelia, respectively, in adults,291 but become colocalized and physically interact with each other to regulate epithelial migration during eyelid closure.291-293 Analogously, SOX9 and KLF5 demarcate HFSCs and EpdSCs, respectively, under adult homeostasis but become coexpressed in wound-activated HFSCs and EpdSCs (Figure 3B). This phenomenon of stem cell plasticity has been referred to as lineage infidelity, where wounded stem cells expand their fates beyond homeostatic patterns, coexpressing otherwise lineage-restricted genes.154 In this case, HFSCs expand fates to regenerate not only hair follicles but also the epidermis during re-epithelialization. Stem cell lineage infidelity is transient in wound repair but sustained in cancer and is functionally required for epithelial regeneration in wounds and malignant progression in SCCs.154 Mechanistically, stem cell fate changes correlate with genome-wide remodelling of the transcriptional and epigenetic landscape. Transcriptional responsiveness to upstream signalling can be differentially dictated based on configurations of in cis elements and in trans regulators.294,295 As HFSCs exit homeostasis and become activated upon injury, core HFSC TFs are decommissioned, and stress TFs are assembled to drive wound-specific epicentres. In both cases, SOX9 is expressed, yet is dependent on different niche signals. Such distinct context dependency at the chromatin level could be reflected by an enhancer switch143,154,296 (Figure 3B). Several speculations may be drawn from the observations of stem cell lineage infidelity. Upstream signalling transductions are specifically induced upon injury. For example, activation of ETS2 and AP-1 results in the ectopic induction of mixed lineage gene expressions, among which the lineage TFs KLF5 and SOX9 may cooperate with ETS2 and AP-1 and form a feedforward circuit to further promote stem cell fate change. Downstream targets may be remodelled to drive fate transition. For instance, KLF5 and SOX9 may be redirected from maintaining homeostatic functions to regulating damage-related cellular pathways that are essential for wound repair and tumorigenesis. Both directions will be worthwhile to further explore and improve our understanding of context-dependent transcriptional and signalling regulations.
One strategy in which squamous TFs regulate gene activation (or repression) and coordinate genome-wide transcriptional regulations is by interacting with epigenetic regulators,297-299 including chromatin binding, modification and remodelling factors (Figure 3C). Loss of histone deacetylases HDAC1 and HDAC2 induced p53 activity along with derepressed p21, p16, Sfn and epidermal hypoplasia,300,301 phenocopying the effects of p63 deficiency.59 Interestingly, suppression of unscheduled cell cycle exits or differentiation, and hence maintaining the stem cell pool, appears to be a common mechanism for many epigenetic regulators in the skin. The skin epidermis lacking EZH2, a component of polycomb repressive complex 2 (PRC2) that catalyses H3K27 methylation, aberrantly induces p16 and undergoes cell cycle arrest.302 Similarly, deletion of chromobox homolog 4 (CBX4, the E3 ligase component of PRC1) induces epidermal senescence.303 Depletion of the maintenance DNA methyltransferase DNMT1, or of ubiquitin-like with PHD and ring finger domains 1 (UHRF1, an E3 ligase that targets DNMT1 to hemi-methylated DNA), leads to aberrant differentiation.304-306 In contrast, the de novo DNA methyltransferases DNMT3a/b are required for epidermal differentiation, and their loss promotes cSCC.307,308 Similarly, HDAC3 governs skin barrier formation by interacting with KLF4, although, interestingly, HDAC3's histone deacetylase activity is dispensable in this particular context.98
Indeed, a reiterative theme arises upon study of squamous lineage TF function. The interactions of these TFs with epigenetic regulators significantly impact epidermal differentiation and cSCC tumorigenesis. The ATP-dependent chromatin remodelling complex BAF (BRG1- or BRM-associated factor) has been shown to regulate chromatin accessibility at key lineage genes in cooperation with p63309 and AP-1.310 For example, p63 induces brahma-related gene 1 (BRG1), a catalytic subunit of the BAF complex, to direct the localization of the epidermal differentiation complex (EDC) locus from the nuclear periphery to the interior for its transcriptional activation.311 An analogous regulation of EDC by p63 has been observed through the special AT-rich sequence binding protein 1 (SATB1),312 another chromatin remodelling factor mediating long-range looping. Meanwhile, p63 is coamplified with the regulatory subunit actin-like protein 6a (ACTL6A) of the BAF complex,313 which prohibits BAF from binding to and activating KLF4, consequently promoting cSCC.314
Furthermore, GRHL3 recruits the trithorax group protein mixed lineage leukaemia (MLL) complex (H3K4 methyltransferases) to facilitate the activation of differentiation genes,315 and notably, both MLL2 and MLL3 are frequently mutated in SCCs.101 The H3K27me3 demethylase Jumonji domain-containing protein 3 (JMJD3) promotes epidermal differentiation by erasing this marker of H3K27me3 upon calcium switch.316 In addition, the H3K9me3 demethylase JMJD2A facilitates the binding of the AP-1 proteins to the promoters of Jun and Fosl1 genes themselves, enhancing AP-1 activity and SCC metastasis.317 Finally, the c-Jun N-terminal kinase (JNK) phosphorylation of JUN (encoded by Jun) released it from HDAC3 suppression,318 and the extracellular signal-regulated kinase (ERK) phosphorylation of Fos-related antigen 1 (Fra-1, encoded by Fosl1) released it from EZH2 suppression to activate EDC genes.319 These observations together illustrate the convergent functions of squamous TFs and epigenetic regulators in governing skin epithelial stem cell maintenance while coordinating stratifications (Figure 3C).
5 ∣. CONCLUSION
Our conceptual advancement of the genetic and developmental principles in skin biology has been instrumental to our understanding of skin diseases. These observations in aggregate illustrate the importance of contextual information including temporal and spatial expression, interacting partners and signalling activities as key aspects of transcriptional regulation in development and diseases. It has become increasingly clear that cancer driver genes rewire existing signalling and transcriptional networks to fuel malignancy, and they do so by hijacking development and regeneration pathways. Investigating these pathways and gene networks will greatly expedite the mechanistic understanding of the context dependency of TFs, thereby revealing cancer-specific vulnerability. Facilitated by rapidly evolving single-cell technology and microscopy, our depicting of stem cell biology in the skin has achieved hitherto unprecedented complexity and depth. We begin to capture signalling events and molecular interactions at high resolution and appreciate intercellular communications at a single-cell level across the tissue and organism. These technical advances are likely the main driver for our understanding of complex skin inflammatory and malignant diseases with regard to cellular crosstalks.
The fascination with transcriptional regulation is further bolstered by the finding that under certain contexts, TFs may sit at the top of a chromatin hierarchy to elicit stem cell plasticity. For example, LEF1 overexpression or bCat activation drives ectopic hair follicle specification and de novo hair follicle tumor formation.320-325 During skin wounding, fibroblasts can be reprogrammed to regenerate the epidermis by a combination of p63, GRHL2, AP2α and MYC.326 In the context of SCCs, in addition to the tissue types frequently giving rise to this cancer type, a few more organs develop squamous-like carcinomas, such as the pancreas and bladder,327-331 suggesting a squamous network in these cancers, whose functional significance and clinical implications await further investigations. The next chapter in skin research is likely to include further advanced applications in regenerative and cancer medicine. The dissection of TF interactions with epigenetic regulators and their signalling crosstalk will facilitate the discovery of druggable targets and appealing therapeutics.
ACKNOWLEDGEMENTS
YG is supported by the NIH K01 career development award 1K01AR072132, CPRIT first-time recruitment award FP00006955, the University of Texas Rising Star award and the University of Texas MD Anderson Cancer Center startup funds. The manuscript was edited by Sarah Bronson, ELS, at the Research Medical Library at MD Anderson.
Abbreviations:
- ACTL6A
actin-related proteins 6a
- BAF
BRG1-or BRM-associated factor
- BRG1
brahma-related gene-1
- cSCC
cutaneous squamous cell carcinoma
- DLL1
delta-1
- DNMT
DNA methyltransferase
- EDA
ectodysplasin A
- EDC
epidermal differentiation complex
- EGFR
epidermal growth factor receptor
- EpdSC
epidermal stem cell
- ETS
e-twenty-six
- FGFR
fibroblast growth factor receptor
- GRH
grainyhead
- GRHL3
grainyhead–like transcription factor 3
- HES1
hairy and enhancer of split-1
- HFSC
hair follicle stem cell
- IkB
inhibitor of NF-κB
- IKKα
IkB kinase alpha
- IRF6
interferon regulatory factor 6
- JAG1
jagged-1
- JMJD
Jumonji domain-containing protein
- JNK
c-Jun N-terminal kinase
- KLF4
Krüppel-like factor 4
- MAPK
mitogen-activated protein kinase
- MLL
mixed lineage leukaemia
- NF-κB
nuclear factor of kappa light polypeptide gene enhancer in B cells
- PRC2
polycomb repressive complex 2
- RAS
rat sarcoma
- RBPJ
recombination signal binding protein for immunoglobulin kappa j region
- RUNX1
runt-related transcription factor 1
- TF
transcription factor
- TGF-β
transforming growth factor-β
- TIMP3
tissue inhibitor of metalloproteinase inhibitor 3
- TNF-α
tumor necrosis factor-alpha
- TPA
12-O-tetradecanoylphorbol-13-acetate
- UHRF1
ubiquitin-like with PHD and ring finger domains 1
- ZNF750
zinc finger protein 750
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- [1].Fuchs E Skin stem cells: rising to the surface. J Cell Biol. 2008;180(2):273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Green KJ, Roth-Carter Q, Niessen CM, Nichols SA. Tracing the evolutionary origin of desmosomes. Curr Biol. 2020;30(10):R535–R543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Eckert RL. Structure, function, and differentiation of the keratinocyte. Physiol Rev. 1989;69(4):1316–1346. [DOI] [PubMed] [Google Scholar]
- [4].Lobitz WC Jr, Dobson RL. Dermatology: the eccrine sweat glands. Annu Rev Med. 1961;12:289–298. [DOI] [PubMed] [Google Scholar]
- [5].Lu C, Fuchs E. Sweat gland progenitors in development, homeostasis, and wound repair. Cold Spring Harb Perspect Med. 2014;4(2):a015222. 10.1101/cshperspect.a015222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stenn KS, Zheng Y, Parimoo S. Phylogeny of the hair follicle: the sebogenic hypothesis. J Invest Dermatol. 2008;128(6):1576–1578. [DOI] [PubMed] [Google Scholar]
- [7].Niemann C, Horsley V. Development and homeostasis of the sebaceous gland. Semin Cell Dev Biol. 2012;23(8):928–936. 10.1016/j.semcdb.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hardy MH. The secret life of the hair follicle. Trends Genet. 1992;8(2):55–61. [DOI] [PubMed] [Google Scholar]
- [9].Paus R, Cotsarelis G. The biology of hair follicles. N Engl J Med. 1999;341(7):491–497. [DOI] [PubMed] [Google Scholar]
- [10].Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102(4):451–461. [DOI] [PubMed] [Google Scholar]
- [11].Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104(2):233–245. [DOI] [PubMed] [Google Scholar]
- [12].Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Morris RJ, Liu Y, Marles L, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22(4):411–417. [DOI] [PubMed] [Google Scholar]
- [14].Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118(5):635–648. [DOI] [PubMed] [Google Scholar]
- [15].Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc Natl Acad Sci USA. 2005;102(41):14677–14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell. 2005;9(6):855–861. [DOI] [PubMed] [Google Scholar]
- [17].Ito M, Liu Y, Yang Z, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11(12):1351–1354. [DOI] [PubMed] [Google Scholar]
- [18].Sennett R, Rendl M. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin Cell Dev Biol. 2012;23(8):917–927. 10.1016/j.semcdb.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shook B, Rivera Gonzalez G, Ebmeier S, Grisotti G, Zwick R, Horsley V. The role of adipocytes in tissue regeneration and stem cell niches. Annu Rev Cell Dev Biol. 2016;32:609–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gur-Cohen S, Yang H, Baksh SC, et al. Stem cell-driven lymphatic remodeling coordinates tissue regeneration. Science. 2019;366(6470):1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li KN, Jain P, He CH, Eun FC, Kang S, Tumbar T. Skin vasculature and hair follicle cross-talking associated with stem cell activation and tissue homeostasis. Elife. 2019;8:e45977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Paus R, Nickoloff BJ, Ito T. A ‘hairy’ privilege. Trends Immunol. 2005;26(1):32–40. [DOI] [PubMed] [Google Scholar]
- [23].Heath WR, Carbone FR. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat Immunol. 2013;14(10):978–985. [DOI] [PubMed] [Google Scholar]
- [24].Ali N, Zirak B, Rodriguez RS, et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell. 2017;169(6):1119–1129.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Woo SH, Lumpkin EA, Patapoutian A. Merkel cells and neurons keep in touch. Trends Cell Biol. 2015;25(2):74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the roles of polycomb repressive complex 2 subunits in the control of skin development. J Invest Dermatol. 2016;136(8):1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307(5710):720–724. [DOI] [PubMed] [Google Scholar]
- [28].Zhang B, Ma S, Rachmin I, et al. Hyperactivation of sympathetic nerves drives depletion of melanocyte stem cells. Nature. 2020;577(7792):676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL. Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell. 2011;8(5):552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shwartz Y, Gonzalez-Celeiro M, Chen CL, et al. Cell types promoting goosebumps form a niche to regulate hair follicle stem cells. Cell. 2020;182(3):578–593.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fujiwara H, Ferreira M, Donati G, et al. The basement membrane of hair follicle stem cells is a muscle cell niche. Cell. 2011;144(4):577–589. 10.1016/j.cell.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Muller-Rover S, Foitzik K, Paus R, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117(1):3–15. [DOI] [PubMed] [Google Scholar]
- [33].Nakamura M, Schneider MR, Schmidt-Ullrich R, Paus R. Mutant laboratory mice with abnormalities in hair follicle morphogenesis, cycling, and/or structure: an update. J Dermatol Sci. 2013;69(1):6–29. [DOI] [PubMed] [Google Scholar]
- [34].Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344(13):975–983. [DOI] [PubMed] [Google Scholar]
- [35].Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. [DOI] [PubMed] [Google Scholar]
- [36].Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9(8):628–638. 10.1038/nrm2455 [DOI] [PubMed] [Google Scholar]
- [37].Ge Y, Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet. 2018;19(5):311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Flores ER, Tsai KY, Crowley D, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416(6880):560–564. [DOI] [PubMed] [Google Scholar]
- [39].Nylander K, Vojtesek B, Nenutil R, et al. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol. 2002;198(4):417–427. [DOI] [PubMed] [Google Scholar]
- [40].Watt FM, Frye M, Benitah SA. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat Rev Cancer. 2008;8(3):234–242. 10.1038/nrc2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Oshimori N, Fuchs E. The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell. 2012;11(6):751–764. 10.1016/j.stem.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li S, Yang J. Ovol proteins: guardians against EMT during epithelial differentiation. Dev Cell. 2014;29(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rognoni E, Walko G. The roles of YAP/TAZ and the Hippo pathway in healthy and diseased skin. Cells. 2019;8(5):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–316. [DOI] [PubMed] [Google Scholar]
- [45].Celli J, Duijf P, Hamel BC, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99(2):143–153. [DOI] [PubMed] [Google Scholar]
- [46].van Bokhoven H, Jung M, Smits AP, et al. Limb mammary syndrome: a new genetic disorder with mammary hypoplasia, ectrodactyly, and other Hand/Foot anomalies maps to human chromosome 3q27. Am J Hum Genet. 1999;64(2):538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. [DOI] [PubMed] [Google Scholar]
- [48].Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. [DOI] [PubMed] [Google Scholar]
- [49].Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol. 1999;113(6):1099–1105. [DOI] [PubMed] [Google Scholar]
- [50].Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci USA. 2001;98(6):3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129(3):523–536. [DOI] [PubMed] [Google Scholar]
- [52].Bian J, Sun Y. p53CP, a putative p53 competing protein that specifically binds to the consensus p53 DNA binding sites: a third member of the p53 family? Proc Natl Acad Sci USA. 1997;94(26):14753–14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zeng X, Levine AJ, Lu H. Non-p53 p53RE binding protein, a human transcription factor functionally analogous to P53. Proc Natl Acad Sci USA. 1998;95(12):6681–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liefer KM, Koster MI, Wang XJ, Yang A, McKeon F, Roop DR. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 2000;60(15):4016–4020. [PubMed] [Google Scholar]
- [55].Yan W, Chen X. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J Biol Chem. 2006;281(12):7856–7862. [DOI] [PubMed] [Google Scholar]
- [56].Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9(1):45–56. [DOI] [PubMed] [Google Scholar]
- [57].Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20(22):3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23(7):2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009;28(13):1904–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19(17):1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wang D, Zhang Z, O'Loughlin E, et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol. 2013;25(10):1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fan X, Wang D, Burgmaier JE, et al. Single cell and open chromatin analysis reveals molecular origin of epidermal cells of the skin. Dev Cell. 2018;47(1):21–37.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dellambra E, Golisano O, Bondanza S, et al. Downregulation of 14-3-3sigma prevents clonal evolution and leads to immortalization of primary human keratinocytes. J Cell Biol. 2000;149(5):1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ilić D, Kanazawa S, Nishizumi H, et al. Skin abnormality in aged fyn−/− fak+/− mice. Carcinogenesis. 1997;18(8):1473–1476. [DOI] [PubMed] [Google Scholar]
- [65].Nguyen BC, Lefort K, Mandinova A, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20(8):1028–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Okuyama R, Ogawa E, Nagoshi H, et al. p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene. 2007;26(31):4478–4488. [DOI] [PubMed] [Google Scholar]
- [67].Yugawa T, Narisawa-Saito M, Yoshimatsu Y, et al. DeltaNp63alpha repression of the Notchl gene supports the proliferative capacity of normal human keratinocytes and cervical cancer cells. Cancer Res. 2010;70(10):4034–4044. [DOI] [PubMed] [Google Scholar]
- [68].Romano RA, Smalley K, Magraw C, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139(4):772–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ihrie RA, Marques MR, Nguyen BT, et al. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120(6):843–856. [DOI] [PubMed] [Google Scholar]
- [70].Beaudry VG, Jiang D, Dusek RL, et al. Loss of the p53/p63 regulated desmosomal protein Perp promotes tumorigenesis. PLoS Genet. 2010;6(10):e1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lopardo T, Lo Iacono N, Marinari B, et al. Claudin-1 is a p63 target gene with a crucial role in epithelial development. PLoS One. 2008;3(7):e2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kurata S, Okuyama T, Osada M, et al. p51/p63 Controls subunit alpha3 of the major epidermis integrin anchoring the stem cells to the niche. J Biol Chem. 2004;279(48):50069–50077. [DOI] [PubMed] [Google Scholar]
- [73].Carroll DK, Carroll JS, Leong CO, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8(6):551–561. [DOI] [PubMed] [Google Scholar]
- [74].Candi E, Rufini A, Terrinoni A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13(6):1037–1047. [DOI] [PubMed] [Google Scholar]
- [75].Koster MI, Dai D, Marinari B, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104(9):3255–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Moretti F, Marinari B, Lo Iacono N, et al. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120(5):1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kondo S, Schutte BC, Richardson RJ, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32(2):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Richardson RJ, Dixon J, Malhotra S, et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38(11):1329–1334. [DOI] [PubMed] [Google Scholar]
- [79].Ingraham CR, Kinoshita A, Kondo S, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet. 2006;38(11):1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Oberbeck N, Pham VC, Webster JD, et al. The RIPK4-IRF6 signalling axis safeguards epidermal differentiation and barrier function. Nature. 2019;574(7777):249–253. [DOI] [PubMed] [Google Scholar]
- [81].Rahimov F, Marazita ML, Visel A, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40(11):1341–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51(2):251–260. [DOI] [PubMed] [Google Scholar]
- [83].Koster MI, Kim S, Huang J, Williams T, Roop DR. TAp63alpha induces AP-2gamma as an early event in epidermal morphogenesis. Dev Biol. 2006;289(1):253–261. [DOI] [PubMed] [Google Scholar]
- [84].Wang X, Bolotin D, Chu DH, Polak L, Williams T, Fuchs E. AP-2alpha: a regulator of EGF receptor signaling and proliferation in skin epidermis. J Cell Biol. 2006;172(3):409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Peyrard-Janvid M, Leslie E, Kousa Y, et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. 2014;94(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bray SJ, Kafatos FC. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 1991;5(9):1672–1683. [DOI] [PubMed] [Google Scholar]
- [87].Ting SB, Wilanowski T, Auden A, et al. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat Med. 2003;9(12):1513–1519. [DOI] [PubMed] [Google Scholar]
- [88].Ting SB, Caddy J, Hislop N, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308(5720):411–413. [DOI] [PubMed] [Google Scholar]
- [89].Gordon WM, Zeller MD, Klein RH, et al. A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. J Clin Invest. 2014;124(12):5205–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Caddy J, Wilanowski T, Darido C, et al. Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev Cell. 2010;19(1):138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Boglev Y, Wilanowski T, Caddy J, et al. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev Biol. 2011;349(2):512–522. [DOI] [PubMed] [Google Scholar]
- [92].Mlacki M, Darido C, Jane SM, Wilanowski T. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One. 2014;9(2):e89247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sen GL, Boxer LD, Webster DE, et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell. 2012;21:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zarnegar BJ, Webster DE, Lopez-Pajares V, et al. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. Am J Hum Genet. 2012;91(3):435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28(18):2013–2026. 10.1101/gad.246579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rubin AJ, Barajas BC, Furlan-Magaril M, et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat Genet. 2017;49(10):1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22(4):356–360. [DOI] [PubMed] [Google Scholar]
- [98].Szigety KM, Liu F, Yuan CY, et al. HDAC3 ensures stepwise epidermal stratification via NCoR/SMRT-reliant mechanisms independent of its histone deacetylase activity. Genes Dev. 2020;34(13-14):973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Birnbaum RY, Zvulunov A, Hallel-Halevy D, et al. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat Genet. 2006;38(7):749–751. [DOI] [PubMed] [Google Scholar]
- [100].Ray-Jones H, Duffus K, McGovern A, et al. Mapping DNA interaction landscapes in psoriasis susceptibility loci highlights KLF4 as a target gene in 9q31. BMC Biol. 2020;18(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Guan Y, Zhang SN, Ma YY, Zhang Y, Zhang YY. Unraveling cancer lineage drivers in squamous cell carcinomas. Pharmacol Ther. 2019;206:107448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18(2):126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S. An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PLoS One. 2009;4(5):e5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].King KE, Ponnamperuma RM, Allen C, et al. The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 2008;68(13):5122–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Yang X, Lu H, Yan B, et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71(10):3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Watanabe H, Ma Q, Peng S, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest. 2014;124(4):1636–1645. 10.1172/JCI71545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Boumahdi S, Driessens G, Lapouge G, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511(7508):246–250. [DOI] [PubMed] [Google Scholar]
- [108].Siegle JM, Basin A, Sastre-Perona A, et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun. 2014;5:4511. 10.1038/ncomms5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sastre-Perona A, Hoang-Phou S, Leitner MC, Okuniewska M, Meehan S, Schober M. De novo PITX1 expression controls bi-stable transcriptional circuits to govern self-renewal and differentiation in squamous cell carcinoma. Cell Stem Cell. 2019;24(3):390–404.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Flores ER, Sengupta S, Miller JB, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. [DOI] [PubMed] [Google Scholar]
- [111].Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66(15):7589–7597. [DOI] [PubMed] [Google Scholar]
- [112].Ehsanian R, Brown M, Lu H, et al. YAP dysregulation by phosphorylation or DeltaNp63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene. 2010;29(46):6160–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133(8):1553–1563. [DOI] [PubMed] [Google Scholar]
- [114].Shalom-Feuerstein R, Lena AM, Zhou H, et al. DeltaNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18(5):887–896. 10.1038/cdd.2010.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Puram SV, Tirosh I, Parikh AS, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171(7):1611–1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Hoadley KA, Yau C, Hinoue T, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(2):291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Campbell JD, Yau C, Bowlby R, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23(1):194–212.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Galang CK, Der CJ, Hauser CA. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene. 1994;9(10):2913–2921. [PubMed] [Google Scholar]
- [119].Coffer P, de Jonge M, Mettouchi A, Binetruy B, Ghysdael J, Kruijer W. junB promoter regulation: Ras mediated transactivation by c-Ets-1 and c-Ets-2. Oncogene. 1994;9(3):911–921. [PubMed] [Google Scholar]
- [120].Karim FD, Urness LD, Thummel CS, et al. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990;4(9):1451–1453. [DOI] [PubMed] [Google Scholar]
- [121].Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Hipskind RA, Rao VN, Mueller CG, Reddy ES, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354(6354):531–534. [DOI] [PubMed] [Google Scholar]
- [123].Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68(3):597–612. [DOI] [PubMed] [Google Scholar]
- [124].Klämbt C The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development. 1993;117(1):163–176. [DOI] [PubMed] [Google Scholar]
- [125].Tan PB, Lackner MR, Kim SK. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell. 1998;93(4):569–580. [DOI] [PubMed] [Google Scholar]
- [126].Wotton D, Ghysdael J, Wang S, Speck NA, Owen MJ. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14(1):840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Sun W, Graves BJ, Speck NA. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69(8):4941–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18(4):2218–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Jayaraman G, Srinivas R, Duggan C, et al. p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem. 1999;274(24):17342–17352. [DOI] [PubMed] [Google Scholar]
- [130].Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9(6):1551–1565. [PubMed] [Google Scholar]
- [131].Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19(55):6443–6454. [DOI] [PubMed] [Google Scholar]
- [132].Yamamoto H, Flannery ML, Kupriyanov S, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12(9):1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73(2):263–278. [DOI] [PubMed] [Google Scholar]
- [134].Mann GB, Fowler KJ, Gabriel A, Nice EC, Williams RL, Dunn AR. Mice with a null mutation of the TGF alpha gene have abnormal skin architecture, wavy hair, and curly whiskers and often develop corneal inflammation. Cell. 1993;73(2):249–261. [DOI] [PubMed] [Google Scholar]
- [135].Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269(5221):234–238. [DOI] [PubMed] [Google Scholar]
- [136].Fischer DF, Gibbs S, van De Putte P, Backendorf C. Interdependent transcription control elements regulate the expression of the SPRR2A gene during keratinocyte terminal differentiation. Mol Cell Biol. 1996;16(10):5365–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lee JH, Jang SI, Yang JM, Markova NG, Steinert PM. The proximal promoter of the human transglutaminase 3 gene. Stratified squamous epithelial-specific expression in cultured cells is mediated by binding of Sp1 and ets transcription factors to a proximal promoter element. J Biol Chem. 1996;271(8):4561–4568. [PubMed] [Google Scholar]
- [138].Andreoli JM, Jang SI, Chung E, Coticchia CM, Steinert PM, Markova NG. The expression of a novel, epithelium-specific ets transcription factor is restricted to the most differentiated layers in the epidermis. Nucleic Acids Res. 1997;25(21):4287–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Oettgen P, Alani RM, Barcinski MA, et al. Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol Cell Biol. 1997;17(8):4419–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hahm KB, Cho K, Lee C, et al. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet. 1999;23(2):222–227. [DOI] [PubMed] [Google Scholar]
- [141].Ries S, Biederer C, Woods D, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103(2):321–330. [DOI] [PubMed] [Google Scholar]
- [142].Ohtani N, Zebedee Z, Huot TJG, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409(6823):1067–1070. [DOI] [PubMed] [Google Scholar]
- [143].Yang H, Schramek D, Adam RC, et al. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. Elife. 2015;4:e10870. 10.7554/eLife.10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Sampath J, Sun D, Kidd VJ, et al. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J Biol Chem. 2001;276(42):39359–39367. [DOI] [PubMed] [Google Scholar]
- [145].Do PM, Varanasi L, Fan S, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26(8):830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Zhu J, Sammons MA, Donahue G, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525(7568):206–211. 10.1038/nature15251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Pourebrahim R, Zhang Y, Liu B, et al. Integrative genome analysis of somatic p53 mutant osteosarcomas identifies Ets2-dependent regulation of small nucleolar RNAs by mutant p53 protein. Genes Dev. 2017;31(18):1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Golub TR, Goga A, Barker GF, et al. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol. 1996;16(8):4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278(5341):1309–1312. [DOI] [PubMed] [Google Scholar]
- [150].Mao X, Miesfeldt S, Yang H, Leiden JM, Thompson CB. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J Biol Chem. 1994;269(27):18216–18222. [PubMed] [Google Scholar]
- [151].Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. [DOI] [PubMed] [Google Scholar]
- [152].Keehn CA, Smoller BR, Morgan MB. Ets-1 immunohistochemical expression in non-melanoma skin carcinoma. J Cutan Pathol. 2004;31(1):8–13. [DOI] [PubMed] [Google Scholar]
- [153].Nagarajan P, Parikh N, Garrett-Sinha LA, Sinha S. Ets1 induces dysplastic changes when expressed in terminally-differentiating squamous epidermal cells. PLoS One. 2009;4(1):e4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Ge Y, Gomez NC, Adam RC, et al. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169(4):636–650.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Colburn NH, Former BF, Nelson KA, Yuspa SH. Tumour promoter induces anchorage independence irreversibly. Nature. 1979;281(5732):589–591. [DOI] [PubMed] [Google Scholar]
- [156].Nishizuka Y The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984;308(5961):693–698. [DOI] [PubMed] [Google Scholar]
- [157].Dlugosz AA, Yuspa SH. Coordinate changes in gene expression which mark the spinous to granular cell transition in epidermis are regulated by protein kinase C. J Cell Biol. 1993;120(1):217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Calautti E, Missero C, Stein PL, Ezzell RM, Dotto GP. fyn tyrosine kinase is involved in keratinocyte differentiation control. Genes Dev. 1995;9(18):2279–2291. [DOI] [PubMed] [Google Scholar]
- [159].Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980;19(1):245–254. [DOI] [PubMed] [Google Scholar]
- [160].Angel P, Imagawa M, Chiu R, et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49(6):729–739. [DOI] [PubMed] [Google Scholar]
- [161].Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49(6):741–752. [DOI] [PubMed] [Google Scholar]
- [162].Bohmann D, Bos T, Admon A, Nishimura T, Vogt P, Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238(4832):1386–1392. [DOI] [PubMed] [Google Scholar]
- [163].Angel P, Allegretto EA, Okino ST, et al. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature. 1988;332(6160):166–171. [DOI] [PubMed] [Google Scholar]
- [164].Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988;54(4):541–552. [DOI] [PubMed] [Google Scholar]
- [165].Sassone-Corsi P, Lamph WW, Kamps M, Verma IM. fos-associated cellular p39 is related to nuclear transcription factor AP-1. Cell. 1988;54(4):553–560. [DOI] [PubMed] [Google Scholar]
- [166].Eckert RL, Crish JF, Banks EB, Welter JF. The epidermis: genes on - genes off. J Invest Dermatol. 1997;109(4):501–509. [DOI] [PubMed] [Google Scholar]
- [167].Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311(5985):433–438. [DOI] [PubMed] [Google Scholar]
- [168].Kruijer W, Cooper JA, Hunter T, Verma IM. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984;312(5996):711–716. [DOI] [PubMed] [Google Scholar]
- [169].Treisman R Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985;42(3):889–902. [DOI] [PubMed] [Google Scholar]
- [170].Prywes R, Roeder RG. Inducible binding of a factor to the c-fos enhancer. Cell. 1986;47(5):777–784. [DOI] [PubMed] [Google Scholar]
- [171].Quantin B, Breathnach R. Epidermal growth factor stimulates transcription of the c-jun proto-oncogene in rat fibroblasts. Nature. 1988;334(6182):538–539. [DOI] [PubMed] [Google Scholar]
- [172].Ryder K, Nathans D. Induction of protooncogene c-jun by serum growth factors. Proc Natl Acad Sci USA. 1988;85(22):8464–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35(3 Pt 2):603–610. [DOI] [PubMed] [Google Scholar]
- [174].Montesano R, Orci L. Tumor-promoting phorbol esters induce angiogenesis in vitro. Cell. 1985;42(2):469–477. [DOI] [PubMed] [Google Scholar]
- [175].Imbra RJ, Karin M. Metallothionein gene expression is regulated by serum factors and activators of protein kinase C. Mol Cell Biol. 1987;7(4):1358–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kerr LD, Holt JT, Matrisian LM. Growth factors regulate transin gene expression by c-fos-dependent and c-fos-independent pathways. Science. 1988;242(4884):1424–1427. [DOI] [PubMed] [Google Scholar]
- [177].Matthews CP, Birkholz AM, Baker AR, et al. Dominant-negative activator protein 1 (TAM67) targets cyclooxygenase-2 and osteopontin under conditions in which it specifically inhibits tumorigenesis. Cancer Res. 2007;67(6):2430–2438. [DOI] [PubMed] [Google Scholar]
- [178].Li G, Gustafson-Brown C, Hanks SK, et al. c-Jun is essential for organization of the epidermal leading edge. Dev Cell. 2003;4(6):865–877. [DOI] [PubMed] [Google Scholar]
- [179].Zenz R, Scheuch H, Martin P, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4(6):879–889. [DOI] [PubMed] [Google Scholar]
- [180].Florin L, Knebel J, Zigrino P, et al. Delayed wound healing and epidermal hyperproliferation in mice lacking JunB in the skin. J Invest Dermatol. 2006;126(4):902–911. [DOI] [PubMed] [Google Scholar]
- [181].Meixner A, Zenz R, Schonthaler HB, et al. Epidermal JunB represses G-CSF transcription and affects haematopoiesis and bone formation. Nat Cell Biol. 2008;10(8):1003–1011. 10.1038/ncb1761 [DOI] [PubMed] [Google Scholar]
- [182].Singh K, Camera E, Krug L, et al. JunB defines functional and structural integrity of the epidermo-pilosebaceous unit in the skin. Nat Commun. 2018;9(1):3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Zenz R, Eferl R, Kenner L, et al. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. 2005;437(7057):369–375. [DOI] [PubMed] [Google Scholar]
- [184].Schonthaler HB, Guinea-Viniegra J, Wculek SK, et al. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity. 2013;39(6):1171–1181. 10.1016/j.immuni.2013.11.011 [DOI] [PubMed] [Google Scholar]
- [185].Guinea-Viniegra J, Jiménez M, Schonthaler HB, et al. Targeting miR-21 to treat psoriasis. Sci Transl Med. 2014;6(225):225re1. 10.1126/scitranslmed.3008089 [DOI] [PubMed] [Google Scholar]
- [186].Ge Y, Zhang L, Nikolova M, Reva B, Fuchs E. Strand-specific in vivo screen of cancer-associated miRNAs unveils a role for miR-21( *) in SCC progression. Nat Cell Biol. 2016;18(1):111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Young MR, Li JJ, Rincón M, et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96(17):9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Behrens A, Jochum W, Sibilia M, Wagner EF. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19(22):2657–2663. [DOI] [PubMed] [Google Scholar]
- [189].Kolev V, Mandinova A, Guinea-Viniegra J, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10(8):902–911. 10.1038/ncb1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Saez E, Rutberg SE, Mueller E, et al. c-fos is required for malignant progression of skin tumors. Cell. 1995;82(5):721–732. [DOI] [PubMed] [Google Scholar]
- [191].Briso EM, Guinea-Viniegra J, Bakiri L, et al. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes Dev. 2013;27(18):1959–1973. 10.1101/gad.223339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Yang J, Meyer M, Müller A-K, et al. Fibroblast growth factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. J Cell Biol. 2010;188(6):935–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Sibilia M, Fleischmann A, Behrens A, et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102(2):211–220. [DOI] [PubMed] [Google Scholar]
- [194].Lichtenberger BM, Tan PK, Niederleithner H, Ferrara N, Petzelbauer P, Sibilia M. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140(2):268–279. 10.1016/j.cell.2009.12.046 [DOI] [PubMed] [Google Scholar]
- [195].Guo L, Yu QC, Fuchs E. Targeting expression of keratinocyte growth factor to keratinocytes elicits striking changes in epithelial differentiation in transgenic mice. EMBO J. 1993;12(3):973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Werner S, Smola H, Liao X, et al. The function of KGF in morphogenesis of epithelium and reepithelialization of wounds. Science. 1994;266(5186):819–822. [DOI] [PubMed] [Google Scholar]
- [197].Rangarajan A, Talora C, Okuyama R, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20(13):3427–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19(12):1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20(21):3022–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10(9):491–500. [DOI] [PubMed] [Google Scholar]
- [201].Sasaki Y, Ishida S, Morimoto I, et al. The p53 family member genes are involved in the Notch signal pathway. J Biol Chem. 2002;277(1):719–724. [DOI] [PubMed] [Google Scholar]
- [202].Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17(10):6057–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Nickoloff BJ, Qin J-Z, Chaturvedi V, Denning MF, Bonish B, Miele L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 2002;9(8):842–855. [DOI] [PubMed] [Google Scholar]
- [204].Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. [DOI] [PubMed] [Google Scholar]
- [205].Crowe R, Henrique D, Ish-Horowicz D, Niswander L. A new role for Notch and Delta in cell fate decisions: patterning the feather array. Development. 1998;125(4):767–775. [DOI] [PubMed] [Google Scholar]
- [206].Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470(7334):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Ezratty EJ, Stokes N, Chai S, Shah A, Williams S, Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145(7):1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Yang H, Adam RC, Ge Y, Hua ZL, Fuchs E. Epithelial-mesenchymal micro-niches govern stem cell lineage choices. Cell. 2017;169(3):483–496.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411–455. [DOI] [PubMed] [Google Scholar]
- [210].Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Nicolas M, Wolfer A, Raj K, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33(3):416–421. [DOI] [PubMed] [Google Scholar]
- [213].Lefort K, Mandinova A, Ostano P, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21(5):562–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Guinea-Viniegra J, Zenz R, Scheuch H, et al. Differentiation-induced skin cancer suppression by FOS, p53, and TACE/ADAM17. J Clin Invest. 2012;122(8):2898–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Quintana RM, Dupuy AJ, Bravo A, et al. A transposon-based analysis of gene mutations related to skin cancer development. J Invest Dermatol. 2013;133(1):239–248. [DOI] [PubMed] [Google Scholar]
- [216].Loganathan SK, Schleicher K, Malik A, et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science. 2020;367(6483):1264–1269. [DOI] [PubMed] [Google Scholar]
- [217].Extance A Alzheimer's failure raises questions about disease-modifying strategies. Nat Rev Drug Discov. 2010;9(10):749–751. [DOI] [PubMed] [Google Scholar]
- [218].Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348(17):1681–1691. [DOI] [PubMed] [Google Scholar]
- [219].Wu X, Nguyen B-C, Dziunycz P, et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature. 2010;465(7296):368–372. 10.1038/nature08996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Mammucari C, di Vignano AT, Sharov AA, et al. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev Cell. 2005;8(5):665–676. [DOI] [PubMed] [Google Scholar]
- [221].Dumortier A, Durham AD, Di Piazza M, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One. 2010;5(2):e9258. 10.1371/journal.pone.0009258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Weber S, Niessen MT, Prox J, et al. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development. 2011;138(3):495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Murthy A, Shao YW, Narala SR, Molyneux SD, Zúñiga-Pflücker JC, Khokha R. Notch activation by the metalloproteinase ADAM17 regulates myeloproliferation and atopic barrier immunity by suppressing epithelial cytokine synthesis. Immunity. 2012;36(1):105–119. [DOI] [PubMed] [Google Scholar]
- [224].Di Piazza M, Nowell CS, Koch U, Durham AD, Radtke F. Loss of cutaneous TSLP-dependent immune responses skews the balance of inflammation from tumor protective to tumor promoting. Cancer Cell. 2012;22(4):479–493. 10.1016/j.ccr.2012.08.016 [DOI] [PubMed] [Google Scholar]
- [225].Demehri S, Turkoz A, Manivasagam S, Yockey LJ, Turkoz M, Kopan R. Elevated epidermal thymic stromal lymphopoietin levels establish an antitumor environment in the skin. Cancer Cell. 2012;22(4):494–505. 10.1016/j.ccr.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27(38):5115–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87(1):13–20. [DOI] [PubMed] [Google Scholar]
- [228].Karin M Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. [DOI] [PubMed] [Google Scholar]
- [229].Lavon I, Goldberg I, Amit S, et al. High susceptibility to bacterial infection, but no liver dysfunction, in mice compromised for hepatocyte NF-kappaB activation. Nat Med. 2000;6(5):573–577. [DOI] [PubMed] [Google Scholar]
- [230].Chen LW, Egan L, Li ZW, Greten FR, Kagnoff MF, Karin M. The two faces of IKK and NF-kappaB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia-reperfusion. Nat Med. 2003;9(5):575–581. [DOI] [PubMed] [Google Scholar]
- [231].Park JM, Greten FR, Wong A, et al. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis-CREB and NF-kappaB as key regulators. Immunity. 2005;23(3):319–329. [DOI] [PubMed] [Google Scholar]
- [232].Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–296. [DOI] [PubMed] [Google Scholar]
- [233].Pikarsky E, Porat RM, Stein I, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–466. [DOI] [PubMed] [Google Scholar]
- [234].Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 2005;121(7):977–990. [DOI] [PubMed] [Google Scholar]
- [235].Ferguson BM, Brockdorff N, Formstone E, Ngyuen T, Kronmiller JE, Zonana J. Cloning of Tabby, the murine homolog of the human EDA gene: evidence for a membrane-associated protein with a short collagenous domain. Hum Mol Genet. 1997;6(9):1589–1594. [DOI] [PubMed] [Google Scholar]
- [236].Srivastava AK, Pispa J, Hartung AJ, et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc Natl Acad Sci USA. 1997;94(24):13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Headon DJ, Overbeek PA. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat Genet. 1999;22(4):370–374. [DOI] [PubMed] [Google Scholar]
- [238].Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 1999;22(4):366–369. [DOI] [PubMed] [Google Scholar]
- [239].Schmidt-Ullrich R, Aebischer T, Hülsken J, Birchmeier W, Klemm U, Scheidereit C. Requirement of NF-kappaB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128(19):3843–3853. [DOI] [PubMed] [Google Scholar]
- [240].Tobin D, van Hogerlinden M, Toftgard R. UVB-induced association of tumor necrosis factor (TNF) receptor 1/TNF receptor-associated factor-2 mediates activation of Rel proteins. Proc Natl Acad Sci USA. 1998;95(2):565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Kato T Jr, Delhase M, Hoffmann A, Karin M. CK2 is a C-terminal IkappaB kinase responsible for NF-kappaB activation during the UV response. Mol Cell. 2003;12(4):829–839. [DOI] [PubMed] [Google Scholar]
- [242].Candi E, Terrinoni A, Rufini A, et al. p63 is upstream of IKK alpha in epidermal development. J Cell Sci. 2006;119(Pt 22):4617–4622. [DOI] [PubMed] [Google Scholar]
- [243].Marinari B, Ballaro C, Koster MI, et al. IKKalpha is a p63 transcriptional target involved in the pathogenesis of ectodermal dysplasias. J Invest Dermatol. 2009;129(1):60–69. [DOI] [PubMed] [Google Scholar]
- [244].Hu Y, Baud V, Delhase M, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284(5412):316–320. [DOI] [PubMed] [Google Scholar]
- [245].Takeda K, Takeuchi O, Tsujimura T, et al. Limb and skin abnormalities in mice lacking IKKalpha. Science. 1999;284(5412):313–316. [DOI] [PubMed] [Google Scholar]
- [246].Li Q, Lu Q, Hwang JY, et al. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 1999;13(10):1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Hu Y, Baud V, Oga T, Kim KI, Yoshida K, Karin M. IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature. 2001;410(6829):710–714. [DOI] [PubMed] [Google Scholar]
- [248].Sil AK, Maeda S, Sano Y, Roop DR, Karin M. IkappaB kinase-alpha acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature. 2004;428(6983):660–664. [DOI] [PubMed] [Google Scholar]
- [249].Senftleben U, Cao Y, Xiao G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293(5534):1495–1499. [DOI] [PubMed] [Google Scholar]
- [250].Cao Y, Bonizzi G, Seagroves TN, et al. IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell. 2001;107(6):763–775. [DOI] [PubMed] [Google Scholar]
- [251].Demicco EG, Kavanagh KT, Romieu-Mourez R, et al. RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland. Mol Cell Biol. 2005;25(22):10136–10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Liu B, Park E, Zhu F, et al. A critical role for I kappaB kinase alpha in the development of human and mouse squamous cell carcinomas. Proc Natl Acad Sci USA. 2006;103(46):17202–17207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Maeda G, Chiba T, Kawashiri S, Satoh T, Imai K. Epigenetic inactivation of IkappaB Kinase-alpha in oral carcinomas and tumor progression. Clin Cancer Res. 2007;13(17):5041–5047. [DOI] [PubMed] [Google Scholar]
- [254].Marinari B, Moretti F, Botti E, et al. The tumor suppressor activity of IKKalpha in stratified epithelia is exerted in part via the TGF-beta antiproliferative pathway. Proc Natl Acad Sci USA. 2008;105(44):17091–17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Descargues P, Sil AK, Sano Y, et al. IKKalpha is a critical coregulator of a Smad4-independent TGFbeta-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci USA. 2008;105(7):2487–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Neri A, Chang CC, Lombardi L, et al. B cell lymphoma-associated chromosomal translocation involves candidate oncogene lyt-10, homologous to NF-kappa B p50. Cell. 1991;67(6):1075–1087. [DOI] [PubMed] [Google Scholar]
- [257].Kitajima I, Shinohara T, Bilakovics J, Brown D, Xu X, Nerenberg M. Ablation of transplanted HTLV-I Tax-transformed tumors in mice by antisense inhibition of NF-kappa B. Science. 1992;258(5089):1792–1795. [DOI] [PubMed] [Google Scholar]
- [258].Seitz CS, Lin Q, Deng H, Khavari PA. Alterations in NF-kappaB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-kappaB. Proc Natl Acad Sci USA. 1998;95(5):2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Makris C, Godfrey VL, Krähn-Senftleben G, et al. Female mice heterozygous for IKK gamma/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol Cell. 2000;5(6):969–979. [DOI] [PubMed] [Google Scholar]
- [260].Zhang JY, Green CL, Tao S, Khavari PA. NF-kappaB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes Dev. 2004;18(1):17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Arnott CH, Scott KA, Moore RJ, et al. Tumour necrosis factor-alpha mediates tumour promotion via a PKC alpha- and AP-1-dependent pathway. Oncogene. 2002;21(31):4728–4738. [DOI] [PubMed] [Google Scholar]
- [262].van Hogerlinden M, Rozell BL, Ahrlund-Richter L, Toftgard R. Squamous cell carcinomas and increased apoptosis in skin with inhibited Rel/nuclear factor-kappaB signaling. Cancer Res. 1999;59(14):3299–3303. [PubMed] [Google Scholar]
- [263].Dajee M, Lazarov M, Zhang JY, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421(6923):639–643. [DOI] [PubMed] [Google Scholar]
- [264].Lind MH, Rozell B, Wallin RP, et al. Tumor necrosis factor receptor 1-mediated signaling is required for skin cancer development induced by NF-kappaB inhibition. Proc Natl Acad Sci USA. 2004;101(14):4972–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Schorpp M, Mallick U, Rahmsdorf HJ, Herrlich P. UV-induced extracellular factor from human fibroblasts communicates the UV response to nonirradiated cells. Cell. 1984;37(3):861–868. [DOI] [PubMed] [Google Scholar]
- [266].Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47(6):921–928. [DOI] [PubMed] [Google Scholar]
- [267].Price MA, Cruzalegui FH, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate Ternary Complex Factors and c-fos transcription in response to UV light. EMBO J. 1996;15(23):6552–6563. [PMC free article] [PubMed] [Google Scholar]
- [268].Ducret C, Maira SM, Dierich A, Wasylyk B. The net repressor is regulated by nuclear export in response to anisomycin, UV, and heat shock. Mol Cell Biol. 1999;19(10):7076–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Treier M, Bohmann D, Mlodzik M. JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell. 1995;83(5):753–760. [DOI] [PubMed] [Google Scholar]
- [270].Whitmarsh AJ, Shore P, Sharrocks AD, Davis RJ. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269(5222):403–407. [DOI] [PubMed] [Google Scholar]
- [271].Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308(5720):381–385. [DOI] [PubMed] [Google Scholar]
- [272].Adiseshaiah P, Peddakama S, Zhang Q, Kalvakolanu DV, Reddy SP. Mitogen regulated induction of FRA-1 proto-oncogene is controlled by the transcription factors binding to both serum and TPA response elements. Oncogene. 2005;24(26):4193–4205. [DOI] [PubMed] [Google Scholar]
- [273].Kim M, McGinnis W. Phosphorylation of Grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc Natl Acad Sci USA. 2011;108(2):650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Huang S, Robinson JB, Deguzman A, Bucana CD, Fidler IJ. Blockade of nuclear factor-kappaB signaling inhibits angiogenesis and tumorigenicity of human ovarian cancer cells by suppressing expression of vascular endothelial growth factor and interleukin 8. Cancer Res. 2000;60(19):5334–5339. [PubMed] [Google Scholar]
- [275].Li R, Pei H, Watson DK, Papas TS. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19(6):745–753. [DOI] [PubMed] [Google Scholar]
- [276].Bond M, Fabunmi RP, Baker AH, Newby AC. Synergistic upregulation of metalloproteinase-9 by growth factors and inflammatory cytokines: an absolute requirement for transcription factor NF-kappa B. FEBS Lett. 1998;435(1):29–34. [DOI] [PubMed] [Google Scholar]
- [277].Lee TL, Yang XP, Yan B, et al. A novel nuclear factor-kappaB gene signature is differentially expressed in head and neck squamous cell carcinomas in association with TP53 status. Clin Cancer Res. 2007;13(19):5680–5691. [DOI] [PubMed] [Google Scholar]
- [278].Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: in search of the ‘epimmunome’. Nat Immunol. 2010;11(8):656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Westfall MD, Joyner AS, Barbieri CE, Livingstone M, Pietenpol JA. Ultraviolet radiation induces phosphorylation and ubiquitin-mediated degradation of DeltaNp63alpha. Cell Cycle. 2005;4(5):710–716. [DOI] [PubMed] [Google Scholar]
- [280].Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15(12):1941–1951. [DOI] [PubMed] [Google Scholar]
- [281].Chatterjee A, Chang X, Sen T, Ravi R, Bedi A, Sidransky D. Regulation of p53 family member isoform DeltaNp63alpha by the nuclear factor-kappaB targeting kinase IkappaB kinase beta. Cancer Res. 2010;70(4):1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Lena AM, Shalom-Feuerstein R, Rivetti P, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15(7):1187–1195. [DOI] [PubMed] [Google Scholar]
- [283].Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing’stemness’. Nature. 2008;452(7184):225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Jackson SJ, Zhang Z, Feng D, et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 2013;140(9):1882–1891. 10.1242/dev.089649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Gille H, Sharrocks AD, Shaw PE. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358(6385):414–417. [DOI] [PubMed] [Google Scholar]
- [286].Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73(2):381–393. [DOI] [PubMed] [Google Scholar]
- [287].Romano RA, Birkaya B, Sinha S. Defining the regulatory elements in the proximal promoter of DeltaNp63 in keratinocytes: potential roles for Sp1/Sp3, NF-Y, and p63. J Invest Dermatol. 2006;126(7):1469–1479. [DOI] [PubMed] [Google Scholar]
- [288].Dotto GP, Gilman MZ, Maruyama M, Weinberg RA. c-myc and c-fos expression in differentiating mouse primary keratinocytes. EMBO J. 1986;5(11):2853–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334(6183):629–631. [DOI] [PubMed] [Google Scholar]
- [290].Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55(5):875–885. [DOI] [PubMed] [Google Scholar]
- [291].Kudryavtseva EI, Sugihara TM, Wang N,et al. Identification and characterization of Grainyhead-like epithelial transactivator (GET-1), a novel mammalian Grainyhead-like factor. Dev Dyn. 2003;226(4):604–617. [DOI] [PubMed] [Google Scholar]
- [292].Yu Z, Lin KK, Bhandari A, et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299(1):122–136. [DOI] [PubMed] [Google Scholar]
- [293].Hislop NR, Caddy J, Ting SB, et al. Grhl3 and Lmo4 play coordinate roles in epidermal migration. Dev Biol. 2008;321(1):263–272. 10.1016/j.ydbio.2008.06.026 [DOI] [PubMed] [Google Scholar]
- [294].Pearson JC, Juarez MT, Kim M, Drivenes Ø, McGinnis W. Multiple transcription factor codes activate epidermal wound-response genes in Drosophila. Proc Natl Acad Sci USA. 2009;106(7):2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Chen AF, Liu AJ, Krishnakumar R, Freimer JW, DeVeale B, Blelloch R. GRHL2-dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell. 2018;23(2):226–238.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Adam RC, Yang H, Rockowitz S, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521(7552):366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Millar SE. Committing to a hairy fate: epigenetic regulation of hair follicle stem cells. Cell Stem Cell. 2011;9(3):183–184. [DOI] [PubMed] [Google Scholar]
- [298].Frye M, Benitah SA. Chromatin regulators in mammalian epidermis. Semin Cell Dev Biol. 2012;23(8):897–905. [DOI] [PubMed] [Google Scholar]
- [299].Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E. Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med. 2014;4(2):a015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].LeBoeuf M, Terrell A, Trivedi S, et al. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19(6):807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Hughes MW, Jiang TX, Lin SJ, et al. Disrupted ectodermal organ morphogenesis in mice with a conditional histone deacetylase 1, 2 deletion in the epidermis. J Invest Dermatol. 2014;134(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Luis NM, Morey L, Mejetta S, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb-dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9(3):233–246. [DOI] [PubMed] [Google Scholar]
- [304].Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463(7280):563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Li J, Jiang TX, Hughes MW, et al. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J Invest Dermatol. 2012;132(12):2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Mulder KW, Wang X, Escriu C, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14(7):753–763. [DOI] [PubMed] [Google Scholar]
- [307].Rinaldi L, Datta D, Serrat J, et al. Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016;19(4):491–501. [DOI] [PubMed] [Google Scholar]
- [308].Rinaldi L, Avgustinova A, Martin M, et al. Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-gamma. Elife. 2017;6:e21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Bao X, Rubin AJ, Qu K, et al. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16(1):284. 10.1186/s13059-015-0840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Vierbuchen T, Ling E, Cowley CJ, et al. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol Cell. 2017;68(6):1067–1082.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Mardaryev AN, Gdula MR, Yarker JL, et al. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development. 2014;141(1):101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Fessing MY, Mardaryev AN, Gdula MR, et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol. 2011;194(6):825–839. 10.1083/jcb.201101148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Saladi SV, Ross K, Karaayvaz M, et al. ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell. 2017;31(1):35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Bao X, Tang J, Lopez-Pajares V, et al. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12(2):193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Hopkin AS, Gordon W, Klein RH, et al. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8(7):e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22(14):1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Ding X, Pan H, Li J, et al. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci Signal.2013;6(273):ra28.1–ra28.13, S0-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Weiss C, Schneider S, Wagner EF, Zhang X, Seto E, Bohmann D. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 2003;22(14): 3686–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Wurm S, Zhang J, Guinea-Viniegra J, et al. Terminal epidermal differentiation is regulated by the interaction of Fra-2/AP-1 with Ezh2 and ERK1/2. Genes Dev. 2015;29(2):144–156. 10.1101/gad.249748.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9(6):700–713. [DOI] [PubMed] [Google Scholar]
- [321].Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95(5):605–614. [DOI] [PubMed] [Google Scholar]
- [322].Van Mater D, Kolligs FT, Dlugosz AA, Fearon ER. Transient activation of beta -catenin signaling in cutaneous keratinocytes is sufficient to trigger the active growth phase of the hair cycle in mice. Genes Dev. 2003;17(10):1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Lo Celso C, Prowse DM, Watt FM. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131(8):1787–1799. [DOI] [PubMed] [Google Scholar]
- [324].Silva-Vargas V, Lo Celso C, Giangreco A, et al. Beta-catenin and Hedgehog signal strength can specify number and location of hair follicles in adult epidermis without recruitment of bulge stem cells. Dev Cell. 2005;9(1):121–131. [DOI] [PubMed] [Google Scholar]
- [325].Zhang Y, Andl T, Yang SH, et al. Activation of beta-catenin signaling programs embryonic epidermis to hair follicle fate. Development. 2008;135(12):2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Kurita M, Araoka T, Hishida T, et al. In vivo reprogramming of wound-resident cells generates skin epithelial tissue. Nature. 2018;561(7722):243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. 10.1016/j.cell.2014.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Cancer Genome Atlas Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47(10): 1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]